Significance
Current methods for CRISPR/Cas9-mediated gene knockout in avian species require isolation, culture, and genome modification of primordial germ cells in vitro for the subsequent injection into the extraembryonic blood vessel of the egg. These processes require highly skilled techniques and the establishment of optimized conditions. In the present study, the adenoviral CRISPR/Cas9 vector, optimized for expression of gRNA and the Cas9 gene in avian cells, was injected into the quail blastoderm in newly laid eggs. The resulting chimeras generated offspring with targeted mutations in the melanophilin gene, resulting in gray plumage of homozygous mutant quail. Implementation of this simplified avian knockout process can accelerate avian knockout studies involved in diverse areas of research and lead to the advancement of future agricultural applications.
Keywords: adenovirus, CRISPR/Cas9, poultry, genome editing, MLPH
Abstract
Zygotes at the 1-cell stage have been genetically modified by microinjecting the CRISPR/Cas9 components for the generation of targeted gene knockout in mammals. In the avian species, genetic modification of the zygote is difficult because its unique reproductive system limits the accessibility of the zygote at the 1-cell stage. To date, only a few CRISPR/Cas9-mediated gene knockouts have been reported using the chicken as a model among avian species, which requires 3 major processes: isolation and culture of primordial germ cells (PGCs), modification of the genome of PGCs in vitro, and injection of the PGCs into the extraembryonic blood vessel at the early embryonic stages when endogenous PGCs migrate through circulation to the genital ridge. In the present study, the adenoviral CRISPR/Cas9 vector was directly injected into the quail blastoderm in newly laid eggs. The resulting chimeras generated offspring with targeted mutations in the melanophilin (MLPH) gene, which is involved in melanosome transportation and feather pigmentation. MLPH homozygous mutant quail exhibited gray plumage, whereas MLPH heterozygous mutants and wild-type quail exhibited dark brown plumage. In addition, the adenoviral vector was not integrated into the genome of knockout quail, and no mutations were detected in potential off-target regions. This method of generating genome-edited poultry is expected to accelerate avian research and has potential applications for developing superior genetic lines for poultry production in the industry.
As a powerful and convenient genome editing technique, the CRISPR/Cas9 system has been used in various areas of scientific research. When conducting studies using food-producing animals, the CRISPR/Cas9 system can be used for the benefit of both the scientific community and the agricultural industry (1). Poultry production is a large portion of the livestock industry worldwide, and poultry can be used as an important biological research model. However, avian species have not been commonly used when conducting research using the CRISPR/Cas9 technique. In mammals, CRISPR/Cas9-mediated genome modification of the zygote has been successfully used to generate targeted gene knockout species (2) because of the easy accessibility of the zygote at the 1-cell stage. However, the same strategy does not apply to the avian species because of its unique reproductive system. In chickens, an oocyte is transported into the oviduct after ovulation, and fertilized in the initial section of the oviduct into which the oocyte is transported, the infundibulum, within 15 min after ovulation; the egg is laid within ∼24 h (3). After fertilization, the 1-cell zygote rapidly proliferates up to ∼60,000 cells at the time of egg laying (4). As a result of rapid cell proliferation of the zygote in the oviduct, it is difficult to access the zygote in vivo at the 1-cell stage for genome editing. In addition, it is challenging to surgically remove, genetically modify, and surgically transplant the zygote attached to the fragile yolk into the oviduct of the recipient. For these reasons, the current method of generating targeted gene knockouts in the avian species consists of 3 processes: isolation and culture of primordial germ cells (PGCs), modification of the genome of PGCs in vitro, and injection of genetically modified PGCs into embryos. For PGC genome modification, several knockout methods have been applied, such as homologous recombination (5, 6), TALENs (transcription activator-like effector nucleases) (7), and CRISPR/Cas9 (8, 9). However, isolation, culture, and genome manipulation of PGCs still require highly skilled expertise and the establishment of optimized conditions. In the present study, a method is described for generation of targeted gene knockout quail by injecting the recombinant adenovirus directly into the quail blastoderm.
Results
Development of the Adenoviral CRISPR/Cas9 System for Quail.
