Abstract
The response of patients with gliomas to alkylating chemotherapy is heterogeneous. However, there are currently no universally accepted predictors of patient response to these agents. We identify the nuclear factor κB (NF-κB) co-regulator B cell CLL/lymphoma 3 (BCL-3) as an independent predictor of response to temozolomide (TMZ) treatment. In glioma patients with tumors that have a methylated O6-methylguanine DNA methyltransferase (MGMT) promoter, high BCL-3 expression was associated with a poor response to TMZ. Mechanistically, BCL-3 promoted a more malignant phenotype by inducing an epithelial-to-mesenchymal transition in glioblastomas through promoter-specific NF-κB dimer exchange. Carbonic anhydrase II (CAII) was identified as a downstream factor promoting BCL-3–mediated resistance to chemotherapy. Experiments in glioma xenograft mouse models demonstrated that the CAII inhibitor acetazolamide enhanced survival of TMZ-treated animals. Our data suggest that BCL-3 might be a useful indicator of glioma response to alkylating chemotherapy and that acetazolamide might be repurposed as a chemosensitizer for treating TMZ-resistant gliomas.
INTRODUCTION
Diffuse gliomas encompass both lower-grade gliomas (LGGs), including grade II and III tumors, and glioblastoma (GBM). Within each grade, subgroups of tumors that have significantly different responses to therapy exist (1, 2). Despite this heterogeneity, virtually all glioma patients receive alkylating chemotherapy as part of their treatment regimen. Although alkylators like temozolomide (TMZ) improve overall patient survival (3), many patients experience minimal benefit from their use. These observations underline the critical need for predictors of response to alkylating therapy.
Glioma cells are distinguished by the presence of recurrent copy number alterations (CNAs) affecting broad chromosomal regions (4). Although these alterations primarily involve driver genes, they can also affect nearby passenger genes not directly involved in malignant progression (5). It has been suggested that targeting such flanking genes might have therapeutic effects (6). However, passenger genes, unlike drivers, have not yet been shown to mediate therapeutic susceptibility in clinical glioma (7).
The best described mechanism of resistance to TMZ involves direct cytotoxic lesion repair by the DNA repair enzyme O6-methylguanine DNA methyltransferase (MGMT) (8). However, even in tumors with low MGMT expression, other pathways have been shown to independently modulate response to therapy and patient outcome (9).
Nuclear factor κB (NF-κB) plays an important role in promoting resistance to DNA-damaging agents in GBM (10, 11). In addition to the five primary NF-κB subunits, p50 (NF-κB1, p105), p52 (NF-κB2, p100), p65 (RelA), RelB, and c-Rel (12), multiple co-regulators also contribute to the overall downstream response. B cell CLL/lymphoma 3 (BCL-3) is an atypical inhibitor κB (IκB) protein that both activates and inhibits NF-κB signaling (13). BCL-3 is a candidate oncoprotein shown to be up-regulated in several malignancies (14, 15) that regulates NF-κB signaling in conjunction with p50- and p52-containing dimers (16-18).
Here, we show that BCL-3 reduced TMZ-induced cytotoxicity in gliomas through the activation of carbonic anhydrase II (CAII). In addition, we found that the CA inhibitor acetazolamide (ACZ) potentiated the effect of TMZ in mouse models of glioma, identifying a potential repurposing strategy for TMZ chemosensitization.
RESULTS
BCL3 is an independent predictor of response to TMZ in GBM
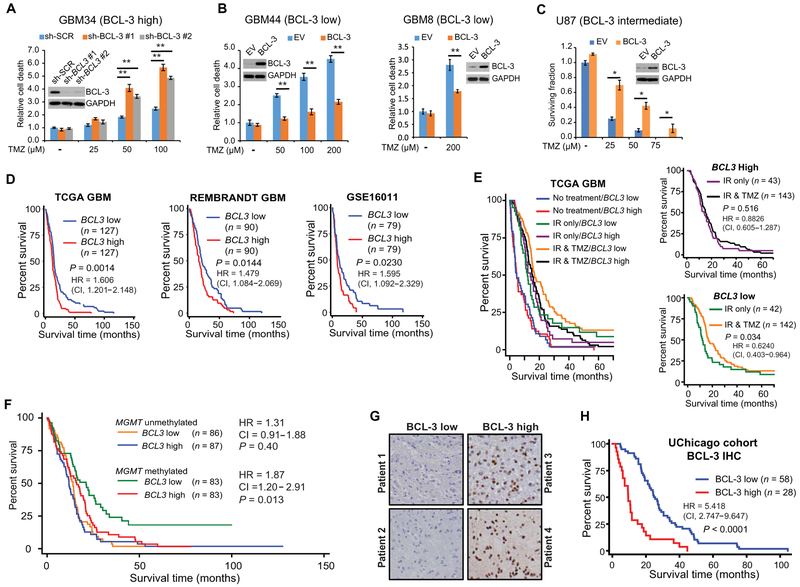
To examine the role of BCL-3 in response to TMZ, we altered its expression in patient-derived glioma stem-like cells (GSCs) (table S1) using vectors expressing human BCL-3 or short hairpins (sh) targeting BCL3 (fig. S1, A and B). Knockdown of BCL3 in GSCs with high basal BCL-3 increased cell death after treatment with TMZ (Fig. 1A and fig. S1C). Conversely, overexpression of BCL-3 in GSCs with low basal BCL-3 decreased TMZ-induced death (Fig. 1B and fig. S1D). Moreover, in U87, U251, and T98 GBM cell lines, knockdown of BCL3 with short interfering RNA (si-RNA) increased the TMZ-induced effect on clonal survival (fig. S1E), whereas overexpression of BCL-3 reduced the efficacy of TMZ (Fig. 1C and fig. S1F). These findings indicate that BCL-3 modulates cytotoxicity induced by TMZ.
