Significance
The 50,000 origins that replicate the human genome are selected from an excess of licensed origins. Firing licensed origins that would otherwise be passively replicated is a simple mechanism to recover DNA replication between stalled replication forks. This plasticity in origin use promotes genome stability if an unknown mechanism prevents a subset of origins from firing during unperturbed DNA replication. We describe ATR and CHK1 kinase signaling that suppresses a CDK1 kinase-dependent phosphorylation on the chromatin protein RIF1. The CDK1 kinase-dependent phosphorylation of RIF1 disrupts its interaction with PP1 phosphatase. Thus, ATR and CHK1 stabilize an interaction between RIF1 and PP1 that counteracts CDC7 and CDK2 kinase signaling at licensed origins. This mechanism limits origin firing during unperturbed DNA replication.
Keywords: replication, ATR, dormant origins, RIF1, CDK1
Abstract
DNA damage-induced signaling by ATR and CHK1 inhibits DNA replication, stabilizes stalled and collapsed replication forks, and mediates the repair of multiple classes of DNA lesions. We and others have shown that ATR kinase inhibitors, three of which are currently undergoing clinical trials, induce excessive origin firing during unperturbed DNA replication, indicating that ATR kinase activity limits replication initiation in the absence of damage. However, the origins impacted and the underlying mechanism(s) have not been described. Here, we show that unperturbed DNA replication is associated with a low level of ATR and CHK1 kinase signaling and that inhibition of this signaling induces dormant origin firing at sites of ongoing replication throughout the S phase. We show that ATR and CHK1 kinase inhibitors induce RIF1 Ser2205 phosphorylation in a CDK1-dependent manner, which disrupts an interaction between RIF1 and PP1 phosphatase. Thus, ATR and CHK1 signaling suppresses CDK1 kinase activity throughout the S phase and stabilizes an interaction between RIF1 and PP1 in replicating cells. PP1 dephosphorylates key CDC7 and CDK2 kinase substrates to inhibit the assembly and activation of the replicative helicase. This mechanism limits origin firing during unperturbed DNA replication in human cells.
The human genome evolved with mechanisms that assemble and activate the replicative helicase to initiate DNA unwinding and DNA replication at 50,000 origins in a spatiotemporal pattern that is broadly conserved from one cell division to the next (1, 2). MCM2-7 hexamers are loaded onto DNA to license origins in G1 phase, and CDC7 and CDK2 kinase activities initiate the assembly of CDC45, MCM2-7 and GINS, as well as the activation of the replicative helicase CMG in S phase (3). The assembly and activation of CMG and the initiation of replication at multiple origins is essential, as the replication of human chromosome 1 by two replisomes synthesizing 1.5 kb/min from a single, central origin would take 50 d (3). Mechanisms that limit origin firing are also essential, as a human epithelial cell contains 200 × 10–18 mol dNTPs (4), 50-fold less than that required to synthesize the 6 × 109-bp genome. The coincident initiation of replication at all licensed origins would rapidly exhaust cellular dNTPs and key replication proteins and stall DNA synthesis (2). While the mechanisms that limit origin firing have not been fully elucidated, it is clear that chromatin structure is among the factors that determine whether origins fire in early or late S phase (5, 6). Key recent findings are that the chromatin factor RIF1 is essential for both the generation of chromatin loops and the spatiotemporal pattern of replication in human cells (7). RIF1 associates with PP1, and this phosphatase activity is proposed to oppose CDC7 and CDK2 kinase activities and prevent the assembly and activation of CMG at licensed origins (8, 9).
The 50,000 origins that replicate the human genome are selected from an 3- to 10-fold excess of licensed origins, with the excess “dormant” origins replicated passively by replication forks emerging from adjacent origins (10, 11). Dormant origins are important for genome stability. If a single replication fork stalls, replication can be recovered by a fork traveling in the opposite direction. If two converging replication forks stall, a simple system to recover the replication lost is to fire an intervening dormant origin. This system requires a mechanism that limits origin firing and generates a pattern that positions dormant origins between active replication forks. Recent computational models of the spatiotemporal pattern of replication in human cells reveal that origin firing in euchromatin and facultative heterochromatin regions can be stochastic and proceed with a domino-like progression, if an unknown mechanism inhibits additional origin firing within 7–120 kb of an origin that fires, a distance that corresponds with the length of chromatin loops (12, 13).
ATR is a DNA-damage-signaling kinase that is activated at single-stranded DNA (ssDNA) exposed by the uncoupling of the CMG helicase from the replicative polymerases at stalled forks and at resected DNA double-strand breaks (14). ATR phosphorylates and activates CHK1 kinase after genotoxic stress, and ATR and CHK1 signaling causes the degradation of CDC25A phosphatase, which prevents dephosphorylation and activation of CDK2 kinase and origin firing in late S phase (15). The ATR and CHK1 signaling that inhibits origin firing in late S phase after genotoxic stress does not inhibit dormant origin firing around stalled replication forks (16). One mechanism that may facilitate dormant origin firing after genotoxic stress is the ATR kinase-dependent phosphorylation of MCM2 Ser108 (17). In Xenopus oocyte extracts, ATR phosphorylates MCM2 after genotoxic stress and phosphorylated MCM2 associates with Plx1 kinase, which opposes CHK1 signaling and facilitates dormant origin firing (18). However, whether this mechanism is present in unperturbed human cells is unclear. Several proteins involved in ATR and CHK1 signaling are required for the assembly and activation of CMG in unperturbed cells (19). TOPBP1 is an allosteric activator of ATR kinase activity (20) that associates with Treslin, and a complex of TOPBP1–Treslin is required for the recruitment of CDC45 into CMG (21). Treslin also associates with CHK1, and the Treslin–CHK1 complex limits origin firing (22). However, the regulation of their interaction has not been studied in detail.
Several groups recently showed that clinical ATR and CHK1 kinase inhibitors induce origin firing in unperturbed human cells with very similar kinetics, revealing the existence of a signaling mechanism that limits origin firing (23–26), which has not been studied to date. Here, we describe an ATR- and CHK1-dependent mechanism that limits origin firing at sites of ongoing replication in unperturbed human cells. We show that ATR and CHK1 kinase activities are temporally associated and essential for the interaction of RIF1 and PP1 at sites of ongoing replication. We show that ATR and CHK1 kinase inhibitors induce CDK1 kinase-mediated RIF1 Ser2205 phosphorylation and that this disrupts an interaction between RIF1 and PP1 phosphatase. Thus, ATR and CHK1 signaling suppresses CDK1 kinase activity throughout S phase and stabilizes an interaction between RIF1 and PP1 at sites of ongoing replication.
Results
ATR Kinase Activity Inhibits the Initiation of Replication at Origins around Active Replication Forks.
ATR and CHK1 kinase inhibitors induce origin firing in unperturbed human cells with very similar kinetics, indicating that an ATR and CHK1 signaling mechanism that limits origin firing exists in the absence of damage (23–26). Previous studies included single-molecule DNA combing that revealed increased origin density in unperturbed cells treated with ATR kinase inhibitors (23–26). However, the population of origins that fire when ATR or CHK1 are inhibited is not known. The possible populations are (i) origins that fire in late S phase; (ii) dormant origins around active replication forks; and (iii) origins that have already fired.