In our previous research, the commercially available mammalian CRISPR/Cas9 vector (lentiCRISPR v2; Addgene) was modified for greater expression of guide RNA (gRNA), and Cas9 in avian cells and the modified CRISPR/Cas9 vector successfully induced multiple insertion/deletion mutations in quail myogenic cells (10). In the vector, a quail 7SK (q7SK) promoter was inserted as a new RNA polymerase III promoter for the expression of gRNA, and a chicken β-actin hybrid (CBh) promoter was used to express the Cas9 gene (10). The melanophilin (MLPH) gene was selected as the target gene because melanophilin functions in melanosome transportation (11) and knockout of the MLPH gene were expected to affect feather pigmentation (12, 13), a modification visible in the phenotype. To knockout all known transcript variants of the Japanese quail MLPH gene, gRNA was designed to match sequences in exon 2, which is a component of all variants (SI Appendix, Fig. S1). To achieve highly acceptable on-target scores and minimal off-target frequencies, potential gRNAs were rigorously preselected using the procedures that are subsequently described. Initially, there was detection of the highly desirable on-target scored gRNAs, using the CRISPR design technique (https://zlab.bio/guide-design-resources). The relatively lesser off-target frequencies of the potential gRNAs were subsequently screened using the BLAST Genome search of the Japanese quail genome sequence (Coturnix japonica 2.0 assembly; GenBank Assembly ID GCA_001577835.1) in the PubMed Genome database (https://www.ncbi.nlm.nih.gov/genome). These optimized gRNA and Cas9 expression cassettes were subsequently integrated into the commercially available adenoviral shuttle vector (Adeno Cas9; Addgene) (14).
Generation of MLPH Mutant Quail.
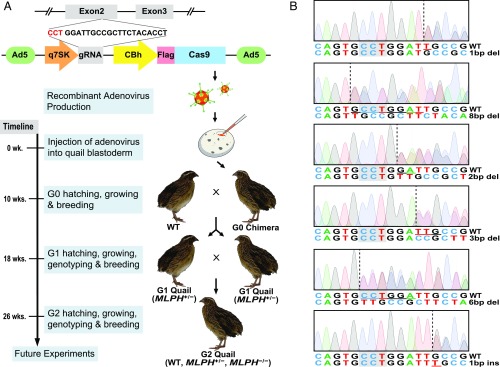
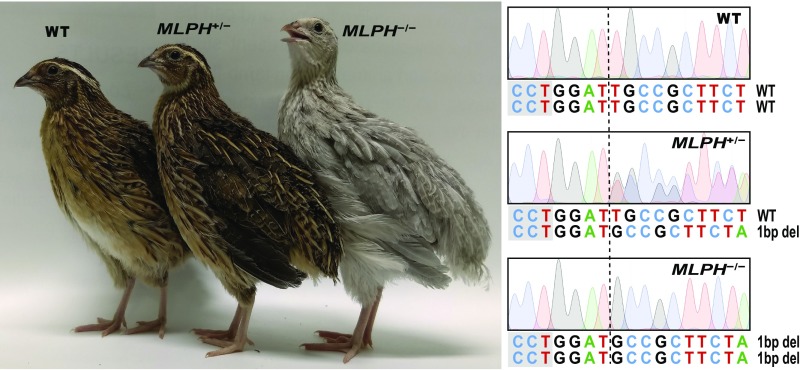
After the production of the recombinant adenovirus type 5 containing the CRISPR/Cas9 vector, the adenovirus was injected into the subgerminal cavity of the quail blastoderm at Eyal-Giladi and Kochav (EGK) (15) stage XI (Fig. 1A). Eleven chimeras (G0) were hatched from 100 injected quail eggs. The germline mutation and transmission of the chimeras were subsequently assessed by breeding chimeric quail with wild-type quail and evaluating the genotype of the offspring (G1) from each line. From the 11 lines, there were 5 chimeric quail that produced MLPH heterozygous mutant (MLPH+/−) quail, resulting in a 45% success rate of generating chimeras with germline transmissible mutations. The efficiencies of germline transmission derived from each chimeric quail ranged from 2.4% to 10% (Table 1). Notably, one chimera produced progeny with 2 different MLPH mutations, indicating there was germline chimerism. A total of 6 different MLPH mutant genotypes were verified using the Sanger sequencing analysis after PCR amplification of the target region (Fig. 1B). To generate MLPH homozygous mutant (MLPH−/−) quail (G2), male and female MLPH+/− quail (G1) were subsequently mated (Fig. 1A). An obvious phenotypic difference was apparent, with MLPH−/− quail exhibiting gray plumage compared with dark brown plumage exhibited by the wild-type and MLPH+/−quail (Fig. 2).
