Significance
Shigella flexneri is an enteroinvasive prokaryote that induces human bacillary dysentery. The delivery of around 30 bacterial effectors inside colonic epithelial cells allows the pathogen to invade, replicate, and move into adjacent cells, hence subverting cellular and immune functions of its host. Intracellular trafficking pathways in eukaryotic cells are essential to regulate cell communication with their environment. Our work shows that two effectors of Shigella flexneri block three main trafficking pathways of its host cell: secretion, recycling, and endocytosis, thereby freezing the exchange through the plasma membrane. As a consequence, Shigella flexneri disorganizes the epithelial cell polarity, disturbs epithelial barrier integrity, and enhances the pathogen capacity to penetrate into the colonic tissue in vivo.
Keywords: bacteria, pathogen, secretion, endocytosis, polarity
Abstract
Intracellular trafficking pathways in eukaryotic cells are essential to maintain organelle identity and structure, and to regulate cell communication with its environment. Shigella flexneri invades and subverts the human colonic epithelium by the injection of virulence factors through a type 3 secretion system (T3SS). In this work, we report the multiple effects of two S. flexneri effectors, IpaJ and VirA, which target small GTPases of the Arf and Rab families, consequently inhibiting several intracellular trafficking pathways. IpaJ and VirA induce large-scale impairment of host protein secretion and block the recycling of surface receptors. Moreover, these two effectors decrease clathrin-dependent and -independent endocytosis. Therefore, S. flexneri infection induces a global blockage of host cell intracellular transport, affecting the exchange between cells and their external environment. The combined action of these effectors disorganizes the epithelial cell polarity, disturbs epithelial barrier integrity, promotes multiple invasion events, and enhances the pathogen capacity to penetrate into the colonic tissue in vivo.
Eukaryotic cells contain a complex array of intracellular membrane-bound compartments, which mediate cell communication with their environment by the bidirectional transport of proteins and lipids between the intracellular and extracellular spaces. This occurs via two main mechanisms: the secretory and the endocytic trafficking pathways. The efficient intracellular transport of molecules is regulated by GTPases of the Arf, Rab, Rho, and dynamin families and is critical to maintain organelle identity and structure. Additionally, the coordination of intracellular trafficking with other pathways regulates vital processes including cell polarity, immunity, signaling, and development, as well as tissue and organ functions (1–3).
Shigella spp. are Gram-negative intracellular bacteria causing bacillary dysentery or shigellosis (4). Shigella invades the colonic epithelium by using a type 3 secretion system that enables the injection of more than 20 virulence factors, the so-called effectors, into the cell (5, 6). These effectors then target multiple cellular functions to promote nonphagocytic uptake, followed by intracellular bacterial replication, cell-to-cell spreading, and subsequently leading to destruction of the colonic epithelium (7, 8). While the enzymatic functions for most effectors have been described and analyzed in cell culture, the mechanisms by which they cooperate with one another to promote infection remains largely unknown. Shigella flexneri induces Golgi apparatus fragmentation and reorganization of the endocytic compartment, leading to a block in secretion and receptor recycling (9). Among the arsenal of injected effectors, two have been specifically implicated in targeting host cell small GTPases essential for Golgi-mediated secretory transport, namely, IpaJ and VirA. IpaJ is a cysteine protease catalyzing the cleavage of myristoylated glycine residues primarily from ADP ribosylation factor (Arf) and Arf-like (Arl) proteins (10, 11). As a consequence, it was shown that IpaJ inhibits STING-mediated activation of the IFN pathway by blocking STING translocation from the endoplasmic reticulum (ER) to ER–Golgi intermediate compartment (ERGIC) (12). Conversely, VirA was reported to impair host cell secretory transport, in addition to inhibiting autophagy (13, 14), by acting as a Rab-GTP activating protein (GAP) with preferential targeting of Rab1, as shown in vitro (13). Although the catalytic activities of these two effectors have been well described, it remains to be elucidated if both act in synergy or independently, and which changes they induce in the intestinal tissue during S. flexneri infection.
In eukaryotes, Arf and Rab protein families work together to regulate intracellular trafficking pathways. However, the exact mechanisms of coordination of action are not yet fully understood. Given that these small GTPases are targeted by both IpaJ and VirA, it raises the question whether these effectors further affect other trafficking pathways in addition to the known secretory transport.
Here, we demonstrate that these two effectors independently block global host cell secretion and concurrently operate to impair receptor recycling. Moreover, we report that IpaJ and VirA decrease receptor-mediated endocytosis. Our results illustrate how S. flexneri “freezes” the invaded host cell by globally interfering on multiple intracellular transport systems, thereby affecting the exchange of molecules between cells and their environment and consequently cell and tissue functions.
Results
S. flexneri Effectors IpaJ and VirA Globally Impair Conventional Secretion.
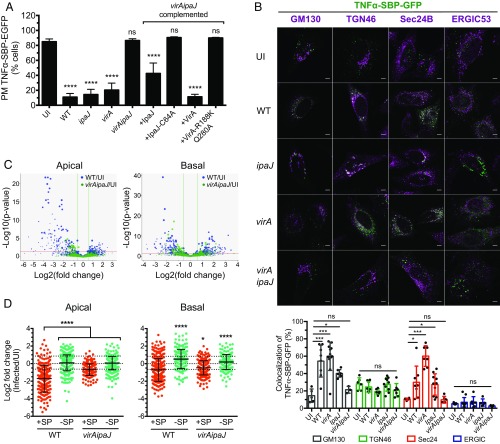
Both IpaJ and VirA S. flexneri effectors have been shown to affect Golgi-mediated transport in host cells (10, 13), raising the question as to whether these effectors operate in synergy or independently. To address this, we first quantified the secretion level of the cytokine TNFα upon infection with wild-type (WT) S. flexneri, or the mutant strains ipaJ, virA, or virAipaJ. We used the retention using selective hooks (RUSH) system (15) to follow the synchronized trafficking of the reporter TNFα-SBP-GFP from the ER to the plasma membrane of epithelial cells (Fig. 1A). Infection with WT S. flexneri blocked 75% of the anterograde trafficking of TNFα-SBP-GFP compared with uninfected cells (UI), in line with previous reports on other cargoes (9, 13). Similar results were obtained when cells were infected with either ipaJ or virA S. flexneri single mutants. However, in cells infected by the virAipaJ double mutant, TNFα-SBP-GFP transport levels were similar to the uninfected condition (Fig. 1A). This differential effect of trafficking during infection by WT, ipaJ, virA, and virAipaJ was not due to impairment in bacterial invasion by the mutant strains (SI Appendix, Fig. S1A). Complementation of the double mutant with either pVirA-myc or pIpaJ-myc was sufficient to restore, at least partially, the secretion inhibitory phenotype obtained with WT bacteria. As expected, complementation with the mutated versions of these effectors in their catalytic sites, pVirA-R188K/Q280A-myc or pIpaJ-C64A-myc (10, 13), did not affect the normal trafficking of TNFα-SBP-GFP (Fig. 1A). Altogether, these results show that each effector, IpaJ and VirA, is sufficient to block anterograde transport of the cargo via their catalytic activities and hence acts largely independently.
Fig. 1.
S. flexneri effectors IpaJ and VirA have a global effect on conventional secretion. (A) IpaJ and VirA effectors block the anterograde transport of the cytokine TNFα. HeLa cells stably expressing the RUSH cargo TNFα-SBP-GFP and the molecular hook streptavidin-KDEL were uninfected (UI) or infected for 1 h with WT-dsRed or the dsRed-expressing mutants ipaJ, virA, virAipaJ, or with virAipaJ complemented with pIpaJ-myc, pIpaJ-C64-myc, pVirA-myc, or pVirA-R188K/Q280A-myc. Cells were then incubated with biotin for 1 h, fixed, and surface-stained for TNFα-SBP-GFP with anti-GFP DyLight 680-conjugated antibody. The arrival of TNFα-SBP-GFP to the plasma membrane (PM) was monitored by FC. Mean ± SD. n = 3. ****P < 0.0001; ns, nonsignificant (one-way ANOVA, Dunnett’s post hoc test, compared with UI). (B) TNFα-SBP-GFP is retained in different subcellular compartments upon S. flexneri infection. HeLa cells stably expressing the RUSH cargo TNFα-SBP-GFP and the molecular hook streptavidin-KDEL were uninfected (UI) or infected for 1 h and incubated with biotin for 1 additional hour. Cells were fixed, permeabilized, and stained with various subcellular markers: anti-GM130 (cis-Golgi), anti-ERGIC53 (ERGIC), anti-Sec24B (ERES), and anti-TGN46 (trans-Golgi network). (Scale bar: 5 µm.) Levels of colocalization between TNFα-SBP-GFP and the different subcellular markers were quantified by SODA plugin in ICY software. Mean ± SD. 5 < n < 12 cells. *P < 0.05; ***P < 0.001 (one-way ANOVA, Dunnett’s post hoc test). (C and D) S. flexneri blocks globally the host conventional secretion. Caco-2/TC7 cells were labeled with SILAC amino acids, grown in Transwell filters, and infected with WT S. flexneri or the mutant virAipaJ. After 4.5 h, the apical and basal media were collected and analyzed by mass spectrometry to determine the relative abundance of secreted proteins in the infected, in comparison with UI, samples. (C) Volcano plots displaying log2 fold changes of secreted proteins from WT or virAipaJ-infected cells in comparison with UI conditions in apical and basal media. (D) Dot plots represent log2 of infected/UI ratio for proteins with (red) and without (green) signal peptide (SP). Mean ± SD. 217 > n > 394 proteins for apical secretome; 194 > n > 281 proteins for basal secretome. *P < 0.05, ****P < 0.0001 [one-way ANOVA, Dunnett’s post hoc test, each condition compared with apical or basal secreted proteins containing signal peptide (+SP)].
