Significance
Phospholipids in the plasma membrane are scrambled between the two leaflets by TMEM16 proteins. Some TMEM16 family members are also present in intracellular membranes, but whether they scramble phospholipids inside cells has not been demonstrated. To address this question, we developed a method to study the distribution of phosphatidylserine (PtdSer) in individual membrane leaflets. Using this method, we found that PtdSer is present in the cytoplasmic leaflet of the endoplasmic reticulum and in the nuclear membrane, and that an increase in intracellular Ca2+ causes the PtdSer in intracellular membranes to be redistributed in a TMEM16K-dependent manner. These results indicate that phospholipids in the endoplasmic reticulum are redistributed by a mechanism similar to that in the plasma membrane.
Keywords: endoplasmic reticulum, nuclear membrane, phosphatidylserine, scramblase, TMEM16K
Abstract
TMEM16K, a membrane protein carrying 10 transmembrane regions, has phospholipid scramblase activity. TMEM16K is localized to intracellular membranes, but whether it actually scrambles phospholipids inside cells has not been demonstrated, due to technical difficulties in studying intracellular lipid distributions. Here, we developed a freeze-fracture electron microscopy method that enabled us to determine the phosphatidylserine (PtdSer) distribution in the individual leaflets of cellular membranes. Using this method, we found that the endoplasmic reticulum (ER) of mammalian cells harbored abundant PtdSer in its cytoplasmic leaflet and much less in the luminal leaflet, whereas the outer and inner nuclear membranes (NMs) had equivalent amounts of PtdSer in both leaflets. The ER and NMs of budding yeast also harbored PtdSer in their cytoplasmic leaflet, but asymmetrical distribution in the ER was not observed. Treating mouse embryonic fibroblasts with the Ca2+ ionophore A23187 compromised the cytoplasmic leaflet-dominant PtdSer asymmetry in the ER and increased PtdSer in the NMs, especially in the nucleoplasmic leaflet of the inner NM. This Ca2+-induced PtdSer redistribution was not observed in TMEM16K-null fibroblasts, but was recovered in these cells by reexpressing TMEM16K. These results indicate that, similar to the plasma membrane, PtdSer in the ER of mammalian cells is predominantly localized to the cytoplasmic leaflet, and that TMEM16K directly or indirectly mediates Ca2+-dependent phospholipid scrambling in the ER.
Phosphatidylserine (PtdSer) is one of the major acidic phospholipids in eukaryotic cells (1, 2) and is involved in various cellular signaling processes (3). PtdSer is exclusively present in the cytoplasmic leaflet of the plasma membrane. However, under some circumstances, such as in activated platelets (4) and in cells undergoing apoptosis (5), the asymmetrical distribution of PtdSer is compromised, and a substantial amount of PtdSer is exposed on the exoplasmic leaflet. We previously showed that TMEM16F, a Ca2+-dependent scramblase, plays a crucial role in the PtdSer exposure on activated platelets (6, 7), and that another scramblase, Xkr8, is activated by caspase-mediated cleavage and promotes the PtdSer exposure on apoptotic cells (8).
In addition to the plasma membrane, several TMEM16 family members are present in intracellular membranes. Among them, the tissue-specifically expressed TMEM16E and ubiquitously expressed TMEM16K have been suggested to have a Ca2+-dependent scrambling activity, based on analyses of their putative scramblase domain (9, 10) and on molecular dynamic simulation experiments (11). More recently, the purified TMEM16K was shown to have scramblase activity in a reconstituted liposome system (12). Nevertheless, whether these molecules actually function as a scramblase inside cells has not been addressed, because the phospholipids in intracellular membranes are not directly accessible and thus are more difficult to study than those in the plasma membrane. However, considering that mutations of TMEM16E and TMEM16K are linked to muscle dystrophy (13–15) and ataxia (16–18), respectively, it is important to understand their cellular functions as thoroughly as possible.
The intracellular PtdSer distribution has mainly been studied by expressing fluorescently tagged PtdSer-binding protein domains in the cytoplasm (19, 20). However, the binding of these molecules to membranes may be affected by factors unrelated to PtdSer (21), and there is uncertainty about the extent to which the observed signal reflects the actual PtdSer content. Moreover, this method does not provide information about PtdSer’s distribution in the luminal leaflet of organelle membranes. Electron microscopy (EM) using ultrathin sections can detect PtdSer in both leaflets in principle (22), but this method cannot distinguish whether PtdSer is present in the luminal or cytoplasmic leaflet, mainly due to section thickness and low membrane contrast.
In the present study, we developed an EM method for analyzing the PtdSer distribution using quick-freeze and freeze-fracture replica labeling (QF-FRL), which can provide information about the two leaflets of a membrane separately (23). Using this method, we found that the endoplasmic reticulum (ER) membrane harbors a large amount of PtdSer in the cytoplasmic leaflet in mouse embryonic fibroblasts (MEFs) and budding yeast. In the ER of MEFs, PtdSer was predominantly localized to the cytoplasmic leaflet, whereas, in the outer and inner nuclear membranes (NMs), the amount of PtdSer was equivalent in the two leaflets. On the other hand, the PtdSer in yeast was symmetrically distributed in both the ER and NMs. When the intracellular Ca2+ concentration was increased in MEFs, the PtdSer asymmetry in the ER was compromised, and PtdSer in the NMs increased, most prominently in the nucleoplasmic leaflet of the inner NM. These Ca2+-induced changes in the PtdSer distribution were TMEM16K-dependent. These results reveal the detailed intracellular PtdSer landscape, and suggest a role of TMEM16K in changing phospholipid distribution in the ER.
Results
PtdSer Labeling by QF-FRL.
The PH domain of evectin-2 (evectin-PH) is reported to specifically bind PtdSer (19, 24). We arranged two of these domains in tandem and fused them to GST (designated as GST-2xPH), and used this construction to label PtdSer (SI Appendix, Fig. S1A). GFP-tagged version of this probe (GFP-2xPH) expressed in live cells distributed to the plasma membrane in yeast and mainly to cytoplasmic structures in MEFs as reported previously (19, 25), but did not show binding to the ER or the NM (SI Appendix, Fig. S1B).