We imaged replication foci pulse-labeled with 5-ethynyl-2′-deoxyuridine (EdU) in early S-phase cells that had been treated with vehicle or ATR kinase inhibitor AZD6738 for 45 min (EdU was added for the last 15 min) using stochastic optical reconstruction microscopy (STORM) (27) and analyzed the spatial pattern of single-molecule localizations using the auto-pair-correlation (auto-PC) function (28). AZD6738 increased the number of fluorophores per focus, resulting in “brighter” EdU foci in both U2OS and BJ-hTERT cells (Fig. 1 A and B), indicating an increase in the amount of DNA synthesized within each focus during the time of treatment. This increase could not result from an increase in replication fork speed, as ATR and CHK1 kinase inhibitors decrease replication fork velocity (23–25). Thus, AZD6738 increases the number of replication forks within each focus, which is consistent with multiple adjacent origins firing close to each other. Similarly, AZD6738 increased the number of proliferating cell nuclear antigen (PCNA) molecules per focus (Fig. 1C). While AZD6738 did not increase the density of EdU foci (number of foci per square micrometer), indicating that new replication factories were not generated (SI Appendix, Fig. S1A), it did increase the density of PCNA foci (SI Appendix, Fig. S1B). This discrepancy is likely a consequence of PCNA foci being smaller and therefore easier to resolve within replication factories than tracks of nascent DNA labeled by EdU. To determine whether ATR kinase inhibitors equally impact DNA synthesis in early and late S phase, we used fluorescence-activated cell sorting (FACS) to quantify the incorporation of EdU. Cells were treated with vehicle or AZD6738 for 30 min, and EdU was then added for 15 min. AZD6738 induced a significant increase of EdU incorporation: 45–50% in U2OS cells and 20% in BJ-hTERT cells (Fig. 1 D–F). We confirmed that the increase in EdU incorporation was due to increased origin firing by pretreating cells with CDC7 kinase inhibitor PHA-767491, which prevented the ATR inhibitor-induced EdU incorporation increase (SI Appendix, Fig. S1 D and E). We compared EdU incorporation in early S-phase (<50% of DNA replicated) and late S-phase (>50% of DNA replicated) cells. The increase in EdU was not significantly different between early and late S-phase cells, showing that the impact of AZD6738 on DNA synthesis is similar in these two populations. These data indicate that ATR inhibition impacts replication throughout the S phase. In addition, we were unable to identify cells with a DNA content greater than 4N after treatment with AZD6738 for 6 h, indicating that ATR inhibition does not induce rereplication in S-phase cells during this time frame (SI Appendix, Fig. S1F).
Fig. 1.
ATR inhibition induces local dormant origin firing throughout the S phase. (A–C) U2OS or BJ-hTERT cells were treated with vehicle or 5 M AZD6738 for 45 min, and EdU was added for the last 15 min of treatment. EdU was detected by click-chemistry reaction, and PCNA was detected with fluorescently tagged antibodies. (A) Representative image of EdU staining is shown. (B and C) Fluorophore per EdU (B) or PCNA (C) focus (FPF) was quantified by STORM imaging and Auto-PC analysis of individual nucleus. One data point displays the average FPF of the foci within one nucleus. The data from three independent experiments are pulled together. t test was used for statistical analyses. ***P < 0.0005; ****P < 0.0001. (D–F) U2OS or BJ-hTERT cells were treated with vehicle or 5 M AZD6738 for 45 min, and EdU was added for the last 15 min of treatment. (D) FACS analysis of EdU/DNA content is shown. (E and F) Fold increase in EdU incorporation after ATR inhibition is quantified from three independent experimental repeats. t test was used for statistical analyses. Cells with 2N-3N DNA content were considered “early S” and 3N-4N “late S.” (G) Repli-seq correlation between log ratio of early to late at LOESS smoothing step of 50-kb windows along the genome of the samples. R, Pearson correlation coefficient. (H) Replication-timing (RT) profiles of U2OS–vehicle and U2OS–AZD6738 for the whole genome and chr7 after LOESS-smoothing step. Data were visualized by using IGV.
To determine whether AZD6738 induces origins that typically fire in the late S phase to fire in early S-phase cells, we analyzed the spatiotemporal pattern of DNA replication before and after AZD6738. We pulse-labeled DNA synthesis with iododeoxyuridine (IdU) for 15 min to identify the replication pattern before treatment, washed the IdU out, and then treated cells with vehicle or AZD6738 for 20 min. We pulse-labeled DNA synthesis during the last 5 min of vehicle or AZD6738 treatment with chlorodeoxyuridine (CldU) (SI Appendix, Fig. S1 G and H). We observed no difference in the spatiotemporal pattern of DNA replication before and after AZD6738 in early or late S-phase U2OS and BJ-hTERT cells, indicating that AZD6738 does not induce late origins to fire in early S-phase cells.
To further confirm the absence of late origin firing in early S-phase cells in response to ATR inhibition, we performed Repli-seq. U2OS cells were treated with vehicle or AZD6738 for 30 min before labeling nascent DNA with bromodeoxyuridine (BrdU) for another 30 min. Cells were sorted based on their DNA content by using FACS. BrdU-labeled DNA was purified from early and late S-phase populations and sequenced, allowing a detailed analysis of replication timing (29). No difference in replication timing between the control and AZD7638-treated samples (Fig. 1 G and H) was observed, confirming that ATR inhibition does not cause immediate late origin firing in early S-phase cells.
Taken together, these data show that ATR kinase signaling inhibits dormant origin firing in unperturbed cells around active replication forks throughout S-phase, without affecting the replication timing program.
ATR and CHK1 Kinase Activities in Unperturbed Cells Are Associated with Replication.
ATR and CHK1 kinase signaling is essential for dormant origin suppression in unperturbed cells. We hypothesized that the cause of ATR kinase activity in unperturbed cells is replication. We treated 293T and U2OS cells with CDC7 kinase inhibitor PHA-767491 for 75 min to block the assembly and activation of CMG. Since, in human cells, the average replicon is 30 kb and replication forks move at 2–3 kb/min, the 75-min incubation with a CDC7 kinase inhibitor is sufficient to allow completion of most ongoing DNA replication. Both CHK1 autophosphorylation of Ser296 and ATR kinase-dependent CHK1 phosphorylation of Ser345 (Fig. 2A) were decreased after CDC7 inhibition (Fig. 2 B and C and SI Appendix, Fig. S2), indicating that ATR and CHK1 kinase activities in 293T and U2OS cells are associated with replication.
Fig. 2.
ATR and CHK1 activities are tightly linked and associated with unperturbed replication. (A) Schematic structure of CHK1 and the phosphorylation sites associated with its activation. (B and C) The 293T (B) or U2OS (C) cells were treated with 5 M AZD6738 and/or 10 M PHA-767491 (CDC7i) for 1 h before the addition of 5 mM HU. Cells were harvested 30 min after the addition of HU; Western blots of total cell lysates are shown. (D) Schematic representation of the experiments in E and F. (E and F) The 293T or BJ-hTERT cells were treated with 5 M AZD6738 or 200 nM UCN01. Inhibitors were added 15 min before or 15 min after irradiation with 10 J/m2 UV, and cells were harvested 1 h after UV treatment; Western blots of total cell lysates are shown. (G) Schematic representation of the experiments in H and I. (H and I) A concentration of 5 M AZD6738 or vehicle was added 30 min after UV, and cells were harvested at the indicated timepoints after UV; Western blots of total cell lysates are shown.
ATR and CHK1 Kinase Activities Are Temporally Associated.