Fig. 1.
Generation of targeted gene knockout quail using adenovirus. (A) Schematic representation of research plan. gRNA of the MLPH gene was selected from exon 2, and the adenoviral CRISPR/Cas9 vector was constructed. Recombinant adenovirus was subsequently produced and injected into the quail blastoderm. Chimeras (G0) were maintained and mated with wild-type (WT) quail to produce G1 quail with a heterozygous genotype of the MLPH gene (MLPH+/−). Male and female G1 MLPH+/− quail were subsequently mated to generate G2 quail with a homozygous genotype of the MLPH gene (MLPH−/−). After production of the recombinant adenovirus, it took a total of 10 wk, 2 wk for egg incubation after virus injection and another 8 wk for sexual maturation, to receive eggs from G0 quail. After G1 quail hatched, the MLPH+/− quail were screened and mated to produce G2 offspring 8 wk later. Fully grown G2 offspring were obtained for further experiments 26 wk after the injection of the adenovirus. (B) Sanger sequencing chromatograms of G1 MLPH+/− quail. Dashed lines indicate the starting point of the mutation, and the deleted or inserted nucleotides are underlined. The PAM sequences are highlighted in gray. Nucleotide sequences are presented in a negative direction.
Table 1.
Efficiency of germline transmission derived from chimeric quail
| Chimeric quail ID | Number of MLPH mutant offspring (%) | Genotype of mutant offspring |
| 9339 | 1/19 (5.3%) | 2 bp deletion |
| 9341 | 2/34 (5.9%) | 6 bp deletion, 1 bp insertion |
| 9343 | 1/10 (10%) | 1 bp deletion |
| 9347 | 1/41 (2.4%) | 8 bp deletion |
| 9348 | 1/36 (2.8%) | 3 bp deletion |
Fig. 2.
Phenotypic and genotypic comparison of G2 offspring. (Left) WT and MLPH+/− quail with dark brown plumage and MLPH−/− quail with gray plumage without any change in plumage pattern. (Right) DNA sequencing chromatograms of MLPH+/− and MLPH−/− quail, which indicate there is a one base pair deletion of one allele and both alleles, respectively. Dashed line indicates the point where one base pair deletion occurred. The PAM sequences are highlighted in gray. Nucleotide sequences are presented in a negative direction.
Analysis of Adenoviral Vector Integration and Off-Target Mutations.
To confirm the adenoviral CRISPR/Cas9 vector transduced the foreign DNA episomally in the nucleus without genome integration, an attempt was made to amplify the viral vector by PCR, using genomic DNAs from all mutant quail lines. As expected, the viral vector was not amplified using PCR, indicating the viral vector was not integrated into the host’s genome (SI Appendix, Fig. S2). Subsequently, the genome was assessed for potential off-target mutations in knockout quail lines. Potential off-target sites were screened on the basis of high homologous scores, using a NCBI Genome BLAST tool, and the 6 closest off-target sites were selected. These off-target sites shared 12–15 matched nucleotides with 11–13 consecutive matched sequences of the 20-base pair gRNA sequences (SI Appendix, Table S1). The PCR amplification of the potential off-target regions using genomic DNAs from every line of mutant quail was conducted. Sanger sequencing of the PCR products did not identify any mutations in the potential off-target regions of the mutant quail lines.
Transduction Assessment of Adenovirus Type 5 in Other Avian Cells.
To explore whether the approaches used in the present study for genome editing can be used in other poultry species, the efficiency of adenovirus-mediated gene expression was examined. In our previous cell culture study, a highly desirable transduction efficiency and gene expression of the adenovirus type 5 in quail and chicken embryonic cells were confirmed (16). In addition, we investigated the transduction efficiency of the adenovirus in turkey embryonic cells (SI Appendix, Fig. S3). Recombinant adenovirus type 5 containing the green fluorescent protein (GFP) gene under a cytomegalovirus promoter was transduced into turkey embryonic muscle cells, and strong expression of GFP was observed.
