Abstract
Objective
The chikungunya virus (CHIKV) is an arthropod-borne Alphavirus transmitted to humans, primarily via Aedes mosquitoes. In Puerto Rico, the first locally transmitted infections were reported in May 2014. Although the virus strain in Puerto Rico is related to the Asian/American lineage, many autochthonous cases have emerged recently in the Caribbean region (including Puerto Rico), raising the question of how CHIKV will evolve and adapt in PR. Taking the role of the envelope glycoprotein (E1) in viral evolution and transmission as a given, we analyzed the genetic diversity of the Puerto Rican (PR) E1 gene sequences and the phylogenetic relationships between those sequences and sequences from other parts of the world.
Materials and Methods
To analyze the overall genetic variation, 772 nucleotide sequences of the E1 gene were obtained from the Virus Pathogen Resource (ViPR). A maximum-likelihood analysis was performed to determine the phylogenetic relationships between the PR sequences and sequences from 48 countries around the world.
Results
The analysis of the E1 gene identified variations at 4 nucleotide positions, which included synonymous and nonsynonymous mutations. In addition, 2 nonsynonymous amino acid changes, T207M and S120L, were unique to the PR CHIKV sequences, and T155I was found to be shared by the PR (n = 3) and Colombia (n = 1) strains.
Conclusions
Our analysis of the E1 gene revealed new molecular signatures in PR CHIKV sequences, 1 of which was also found in Colombia. While studies have shown possible relationships between T98A and A226V with viral adaptation and spread, no other PR sequence contained these vector-adaptive mutations. Thus, constant monitoring of the virus remains an essential factor in the establishment of control strategies to track viral spread.
Keywords: chikungunya virus, evolution, envelope glycoprotein 1
Article Summary Line
While no mutations contributing to the increased transmissibility of Aedes albopictus were detected, phylogenetic analysis of the chikungunya virus (CHIKV) envelope glycoprotein E1 in the Puerto Rican strain of the virus revealed unique E1 mutations.
Resumen
Objetivo
El virus chikungunya (CHIKV) es un Alphavirus transmitido a los humanos principalmente a través de los mosquitos Aedes. En Puerto Rico, las primeras infecciones transmitidas localmente se notificaron en mayo de 2014. Aunque el virus en Puerto Rico está relacionado con el linaje asiático/americano, muchos casos autóctonos han surgido recientemente en la región del Caribe, lo que plantea la pregunta: ¿Cómo CHIKV evolucionará y se adaptará en Puerto Rico? Dado el papel de la glicoproteína de la envoltura (E1) en la evolución y transmisión del virus, analizamos la diversidad genética y las relaciones filogenéticas entre las secuencias del gen E1 en Puerto Rico y otras partes del mundo.
Materiales y métodos
Para analizar la variación genética, se obtuvieron 772 secuencias de nucleótidos del gen E1 de la base de datos “Virus Pathogen Resource (ViPR)”. Se realizó un análisis de máxima verosimilitud para observar las relaciones filogenéticas entre las secuencias de Puerto Rico y las secuencias de cuarenta y ocho países alrededor del mundo.
Resultados
Análisis del gen E1 identificó variantes en cuatro posiciones de nucleótidos, que incluyen mutaciones sinónimas y no sinónimas. Además, dos cambios de aminoácidos no sinónimos, T207M y S120L, son únicos en las secuencias de PR, y se encontró que T155I se compartió entre las cepas PR (n = 3) y Colombia (n = 1).
Conclusiones
Nuestro análisis del gen E1 reveló nuevas marcas moleculares en las secuencias de Puerto Rico, una de las cuales también se encontró en Colombia. Si bien los estudios han demostrado posibles relaciones entre T98A y A226V con la adaptación y propagación viral, ninguna secuencia de Puerto Rico contenía estas mutaciones adaptativas a los vectores. Por lo tanto, el monitoreo constante del virus sigue siendo un factor importante en el establecimiento de estrategias de control para monitorear la propagación viral.
