Significance
The sympathetic nervous system plays a critical role in stimulating heart rate and contractility to increase cardiac output and thereby insure adequate tissue O2 delivery. Altered sympathetic output contributes to cardiac pathologies, such as sudden cardiac death and heart failure. Our data provide insight into the role of the transcription factor hypoxia-inducible factor 1α (HIF-1α) in the development of sympathetic ganglion neurons and cardiac sympathetic innervation. We found that genetic deletion of HIF-1α resulted in increased cell death and decreased proliferation of neuronal progenitors of the sympathetic system. These findings suggest that dysregulated HIF-1α expression may contribute to cardiac dysfunction and disease associated with defects in the cardiac sympathetic system by affecting the function and survival of sympathetic neurons.
Keywords: sympathetic neurons, cardiac innervation, tyrosine hydroxylase, hypoxia, coronary artery branching
Abstract
The molecular mechanisms regulating sympathetic innervation of the heart during embryogenesis and its importance for cardiac development and function remain to be fully elucidated. We generated mice in which conditional knockout (CKO) of the Hif1a gene encoding the transcription factor hypoxia-inducible factor 1α (HIF-1α) is mediated by an Islet1-Cre transgene expressed in the cardiac outflow tract, right ventricle and atrium, pharyngeal mesoderm, peripheral neurons, and hindlimbs. These Hif1aCKO mice demonstrate significantly decreased perinatal survival and impaired left ventricular function. The absence of HIF-1α impaired the survival and proliferation of preganglionic and postganglionic neurons of the sympathetic system, respectively. These defects resulted in hypoplasia of the sympathetic ganglion chain and decreased sympathetic innervation of the Hif1aCKO heart, which was associated with decreased cardiac contractility. The number of chromaffin cells in the adrenal medulla was also decreased, indicating a broad dependence on HIF-1α for development of the sympathetic nervous system.
The heart is innervated via the autonomic nervous system, which comprises sympathetic and parasympathetic nerves that exert antagonistic effects on cardiac function. Disruption of sympathetic–parasympathetic balance in the heart is central to the pathophysiology and progression of many types of heart disease. Sympathetic innervation regulates cardiac function by increasing heart rate, conduction velocity, and myocardial contractility (1). Changes in cardiac innervation pattern and activity of the sympathetic nervous system are implicated in many cardiac pathologies, ranging from sudden infant death syndrome (2) to such common diseases of adulthood as hypertension, myocardial ischemia, cardiac arrhythmias, sudden cardiac death, and heart failure (3).
The sympathetic nervous system is composed of preganglionic neurons residing in the spinal cord (the preganglionic motor column) and postganglionic neurons forming the sympathetic ganglion chain. Postganglionic sympathetic neurons that innervate the heart are generated from trunk neural crest cells, which first migrate and form the primary sympathetic ganglia (PSG) chain near the dorsal aorta, starting at embryonic day (E) 10.5 in the mouse (4, 5). Sympathetic neuroblasts express markers associated with mature sympathetic neurons, such as tyrosine hydroxylase (TH), but still undergo proliferation (6). TH is the first enzyme in the catecholamine biosynthetic pathway that produces norepinephrine, the primary neurotransmitter released from postganglionic sympathetic neurons (7). In the second migration step, sympathetic progenitors migrate away from the dorsal aorta to form secondary sympathetic ganglia at E13.5 and undergo final differentiation into sympathetic neurons with exit from the cell cycle that peaks at E14.5 (6).
The differentiation of sympathetic neurons is induced by signaling molecules of the bone morphogenetic protein family secreted by the dorsal aorta, followed by expression of a variety of transcription factors that initiate noradrenergic and neuronal differentiation, including PHOX2A, PHOX2B, GATA2, GATA3, INSM1, HAND2, ASCL1, SOX4, and SOX11 (5, 8). Another transcription factor required for the development of sympathetic progenitors in the PSG is the LIM homeodomain protein Islet-1 (ISL1), which acts at both early and late stages of sympathetic neurogenesis (9, 10).
Postganglionic sympathetic neurons innervating the heart are located primarily in the stellate ganglion, with minor contributions from the middle cervical and upper thoracic paravertebral sympathetic ganglia (11, 12). Cardiac sympathetic fibers originate in sympathetic ganglia, extend along large-diameter coronary vessels into the underlying myocardium, and end as sympathetic nerve terminals reaching the endocardium (13). The density of sympathetic innervation is highest in the subepicardium and central conduction system, which consists of the sinoatrial node (SAN), atrioventricular node, and bundle of His (1), and sympathetic innervation gradually decreases from the atria to the ventricles and from the base to the apex of the heart (13).
Tissue hypoxia plays important roles in the recruitment of neural crest cells, sympathetic axon guidance, and patterning of cardiac innervation (13, 14). The master regulator of transcriptional responses to decreased O2 availability is hypoxia-inducible factor 1 (HIF-1), a heterodimeric protein consisting of an O2-regulated HIF-1α subunit and a constitutively expressed HIF-1β subunit. Germline knockout of the Hif1a gene results in mouse embryos with profound cardiac, vascular, and neural tube defects and developmental arrest at E8.5 (15, 16). Severe placental and vascular defects, as well as the resulting early embryonic lethality of germline Hif1a knockout mice, preclude an in-depth investigation of the mechanisms by which HIF-1 contributes to heart development and function.
Conditional knockout (CKO) by deletion of a floxed Hif1a exon 2 (Hif1aloxP/loxP) (17) under the control of several CRE driver lines (18–20) has provided insight into the role of HIF-1α in later stages of cardiogenesis, but no previous studies have investigated whether Hif1a is required for cardiac sympathetic innervation. Although TH expression is regulated by HIF-1 in certain cell types (21, 22), whether HIF-1α is required for the development of sympathetic neurons is unclear. In this study, we generated Hif1aCKO mice by mating Hif1aloxP/loxP and Isl1-Cre mice. Lineage studies have demonstrated Isl1-Cre expression in progenitors of the second heart field, pharyngeal mesoderm, hindlimbs, spinal motor neurons, cranial ganglia, dorsal root ganglia, and sympathetic chain (23, 24). Our analysis of mice with Hif1aCKO in ISL1-expressing cells has revealed a key role for HIF-1α in sympathetic innervation of the heart and development of sympathetic ganglia.
Results
Loss of HIF-1α in ISL1+ Cells Leads to Neonatal Mortality and Impaired Cardiac Function.
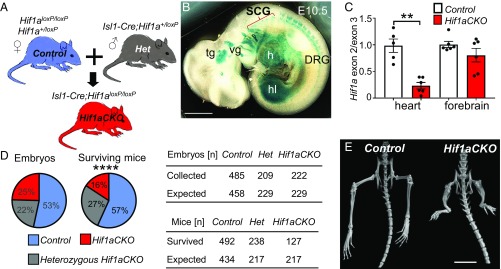
Embryonic lethality of germline Hif1a knockout mice by E10.5 (15, 16) precludes analysis of HIF-1α–dependent events at later developmental stages. To address this limitation, we disrupted the HIF-1α coding sequence in Hif1aloxP/loxP mice (17) by expression of an Isl1-Cre transgene (Fig. 1A) that has been used extensively to study cardiovascular and nervous system development (24–29). To identify tissues with Isl1-directed expression of CRE recombinase, we crossed Isl1-Cre mice with Rosa26 reporter mice, which express β-galactosidase only in the presence of CRE. Descendants of cells expressing the Isl1-Cre transgene were detected mainly in the heart, hindlimbs, cranial ganglia, sympathetic ganglion chain, and dorsal root ganglia at E10.5 (Fig. 1B), similar to previous reports (24). The CRE-mediated excision of Hif1a exon 2, which encodes the basic helix-loop-helix DNA-binding domain of HIF-1α in Hif1aloxP/loxP;Isl1-Cre (Hif1aCKO) mice was very efficient, with quantitative real-time PCR (qPCR) analysis of genomic DNA revealing 77% elimination of exon 2 in the heart, whereas exon 2 copy number was not significantly different in the forebrains of control and Hif1aCKO mice at E10.5 (Fig. 1C).
Fig. 1.
Decreased neonatal survival and hindlimb malformations in Hif1aCKO mice. (A) The Isl1-Cre transgene was transmitted paternally to eliminate any potential influence of maternal HIF-1α haploinsufficiency on the developing embryos. (B) Whole-mount X-gal staining of E10.5 Isl1-Cre;RosaR26R embryos revealed β-galactosidase expression in the dorsal root ganglia (DRG) and sympathetic cervical ganglia (SCG), trigeminal ganglion (tg), vestibular ganglion (vg), heart (h), and hindlimb (hl). (Scale bar: 100 μm.) (C) qPCR analysis of exon 2 (normalized to exon 3) sequences in genomic DNA isolated from heart and forebrain of Hif1aCKO and control embryos at E10.5. The data are presented as mean ± SEM (n = 6 embryos per group). **P < 0.01, Student’s t test. (D, Left) The genotype distribution of embryos and surviving mice. (D, Right) Observed distribution of genotypes of embryos collected between E7.5 and E17.5 according to the expected Mendelian ratio (Top). In contrast, the observed genotype distribution of surviving mice at 3 wk after birth shows a decreased number of Hif1aCKO mice (Bottom). ****P < 0.0001, χ2 test. (E) Micro-CT scan shows hindlimb malformations with reduced tibia, femur, and pelvis lengths in adult Hif1aCKO mice. (Scale bar: 10 mm.)