Discussion
The present study reports the development of an effective approach for the CRISPR/Cas9-mediated targeted gene knockout in the avian species without using in vitro genetic modification of PGCs. Instead of modifying PGCs in vitro, the recombinant adenovirus type 5 containing the optimized CRISPR/Cas9 vector for quail cells was directly injected into the EGK stage XI quail blastoderm in the newly laid eggs (17). At the time of injection, there are only 2–3 PGCs (18) among 32,500–46,250 cells in the quail blastoderm (17), and the PGCs rapidly proliferate up to more than 100 cells within 18 h of egg incubation (18). The influence of PGC proliferation during the early development of the blastoderm, in addition to the rate and timing of viral infection, may have affected the germline transmission efficiencies (2.4–10%) among knockout quail lines in the present study. The genotypes of G2 progeny exhibited Mendelian inheritance patterns, indicating the MLPH knockout did not affect viability of the embryos. From all mutant lines, MLPH−/− quail exhibited gray plumage; however, MLPH+/− quail exhibited dark brown plumage similar to wild-type quail. The gray plumage color of MLPH−/− quail clearly demonstrates the phenotype of the autosomal recessive inheritance.
In addition, there were no differences in body weight among male quail from wild-type, MLPH+/−, and MLPH−/− quail genotypes when the body weight of male quail plateaued at 7 wk of age (19). The body weight of female quail was not assessed because the timing of daily egg production caused a significant fluctuation in body weight (20). Furthermore, mating of MLPH−/− pairs resulted in the production of MLPH−/− offspring. These data indicate the knockout of the MLPH gene does not affect the growth and reproductive capacity of MLPH−/− quail.
The lentiviral vector has been primarily used for the generation of transgenic birds (21–26) because of its effectiveness to integrate transgenes into the host’s genome (27), and as a consequence, transfer transgenes onto offspring of the subsequent generation. However, this particular advantage of the lentivirus for the generation of transgenic birds may not be desirable for the generation of the CRISPR/Cas9-mediated gene knockout birds because integration of the CRISPR/Cas9 vector would result in a sustained expression of gRNA and Cas9 and can result in an increased probability of off-target mutations. In support of this, results from a recent study demonstrated that the integrase-competent lentiviral vector resulted in more off-target mutations compared with the use of the integrase-deficient lentiviral vector in the human embryonic kidney (HEK293T) cells (28). Therefore, the replication-defective adenovirus type 5 (27) was selected as a delivery system for gRNA and Cas9 expression cassettes because of the advantage of episomal transduction of the nonintegrating viral vector (27, 29), and the transient expression of transgenes resulting from clearance of the adenoviral vector as a result of the host’s innate immune system (30, 31). In addition, nonintegrated foreign DNA is diluted and not propagated, as cells proliferate rapidly (29) in the blastodermal developmental stage. Considering these characteristics of adenovirus-mediated gene delivery, the adenoviral vector may be a more desirable delivery system that prevents potential off-target mutations because of temporary expression of gRNA and Cas9.
The present study has established a convenient knockout system for use in avian species by directly injecting the adenoviral CRISPR/Cas9 vector into the EGK stage XI quail blastoderm without in vitro processes for genome modification of PGCs. Therefore, this system is potentially applicable in the avian species from which isolating, maintaining, and genetically modifying PGCs in culture has not been successful. In addition, the blastoderm is durable and wide enough to be injected with the adenoviral vector. Furthermore, precise incubation time is not required because quail eggs contain the EGK stage XI blastoderm after oviposition (17). When newly laid chicken eggs are incubated, PGCs migrate from the central area to the genital crescent within the embryo, and subsequently migrate toward the genital ridge through the extraembryonic blood vessel (32). At Hamburger and Hamilton (33) stage 13–15, when PGCs migrate through circulation (32), a lentiviral vector (34) or transfection reagent (35) was injected into the extraembryonic blood vessel to generate transgenic birds. These results suggest that migrating PGCs might be permissive for adenovirus-mediated genome modification. However, our method may not be applicable for gene knockin of the target region of the avian genome, which can be accomplished by inducing gene knockin in cultured PGCs and injecting the knockin cells into the extraembryonic blood vessel at Hamburger and Hamilton stage 13–15 (36). Therefore, both conventional avian genome modification using cultured PGCs and our avian knockout method using adenovirus can be considered for future studies, depending on the availability of established methods for PGC isolation and culture, and types of genome modifications (knockout or knockin).