Introduction
The chikungunya virus (CHIKV) is an arthropod-borne virus (arbovirus) that is transmitted to humans primarily via the bite of an infected Aedes species mosquito (1, 2). The virus is a member of the genus Alphavirus; many alphaviruses are important human pathogens (3, 4). The CHIKV infection is characterized by acute febrile illness and severe polyarthralgia that can persist for a long time (5, 6). The first reported case was in the early 1950s, on the border of Tanzania and Mozambique (the Makonde Plateau) (7). In the Caribbean, the first reported case was in Saint Martin (2013), which is a part of the French Antilles (8). Immediately, further cases were identified in other countries in that area (9, 10). The principal vector in the region is Aedes aegypti. However, Aedes albopictus is also present in some Caribbean countries (11, 12). In May 2014, the first laboratory-positive chikungunya case was reported in Puerto Rico (13). The CHIKV fever expanded rapidly, and 4,465 confirmed and 30,247 suspected cases were reported during that year (14, 15). The expansion of the virus that was observed during 2014 and 2015 was typical of a new pathogen in an immunologically naive population (16). Currently, the number of new cases has dropped in the last 2 years, due, in part, to increased efforts by the government to eradicate the vector (Aedes aegypti), the use of repellent, and the implementation of educational strategies (13, 17).
The CHIKV genome is composed of 11.8 kb of single-stranded, positive-sense RNA. It codes for 4 non-structural and 5 structural proteins and includes the E1 envelope glycoprotein (18). The gene that encodes this protein is composed of approximately 1,300 nucleotides. The E1 glycoprotein is a significant participant in the virion attachment to the host cell and in the fusion of the virus membrane with the host endosome membrane (19). CHIKV has 3 main genotypes—Asian, East/Central/South African, and West African—which can be distinguished by differences in the E1 glycoprotein gene (20). The CHIKV in Puerto Rico is related to the Asian/Pacific and American strains; however, many autochthonous cases have emerged recently in the Caribbean and South America (21, 22). Single mutations in the E1 gene can potentiate the capacity of the vector to transmit the CHIKV (5, 23). The A226V mutation promotes increased infectivity of the midgut, dissemination to the salivary glands, and transmission of the virus by Aedes albopictus mosquitoes (23–25). The CHIKV with the A226V mutation was responsible for several outbreaks, worldwide (26–29).
Although the CHIKV in Puerto Rico is related to the Asian/American lineage, many autochthonous cases have emerged recently in the Caribbean region, raising the question of how CHIKV will evolve and adapt in Puerto Rico. Our current study wanted to evaluate the hypothesis that during the CHIKV epidemic in Puerto Rico, unique sequences emerged. Taking the role of the envelope glycoprotein (E1) in viral evolution and transmission as a given, we analyzed the E1 envelope glycoprotein partial nucleotide sequences from Puerto Rico, which were available in the Virus Pathogen Resource (ViPR). This analysis can help us to identify the possible autochthonous cases, which could be the result of a mutation that enhances viral fitness and replication capacity in the mosquito and human cells.
Materials and Methods
Ethics Approval
The protocol underwent review at the Institutional Review Board of the Ponce Medical School Foundation and was determined to be exempt from the federal policy for the protection of human subjects, as the data are publicly available from the ViPR, a National Institute of Allergy and Infectious Diseases (NIAID)-sponsored database and analysis resource that supports the research of viral pathogens in categories A, B, and C.