CHK1 kinase activity is ATR kinase-dependent after genotoxic stress (30) and ATR and CHK1 kinase inhibitors induce origin firing with similar kinetics (24). We hypothesized that ATR kinase-dependent CHK1 activity is rapidly reversible and that ATR kinase inhibitors cause a rapid inactivation of CHK1 kinase. To address this possibility, we examined the ATR kinase-dependent phosphorylation of CHK1 Ser345 and CHK1 kinase autophosphorylation of Ser296. We confirmed that while ATR kinase inhibition blocked UV-irradiation-induced CHK1 phosphorylation on both Ser345 and Ser296, the impact of CHK1 kinase inhibition was limited to blocking CHK1 autophosphorylation of Ser296 and not ATR kinase-dependent CHK1 phosphorylation of Ser345 (Fig. 2 D–F). To determine whether ATR kinase activity is required to maintain active CHK1, we UV-treated cells and added AZD6738 30 min after irradiation, at which time both phosphorylations had been induced. Within 10 min of treatment with AZD6738, both CHK1 autophosphorylation of Ser296 and ATR kinase-dependent CHK1 phosphorylation of Ser345 were reversed, indicating that CHK1 was inactivated very quickly after ATR kinase inhibition (Fig. 2 G–I). Thus, CHK1 kinase activity is ATR kinase-dependent, and the two are temporally and sequentially associated. These data show that CHK1 kinase activity is highly dynamic and that CHK1 kinase activity requires continuous ATR kinase activity. This is an important conclusion that has not been demonstrated previously.
Spatiotemporal Analyses of CHK1 Kinase Activity.
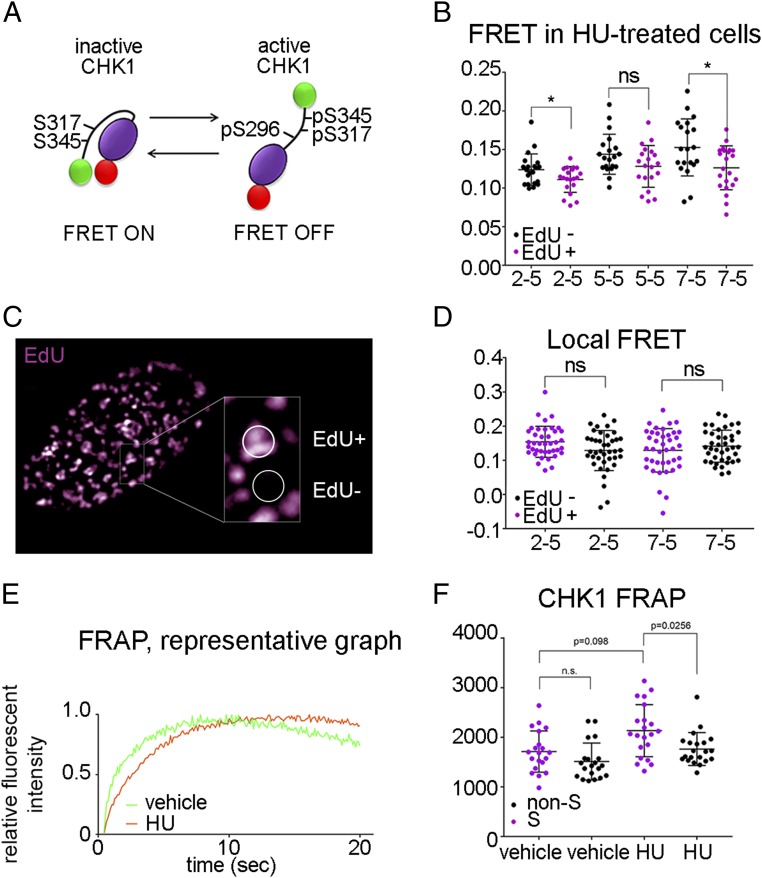
The C-terminal regulatory domain of CHK1 interacts with its N-terminal kinase domain, and this intramolecular interaction physically inhibits kinase activity in vitro (31, 32). ATR kinase-dependent phosphorylations of CHK1 Ser317 and Ser345 induce a conformational change from closed inactive to open active CHK1 kinase (32–40). We examined CHK1 activation using an intramolecular fluorescence resonance energy transfer (FRET) reporter of this conformational change. We generated a FRET construct, similar to that described, in which the two fluorophores are in close apposition in “closed conformation” inactive CHK1 allowing a FRET signal, and the two fluorophores do not interact in “open conformation” active CHK1 allowing no FRET between the two fluorophores (41). We fused mCherry to the N terminus and mNeon to the C terminus of CHK1 (Fig. 3A). We generated three mCherry–CHK1–mNeon constructs with different linker lengths: two amino acids between mCherry and CHK1 and five amino acids between CHK1 and mNeon (2-5), as well as 5-5 and 7-5, and transfected these plasmids into U2OS cells. These three recombinant CHK1 proteins were stably expressed and active in cells (SI Appendix, Fig. S3A). To determine whether these recombinant CHK1 proteins were FRET reporters of CHK1 kinase activity, we labeled ongoing replication with EdU and then induced replication stress by treating cells with hydroxyurea (HU) for 30 min. We anticipated increased CHK1 kinase activity and reduced FRET in the EdU-positive cells treated with HU, and no change in CHK1 kinase activity and FRET in the nonreplicating cells. FRET was determined by measuring the increase in donor fluorescence after photobleaching the acceptor (SI Appendix, Fig. S3B). We observed increased CHK1 kinase activity and reduced FRET in the “2-5” and “7-5” recombinant CHK1 proteins in EdU-positive cells after HU (Fig. 3B).
Fig. 3.
Spatiotemporal analyses of CHK1 kinase activity. (A) Schematic structure of the CHK1–FRET construct and its conformational change after CHK1 activation. (B) U2OS cells were transfected with mCherry–CHK1–mNeon constructs with different linker lengths. At 48 h after transfection, ongoing replication was labeled with EdU for 15 min, followed by HU treatment (5 mM, 30 min) to activate ATR and CHK1. Cells were fixed, and EdU was detected by using click chemistry. Cells were mounted, and FRET was probed by acceptor photobleaching. The relative increase in donor fluorescence is shown. t test was used for statistical analyses. *P < 0.05. ns, not significant. (C and D) U2OS cells were transfected with mCherry–CHK1–mNeon constructs with different linker lengths. At 48 h after transfections, ongoing replication was labeled with EdU for 15 min, cells were fixed, and EdU was detected by using click chemistry. Cells were mounted, and FRET was probed by acceptor photobleaching. The relative increase in donor fluorescence within EdU foci vs. EdU-negative areas of the replicating nuclei is shown. One-way ANOVA was used for statistical analyses. (E and F) U2OS cells were transfected with mCherry–CHK1–mNeon 7-5 construct and vector expressing mCerulean–PCNA. FRAP experiments were performed as described in Materials and Methods. HU (5 mM)-treated cells were imaged between 30 and 60 min of HU treatment. (E) Representative graph of the relative fluorescent intensity normalized to the maximum intensity after recovery in replicating cells. (F) Time of half-recovery was quantified. One-way ANOVA was used for statistical analyses.