Hif1aCKO embryos were recovered at expected Mendelian ratios at all embryonic days examined in utero (Fig. 1D and SI Appendix, Table S1). We analyzed cardiac development, including the outflow tract and pharyngeal arch arteries, by histological analysis of E14.5 and E15.5 embryos. Hif1aCKO hearts (n = 11) were indistinguishable from control (Hif1a+/loxP or Hif1aloxP/loxP) hearts (n = 7) with no gross structural abnormalities. Thus, Hif1a deletion in ISL1+ cardiac progenitor cells did not affect overall heart development. However, Hif1aCKO mice had significantly increased neonatal mortality (P < 0.0001, χ2 test), with 40% fewer Hif1aCKO mice identified at the time of genotyping (3 wk) compared with Hif1a+/loxP;Isl1-Cre mice (Fig. 1D). Examination of surviving adult Hif1aCKO mice revealed major malformations of the proximal and distal hindlimbs (Fig. 1E and SI Appendix, Fig. S1). Conditional inactivation of HIF-1α during limb development using Prx1-Cre caused malformation of both forelimbs and hindlimbs (30).
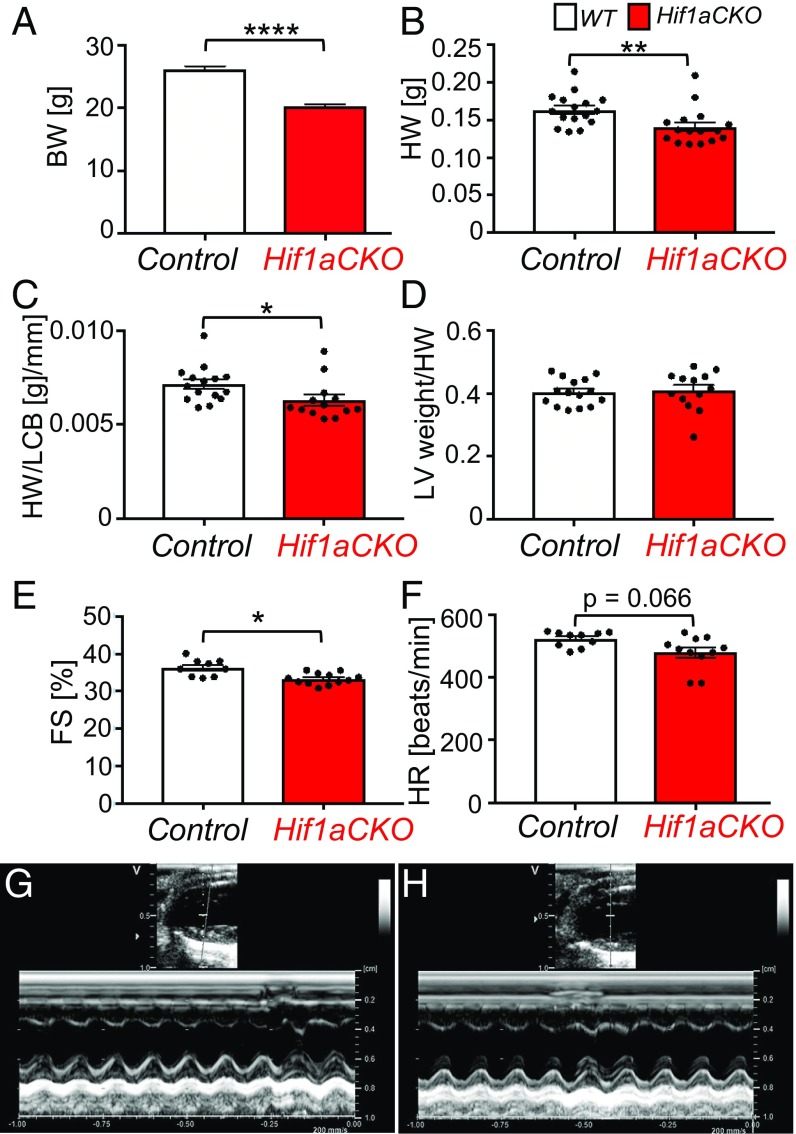
Due to gender differences in cardiac electrophysiological properties (31), we focused our functional analyses on male mice. Since the body weight of Hif1aCKO mice was decreased (Fig. 2A), as was heart weight (HW) (Fig. 2B), we adjusted HW using the length of the cranial base (LCB) measured by micro-computed tomography (CT) (Fig. 2C and SI Appendix, Fig. S2 and Table S2). The HW of the surviving Hif1aCKO mice was modestly decreased compared with age-matched controls (Fig. 2B), but the ratio of left ventricular (LV) weight to HW was unaffected (Fig. 2D). Although Hif1aCKO mice had no detectable structural abnormalities of the cardiac chambers or valves, analysis of adult males using echocardiography revealed significantly impaired LV function, with fractional shortening of 33.3 ± 0.4%, compared with 37.0 ± 0.9% in control littermates (Fig. 2 E, G, and H and SI Appendix, Table S3). Observed differences in heart rate were not statistically significant (Fig. 2F and SI Appendix, Table S3).
Fig. 2.
Impaired LV function in Hif1aCKO mice. (A) Body weight (BW) was decreased in adult Hif1aCKO mice (n = 37) compared with controls (n = 40). (B and C) Both HW and HW relative to the LCB (HW/LCB) were decreased in Hif1aCKO mice (n = 13) compared with controls (n = 15). (D) The ratio of LV weight to HW (LV/HW) was not significantly different between genotypes. (E and F) Echocardiographic analyses of adult (3-mo-old) males revealed significantly decreased fractional shortening (FS) but no significant difference in heart rate (HR) in Hif1aCKO mice (n = 12) compared with control mice (n = 10). (G and H) Representative M-mode echocardiography (long-axis view of LV) in control (G) and Hif1aCKO (H) mice. All numerical data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ****P < 0.0001, Student’s t test.
Impaired Sympathetic Innervation in Hif1aCKO Hearts.
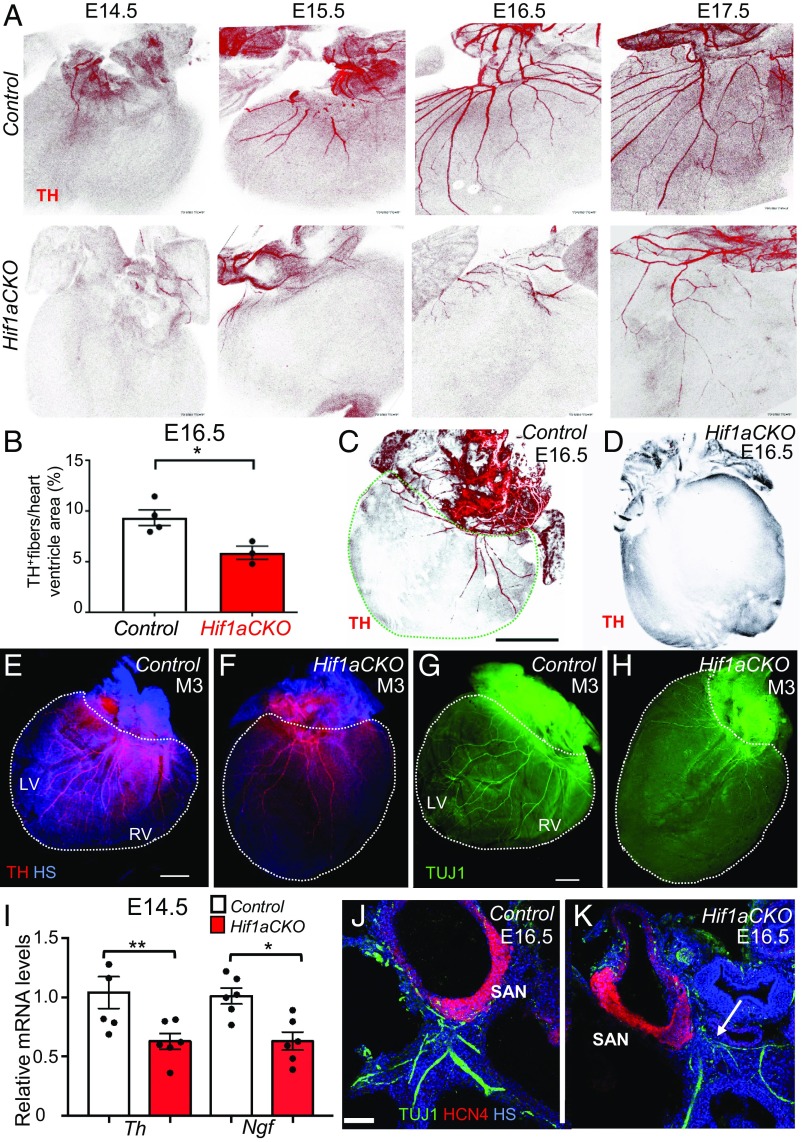
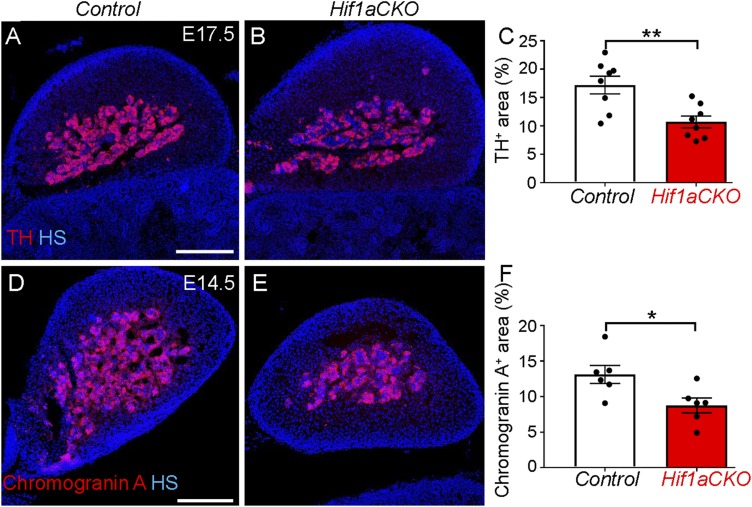
Immunohistochemical staining of TH revealed significantly impaired sympathetic innervation in the hearts of Hif1aCKO mice. At E14.5, TH+ sympathetic axons were found on the heart surface and in an area adjacent to the sinus venosus, reaching the base of the ventricles in control hearts, whereas in Hif1aCKO hearts, TH+ axons were significantly reduced in number and length and were restricted to the area of the sinus venosus (Fig. 3A). Between E14.5 and E17.5, sympathetic axons extended to and branched in the dorsal subepicardium of control hearts, whereas expansion of sympathetic innervation in the Hif1aCKO hearts was attenuated. In comparison with control littermates, sympathetic innervation was significantly decreased in E16.5 Hif1aCKO hearts (Fig. 3B). In contrast to the abundant TH+ neurons in control hearts (Fig. 3C), considerable variation in sympathetic innervation of Hif1aCKO hearts was observed, ranging from a 30% reduction to a complete absence of TH+ neurons (Fig. 3D). Decreased sympathetic innervation was also detected in the hearts of surviving 3-mo-old Hif1aCKO mice, as shown by anti-TH and anti-TUJ1 immunofluorescence (Fig. 3 E–H). Consistent with the reduced innervation of Hif1aCKO hearts at E14.5, reverse-transcription and qPCR revealed decreased levels of mRNA encoding TH as well as nerve growth factor (NGF) (Fig. 3I), which is required for terminal sympathetic innervation of target tissues (32).