Sequence Data and Analysis
At the time of the study, a total of 2,055 CHIKV E1 gene nucleotide sequences were obtained from the ViPR database. Of these sequences, a total of 772 CHIKV E1 gene sequences from 50 countries, including 42 sequences from Puerto Rico, were included in the analysis (30). In choosing non-PR sequences, only those that covered the same region and 696-nucleotide length as the PR sequences were selected. The sequences included in the analysis started at position 10096 and ended at position 10792, relative to the CHIKV isolate MY/06/37348 (Malaysia, 2006) reference sequence (accession number: FN295483) (31, 32). This fragment extends from position 142 to position 837 of the E1 glycoprotein gene sequence from the MY/06/37348 reference sequence. Positions with less than 95% site coverage were eliminated from the analysis. Nucleotide sequences were input in FASTA, which format was also used for the output data. Sequence edition and sorting were performed using BioEdit (v.7.2.5), a sequence alignment editor (33). The multiple sequence alignment software MAFFT (v. 7.304b) was used for similarity-based methods, using the default penalties in which the software automatically selected the appropriate strategy from L-INS-i, FFT-NS-i, or FFT-NS-2, according to data size (30). Phylogenetic relationships were analyzed with MEGA software (v.6), using the maximum-likelihood method (34). The Tamura-Nei model was used to perform the analysis (35). To construct the initial tree for the heuristic search, we applied the neighbor-joining method to a pairwise distance matrix that had been computed using the maximum composite likelihood approach (34). Bootstrap analysis was performed on 50 replicates, and those with bootstrap values greater than 70 were indicated at the nodes. Nodes where no value was reported were not supported at this level. Evolutionary rate differences among sites were obtained using a discrete gamma distribution (5 categories [+G, parameter = 0.4311]). The CHIKV genotyping details were obtained using the Dengue, Zika & Chikungunya Viruses Typing Tool (v. 1.0, 2015), a tool designed to identify viral genotypes using BLAST and phylogenetic methods (36). The software tool Highlighter was used to highlight mismatches, matches, transition and transversion mutations, and silent and non-silent mutations in the nucleotide sequence alignments (37).
Statistical Analysis