To determine whether CHK1 kinase activity is localized at sites of ongoing DNA replication during unperturbed S-phase, we transfected U2OS cells with the 2-5 and 2-7 constructs. We pulse-labeled replication with a 15-min EdU treatment and compared FRET at EdU-positive replication foci with FRET at the EdU-negative (nonreplicating) areas in S-phase cells (Fig. 3C). We hypothesized that CHK1 kinase activity localized around replication forks emerging from fired origins might inhibit origin firing in the vicinity. We observed no significant difference in CHK1 kinase activity, as measured by FRET, between replication foci and nonreplicating areas in S-phase nuclei (Fig. 3D). Since CHK1 kinase activity requires continuous ATR kinase activity, and ATR kinase activity is associated with origin firing, these data suggest that CHK1 kinase activity diffuses very rapidly in S-phase nuclei.
To determine the diffusion coefficient of inactive and active CHK1, we used a stable clonal U2OS cell line expressing 7-5 mCherry–CHK1–mNeon. Transient expression of mCerulean–PCNA was used to identify S-phase cells (SI Appendix, Fig. S3C). We photobleached mCherry in mCherry–CHK1–mNeon in 50% of the nucleus of live cells and measured the time to recover half of maximum fluorescent intensity (fluorescence recovery after photobleaching; FRAP). We observed no significant difference in CHK1 diffusion in replicating and nonreplicating cells. We observed decreased CHK1 diffusion in S-phase cells treated with HU, which induces stalled replication forks and CHK1 activation, compared with G1- and G2-phase cells treated with HU, as well as unperturbed S-phase cells (Fig. 3 E and F). These data show that active CHK1 has a lower diffusion coefficient than inactive CHK1. Since ATR is bound in complexes at structures that increase its activity, and ATR and CHK1 activities are temporally associated, the reduced mobility of active CHK1 may create increased concentrations of CHK1 kinase activity in the vicinity of active replication.
ATR and CHK1 Signaling Is Essential for the Association of RIF1 and PP1.
Our published data (24) show that ATR and CHK1 activity inhibits origin firing by opposing CDC7 kinase activity. The mechanism that connects CHK1 and CDC7 is not known. Recently, two groups (8, 9) reported that an association between RIF1 and PP1 inhibits origin firing in S-phase cells. In the proposed models, PP1 phosphatase opposes CDC7 kinase-dependent phosphorylations preventing origin firing. Since the phenotype induced by ATR and CHK1 kinase inhibitors is similar to the phenotype observed when the association of RIF1 and PP1 is disrupted (9), we hypothesized that the RIF1–PP1 complex is regulated by ATR and CHK1 signaling.
We examined the association of RIF1 and PP1alpha using the proximity ligation assay (PLA). Cells were treated with vehicle or AZD6738 for 1 h and pulse-labeled with EdU to identify S-phase cells. RIF1–PP1alpha PLA foci in EdU-positive cells were enumerated. AZD6738 significantly decreased the number of RIF1–PP1alpha PLA foci in S-phase U2OS and BJ-hTERT cells (Fig. 4 A and B and SI Appendix, Fig. S4 A and B).
Fig. 4.
The RIF1–PP1alpha interaction is disrupted by ATR and CHK1 inhibitors. (A) Schematic representation of RIF1–PP1 PLA principle. (B) U2OS were treated with vehicle or 5 M AZD6738 for 60 min, and EdU was added for the last 15 min of treatment. The quantification of PLA data in the EdU-positive (S-phase) cells is shown (the number of PLA foci per nucleus). One-way ANOVA was used for statistical analyses. ****P < 0.0001. (C and D) The 293T cells were transfected with a plasmid expressing RIF1–GFP. At 48 h later, cells were treated with 5 M vehicle or the indicated concentrations of AZD6738 or 200 nM CHK1 inhibitor AZD7762 for 1 h. RIF1 was immunoprecipitated by using GFP-Trap beads. Western blot analyses of RIF1 and PP1alpha in the immunoprecipitation samples are shown.
We immunoprecipitated GFP-tagged RIF1 from 293T cells treated with vehicle or AZD6738. ATR kinase inhibition decreased the amount of PP1alpha that coimmunoprecipitated with RIF1 (Fig. 4C). The association of RIF1 and PP1alpha was not mediated by DNA (SI Appendix, Fig. S4C) and was disrupted by both ATR and CHK1 kinase inhibitors (Fig. 4D). The levels of both RIF1 and PP1 proteins in the total cell lysates were unaffected by ATR or CHK1 inhibition (SI Appendix, Fig. S4D). Together, our data showed that stable RIF–PP1 association required continuous ATR and CHK1 activities.
RIF1 Is Phosphorylated on S2205 in Response to ATR Inhibition to Disrupt the RIF1–PP1 Interaction.
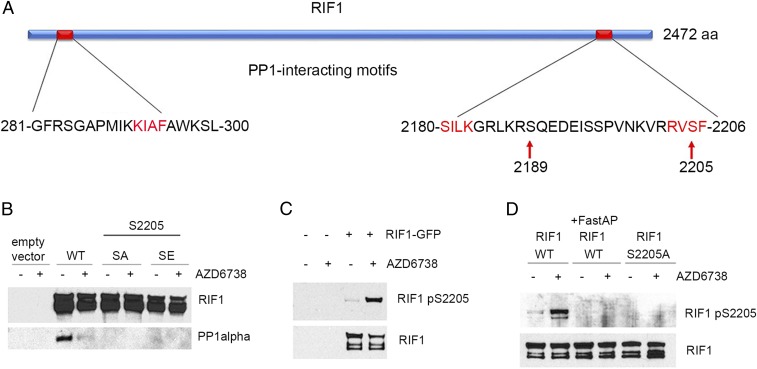
Since the association of RIF1 and PP1alpha was disrupted by ATR and CHK1 inhibitors, we examined the sequence of RIF1 for phosphorylation sites that might mediate the interaction with PP1. We identified two known phosphorylation sites in the domain of RIF1 that is known to associate with PP1. RIF1 Ser2205 was identified as a CHK1-dependent phosphorylation site (42), and RIF1 Ser2189 is followed by glutamine generating a consensus sequence (SQ) for ATR kinase phosphorylation (Fig. 5A). We mutated RIF1 Ser2205 to alanine (RIF1 S2205A), an amino acid that cannot be phosphorylated, and to glutamic acid (RIF1 S2205E), which mimics serine phosphorylation, and determined the impact of these mutations on the association of RIF1 and PP1alpha. Both RIF1 S2205A and RIF1 S2205E disrupted the association of RIF1 and PP1alpha, revealing a critical role for RIF1 Ser2205 in this association, but providing no insight as to how the phosphorylation of RIF1 Ser2205 impacts the association (Fig. 5B). Mutations in RIF1 Ser2189 did not impact the association of RIF1 and PP1alpha (SI Appendix, Fig. S5A).
Fig. 5.
ATR and CHK1 inhibitors induce phosphorylation of RIF1 on Ser2205. (A) Schematic representation of RIF1 and its PP1-interacting motifs (red font). Phosphorylation sites are shown with arrows. (B–D) The 293T cells were transfected with a plasmid expressing wild-type (WT) RIF1–GFP or S2205A (SA) or S2205E (SE) RIF1 mutants. At 48 h later, cells were treated with vehicle or 5 M AZD6738, followed by RIF1–GFP immunoprecipitation. (B) RIF1 and PP1alpha Western blots are shown. (C and D) RIF1 and RIF1 pSer2205 Western blots are shown. (D) Indicated samples were treated with FastAP phosphatase for 30 min after the immunoprecipitation.