To determine in which subcellular compartment TNFα-SBP-GFP was retained upon S. flexneri infection, we utilized immunostaining of various subcellular compartments after 1 h of synchronized trafficking and quantified the percentage of colocalization using a statistical object-based method (Fig. 1B). In uninfected and virAipaJ-infected cells, TNFα-SBP-GFP was mostly at plasma membrane (Fig. 1A), but the minor intracellular pool was mostly colocalized with trans-Golgi network (TGN) labeled with TGN46 (28–21%, respectively) and with cis-Golgi remnants (GM130-positive compartment, 14–20%) (Fig. 1B). By contrast, in WT-infected cells TNFα-SBP-GFP was mostly intracellular (Fig. 1A) and colocalized with GM130 (54%), Sec24B (29%), a marker of ER exit sites (ERES), and with TGN46 (24%). This reveals that S. flexneri WT infection blocked the Golgi-mediated transport at different stages, mainly at ERES and at cis-Golgi. Interestingly, infection with virA or ipaJ mutants resulted in a phenotype similar to the WT strain except that, in absence of IpaJ, TNFα-SBP-GFP was strongly colocalized with Sec24 (60%, Fig. 1B). These data suggest that the retention in the Sec24B-positive compartment was due to the action of IpaJ. We could not observe colocalization of TNFα-SBP-GFP with ERGIC53, a marker of the ERGIC, in any of the conditions tested (Fig. 1B). Taken together, our data show that IpaJ and VirA block the anterograde trafficking at multiple stages, confirming their independent action in blocking host cell secretion.
To investigate the global effects of S. flexneri infection on the secretory transport in a polarized cell system, we used the stable isotope labeling with amino acids in cell culture (SILAC) technology coupled to liquid chromatography–tandem mass spectrometry (LC-MS/MS) (16). Polarized Caco-2/TC7 cells labeled with “medium” amino acids were uninfected, and cells labeled with “heavy” amino acids were infected with either WT S. flexneri or the mutant virAipaJ. After 5 h of infection, the SILAC-labeled secretome-containing supernatants from the apical and basal side were collected and treated for mass spectrometry analysis to determine the relative abundance of peptides between infected and uninfected samples (SI Appendix, Fig. S1B). Fold change-based gene ontology (GO) enrichment analysis showed that apical and basal secretome proteins were more than four-times enriched in extracellular proteins in comparison with the human theoretical proteome, hence revealing the good quality of our analysis. We quantified 148 proteins in the apical and 136 in the basal supernatants that were differentially secreted in WT S. flexneri-infected cells compared with UI cells, from which 125 (84%) and 79 (58%) proteins, respectively, were less secreted (Fig. 1C and SI Appendix, Table S1). This secretion impairment was independent of global changes in the proteome of infected cells (SI Appendix, Fig. S1C). In addition, tight junctions and cell viability were not affected during the secretome collection (SI Appendix, Fig. S1 D–F). Among the differentially secreted proteins, 116 (92.8%) of the apical and 64 (81%) of the basal proteins contained predicted cleavable signal peptides indicating targets of the conventional secretory pathway (Fig. 1D). In contrast, analysis of the same proteins from the virAipaJ-infected samples revealed only a marginal impairment on the secretion of signal-peptide–containing proteins; indeed, the mean fold change was less striking than for the WT-infected secretomes (Fig. 1D and SI Appendix, Table S1). Taken together, these results indicate that S. flexneri infection affects the conventional secretory pathway in a global manner, and that this is mediated by the action of both IpaJ and VirA effectors.
Among the proteins less secreted by WT-infected cells, the most represented class were hydrolytic enzymes, like proteases (cathepsin D, carboxypeptidase M) and enzyme regulators. However, we also found cell receptors (LDLR, HGFR), lipid transporters (apolipoproteins), proteins related to cell adhesion and extracellular matrix (ECM) (laminins), and immunomodulators (IL6ST) (SI Appendix, Table S1). The secretion inhibition of this wide array of proteins may have severe consequences on cell polarity and intestinal tissue organization and functions.
S. flexneri Effectors IpaJ and VirA both Block the Host Cell Recycling Pathway.
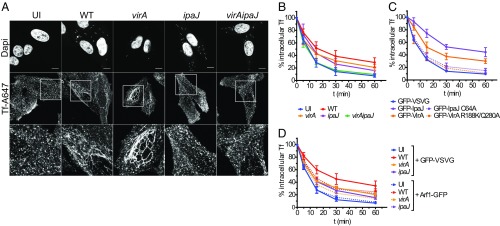
As IpaJ and VirA effectors are targeting small GTPases from the Arf and Rab families, which are important for normal endosomal functioning (1, 3), we further investigated whether they were also implicated in the previously described reorganization of the endosomal compartment and impairment of transferrin (Tf) receptor (TfR) recycling (9). We loaded Hep2 cells with fluorescent Tf coupled to Alexa Fluor 647 (Tf-AF647), before infection with WT, virA, ipaJ, or virAipaJ strains (Fig. 2A). In line with what was previously reported (9), after 1 h of infection with WT S. flexneri, we observed an extensive formation of membrane tubules labeled with Tf-AF647, which resembled the phenotype induced by the drug brefeldin A (BFA) (SI Appendix, Fig. S2A) (17). A comparable, tubular phenotype was observed when cells were infected with the virA mutant strain. In contrast, in cells infected with either the ipaJ or virAipaJ mutant, Tf-AF647 was located in punctate vesicles throughout the cytoplasm, resembling the uninfected control (Fig. 2A). These findings demonstrate that IpaJ is involved in endosomal compartment tubulation. As shown in the case of BFA-treated cells (17, 18) (SI Appendix, Fig. S2B), the formation of endosomal tubules is not necessarily linked to a striking recycling impairment. We therefore assessed TfR recycling kinetics by pulse-chase experiments. Here, cells were loaded with Tf-AF647, uninfected or infected for 1 h with the GFP-expressing WT or mutant S. flexneri strains, and then chased with the unlabeled holo-Tf for different time points before analysis by flow cytometry (FC) (Fig. 2B and SI Appendix, Fig. S2D). We observed a notable reduction of Tf-AF647 recycling in WT-infected cells, whereby 50% of the internalized Tf-AF647 recycled back to the plasma membrane in 17 min, compared with only 9 min for uninfected cells. In addition to the observed time delay induced by the WT strain, Tf recycling was also blocked, as indicated by 29% retention of Tf-AF647 at the 60-min kinetic end-point. When cells were infected with the ipaJ or virA mutant, an intermediate phenotype was observed while the virAipaJ double mutant had normal levels of Tf recycling similarly to uninfected conditions (Fig. 2B and SI Appendix, Fig. S2D). These results demonstrate that a combined action of these two effectors is required to efficiently block receptor recycling in the host cell. Furthermore, we confirmed that the inhibitory effect of IpaJ and VirA on receptor recycling is due to their catalytic activities. While their ectopic expression in Hep2 cells dramatically decreased the Tf recycling rate (Fig. 2C and SI Appendix, Fig. S2E), control levels of Tf recycling were obtained when the VirA-R188K/Q280A or IpaJ-C64A mutated versions were expressed (Fig. 2C and SI Appendix, Fig. S2E). To gain further insights into the mechanisms at play, we overexpressed one of the targets of IpaJ, the small GTPase Arf1 fused to GFP, before infection with dsRed-expressing bacterial strains (Fig. 2D and SI Appendix, Fig. S2F). In uninfected cells, the Tf recycling rate remained unchanged despite overexpression of either Arf1-GFP or GFP-VSVG, which served as a transfection control (Fig. 2D and SI Appendix, Fig. S2 C–F). Interestingly, the rate of Tf recycling was faster in cells infected by the IpaJ-expressing strains, WT and virA, and overexpressing Arf1-GFP (Fig. 2D and SI Appendix, Fig. S2F). In contrast, in ipaJ-infected cells, the recycling kinetics remained unchanged by Arf1 overexpression. These results strongly suggest that IpaJ slows down Tf recycling by targeting Arf1. Overall, we demonstrate that the two S. flexneri effectors, IpaJ and VirA, present catalytic activities that together strongly inhibit two major trafficking pathways, secretion and recycling, that are implicated in the delivery of molecules to the cellular surface.
Fig. 2.