The chi-square test of independence was used for comparative statistical analyses of nucleotide and amino acid sequence data to identify correlations between sequence variations and distribution. A p-value of less than or equal to 0.05 was considered statistically significant.
Availability of Data
The sequences detailed in this report are available from GenBank. The information regarding the accession numbers will be provided upon request.
Results
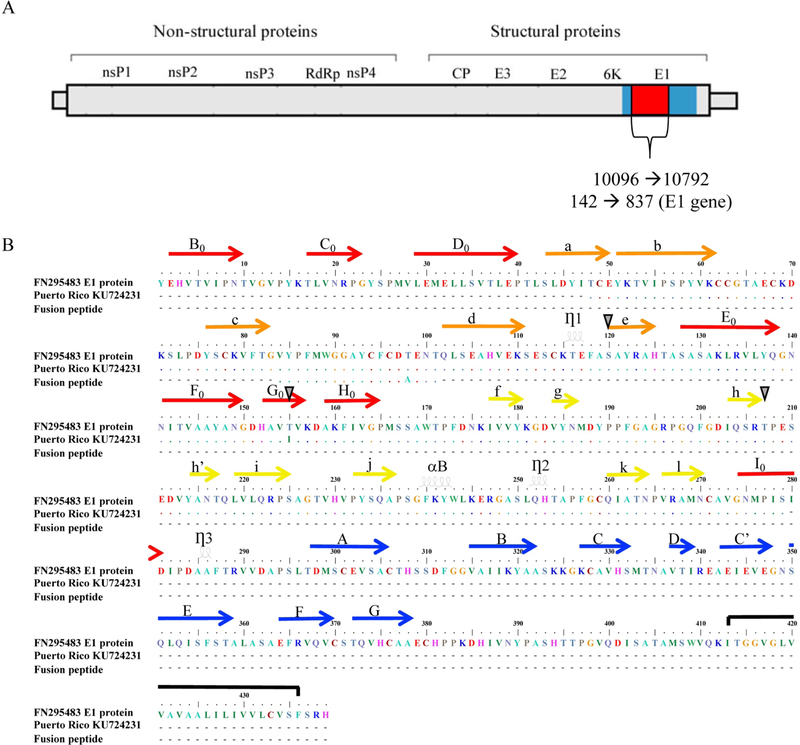
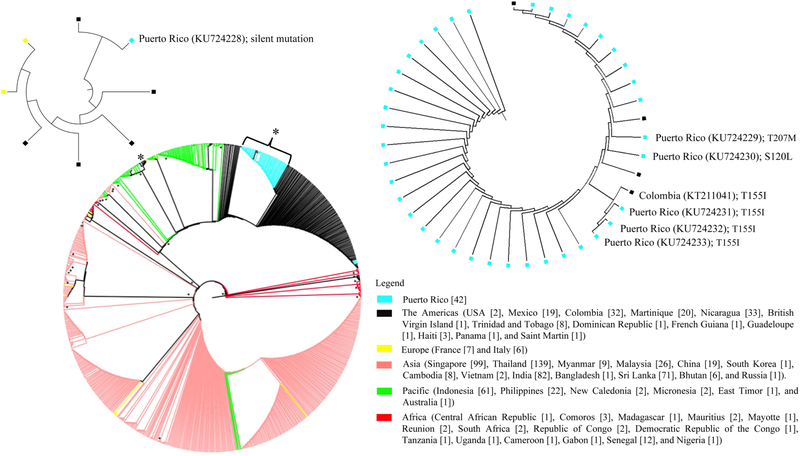
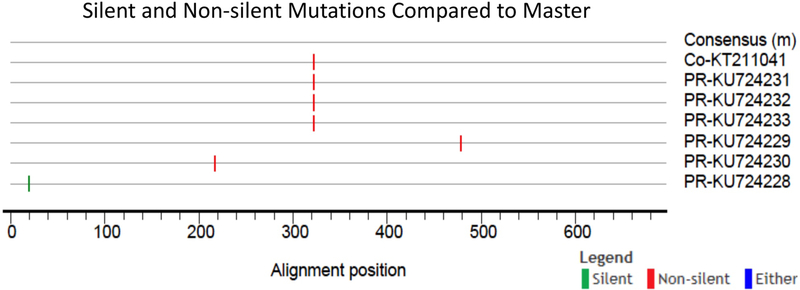
The CHIKV envelope sequence analyzed included 696 nucleotides of the total 1317 nucleotides from the complete E1 gene (Figure 1, panel A). The deduced amino acid sequence of the E1 glycoprotein region under study comprised domains I and II (Figure 1, panel B). The sequence analysis of the E1 gene using the Dengue, Zika & Chikungunya Virus Typing Tool revealed that the sequences from Puerto Rico belong to the Asian genotype, Caribbean clade (data not shown). PR sequences (bootstrap value = 99%) are similar to Asian/Pacific (Thailand, Malaysia, Singapore, Indonesia, and Philippines, among others), Caribbean (the Dominican Republic, Haiti, the British Virgin Islands, Martinique, and Saint Martin), Central American (Mexico, Nicaragua, and Panama), and South American (Colombia, Brazil, and French Guiana) sequences, suggesting a single introduction. Phylogenetic analysis revealed that the PR sequences clustered with countries in the Americas (Figure 2). The analysis of the E1 envelope glycoprotein gene identified variations at 4 nucleotides positions, compared to the to the consensus sequence from the Americas (Figure 3). Six PR sequences were different from the consensus sequence. Changes included both synonymous (n = 1) and nonsynonymous mutations (n = 5) (Figure 3). The nonsynonymous mutations were located within domains I and II of the E1 glycoprotein (Figure 1, panel B). Two nonsynonymous mutations, S120L and T207M, were unique to PR sequences. One nonsynonymous mutation, T155I, was found to be unique to Puerto Rico (n = 3) and Colombia (CO; n = 1). The PR sequences with the mutations clustered together with sequences from the Americas and the PR sequences that shared the mutation with CO were in the same node (Figure 2). We ran a chi-square analysis on the distribution of the changes at S120L, T155I, and T207M and compared PR sequences (group 1, n = 42) to sequences from around the world (group 2, n = 730). The analysis revealed that the changes at these positions had a statistically significant non-random distribution between the specified groups (Table 1).
Figure 1.