Fig. 3.
Impaired sympathetic innervation of Hif1aCKO hearts. (A) Immunohistochemical staining of TH in posterior view of hearts from control (Upper) and Hif1aCKO (Lower) littermate embryos between E14.5 and E17.5. (B) Quantification of TH+ fibers per ventricle area in E16.5 control and Hif1aCKO hearts (n = 4 each). *P < 0.05, Student’s t test. (C and D) Posterior view of E16.5 control heart (C) and Hif1aCKO heart with no detectable TH+ fibers (D). In C, the dashed green line demarcates the ventricular area of the heart. (Scale bar: 500 μm.) (E–H) Sympathetic innervation of hearts from 3-mo-old male mice (M3) was analyzed by anti-TH (E and F) and anti-TUJ1 (G and H) immunohistochemistry. The posterior view is shown, and dotted lines encompass the left ventricle (LV) and right ventricle (RV) of the heart. (Scale bar: 1 mm.) (I) Relative Th and Ngf mRNA levels were measured by reverse-transcription and qPCR, using mRNA extracted from the whole heart at E14.5, when the sympathetic neurons reach the heart and start to innervate the ventricles (n = 8 hearts per genotype). *P < 0.05, **P < 0.01, Student’s t test. (J and K) Expression of HCN4, a marker for the cardiac conduction system, in E16.5 heart sections. HCN4+ area of the SAN in the Hif1aCKO embryo is comparable to that in the control embryo at E16.5, but TUJ1-labeled nerve fibers in proximity of the SAN are reduced, thinner, and more disorganized in the Hif1aCKO (arrow) compared with control heart. (Scale bar: 100 μm.) HS, Hoechst stain. All numerical data are presented as mean ± SEM.
Since ISL1 is expressed in the SAN during the development of cardiac pacemaker cells (33), we investigated formation of the SAN in Hif1aCKO hearts. Hyperpolarization-activated nucleotide-gated cation channel 4 (HCN4) is a well-established marker for slow-conducting components of the cardiac conduction system with pacemaker potential (34). At E16.5, formation of the SAN was comparable between genotypes, although innervation in the proximity of the SAN was reduced, thinner, and more disorganized in Hif1aCKO hearts compared with control hearts (Fig. 3 J and K).
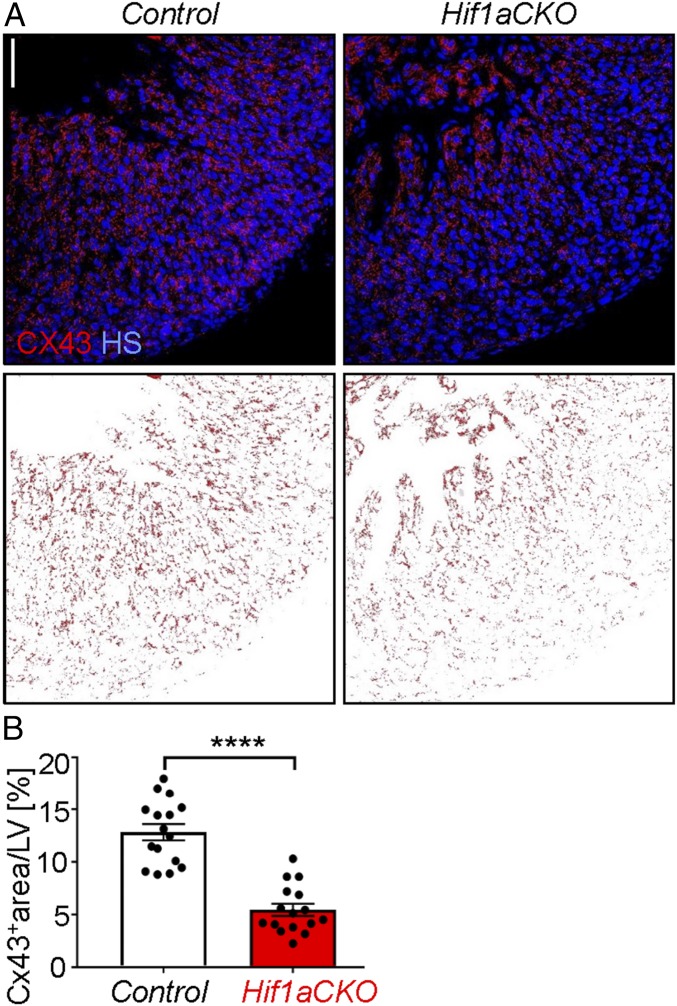
The gap junction alpha 1 (GJA1) protein, also known as connexin 43 (CX43), plays a key role in myocardial conduction, and decreased CX43 expression is associated with decreased conduction properties in a heart failure model (35). Furthermore, HIF-1 binds directly to the GJA1 gene and activates its transcription in hypoxic melanoma cells (36). Immunohistochemistry (Fig. 4 A, Upper) and image analysis (Fig. 4 A, Lower) revealed significantly decreased CX43 protein levels in the compact myocardium of the LV in Hif1aCKO hearts compared with controls (Fig. 4B).
Fig. 4.
Decreased connexin 43 expression in Hif1aCKO hearts at E16.5. (A) CX43 immunohistochemistry was performed (Upper), and the CX43+ area was captured using ImageJ (Lower). (Scale bar: 50 µm.) (B) CX43 staining was quantified using ImageJ, and the CX43+ area was expressed as percentage of total cross-sectional area of compact myocardium of LV in control and Hif1aCKO littermates (n = 3 embryos per genotype and 4–6 fields per LV). ****P < 0.0001, Student’s t test. The data are presented as mean ± SEM.
Aberrant Coronary Artery Architecture in Hif1aCKO Hearts.