To further investigate the role of RIF1 pSer2205 in association of RIF1 and PP1alpha, we generated an antibody that only recognizes RIF1 when it is phosphorylated on Ser2205. The phosphorylation of RIF1 Ser2205 was greatly increased in cells treated with AZD6738 (Fig. 5C), two other ATR inhibitors (Ve822 and ETP46464), and two CHK1 inhibitors (AZD7762 and UCN01) (SI Appendix, Fig. S5 C and D). The selectivity of this anti-RIF1 pSer2205 antibody was confirmed by immunoblotting and dot blot (Fig. 5D and SI Appendix, Fig. S5B). The anti-RIF1 pSer2205 antibody recognized RIF1 pSer2205, but not RIF1 S2205A mutant, and not RIF1 treated with alkaline phosphatase after the immunoprecipitation (IP) (Fig. 5D). Consistent with our hypothesis, during the course of this work, it was shown that a RIF1 peptide containing pSer2205 was unable to bind PP1alpha in vitro (43). Taken together, these data suggest that ATR and CHK1 kinase inhibitors induce the phosphorylation of RIF1 Ser2205, and this modification disrupts the association of RIF1 and PP1alpha around active replication forks where it opposes CDC7 and CDK2 kinase activities and origin firing.
ATR Inhibitor-Induced Dormant Origin Firing Is CDK1-Dependent.
To identify the kinase that phosphorylates RIF1 Ser2205 after ATR inhibition, we investigated Aurora B (43) and PKA (44) kinases that have been reported to phosphorylate this site; CKII, which phosphorylates a consensus sequence that includes the sequence at RIF1 Ser2205; and CDC7 and CDK2 that are essential for the assembly and activation of the replicative helicase. Inhibitors of these kinases did not prevent ATR inhibitor-induced RIF1 pSer2205 (SI Appendix, Fig. S6).
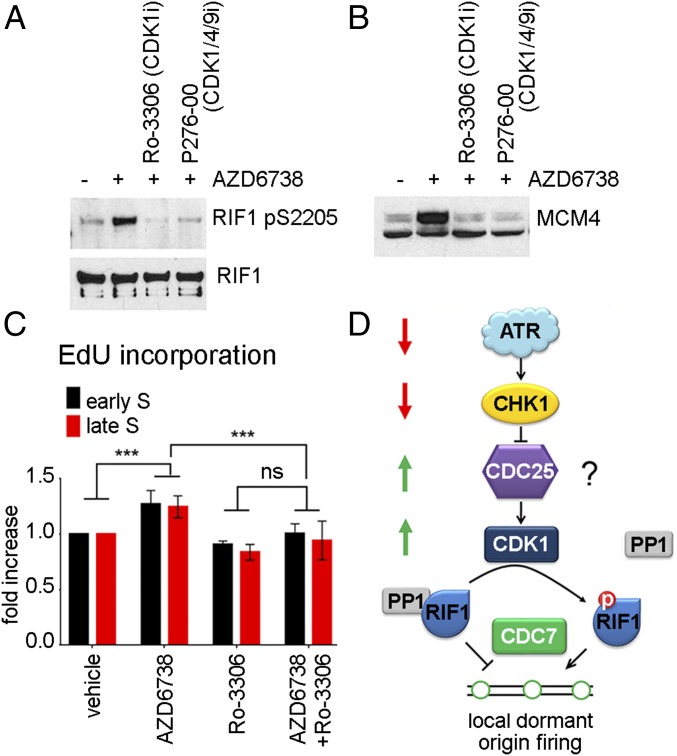
Recently, ATR kinase was reported to control the S/G2 transition by preventing CDK1-dependent phosphorylation events in S-phase cells (45). We used CDK1 and CDK1/4/9 inhibitors to determine whether CDK1 plays a role in ATR inhibitor-induced origin firing. CDK1 inhibition blocked ATR inhibitor-induced RIF1 pSer2205 (Fig. 6A), MCM4 hyperphosphorylation on chromatin (Fig. 6B), and an increase in EdU incorporation (Fig. 6C). We conclude that ATR inhibitor-induced RIF1 phosphorylation is CDK1 kinase-mediated.
Fig. 6.
ATR inhibitor-induced increase in origin firing is CDK1-mediated. (A) The 293T cells were transfected with a plasmid expressing RIF1–GFP. At 48 h later, cells were pretreated with 5 M CDK1 inhibitor Ro-3306 or 5 M CDK1/4/9 inhibitor P276-00 for 15 min before adding 5 M AZD6738 for 1 h, followed by RIF1–GFP immunoprecipitation. RIF1 and RIF1 pSer2205 Western blots are shown. (B) The 293T cells were pretreated with 5 M CDK1 inhibitor Ro-3306 or 5 M CDK1/4/9 inhibitor P276-00 for 15 min before adding 5 M AZD6738 for 30 min. Western blot of the nuclease-insoluble chromatin fraction is shown. (C) U2OS cells were pretreated with 5 M CDK1 inhibitor Ro-3306 for 15 min before adding 5 M AZD6738 for 30 min. EdU was added for the last 15 min of treatment. EdU incorporation and DNA content were analyzed by FACS. Fold increase in EdU incorporation after ATR inhibition was quantified from 3 independent experimental repeats. Cells with 2N-3N DNA content were considered “early S,” and 3N-4N were “late S.” Two-way ANOVA test was used for statistical analyses. ***P < 0.0005; ns, not significant. (D) Proposed mechanism that limits origin firing during unperturbed replication.
Discussion
Recent computational models of the replication timing program that accurately predict the spatiotemporal pattern of replication through the S phase require an unknown mechanism that inhibits additional origin firing within 7–120 kb of an origin that fires (13). Since the chromatin factor RIF1 is essential for the generation of chromatin loops that are 7–120 kb in length, it is a candidate platform for this unknown mechanism (12, 13). RIF1 is required for the spatiotemporal pattern of replication in human cells (7). RIF1 also associates with PP1 phosphatase, which opposes CDC7 and CDK2 kinase activities and prevents the assembly and activation of CMG at licensed origins (8, 9). The work presented here advances these findings by showing that ATR and CHK1 signaling limits origin firing at sites of ongoing DNA replication in unperturbed cells. ATR and CHK1 kinase activities stabilize an interaction between RIF1 and PP1 by preventing CDK1 activation and the phosphorylation of RIF1 on Ser2205 during unperturbed DNA replication. ATR and CHK1 kinase inhibitors induce dormant origin firing through CDK1 kinase-mediated phosphorylation of RIF1 Ser2205 (Fig. 6) Our FRET-based CHK1 activity reporter was not sensitive enough to show CHK1 kinase activity localized around active replication forks, perhaps because of the limitations of the FRET reporters generated to date. However, since ATR is associated in complex protein machines at DNA lesions and structures that increase ATR kinase activity, and ATR and CHK1 activities are temporally associated, the reduced mobility of active CHK1 may create increased concentrations of CHK1 kinase activity in the vicinity of active replication forks during unperturbed DNA replication.
We show that during unperturbed replication, a low number of ATR and CHK1 molecules are active, and ATR and CHK1 signaling limits the assembly and activation of the replicative helicase in regions of ongoing DNA replication. How is ATR and CHK1 signaling activated? ATR and CHK1 signaling could be initiated by the ssDNA that is generated by CMG helicase activity and between unligated Okazaki fragments. TOPBP1 is an allosteric activator of ATR kinase activity (20) that associates with Treslin, and a complex of TOPBP1–Treslin is required for the assembly of CDC45 into CMG (21). ATR and CHK1 signaling could be initiated by TOPBP1 that is associated with CMG. In agreement with this hypothesis, the ATR-activating domain of TOPBP1 was recently found to play a role in origin suppression during the S phase (46). Recent findings by Saldivar et al. (45) document ATR signaling suppressing CDK1 activity in the S phase and specifically at the S/G2 transition in unperturbed cells. The ATR signaling that suppresses CDK1 signaling at the S/G2 transition was shown to be dependent on ETAA1, a second allosteric activator of ATR kinase activity, in unperturbed cells (45). Further investigation is needed to establish the roles of different mechanisms of ATR activation during unperturbed replication. Our work advances the role of ATR signaling suppressing CDK1 activity in the S phase in a mechanism that limits origin firing during unperturbed DNA replication.
