Abstract
Bmp2 is known to play an essential role in the initiation of fracture healing via periosteal activation. Specifically, activation and subsequent differentiation of periosteal progenitor cells requires Bmp2 signaling for activation of the osteo-chondrogenic pathway. Here, we explored the interactive transcriptional gene-gene interplays between Bmp2 and 150 known candidate genes during fracture repair. We constructed the interactive Bmp2 signaling pathways in vivo, by comparing gene expression levels prior and 24 hours post femur fracture, in presence (wild type) and in absence of Bmp2 (Bmp2c/c;Prx1::cre limb-specific conditional knockout). Twenty-six differentially expressed genes (pre- vs. post-fracture), which demonstrated high correlations within each experimental condition, were used to construct the co-expression networks. Topological dynamic shifts across different co-expression networks characterized the 26 differentially expressed genes as non-redundant focal linking hubs, redundant connecting hubs, periphery genes, or non-existent. Top-ranked up- or down-regulated genes were identified and discussed. Protein-protein interactions in public databases support our findings. Thus, the co-expression networks from this study can be used for future experimental hypotheses.
Keywords: Bmp2, fracture healing, co-expression, differential gene expression, conditional knockout, networks
1. Introduction
Since its identification as a key regulator of bone formation(1), bone morphogenetic protein 2 (Bmp2) has been shown to induce osteoblastic differentiation in vitro(2) and in vivo(3), and to be clinically effective in bone regenerative therapy(4). Specifically, Bmp2 expression is essential to initiate fracture healing(5) and regulate embryonic patterning(6), and is an indispensable multifunctional regulator of vertebrate development(7, 8). Polymorphisms of the human Bmp2 gene have been linked to osteoporosis(9–12) and osteoarthritis(13, 14). Thus, Bmp2 plays a crucial role in biological processes associated with bone formation, homeostasis, and regeneration.
Evidence has shown that selective genes in the Wnt and TGF-beta signaling pathways are active in bone fracture healing (15). Our goal is to explore the gene-gene interplays between Bmp2 and a list of 150 candidate genes during the bone fracture healing process. To do so, we compared gene expression in fractured femurs of animals expressing Bmp2 (wild type; denoted as WT) vs. those fractured femurs of animals not expressing Bmp2 (Bmp2c/c;Prx1::cre limb-specific conditional knockout; denoted as cKO)(16). The 150 candidate genes, selected for their roles in bone development, are enriched in such pathways as Wnt, Bmp, PTH and TGF-beta signaling. Details are provided in Suppl. Table 1.
Previous studies using a mouse femur fracture(5) model found maximum Bmp2 expression at 24 hours post-fracture. Therefore, in this report, gene expression profiles were obtained and compared at two time points – before fracture and 24 hours post-fracture. We demonstrated that markers for chondrogenesis and osteogenesis were absent at 24 hours post-fracture in Bmp2 cKO mice. This is consistent with human studies that examined co-expression of BMPs in non-unions(17). We further investigated differential gene expression signatures (DGEs) and their correlation strength in the Bmp2 WT vs. cKO groups. Interplays among the DGEs were characterized with four co-expression networks. The identified changes in topological characteristics and the associated correlation strength between Bmp2 and DGEs provided an in-depth dynamic understanding of Bmp2 networks during fracture repair.
2. Materials and Methods
2.1. Experimental Design
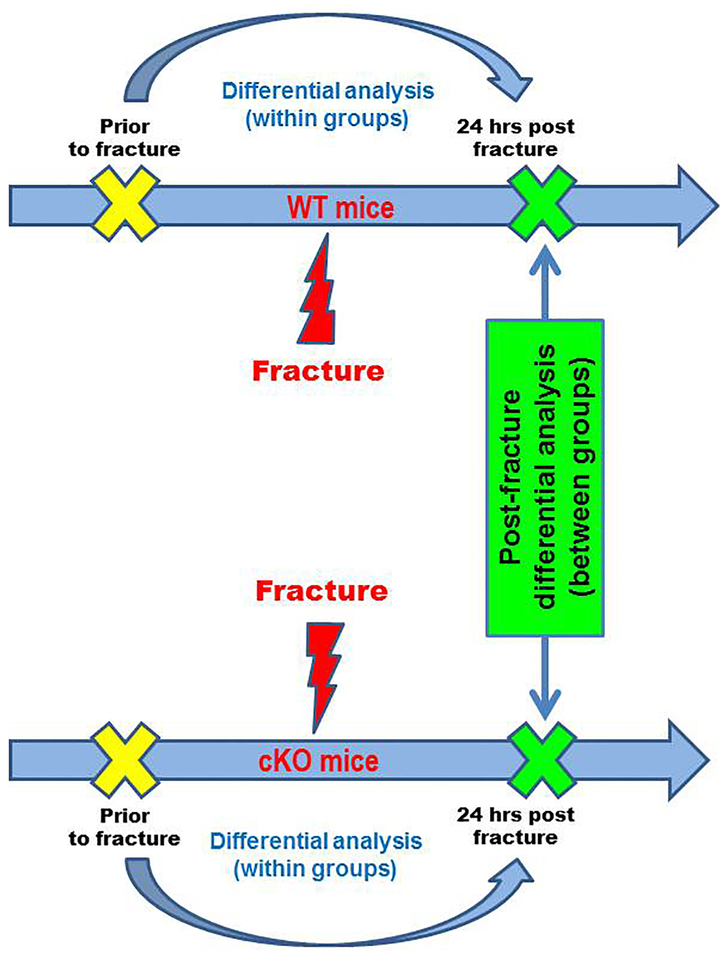
Our research design follows the flowchart shown in Figure 1. In the WT group, 5 animals were sacrificed post-fracture vs. 6 pre-fracture, leading to 30 combinatory differences in gene expression activity between post- vs. pre-fracture (denoted WT Δpost-pre). Similarly, in the cKO group, we calculated 36 combinatory differences of gene activity by comparing 6 animals post-fracture vs. 6 pre-fracture (denoted cKO Δpost-pre). Student t-test was used to compare gene activities between two groups (WT vs. cKO) at the baseline before fracture, 24 hours post-fracture, as well as combinatory differences in gene expression activity (WT Δpost-pre vs. cKO Δpost-pre; with Bonferroni correction) (18). Analysis of Variances (ANOVA) was performed for statistical analyses among four groups.