Both S. flexneri IpaJ and VirA affect TfR recycling. (A) IpaJ induces endosomal tubulation. Hep2 cells loaded with Tf-AF647 were uninfected (UI) or infected for 1 h with WT, virA, ipaJ, or virAipaJ strains, fixed, and stained with DAPI (nuclei and bacteria). (Scale bar: 10 µm.) (B–D) Tf recycling kinetics monitored by FC. Hep2 cells nontransfected or transfected with the indicated plasmids were loaded with Tf-AF647 and were left UI or infected for 1 h with the indicated S. flexneri strains. Cells were then chased with unlabeled holo-Tf, and the loss of intracellular Tf-AF647 fluorescence was monitored over time. (B) IpaJ and VirA block Tf recycling. Cells were UI or infected with WT, virA, ipaJ, or virAipaJ strains followed by Tf recycling kinetic. (C) Tf recycling is inhibited by IpaJ and VirA catalytic activities. Cells were transfected with GFP-VSVG (control), GFP-VirA, GFP-IpaJ, or their mutated versions GFP-VirA-R188K/Q280A and GFP-IpaJ-C64A. Tf recycling kinetic was performed 24 h posttransfection. (D) Arf1-GFP overexpression partially recovers Tf recycling. Cells were transfected either with GFP-VSVG (control) or Arf1-GFP, loaded with Tf-AF647, UI or infected with WT, virA or ipaJ strains followed by Tf recycling. Means ± SD from at least three independent experiments are shown (statistical tests in SI Appendix, Fig. S2 D–F).
S. flexneri Infection Affects Different Host Endocytic Pathways.
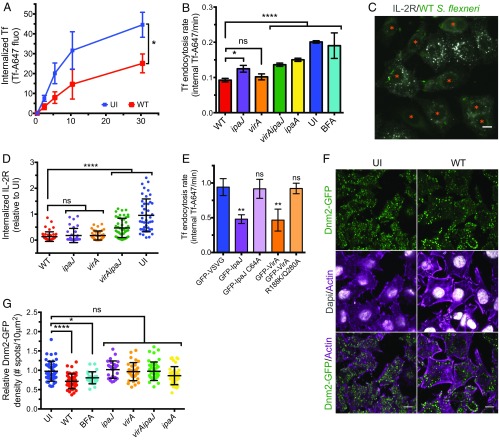
Next, we investigated whether S. flexneri directly blocked the endocytic pathways. In the first instance, we aimed to test S. flexneri infection on clathrin-dependent endocytosis (CDE). To do this, we performed Tf uptake experiments using Tf, a bona fide cargo of CDE (19). Hep2 cells were either left uninfected or were infected for 90 min with GFP-WT S. flexneri, before incubation with Tf-AF647 at 37 °C for different time points, and followed by FC analysis. In WT-infected cells, we observed a 54% reduction in the endocytosis rate, as well as a significant inhibition of the total Tf uptake at the 30 min end-point (Fig. 3 A and B), which was independent of the surface expression of TfR (SI Appendix, Fig. S3A). This defect on Tf uptake was also observed in WT-infected polarized Caco-2/TC7 cells, after basolateral incubation with fluorescent Tf (SI Appendix, Fig. S3 C and D). To test whether other endocytic pathways were affected by S. flexneri infection, we assayed for IL-2 receptor (IL-2R) uptake, a well-described marker of a clathrin-independent endocytosis (CIE) pathway (20). Hep2β cells, stably expressing the IL-2Rβ chain, were left uninfected or infected with WT S. flexneri strain for 90 min and then incubated with an anti–IL-2Rβ chain antibody coupled to Cy3 for 15 min, fixed, and analyzed by fluorescent microscopy (21) (Fig. 3C). We observed a dramatic reduction in IL-2Rβ uptake in WT-infected cells after 15 min of endocytosis compared with uninfected cells (Fig. 3D). These results indicate that WT S. flexneri reduces both clathrin-dependent and -independent endocytosis pathways.
Fig. 3.
S. flexneri affects clathrin-dependent and -independent endocytosis. (A) Tf uptake is decreased upon WT infection. Hep2 cells were left uninfected (UI) or infected with the WT strain for 30 min, and then incubated for different time points with Tf-AF647 at 37 °C. After an acidic wash, the total internal Tf-AF647 fluorescence was quantified by FC. Mean ± SD. n = 4. *P < 0.05 (unpaired two-tailed Welch’s t test performed on area under the curve). (B) Different S. flexneri effectors decrease the Tf endocytosis rate. Hep2 cells were left UI or infected with GFP-expressing WT or mutant strains for 90 min, before monitoring by FC the Tf-AF647 uptake over time. Mean ± SD. n 3. (C and D) IL-2R endocytosis is decreased upon WT S. flexneri infection. (C) UI or WT-infected Hep2β cells were incubated with an anti–IL-2Rβ-Cy3 antibody for 15 min at 37 °C, fixed, and subjected to fluorescent microscopy. (Scale bar: 10 µm.) (D) Quantification of IL-2R endocytosis. Cells either UI or infected by WT, ipaJ, virA, and virAipaJ S. flexneri strains for 90 min were incubated with an anti–IL-2Rβ-Cy3 antibody for 30 min at 37 °C, fixed, subjected to confocal fluorescence microscopy, and analyzed by quantifying the fluorescence intensity of IL-2R–positive vesicles per cell. Mean ± SD. 35 < n < 48 cells. (E) Tf endocytosis is inhibited by IpaJ and VirA catalytic activities. Cells were transfected with GFP-VSVG (control), GFP-VirA, GFP-IpaJ, or their mutated versions GFP-VirA-R188K/Q280A and GFP-IpaJ-C64A. Tf endocytosis kinetic was performed 24 h posttransfection. Mean ± SD. n = 3. (F and G) S. flexneri decreases Dnm2-GFP density at plasma membrane. Hep2β Dnm2-GFP cells were UI, treated with BFA for 30 min, or infected with WT or mutant strains for 90 min. Cells were fixed and labeled with rabbit anti-LPS followed by anti–rabbit-A405 and phalloidin-A647 before confocal (F) or TIRF (G) imaging. (Scale bar: 10 µm.) (G) Quantification of Dnm2-GFP density (number of Dnm2-GFP spots/area) at plasma membrane from TIRF microscopy images. Mean ± SD. 27 < n < 45 cells pooled from two independent experiments. (B, D, E, and G) ns, nonsignificant; *P < 0.05; **P < 0.01; ****P < 0.0001 (one-way ANOVA, Dunnett’s post hoc test).
As IpaJ and VirA effectors target key players in regulating intracellular transport, we asked whether they had an impact on this inhibition of endocytosis. Thus, we infected cells with S. flexneri WT, virA, ipaJ, and virAipaJ strains and performed Tf-AF647 uptake kinetics. We observed a partial recovery of Tf uptake when infecting cells with the three mutant strains (Fig. 3B), indicating that both IpaJ and VirA decrease endocytosis. This IpaJ/VirA-dependent Tf endocytosis inhibition is stronger than BFA, which does not induce significant changes in contrast to uninfected conditions (Fig. 3B), as previously reported (17, 22). In addition, we confirmed that the inhibitory effect of IpaJ and VirA on endocytosis was due to their catalytic activities, as their ectopic expression in Hep2 cells considerably decreased Tf uptake but not when their catalytic-inactive forms were expressed (Fig. 3E). To determine whether other effectors were involved in the Tf uptake impairment, we next tested a panel of S. flexneri mutants lacking effectors with different cellular targets (Fig. 3B and SI Appendix, Fig. S3B). Interestingly, we also found a partial recovery of Tf uptake in cells infected with the ipaA mutant, which lacks the vinculin-binding protein IpaA (Fig. 3B) (23). Again, the differences in Tf uptake rate between mutant strains and WT could not be explained by differences of TfR surface expression (SI Appendix, Fig. S3A). Overall, these results demonstrate that three S. flexneri effectors, IpaJ, VirA, and IpaA, inhibit endocytosis in the host cell.
To gain more insight into the mechanisms by which S. flexneri affects CDE and CIE, we looked at dynamin 2 (Dnm2), an enzyme implicated in both pathways and involved in the pinching off of vesicles from the plasma membrane (24). To this end, we left uninfected or infected Hep2β Dnm2-GFP genome-edited cells (25) with WT S. flexneri and analyzed the Dnm2-GFP distribution at plasma membrane by total internal reflection fluorescence (TIRF) microscopy (Fig. 3 F and G). Upon WT infection, we observed a 27% reduction of Dnm2-GFP at the plasma membrane (Fig. 3F), which was quantified by image analysis (Fig. 3G). This reduction was not due to a decrease of total Dnm2-GFP levels (SI Appendix, Fig. S3E), but likely linked to a reduced recruitment of Dnm2-GFP to the plasma membrane and hence to endocytic sites. Similar results were obtained when we analyzed the clathrin light chain A (CLC) behavior in Hep2β CLC-GFP genome-edited cells (25) (SI Appendix, Fig. S3 F and G). Interestingly, this phenotype was correlated with IpaJ and VirA expression since cells infected with either the single or the double virAipaJ mutants recovered the normal Dnm2-GFP and CLC-GFP recruitment at the plasma membrane (Fig. 3G and SI Appendix, Fig. S3G). In addition, BFA-treated cells also showed a reduction in Dnm2-GFP at the plasma membrane, although smaller than upon infection with WT bacteria (Fig. 3G). In contrast, IpaA led to a marginal effect on Dnm2-GFP distribution (Fig. 3G). These results indicate that Dnm2, a key host factor involved in both CDE and CIE, is affected by S. flexneri, explaining the observed reduction in the uptake of both Tf and IL-2R upon infection. Altogether, our results show how IpaJ and VirA effectors induce multifactorial defects on the general intracellular trafficking of host cells.