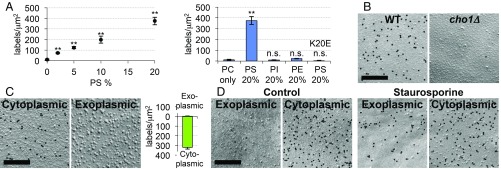
To confirm the specificity of our detection procedure, we first showed that GST-2xPH specifically bound to freeze-fracture replicas of liposomes containing PtdSer, and not to those without PtdSer (Fig. 1A and SI Appendix, Fig. S2A). The labeling intensity in the liposome replica increased roughly in proportion to the PtdSer content up to 20% (Fig. 1A), and the presence of 20% phosphatidylethanolamine in the PtdSer-containing liposomes had no significant effect on the labeling for PtdSer (SI Appendix, Fig. S2B). Second, a mutant of evectin-PH [GST-2xPH(K20E)], in which Lys-20 in the PtdSer-recognition site was replaced by Glu (19), did not bind to the PtdSer-containing liposomes (Fig. 1A). Third, GST-2xPH only negligibly labeled cho1Δ yeast, which lacks PtdSer (26) (Fig. 1B and SI Appendix, Fig. S2C).
Fig. 1.
Validation of QF-FRL for PtdSer detection. (A) Liposomes were prepared from phosphatidylcholine (PC) alone, PC and 2 to 20% PtdSer (PS), PC and 20% phosphatidylethanolamine (PE), and PC and 20% phosphatidylinositol (PI). Freeze-fracture replicas were prepared from these liposomes, and labeled with GST-2xPH or GST-2xPH(K20E). The density of the gold particles (gold particles per square micrometer) is shown. The data are the mean ± SEM of three independent experiments (n = 100 for each experiment); n.s., not significant. Difference from the result for the PC-only liposomes was examined by Student’s t test. **P < 0.01. The EM pictures are shown in SI Appendix, Fig. S2A. (B) PtdSer-labeling of the cytoplasmic leaflet of the plasma membrane of wild-type but not of cho1Δ yeast. (Scale bar, 0.2 μm.) See SI Appendix, Fig. S2C for larger areas, and Fig. 2B for quantification. (C) (Left and Center) Labeling of a human red blood cell membrane with GST-2xPH. (Scale bar, 0.2 μm.) See SI Appendix, Fig. S2D for larger areas. (Right) Three independent experiments were performed, and the mean labeling intensity (gold particles per square micrometer) in the exoplasmic and cytoplasmic leaflets are shown ± SEM (n > 30). (D) Localization of PtdSer in the exoplasmic leaflet of the plasma membrane in apoptotic cells. MEFs were untreated or treated with 0.2 μM staurosporine for 6 h, and stained with GST-2xPH. (Scale bar, 0.2 μm.)
We then used GST-2xPH to label human erythrocyte plasma membranes, which showed predominant labeling in their cytoplasmic leaflet (Fig. 1C and SI Appendix, Fig. S2D), confirming that it detected the known asymmetrical distribution of PtdSer (27). The plasma membrane of MEFs under normal culture conditions also showed the cytoplasmic leaflet-dominant PtdSer asymmetry, while, in cells undergoing apoptosis, the labeling in the exoplasmic leaflet increased (Fig. 1D). In cellular samples, access of the labeling probe to PtdSer could be hindered by integral membrane proteins retained in the replica. However, pretreatment of the replicas with proteinase K did not change the PtdSer labeling intensity significantly (SI Appendix, Fig. S2E), indicating that the effect of proteins was negligible. We thus concluded that the use of GST-2xPH in QF-FRL would be reliable for determining the intracellular distribution of PtdSer.
PtdSer Distribution in Intracellular Membranes.
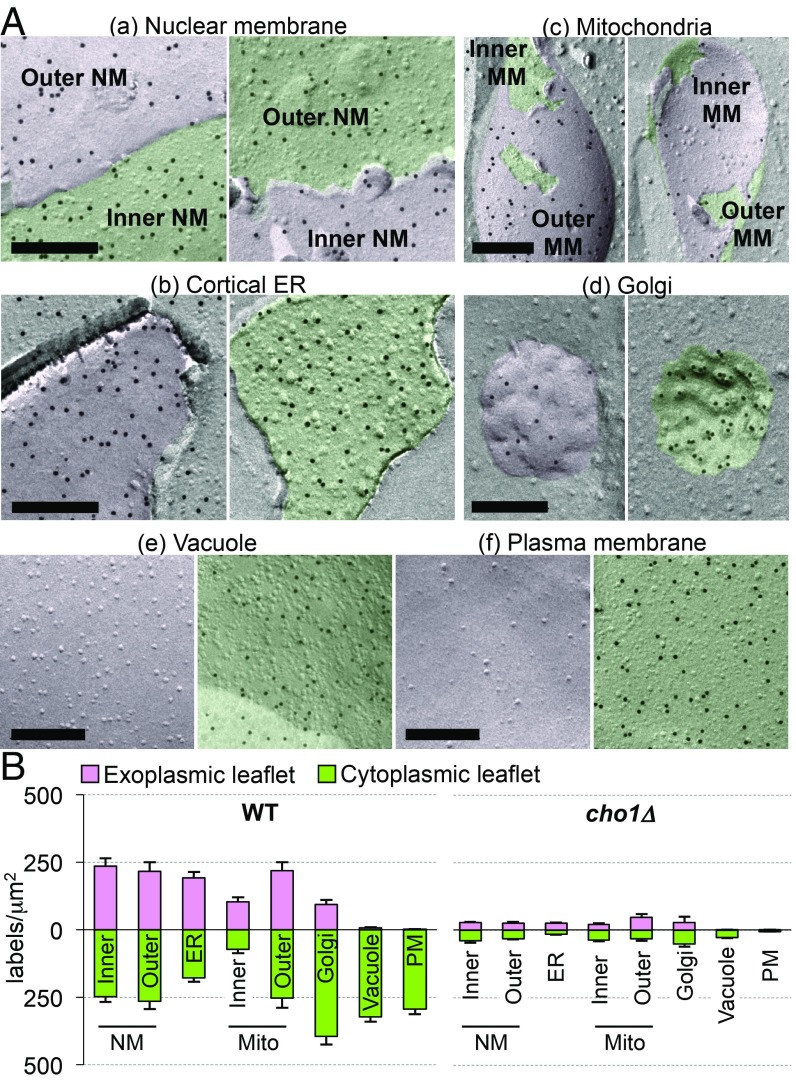
Using the QF-FRL method validated above, we examined the PtdSer distribution in the intracellular membranes of budding yeast and MEFs. Yeast has a simple cellular structure, and most of its organelles can be readily identified morphologically in freeze-fracture EM (28, 29). The cytoplasmic (nucleoplasmic in the inner NM) and exoplasmic (luminal) leaflets of the NMs and the ER were labeled with GST-2xPH with comparable intensities (Fig. 2 A, a and b and SI Appendix, Fig. S3A). The labeling intensity of the two leaflets in the cortical ER was similar between the leaflet facing the plasma membrane and