Figure 1. Flow Chart of Experimental Design and Differential Expression Analysis.
Genes with induced or suppressed expression activities were identified within the WT and the cKO mice. Post-fracture transcriptional variations were compared between the two groups and corrected for multiple testing.
2.2. Animal Experiment and Humane Endpoints
All experiments were conducted in compliance with the Guide for the Care and Use of Laboratory Animals and were approved by the Harvard Medical Area Institutional Animal Care and Use Committee. Mice carrying floxed Bmp2 alleles (Bmp2c/c) with C57BL/6 background (16) were crossed with heterozygous Prx1::cre mice with C57BL/6 background (19) to obtain the following littermates: 1) Bmp2c/c mice (WT); 2) Bmp2c/c;Prx1::cre (cKO) mice. Mice were born at the expected Mendelian ratios. Tail biopsies were collected for genotyping by PCR as described by Tsuji et al. (16) Four groups of 8–10 weeks old mice (un-fractured WT, fractured WT, un-fractured cKO, fractured cKO) were created by randomly distributing 11 males and 12 female mice in each group (n=5–6). Using a method previously described (20), unilateral fractures were produced in the right femurs of the fractured-group mice. X-rays were taken using Micro50 (Microfocus Imaging, Faxitron Bioptics LLC, Tucson, AZ, USA) at 50 kV for 100 s, to ensure that the fracture sites were consistently located in the same central area of the diaphysis.
2.3. RNA Data Processing
24 hours after fracture, the two diaphysal fractured segments of each femur were harvested. Segments of 1 mm in length (from the fracture point) were collected. Care was taken to collect each bone segment with intact periosteal tissue. In samples without fractures, two mm of diaphysal bone, inclusive of the periosteum, were harvested from the corresponding areas. Total RNA was isolated from the tissue using a commercially available RNA isolation kit (TRIzol, Invitrogen). Multiple assessments of isolated RNA quality were performed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., USA), UV spectroscopy at 260 nm and 280 nm absorbance (A260/A280), gel electrophoresis, and by RT-PCR detection of three housekeeping genes (actin-β, TBP and TUBB5). Double-stranded DNA synthesis, biotin-labeled cRNA synthesis, and cRNA fragmentation were conducted as described by the nanoString nCounter® Analysis system and the associated analysis protocol (Seattle, WA). (21)
2.4. nanoString nCounter Data Normalization and Analysis
Raw data from the nanoString nCounter were normalized at three levels as recommended(21) using the R software nanoStringNorm package(22). Specifically, we used the geometric means of the positive controls, applied a stringent background correction of the mean plus 2 standard deviations, and chose TBP and TUBB5 as the housekeeping genes. To avoid problems with zero values (23), we further added 1 to the normalized data in log2 scale. We first calculated all combinatory differences in gene expression activity post- vs. pre-fracture within the WT and cKO groups (WT Δpost-pre and cKO Δpost-pre), respectively. Then, we used Student’s t-test to compare gene activities between two groups (WT vs. cKO) at the baseline before fracture, 24 hours post-fracture, as well as for WT Δpost-pre vs. cKO Δpost-pre. Analysis of Variances (ANOVA) was performed for statistical analyses among four groups. Statistical significance was set at p<0.05; except for WT Δpost-pre vs. cKO Δpost-pre that genes were considered significant if p value was less than the Bonferroni corrected p values (p < 0.05/36)(18). Differential gene expression profiles among the significant genes were displayed in the networks as red for up-regulation and green for down-regulation (medians of WT Δpost-pre and cKO Δpost-pre).
To validate the gene expression values obtained in vivo with the nanoString nCounter Analysis, we performed an in vitro quantitative RT-PCR for 10 randomly selected genes (among the 150 candidate genes studied in vivo) in mouse mesenchymal stem cells treated with Bmp2 (data not shown).
2.5. Functional Annotation
We characterized the functional enrichment of differentially expressed genes (DEGs) using the Database for Annotation, Visualization and Integrated Discovery(24) (DAVID; http://david.abcc.ncifcrf.gov/). GenBank accession numbers of the complete 150 genes as well as the 26 DEGs were uploaded as input for DAVID. Enriched biological processes were generated by an established algorithm(24). Functional clusters were sorted by the Enrichment scores (ES) output from the DAVID. Enrichment Score was developed based on the geometric means of the EASE scores (modified Fisher Exact tests) associated with the enriched annotation terms that belong to this gene group.
2.6. Co-expression Analyses and Networks Construction
Network models were constructed based on previously published protocol(25). In brief, co-expression Pearson Correlation Coefficients (PCCs) of every possible gene-gene correlation among the 26 DEGs were calculated using expression profiles in each of the four conditions: WT pre-fracture (n=6), WT post-fracture (n=5), cKO pre-fracture (n=6), and cKO post-fracture (n=6). Significant (p<0.05) absolute values of PCCs of 0.8 or greater for co-expression were set as the cut-off threshold. When significant correlated co-expression occurred between any gene-pair among the 26 DEGs in each experimental condition, a connecting link between the gene-pair was plotted in the network.
2.7. Network Topological Analyses
Using previously developed algorithm(26), we categorized each of the 26 DEGs based on their position and inter-relations with other genes (topology) to infer their potential dynamic biological roles across the four co-expression networks. Using this concept of connectivity, we evaluated the functional significance of each gene. In a connected network, a gene is defined as a hub when connected with two or more genes (otherwise it is periphery with only one connection). A hub was then classified as redundant or non-redundant based on whether a network will breakdown or dis-integrate into separate groups with its removal. Networks will break down into smaller disconnected parts with the removal of non-redundant focal linking hub from the originally connected networks (i.e. inter-modular hubs). Focality score of topological significance was calculated as previously described(26) for the non-redundant focal linking hubs. The higher the focality score, the more disruption of information is created by removal of such non-redundant focal linking hubs. On the other hand, alternative routes exist to maintain the integrity of networks if a redundant connecting hub is removed (i.e. intra-modular hubs). Number of connections represents as estimates of topological significance for both kinds of hubs. However, the number of connections is the only kind of estimate for redundant connecting (intra-modular) hubs. Lastly, a periphery gene can be considered as the start (initiating point) or the exit point in the networks. Less gene-gene interactive context is suggested for the periphery genes.