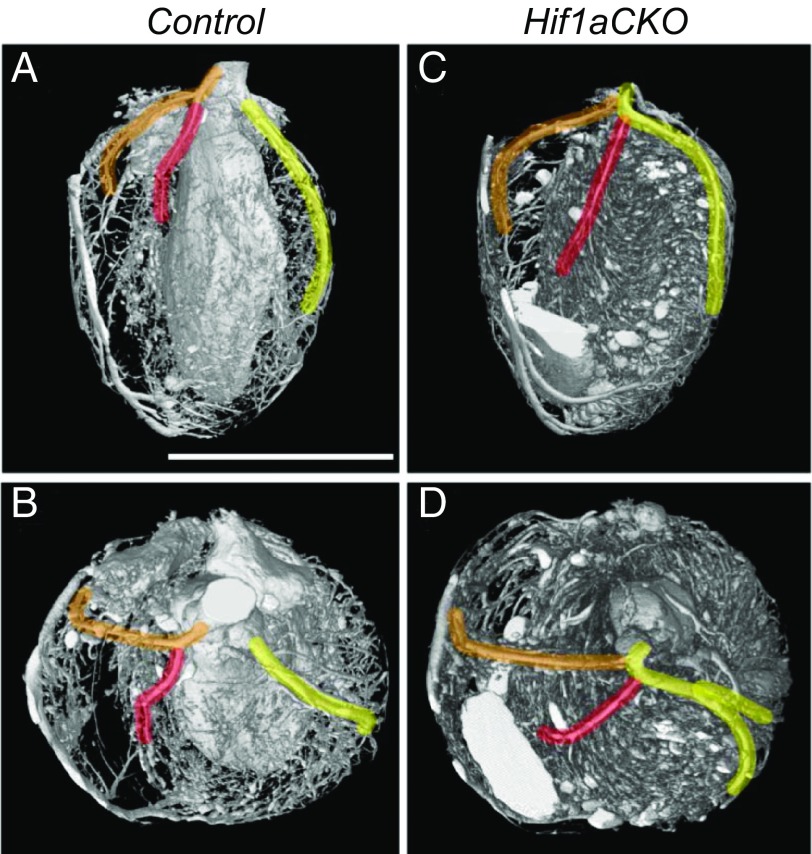
The physical proximity and similar branching pattern of the coronary vasculature and peripheral nerves imply coordinated development of these networks. Since sympathetic innervation was impaired in Hif1aCKO hearts, we investigated coronary vascular architecture using Microfil injection (Flow Tech) and micro-CT. Quantification of the branching pattern (i.e., frequency and distance of identifiable branches from the left coronary artery) did not reveal any significant differences between control and Hif1aCKO adult mice (SI Appendix, Table S4). As described previously (37), the most common coronary architecture was the presence of two independent coronary arteries originating from the left and right aortic sinuses, with the septal artery originating from the right coronary artery (Fig. 5 A and B). However, in 2 out of 13 Hif1aCKO hearts, we detected a major anomaly in which a single coronary artery arose from the right aortic sinus, from which the left coronary and septal arteries then originated (Fig. 5 C and D). This type of coronary artery anomaly, which was observed in none of 15 control hearts, is considered a pathological malformation in humans (37).
Fig. 5.
Main coronary artery anomalies in Hif1aCKO hearts. Micro-CT visualization of coronary artery branching patterns in 2-mo-old control (A and B) and Hif1aCKO (C and D) mice. Anterior aspect (A and C) and top view (B and D) are shown. Arteries are highlighted from their origin: right coronary artery (RCA; orange), left coronary artery (LCA; yellow), and septal artery (SA; red). The classic arrangement with two independent coronary arteries, the RCA and LCA, arising separately from the aorta and the SA arising from the RCA was observed in 15 out of 15 control hearts (A and B). An anomalous single coronary artery arising from the aorta with subsequent division of the RCA, SA, and LCA was observed in 2 out of 13 Hif1aCKO hearts (C and D). (Scale bar: 5 mm.)
HIF-1α Is Required for Development of Sympathetic Ganglia.
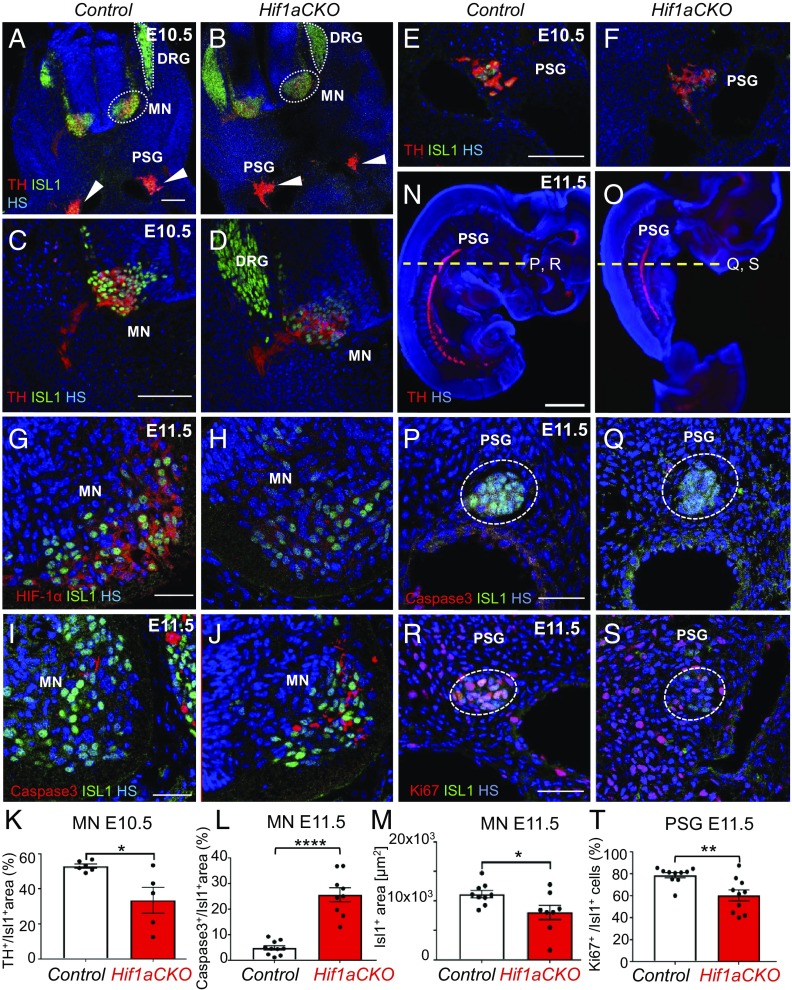
Based on our finding of compromised sympathetic innervation of the heart in Hif1aCKO mice, we assessed the development of the sympathetic nervous system more broadly (Fig. 6). ISL1 expression is initiated in the developing sympathetic progenitors at E9.5 and is maintained in sympathetic ganglia until E16.5 (9). ISL1 is also expressed in developing preganglionic motor neurons of the ventral spinal cord (38). We first analyzed the expression of ISL1 and TH, the marker of sympathetic lineage cells, at E10.5. TH expression was reduced in preganglionic motor neurons and their axons within the thoracic spinal cord of Hif1aCKO mice compared with control littermates (Fig. 6 A–D). The TH+ domain was significantly decreased in motor neuron columns of Hif1aCKO embryos (Fig. 6K). TH and ISL1 expression appeared to be decreased in neurons of developing PSG of Hif1aCKO mice compared with controls (Fig. 6 E and F). At E11.5, HIF-1α expression was almost completely absent in thoracic motor columns of Hif1aCKO embryos compared with control littermates (Fig. 6 G and H).
Fig. 6.
Abnormal development of sympathetic neurons in Hif1aCKO embryos. (A and B) Cross-section images of E10.5 embryos showing decreased expression of TH (red) in developing neurons of the sympathetic system in Hif1aCKO embryos compared with control embryos. ISL1+ neurons (green) were detected in the dorsal root ganglia (DRG), in developing motor neuron populations (including preganglionic motor columns of the sympathetic system) within the ventral neural tube (MN), and in forming PSG (arrowheads). TH expression was detected in the MN, sympathetic axons, and PSG neurons. HS is shown in blue. (C–F) Higher magnification images show TH expression in the MN (C and D) and PSG (E and F). (G and H) HIF-1α expression (red) was decreased in the MN of Hif1aCKO embryos compared with control embryos. (I, J, L, and M) Cleaved caspase-3 immunostaining shows significantly increased apoptosis and decreased ISL1+ area in the MN of Hif1aCKO embryos at E11.5 (n = 5 embryos per genotype and 2 ganglia per embryo). ****P < 0.0001, Mann–Whitney U test; *P < 0.05, Student’s t test. (K) Quantification of TH+ area as a percentage of the total ISL1+ area in the MN (n = 3 embryos per genotype and two sections per embryo). *P < 0.05, Student’s t test. (N and O) Sagittal sections of E11.5 embryos were analyzed by TH immunostaining to identify the PSG. The dotted yellow line indicates the plane of cross-sectioning for the images shown in P–S. (P and Q) Cross-sections of E11.5 embryos show no apoptotic cells in PSG neurons of control or Hif1aCKO embryos (n = 5 embryos per genotype analyzed). (R and S) Immunostaining for proliferating cell nuclear antigen Ki67 (red) in PSG neurons. (T) Quantification of proliferating neurons in PSG of control and Hif1aCKO embryos (n = 5 embryos per genotype and 2 ganglia per embryo). **P < 0.01, Student’s t test. (Scale bars: 100 μm for A–F, 50 μm for G–J and P–S, and 1,000 μm for N and O.) All numerical data are presented as mean ± SEM.