A schematic representation of the CHIKV genome and the E1 glycoprotein. Panel A: The position of the CHIKV full-length E1 gene and the 696 nucleotides analyzed in this study are denoted in reference to the complete genome. Panel B: The top line represents the CHIKV isolate MY/06/37348 (Malaysia, 2006) reference sequence FN295483. The second line is the PR sequence KU724231, which includes the mutation T155I as an example. The third line represents the E1 fusion peptide. The domains are identified according to Roussel et al. (52). The arrows represent the b sheets within each domain. Red arrows indicate E1 domain I; yellow arrows indicate domain II; and blue arrows indicate domain III. Orange arrows indicate domain II regions that are located within domain I. The loops indicate the a helices. The black line shows the transmembrane region. Sites of unique mutations detected in the sequences from PR are denoted with a grey triangle.
Figure 2.
The evolutionary history was inferred using the maximum likelihood method based on the Tamura-Nei model. The bootstrap consensus tree was inferred from 50 replicates. The study involved 772 nucleotide sequences. Evolutionary analyses were conducted in MEGA v.6. The colors indicate world region and the bullets (•) indicate the branch with a bootstrap value > 0.70. The number of sequences per country is shown in brackets. The asterisks (*) denote the region in the phylogenetic tree that contains the PR sequences with the mutations. These regions are provided as inserts for better visualization of the relationship of PR sequences to nearby sequences.
Figure 3.
Highlighter plot traces mismatched mutations as compared to the Asian/American consensus sequence. Silent (n = 1, green) and non-silent mutations (n = 5, red) are in 4 positions of the gene. One of the nonsynonymous mutations was found to be unique to the PR (n = 3) and CO (n = 1) strains.
Table 1.
Positions within the multiple sequence alignment that significantly differ from the expected (random) distribution
| Position* | Chi-square† | p-value | Degrees of Freedom | Residue Diversity |
|---|---|---|---|---|
| 10313 (S120L) | 17.416 | 5.80E-04 | 3 | Group 1 (41 C, 1 T) Group 2 (730 C) |
| 10418 (T155I) | 37.835 | 3.06E-08 | 3 | Group 1 (39 C, 3 T) Group 2 (729 C, 1 T) |
| 10574 (T207M) | 17.416 | 5.80E-04 | 3 | Group 1 (41 C, 1 T) Group 2 (730 C) |
The positions are in relation to the CHIKV isolate MY/06/37348 (Malaysia, 2006) reference sequence (accession number: FN295483), which start at position 10096 and ends at position 10792.
The chi-square test of independence was performed with the Metadata-driven Comparative Analysis Tool for Sequences (meta-CATS) (51).
Changes from the consensus sequences are in bold.
Discussion
The E1 glycoprotein is a fusion peptide and is one of the significant determinants of the evolutionary adaptation and dissemination of CHIKV (22, 38, 39). Recent studies have shown a possible relationship between nonsynonymous mutations and worldwide virus adaptation and spread. In the 2005 Indian Ocean outbreak (Reunion Island), Aedes albopictus was the primary vector responsible for the transmission of CHIKV (40–42). Molecular analysis identified an alanine to valine mutation at position 226 in the E1 glycoprotein (A226V). This E1-A226V mutation influenced the fitness of the virus in Aedes albopictus and affected the vector specificity and the duration of the epidemic (23, 28, 31). This mutation was related to the modulation of cholesterol in the cellular membrane. In the Semliki Forest virus (SFV), the fusion and exit of the virus is cholesterol-dependent (23, 43).
The mutation E1-A226V, which contributes to increased virus transmissibility by Aedes albopictus, was not detected in the E1 gene of the PR sequences included in this study. Its absence may in part be due to the CHIKV vector in Puerto Rico, Aedes aegypti. While the E1-A226V mutation has not been shown to be associated with Aedes aegypti, a single mutation can modify viral infectivity for a specific vector and expand the epidemic in a region that lacks the typical vector (23, 44). CHIKV sequence analysis in India showed 2 single nucleotide mutations that increased the virus’s capacity to be transmitted by the Aedes aegypti mosquito (45). Even though the Aedes aegypti is the principal vector on the island, Aedes albopictus mosquitoes can also be found, albeit with less frequency (46). Thus, constant monitoring of the vector and virus is essential to assess the presence of mutations that may affect CHIKV transmissibility and pathogenesis.