the one facing the cell interior (SI Appendix, Fig. S3B). The two leaflets in the cytoplasmic ER were labeled similarly (SI Appendix, Fig. S3B). The two leaflets of the outer and inner mitochondrial membranes were also labeled with GST-2xPH with similar densities, although relatively low labeling was consistently observed in the inner membrane (Fig. 2 A, c). In contrast, the cytoplasmic leaflets of the Golgi, the vacuole, and the plasma membrane were labeled much more densely than their luminal counterpart (Fig. 2 A, d–f and SI Appendix, Fig. S3A). Proteinase K treatment of the replica did not affect the labeling intensity (SI Appendix, Fig. S3C). In cho1Δ, the labeling of the intracellular membranes was essentially negligible (Fig. 2B and SI Appendix, Fig. S3D), reconfirming the labeling specificity of the method. These analyses indicated that PtdSer was distributed at similar densities in the plasma membrane and the vacuole in yeast, which appears to be different from the reported biochemical analysis, which showed that PtdSer in the plasma membrane occupies a significantly larger proportion of phospholipids than in the vacuole (30) (SI Appendix, Table S1). However, the two results may agree well if we consider that our method detects the distribution density of PtdSer in the membrane, while the previous study measured only the relative PtdSer amount in phospholipids and did not count other lipids and proteins, which exist more abundantly in the plasma membrane than in the vacuole (SI Appendix, Table S1 and Fig. S3E).
Fig. 2.
PtdSer distribution in yeast. (A) PtdSer labeling in budding yeast. Yeast cells were analyzed by QF-FRL. Staining profiles with GST-2xPH for the NMs (a), cortical ER (b), mitochondria (Mito) (c), Golgi (d), vacuole (e), and plasma membrane (PM) (f) are shown. The cytoplasmic leaflet (and the leaflets facing the nucleoplasm and the mitochondrial matrix) and the exoplasmic leaflet are colored in green and pink, respectively. (Scale bars, 0.2 μm.) Wider areas of the NMs, cortical ER, and vacuole are shown in SI Appendix, Fig. S3A. (B) The density of PtdSer labeling (gold particles per square micrometer) measured from electron micrographs of wild-type and cho1Δ yeast. Mean ± SEM values obtained from three independent experiments are shown; n > 30.
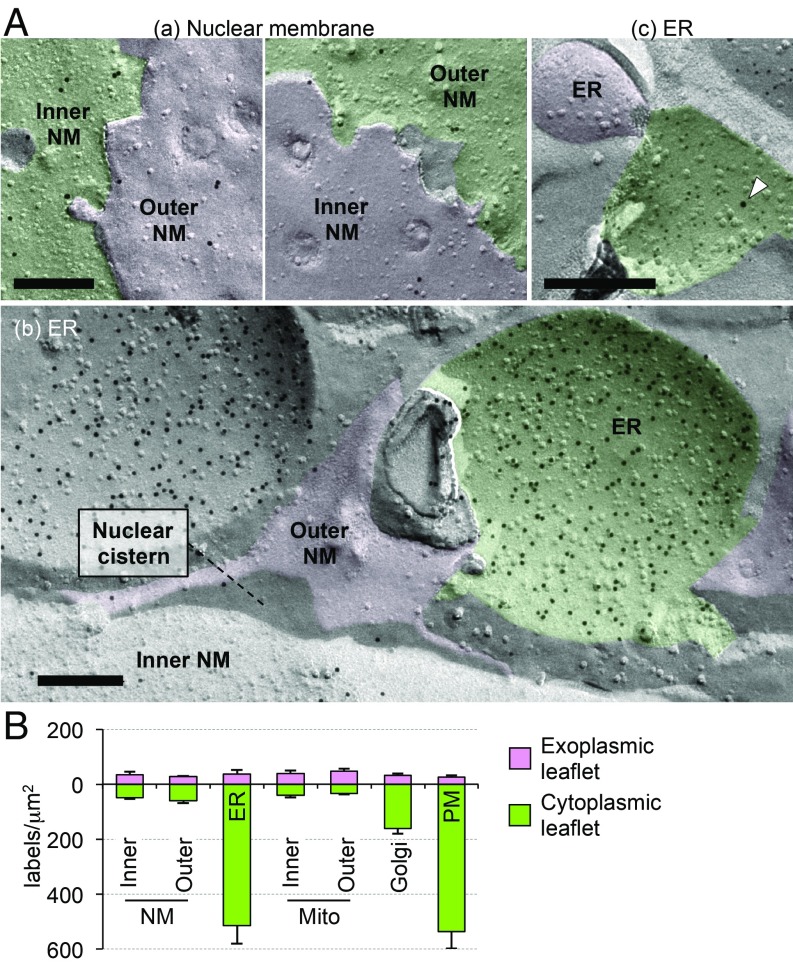
We next examined the distribution of PtdSer in MEFs. As shown in Fig. 3 A, a, PtdSer was present in both leaflets of the outer and inner NM with similar low densities. The ER was recognized as a membrane continuous with the outer NM (Fig. 3 A, b), or as tubular or sheet-like structures with a narrow lumen, which were labeled with anti-cytochrome P450 reductase (POR) (SI Appendix, Fig. S4A). PtdSer was labeled intensely in the cytoplasmic leaflet but not in the luminal leaflet of these ER membranes (Fig. 3 A, b and SI Appendix, Fig. S4B). The ER labeling was observed similarly in proteinase K-treated replicas (SI Appendix, Fig. S4C), and confirmed by double labeling for PtdSer and POR (Fig. 3 A, c and SI Appendix, Fig. S4D). The cytoplasmic leaflet-dominant PtdSer labeling was also observed in the ER of McA-RH7777 cells, a rat liver hepatoma cell line (SI Appendix, Fig. S4E). Symmetrical labeling in the outer and inner mitochondrial membranes and cytoplasmic leaflet-dominant labeling in the Golgi and plasma membrane were also observed, similar to the findings in yeast (SI Appendix, Fig. S4F). The recycling endosome, which was identified by anti-Rab11 labeling, was densely labeled for PtdSer, as reported previously (SI Appendix, Fig. S4G) (19). GST-2xPH(K20E) showed only negligible labeling, verifying the specificity of the results in MEFs (SI Appendix, Fig. S4H). The quantified PtdSer-labeling density confirmed that, similar to the plasma membrane, PtdSer was predominantly distributed in the cytoplasmic leaflet of the ER in MEFs (Fig. 3B).