2.8. Mouse Protein-Protein Interactions in the Public Databases
We downloaded protein-protein interactions data (PPIs) from the NCBI Gene FTP database and extracted those specific to mice. Extracted PPIs related to the 150 candidate genes and the 26 DEGs signature were provided in Suppl. Table 6.
3. Results
Functional annotations(24) of the selected 150 genes (Suppl. Table 1) indicated a higher enrichment in signal peptides and secreted extracellular glycoproteins (95 genes; ES=30.05). A cluster of 30 genes, enriched in TGF-beta signaling pathway, consisted of Smad proteins, BMP receptor binding and growth factor activities (ES=14.13). Another cluster of 25 genes were enriched in the Wnt signaling pathway (ES=13.12). Please refer to Suppl. Table 1 for additional details of functional annotations.
3.1. Identifying Differential Expression Gene Signature (26 genes)
By comparing the post-fracture variations between WT vs. cKO groups (WT Δpost-pre vs. cKO Δpost-pre), we identified 26 genes demonstrating differential expression patterns during the fracture healing. The group comparisons, the ANOVA testing, and gene activities before and after fracture were plotted for each gene (see Suppl. Table 2; Suppl. Fig. S8–S33 and page 34~184 of supplemental material).
Function annotations indicated that 17 out of these 26 genes consisted of glycoproteins and signal peptides, with many of them secreted in the extracellular region and containing disulfide bonds (ES =6.16). A second cluster of 6 genes involved in regulating pluripotency of stem cells and osteoblast differentiation (ES = 3.39). A third cluster of 5 genes were associated with regulation of protein phosphorylation and angiogenesis (ES = 3.29). Lastly, 6 genes were mapped in the Wnt signaling pathway and extracellular matrix (ES = 3.16). Please see Table 1 for details.
Table 1.
NIH DAVID Functional Annotation Clusters for 26 genes
| Cluster % of list | Biological Terms | Accession | Enrichment Score |
|---|---|---|---|
| #1 17/26 (65.4%) |
GO:0005615~extracellular space Glycoprotein signal peptide GO:0005576~extracellular region Disulfide bond glycosylation site:N-linked (GlcNAc...) |
MMP9, Postn, Pparg, Tnn, Bmp2, Igf1r, Wnt2b, Eng1, MCAM, Dkk1, Ctsk, Tnn, Notch2, Adipoq, Wnt7b, Tnfrsf1a, Sost | 6.16 |
| #2 8/26 (30.8%) |
mmu04550:Signaling pathways regulating pluripotency of stem cells mmu05200:Pathways in cancer GO:0045669~positive regulation of osteoblast differentiation GO:0045165~cell fate commitment |
MMP9, Pparg, Bmp2, Igf1r, Wnt7b, Wnt2b, Cebpa, Fgf2 | 3.39 |
| #3 5/26 (19.2%) |
GO:0001934~positive regulation of protein phosphorylation GO:0045766~positive regulation of angiogenesis GO:0010628~positive regulation of gene expression |
MMP9, Bmp2, Adipoq, Fgf2, Eng1 | 3.29 |
| #4 6/26 (23.1%) |
GO:0005578~proteinaceous extracellular matrix GO:0016055~Wnt signaling pathway mmu04310:Wnt signaling pathway GO:0031012~extracellular matrix |
MMP9, Postn, Tnn, Wnt7b, Wnt2b, Sost | 3.16 |
3.2. Post-fracture Expression Variation in the Wild Type (WT) Group
In WT, the medians of post-fracture expression variation ranged from a maximum of 6.37 (Myod) to a minimum of −6.79 (Adipoq), on the log scale. We noted significant induction of Fgf2, MCAM, PTHrP and Bmp2 after fracture, with reduction of Smad8, Wnt2b, Wnt7b, Sost and Adipoq. Of note, suppression of Wnt2b and Wnt7b was found only in the WT group. Please see Table 2 for details of expression variation and the topological role shift in the pre- vs. post-fracture networks. Additional information is provided in Suppl. Tables 2&3.
Table 2.
Differentially Expressed Genes in the wild type (WT) and knockout (cKO) groups
| Wild Type (WT) Group | Knockout (cKO) Group | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Median (Range) ΔWT post-pre fracture | Rank | Network Pre → Post | Network Pre → Post | Rank | Median (Range) ΔcKO post-pre fracture | Gene |
| Myod | 6.37 (5.89) | Up 1 | P → Hc (2) | Hc (2) → Hc (3) | Up 1 | 5.41 (9.38) | Wnt7b |
| FGF2 | 2.80 (2.71) | Up 2 | N → Hc (4) | N → Hc (6) | Up 2 | 3.69 (5.94) | Myod |
| MCAM | 2.66 (2.19) | Up 3 | Hf (2) → Hc (3) | Hc (3) → P | Up 3 | 2.92 (7.20) | Tbx4 |
| PTHrP | 2.52 (8.66) | Up 4 | Hf (5)[8.5] → Hc(5) | Hc (5) → Hc (6) | Up 4 | 2.32 (1.96) | MCAM |
| Bmp2 | 2.44 (4.42) | Up 5 | P → N | Hc(4) →Hf(10)[17] | Up 5 | 2.15 (2.40) | Eng1 |
| Smad8 | 0 (8.66) | Dn 5 | Hf (7)[22.5] → Hc(2) | P → N | Dn 5 | −0.60 (2.77) | Ctsk |
| Wnt2b | 0 (0.004) | Dn 4 | Hc (3) → N | N → P | Dn 4 | −1.18 (7.10) | Bmp2 |
| Wnt7b | 0 (4.31) | Dn 3 | Hc (3) → N | Hc(3)→Hf(3)[11.67] | Dn 3 | −1.92 (8.80) | Adipoq |
| Sost | −0.48 (5.02) | Dn 2 | Hc (6) → Hc (4) | Hf(2)[8.5] → Hc (6) | Dn 2 | −3.27 (8.88) | Sost |
| Adipoq | −6.79 (6.13) | Dn 1 | P → Hc (3) | P → N | Dn 1 | −3.64 (5.62) | Smad8 |
Note: Expression levels were log2 transformed.