The finding that 2 amino acid substitutions were unique to PR CHIKV sequences and that 1 mutation was detected only in PR (n = 3) and CO sequences (n = 1) (compared to sequences from around the world) suggests that autochthonous cases are emerging in Puerto Rico. Previous studies have shown that single amino acid changes in the E1 glycoprotein can affect how the virus fuses with the target cells (47). While at this time the implication of these mutations remains unclear, the 3 mutations are changes from small, non-polar amino acids to hydrophobic amino acids, similar to mutation A226V. Whether these mutations have the potential to alter E1’s biological activity, including fusogenicity, remains to be elucidated. Additional epidemiological information will help us to better understand the role of these amino acid substitutions in CHIKV pathogenesis. Since these mutations are unique to the PR CHIKV sequences, we suggest that these mutations may represent a signature from the island. However, additional research regarding CHIKV evolution in Puerto Rico is necessary to confirm this. In addition, the virus gene flow between Puerto Rico and CO should be monitored to understand the role of CHIKV mutations in disease severity and chronicity in the 2 countries.
Recent data suggest that around 25% of the blood donors in Puerto Rico acquired CHIKV during the 2014 epidemic, meaning that approximately 800,000 individuals in this population were infected (16). Nevertheless, the number of new cases appears to have dropped in the last few years, possibly as a result of the establishment of strategies for vector control (Aedes aegypti) and an increased awareness of mosquito-borne diseases. However, according to ArboNET, in 2016, 100% of the transmitted cases in the United States and its territories occurred in Puerto Rico (48). Puerto Rico is a United States Commonwealth, and because of the high level of travel between Puerto Rico and the United States (49), how the virus evolves and adapts to new hosts remains a crucial issue under study. After the first case was identified, the expansion of CHIKV in Puerto Rico decreased dramatically (50), likely resulting from long-lasting immunity to the virus. However, the viral mutation may lead to the re-emergence of chikungunya in Puerto Rico, and thus it is essential to carefully monitor autochthonous transmission for the emergence of mutations that might alter CHIKV transmissibility and pathogenesis.
Acknowledgments
The study was supported by the Research Centers in Minority Institutions AIDS Research Infrastructure Program (National Institute on Minority Health and Health Disparities: 5G12MD007579-32) and by the Research Initiative for Scientific Enhancement (RISE) Program (R25GM82406) of the National Institutes of Health. We would like to thank the personnel of the AIDS Research Infrastructure Program for their insightful discussions.
Funding sources: NIMHD RCMI 5G12MD007579-32; NIGMS RISE R25GM82406
Footnotes
Disclosure Statement
The authors have no conflict of interest to disclose.
References
- 1.Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis 2007;7(5):319–327. [DOI] [PubMed] [Google Scholar]
- 2.Thiberville SD, Boisson V, Gaudart J, Simon F, Flahault A, de Lamballerie X. Chikungunya fever: a clinical and virological investigation of outpatients on Reunion Island, South-West Indian Ocean. PLoS Negl Trop Dis 2013;7(1):e2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol 1998;72(l):73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mlera L, Melik W, Bloom ME. The role of viral persistence in flavivirus biology. Pathog Dis 2014;71(2):137–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuffenecker I, Iteman I, Michault A, et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med 2006;3(7):e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohan A, Kiran DH, Manohar IC, Kumar DP. Epidemiology, clinical manifestations, and diagnosis of Chikungunya fever: lessons learned from the re-emerging epidemic. Indian J Dermatol 2010;55(1):54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross RW. The Newala epidemic. III. The virus: isolation, pathogenic properties and relationship to the epidemic. J Hyg (Lond) 1956;54(2):177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cauchemez S, Ledrans M, Poletto C, et al. Local and regional spread of chikungunya fever in the Americas. Euro Surveill 2014;19(28):20854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chikungunya--coming to America. Lancet 2014;383(9916):488. [DOI] [PubMed] [Google Scholar]