Direct injection of the adenoviral CRISPR/Cas9 vector into the blastoderm provides a rapid way to generate knockout birds by avoiding the need for PGC culture and genome modification in vitro before injection into the embryo. In addition, this method can be broadly applied to other poultry species because of the high transduction efficiency of the adenovirus into other avian cells, including the chicken (16) and turkey (SI Appendix, Fig. S3). Furthermore, episomal transduction and transient gene expression of the adenovirus (27, 29) can provide an efficient and safe approach to generate targeted gene knockout in birds with a minimal risk for off-target mutations. The method used in the present study allows avian knockout studies in broad research areas to be conducted for the advancement of developing more productive genomic lines of poultry.
Materials and Methods
Animal Care.
All experimental procedures and the animal care protocol were approved by the Institutional Animal Care and Use Committee at The Ohio State University (Protocol 2015A00000013-R1). Experimental quail were maintained at The Ohio State University Poultry Facilities in Columbus and Wooster, Ohio, until birds were killed at 5 mo old via CO2 inhalation.
Adenoviral Shuttle Vector Construction.
Previously, an optimized CRISPR/Cas9 vector was generated for the expression of MLPH gRNA and Cas9 under the regulation of a quail 7SK promoter and CBh promoter, respectively, by customizing a commercially available lentiviral CRISPR/Cas9 vector (lentiCRISPR v2, Addgene) (10). The gRNA and Cas9 expression cassettes were moved to a commercially available adenoviral shuttle vector (Adeno Cas9; Addgene) (14). This was conducted by replacing a human U6 promoter-gRNA-CBh fragment in the adenoviral shuttle vector with the quail 7SK promoter-MLPH gRNA-CBh fragment using XhoI and AgeI restriction enzyme sites. After confirming the insertion of the gRNA and Cas9 expression cassettes into the shuttle vector by Sanger sequencing at The Ohio State University Comprehensive Cancer Center, the confirmed adenoviral shuttle vector was used for production and concentration of the recombinant adenovirus type 5 (ViraQuest Inc). The final titer of the recombinant adenovirus was 3.0 × 1010 PFU/mL.
Egg Injection of the Adenovirus.
Wild-type Japanese quail eggs were sanitized using 70% ethanol and positioned upside down for 2 h and laterally for another 2 h to position quail blastoderm on the lateral apex of the eggs. Microneedles were prepared by sharpening and grinding 50 µL microcapillary tubes (Drummond), using a micropipette puller (PC-100, Narishige) and a micropipette grinder (EG-44, Narishige). The microneedle was attached to a microinjector (MINJ-PD, Tritech Research). The inner portion of the microneedle was sanitized with 70% ethanol and subsequently washed with PBS before loading the adenovirus particle into the microneedle. A small opening was subsequently constructed on the lateral apex of the egg shell, using fine-tip tweezers, and 2 µL adenovirus was injected into the middle of the quail blastoderm through the constructed opening under the visual support area of a stereomicroscope (SZ61, Olympus). The eggs were sealed with a paraffin film, and the pointed end of the eggs was placed down and incubated in an automatic egg incubator (Type 65Hs, Masalles) with a temperature of 37.5 °C and 60% relative humidity. The eggs were placed on rotating trays for 14 d and then transferred to hatching trays for 3 d until hatching occurred. After feathers of newly hatched chicks were dried in the incubator, the chicks were tagged and transferred to brooder cages. A total of 11 potential chimeras (G0) were maintained until sexually mature, and mated with wild-type quail in separate breeding cages.
Genomic DNA Extraction.