Fig. 3.
The intracellular PtdSer distribution in MEFs. (A) Distribution of PtdSer in the NMs (a) and the ER (b and c). The cytoplasmic and exoplasmic leaflets are colored in green and pink, respectively. In b, the ER was identified by its membrane continuity with the outer NM. In c, small (6 nm) and large (12 nm; arrowhead) colloidal gold particles label PtdSer and POR, respectively. (Scale bars, 0.2 μm.) Another example is shown in SI Appendix, Fig. S4D. (B) The density of PtdSer labeling (gold particles per square micrometer) measured from electron micrographs of MEFs. Mean ± SEM values obtained from three independent experiments are shown; n > 30.
Redistribution of PtdSer in the ER and Nuclear Membrane upon Ca2 Ionophore Treatment.
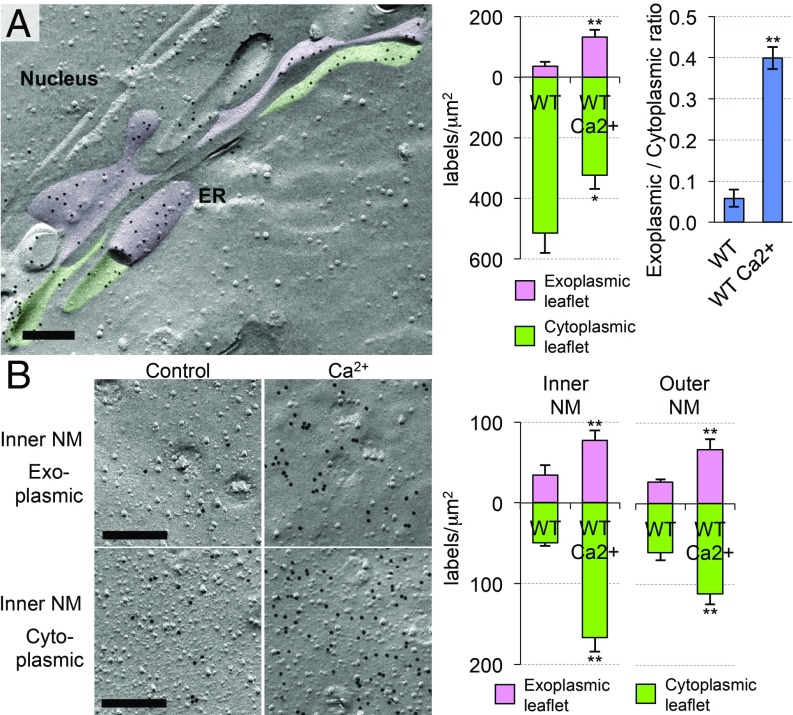
An increase of the intracellular Ca2+ concentration induces phospholipid scrambling in the plasma membrane by activating TMEM16F (7). However, whether similar scrambling occurs in intracellular membranes has not been known. To address this question, we treated MEFs with a Ca2+ ionophore, A23187, and examined the PtdSer distribution in their intracellular membranes. Remarkably, the A23187 treatment dramatically altered the PtdSer distribution in the ER and NMs (Fig. 4 and SI Appendix, Fig. S5A). That is, PtdSer in the ER decreased in the cytoplasmic leaflet, whereas it increased in the luminal leaflet, making the PtdSer asymmetry less severe than that in the untreated MEFs. In the NMs, the A23187 treatment significantly increased the PtdSer in both leaflets, most prominently in the nucleoplasmic leaflet of the inner NM (Fig. 4B). The A23187 treatment of McA-RH7777 cells induced similar changes in the PtdSer distribution in the ER and NMs (SI Appendix, Fig. S5B).
Fig. 4.
Ca2+-induced redistribution of PtdSer in the ER and NMs. MEFs were treated with 3 μM A23187 for 10 min at 37 °C, and the PtdSer distribution in the (A) ER and (B) NMs was examined by QF-FRL. (Scale bars, 0.2 μm.) (Right) The density of the PtdSer labeling in control (WT) and A23187-treated MEFs (WT Ca2+) is shown. The data for the untreated sample are the same as in Fig. 3B. Mean ± SEM values obtained from three independent experiments are shown (n > 30). The data were statistically analyzed by Mann−Whitney U test. *P < 0.05; **P < 0.01. (Upper Right) The ratio of the PtdSer labeling density in the exoplasmic leaflet to that in the cytoplasmic leaflet in the ER is also shown. The data were statistically analyzed by Student’s t test. **P < 0.01.