- 10.Mowatt L, Jackson ST. Chikungunya in the Caribbean: An Epidemic in the Making. Infect Dis Ther 2014;3(2):63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffey LL, Failloux AB, Weaver SC. Chikungunya virus-vector interactions. Viruses 2014;6(11):4628–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brathwaite Dick O, San Martin JL, Montoya RH, del Diego J, Zambrano B, Dayan GH. The history of dengue outbreaks in the Americas. Am J Trop Med Hyg 2012;87(4):584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenzi OD, Major C, Acevedo V, et al. Reduced Incidence of Chikungunya Virus Infection in Communities with Ongoing Aedes Aegypti Mosquito Trap Intervention Studies - Salinas and Guayama, Puerto Rico, November 2015-February 2016. MMWR Morb Mortal Wkly Rep. 2016;65(18):479–480. [DOI] [PubMed] [Google Scholar]
- 14.Fischer M, Staples JE; Arboviral Diseases Branch, National Center for Emerging and Zoonotic Infectious Diseases, CDC. Notes from the field: chikungunya virus spreads in the Americas - Caribbean and South America, 2013–2014. MMWR Morb Mortal Wkly Rep 2014;63(22):500–501. [PMC free article] [PubMed] [Google Scholar]
- 15.van Haastregt JC, Zijlstra GA, van Rossum E, van Eijk JT, Kempen GI. Feelings of anxiety and symptoms of depression in community-living older persons who avoid activity for fear of falling. Am J Geriatr Psychiatry 2008;16(3):186–193. [DOI] [PubMed] [Google Scholar]
- 16.Simmons G, Bres V, Lu K, et al. High Incidence of Chikungunya Virus and Frequency of Viremic Blood Donations during Epidemic, Puerto Rico, USA, 2014. Emerg Infect Dis 2016;22(7):1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention (CDC). Dengue in Puerto Rico. December 14, 2015. Available at: https://www.cdc.gov/dengue/about/inpuerto.html. Accessed January 15, 2017.
- 18.Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58(3):491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng X, Mukhopadhyay S, Brooks CL 3rd. Residue-level resolution of alphavirus envelope protein interactions in pH-dependent fusion. Proc Natl Acad Sci U S A 2015;112(7):2034–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powers AM, Brault AC, Tesh RB, Weaver SC. Re-emergence of Chikungunya and O’nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol 2000;81(Pt 2):471–479. [DOI] [PubMed] [Google Scholar]
- 21.Perti T, Lucero-Obusan CA, Schirmer PL, Winters MA, Holodniy M. Chikungunya Fever Cases Identified in the Veterans Health Administration System, 2014. PLoS Negl Trop Dis 2016;10(5):e0004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodas JD, Kautz T, Camacho E, et al. Genetic Characterization of Northwestern Colombian Chikungunya Virus Strains from the 2014–2015 Epidemic. Am J Trop Med Hyg 2016;95(3):639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 2007;3(12):e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Souza TM, Azeredo EL, Badolato-Correa J, et al. First Report of the East-Central South African Genotype of Chikungunya Virus in Rio de Janeiro, Brazil. PLoS Curr 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thiboutot MM, Kannan S, Kawalekar OU, et al. Chikungunya: a potentially emerging epidemic? PLoS Negl Trop Dis 2010;4(4):e623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vazeille M, Moutailler S, Coudrier D, et al. Two Chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS One 2007;2(11):e1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pages F, Peyrefitte CN, Mve MT, et al. Aedes albopictus mosquito: the main vector of the 2007 Chikungunya outbreak in Gabon. PLoS One 2009;4(3):e4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar NP, Joseph R, Kamaraj T, Jambulingam P. A226V mutation in virus during the 2007 chikungunya outbreak in Kerala, India. J Gen Virol 2008;89(Pt 8):1945–1948. [DOI] [PubMed] [Google Scholar]
- 29.Rianthavorn P, Prianantathavorn K, Wuttirattanakowit N, Theamboonlers A, Poovorawan Y. An outbreak of chikungunya in southern Thailand from 2008 to 2009 caused by African strains with A226V mutation. Int J Infect Dis 2010;14 Suppl 3:e161–e165. [DOI] [PubMed] [Google Scholar]