Abbreviation used: Up, up-regulated; Dn, down-regulated; P, periphery gene; Hc (degree), redundant connecting hub (number of neighboring genes); Hf, non-redundant focal linking hub (number of neighboring genes)[estimated focality scores]; N, non-existent in networks.
Italic: Bmp2 and Wnt7b had opposite differential expression profiles between WT vs. KO groups.
Bold: Myod, MCAM, Smad8, Sost and Adipoq had similar direction in both WT and KO groups.
3.3. Post-fracture Expression Variation in the Knockout (cKO) Group
Bmp2 was found down-regulated in both pre- and post-fracture in the cKO mice, confirming the validity of our experimental design. Relative to WT, post-fracture variations in the cKO group showed that Myod and MCAM were similarly up-regulated and that Adipoq, Smad8 and Sost were similarly down-regulated. A few differences occurred in the post-fracture expression variations between WT and cKO: (1) Wnt7b was highly induced in the cKO post-fracture, whereas it was maintained at minimal expression levels in the WT; (2) a significant induction of Tbx4 and Eng1 was detected in the cKO, when compared to WT; and (3) Bmp2 and Ctsk were significantly down-regulated in the cKO. Please see Table 2 for details.
3.4. Topological Gene-Gene Interplays in Co-expression Networks
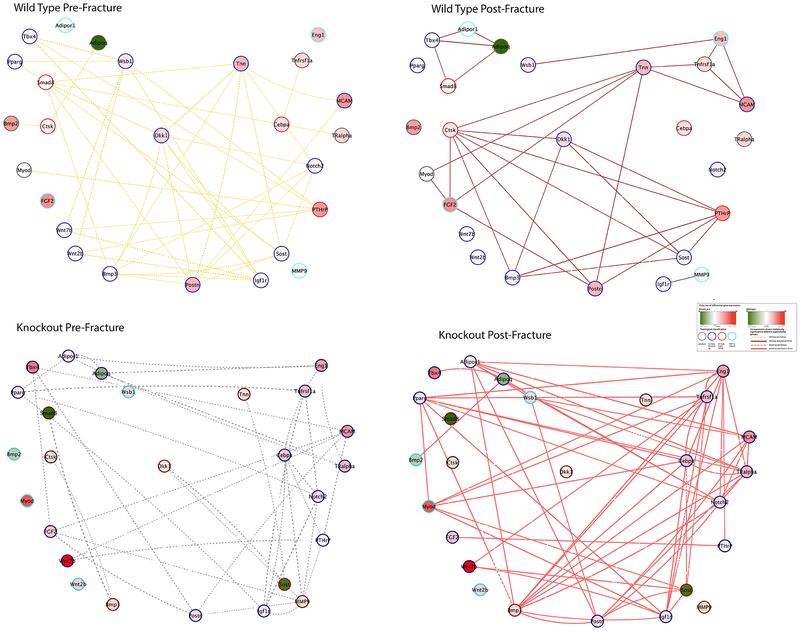
After analyzing the post-fracture expression variations, we constructed co-expression networks in each of the experimental condition to characterize the underlying gene-gene interplays. A total of four co-expression networks were compiled using the expression values of the 26 DEGs in each experimental group (WT pre-fracture, WT post-fracture, cKO pre-fracture and cKO post-fracture; Figure 2). For each gene, four dynamic topological roles and gene-gene interplays were provided. Accordingly, a gene can be categorized into the following membership/role groups based on the network topology (25, 26): (1) non-redundant focal linking hub (denoted as Hf); (2) redundant connecting hub, denoted as Hc; (3) periphery gene, denoted as P; or (4) non-existent, i.e. not correlated with other genes among the 26 genes, denoted as N. See additional information provided in Suppl. Tables 3&4.
Figure 2. Differential Co-Expression Networks: panels for WT pre-fracture (upper-right), WT post-fracture (upper-left), cKO pre-fracture (lower-right) and cKO post-fracture (lower-left).
Node coloring indicates gene activity after fracture – red for induced, and green, suppressed; node border coloring indicates topological membership/role classification; and edge coloring is based on experimental condition. All information is illustrated in the “inset”.
3.5. Comparison of Co-expression networks in the WT Group
For the WT group, pre- (dotted yellow line) vs. post-fracture (solid crimson line) networks were presented in Figure 2 (upper panels). Cyan node border indicated no connection in the pre-fracture network. In the WT pre-fracture network, 22 out of 26 genes demonstrated significant co-expression before fracture. After fracture, only 19 genes showed co-expression. Bmp2 is a periphery gene in the WT pre-fracture networks, simulating an initiating position that connects to Ctsk and then to Adipoq. After fracture, Bmp2 lost connection with Ctsk, but both Ctsk, and Adipoq acquired additional connections to other genes in the network. More specifically, after fracture, Ctsk linked to such up-regulated genes as Fgf2 and PTHrP.
In the pre-fracture network, MCAM (ranked as 3rd most up-regulated gene) occupied a non-redundant focal linking hub (neighbor genes: Postn and Tnn). This position changed to a redundant connecting hub (neighbor genes: Eng1, Tnfrsf1a and Tnn) after fracture (Table 1; Figure 2). Although both MCAM and Tnn demonstrated up-regulated post-fracture expression variations, the co-expression coefficients between MCAM and Tnn changed from 0.85 before fracture to −0.91 after fracture, indicating that compared to Tnn, the MCAM post-fracture expression level was much higher. Please see Figure Suppl. Tables S3–5, and Suppl. Fig. S8–33 for more details of all co-expression links.
We noted that Smad8, PTHrP, Ctsk and Cebpa played important non-redundant linking positions in the pre-fracture networks (see numbers of connections in Table 3). Smad8 had 7 neighbor genes prior to fracture but linked only to Adipoq and Tbx4 after fracture, whereas PTHrP maintained 5 connections (although different from those in the pre-fracture networks) and Ctsk gained 5 additional connections (Figure 2). Differently, Cebpa lost all connections after fracture. In addition, Tnn and Eng1 became topologically critical as non-redundant focal linking hubs after fracture.