IpaJ and VirA Disorganize Cell Polarity and Colonic Tissue Structure.
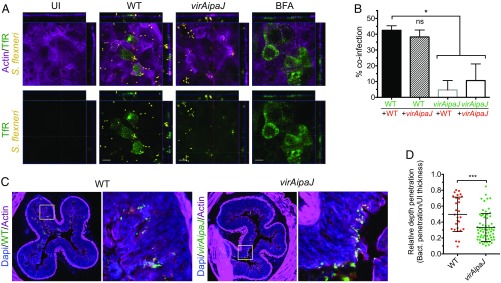
According to our results, IpaJ and VirA are responsible for blocking three intracellular trafficking pathways: secretion, recycling, and endocytosis. We therefore asked whether this vesicular trafficking blockage affected the maintenance of cell polarity. Therefore, we infected polarized Caco-2/TC7 cells and stained the surface TfR, which is usually expressed in the basolateral domain of polarized cells. We observed a pool of the TfR localizing at the apical cell domain upon WT S. flexneri infection, similarly to what has been previously reported in BFA-treated cells (26) (Fig. 4A). On the contrary, the TfR did not localize at the apical domain of uninfected or virAipaJ-infected cells (Fig. 4A). These results indicate that IpaJ and VirA lead to a loss in the polarized transport of TfR, hence highlighting the S. flexneri-mediated disorganization of epithelial polarity.
Fig. 4.
S. flexneri invasion disrupts cell polarity and colonic epithelium. (A) S. flexneri delocalizes the TfR to the apical domain of polarized Caco-2/TC7 cells. Cells were left uninfected (UI), infected with the WT or virAipaJ strains for 5 h, or treated with BFA for 30 min, before fixation without permeabilization and subsequent apical surface staining with an anti-TfR (OKT9) recognizing the extracellular domain of the receptor. (Scale bars: 10 µm.) (B) IpaJ and VirA enhance reinfection of polarized cells. Caco-2/TC7 cells were primo-infected with GFP-expressing WT or virAipaJ strains (green) for 4 h, and then reinfected with the same strains expressing dsRed for 2 more hours (red). The percentage of coinfection was quantified by determining the percentage of primo-infected cells (GFP positive) that were reinfected with dsRed-expressing bacteria. Mean ± SD. n = 4. ns, nonsignificant; *P < 0.05 (Kruskal–Wallis test followed by Dunn’s post hoc test). (C and D) S. flexneri WT has a deeper penetration than virAipaJ mutant into the guinea pig colonic tissue. (C) Colonic tissue from animals infected with GFP-expressing WT or virAipaJ mutant, 8 h postinfection, were stained with phalloidin-A647 (actin) and DAPI (nuclei). (D) The relative depth penetration of both strains into the colonic tissue was measured by their penetration from the epithelial surface compared with the total epithelial mucosa thickness at 4 h postinfection. Mean ± SD. 28 < n < 48 focus of infection pooled from two biological replicates. ***P < 0.001 (Mann–Whitney test).
We next asked whether these two effectors, by disrupting cell polarity, promoted new invasion events in a second round of bacterial infection (Fig. 4B and SI Appendix, Fig. S4A). For that, we first infected cells with either GFP-expressing WT or virAipaJ strains, and then with the same strains but expressing dsRed. We then quantified the percentage of cells coinfected by the GFP and dsRed strains (Fig. 4B and SI Appendix, Fig. S4A). We observed that cells primo-infected with the WT strain were more prone to be reinfected with either the WT or the virAipaJ strain when comparing with virAipaJ primo-infected cells, which were poorly reinfected (Fig. 4B). This result indicates that IpaJ and VirA favor multiple invasion events, thereby enhancing the efficiency of infection.
Next, we used an in vivo model of infection to analyze whether the trafficking impairment caused by IpaJ and VirA induced changes in the colonic epithelial structure and function. We infected guinea pigs intrarectally (27, 28) with S. flexneri WT or the double-mutant virAipaJ. First, we observed a compact Golgi structure in colonocytes from virAipaJ-infected animals compared with a fragmented Golgi apparatus observed in WT-infected animals, confirming the combined action of IpaJ and VirA on disrupting Golgi structure in vivo (SI Appendix, Fig. S4B). We then analyzed the relative depth penetration of WT and virAipaJ strains from the epithelial surface within the colonic tissue (Fig. 4 C and D and SI Appendix, Fig. S4B). We observed and quantified a 40% decrease in the penetration depth of the virAipaJ mutant in contrast to the WT strain. This difference cannot be explained by a spreading defect in the virAipaJ mutant, as the plaques formed on a cell monolayer by the virAipaJ mutant after 48 h of infection are only 3% smaller than the ones formed by the WT strain (SI Appendix, Fig. S4C). Overall, these results indicate that IpaJ and VirA, by the induction of a general trafficking impairment, are critical for intestinal epithelial invasion in vivo.
Discussion
In this study, we showed how two S. flexneri effectors, IpaJ and VirA, are necessary and sufficient to block several key intracellular trafficking pathways in invaded cells, inducing a “frozen” state in which cells are no longer able to exchange molecules with their environment. These results were observed in vitro both in nonpolarized and polarized cells. Moreover, we were able to show in vivo the impact of some of the functions of these effectors on the efficient intestinal invasion by the bacteria.
Our work confirmed, in the context of cellular S. flexneri infection, the secretion blockage described when overexpressing either of the effectors, IpaJ or VirA (10, 13). This is not trivial as EspG, for instance, the enterohemorrhagic and enteropathogenic Escherichia coli (EHEC/EPEC) homolog of Shigella VirA, does not affect the secretory transport during EHEC infection (29), but only during ectopic overexpression. This suggests that Rab1 is not targeted in vivo by EHEC EspG and illustrates how the overexpression of certain virulence factors might induce phenotypes that are not observed during natural infection. However, the fact that VirA blocks the secretory transport in S. flexneri-infected cells strongly suggests that Rab1 is a VirA substrate but does not exclude the possibility that other Rabs might be targeted by this effector. In line with this, we demonstrated that VirA, in combination with IpaJ, impairs the normal recycling of cell receptors. VirA, in contrast to EPEC/EHEC EspG, was shown in vitro to have a broader range of specificity toward Rab GTPases (13). This strongly suggests that endosomal Rabs, such as Rab11, Rab35, or Rab22 (30), are targeted and inactivated by VirA, explaining part of the recycling impairment. Interestingly, it was reported that EspG reduces surface receptor levels and receptor recycling in EHEC-infected cells (29), and this is due to the modulation of an Arf6:Rab35 signaling axis (31). Moreover, EspG was shown to interact with Arf GTPases and PAK (13, 32). However, unlike in EHEC infection, a scaffolding role of VirA modulating Rab-Arf signaling has not been described so far for S. flexneri. On the other hand, our results show that IpaJ and VirA work in concert to induce an additive defect in receptor recycling. Although IpaJ activity slows down Tf recycling by targeting Arf1, this might also happen via the inactivation of other Arf family members (11, 33). Altogether, our work reveals that, in the case of S. flexneri infection, both IpaJ and VirA are necessary to induce strong recycling inhibition by acting on Arf GTPases and possibly on Rab GTPases.
The targeting of the endosomal compartments by these two effectors has consequences not only restricted to the surface receptor recycling, but also to an endocytosis blockage. How IpaJ and VirA reduce Tf uptake, as well as dynamin 2 and clathrin recruitment to endocytic sites at the plasma membrane, remains a key question. One hypothesis is that the concerted action of these two effectors on the secretory and recycling pathways will induce downstream alterations on the endocytic route, as the intracellular trafficking routes are intimately interlinked. Indeed, these two S. flexneri effectors target two families of proteins that are key regulators of many intracellular trafficking events, possibly explaining the endocytosis impairment as an indirect consequence of their activities on small GTPases. One possibility suggested by our results is that impairment of exocytosis pathways by IpaJ and VirA blocks the recruitment of dynamin 2 to the plasma membrane. Interestingly, this inhibition is also observed upon BFA treatment, which targets only Arf proteins. However, the effect of BFA being less strong than Shigella infection might explain why this drug does not significantly decrease Tf endocytosis and further suggests that the combined action of IpaJ and VirA on Arf and Rab proteins is needed to strongly affect endocytosis. Moreover, it was also reported that dynamin 2 is recruited to the budding sites of recycling endosomes, as well as to the TGN (34, 35), possibly explaining the decreased dynamin 2 recruitment upon S. flexneri infection. The bacterium might also affect endocytosis by targeting actin, an important factor for dynamin 2 recruitment (36, 37), since it possesses several effectors modulating actin polymerization, such as IpaC, IcsA, and IpaA (38). In agreement with this, our results demonstrated that IpaA, a vinculin-binding protein involved in actin reorganization, partially inhibits endocytosis. Moreover, the plasma membrane tension, which is regulated by actin polymerization, was shown to affect endocytosis dynamics in polarized cells (39). Thus, by targeting actin dynamic regulators, S. flexneri might modulate the plasma membrane tension, thereby inhibiting the endocytosis rate of the host cell.