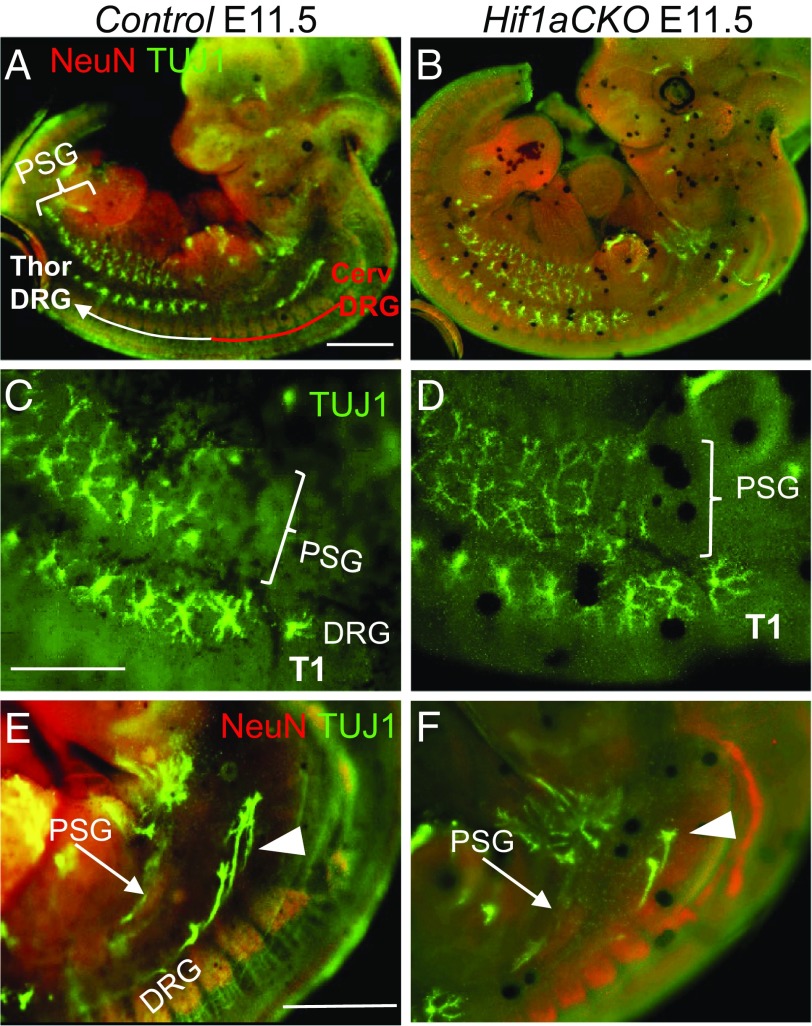
To determine whether the impaired development of sympathetic ganglia was due to altered proliferation or survival, we first performed immunostaining for cleaved caspase 3, which revealed significantly increased apoptosis of preganglionic neurons in motor columns of E11.5 Hif1aCKO embryos (Fig. 6 I, J, and K). The ISL1+ domain was significantly decreased in Hif1aCKO motor columns compared with controls (Fig. 6M). Since developing preganglionic neurons and postganglionic sympathetic neurons interact extensively (39), increased death of preganglionic neurons may influence the formation of sympathetic ganglia. In contrast to increased apoptosis of neurons in the neural tube, no caspase 3-positive cells were observed in the PSG of Hif1aCKO or control embryos at E11.5 (Fig. 6 P and Q). However, Ki67 immunostaining revealed significantly attenuated proliferation of sympathetic neurons in the developing ganglia in Hif1aCKO embryos at E11.5 (Fig. 6 R–T), when most sympathetic neurons have reentered the cell cycle until their terminal differentiation (6). Taken together, these data suggest that HIF-1α is required for proliferation of progenitors of the sympathetic ganglia and for survival of preganglionic neurons in the developing thoracic spinal cord. Consistent with these changes, axon projections in the cervical and upper thoracic area were diminished in Hif1aCKO embryos compared with control embryos at E11.5 (Fig. 7). Axonal bundles of sympathetic neurons also appeared thinner in Hif1aCKO embryos (Fig. 7 C and D).
Fig. 7.
HIF-1α is required for normal axon growth. (A and B) Whole-mount immunostaining of nerve fibers with anti-TUJ1 (green) shows decreased innervation in the cervical area of the DRG and developing PSG in Hif1aCKO embryos compared with control embryos at E11.5. Neurons are labeled with anti-NeuN (red). The cervical (red line) and thoracic (white line) DRG are indicated in A. (C–F) Higher-magnification images of the upper thoracic region show thinner axonal bundles of neurons in PSG (arrows in E and F) and decreased numbers of DRG axons (arrowheads in E and F) in Hif1aCKO embryos compared with control embryos. Thoracic T1 vertebra are indicated in C and D. (Scale bars: 1,000 μm for A, B, E, and F; 500 μm for C and D)
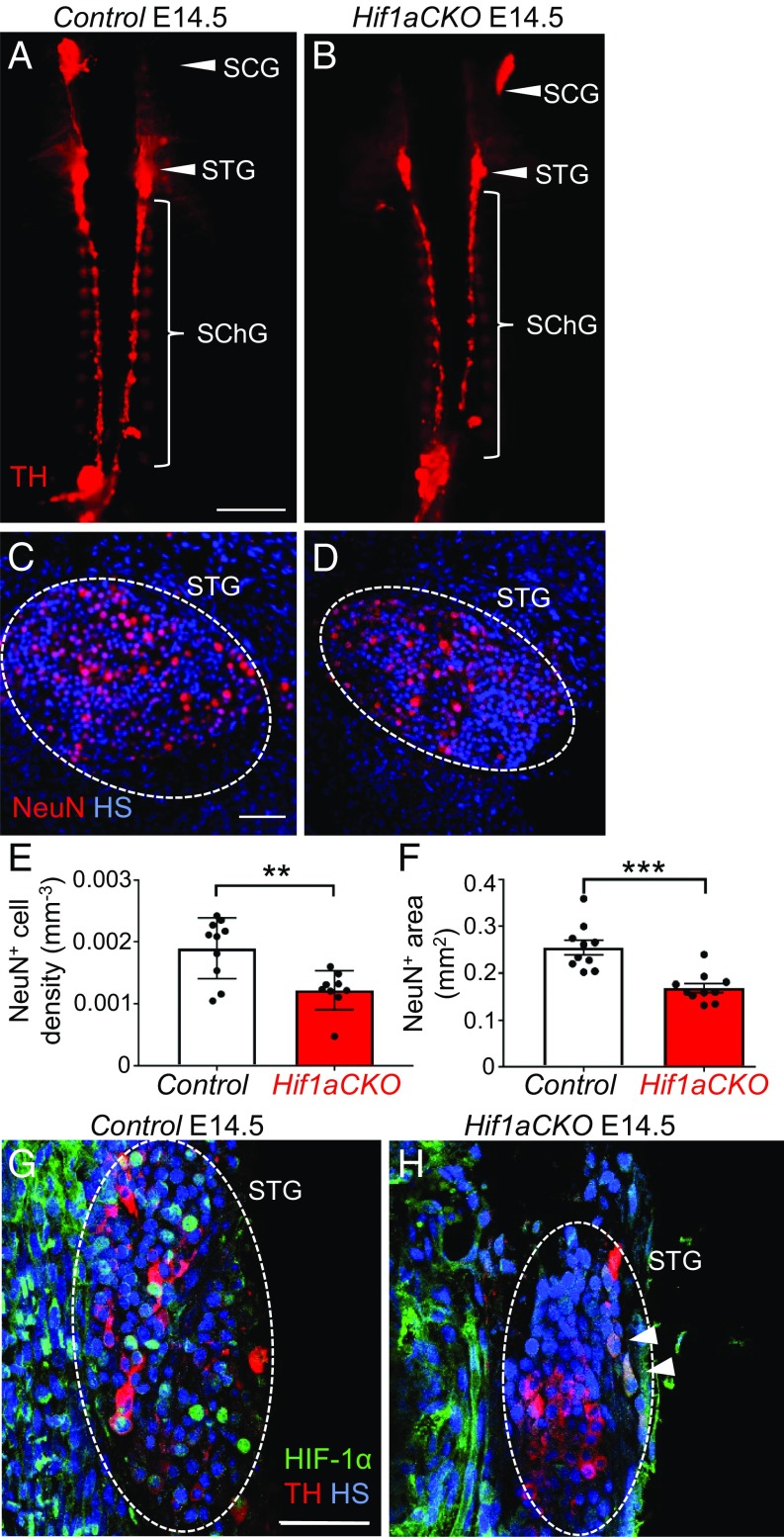
We next investigated formation of the secondary sympathetic ganglia. Hypoplasia of PSG was accompanied by decreased overall size of the secondary sympathetic chain ganglia in Hif1aCKO embryos. The superior cervical ganglia, stellate ganglia, and thoracic sympathetic chain ganglia of Hif1aCKO embryos were noticeably smaller than those of littermate controls at E14.5 as visualized by TH immunostaining (Fig. 8 A and B). We focused our analyses on the stellate ganglia, where the overwhelming majority of cardiac nerve fibers originate (12). To assess the functional state of neurons in the stellate ganglia, we evaluated the expression of neuronal nuclear antigen (NeuN), a differentiation marker of mature postmitotic neurons. The number of NeuN+ neurons in the stellate ganglia of Hif1aCKO embryos was significantly decreased (Fig. 8 C–F). Using NeuN whole-mount staining, we determined that Hif1aCKO ganglia were 34% smaller compared with those of littermate controls at E14.5 (Fig. 8F and SI Appendix, Fig. S3). At E14.5, strong nuclear HIF-1α was detected in neurons of the stellate ganglia in control embryos (Fig. 8G), whereas HIF-1α expression was reduced and localized to the cytoplasm of TH+ neurons in Hif1aCKO embryos (arrowheads in Fig. 8H). The cross-sectional area of the stellate ganglia (dotted ovals in Fig. 8 G and H) was decreased in Hif1aCKO embryos. Thus, the loss of HIF-1α expression and differentiated neurons in sympathetic chain ganglia correlates with the attenuated sympathetic innervation of Hif1aCKO hearts and impaired development of secondary sympathetic ganglia at E14.5.