TMEM16K-Dependent Ca2+-Induced Redistribution of PtdSer.
Among the TMEM16 family proteins that have a domain capable of scrambling phospholipids, TMEM16E and TMEM16K are localized to intracellular membranes (9, 11), and MEFs expressed only TMEM16K (SI Appendix, Fig. S6A). To examine whether TMEM16K was involved in the Ca2+-induced PtdSer redistribution, TMEM16K−/− MEFs were generated using the CRISPR/Cas9 system (SI Appendix, Fig. S6B).
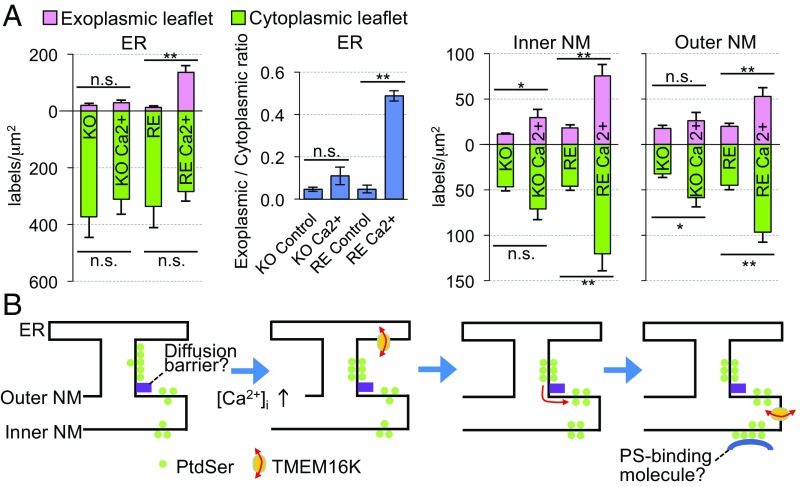
The PtdSer distribution in TMEM16K-null MEFs was similar to that in the wild-type MEFs under the untreated condition. However, unlike the wild-type cells, the A23187 treatment of TMEM16K−/− MEFs did not cause a significant change in the PtdSer distribution in the ER or NMs (Fig. 5A). This lack of change was rescued by reintroducing TMEM16K into the TMEM16K−/− MEFs (SI Appendix, Fig. S6B). Specifically, Flag-tagged TMEM16K introduced into TMEM16K−/− MEFs was localized to the ER and NMs (SI Appendix, Fig. S6C), and A23187 treatment induced the redistribution of PtdSer in these membranes (Fig. 5A and SI Appendix, Fig. S6D). Since intracellular Ca2+ concentration increased similarly in these cell lines upon the A23187 treatment, monitored with Fluo-4 (SI Appendix, Fig. S6E), these results indicated that TMEM16K played an important role in the Ca2+-induced PtdSer redistribution in the ER and the NM.
Fig. 5.
Critical role of TMEM16K in the Ca2+-induced redistribution of PtdSer. (A) TMEM16K−/−MEFs (KO) and their transformants expressing TMEM16K (RE) were untreated or treated with A23187 (Ca2+). The PtdSer distribution in the ER and NMs was examined by QF-FRL, and the PtdSer-labeling densities in the exoplasmic and cytoplasmic leaflets are shown. Mean ± SEM values obtained from three independent experiments (n > 30) are shown. The data were statistically analyzed by Mann−Whitney U test. *P < 0.05; **P < 0.01. The ratio of the ER PtdSer labeling density in the exoplasmic leaflet to that in the cytoplasmic leaflet is also presented. The data were statistically analyzed by Student’s t test. (B) A putative mechanism for the TMEM16K-mediated Ca2+-induced PtdSer redistribution in the ER and NMs. See Discussion for details.
Discussion
In both budding yeast and mammalian cells, PtdSer is synthesized in the ER (31, 32), and is thought to be incorporated into the cytoplasmic leaflet of the ER membrane. However, since fluorescent biosensors for PtdSer do not bind to the cytoplasmic leaflet of the ER (19, 20), it was speculated that the de novo synthesized PtdSer is quickly transported to the luminal leaflet (33). In contrast to this scenario, we could detect PtdSer in both leaflets of the ER membrane in yeast and mammalian cells. In particular, PtdSer was found to be more abundant in the cytoplasmic leaflet than in the luminal leaflet of the ER in MEFs. The density of colloidal gold labels in QF-FRL may not be strictly proportional to the actual PtdSer content (23), and difficulty exists in accurately estimating the area of curved membrane in electron micrographs. Nevertheless, it was evident that the cytoplasmic leaflet of the ER contained a substantial amount of PtdSer. On the other hand, it is not clear why the existing fluorescent biosensors do not bind to PtdSer in the cytoplasmic leaflets of the ER and the NM. It is possible that coincident detection, which underlies the variable behavior of phosphoinositide biosensors (34), may also occur for PtdSer biosensors. It is also possible that the PtdSer in the cytoplasmic leaflet of the ER is occupied by PtdSer-binding proteins. In this regard, it is noteworthy that, in the QF-FRL procedure, the membranes were pretreated with sodium dodecyl sulfate (SDS) before labeling. This methodological issue needs to be explored further.
The results obtained by QF-FRL indicate that we should revise the current view of the intracellular PtdSer distribution and PtdSer-related cellular functions in mammalian cells. For example, the presence of PtdSer in the cytoplasmic leaflet of the ER may be more reasonable than its absence with respect to the countertransport of PtdSer and phosphatidylinositol 4-phosphate at the ER–plasma membrane contact site (35, 36). Our results also indicated that the cytoplasmic leaflet-dominant asymmetric distribution of PtdSer already exists in the ER, and thus specific flippases in the post-ER compartment may not create the plasmalemmal PtdSer asymmetry from scratch. The detailed mechanism by which PtdSer is enriched and maintained in the cytoplasmic leaflet of the ER in mammalian cells is a topic for future study.