Finally, the global secretion inhibition determined by our proteomic approach revealed a list of less secreted proteins that participate in cell polarity establishment and differentiation (40), and their modulation by S. flexneri might perturb the integrity of the intestinal barrier. Candidates among such proteins are those regulating cell adhesion, ECM composition, and lipid transport. In addition, the impairment in the secretion of immunomodulators might affect the host immune response. Moreover, our in vitro data show that IpaJ and VirA subvert the epithelial polarity and promote multiple invasion events, and we have preliminary data suggesting that they might regulate intercellular junctions’ stability. These results are in total agreement with our in vivo data showing that IpaJ and VirA induce a deeper S. flexneri penetration into the colonic tissue.
Overall, our work shows how S. flexneri IpaJ and VirA effectors coordinate and modulate the host cell intracellular trafficking, leading to the subversion of the infected cells and tissue that will result in more efficient bacterial invasion.
Materials and Methods
Bacterial Strains.
Shigella flexneri 5a strain M90T, harboring a streptomycin resistance mutation (41), was used as the WT strain. All of the mutants from this study were generated from the WT strain. All of the mutant strains used in this study were part of a S. flexneri mutant collection (42), except the following: ipgD (43), virAipaJ, ospE1E2, and ospC1C2C3. Tetracycline resistance cassette was removed from virA strain to avoid reduction in IcsA protein levels by FLP-FRT recombination using the pCP20 plasmid. Frozen bacterial stocks were streaked onto trypticase soy agar (TSA) plates containing 0.1% (wt/vol) Congo red (CR) and grown at 37 °C overnight. Plates were kept at 4 °C for up to 2 wk.
Cell Lines.
Hep2 cells (HeLa derivative) and its derivatives expressing the β-chain of IL-2R, Hep2β, were grown in DMEM (1 g/L) (Gibco) supplemented with 10% heat-inactivated FCS (HI-FCS) (Eurobio) at 5% CO2 at 37 °C (44), in the presence of 1 mg/mL G418 (Sigma) for Hep2β cells. CRISPR-Cas9 genome edited Hep2β Dnm2-GFP and Hep2β CLC-GFP cells (25), and the RUSH stable HeLa cell line expressing the ER molecular hook streptavidin-KDEL and the cargo SBP-EGFP-TNFα (45) were described previously. Hep2 and HeLa cells are nonpolarized cells and were always cultured to 70% confluence before infection experiments. Caco-2/TC7 cells (a clone of Caco-2 cells, human colorectal adenocarcinoma origin) were grown in DMEM (1 g/L) supplemented with 20% HI-FCS, GlutaMAX, and nonessential amino acids (Gibco). For complete polarization and differentiation of Caco-2/TC7 cells, 2 × 105 cells/cm2 cells were seeded into 12-well or 6-well Transwell inserts (pore size, 0.4 µm; Corning) and cultured for 18–21 d at 10% CO2 at 37 °C; fresh media was added triweekly, except for plaque assay experiments, where cells were cultured on plastic six-well plates. Transepithelial electrical resistance was measured using a Millicell-ERS volt-ohm meter (Millipore). Dextran permeability was assessed by adding 70-kDa FITC-dextran (200 µg/mL) (Sigma) in Ringer’s buffer to the apical compartment of Transwell inserts (uninfected or infected in Ringer’s solution: 155 mM NaCl, 3 mM KCl, 3 mM NaH2PO4, 5 mM Hepes 10 mM glucose, pH 7.0, with 2 mM CaCl2, 1 mM MgCl2) after a 15-min preincubation in Ringer’s buffer or in calcium-free Ringer’s (containing 10 mM glucose). Fluorescence pass-through to the basal compartment medium was measured with an Infinite M200 Pro multimode plate reader (TECAN). Lactate dehydrogenase (LDH) was quantified with the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega).
Plasmids.
Bacteria transformed with plasmids coding for the Escherichia coli AfaE adhesin (46), or GFP (pFPV 25.1) (47), dsRed (48), or mCherry fluorescent proteins, were used as indicated. pTRIO-mCherry plasmid was generated by inserting mCherry CDS in XmaI–NheI sites of pTRIO plasmid, which is the basic backbone of the pTSAR plasmids series (49). The CDS of ipaJ was cloned through EcoRI–BamHI into pSU2.1tt plasmid (49), giving the pSU2.1tt-IpaJ-Myc plasmid. The catalytic site variant was generated by PCR-based mutagenesis introducing the C64A mutation into ipaJ (plasmid pSU2.1tt-IpaJ-C64A-Myc). ipaJ and virAipaJ mutant strains were complemented with pSU2.1tt-IpaJ-Myc or pSU2.1tt-IpaJ-C64A-Myc plasmids. pSU2.1tt-VirA-Myc or pSU2.1tt-VirA-RQ-Myc plasmids (14) were used to complement virAipaJ or virA strains. For the ectopic expression of GFP-VirA, GFP-IpaJ and their mutated variants in mammalian cells, the CDS of ipaJ and virA were cloned through HindIII–KpnI into pEGFP-C1 plasmid.
Antibodies and Reagents.
The following primary antibodies were used: rabbit anti-S. flexneri 5a M90T LPS (1:300), mouse anti-GM130 (1:200) (BD; 610823), rabbit anti-GM130 (1:100) (Abcam; ab52649), mouse anti-GFP DyLight 680 (1:1,000) (Rockland; 600-144-215), mouse anti-ERGIC53 (1:100) (Sigma; SAB4200585), rabbit anti-Sec24b (1:100) (D7D6S) (Cell Signaling Technology; 12042), sheep anti-TGN46 (1:100) (Bio-Rad; AHP500GT), purified mouse mAb OKT9 (anti-TfR) (1:100), goat anti-dynamin-2 (1:500) (Santa Cruz; sc-6400), rabbit anti-clathrin LCA (H-55) (1:500) (Santa Cruz; sc-28276), rabbit anti-GFP (1:1,000) (Rockland; 600-401-215), rabbit anti-actin (1:5,000) (Sigma; A2066), and chicken anti-GFP (1:1,000) (Abcam; ab13970). All of the secondary antibodies were from Molecular Probes and used at 1:500 dilution; FITC-dextran 70 (Sigma; 46945); DAPI (1 µg/mL) (Sigma); Phalloidin-AF647 (1:100) (A22287; Molecular Probes); BFA (Sigma; B7651) was used at 1 µg/mL.
Bacterial Infections of Cultured Cells.
Hep2 and Hep2β cells were plated the day before the experiment onto 12-mm coverslips at a density of 0.4 × 105 cells/cm2 for immunofluorescence, or on glass-bottom dishes (MatTek) for TIRF microscopy experiments, and at a density of 0.4 × 105 cells/cm2 on six-well plates for FC or Western blot experiments. Bacterial cultures were prepared by picking a single colony from each strain from TSA-CR plates and grown in 8 mL trypticase soy broth (TSB) supplemented with the appropriate antibiotics (ampicillin, 100 µg/mL; chloramphenicol, 10 µg/mL) in a shaking incubator overnight at 30 °C. Bacteria were subcultured in fresh 8 mL of TSB at 37 °C until OD600 0.8–1.0, pelleted, washed in PBS, and coated with poly-l-lysine (molecular weight, 70,000–100,000; Sigma) 10 µg/mL in PBS for 10 min, washed twice with PBS, and resuspended in the infection medium (DMEM supplemented with 20 mM Hepes) to the adequate multiplicity of infection (MOI). Coated bacteria were added to the cells and allowed to adhere for 15 min at room temperature (RT), and then incubated at 37 °C in a CO2 incubator or in a water bath when short time of infection or short kinetics was performed. For infections of Caco-2/TC7 cells grown in Transwell filters, AfaE-expressing strains were cultured as described, pelleted, washed with PBS, and resuspended in infection medium to the adequate MOI. Apical and basal chambers were washed twice with warm DMEM–Hepes, and bacteria were added to the apical chamber at a MOI of 75, incubated for 15 min at RT, and switched to a 37 °C CO2 incubator. After 30 min, the medium was aspirated and replaced for fresh DMEM–Hepes supplemented with gentamicin at 50 µg/mL and incubated for the indicated additional time.
For plaque assays, Caco-2/TC7 cell monolayers were cultured on plastic six-well plates for 3 wk, infected at a MOI of 5 for 2 h, and washed, followed by the addition of a 0.5% agarose overlay containing 50 µg/mL gentamicin in culture medium. Forty-eight hours later, cells were fixed with ethanol and stained with Giemsa R solution. Plaque sizes were quantified using Fiji (ImageJ) software.
RUSH Assay to Assess Anterograde Trafficking.