Fig. 8.
The growth and maturation of secondary sympathetic ganglia are impaired in E14.5 Hif1aCKO embryos. (A and B) Whole-mount TH immunostaining was performed to detect the secondary chain of sympathetic ganglia, which are smaller in Hif1aCKO mice compared with control littermates. Arrowheads indicate the superior cervical (SCG) and stellate (STG) ganglia; bracket indicates the thoracic chain of sympathetic ganglia (SChG). (Scale bar: 1,000 μm.) (C and D) Immunostaining for NeuN (a marker for differentiated neurons; red) shows reduced neuronal density in the STG of Hif1aCKO compared with control embryos (n = 5 embryos per genotype and 2 ganglia per embryo). **P < 0.01, Student’s t test. (Scale bar: 50 μm.) (E and F) Morphometry of sympathetic ganglia (the STG and three upper ganglia of the thoracic sympathetic chain) by whole-mount NeuN immunostaining of the spinal cord (n = 5 embryos per genotype, left and right sympathetic chain ganglia per embryo). ***P < 0.0005, Student’s t test. (G and H) Coronal sections of embryos immunolabeled with anti-TH (red) and anti–HIF-1α (green) show a smaller STG area (dotted oval) and lower HIF-1α expression in Hif1aCKO embryos. Strong HIF-1α expression was detected in nuclei of TH+ neurons in control embryos (G) but only in the cytoplasm of a few TH+ neurons in Hif1aCKO embryos (H, arrowheads). (Scale bar: 50 μm.) All numerical data are presented as mean ± SEM.
Development of Adrenal Chromaffin Cells Is Impaired in Hif1aCKO Embryos.
Since sympathetic neurons and neuroendocrine chromaffin cells in the adrenal medulla derive from a common catecholaminergic sympathoadrenal progenitor (9), we analyzed the development of adrenal chromaffin cells in Hif1aCKO embryos. We first analyzed TH expression in the adrenal medulla at E17.5, and found that TH expression in the adrenal medulla of Hif1aCKO embryos was only 55% of that of control littermates (Fig. 9 A–C). We next analyzed expression of chromogranin A, a secretory protein located in chromaffin vesicles that is costored and coreleased with catecholamines, in the adrenal medulla. The chromogranin A expression domain in the adrenal gland was also decreased in Hif1aCKO compared with control littermates (Fig. 9 D–F). These findings show that HIF-1α deficiency affects the development of chromaffin cells as well as sympathetic neurons.
Fig. 9.
Decreased number of adrenal chromaffin cells in Hif1aCKO embryos. (A and B) Cross-sections through the adrenal glands of Hif1aCKO and control embryos at E17.5 were stained with anti-TH antibody (red). (C) TH+ area was quantified using ImageJ software and expressed as percentage of total adrenal gland area (n = 4 embryos per genotype and two sections per embryo). **P < 0.005, Student’s t test. (D and E) Chromogranin A expression in cross-sections of the adrenal glands of control and Hif1aCKO E14.5 embryos was detected by immunostaining. (F) Chromogranin A+ area was quantified using ImageJ and expressed as a percentage of the total adrenal gland area (n = 3 embryos per genotype and two sections per embryo). *P < 0.05, Student’s t test. (Scale bar: 200 μm.) All numerical data are presented as mean ± SEM.
Discussion
In this study, we generated conditional deletion of the Hif1a gene to bypass the early embryonic lethality associated with germline Hif1a knockout (15, 16). We found that mice selectively lacking HIF-1α in ISL1+ cells have a profound deficit in sympathetic innervation of the heart and impaired development of sympathetic ganglia and chromaffin cells. Thus, HIF-1α is essential for the development of neuronal and endocrine sympathoadrenal lineage derivatives.
HIF-1α is required for physiological responses to hypoxia and plays critical roles in the pathophysiology of cancer, cardiovascular disease, and other common causes of mortality (40, 41). During development, HIF-1α is required for circulatory system development and O2 homeostasis (15, 40, 42). Mice lacking Hif1a expression die at midgestation due to profound cardiovascular and neural tube defects (15, 16). We previously reported that global Hif1a haploinsufficiency in combination with an adverse diabetic environment in utero is associated with an increased incidence of congenital cardiac defects and impaired cardiac function (43, 44). In the present study, we used an Isl1-Cre transgene to generate selective HIF-1α deficiency. ISL1 is expressed in progenitors of the cardiac outflow tract, right ventricle and atria (23), hindlimbs, and all peripheral neurons (45). ISL1 regulates both early and late development of the sympathetic nervous system, including axon growth as well as neuron proliferation and maturation (10).
Isl1-Cre–mediated excision of Hif1a exon 2 did not affect heart development at a gross morphological level, as the chambers and outflow tract were comparable between control and Hif1aCKO embryos. Thus, the disruption of Hif1a in ISL1+ cardiac progenitor cells did not affect cardiac morphogenesis. This conclusion is consistent with the recent finding that ISL1+ cardiac progenitor cells proliferate in a normoxic niche with low HIF-1α expression (46). Conversely, HIF-1α forms a repressor complex to silence Isl1 expression under pathological hypoxia, leading to congenital cardiac defects (46).
In attempting to understand how HIF-1α loss of function in ISL1+ cells influences neonatal survival and cardiac function of Hif1aCKO mice, we uncovered an essential role for HIF-1α in the development of cardiac sympathetic neurons. We observed significantly impaired sympathetic innervation of Hif1aCKO hearts compared with littermate control hearts at E14.5. Expression of the Th gene, a direct target of HIF-1 (21), was decreased in the heart at E14.5, when sympathetic nerve terminals reach the heart and begin to innervate the ventricles. The expression of mRNA encoding NGF, which is required for cardiac sympathetic innervation and axon growth (32, 47, 48), was also reduced in E14.5 Hif1aCKO hearts. We cannot rule out the possibility that these changes in gene expression represent the consequence, rather than the cause, of decreased sympathetic innervation or reflect an indirect effect of HIF-1α loss of function. We also cannot exclude the possibility that impaired production of neurotrophic factors by cardiac myocytes might contribute to impaired cardiac innervation. However, the impaired development of sympathetic chain ganglia suggests direct effects of HIF-1α deficiency on sympathetic neurons.
In addition to the impaired sympathetic innervation, CX43 expression was significantly decreased in Hif1aCKO hearts. CX43 is an integral membrane protein and the major gap junction protein that contributes to the electrical coupling of ventricular cardiomyocytes (49). A single nucleotide polymorphism in the GJA1 gene encoding CX43 is associated with arrhythmias and sudden cardiac death (50). CX43 is a key mediator of cardiomyocyte protection during hypoxia (51), and CX43 expression is directly regulated by HIF-1 in melanoma cells (36). Decreased CX43 expression in the LV of Hif1aCKO mice may have contributed to the impaired contractility detected on echocardiography and may have increased the risk of arrhythmias and sudden death. Isl1 expression has been documented in cardiac pacemaker cells (33, 52), providing another potential mechanism for arrhythmias in Hif1aCKO mice.
It is interesting that increased neonatal mortality was reported in mice with germline deletion of Th, and that, as in the case of Hif1aCKO mice, no pathological abnormalities were found in Th−/− mice that could definitively establish the primary cause of death (53). It is possible that the neonatal mortality of Hif1aCKO mice is due to reduced catecholamine production or defective sympathetic innervation of other tissues, such as the lungs, because a catecholamine surge at birth is critical to maintain euglycemia (53) and trigger the pulmonary fluid absorption necessary to increase lung compliance and pulmonary function (54). A number of additional mechanisms underlying sudden infant death syndrome have been proposed, including inborn errors of metabolism, respiratory dysfunction, cardiorespiratory instability, and cardiac arrhythmias (55). Finally, the hindlimb defects of Hif1aCKO mice might impair suckling and thereby contribute to neonatal mortality, but conditional inactivation of Hif1a in the limb bud mesenchyme using Prx1-Cre resulted in severe forelimb and hindlimb shortening without any effect on neonatal survival (30).