We propose that the ATR and CHK1 kinase signaling that limits origin firing during unperturbed DNA replication is the essential ATR kinase signaling mechanism. We propose that ATR and CHK1 kinase “dormant origin signaling” remains constitutively active during replication stress and after activation of the replicative checkpoint. This ATR and CHK1 dormant origin signaling stabilizes an interaction between RIF1 and PP1 by suppressing CDK1 kinase-mediated phosphorylation of RIF1 Ser2205 and the assembly and activation of the replication fork at the majority of, but not all, licensed origins in regions of ongoing DNA replication. Following replication stress and activation of the replication checkpoint, reduced replication fork velocity and stalled and collapsed replication forks increase the half-life of licensed origins in regions of active replication as their passive replication is delayed, and this allows those that are not suppressed by the ATR–RIF1 pathway to fire. In addition, CHK1 is degraded after replication stress, and this would reduce the ATR and CHK1 kinase signaling that suppresses CDK1 (47). In this system, the failsafe is origins fire in regions of ongoing replication, and genome stability is promoted.
In our experiments, we used both cancer (U2OS) and normal (BJ-hTERT) cell lines, and the mechanisms we describe here are common to both cell lines. However, when we quantified the increase in EdU incorporation induced by ATR kinase inhibitor, the increase in U2OS cells was 45–50%, while the increase in BJ-hTERT fibroblasts was 20% (Fig. 1 E and F). This is an important difference between cancer and normal cells, probably indicating differences in the availability of dormant origins or sensitivity to replication stress or ATR kinase inhibition. Cancer cells tend to be more prone to replication stress and may therefore be more dependent on dormant origins to complete replication. It has been demonstrated that origin use may vary between cell lines and cell types (48). As of yet, the numbers of licensed origins have not been compared between normal and cancer cells. If normal cells have fewer dormant origins, they may be less sensitive to ATR kinase inhibitors as a single agent, and this difference could be exploited to generate a therapeutic window for cancer treatment.
In summary, we define essential ATR and CHK1 kinase dormant origin signaling and describe a mechanism that limits origin firing during unperturbed DNA replication in human cells.
Materials and Methods
Cell Lines.
HEK293T cells and BJ-hTERT fibroblasts were cultured in DMEM and U2OS in RPMI medium containing 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin (Lonza). All of the cell lines were purchased from ATCC. Cells were routinely tested for mycoplasma every 6 mo.
Antibodies and Chemicals.
Antibodies were as follows: MCM4 (Cell Signaling, catalog no. 3228) (used at 1:1,000), CHK1 (Cell Signaling, catalog no. 2360) (used at 1:1,000), CHK1 pS345 (Cell Signaling, catalog no. 2348) (used at 1;1000), CHK1 pS296 (Cell Signaling, catalog no. 2349) (used at 1:1,000), GAPDH (Abcam, catalog no. 8245) (used at 1:10,000), IdU (Becton Dickinson, catalog no. 347580) [used 1:300 for immunofluorescence (IF)], CldU (Abcam, catalog no. 6326) (used 1:100 for IF), PCNA (Abcam, catalog no. ab201674) (used 1:1,000 for STORM), RIF1 [Bethyl, catalog no. A300-568A; used 1:1,000 for Western blot (WB) and 1:500 for PLA], PP1alpha (Thermo, catalog no. 43-8100, used 1:1,000 for PLA; Cell Signaling, catalog no. 2582, used 1:500 for WB). RIF1 pSer2205 antibody was custom-made by Bethyl by injecting rabbits with KVRRV(pS)FADPI peptide and subsequent purifications. Other materials included ATR inhibitor AZD6738 (AstraZeneca); CHK1 inhibitor UCN01 (Sigma); CHK1 inhibitor AZD7762 (AstraZeneca); CDC7 inhibitor PHA-767491, Aurora B inhibitor AZD1152, PKA inhibitor H89, CK2 inhibitor CX-4945, CDK1 inhibitor Ro-3306, and CDK1/4/9 inhibitor P276-00 (all from Selleckchem); CDK2i CVT-313 (Santa Cruz); EdU (Invitrogen); IdU (Sigma); and CldU (MP Biomedical).
Immunoprecipitations and Immunoblotting.
Cells were lysed in 50 mM TrisHCl (pH 7.5), 150 mM NaCl, 50 mM NaF, 0.5% Tween-20, 1% Nonidet P-40, and protease inhibitors for 20 min on ice. Lysates were cleared by centrifugation, and soluble protein was used for immunoprecipitation or mixed with 2 Laemmli Sample Buffer (Bio-Rad) and incubated for 7 min at 96 °C and analyzed by Western blot. For immunoprecipitation, protein extracts were incubated with GFP-Trap beads (ChromoTek) at 4 °C for 120 min. Beads were washed five times with lysis buffer and incubated with 2 Laemmli Sample Buffer (Bio-Rad) for 7 min at 96 °C. In case of phosphatase treatment, washed beads after IP were incubated with 10 U of FastAP (Fermentas) in 1 FastAP buffer at 37 °C for 30 min, pelleted, and incubated with Laemmli Sample Buffer. Proteins were resolved in 4–12% Bis-Tris or 3–8% Tris–acetate gels (Life Technologies), transferred to 0.45-m nitrocellulose membrane (Bio-Rad), and immunoblotted.
Purification of the Nuclease Insoluble Chromatin Fraction.
Cells were lysed in buffer containing 50 mM TrisHCl (pH 7.5), 150 mM NaCl, 50 mM NaF, 0.5% Tween-20, 1% Nonidet P-40, and protease inhibitors for 20 min on ice. Chromatin was pelleted by centrifugation and suspended in 150 mM Hepes (pH 7.9), 1.5 mM MgCl2, 10% glycerol, 150 mM potassium acetate, and protease inhibitors containing universal nuclease for cell lysis (ThermoFisher, catalog no. 88700) and incubated for 10 min at 37° on the shaker. Nuclease-insoluble chromatin was pelleted by centrifugation, washed with water, and dissolved in Laemmli Sample Buffer.
FRET/Photobleaching.
FRET data were acquired on a Nikon A1 confocal system by using an Apo 60 oil [numerical aperture (NA) 1.4] objective. mNeon was excited at 488 nm, and emission was collected by using a 525/50 bandpass filter. mCherry was excited at 561 nm, and emission was collected by using a 595/50 bandpass filter. FRET efficiency was calculated by enhanced donor fluorescence after acceptor photobleaching [EF = (Ipre – Ipost) × 100/Ipre]. Photobleaching was performed by using a 561-nm laser at 100% for four frames.
FRAP.