RUSH stable HeLa cell line expressing the ER molecular hook streptavidin-KDEL and the cargo SBP-EGFP-TNFα (45) were uninfected or infected with the indicated S. flexneri strain expressing mCherry fluorescent protein. After 15 min at RT and 45 min at 37 °C, the medium was replaced by medium containing 40 µM biotin (Sigma) to initiate the cargo transport from the ER and incubated at 37 °C for an additional 60 min. Cells were washed with ice-cold PBS, fixed in PFA (4%)–PBS for 20 min on ice, and stained for 1 h at RT with an anti-GFP DyLight 680 antibody (Rockland) to detect the surface-arrived SBP-EGFP-TNFα. After harvesting, surface anti-GFP DyLight 680 fluorescence was measured by FC (MoFlo Astrios EQ, Beckman) in the infected population of cells (mCherry positive). Results are the mean expressed as the percentage of total (uninfected or infected) cells expressing the RUSH cargo at their surface. Alternatively, cells grown on coverslips were infected as before, and subsequent immunofluorescences were performed as indicated in the figures.
SILAC Labeling, Infections, Sample Collection, and Preparation.
Human Caco-2/TC7 cells were cultured for six passages in SILAC DMEM flex media deficient for l-arginine and l-lysine (Gibco) with 20% heat inactivated dialyzed FBS (dFBS) (Thermo Fisher), 1 g/L glucose (Sigma), GlutaMAX 1×, nonessential amino acids, 10 U/mL penicillin/streptomycin (all from Gibco) supplemented with either 13C6,15N4 l-arginine-HCl and 13C6,15N2 l-lysine-2HCl (Heavy media) or with 13C6 l-arginine-HCl and 4,4,5,5-D4 l-lysine-2HCl (Medium media) (arginine at 84 mg/mL and lysine at 146 mg/mL; Thermo Fisher Scientific). The stable isotope labeling was confirmed by LC-MS/MS after protein in-gel separation and digestion of blue bands. Labeled cells were seeded on six-well Transwell inserts and cultured for 21 d in Heavy or Medium SILAC medium changing the medium three times per week.
For secretome collection, 24 h before infections, cells were washed with DMEM for SILAC with 20 mM Hepes, and medium was replaced with either Medium or Heavy SILAC media but containing 2% dFBS. Cells were uninfected or infected with AfaE-expressing WT or virAipaJ strains. To that end, apical and basal chambers of Transwell inserts were washed three times with DMEM for SILAC/Hepes, Medium SILAC medium was added to uninfected cells, and bacteria were added in the apical chamber at an MOI of 75 in Heavy SILAC medium without FBS or antibiotics. After 15 min at RT and 30 min at 37 °C, cells were washed three times with DMEM for SILAC/Hepes, 1.5 mL of FBS-free Medium or Heavy SILAC media with 50 µg/mL gentamicin were added at either apical or basal Transwell chambers, and cells were further incubated at 37 °C for 4.5 h. Apical and basal secretomes were collected (total volume: 3.6 mL/condition), UI and infected samples mixed in a 1:1 ratio, centrifuged (200 × g, 5 min), filtered with 0.22-µm syringe filters (Minisart; Sartorius Stedim Biotech S.A.), and snap-frozen in liquid N2. For proteome analysis, cells were lysed in RIPA buffer containing protease inhibition mixture, and samples were mixed at a protein stoichiometry ratio of UI:infected 1:1. Samples were kept at −80 °C until use. Before trypsin digestion, uninfected and infected secretomes mixing at a 1:1 ratio was done according to cell number in Transwells from which the secretome was prepared (total volume, 3.6 mL), and then concentrated to 500 μL on Amicon Ultra-15, 10,000 molecular weight cutoff centrifugation filter units (Millipore). For the filter-aided sample preparation protocol, a total of 60 mg of urea and 16 μL of M DTT were added to 500 μL of concentrated secretome; the solution was mixed on a Nanosep (10 KDa, Pall) device and was incubated at 57 °C for 15 min. The mixture was spun down and was washed two times with 500 μL of 2 M urea in 0.1 Tris/HCl, pH 8.5. A total of 100 μL of 0.05 M iodoacetamide was added and was incubated for 30 min at RT in the dark. Two washes with 25 mM ammonium bicarbonate were performed, and the secretome was digested with 5 μg of trypsin/LysC (Promega) for 4 h at 37 °C. The digested peptides were collected by centrifugation, and the filtrate was dried in a vacuum concentrator at room temperature and was redissolved in solvent A (2% acetonitrile, 0.1% formic acid). Peptides were then subjected to LC/MS analysis. For proteome analysis, mixed proteins lysates were separated on 10% SDS/PAGE (Thermo Fisher Scientific) and were digested in-gel with trypsin/LysC (Promega) as described in standard protocols. Extracted peptides were dried in a vacuum concentrator at RT and were redissolved in solvent A (2% MeCN, 0.1% HCO2H) before LC/MS analysis.
LC-MS/MS Analysis.
For the analysis of cell lysates, peptides were separated by reverse-phase chromatography by using an UltiMate 3000 RSLCnano system coupled to an Orbitrap Fusion mass spectrometer (Q-OT-qIT; Thermo Fisher Scientific). Samples were loaded on a nanoViper C18 µ-precolumn (75 µm × 2 cm; Acclaim PepMap; Thermo Scientific) at 5 µL/min of solvent A. After a desalting of 8 min, the precolumn was switched on the C18 column (75 µm × 50 cm; 3 µm, 100 Å; Acclaim PepMap; Thermo Scientific) equilibrated in solvent A. Bound peptides were eluted using a 168-min four-step linear gradient [from 5 to 6% (vol/vol) in 1 min, 6 to 9% in 18 min, 9 to 30% in 132 min, and 30 to 40% in 9 min] of solvent B (100% MeCN, 0.085% HCO2H) at 60 °C and a 300 nL/min flow rate.
For the analysis of secretome samples, peptides were loaded on a C18 µ-precolumn (Thermo Scientific) at 20 µL/min in solvent A. After a desalting step for 3 min, the precolumn was switched on the C18 column (Thermo Scientific) equilibrated in solvent A, and peptides were eluted with a 215-min two-step linear gradient of solvent B [from 5 to 20% (vol/vol) in 147 min and 20 to 40% in 65 min].
We acquired Survey MS scans at a resolution set to a value of 120,000, with a mass range of m/z 400–1,500 and a 4 × 105 ion count target. Tandem MS was performed by isolation at 1.6 Th with the quadrupole, HCD fragmentation with normalized collision energy of 28, and rapid-scan MS analysis in the ion trap. The MS2 ion count target was set to 1 × 104, and only those precursors with charge state from two to seven were sampled for MS2 acquisition. The instrument was run in top speed mode with 3-s cycles.
LC-MS/MS Data Processing and Protein Identification.
Data were acquired using the Xcalibur software (version 3.0), and the resulting spectra were interrogated by Sequest HT through Thermo Scientific Proteome Discoverer (version 2.1) with the SwissProt human database (012016). The mass tolerances in MS and MS/MS were set to 10 ppm and 0.6 Da, respectively. We set carbamidomethyl cysteine, oxidation of methionine, N-terminal acetylation, heavy 13C615N2-lysine (Lys8) and 13C615N4-arginine (Arg10), medium 2H4-lysine (Lys4) and 13C6-arginine (Arg6) as variable modifications. We set specificity of trypsin digestion and allowed two missed cleavage sites.
The resulting files were further processed by using myProMS (version 3.5) (50). The Sequest HT target and decoy search result were validated at 1% false-discovery rate with Percolator. For SILAC-based protein quantification, peptides extracted ion chromatograms (XICs) were retrieved from Thermo Scientific Proteome Discoverer or computed with MassChroQ, version 1.2.1 (51). Global MAD normalization or not was applied on the total signal to correct the XICs for each biological replicate. Protein ratios were computed as the geometrical mean of related peptides. To estimate ratio significance, a t test was performed with the R package limma (52) and the false-discovery rate has been controlled thanks to the Benjamini–Hochberg procedure (53) with a threshold set to 0.05.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (54) partner repository with the dataset identifier PXD012291.
Bioinformatics Analysis.
myProMS (version 3.5) (50) was used to analyze SILAC results. Proteins from infected secretomes were considered as differentially secreted from uninfected secretomes when showing a fold change above or less than 2, P < 0.05 and at least three detected peptides. Fold change-based GO enrichment analysis was performed as in ref. 55. SignalP 4.0 (56) was used to determine which proteins contained a signal peptide (predicted or confirmed). SecretomeP 2.0 (57) was used to detect nonconventional secretion; proteins above a cutoff of 0.5 were considered as secreted proteins. GO terms (58) were extracted from PANTHER (59) and UniprotKB.
Tf Recycling and Endocytosis.
For Tf recycling experiments, Hep2 cells cultured in six-well plates were incubated at RT with DMEM–Hepes (UI) or the indicated bacterial strains expressing GFP or dsRed proteins at an MOI of 100. After 15 min of infection, human Tf coupled to Alexa Fluor 647 (Tf-AF647) (Invitrogen) was added at 0.5 µg/mL in DMEM–Hepes–0.1% BSA at 37 °C. After 30 min of incubation to allow Tf-AF647 endocytosis, surface Tf-AF647 was removed by treating cells with ice-cold sodium acetate (20 mM pH 3.0) for 3 min at 4 °C before neutralization with DMEM–Hepes (pH 10). Cells were incubated back at 37 °C in DMEM–Hepes–0.1% BSA supplemented with 50 µg/mL human nonfluorescent holo-transferrin to perform a time course. Cells were scraped gently and transferred into ice-cold PBS for FC analysis.