The present study also revealed that the PtdSer distribution is different between yeast and mammalian cells, but this may not be surprising in light of their known differences in PtdSer’s synthesis, decarboxylation, and transport: 1) PtdSer in mammals is synthesized by a base exchange of the polar head group of phosphatidylcholine or phosphatidylethanolamine to serine (37), whereas PtdSer in yeast is generated from CDP-diacylglycerol and serine (38); 2) Ca2+ stimulates PtdSer synthases in mammals (39), but inhibits it in yeast (38); 3) PtdSer decarboxylase is present only in mitochondria in mammalian cells (40), whereas yeast has two different enzymes, which are distributed in mitochondria, the ER, the Golgi, and vacuoles (30, 41, 42); and 4) PtdSer export from the mitochondria-associated membrane requires adenosine 5′-triphosphate in mammals (43), but not in yeast (44). Furthermore, Ist2, the only yeast TMEM16 homolog, lacks phospholipid scrambling activity in vitro (45), and we found that deleting the ist2 gene did not affect the PtdSer distribution (SI Appendix, Fig. S7). Microsomal membranes show a constitutive phospholipid scrambling activity (46), but the molecule(s) executing the activity and how the mechanism operates in the ER in vivo remain unknown. The present results suggest that TMEM16K has a regulatory role, thus imparting mammalian cells with an elaborate mechanism for controlling PtdSer distribution in a Ca2+-dependent manner. Further analyses are needed to determine the functional importance of the Ca2+-induced PtdSer redistribution in the intracellular membranes of mammalian cells.
Using the QF-FRL method, we showed here that an increase in intracellular Ca2+ in MEFs significantly reduced the PtdSer asymmetry in the ER, only when TMEM16K was present. In light of the scrambling activity shown with the putative scramblase domain of TMEM16K (9) and the purified protein (12), these results suggest that TMEM16K may cause the PtdSer redistribution in the ER through scrambling. It is notable that, in untreated MEFs, the PtdSer density in the cytoplasmic leaflet was much higher in the ER than in the NM despite their continuity, whereas the PtdSer distribution in the luminal leaflet was equivalent between these two membranes. This finding suggests that PtdSer in the cytoplasmic leaflet may not freely move from the ER to the outer NM, due to either a barrier between them or sequestration to the ER membrane (Fig. 5B). We speculate that, once PtdSer is increased in the luminal leaflet of the ER by TMEM16K-mediated process, it diffuses to the NM, and then flips to the cytoplasmic/nucleoplasmic leaflets in the NMs. The inactivation of the barrier (or the sequestration) may also be caused by a high concentration of Ca2+. The prominent increase in PtdSer in the nucleoplasmic leaflet of the inner NM may have been caused by some mechanism that sequestered PtdSer in that location, such as a PtdSer-binding protein in the nucleoplasm (Fig. 5B).
Mutations in the TMEM16K gene cause autosomal recessive spinocerebellar ataxia 10 (16–18). Recently, Tsuchiya et al. (47) reported that an increase in PtdSer in the extracellular leaflet of the myoblast plasma membrane down-regulates the function of a Ca2+ channel, PIEZO1. Similarly, the enzymatic activity and/or cellular localization of proteins are often regulated by their binding to specific phospholipids (48, 49). Thus, it is tempting to speculate that a deficiency in phospholipid scrambling due to TMEM16K dysfunction might affect ER protein functions, causing cellular defects such as the abnormal Ca2+ signaling observed in TMEM16K mutants (50). In addition, since Axs, a Drosophila TMEM16K homolog (51), is essential for meiotic spindle formation (52), phospholipid scrambling in the NMs may also play a role in spindle formation or function.
Materials and Methods
Cells.
The wild-type Saccharomyces cerevisiae (strain, SEY6210) (53), cho1Δ (YSC1021-98804848, Open Biosystems), and ist2Δ (YBR086C) were cultured in YPD medium (1% yeast extract, 2% polypeptone, and 2% glucose), except that the medium for cho1Δ was supplemented with 1 mM ethanolamine. These strains were used at a logarithmic growth phase.
MEFs were established by transforming fibroblasts from E13.5 mouse embryos with SV40 T antigen. The experiment was approved by the Animal Experimentation Committee of Nagoya University Graduate School of Medicine (Approval ID: 28086). MEFs and rat McA-RH7777 cells (American Type Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS) and antibiotics. To increase intracellular Ca2+, cells were treated at 37 °C for 10 min with 3 μM A23187 (Sigma) in Hanks’ balanced salt solution (Sigma) containing 2.5 mM CaCl2.
Gene Knockout by the CRISPR/Cas9 System.
The TMEM16K gene was disrupted in MEFs as described (54). The sequence for the guide RNA (AATGTACGGACGCTGCCATG) was selected using a web-based prediction tool (Crispr direct; https://crispr.dbcls.jp). The guide RNA was generated from a custom-made DNA template (Thermo Fisher Scientific) using the MEGAshortscript T7 Transcription KIT (Thermo Fisher Scientific), and transfected into MEFs along with the GeneArt CRISPR Nuclease mRNA using the Lipofectamine MessengerMax reagent (Thermo Fisher Scientific). Cell colonies were picked and expanded, and the gene disruption was verified by genomic DNA sequencing and by Western blotting with an anti-TMEM16K antibody (Ab).
Quick-Freezing and Freeze-Fracture.
For yeast, a copper EM grid (200 mesh, Nisshin EM) was immersed in cell pellet, sandwiched between a flat aluminum disk (Engineering Office M. Wohlwend, Sennwald, Switzerland) and a thin copper foil (20 μm thick; Nilaco), and frozen using an HPM 010 high-pressure freezing machine (Leica) according to the manufacturer’s instructions. The liposome solutions were processed similarly. Adherent mammalian cells were either frozen either after culturing on a piece of gold foil (20 μm thick; Nilaco) or after detached from the substrate by a brief treatment with trypsin and ethylenediaminetetraacetic acid (EDTA) and pelleted in CO2-Independent Medium (Thermo Fisher Scientific) as described for yeast. The frozen specimens were transferred to a cold stage of a BAF 400 apparatus (Balzers) and freeze-fractured at −115 °C to −105 °C under a vacuum of ∼1 × 10−6 mbar. Replicas were made by the electron beam evaporation of carbon (2 nm to 5 nm thick), followed by platinum/carbon (Pt/C) (2 nm thick), and then by carbon (20 nm thick) as described previously (55).