Within the central nervous system, blood vessels modulate neurogenesis and neuronal migration (56), and neurons reciprocally regulate angiogenesis by expressing angiogenic factors (57). Similarly, the development and function of peripheral innervation and vasculature in the heart depend on precise communication between the nervous and vascular systems (13). Based on our finding of impaired sympathetic innervation, as well as on genetic and toxicologic data linking HIF-1 to coronary development (58, 59), we analyzed the arrangement of coronary arteries. Notably, in 15% of Hif1aCKO hearts, we observed a single coronary artery (from the right aortic sinus), which is considered a major anomaly in human pathology associated with myocardial ischemia (37). We interpreted this as a defect in remodeling of the periaortic plexus, from which two coronary arteries normally form, one penetrating to the left aortic sinus and the other to the right aortic sinus. Major coronary anomalies could be the cause of death in a proportion of the Hif1aCKO mice. Further studies are needed to identify the major defects in cardiac vascularization or innervation (e.g., Fig. 3D) that are found in late-stage Hif1aCKO fetuses more frequently than in surviving Hif1aCKO neonates and thus are likely to contribute to neonatal mortality.
Multiple transcription factors drive the development of sympathetic neurons (4, 5), and our results demonstrate that HIF-1α is involved in this process. We found that HIF-1α loss of function resulted in significantly decreased TH expression and increased cell death in neurons of the preganglionic motor columns. Furthermore, reduced proliferation of sympathetic progenitors in the PSG impaired the development of sympathetic chain ganglia and sympathetic innervation of the heart, demonstrating a continued requirement for HIF-1α at later stages of sympathetic neuron differentiation and maturation. As a master regulator of responses to hypoxia, HIF-1 signaling directly or indirectly targets hundreds of genes (40). HIF-1 has been shown in different cell types to regulate genes involved in a regulatory network of early sympathetic neuron development, for example, bone morphogenetic protein signaling (60), Hand1 and Hand2 (16), Th (21), and Sox4 (61). In addition, HIF-1 is an important regulator of a key angiogenic factor, vascular endothelial growth factor (VEGF), which is essential for the regulation of axon growth (62). Nerve-derived VEGF is required for arterial differentiation (63), and decreased VEGF levels cause neuron degeneration (64).
The present study has uncovered a requirement for HIF-1α in cardiac sympathetic neurogenesis, whereas germline knockout of the Phd3 gene causes abnormal adrenal and sympathetic nervous system development due to HIF-2α overexpression (65). HIF-1α is required for sympathetic nervous system activation mediated by the carotid body, and its action is opposed by HIF-2α (66). Our findings raise the intriguing possibility that in addition to its critical role in the development of sympathetic neurons, HIF-1α may play a role in various sympathetic abnormalities underlying cardiac pathologies, such as sudden cardiac death and heart failure.
Materials and Methods
Expanded descriptions of our animal models and methodology are provided SI Appendix, Materials and Methods.
Animals.
Experiments were conducted according to protocols approved by the Animal Care and Use Committee of the Institute of Molecular Genetics, Czech Academy of Sciences. All experiments were performed with littermates (males and females) cross-bred from two transgenic mouse lines: floxed Hif1atm3Rsjo with exon 2 of the Hif1a gene flanked by loxP sites (Hif1aloxP/loxP) (17) and Isl1-Cre (Isl1tm1(cre)Sev/+) (24). Hif1a+/loxP or Hif1aloxP/loxP mice served as controls. Control and heterozygous Isl1-Cre;Hif1a+/loxP mice were born and survived according to the expected Mendelian ratio of genotypes. To determine the efficiency of CRE‐mediated recombination, Isl1-Cre mice were bred with Rosa26 (Gt(ROSA)26Sortm1Sor) reporter mice. Phenotyping and data analysis were performed by investigators blinded to the genotypes of the mice.
Detection of Hif1a Exon 2 Deletion.
DNA was isolated from embryonic hearts and forebrains at E10.5. Primers for exon 2 and exon 3 of the Hif1a gene were used for qPCR as described previously (67). The difference (Δ) between the quantification cycle (Cq) of exon 2 and Cq of exon 3 was determined for each sample, and ΔCq values were compared between groups of Hif1aCKO and control mice using an unpaired two-tailed t test (GraphPad Prism 7). The relative amount of exon 2 DNA was calculated, based on the following formula: ΔΔCq = ΔCq of Hif1aCKO –ΔCq of control (68).
Echocardiography.
Echocardiographic evaluation of geometrical and functional LV parameters was performed using the GE Vivid 7 Dimension (GE Vingmed Ultrasound) with 12-MHz linear matrix probe M12L. The diastolic and systolic dimensions of the LV were measured in 12-wk-old mice as described previously (43).
Coronary Artery Visualization by Micro-CT Imaging.
Hearts perfused with polymerized yellow Microfil (Flow Tech) were prepared as described previously (69) and processed by micro-CT scanning (SkyScan 1272; Bruker). Projection images were reconstructed using NRecon SW (Bruker). 3D visualizations were achieved with CTVox SW (Bruker). For some specimens, CTAnalyser SW (Bruker) was applied for removal of excess extravascular contrast that limited image clarity. Branching of coronary arteries was evaluated using 3D visualization, and distances between branches of the left coronary artery were measured in 3D.
Cranial Morphometry.
Whole mouse cadavers were scanned on an Albira MikroPET/CT scanner (Bruker), and micro-CT images were used for measurement of the LCB skull dimension, the distance between the anterior point of premaxilla and occipital condyles. Skulls were reoriented in 3D using DataViewer SW (Bruker) to reach the same position for all specimens. Distances (LCB) were measured in the sagittal plane of 3D visualization.
Statistical Methods.
Data are presented as mean ± SEM. Differences between two groups were analyzed by an unpaired two-tailed t test. The Mann–Whitney U test was used when Gaussian distribution could not be assumed. Differences between groups were considered significant at P < 0.05. All statistical tests were performed using GraphPad Prism 7. Sample sizes and individual statistical results for all analyses are provided in the figure legends and tables.
Supplementary Material
Acknowledgments
We thank the Imaging Methods Core Facility at Biotechnology and Biomedicine Centre of the Academy of Sciences and Charles University (BIOCEV) and micro-CT at the First Faculty of Medicine (CZ.1.05/41.00/16.0346). We also thank A. Pavlinek for manuscript editing and Nanduri Prabhakar (University of Chicago) for helpful discussions and presubmission manuscript review. This research was supported by Czech Science Foundation Grants 17-04719S and 19-07378S (to G.P.); institutional support of Czech Academy of Sciences Grant RVO 86652036 (to G.P.); European Regional Development Fund Grant BIOCEV CZ.1.05/1.1.00/02.0109 (to G.P.); and Czech Ministry of Education, Youth and Sports [Grants Progres Q29 (to M.B.) and Progres Q38 (to D.S.), European Cooperation in Science and Technology (INTER-COST) Award LTC17023, and Czech Bio-Imaging Award LM2015062].
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903510116/-/DCSupplemental.