Table 3.
Critical hub genes
| Wild Type Group Networks | Same Gene in the Knockout Group Networks | |||||
|---|---|---|---|---|---|---|
| Hf Gene | Median (Range) | Rank | Pre → Post | Pre → Post | Rank | Median (Range) |
| Smad8 | 0 (8.66) | Dn 5 | Hf (7) [22.5] → Hc (2) | P → N | Dn 1 | −3.64 (5.62) |
| Cebpa | 0.6 (0.94) | -- | Hf (3) [11] → N | Hc (5) → Hc (8) | -- | 1.43 (2.83) |
| PTHrP | 2.52 (8.66) | Up 4 | Hf (5) [8.5] → Hc (5) | Hc (4) → Hf (2) [9] | -- | 0.02 (6.72) |
| Ctsk | 0.32 (3.92) | -- | Hf (2) [0.5] → Hc (7) | P → N | Dn 5 | −0.60 (2.77) |
| Tnn | 1.78 (4.22) | -- | Hc (7) → Hf (6) [16] | P → N | -- | 0.85 (3.32) |
| Eng1 | 1.55 (1.60) | -- | N →Hf (3) [5.5] | Hc (4) → Hf (10) [17] | Up 5 | 2.15 (2.40) |
| Same Gene in the Wild Type Group Networks | Knockout Group Networks | |||||
| Hf Gene | Median (Range) | Rank | Pre → Post | Pre → Post | Rank | Median (Range) |
| MMP9 | 0.20 (1.82) | -- | N → P | Hf (7) [22.5] → N | -- | −0.54 (2.94) |
| Dkk1 | 0.93 (5.78) | -- | Hc (7) → Hc (4) | Hf (2) [16] → N | -- | −0.33 (5.23) |
| Sost | −0.48 (5.02) | Dn 2 | Hc (6) → Hc (4) | Hf (2) [8.5] → Hc (6) | Dn 4 | −3.27 (8.88) |
| Bmp3 | 0.10 (3.44) | -- | Hc (7) → Hc (4) | Hf (2) [0.5] → Hc (10) | -- | 0.89 (3.10) |
| Notch2 | 0.31 (1.55) | -- | Hc (5) → N | Hc (5) → Hf (2) [24] | -- | 0.79 (3.56) |
| Eng1 | 1.55 (1.60) | -- | N →Hf (3) [5.5] | Hc (4) → Hf (10) [17] | Up 5 | 2.15 (2.40) |
| Adipoq | −6.79 (6.13) | Dn 1 | P → Hc (3) | Hc (3) → Hf (3) [11.67] | Dn 3 | −1.92 (8.80) |
| PTHrP | 2.52 (8.66) | Up 4 | Hf (5) [8.5] → Hc(5) | Hc (4) → Hf (2) [9] | -- | 0.02 (6.72) |
Abbreviation used: Up, up-regulated; Dn, down-regulated; P, periphery gene; Hc (degree), redundant connecting hub (number of neighboring genes); Hf, non-redundant focal linking hub (number of neighboring genes)[estimated focality scores]; N, non-existent in networks.
3.6. Comparison of Co-expression Networks in the Knockout Group
For the cKO group, pre- (dotted gray line) vs. post-fracture (solid rose line) networks were shown in Figure 2 (lower panels). Twenty-two out of 26 genes demonstrated significant correlation before fracture. Bmp2, Myod, Wnt2b and Wsb1 did not show up in the knockout pre-fracture networks (node border colored cyan). After fracture (Figure 2 lower-right), the network consisted of 20 genes, with six genes without connections (Ctsk, Dkk1, MMP9, Smad8, Tnn, Wnt2b). With the exception for Wnt2b, these genes were all highly connected in the WT post-fracture network.
Consistent with its significant low level of expression, Bmp2 did not connect with other genes in the pre-fracture networks. After fracture, Bmp2 presented with even lower levels of expression but gained a connection with Adipoq, a gene that became extremely suppressed after fracture. Compared with the WT group, two differences in topological role shift were noted: (1) Dkk1, Sost and Bmp3 were located at significant non-redundant linking positions in the pre-fracture network, indicating that these inhibitors of osteo-differentiation were substantially present in the cKO mice. After fracture, Sost and Bmp3 remained highly connected with Sost having 6 connections and Bmp3 10 connections. This was in contrast from that observed in the WT post-fracture networks where Sost and Bmp3 presented with only 4 connections each; (2) Notch2 changed from a redundant connecting hub to a critical non-redundant focal linking hub in cKO post-fracture network, whereas it was non-existent in the WT. Interestingly, Eng1 and PTHrP demonstrated similar topological importance both in the cKO and WT networks.
4. Discussion
Network topological approach to examine Bmp2 activity in vivo during fracture healing, provided us with a dynamic perspective of post-fracture transcriptional variations. By examining changes in topological membership in the co-expression networks under each condition (WT pre-, WT post-, cKO pre-, cKO post-fracture), an integrative view of the 26 DEGs was presented. The identification of the gene-gene interplays helped us recognizing potential Bmp2 target genes such as Sost, Myod, PTHrP, Fgf2, Dkk1, Postn, Tnn, Ctsk, MMP9, Wnt7b, Adipoq, Tbx4, and Wnt2b. As of today, no experimentally validated protein-protein interactions study of BMP2 is present in the NCBI database or in current literature; little information about BMP2 co-expression has been published in PubMed (Suppl. Tables S2&6). Thus, the present work provides an alternative approach to identify potential targets for BMP2. Further independent analyses will help verify the proposed correlations.
Several findings merit discussion. First, we found that Fgf2 was not present in the WT pre-fracture network (Figure 2, upper left panel), but demonstrated connections with Ctsk, Myod, Postn and Tnn in the WT post-fracture network. This result is, at least in part, confirmed by the studies of Downey et al., who reported Fgf2 induction of bone formation via Ctsk(27). In contrast, in the cKO group, Fgf2 was connected with MCAM, MMP9, Postn and Tbx4 before fracture and lost connections with all except PTHrP post-fracture. In addition, our results indicate that MCAM, Postn and Tbx4 were up-regulated in cKO mice pre-fracture compared with the WT. We suspect that absence of BMP2 in cKO mice may correlate with overexpression of these genes, which may influence the homeostasis of bone in the cKO mice.
