PLANT BIOLOGY Correction for “Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway,” by Benoît Derrien, Nicolas Baumberger, Mikhail Schepetilnikov, Corrado Viotti, Julia De Cillia, Véronique Ziegler-Graff, Erika Isono, Karin Schumacher, and Pascal Genschik, which was first published September 10, 2012; 10.1073/pnas.1209487109 (Proc. Natl. Acad. Sci. U.S.A. 109, 15942–15946).
The authors wish to note the following: “Corrections are necessary for Fig. 1 panel E and Fig. 4 panel A, as described below.
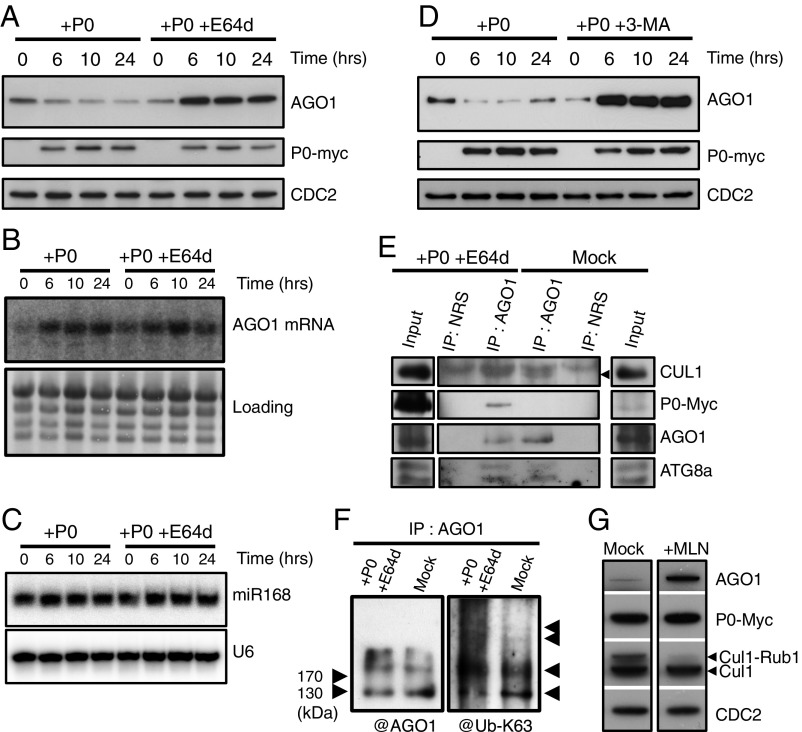
Fig. 1.
P0-mediated degradation of AGO1 is blocked by autophagy inhibitors. AGO1 degradation kinetics were performed on 7-d-old XVE-P0BW-myc seedlings treated with β-estradiol (5 μM) for P0-myc induction. Autophagy was inhibited in its last steps using E64d (20 μM) (A) and AGO1, P0-myc, and CDC2 (loading control) protein accumulation levels were assayed by Western blot on a 24-h period. In a similar manner, AGO1 mRNA (B) and miR168 accumulation (C) was assayed by Northern blot analyses along P0-mediated degradation of AGO1 in presence or in absence of E64d (20 μM). Loading controls are methylene blue staining of the membrane for mRNA and U6 for small RNA blots. (D) Autophagy was inhibited in its first steps using the specific PI-3-kinase class III inhibitor 3-MA (5 mM) and AGO1, P0-myc, and CDC2 (loading control) protein accumulation levels were assayed by Western blot on a 24-h period. (E) Coimmunoprecipitation of AGO1 and SCFP0. XVE-P0BW-myc seedlings were treated with β-estradiol (5 μM) for P0-myc induction and E64d (20 μM) for at least 6 h before protein extraction. Plant extract were immunoprecipitated with an anti-AGO1 antibody and with normal rabbit serum (NRS). IP fractions were submitted to Western blot analysis using antibodies raised against the myc tag for P0 detection and against CUL1, AGO1, and ATG8a. The arrow indicates CUL1. (F) Ubiquitylation status of AGO1 was determined by Western blot analysis of IP fractions using an antibody specifically raised against K63-Ub. (G) Inhibition of SCF activity prevents P0-mediated degradation of AGO1. Seven-day-old XVE-P0BW-myc seedlings were pretreated with MLN-4924 (25 μM) for 3 h before P0-myc induction with β-estradiol (5 μM). The accumulation level of AGO1, P0-myc, CUL1, and CDC2 (loading control) was assayed by Western blot 24 h after P0 induction. Anti-CUL1 antibody detects two bands, the upper one corresponding to the NEDD8/RUB1-modified form of CUL1.