For Tf endocytosis, Hep2 cells were uninfected or infected with GFP-expressing relevant strains as previously described. After 60 min of infection at an MOI of 10, cells were washed twice with warm DMEM–Hepes and incubated for an additional 30 min in the same medium containing 0.1% BSA and gentamicin (50 µg/mL) to kill extracellular bacteria. Tf uptake time course was performed by incubating cells in a water bath at 37 °C with DMEM–Hepes–0.1% BSA in the presence of 0.5 µg/mL Tf-AF647 for different time points, followed by an acid stripping of remaining membrane-associated Tf-AF647.
To measure surface TfR levels, UI or infected cells were incubated at 4 °C in the presence of Tf-AF647 for 30 min. Cells were acid washed or untreated, and the total surface Tf-AF647 fluorescence was quantified by FC.
Where indicated, cells were transfected with the relevant plasmids the day before the Tf recycling or endocytosis experiments by electroporation (10 µg of DNA/4.5 × 106 cells) or using Lipofectamine 3000 (Thermo Fisher Scientific).
The geometrical mean fluorescence of intracellular Tf-AF647 was measured in gated living cells (using DAPI or Live/Dead Fixable Violet Cell Stain Kit; L34955; Molecular Probes) by FC in a BD FACSCanto II Flow Cytometer (BD Biosciences) or an Attune NxT Flow Cytometer (Thermo Fisher Scientific). Tf recycling results were expressed as the percentage of the remaining intracellular Tf-AF647 at each time point with respect to time 0 of recycling. Tf recycling rate was determined from the slope of ln2 data during the first 30 min of kinetics. Tf endocytosis results were expressed as Tf endocytosis rate, calculated as the internal Tf-AF647 fluorescence over time, during the first 10 min of kinetics.
IL-2Rβ Endocytosis.
Hep2β cells seeded on 12-mm coverslips were uninfected or infected with GFP-expressing bacteria. After 90 min of infection, a time course of IL-2Rβ endocytosis was performed by incubating cells with an anti–IL-2Rβ antibody (mouse Ab 561) conjugated to Cy3 (1:1,000) (44) at 37 °C for different time points. Cells were fixed, permeabilized, and stained with HCS CellMask Blue Stain (Molecular Probes) and phalloidin-AF647 or Tf-AF647. Imaging was performed by TIRF microscopy. IL-2Rβ endocytosis was quantified with ICY software as described in ref. 21.
Animals, Infections, and Sample Preparation.
Guinea pigs were infected according to previously described protocols (27, 28). Female specific-pathogen–free Hartley guinea pigs (120–250 g) were purchased from Charles River Laboratories, maintained in animal care facilities of Institut Pasteur, and provided with food and water ad libitum. Animal experiments were carried out under approval by the “Use Committee of Institut Pasteur and by the French Ministry of Agriculture no. 2013-0113.” Briefly, animals were anesthetized intraperitoneally using a mixture of ketamine (100 mg/kg; Merial) and xylazine hydrochloride (10 mg/kg; Bayer) before intrarectal inoculation of S. flexneri strains at 5 × 1010 cfu per 200 μL. Animals were killed at 4 and 8 h postchallenge. The distal 10 cm of colon was harvested and fixed overnight in 4% (vol/vol) PFA in PBS, and incubated in PBS–glycine (100 mM) for 30 min to quench the PFA. Tissues were then immersed successively in 15% and 30% (wt/vol) sucrose at 4 °C overnight. Tissues were cut and embedded in Tissue-Tek OCT compound (Sakura) using a flash-freeze protocol and frozen at −80 °C.
Immunofluorescence.
Hep2 and Hep2β cells were fixed in PFA (4%) sucrose (4%) in PBS for 20 min and quenched with NH4Cl (50 mM) for 10 min. Permeabilization, blocking, incubations, and washes were done with PBS BSA (0.1%) saponin (0.05%). Polarized Caco-2/TC7 cells were washed, fixed, and quenched preparing solutions in PBS with Ca2+ and Mg2+ (Gibco). Permeabilization was done in PBS Ca2+ Mg2+ gelatin (0.2%), saponin (0.075%) for 1 h at RT, primary antibodies incubated for 90 min at RT or overnight at 4 °C, secondary antibodies for 1 h at RT, together with DAPI (1 µg/mL) and phalloidin coupled to Alexa Fluor (Molecular Probes) if indicated.
For tissue sections, immunofluorescence samples were prepared as follows: 10-μm-thick transversal colon sections were permeabilized in PBS 0.5% Triton X-100 for 30 min, blocked in PBS 1% BSA for 30 min at RT, and incubated overnight at 4 °C with the indicated primary antibodies, together with phalloidin Alexa Fluor 647 (1:100) diluted in PBS, 0.1% Triton X-100, and 1% BSA. Sections were then washed with PBS and stained for 1–2 h at RT with Alexa Fluor 568 goat anti-mouse or anti-rabbit (A11031 and A11036; Molecular Probes), followed by incubation with DAPI (1 µg/mL) for 10 min at RT. Samples were washed with PBS before mounting.
ProLong Gold Antifade (Molecular Probes) was used as mounting medium.
Image Acquisition.
The following equipment was used for image acquisition: a LSM700 inverted laser-scanning confocal microscope (Zeiss), with a 40×/1.4 oil immersion or a 63×/1.4 oil immersion objective (Zeiss); an Axio Observer.Z1 microscope (Zeiss) equipped with a swept field confocal Opterra system (Bruker) and an Evolve 512 Delta EMCCD camera (Photometrics), using a 63× PlanAPO-CHROMAT oil immersion/1.4 N.A. objective (Zeiss); a slide scanner Axio Scan.Z1 (Zeiss), using a 40× dry objective; and an inverted confocal microscope LSM 780 Elyra PS.1 (Zeiss), using an alpha Plan Apo 100×/1.46 N.A. oil objective (Zeiss) (TIRF imaging).
Image Analysis and Quantification.
Microscopy images were processed and quantified with Fiji (ImageJ) (60), ICY software (61) (http://icy.bioimageanalysis.org), or Zen (Zeiss). Colocalization quantification was performed on confocal images using the Statistical Object Distance Analysis (SODA) plugin in ICY software described in ref. 62. IL-2Rβ endocytosis was quantified with ICY software using “HK-Means” and “Active Contours” plugins to automatically detect cell boundaries and “Spot Detector” plugin to measure the number of the IL-2Rβ spots within the detected cells. The total intensity of IL-2Rβ spots was normalized to the mean value of the uninfected conditions. Quantification of Dnm2 or CLC density at plasma membrane from TIRF images was performed using ICY Spot Detector plugin to quantify the number of spots per cell area. Quantification of Tf-A647 uptake by Caco-2/TC7 cells was performed with Fiji, by quantifying the total fluorescence per cell from 10 slices of 0.8 µm from a z-stack. Tissue images were acquired with 40× objectives, and subsequent stitching was performed using Zen software to build the mosaic images. Quantification of bacteria depth penetration into the intestinal mucosa was done from images illustrated in SI Appendix, Fig. S4C with Fiji, by measuring the maximum bacterial distance of penetration from the epithelium surface, with respect to the total mucosal thickness.
Data Presentation and Statistical Analysis.
Prism 6.0 (GraphPad Software) was used to perform statistical analysis. Results are represented as mean ± SD, except as otherwise indicated. The following statistical tests were used: Welch’s t test performed as unpaired two-tailed analysis; one-way ANOVA followed by Dunnett’s or Tukey’s post hoc multiple-comparison tests; Mann–Whitney; and Kruskal–Wallis followed by Dunn’s post hoc test. P < 0.05 was considered significant for all analyses.
Fiji and Zen (Zeiss) were used to process microscopy images. Inkscape software (https://inkscape.org/) was used for assembling figures.
Supplementary Material
Acknowledgments
We are very grateful to Pierre-Henri Commere (Flow Cytometry Platform, Institut Pasteur) for technical help with the FACS MoFlo Astrios EQ, Gaëlle Boncompain and Frank Perez for RUSH reagents, Laurie Pinaud and Claude Parsot for sharing reagents and helpful discussion, and Katja Brunner for critical reading of the manuscript. We thank Photonic BioImaging (Imagopole) platform of Institut Pasteur for microscope maintenance and technical help. This project was funded by European Research Council Advanced Grants 232798 and 339579 to P.J.S., Fondation pour la Recherche Médicale Grant SPF20121226366 to M.L.F., Transversal Research Program 22-16 grant to A.G., and “Région Ile-de-France” and Fondation pour la Recherche Médicale grants to D.L. L.S. is part of the Pasteur Paris University International PhD Program and has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under Marie Sklodowska-Curie Grant Agreement 665807.
Footnotes
The authors declare no conflict of interest.
Data deposition: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (dataset identifier PXD012291).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902922116/-/DCSupplemental.