Eggs from 11 G0 chimeric quail lines were collected daily and stored in a cooler (20 °C) until being placed in the egg incubator at weekly intervals. After G1 chicks were hatched, the chicks were tagged and transferred into brooder cages. When G1 quail were 3 wk old, blood was extracted from the brachial vein of the wing, using a Monoject 1/2 mL insulin syringe (Covidien). To obtain genomic DNA from blood, 400 µL of cell lysis solution (200 mM NaCl, 50 mM Tris⋅Cl, 10 mM EDTA, 1% SDS at pH 8.0) containing proteinase K solution (0.1 mg/mL, Invitrogen) was added to 3 µL blood, and the blood was incubated at 55 °C in a heat block for at least 4 h. Subsequently, 400 µL phenol:chloroform:isoamyl alcohol (25:24:1) (Sigma) was added, the mixture was vortexed vigorously, and the mixture was centrifuged at 13,000 × g for 10 min. After the clear supernatant was separated, 240 µL of the supernatant was transferred carefully to a different 1.5-mL tube, and 80 µL ammonium acetate solution (Sigma) and 240 µL 2-Propanol (Fisher Scientific) were added to precipitate genomic DNA. The mixture was inverted 10 times and centrifuged at 13,000 × g for 5 min. After removal of the supernatant, a pellet was washed with 300 µL 70% ethanol, followed by centrifuging and drying. The dried pellet was dissolved with 30 µL TE buffer containing RNase A (10 µg/mL, Qiagen).
Detection of Mutation in the MLPH Gene.
On the basis of the Japanese quail genome sequence (Coturnix japonica 2.0 assembly; GenBank Assembly ID GCA_001577835.1), 2 primer sets were designed for PCR amplification of the MLPH gRNA locus (SI Appendix, Table S2). The Taq DNA Polymerase (New England Biolabs) was used for PCR amplification, and PCR was performed with an initial incubation at 95 °C for 90 s, followed by 40 cycles at 95 °C for 30 s, 54 °C for 40 s, and 68 °C for 60 s. The final extension occurred at 68 °C for 5 min. The reaction mixture without genomic DNA was used as a negative control. Using an agarose gel electrophoresis system (Bio-Rad), PCR products were processed at 140 v for 20 min to separate the bands. The bands were extracted using a QIAquick Gel Extraction Kit (Qiagen), according to the manufacturer’s protocol, and the extracted PCR samples were sent to The Ohio State University Comprehensive Cancer Center for Sanger sequencing. After sequencing analysis, MLPH+/− quail were maintained and mated with other MLPH+/− quail to produce MLPH−/− offspring. Eggs from MLPH+/− parents were collected daily, labeled, and stored in a cooler. Every week, the eggs were transferred to the egg incubator and newly hatched chicks were tagged and placed in brooder cages. Genomic DNA from blood was extracted and the target region in the MLPH gene was analyzed after PCR amplification, using Sanger sequencing to confirm the MLPH−/− genotype.
Analysis of Off-Target Mutation and Chromosomal Integration of the Viral Vector.
A BLAST Genome search in the PubMed Genome database (https://www.ncbi.nlm.nih.gov/genome) of 23 nucleotide sequences containing the MLPH gRNA and protospacer adjacent motif (PAM) motif, 5′-NGG-3′, for the Japanese quail genome sequence was performed to identify potential off-target sites. Among candidates having PAM sequences, -5′-NGG-3′, 4 sites with the highest homology scores and 2 sites with the second highest homology scores were selected (SI Appendix, Table S1). Primers for each of the off-target regions were designed (SI Appendix, Table S2), and PCR amplification of 6 off-target regions was conducted, using genomic DNA of 6 MLPH+/− quail lines. The PCR was conducted with an initial incubation at 95 °C for 90 s, followed by 40 cycles at 95 °C for 40 s, 56 °C for 40 s, and 68 °C for 30 s. The final extension was performed at 68 °C for 5 min. The sequence analysis of PCR products was subsequently performed. In addition, integration of the viral vector sequence into the chromosomes of all lines of MLPH+/− quail (G1) was analyzed using PCR amplification. Primers were designed to be located at the 3′ end portion of the CBh promoter and the 5′ end portion of the Cas9 sequence (SI Appendix, Table S2). Furthermore, the first exon of the myostatin gene was amplified as a positive control for integrity of extracted genomic DNA from each sample.
Cell Culture.