FRAP data were acquired on a Leica SP8 confocal system by using an Apo 93 glycerolC (NA 1.3) objective, a white light laser, and an acousto-optic beam splitter. mCherry was excited at 585 nm, and emission was collected from 599 to 702 nm. A galvo scanner was used to collect 9.25 frames per second. Four continuous time points were collected, followed by photobleaching in a region of interest covering approximately half of the nucleus using 100% power at 592 nm, followed by 27 s (250 frames) of postbleaching imaging. Half recovery was calculated using Nikon Elements (Version 5.10).
EdU FACS.
Cells were treated with 10 M EdU for 10 min, trypsinized, washed with PBS, and fixed with cold 70% ethanol on ice for 30 min to overnight. Cells were washed with PBS, and EdU staining was performed by using the EdU Click-iT kit (Thermofisher, catalog no. C10632), according to manufacturer’s instructions. For DNA staining, we used FxCycleTM Far Red Stain (Thermofisher, catalog no. F10348). Flow cytometry was performed by using an Accuri C6 flow cytometer, and data were analyzed by using Accuri C6 software or FlowJo V10 software.
Plasmids.
pDEST pcDNA5-FRT/TO-eGFP-RIF1 was a gift from Daniel Durocher, The Lunenfeld-Tanenbaum Research Institute (Addgene plasmid no. 52506) (49). pcDNA4-CHK1-Flag was a gift from Aziz Sancar, University of North Carolina (Addgene plasmid no. 22894) (50). mCerulean-PCNA-19-SV40NLS-4 was a gift from Michael Davidson, Florida State University (Addgene plasmid no. 55386).
IF, EdU Staining, and PLA.
For IF staining, cells were plated on eight-well glass slides (Falcon). After treatments, cells were washed with PBS, fixed with 4% formaldehyde for 10 min at room temperature, permeabilized with 0.5% Triton X-100 in PBS for 20 min, blocked by 3% BSA in PBS with 0.1% Triton X-100 for 1 h, and incubated with primary antibodies overnight at 4 °C. Slides were washed with 0.5% Triton X-100 in PBS and incubated with secondary antibodies for 1 h in the dark. After washes with 0.1% Triton X-100 in PBS, slides were mounted with Prolong Diamond Antifade Mountant with DAPI (Thermofisher, catalog no. P36962).
In case of IdU/CldU IF, cells were fixed with 70% ethanol for 10 min on ice, followed by 90% methanol for 5 min on ice, then washed twice with PBS. DNA was denatured by incubation with 1.5 N HCl for 40 min at room temperature. Cells were washed with PBS, incubated with 0.5% Tween-20 in PBS for 5 min, and blocked for 30 min with NGS buffer (5% normal goat serum, 0.5% Tween-20, and 0.1% BSA in PBS). Slides were incubated with primary antibodies diluted in NGS buffer overnight at 4 °C. After washes with 0.5% Tween-20 in PBS and high salt buffer (250 mM NaCl, 0.2% Tween-20, and 0.2% Nonidet P-40 in PBS), 15 min each, cells were reblocked for 20 min with NGS buffer and incubated with secondary antibodies labeled with fluorophores for 1 h at room temperature. After washing with 0.5% Tween-20 in PBS, cells were mounted as described above.
In case of EdU staining, cells were labeled with 10 M EdU for 15–30 min. Click-iTTM Plus Alexa FluorTM 647 or 488 Picolyl Azide Toolkits (Thermofisher, catalog nos. C10643 and C10641) were used as directed by the manufacturer after IF antibodies’ staining but before mounting.
PLA was performed by using the DuoLink kit (Sigma), according to manufacturer’s instructions. The concentrations of primary and secondary antibodies were adjusted by analyzing single-antibody controls, that could not have more than one or two foci per nucleus.
Repli-Seq.
U2OS cells were treated with vehicle or 5 M AZD6738 for 30 min before labeling nascent DNA for 30 min with 10 M EdU. Cells were fixed with 70% ethanol, stained with propidium iodide (Thermofisher, catalog no. F10797), sorted based on DNA content by using Beckman Coulter MoFlo Astrios into early S (2N-3N) and late S (3N-4N). DNA extraction and library prep was performed essentially as described (29), with modifications in the BrdU immunoprecipitation. Anti-BrdU antibody (BD, catalog no. 555627) was used in combination with bridging antibody for mouse IgG (Active Motif, catalog no. 53017) and protein A/G PLUS agarose (sc-2003). NEBNext kit no. 7370 was used for library preparation. Sequencing was performed by GENEWIZ using Illumina HiSeq. Raw Repli-seq reads were trimmed and filtered for quality by using Trim Galore. Reads were aligned using bowtie2 (51) against GRCh38(hg38). Genome-wide replication-timing profiles were constructed, scaled, and pooled for analysis as described (29). Briefly, Log2 ratios of early vs. late read counts were calculated for 50-kb nonoverlapping windows and locally estimated scatterplot smoothing (LOESS) smoothed at 300-kb windows. Data were visualized by using IGV (52). Repli-seq data have been deposited in the Gene Expression Omnibus database (accession no. GSE131018) (53).
STORM Imaging.
STORM imaging was performed on a custom-built optical imaging platform based on a Leica DMI 300 inverse microscopy. Briefly, a 639-nm laser (UltraLaser, catalog no. MRL-FN-639-800) was collimated into an HCX APO TIRF Objective (Zeiss, 63, NA = 1.47) and was adjusted to a Highly Inclined and Laminated Optical sheet illumination mode with of 1.5 kW/cm2 at sample illumination area. The emitted photons from the sample-labeling fluorophores were collected through the same objective, further expended by a 2 lens tube, and filtered by a single-band pass filter (Semrock, catalog no. FF01-676/37). A scientific complementary metal–oxide–semiconductor camera (Photometrics, Prime 95B) was used to convert collected photons to analog signals. All raw image acquisition was carried out at 33 Hz and 2,000 frames.
To precisely localize each collected single-molecule Point Spread Function, a 2D Gaussian distribution was used via the maximum-likelihood estimation (MLE). In details, each frame of the collected raw image stack was first box-smoothed with a box size of 4 full-width at half-maximum of a 2D Gaussian profile. We note that the smoothing was weighted by the inverse of the variance of the read-out noise of each pixel. The local maximum after smoothing was then picked and submitted to MLE 2D Gaussian fitting. The likelihood function at each pixel was constructed by convolving the Poisson distribution of the shot noise governed by the photons emitted from a fluorophore nearby and a Gaussian modeled read-out noise of the pixel itself (51). The MLE 2D Gaussian regression was performed via the Gauss–Newton algorithm, and the precision of fitting results was estimated by Cramér–Rao lower bound. The distribution of the fitting precisions was then fitted into a skew-Gaussian distribution. The fitted center was used as the average localization precision, and localization events appeared in consecutive frames within 2.5 such averaged localization precision were averaged into one blinking event. The localization results were then constructed into the final result table that records the localization coordinates of each fluorophore blinking event.
STORM Auto-PC Analyses.
Please see SI Appendix.
Dot Blot.
Primary antibody was preincubated overnight with excess of unmodified (VRRVSFADPI) or phosphorylated (KVRRV(pS)FADPI) RIF1 peptide (Bethyl). A quantity of 32, 64, and 128 g of unmodified or phosphorylated RIF1 peptide was loaded onto a nitrocellulose membrane. The membrane was stained with FastGreen stain to assess the loading, washed, dried, blocked in 5% nonfat milk, and incubated for 1 h with primary antipSer2205 RIF1 antibody (1:2,500 dilution). The membrane was washed and probed with secondary antibody (1:4,000, goat anti-rabbit) for 30 min, followed by washes and visualization with LumiGLO Cell Signaling (catalog no. 7003).