Thawed replicas were treated with 2.5% SDS in 0.1 M Tris⋅HCl (pH 8.0) at 60 °C overnight. The yeast cell walls were removed by treating the replicas for 2 h at 30 °C with 0.5% Westase (Takara Bio) in McIlvain citrate-phosphate buffer (pH 6.0) containing 10 mM EDTA, 30% FCS, and a protease inhibitor mixture (Nacalai), followed by treatment at 60 °C overnight with 2.5% SDS. The cleaned replicas were stored at −20 °C in 0.1 M Tris⋅HCl buffer (pH 8.0) containing 50% glycerol.
Labeling of Freeze-Fracture Replicas.
Freeze-fracture replicas were washed with phosphate-buffered saline (PBS) containing 0.1% Triton X-100 (PBST). After blocking nonspecific sites with a mixture of 3% bovine serum albumin (BSA) and 2% cold fish gelatin in 0.1 M sodium phosphate buffer (pH 6.0) (PB), the samples were incubated at 4 °C overnight with 0.15 μg/mL GST-2xPH or GST-2xPH(K20E) in 0.1 M PB containing 1% BSA and 1% cold fish gelatin. The samples were then successively incubated in PBS containing 1% BSA at 37 °C for 30 min each with a rabbit anti-GST Ab (2.5 μg/mL) and with 50-fold diluted colloidal gold (10 nm)-conjugated Protein A (PAG10; The University Medical Center Utrecht).
For the double labeling of PtdSer and either POR or Rab11, PtdSer was labeled by sequentially incubating the samples with GST-2xPH, 10 μg/mL mouse anti-GST Ab, and 20-fold diluted colloidal gold (6 nm)-conjugated goat anti-mouse IgG, whereas POR and Rab11 were labeled by a rabbit Ab against POR (10 μg/mL) or against Rab11 (2.5 μg/mL), followed by incubation with 20-fold diluted colloidal gold (12 nm)-conjugated goat anti-rabbit Ab.
The labeled replicas were picked up on formvar-coated EM grids and observed with a JEM-1011 electron microscope (JEOL) operated at 100 kV. Digital images were captured by a charge-coupled device camera (Gatan) and subjected to further analysis.
Statistical Analysis.
To obtain the labeling density, the number of colloidal gold particles was counted manually, and the area was measured using ImageJ (NIH). Statistical differences between samples were examined using the Student’s t test or Mann−Whitney U test. More than 10 areas were chosen randomly for each membrane leaflet in each experiment, and more than three independent experiments were performed.
Detailed materials and methods are available in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank T. Kitamura (Institute of Medical Science, University of Tokyo) for the PLAT-E cells and pMXs-IP vector, and technical staff in the Division for Medical Research Engineering, Nagoya University Graduate School of Medicine, for technical support. This study was supported by Grants-in-Aid for Scientific Research from the Japan Society of the Promotion of Science to T.Tsuji (17K15544), K.S. (16H01360 and 17H05506), S.N. (15H05785), and T.F. (15H05902 and 18H04023).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1822025116/-/DCSupplemental.
References
- 1.Holthuis J. C., Menon A. K., Lipid landscapes and pipelines in membrane homeostasis. Nature 510, 48–57 (2014). [DOI] [PubMed] [Google Scholar]
- 2.van Meer G., Voelker D. R., Feigenson G. W., Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kay J. G., Grinstein S., Phosphatidylserine-mediated cellular signaling. Adv. Exp. Med. Biol. 991, 177–193 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Bevers E. M., Comfurius P., van Rijn J. L., Hemker H. C., Zwaal R. F., Generation of prothrombin-converting activity and the exposure of phosphatidylserine at the outer surface of platelets. Eur. J. Biochem. 122, 429–436 (1982). [DOI] [PubMed] [Google Scholar]
- 5.Martin S. J., et al. , Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: Inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 182, 1545–1556 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki J., et al. , Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J. Biol. Chem. 288, 13305–13316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki J., Umeda M., Sims P. J., Nagata S., Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468, 834–838 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Suzuki J., Denning D. P., Imanishi E., Horvitz H. R., Nagata S., Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 341, 403–406 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Gyobu S., Ishihara K., Suzuki J., Segawa K., Nagata S., Characterization of the scrambling domain of the TMEM16 family. Proc. Natl. Acad. Sci. U.S.A. 114, 6274–6279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gyobu S., et al. , A role of TMEM16E carrying a scrambling domain in sperm motility. Mol. Cell. Biol. 36, 645–659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bethel N. P., Grabe M., Atomistic insight into lipid translocation by a TMEM16 scramblase. Proc. Natl. Acad. Sci. U.S.A. 113, 14049–14054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bushell S. R., et al. , The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K. bioRχiv 10.1101/447417 (12 December 2018). [DOI] [PMC free article] [PubMed]
- 13.Bolduc V., et al. , Recessive mutations in the putative calcium-activated chloride channel Anoctamin 5 cause proximal LGMD2L and distal MMD3 muscular dystrophies. Am. J. Hum. Genet. 86, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hicks D., et al. , A founder mutation in Anoctamin 5 is a major cause of limb-girdle muscular dystrophy. Brain 134, 171–182 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahjneh I., et al. , A new distal myopathy with mutation in anoctamin 5. Neuromuscul. Disord. 20, 791–795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balreira A., et al. , ANO10 mutations cause ataxia and coenzyme Q10 deficiency. J. Neurol. 261, 2192–2198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renaud M., et al. , Autosomal recessive cerebellar ataxia type 3 due to ANO10 mutations: Delineation and genotype-phenotype correlation study. JAMA Neurol. 71, 1305–1310 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Vermeer S., et al. , Targeted next-generation sequencing of a 12.5 Mb homozygous region reveals ANO10 mutations in patients with autosomal-recessive cerebellar ataxia. Am. J. Hum. Genet. 