Pectoralis major muscle tissues were collected from embryonic turkeys at embryonic day 11. The embryonic tissues were finely minced in PBS, using an autoclaved razor blade, and dissociated with 0.05% trypsin-EDTA (Invitrogen) for 5 min at room temperature. The dissociated cells were filtered through a 100-µm cell strainer (BD Biosciences) and centrifuged to remove trypsin-EDTA. After disrupting the cell pellet, cells were seeded in 35-mm tissue culture plates containing growth media, including DMEM (GIBCO), 10% FBS (GIBCO), penicillin (100 U/mL), and streptomycin sulfate (100 g/mL; GIBCO). Before cell culture, the plates were coated with 0.01% calf skin collagen for 30 min at 37 °C and washed twice with PBS.
Feasibility Assessment of Adenovirus Type 5 in Primary Turkey Cells.
To assess the feasibility of the adenovirus type 5 in primary turkey myocytes, a commercially available empty adenoviral shuttle vector containing GFP under the control of the cytomegalovirus promoter (pAdTrack-cytomegalovirus; ATCC) was used. Homologous recombination of the shuttle vector with the adenoviral backbone, pAdEasy-1 (ATCC), was conducted using Escherichia. coli BJ5183 (Stratagene), as described in a previous report (37). Recombinant adenoviral plasmids were transfected into AD-293 cells (Stratagene) after screening plasmids with correct homologous recombination, as described in a previous report (37). The cytopathic effect assay was used to analyze titer of the recombinant adenovirus, using procedures that have been previously published (38). The final titer of the recombinant adenovirus was 1.6 × 1011 pfu/mL, and the virus was transduced into primary turkey myocytes (multiplicity of infection, 7,000). After overnight incubation of the myocytes with the adenovirus, the media was changed to fresh growth media and GFP gene expression was determined 3 d after transduction using an inverted fluorescent microscope at 100× or 200× magnifications (Olympus IX50, Olympus).
Supplementary Material
Acknowledgments
We thank Jonah Perkins for quail management, Jonghyun Shin for adenoviral transduction of turkey myocytes, and Taewan Kim for illustration. This research was supported in part by the U.S. Department of Agriculture National Institute of Food and Agriculture Grant (Project No. 2016-08413), and the Ohio Agricultural Research and Development Center Research SEEDS funding (Project No. 2017-069). We acknowledge the Addgene company and Dr. Andrea Ventura for supplying us with the Adeno Cas9 (#64072) plasmid.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903230116/-/DCSupplemental.
References
- 1.Lamas-Toranzo I., et al. , CRISPR is knocking on barn door. Reprod. Domest. Anim. 52 (suppl. 4), 39–47 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Hsu P. D., Lander E. S., Zhang F., Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sang H., Prospects for transgenesis in the chick. Mech. Dev. 121, 1179–1186 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Pokhrel N., Ben-Tal Cohen E., Genin O., Sela-Donenfeld D., Cinnamon Y., Cellular and morphological characterization of blastoderms from freshly laid broiler eggs. Poult. Sci. 96, 4399–4408 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Schusser B., et al. , Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc. Natl. Acad. Sci. U.S.A. 110, 20170–20175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimitrov L., et al. , Germline gene editing in chickens by efficient CRISPR-mediated homologous recombination in primordial germ cells. PLoS One 11, e0154303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park T. S., Lee H. J., Kim K. H., Kim J.-S., Han J. Y., Targeted gene knockout in chickens mediated by TALENs. Proc. Natl. Acad. Sci. U.S.A. 111, 12716–12721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oishi I., Yoshii K., Miyahara D., Kagami H., Tagami T., Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci. Rep. 6, 23980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park T. S., Park J., Lee J. H., Park J.-W., Park B.-C., Disruption of G0/G1 switch gene 2 (G0S2) reduced abdominal fat deposition and altered fatty acid composition in chicken. FASEB J. 33, 1188–1198 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Ahn J., et al. , Targeted genome editing in a quail cell line using a customized CRISPR/Cas9 system. Poult. Sci. 96, 1445–1450 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Matesic L. E., et al. , Mutations in Mlph, encoding a member of the Rab effector family, cause the melanosome transport defects observed in leaden mice. Proc. Natl. Acad. Sci. U.S.A. 98, 10238–10243 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaez M., et al. , A single point-mutation within the melanophilin gene causes the lavender plumage colour dilution phenotype in the chicken. BMC Genet. 9, 7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bed’hom B., et al. , The lavender plumage colour in Japanese quail is associated with a complex mutation in the region of MLPH that is related to differences in growth, feed consumption and body temperature. BMC Genomics 13, 442 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maddalo D., et al. , In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516, 423–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eyal-Giladi H., Kochav S., From cleavage to primitive streak formation: A complementary normal table and a new look at the first stages of the development of the chick. I. General morphology. Dev. Biol. 49, 321–337 (1976). [DOI] [PubMed] [Google Scholar]