References
- 1.Kimura K., Ieda M., Fukuda K., Development, maturation, and transdifferentiation of cardiac sympathetic nerves. Circ. Res. 110, 325–336 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Leistner H. L., et al. , Heart rate and heart rate variability during sleep in aborted sudden infant death syndrome. J. Pediatr. 97, 51–55 (1980). [DOI] [PubMed] [Google Scholar]
- 3.Vaseghi M., Shivkumar K., The role of the autonomic nervous system in sudden cardiac death. Prog. Cardiovasc. Dis. 50, 404–419 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rohrer H., Transcriptional control of differentiation and neurogenesis in autonomic ganglia. Eur. J. Neurosci. 34, 1563–1573 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Huber K., The sympathoadrenal cell lineage: Specification, diversification, and new perspectives. Dev. Biol. 298, 335–343 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Gonsalvez D. G., et al. , Proliferation and cell cycle dynamics in the developing stellate ganglion. J. Neurosci. 33, 5969–5979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kameda Y., Signaling molecules and transcription factors involved in the development of the sympathetic nervous system, with special emphasis on the superior cervical ganglion. Cell Tissue Res. 357, 527–548 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Potzner M. R., et al. , Sequential requirement of Sox4 and Sox11 during development of the sympathetic nervous system. Development 137, 775–784 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huber K., et al. , The LIM-homeodomain transcription factor Islet-1 is required for the development of sympathetic neurons and adrenal chromaffin cells. Dev. Biol. 380, 286–298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Q., et al. , Temporal requirements for ISL1 in sympathetic neuron proliferation, differentiation, and diversification. Cell Death Dis. 9, 247 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irie T., et al. , Cardiac sympathetic innervation via middle cervical and stellate ganglia and antiarrhythmic mechanism of bilateral stellectomy. Am. J. Physiol. Heart Circ. Physiol. 312, H392–H405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardini B. J., Lund D. D., Schmid P. G., Organization of the sympathetic postganglionic innervation of the rat heart. J. Auton. Nerv. Syst. 28, 193–201 (1989). [DOI] [PubMed] [Google Scholar]
- 13.Nam J., et al. , Coronary veins determine the pattern of sympathetic innervation in the developing heart. Development 140, 1475–1485 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H., et al. , Role of VEGF and tissue hypoxia in patterning of neural and vascular cells recruited to the embryonic heart. Dev. Dyn. 238, 2760–2769 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer N. V., et al. , Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 12, 149–162 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Compernolle V., et al. , Cardia bifida, defective heart development and abnormal neural crest migration in embryos lacking hypoxia-inducible factor-1α. Cardiovasc. Res. 60, 569–579 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Ryan H. E., et al. , Hypoxia-inducible factor-1α is a positive factor in solid tumor growth. Cancer Res. 60, 4010–4015 (2000). [PubMed] [Google Scholar]
- 18.Krishnan J., et al. , Essential role of developmentally activated hypoxia-inducible factor 1α for cardiac morphogenesis and function. Circ. Res. 103, 1139–1146 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Huang Y., et al. , Cardiac myocyte-specific HIF-1α deletion alters vascularization, energy availability, calcium flux, and contractility in the normoxic heart. FASEB J. 18, 1138–1140 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Guimarães-Camboa N., et al. , HIF-1α represses cell stress pathways to allow proliferation of hypoxic fetal cardiomyocytes. Dev. Cell 33, 507–521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnell P. O., et al. , Regulation of tyrosine hydroxylase promoter activity by the von Hippel-Lindau tumor suppressor protein and hypoxia-inducible transcription factors. J. Neurochem. 85, 483–491 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Milosevic J., et al. , Lack of hypoxia-inducible factor-1 alpha impairs midbrain neural precursor cells involving vascular endothelial growth factor signaling. J. Neurosci. 27, 412–421 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai C. L., et al. , Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 5, 877–889 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L., et al. , Isl1Cre reveals a common Bmp pathway in heart and limb development. Development 133, 1575–1585 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dvorakova M., et al. , Incomplete and delayed Sox2 deletion defines residual ear neurosensory development and maintenance. Sci. Rep. 6, 38253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin L., et al. , β-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 9313–9318 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer I., et al. , A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron 49, 379–393 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Peng T., et al. , Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature 500, 589–592 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macova I., et al. , Neurod1 is essential for the primary tonotopic organization and related auditory information processing in the midbrain. J. Neurosci. 39, 984–1004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Provot S., et al. , Hif-1α regulates differentiation of limb bud mesenchyme and joint development. J. Cell Biol. 177, 451–464 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendelsohn M. E., Karas R. H., Molecular and cellular basis of cardiovascular gender differences. Science 308, 1583–1587 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Glebova N. O., Ginty D. D., Heterogeneous requirement of NGF for sympathetic target innervation in vivo. J. Neurosci. 24, 743–751 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang X., et al. , Transcription factor ISL1 is essential for pacemaker development and function. J. Clin. Invest. 125, 3256–3268 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang X., et al. , HCN4 dynamically marks the first heart field and conduction system precursors. Circ. Res. 113, 399–407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sedmera D., et al. , Changes in myocardial composition and conduction properties in rat heart failure model induced by chronic volume overload. Front. Physiol. 7, 367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tittarelli A., Janji B., Van Moer K., Noman M. Z., Chouaib S., The selective degradation of synaptic connexin 43 protein by hypoxia-induced autophagy impairs natural killer cell-mediated tumor cell killing. J. Biol. Chem. 290, 23670–23679 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pérez-Pomares J. M., et al. , Congenital coronary artery anomalies: A bridge from embryology to anatomy and pathophysiology. A position statement of the Development, Anatomy, and Pathology ESC Working Group. Cardiovasc. Res. 109, 204–216 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Pfaff S. L., Mendelsohn M., Stewart C. L., Edlund T., Jessell T. M., Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell 84, 309–320 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Kasemeier-Kulesa J. C., Morrison J. A., Lefcort F., Kulesa P. M., TrkB/BDNF signalling patterns the sympathetic nervous system. Nat. Commun. 6, 8281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Semenza G. L., Oxygen sensing, homeostasis, and disease. N. Engl. J. Med. 365, 537–547 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Semenza G. L., Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cerychova R., Pavlinkova G., HIF-1, metabolism, and diabetes in the embryonic and adult heart. Front. Endocrinol. (Lausanne) 9, 460 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerychova R., et al. , Adverse effects of Hif1a mutation and maternal diabetes on the offspring heart. Cardiovasc. Diabetol. 17, 68 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bohuslavova R., Skvorova L., Sedmera D., Semenza G. L., Pavlinkova G., Increased susceptibility of HIF-1α heterozygous-null mice to cardiovascular malformations associated with maternal diabetes. J. Mol. Cell. Cardiol. 60, 129–141 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Sun Y., et al. , A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat. Neurosci. 11, 1283–1293 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan X., et al. , Disruption of spatiotemporal hypoxic signaling causes congenital heart disease in mice. J. Clin. Invest. 127, 2235–2248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Francis N., et al. , NT-3, like NGF, is required for survival of sympathetic neurons, but not their precursors. Dev. Biol. 210, 411–427 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Hassankhani A., et al. , Overexpression of NGF within the heart of transgenic mice causes hyperinnervation, cardiac enlargement, and hyperplasia of ectopic cells. Dev. Biol. 169, 309–321 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Schulz R., et al. , Connexin 43 is an emerging therapeutic target in ischemia/reperfusion injury, cardioprotection and neuroprotection. Pharmacol. Ther. 153, 90–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deo R., Albert C. M., Epidemiology and genetics of sudden cardiac death. Circulation 125, 620–637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waza A. A., Andrabi K., Hussain M. U., Protein kinase C (PKC)-mediated interaction between conexin43 (Cx43) and K+(ATP) channel subunit (Kir6.1) in cardiomyocyte mitochondria: Implications in cytoprotection against hypoxia-induced cell apoptosis. Cell. Signal. 26, 1909–1917 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Sun Y., et al. , Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev. Biol. 304, 286–296 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi K., et al. , Targeted disruption of the tyrosine hydroxylase locus results in severe catecholamine depletion and perinatal lethality in mice. J. Biol. Chem. 270, 27235–27243 (1995). [DOI] [PubMed] [Google Scholar]
- 54.Faxelius G., Hägnevik K., Lagercrantz H., Lundell B., Irestedt L., Catecholamine surge and lung function after delivery. Arch. Dis. Child. 58, 262–266 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwartz P. J., et al. , Prolongation of the QT interval and the sudden infant death syndrome. N. Engl. J. Med. 338, 1709–1714 (1998). [DOI] [PubMed] [Google Scholar]
- 56.Le Magueresse C., et al. , Subventricular zone-derived neuroblasts use vasculature as a scaffold to migrate radially to the cortex in neonatal mice. Cereb. Cortex 22, 2285–2296 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Himmels P., et al. , Motor neurons control blood vessel patterning in the developing spinal cord. Nat. Commun. 8, 14583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duran J., et al. , The HIF1A C85T single nucleotide polymorphism influences the number of branches of the human coronary tree. Cardiology 121, 156–159 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Wikenheiser J., et al. , Altering HIF-1α through 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) exposure affects coronary vessel development. Cardiovasc. Toxicol. 13, 161–167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pistollato F., et al. , Hypoxia and HIF1α repress the differentiative effects of BMPs in high-grade glioma. Stem Cells 27, 7–17 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Manalo D. J., et al. , Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 105, 659–669 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Ruiz de Almodovar C., et al. , VEGF mediates commissural axon chemoattraction through its receptor Flk1. Neuron 70, 966–978 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mukouyama Y. S., Gerber H. P., Ferrara N., Gu C., Anderson D. J., Peripheral nerve-derived VEGF promotes arterial differentiation via neuropilin 1-mediated positive feedback. Development 132, 941–952 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Oosthuyse B., et al. , Deletion of the hypoxia-response element in the vascular endothelial growth factor promoter causes motor neuron degeneration. Nat. Genet. 28, 131–138 (2001). [DOI] [PubMed] [Google Scholar]
- 65.Bishop T., et al. , Abnormal sympathoadrenal development and systemic hypotension in PHD3−/− mice. Mol. Cell. Biol. 28, 3386–3400 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Semenza G. L., Prabhakar N. R., The role of hypoxia-inducible factors in carotid body (patho) physiology. J. Physiol. 596, 2977–2983 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarkar K., et al. , Hypoxia-inducible factor 1 transcriptional activity in endothelial cells is required for acute phase cardioprotection induced by ischemic preconditioning. Proc. Natl. Acad. Sci. U.S.A. 109, 10504–10509 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kolesová H., Bartoš M., Hsieh W. C., Olejníčková V., Sedmera D., Novel approaches to study coronary vasculature development in mice. Dev. Dyn. 247, 1018–1027 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.