Secondly, a group of 5 genes (Ctsk, Dkk1, MMP9, Smad8, and Tnn) were expressed in the WT post-fracture network but were not expressed in the cKO group (see Figure 2, upper and lower right panels). The membership of Smad8 changed from a focal linking hub in WT pre-fracture (focality score 22.5; 7 connections) to a redundant hub linked to only Tbx4 and Adipoq in WT post-fracture (with Tbx4 up-regulated and Adipoq down-regulated; see Suppl. Fig. S8 and S27). In the cKO group, Smad8 was a periphery gene connected to Bmp3 before fracture and became absent in the cKO post-fracture network. This may imply that Bmp3, being a well-known inhibitor of Bmp signaling(28, 29), has a role in the alteration of the bone homeostasis in the cKO group, suggesting that lower levels of Bmp2 expression may be responsible for Bmp3 up-regulation. Ctsk changed from a non-redundant focal linking hub in the WT pre-fracture network (focality score 0.5; 2 connections of Bmp2 and Adipoq) to a redundant connecting hub having 7 neighbor genes in the WT post-fracure network. In the cKO group, we noticed co-expression of Bmp3 and Ctsk pre-fracture and absence of Ctsk post-fracture, which may imply a role of Ctsk in bone homeostasis under the cKO condition. MMP9 was absent in the WT pre-fracture network but later connected with only Igf1r in the WT post-fracture network. In contrast, in the cKO group, MMP9 changed from a focal linking hub (focality score 22.5; 7 connections) in cKO pre-fracture network to absent in cKO post-fracture network. Co-expressions of MMP9 in the cKO pre-fracture network with Notch2, Postn, Pthrp, Cebpa, Dkk1, Fgf2, Igf1r might be responsible for the inability of bone healing in the cKO group, which could be related to the possible inhibiting role of these 7 genes in the osteo-differentiation pathways. Dkk1, a known inhibitor of the canonical Wnt signaling pathway, played an essential role in the WT networks (pre-fracture 7 neighboring genes – Igf1r, Notch2, Postn, Smad8, Sost, Tnn and Bmp3; post-fracture 4 neighboring genes, PTHrP, Sost, Bmp3 and Ctsk). However, in absence of Bmp2 (cKO group) Dkk1 only correlated with MMP9 and Sost in the cKO pre-fracture network and became absent in cKO post-fracture. We suspect a central role of Dkk1 in fracture healing, which highlights the crosstalk between Wnt and Bmp pathways. Indeed, we previously reported that Dkk1 haplo-insufficiency does not rescue the inability to heal fracture in Bmp2 cKO mice, further implying that Bmp signaling is required for the Wnt-mediated bone anabolic activity(20). In addition, we extracted mouse protein-protein interactions (PPIs) from the NCBI Gene database and found that Dkk1 has multiple interactions with proteins that were included in our co-expression networks (see for instance Dkk1 interacting with Lrp6 and Sost in the PPIs; Suppl. Table S6) (30–32). We acknowledge that the information can only serve as indirect support for our finding and propose further experimental validations.
Third, Endoglin, a co-receptor for Bmp2 signaling (33), was very active in both the WT and cKO groups. We speculate that the focal linking position of Eng1 (hence its biological activity) in the cKO post-fracture network indicates an ineffective role of this receptor in absence of Bmp2 expression. In fact, Endoglin was found to mediate Bmp2/Bmp4 signaling during yolk sac hematopoietic development(34) as well as being involved in Bmp2-induced osteoblastic differentiation independently of Smad1/5/8 phosphorylation(35). Fourth, Bmp2 is known to stimulate expression of Tenascin (Tnn) (36, 37). Our findings showed that in WT networks, Tnn was a high-connected redundant hub before fracture (7 neighbor genes: Bmp3, Dkk1, MCAM, Notch2, Postn, Smad8 and Sost) then became a critical non-redundant focal linking hub after fracture (connected to Ctsk, Fgf2, MCAM, Myod, Postn, Tnfrsf1a). In the cKO group, Tnn was a periphery gene before fracture and became absent after fracture. This information may provide a stronger foundation to further explore the possible interplay between Bmp2 and Tnn.
Several limitations of this study are worth mentioning. First, given the nature of co-expression analyses, direct or causal relationships cannot be inferred among the genes in the networks. Secondly, in our models Bmp2 was conditionally knockout in a limb-specific manner and therefore our finding may not be generalizable to other tissues and organs. Additionally, we acknowledge that by utilizing a Prx1-cre mouse deleter we have not been able to inactivate Bmp2 in other areas such as vascular tissue. Therefore, this analysis presents with the limitations of evaluating genes that depend on Bmp2 when it is expressed in tissue where Prx1 is or was active. Third, our results were confined to the examined candidate genes (150 genes) and a systematic genome-wide approach will certainly provide additional information. Fourth, while Bmp2 seems to have a periosteal activation function (16), the tissue samples collected in our experiments contained cortical bone as well as bone marrow and therefore our gene expression analysis was not limited to the periosteum. Nevertheless, the discovery of the molecular mechanisms associated with Bmp2 activation may shed light on the role of Bmp2 in bone regeneration.
In summary, our investigation used a candidate genes approach(38) to find Bmp2 downstream gene-gene interplays in the fracture healing process in vivo. We identified 26 differentially expressed genes by comparing the post-fracture gene expression variation in the WT vs. cKO groups. We provide an analytical view of the dynamic topological role shifts among the co-expressed genes under the conditional Bmp2 inactivation. Our results can be useful to identify potential functional interplays between genes to be tested in future studies for characterizing the molecular mechanisms of Bmp2 signaling.
Supplementary Material
Supplementary Table 1 Details of 150 candidate genes and DAVID output
Supplementary Table 2 Details of 26 differentially expressed genes
Supplementary Table 3 Details of topological roles of 26 differentially expressed genes among different networks
Supplementary Table 4 & 5 Details of co-expression gene activities among different networks.