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R01 CA204173 (to C.J.B.) and R01 GM108119 (to E.R.); American Cancer Society Grant 130304-RSG-16-241-01-DMC (to E.R.); V Foundation for Cancer Research Grant D2018-020 (to E.R.), and National Cancer Institute R00 Award CA207871(to H.U.O.). This project used the Hillman Cell and Tissue Imaging Facility and Hillman Flow Cytometry Facility that were supported in part by Award P30CA047904.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Repli-seq data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE131018).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903418116/-/DCSupplemental.
References
- 1.Cayrou C., et al. , Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 21, 1438–1449 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chagin V. O., et al. , 4D visualization of replication foci in mammalian cells corresponding to individual replicons. Nat. Commun. 7, 11231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Donnell M., Langston L., Stillman B., Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harbor Perspect. Biol. 5, a010108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferraro P., Franzolin E., Pontarin G., Reichard P., Bianchi V., Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 38, e85 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon J. R., et al. , Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pope B. D., et al. , Topologically associating domains are stable units of replication-timing regulation. Nature 515, 402–405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamazaki S., et al. , Rif1 regulates the replication timing domains on the human genome. EMBO J. 31, 3667–3677 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alver R. C., Chadha G. S., Gillespie P. J., Blow J. J., Reversal of DDK-mediated MCM phosphorylation by Rif1-PP1 regulates replication initiation and replisome stability independently of ATR/Chk1. Cell Rep. 18, 2508–2520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiraga S. I., et al. , Human RIF1 and protein phosphatase 1 stimulate DNA replication origin licensing but suppress origin activation. EMBO Rep. 18, 403–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burkhart R., et al. , Interactions of human nuclear proteins P1Mcm3 and P1Cdc46. Eur. J. Biochem. 228, 431–438 (1995). [PubMed] [Google Scholar]
- 11.Ibarra A., Schwob E., Mendez J., Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl. Acad. Sci. U.S.A. 105, 8956–8961 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gindin Y., Valenzuela M. S., Aladjem M. I., Meltzer P. S., Bilke S., A chromatin structure-based model accurately predicts DNA replication timing in human cells. Mol. Syst. Biol. 10, 722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lob D., et al. , 3D replicon distributions arise from stochastic initiation and domino-like DNA replication progression. Nat. Commun. 7, 11207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou L., Elledge S. J., Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Sorensen C. S., et al. , Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25a. Cancer Cell 3, 247–258 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Ge X. Q., Blow J. J., Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J. Cell Biol. 191, 1285–1297 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortez D., Glick G., Elledge S. J., Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 101, 10078–10083 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trenz K., Errico A., Costanzo V., Plx1 is required for chromosomal DNA replication under stressful conditions. EMBO J. 27, 876–885 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J., Gold D. A., Shevchenko A., Shevchenko A., Dunphy W. G., Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Mol. Biol. Cell 16, 5269–5282 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumagai A., Lee J., Yoo H. Y., Dunphy W. G., TopBP1 activates the ATR-ATRIP complex. Cell 124, 943–955 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Kumagai A., Shevchenko A., Shevchenko A., Dunphy W. G., Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 140, 349–359 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo C., et al. , Interaction of Chk1 with Treslin negatively regulates the initiation of chromosomal DNA replication. Mol. Cell 57, 492–505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couch F. B., et al. , ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 27, 1610–1623 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moiseeva T., et al. , ATR kinase inhibition induces unscheduled origin firing through a Cdc7-dependent association between GINS and And-1. Nat. Commun. 8, 1392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petermann E., Woodcock M., Helleday T., Chk1 promotes replication fork progression by controlling replication initiation. Proc. Natl. Acad. Sci. U.S.A. 107, 16090–16095 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Syljuasen R. G., et al. , Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell Biol. 25, 3553–3562 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rust M. J., Bates M., Zhuang X., Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sengupta P., et al. , Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat. Methods 8, 969–975 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchal C., et al. , Genome-wide analysis of replication timing by next-generation sequencing with E/L Repli-seq. Nat. Protoc. 13, 819–839 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q., et al. , Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 14, 1448–1459 (2000). [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao H., Piwnica-Worms H., ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell Biol. 21, 4129–4139 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen P., et al. , The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: Implications for Chk1 regulation. Cell 100, 681–692 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Walker M., Black E. J., Oehler V., Gillespie D. A., Scott M. T., Chk1 C-terminal regulatory phosphorylation mediates checkpoint activation by de-repression of Chk1 catalytic activity. Oncogene 28, 2314–2323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira E., Chen Y., Sanchez Y., Conserved ATRMec1 phosphorylation-independent activation of Chk1 by single amino acid substitution in the GD domain. Cell Cycle 8, 1788–1793 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., Han X., Zhang Y., Autoregulatory mechanisms of phosphorylation of checkpoint kinase 1. Cancer Res. 72, 3786–3794 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S. X., Dunphy W. G., Activation of Cenopus Chk1 by mutagenesis of threonine-377. FEBS Lett. 487, 277–281 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Capasso H., et al. , Phosphorylation activates Chk1 and is required for checkpoint-mediated cell cycle arrest. J. Cell Sci. 115, 4555–4564 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Wilsker D., Petermann E., Helleday T., Bunz F., Essential function of Chk1 can be uncoupled from DNA damage checkpoint and replication control. Proc. Natl. Acad. Sci. U.S.A. 105, 20752–20757 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niida H., Katsuno Y., Banerjee B., Hande M. P., Nakanishi M., Specific role of Chk1 phosphorylations in cell survival and checkpoint activation. Mol. Cell Biol. 27, 2572–2581 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong E. Y., et al. , KA1-targeted regulatory domain mutations activate Chk1 in the absence of DNA damage. Sci. Rep. 5, 10856 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han X., et al. , Conformational change of human checkpoint kinase 1 (Chk1) induced by DNA damage. J. Biol. Chem. 291, 12951–12959 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blasius M., et al. , A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 12, R78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nasa I., Rusin S. F., Kettenbach A. N., Moorhead G. B., Aurora B opposes PP1 function in mitosis by phosphorylating the conserved PP1-binding RVxF motif in PP1 regulatory proteins. Sci. Signal. 11, eaai8669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giansanti P., Stokes M. P., Silva J. C., Scholten A., Heck A. J., Interrogating cAMP-dependent kinase signaling in Jurkat T cells via a protein kinase a targeted immune-precipitation phosphoproteomics approach. Mol. Cell Proteomics 12, 3350–3359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saldivar J. C., et al. , An intrinsic S/G2 checkpoint enforced by ATR. Science 361, 806–810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sokka M., Koalick D., Hemmerich P., Syvaoja J. E., Pospiech H., The ATR-activation domain of TopBP1 is required for the suppression of origin firing during the S phase. Int. J. Mol. Sci. 19, 2376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y. W., et al. , The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol. Cell 35, 442–453 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petryk N., et al. , Replication landscape of the human genome. Nat. Commun. 7, 10208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Escribano-Diaz C., et al. , A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49, 872–883 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Unsal-Kacmaz K., et al. , The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol. Cell Biol. 27, 3131–3142 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B., Salzberg S. L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson J. T., et al. , Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moiseeva T. N., Qian C., Sugitani N., Osmanbeyoglu H. U., Bakkenist C. J., The effect of ATR inhibition on the replication timing program. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131018. Deposited 10 May 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.