References
- 1.Donaldson J. G., Jackson C. L., ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12, 362–375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr F. A., Review series: Rab GTPases and membrane identity: Causal or inconsequential? J. Cell Biol. 202, 191–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutagalung A. H., Novick P. J., Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91, 119–149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phalipon A., Sansonetti P. J., Shigella’s ways of manipulating the host intestinal innate and adaptive immune system: A tool box for survival? Immunol. Cell Biol. 85, 119–129 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Galán J. E., Lara-Tejero M., Marlovits T. C., Wagner S., Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68, 415–438 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroeder G. N., Hilbi H., Molecular pathogenesis of Shigella spp.: Controlling host cell signaling, invasion, and death by type III secretion. Clin. Microbiol. Rev. 21, 134–156 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashida H., Mimuro H., Sasakawa C., Shigella manipulates host immune responses by delivering effector proteins with specific roles. Front. Immunol. 6, 219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsot C., Shigella type III secretion effectors: How, where, when, for what purposes? Curr. Opin. Microbiol. 12, 110–116 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Mounier J., et al. , Shigella effector IpaB-induced cholesterol relocation disrupts the Golgi complex and recycling network to inhibit host cell secretion. Cell Host Microbe 12, 381–389 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Burnaevskiy N., et al. , Proteolytic elimination of N-myristoyl modifications by the Shigella virulence factor IpaJ. Nature 496, 106–109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burnaevskiy N., Peng T., Reddick L. E., Hang H. C., Alto N. M., Myristoylome profiling reveals a concerted mechanism of ARF GTPase deacylation by the bacterial protease IpaJ. Mol. Cell 58, 110–122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobbs N., et al. , STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe 18, 157–168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong N., et al. , Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell 150, 1029–1041 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Campbell-Valois F. X., Sachse M., Sansonetti P. J., Parsot C., Escape of actively secreting Shigella flexneri from ATG8/LC3-Positive vacuoles formed during cell-to-cell spread is facilitated by IcsB and VirA. MBio 6, e02567-e14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boncompain G., et al. , Synchronization of secretory protein traffic in populations of cells. Nat. Methods 9, 493–498 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Ong S.-E., et al. , Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Lippincott-Schwartz J., et al. , Brefeldin A’s effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell 67, 601–616 (1991). [DOI] [PubMed] [Google Scholar]
- 18.de Figueiredo P., et al. , Inhibition of transferrin recycling and endosome tubulation by phospholipase A2 antagonists. J. Biol. Chem. 276, 47361–47370 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Robinson M. S., Forty years of clathrin-coated vesicles. Traffic 16, 1210–1238 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Lamaze C., et al. , Interleukin 2 receptors and detergent-resistant membrane domains define a clathrin-independent endocytic pathway. Mol. Cell 7, 661–671 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Basquin C., et al. , Membrane protrusion powers clathrin-independent endocytosis of interleukin-2 receptor. EMBO J. 34, 2147–2161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunziker W., Whitney J. A., Mellman I., Selective inhibition of transcytosis by brefeldin A in MDCK cells. Cell 67, 617–627 (1991). [DOI] [PubMed] [Google Scholar]
- 23.Tran Van Nhieu G., Ben-Ze’ev A., Sansonetti P. J., Modulation of bacterial entry into epithelial cells by association between vinculin and the Shigella IpaA invasin. EMBO J. 16, 2717–2729 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antonny B., et al. , Membrane fission by dynamin: What we know and what we need to know. EMBO J. 35, 2270–2284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertot L., et al. , Quantitative and statistical study of the dynamics of clathrin-dependent and -independent endocytosis reveal a differential role of endophilin A2. Cell Rep. 22, 1574–1588 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Wang E., Pennington J. G., Goldenring J. R., Hunziker W., Dunn K. W., Brefeldin A rapidly disrupts plasma membrane polarity by blocking polar sorting in common endosomes of MDCK cells. J Cell Sci. 114, 3309–3321 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Arena E. T., et al. , Bioimage analysis of Shigella infection reveals targeting of colonic crypts. Proc. Natl. Acad. Sci. U.S.A. 112, E3282–E3290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shim D.-H., et al. , New animal model of shigellosis in the guinea pig: Its usefulness for protective efficacy studies. J. Immunol. 178, 2476–2482 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Clements A., Stoneham C. A., Furniss R. C. D., Frankel G., Enterohaemorrhagic Escherichia coli inhibits recycling endosome function and trafficking of surface receptors. Cell. Microbiol. 16, 1693–1705 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant B. D., Donaldson J. G., Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10, 597–608 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furniss R. C. D., Slater S., Frankel G., Clements A., Enterohaemorrhagic E. coli modulates an ARF6:Rab35 signaling axis to prevent recycling endosome maturation during infection. J. Mol. Biol. 428, 3399–3407 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selyunin A. S., et al. , The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature 469, 107–111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volpicelli-Daley L. A., Li Y., Zhang C., Kahn R. A., Isoform-selective effects of the depletion of ADP-ribosylation factors 1–5 on membrane traffic. Mol. Biol. Cell. 16, 4495–4508 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Dam E. M., Stoorvogel W., Dynamin-dependent transferrin receptor recycling by endosome-derived clathrin-coated vesicles. Mol. Biol. Cell 13, 169–182 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao H., et al. , Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat. Cell Biol. 7, 483–492 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Taylor M. J., Lampe M., Merrifield C. J., A feedback loop between dynamin and actin recruitment during clathrin-mediated endocytosis. PLoS Biol. 10, e1001302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grassart A., et al. , Actin and dynamin2 dynamics and interplay during clathrin-mediated endocytosis. J. Cell Biol. 205, 721–735 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valencia-Gallardo C. M., Carayol N., Tran Van Nhieu G., Cytoskeletal mechanics during Shigella invasion and dissemination in epithelial cells. Cell. Microbiol. 17, 174–182 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Boulant S., Kural C., Zeeh J. C., Ubelmann F., Kirchhausen T., Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat. Cell Biol. 13, 1124–1131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.García-Lorenzo A., Rodríguez-Piñeiro A. M., Rodríguez-Berrocal F. J., Cadena M. P., Martínez-Zorzano V. S., Changes on the Caco-2 secretome through differentiation analyzed by 2-D differential in-gel electrophoresis (DIGE). Int. J. Mol. Sci. 13, 14401–14420 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allaoui A., Sansonetti P. J., Parsot C., MxiJ, a lipoprotein involved in secretion of Shigella Ipa invasins, is homologous to YscJ, a secretion factor of the Yersinia Yop proteins. J. Bacteriol. 174, 7661–7669 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sidik S., et al. , A Shigella flexneri virulence plasmid encoded factor controls production of outer membrane vesicles. G3 (Bethesda) 4, 2493–2503 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allaoui A., Ménard R., Sansonetti P. J., Parsot C., Characterization of the Shigella flexneri ipgD and ipgF genes, which are located in the proximal part of the mxi locus. Infect. Immun. 61, 1707–1714 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grassart A., Dujeancourt A., Lazarow P. B., Dautry-Varsat A., Sauvonnet N., Clathrin-independent endocytosis used by the IL-2 receptor is regulated by Rac1, Pak1 and Pak2. EMBO Rep. 9, 356–362 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fourriere L., Divoux S., Roceri M., Perez F., Boncompain G., Microtubule-independent secretion requires functional maturation of Golgi elements. J. Cell Sci. 129, 3238–3250 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Labigne-Roussel A. F., Lark D., Schoolnik G., Falkow S., Cloning and expression of an afimbrial adhesin (AFA-I) responsible for P blood group-independent, mannose-resistant hemagglutination from a pyelonephritic Escherichia coli strain. Infect. Immun. 46, 251–259 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valdivia R. H., Falkow S., Bacterial genetics by flow cytometry: Rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22, 367–378 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Sörensen M., et al. , Rapidly maturing red fluorescent protein variants with strongly enhanced brightness in bacteria. FEBS Lett. 552, 110–114 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Campbell-Valois F.-X., et al. , A fluorescent reporter reveals on/off regulation of the Shigella type III secretion apparatus during entry and cell-to-cell spread. Cell Host Microbe 15, 177–189 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Poullet P., Carpentier S., Barillot E., myProMS, a web server for management and validation of mass spectrometry-based proteomic data. Proteomics 7, 2553–2556 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Valot B., Langella O., Nano E., Zivy M., MassChroQ: A versatile tool for mass spectrometry quantification. Proteomics 11, 3572–3577 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Ritchie M. E., et al. , Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 54.Vizcaíno J. A., et al. , 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kowal J., et al. , Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U.S.A. 113, E968–E977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petersen T. N., Brunak S., von Heijne G., Nielsen H., SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Bendtsen J. D., Jensen L. J., Blom N., Von Heijne G., Brunak S., Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17, 349–356 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Ashburner M., et al. ; The Gene Ontology Consortium , Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas P. D., et al. , PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Chaumont F., et al. , Icy: An open bioimage informatics platform for extended reproducible research. Nat. Methods 9, 690–696 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Lagache T., et al. , Mapping molecular assemblies with fluorescence microscopy and object-based spatial statistics. Nat. Commun. 9, 698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.