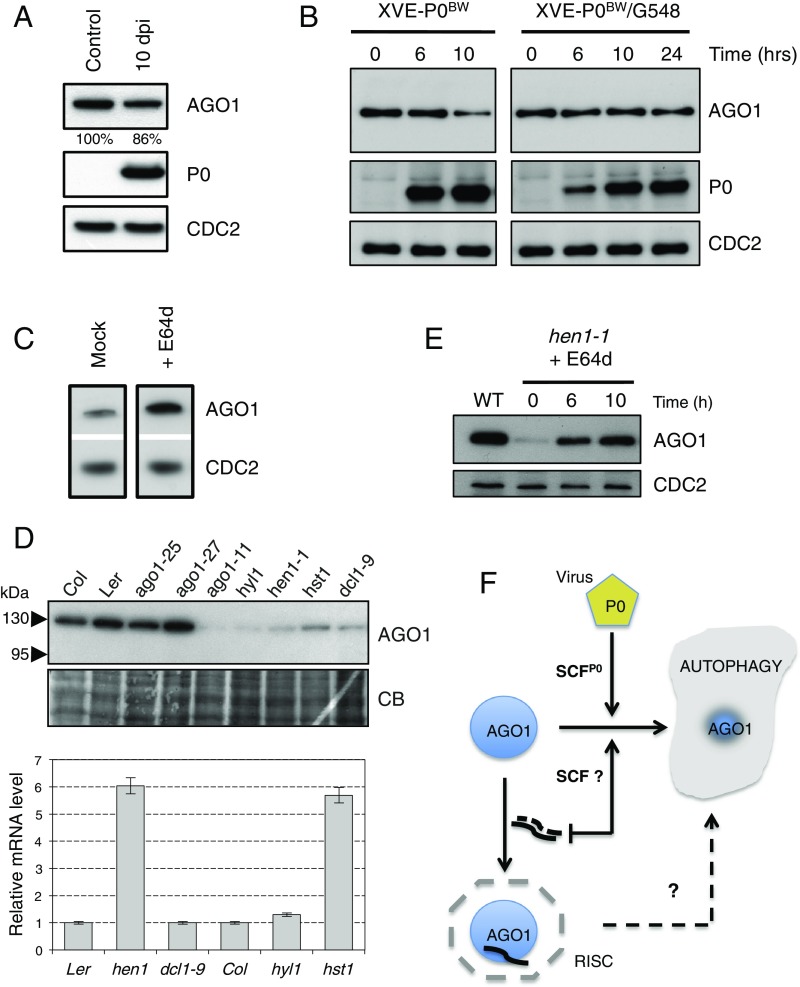
Fig. 4.
P0-mediated degradation of AGO1 is compromised in amsh3-1 mutant and in TOR-overexpressing plants and the endogenous pathway for AGO1 degradation also relies on autophagy. (A) Comparison of AGO1 protein accumulation level between uninduced 10-d–old XVE-P0BW-myc/amsh3-1 seedlings (control) and P0-induced 10-d-old XVE-P0BW-myc/amsh3-1 seedlings. For P0 induction, plants have been germinated and grown for 10 d on MS-agar dishes supplemented with 10 μM β-estradiol [10 d passed on induction (dpi)]. (B) AGO1 degradation kinetics performed on 7-d old XVE-P0BW-myc and XVE-P0BW-myc/G548 seedlings treated with β-estradiol (5 μM) for P0-myc induction. Because P0 induction is delayed in the XVE-P0BW-myc/G548 line, we extended this kinetic to 24 h. AGO1, P0-myc, and CDC2 (loading control) protein contents were assayed by Western blot. (C) E64d treatment (20 μM for 24 h) on wild-type Col-0 seedling leads to a higher accumulation of AGO1 protein. (D) Mutants affected in miRNA maturation and production pathways show reduced level of AGO1. (Upper) AGO1 protein accumulation in mutants and wild-type controls assayed by Western blot using the anti-AGO1 antibody. Coomassie blue staining is given as loading control. (Lower) AGO1 mRNA accumulation in each mutant assayed by quantitative RT-PCR. (E) The hen1-1 seedlings treated with E64d (20 μM) show AGO1 protein reaccumulation. AGO1 and CDC2 (loading control) protein contents were assayed by Western blot at the indicated time point. (F) Model for AGO1 turnover in a viral and nonviral context.
“Fig. 1E: This panel displays issues: (1) The AGO1 blot presented in Fig. 1E is the same as in Fig. 1F and the AGO1 image was inadvertently flipped during image assembly. (2) IPs with the normal rabbit serum (NRS) should be on the same blots (with same exposures) as the AGO1-IPs, which was not the case. The blots for inputs (CUL1, P0-myc, and ATG8a) were deposited on separate gels, as their signal was too strong. Therefore, we corrected Fig. 1E by mounting it in the appropriate way and including the AGO1 blot corresponding to this specific IP.
“Fig. 4A: The control lane (Lane 1) was duplicated to better appreciate the differences in AGO1 protein levels between induced and noninduced conditions. We presented a 10-d induction of P0 because the standard treatment of 24 h of P0 induction is less efficient in the amsh3-1 mutant background. To alleviate any doubt about the antagonizing effect of the amsh3-1 mutation on AGO1 degradation by P0, a long P0 induction with a significant amount of P0 protein accumulation has been added. The short-time induction was removed from Fig. 4A as not essential for the conclusion of this experiment. In addition, the legend of Fig. 4A was amended to clearly explain how the samples were produced.
“The original results and conclusions are unaffected by these corrections. B.D. takes responsibility for the inappropriate figure assembly. Corresponding author P.G. takes responsibility for not reviewing the data sufficiently and apologizes for not detecting these errors before publication.” The corrected Fig. 1 and Fig. 4 appear below with their respective corrected legends.