87, 813–819 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uchida Y., et al. , Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc. Natl. Acad. Sci. U.S.A. 108, 15846–15851 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeung T., et al. , Membrane phosphatidylserine regulates surface charge and protein localization. Science 319, 210–213 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Kay J. G., Koivusalo M., Ma X., Wohland T., Grinstein S., Phosphatidylserine dynamics in cellular membranes. Mol. Biol. Cell 23, 2198–2212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fairn G. D., et al. , High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J. Cell Biol. 194, 257–275 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takatori S., Mesman R., Fujimoto T., Microscopic methods to observe the distribution of lipids in the cellular membrane. Biochemistry 53, 639–653 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Lee S., et al. , Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J. 34, 669–688 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsudaira T., et al. , Endosomal phosphatidylserine is critical for the YAP signalling pathway in proliferating cells. Nat. Commun. 8, 1246 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letts V. A., Klig L. S., Bae-Lee M., Carman G. M., Henry S. A., Isolation of the yeast structural gene for the membrane-associated enzyme phosphatidylserine synthase. Proc. Natl. Acad. Sci. U.S.A. 80, 7279–7283 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zachowski A., Phospholipids in animal eukaryotic membranes: Transverse asymmetry and movement. Biochem. J. 294, 1–14 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng J., et al. , Yeast and mammalian autophagosomes exhibit distinct phosphatidylinositol 3-phosphate asymmetries. Nat. Commun. 5, 3207 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Iyoshi S., et al. , Asymmetrical distribution of choline phospholipids revealed by click chemistry and freeze-fracture electron microscopy. ACS Chem. Biol. 9, 2217–2222 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Zinser E., et al. , Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 173, 2026–2034 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuge O., Nishijima M., Akamatsu Y., A Chinese hamster cDNA encoding a protein essential for phosphatidylserine synthase I activity. J. Biol. Chem. 266, 24184–24189 (1991). [PubMed] [Google Scholar]
- 32.Kuge O., Saito K., Nishijima M., Cloning of a Chinese hamster ovary (CHO) cDNA encoding phosphatidylserine synthase (PSS) II, overexpression of which suppresses the phosphatidylserine biosynthetic defect of a PSS I-lacking mutant of CHO-K1 cells. J. Biol. Chem. 272, 19133–19139 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Hankins H. M., Baldridge R. D., Xu P., Graham T. R., Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic 16, 35–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammond G. R., Balla T., Polyphosphoinositide binding domains: Key to inositol lipid biology. Biochim. Biophys. Acta 1851, 746–758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung J., et al. , INTRACELLULAR TRANSPORT. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science 349, 428–432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moser von Filseck J., et al. , INTRACELLULAR TRANSPORT. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science 349, 432–436 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Kuge O., Nishijima M., Akamatsu Y., Phosphatidylserine biosynthesis in cultured Chinese hamster ovary cells. III. Genetic evidence for utilization of phosphatidylcholine and phosphatidylethanolamine as precursors. J. Biol. Chem. 261, 5795–5798 (1986). [PubMed] [Google Scholar]
- 38.Yamashita S., Nikawa J., Phosphatidylserine synthase from yeast. Biochim. Biophys. Acta 1348, 228–235 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Tomohiro S., Kawaguti A., Kawabe Y., Kitada S., Kuge O., Purification and characterization of human phosphatidylserine synthases 1 and 2. Biochem. J. 418, 421–429 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Zborowski J., Dygas A., Wojtczak L., Phosphatidylserine decarboxylase is located on the external side of the inner mitochondrial membrane. FEBS Lett. 157, 179–182 (1983). [DOI] [PubMed] [Google Scholar]
- 41.Friedman J. R., et al. , Lipid homeostasis is maintained by dual targeting of the mitochondrial PE biosynthesis enzyme to the ER. Dev. Cell 44, 261–270.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trotter P. J., Voelker D. R., Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270, 6062–6070 (1995). [DOI] [PubMed] [Google Scholar]
- 43.Voelker D. R., Phosphatidylserine translocation to the mitochondrion is an ATP-dependent process in permeabilized animal cells. Proc. Natl. Acad. Sci. U.S.A. 86, 9921–9925 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Achleitner G., et al. , Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur. J. Biochem. 264, 545–553 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Malvezzi M., et al. , Ca2+-dependent phospholipid scrambling by a reconstituted TMEM16 ion channel. Nat. Commun. 4, 2367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pomorski T. G., Menon A. K., Lipid somersaults: Uncovering the mechanisms of protein-mediated lipid flipping. Prog. Lipid Res. 64, 69–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsuchiya M., et al. , Cell surface flip-flop of phosphatidylserine is critical for PIEZO1-mediated myotube formation. Nat. Commun. 9, 2049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Mark V. A., et al. , Phospholipid flippases attenuate LPS-induced TLR4 signaling by mediating endocytic retrieval of Toll-like receptor 4. Cell. Mol. Life Sci. 74, 715–730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verkleij A. J., Post J. A., Membrane phospholipid asymmetry and signal transduction. J. Membr. Biol. 178, 1–10 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Schreiber R., et al. , Anoctamins support calcium-dependent chloride secretion by facilitating calcium signaling in adult mouse intestine. Pflugers Arch. 467, 1203–1213 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Falzone M. E., Malvezzi M., Lee B. C., Accardi A., Known structures and unknown mechanisms of TMEM16 scramblases and channels. J. Gen. Physiol. 150, 933–947 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kramer J., Hawley R. S., The spindle-associated transmembrane protein Axs identifies a membranous structure ensheathing the meiotic spindle. Nat. Cell Biol. 5, 261–263 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D., Protein sorting in Saccharomyces cerevisiae: Isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936–4948 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujita A., Cheng J., Fujimoto T., Quantitative electron microscopy for the nanoscale analysis of membrane lipid distribution. Nat. Protoc. 5, 661–669 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.