Supplementary Table 6 Details of mouse protein-protein interactions of the 150 candidate genes and the 26 differentially expressed genes
Fig S1-S6 differential co-expression networks
Fig S7 heatmap of transcriptional expression of 26 differentially expressed genes
Fig S8-S33 streamline presentations of differentially co-expression (by each gene)
Page 34 ~184 Bar graph and line graph of group comparisons with p-values (for 150 candidate genes, one page per gene)
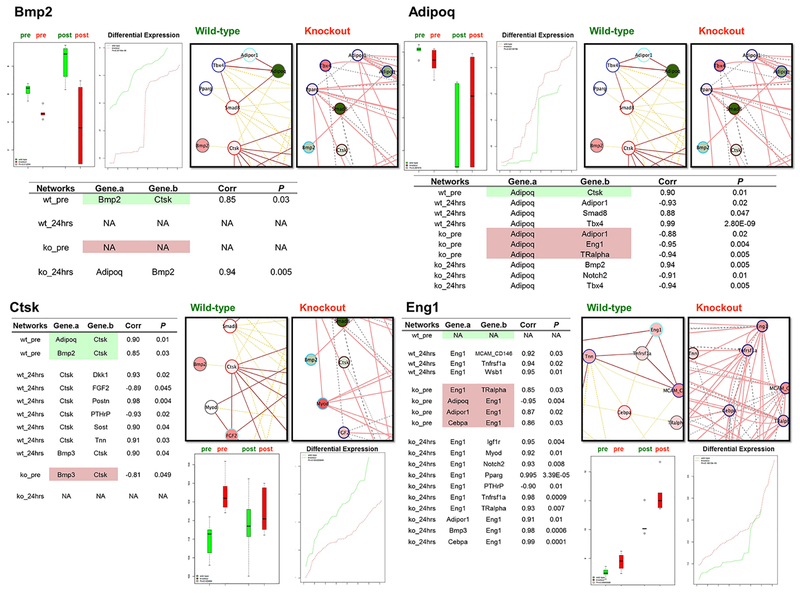
Figure 3. Streamline Presentations of Differentially Co-Expressed Genes (Bmp2, Adipoq, Ctsk, and Eng1).
Expression levels were plotted in bar charts. Green boxes: WT group; red boxes, knockout group. The network illustration is similar as in Figure 2 (pre-fracture and post-fracture merged) with smaller snapshots of the gene of interest and the connected genes. Node coloring indicates gene activities after fracture – red, induced; green, suppressed; Node border: topological gene grouping; edge coloring: experimental groups. See Figure 2 “inset”. Table of co-expression gene activities in each experimental condition is provided (please see Fig. S7–S32 for the complete list of the 26 DEGs).
Acknowledgments
Dr. Yu is currently supported by the National Institute of Dental and Craniofacial Research (1K23DE026804–01A1).
This project was supported by grant # 560_2008 to Giuseppe Intini (ITI International Team for Implantology), and in part by grant # K99/R00 DE021069 to Dr. Giuseppe Intini (NIH/NIDCR) and grant # R01AR055904 to Dr. Vicki Rosen (NIH/NIAMS) while Dr. Intini was a postdoctoral fellow in Dr. Rosen’s laboratory at the Harvard School of Dental Medicine. Authors thank Dr. Vicki Rosen for releasing the data and permitting publication.
Footnotes
Disclosure
All authors declare no conflict of interest.
References
- 1.Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, et al. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242(4885):1528–34. [DOI] [PubMed] [Google Scholar]
- 2.He X, Dziak R, Yuan X, Mao K, Genco R, Swihart M, et al. BMP2 genetically engineered MSCs and EPCs promote vascularized bone regeneration in rat critical-sized calvarial bone defects. PLoS One. 2013;8(4):e60473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Z, Yuan X, Fernandes G, Dziak R, Ionita CN, Li C, et al. The combination of nano-calcium sulfate/platelet rich plasma gel scaffold with BMP2 gene-modified mesenchymal stem cells promotes bone regeneration in rat critical-sized calvarial defects. Stem Cell Res Ther. 2017;8(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Termaat MF, Den Boer FC, Bakker FC, Patka P, Haarman HJ. Bone morphogenetic proteins. Development and clinical efficacy in the treatment of fractures and bone defects. J Bone Joint Surg Am. 2005;87(6):1367–78. [DOI] [PubMed] [Google Scholar]
- 5.Cho TJ, Gerstenfeld LC, Einhorn TA. Differential temporal expression of members of the transforming growth factor beta superfamily during murine fracture healing. J Bone Miner Res. 2002;17(3):513–20. [DOI] [PubMed] [Google Scholar]
- 6.Kishigami S, Mishina Y. BMP signaling and early embryonic patterning. Cytokine & growth factor reviews. 2005;16(3):265–78. [DOI] [PubMed] [Google Scholar]
- 7.Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes & development. 1996;10(13):1580–94. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Bradley A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development (Cambridge, England). 1996;122(10):2977–86. [DOI] [PubMed] [Google Scholar]
- 9.Styrkarsdottir U, Cazier JB, Kong A, Rolfsson O, Larsen H, Bjarnadottir E, et al. Linkage of osteoporosis to chromosome 20p12 and association to BMP2. PLoS biology. 2003;1(3):E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reneland RH, Mah S, Kammerer S, Hoyal CR, Marnellos G, Wilson SG, et al. Association between a variation in the phosphodiesterase 4D gene and bone mineral density. BMC medical genetics. 2005;6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong DH, Shen H, Zhao LJ, Xiao P, Yang TL, Guo Y, et al. Robust and comprehensive analysis of 20 osteoporosis candidate genes by very high-density single-nucleotide polymorphism screen among 405 white nuclear families identified significant association and gene-gene interaction. J Bone Miner Res. 2006;21(11):1678–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fritz DT, Jiang S, Xu J, Rogers MB. A polymorphism in a conserved posttranscriptional regulatory motif alters bone morphogenetic protein 2 (BMP2) RNA:protein interactions. Molecular endocrinology (Baltimore, Md. 2006;20(7):1574–86. [DOI] [PubMed] [Google Scholar]
- 13.Valdes AM, Hart DJ, Jones KA, Surdulescu G, Swarbrick P, Doyle DV, et al. Association study of candidate genes for the prevalence and progression of knee osteoarthritis. Arthritis and rheumatism. 2004;50(8):2497–507. [DOI] [PubMed] [Google Scholar]
- 14.Valdes AM, Van Oene M, Hart DJ, Surdulescu GL, Loughlin J, Doherty M, et al. Reproducible genetic associations between candidate genes and clinical knee osteoarthritis in men and women. Arthritis and rheumatism. 2006;54(2):533–9. [DOI] [PubMed] [Google Scholar]
- 15.Majidinia M, Sadeghpour A, Yousefi B. The roles of signaling pathways in bone repair and regeneration. J Cell Physiol. 2017. [DOI] [PubMed] [Google Scholar]
- 16.Tsuji K, Bandyopadhyay A, Harfe BD, Cox K, Kakar S, Gerstenfeld L, et al. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nature genetics. 2006;38(12):1424–9. [DOI] [PubMed] [Google Scholar]