- 30.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013;30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sam IC, Chan YF, Chan SY, et al. Chikungunya virus of Asian and Central/East African genotypes in Malaysia. J Clin Virol 2009;46(2):180–183. [DOI] [PubMed] [Google Scholar]
- 32.Sam IC, Loong SK, Michael JC, et al. Genotypic and phenotypic characterization of Chikungunya virus of different genotypes from Malaysia. PLoS One 2012;7(11):e50476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf) 1999;41:95–98. [Google Scholar]
- 34.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013;30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993;10(3):512–526. [DOI] [PubMed] [Google Scholar]
- 36.Dengue Zika, Chikungunya & Yellow Fever Viruses Typing Tool [computer program].Version 1. Durban, South Africa: KRISP Group, Nelson R Mandela School of Medicine, UKZN; 2015. Available at: http://www.bioafrica.net/software.php. Accessed January 15, 2017. [Google Scholar]
- 37.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 2008;105(21):7552–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voss JE, Vaney MC, Duquerroy S, et al. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 2010;468(7324):709–712. [DOI] [PubMed] [Google Scholar]
- 39.Weaver SC, Lecuit M. Chikungunya virus and the global spread of a mosquito-borne disease. N Engl J Med 2015;372(13):1231–1239. [DOI] [PubMed] [Google Scholar]
- 40.Arias-Goeta C, Mousson L, Rougeon F, Failloux AB. Dissemination and transmission of the E1–226V variant of chikungunya virus in Aedes albopictus are controlled at the midgut barrier level. PLoS One 2013;8(2):e57548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Lamballerie X, Leroy E, Charrel RN, Ttsetsarkin K, Higgs S, Gould EA. Chikungunya virus adapts to tiger mosquito via evolutionary convergence: a sign of things to come? Virol J 2008;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bagny L, Delatte H, Quilici S, Fontenille D. Progressive decrease in Aedes aegypti distribution in Reunion Island since the 1900s. J Med Entomol 2009;46(6):1541–1545. [DOI] [PubMed] [Google Scholar]
- 43.Vashishtha M, Phalen T, Marquardt MT, Ryu JS, Ng AC, Kielian M. A single point mutation controls the cholesterol dependence of Semliki Forest virus entry and exit. J Cell Biol. 1998;140(1):91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arankalle VA, Shrivastava S, Cherian S, et al. Genetic divergence of Chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. J Gen Virol 2007;88(Pt 7):1967–1976. [DOI] [PubMed] [Google Scholar]
- 45.Agarwal A, Sharma AK, Sukumaran D, Parida M, Dash PK. Two novel epistatic mutations (E1:K211E and E2:V264A) in structural proteins of Chikungunya virus enhance fitness in Aedes aegypti. Virology 2016;497:59–68. [DOI] [PubMed] [Google Scholar]
- 46.Kraemer MU, Sinka ME, Duda KA, et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife 2015;4:e08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuo SC, Chen YJ, Wang YM, et al. Cell-based analysis of Chikungunya virus E1 protein in membrane fusion. J Biomed Sci 2012;19:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Centers for Disease Control. CDC. Laboratory-confirmed chikungunya virus disease cases reported to ArboNET by state or territory — United States, 2016. 2016. Available at: https://www.cdc.gov/chikungunya/pdfs/2016/2016Map-final-508.pdf. Accessed January 15, 2017.
- 49.Khan K, Bogoch I, Brownstein JS, et al. Assessing the origin of and potential for international spread of chikungunya virus from the Caribbean. PLoS Curr 2014;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puerto Rico Department of Health. Weekly Case Surveillance Report of Chikungunya. Available at: http://www.salud.gov.pr/. Accessed January 15, 2017.
- 51.Pickett BE, Liu M, Sadat EL, et al. Metadata-driven comparative analysis tool for sequences (meta-CATS): an automated process for identifying significant sequence variations that correlate with virus attributes. Virology 2013;447(1–2):45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roussel A, Lescar J, Vaney MC, Wengler G, Wengler G, Rey FA. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure 2006;14(1):75–86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences detailed in this report are available from GenBank. The information regarding the accession numbers will be provided upon request.