- 16.Shin J., et al. , Technical note: A gene delivery system in the embryonic cells of avian species using a human adenoviral vector. J. Anim. Sci. 87, 2791–2795 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Stepińska U., Olszańska B., Cell multiplication and blastoderm development in relation to egg envelope formation during uterine development of quail (Coturnix coturnix japonica) embryo. J. Exp. Zool. 228, 505–510 (1983). [Google Scholar]
- 18.Pardanaud L., Buck C., Dieterlen-Lièvre F., Early germ cell segregation and distribution in the quail blastodisc. Cell Differ. 22, 47–59 (1987). [DOI] [PubMed] [Google Scholar]
- 19.Choi Y. M., Suh Y., Shin S., Lee K., Skeletal muscle characterization of Japanese quail line selectively bred for lower body weight as an avian model of delayed muscle growth with hypoplasia. PLoS One 9, e95932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang S., et al. , Loss of fat with increased adipose triglyceride lipase-mediated lipolysis in adipose tissue during laying stages in quail. Lipids 48, 13–21 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Shin S., Choi Y. M., Han J. Y., Lee K., Inhibition of lipolysis in the novel transgenic quail model overexpressing G0/G1 switch gene 2 in the adipose tissue during feed restriction. PLoS One 9, e100905 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao D., et al. , Expression of recombinant human lysozyme in egg whites of transgenic hens. PLoS One 10, e0118626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn J., et al. , Identification of the avian RBP7 gene as a new adipose-specific gene and RBP7 promoter-driven GFP expression in adipose tissue of transgenic quail. PLoS One 10, e0124768 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu T., et al. , Oviduct-specific expression of human neutrophil defensin 4 in lentivirally generated transgenic chickens. PLoS One 10, e0127922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen P. R., et al. , Overexpression of G0/G1 switch gene 2 in adipose tissue of transgenic quail inhibits lipolysis associated with egg laying. Int. J. Mol. Sci. 17, 384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodfint R. M., et al. , Identification of the MUC2 promoter as a strong promoter for intestinal gene expression through generation of transgenic quail expressing GFP in gut epithelial cells. Int. J. Mol. Sci. 18, 196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee C. S., et al. , Adenovirus-mediated gene delivery: Potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 4, 43–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortinski P. I., O’Donovan B., Dong X., Kantor B., Integrase-deficient lentiviral vector as an all-in-one platform for highly efficient CRISPR/Cas9-mediated gene editing. Mol. Ther. Methods Clin. Dev. 5, 153–164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Athanasopoulos T., Munye M. M., Yáñez-Muñoz R. J., Nonintegrating gene therapy vectors. Hematol. Oncol. Clin. North Am. 31, 753–770 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Verma I. M., Somia N., Gene therapy–Promises, problems and prospects. Nature 389, 239–242 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Nayak S., Herzog R. W., Progress and prospects: Immune responses to viral vectors. Gene Ther. 17, 295–304 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang K. S., et al. , Spatial and temporal action of chicken primordial germ cells during initial migration. Reproduction 149, 179–187 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Hamburger V., Hamilton H. L., A series of normal stages in the development of the chick embryo. J. Morphol. 88, 49–92 (1951). [PubMed] [Google Scholar]
- 34.Sun P., et al. , Transgenic chimera quail production by microinjecting lentiviral vector into the blood vessel of the early embryo. Anim. Sci. J. 83, 291–298 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Tyack S. G., et al. , A new method for producing transgenic birds via direct in vivo transfection of primordial germ cells. Transgenic Res. 22, 1257–1264 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Oishi I., Yoshii K., Miyahara D., Tagami T., Efficient production of human interferon beta in the white of eggs from ovalbumin gene-targeted hens. Sci. Rep. 8, 10203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He T.-C., et al. , A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee K., et al. , Expression of beta-galactosidase and pig leptin gene in vitro by recombinant adenovirus. Anim. Biotechnol. 10, 37–48 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.