- 17.Kloen P, Lauzier D, Hamdy RC. Co-expression of BMPs and BMP-inhibitors in human fractures and non-unions. Bone. 2012;51(1):59–68. [DOI] [PubMed] [Google Scholar]
- 18.Dunn OJ. Multiple Comparisons among Means. J Am Stat Assoc. 1961;56(293):52-&. [Google Scholar]
- 19.Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33(2):77–80. [DOI] [PubMed] [Google Scholar]
- 20.Intini G, Nyman JS. Dkk1 haploinsufficiency requires expression of Bmp2 for bone anabolic activity. Bone. 2015;75:151–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature biotechnology. 2008;26(3):317–25. [DOI] [PubMed] [Google Scholar]
- 22.Waggott D, Chu K, Yin S, Wouters BG, Liu FF, Boutros PC. NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics. 2012;28(11):1546–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suehara Y, Arcila M, Wang L, Hasanovic A, Ang D, Ito T, et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res. 2012;18(24):6599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4(5):P3. [PubMed] [Google Scholar]
- 25.Yu YH, Chiou GY, Huang PI, Lo WL, Wang CY, Lu KH, et al. Network biology of tumor stem-like cells identified a regulatory role of CBX5 in lung cancer. Sci Rep. 2012;2:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu YH, Kuo HK, Chang KW. The evolving transcriptome of head and neck squamous cell carcinoma: a systematic review. PLoS One. 2008;3(9):e3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Downey ME, Holliday LS, Aguirre JI, Wronski TJ. In vitro and in vivo evidence for stimulation of bone resorption by an EP4 receptor agonist and basic fibroblast growth factor: Implications for their efficacy as bone anabolic agents. Bone. 2009;44(2):266–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamer LW, Ho V, Cox K, Rosen V. Expression and function of BMP3 during chick limb development. Dev Dyn. 2008;237(6):1691–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gamer LW, Cox K, Carlo JM, Rosen V. Overexpression of BMP3 in the developing skeleton alters endochondral bone formation resulting in spontaneous rib fractures. Dev Dyn. 2009;238(9):2374–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter M, Chen X, Slowinska B, Minnerath S, Glickstein S, Shi L, et al. Crooked tail (Cd) model of human folate-responsive neural tube defects is mutated in Wnt coreceptor lipoprotein receptor-related protein 6. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(36):12843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubota T, Michigami T, Sakaguchi N, Kokubu C, Suzuki A, Namba N, et al. Lrp6 hypomorphic mutation affects bone mass through bone resorption in mice and impairs interaction with Mesd. J Bone Miner Res. 2008;23(10):1661–71. [DOI] [PubMed] [Google Scholar]
- 32.Ellies DL, Viviano B, McCarthy J, Rey JP, Itasaki N, Saunders S, et al. Bone density ligand, Sclerostin, directly interacts with LRP5 but not LRP5G171V to modulate Wnt activity. J Bone Miner Res. 2006;21(11):1738–49. [DOI] [PubMed] [Google Scholar]
- 33.Barbara NP, Wrana JL, Letarte M. Endoglin is an accessory protein that interacts with the signaling receptor complex of multiple members of the transforming growth factor-beta superfamily. The Journal of biological chemistry. 1999;274(2):584–94. [DOI] [PubMed] [Google Scholar]
- 34.Borges L, Iacovino M, Koyano-Nakagawa N, Baik J, Garry DJ, Kyba M, et al. Expression levels of endoglin distinctively identify hematopoietic and endothelial progeny at different stages of yolk sac hematopoiesis. Stem Cells. 2013;31(9):1893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishibashi O, Ikegame M, Takizawa F, Yoshizawa T, Moksed MA, Iizawa F, et al. Endoglin is involved in BMP-2-induced osteogenic differentiation of periodontal ligament cells through a pathway independent of Smad-1/5/8 phosphorylation. J Cell Physiol. 2010;222(2):465–73. [DOI] [PubMed] [Google Scholar]
- 36.Tucker RP, Chiquet-Ehrismann R. The regulation of tenascin expression by tissue microenvironments. Biochim Biophys Acta. 2009;1793(5):888–92. [DOI] [PubMed] [Google Scholar]
- 37.Scherberich A, Tucker RP, Degen M, Brown-Luedi M, Andres AC, Chiquet-Ehrismann R. Tenascin-W is found in malignant mammary tumors, promotes alpha8 integrin-dependent motility and requires p38MAPK activity for BMP-2 and TNF-alpha induced expression in vitro. Oncogene. 2005;24(9):1525–32. [DOI] [PubMed] [Google Scholar]
- 38.Dryja TP. Human genetics. Deficiencies in sight with the candidate gene approach. Nature. 1990;347(6294):614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 Details of 150 candidate genes and DAVID output
Supplementary Table 2 Details of 26 differentially expressed genes
Supplementary Table 3 Details of topological roles of 26 differentially expressed genes among different networks
Supplementary Table 4 & 5 Details of co-expression gene activities among different networks.
Supplementary Table 6 Details of mouse protein-protein interactions of the 150 candidate genes and the 26 differentially expressed genes
Fig S1-S6 differential co-expression networks
Fig S7 heatmap of transcriptional expression of 26 differentially expressed genes
Fig S8-S33 streamline presentations of differentially co-expression (by each gene)
Page 34 ~184 Bar graph and line graph of group comparisons with p-values (for 150 candidate genes, one page per gene)