Fig. 1. BCL3 predicts response to alkylating chemotherapy in GBM.
(A and B) Trypan blue assays in GSCs expressing the indicated construct treated with TMZ (72 hours). Data show fold change in percent dead cells relative to cells expressing a scrambled RNA sequence (sh-SCR), or empty vector (EV), treated with vehicle, ± SD (n = 3). Inset: Immunoblotting (IB) with anti–BCL-3. (C) Clonogenic assay in U87 cells stably expressing BCL-3 or EV treated with vehicle or TMZ. Data show mean value relative to vehicle ± SD (n = 3). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (D to F) Kaplan-Meier overall survival curves in GBM patients. (D) Survival based on median mRNA expression (REMBRANDT and GSE16011) and upper and lower quartile (TCGA). (E) TCGA GBM patients separated by median BCL3 expression and treatment modality. n = 490 patients total: no treatment, IR alone, and IR + TMZ; inset graphs: IR versus IR + TMZ in patients with high or low BCL3 mRNA (median cutoff). (F) TCGA GBM patients separated by median MGMT promoter methylation and median BCL3 expression. (G) Representative IHC staining for BCL-3 in GBM. (H) Survival curves in GBM patients separated by BCL-3 IHC staining. Kaplan-Meier curves analyzed by the log-rank test. *P < 0.05 and **P < 0.01.
Next, to investigate BCL-3 in clinical GBM, we examined four independent expression databases. In these data sets, patients with high BCL3 expression had reduced survival compared to patients with low BCL3 expression (Fig. 1D, fig. S1G, and table S2). BCL3 expression was significantly associated with survival on multivariate analysis, taking the primary GBM prognostic factors into consideration [hazard ratio (HR), 1.455; P = 0.017; Table 1]. The expression of none of the primary NF-κB subunits correlated with survival (fig. S1H). Similarly, NFKBIA, the gene encoding IκBα, a potential tumor suppressor in GBM (19), also had no prognostic value on its own (fig. S1I). These results indicate that BCL3 expression plays an independent role in GBM patient outcome.
Table 1. Cox regression analysis in TCGA GBM patients.
| Covariate | Univariate regression | Multivariate regression | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| BCL3 expression | 1.408 | 1.131–1.752 | 0.002 | 1.455 | 1.069–1.981 | 0.017 |
| MGMT methylation | 0.624 | 0.482–0.808 | 0.000 | 0.754 | 0.580–0.981 | 0.035 |
| IDH1 mutation | 0.232 | 0.075–0.724 | 0.012 | 0.490 | 0.153–1.573 | 0.231 |
| Radiotherapy | 0.357 | 0.287–0.445 | 0.000 | 0.424 | 0.307–0.585 | 0.000 |
| Chemotherapy | 0.576 | 0.471–0.705 | 0.000 | 0.650 | 0.492–0.860 | 0.003 |
| Procurement method | 1.155 | 0.864–1.543 | 0.330 | 1.055 | 0.734–1.516 | 0.773 |
| Age | 1.035 | 1.027–1.044 | 0.000 | 1.027 | 1.016–1.039 | 0.000 |
To understand whether BCL3 is specifically relevant in the setting of alkylation damage, we looked at TCGA to analyze whether BCL3 expression affected survival of GBM patients who were untreated, treated with ionizing radiation (IR) alone, or treated with IR in combination with TMZ. BCL3 expression did not segregate GBM patients into survival groups if they were untreated or treated with IR alone (Fig. 1E and fig. S1J). Consistent with this latter finding, overexpression of BCL-3 in GBM cells did not alter clonal survival in response to IR (fig. S1K). However, when TMZ was added to IR, patients with low BCL3 expression survived significantly longer than those with high expression (P = 0.0084; Fig. 1E and fig. S1J). In patients treated with TMZ, BCL3 expression remained associated with survival on multivariate analysis, taking into account MGMT promoter methylation, the primary factor associated with resistance to alkylating chemotherapy in GBM (table S3) (8). Moreover, our analysis revealed that addition of TMZ improved survival only in tumors with low BCL3 expression (Fig. 1E, inset graphs). To validate this role of BCL3, we examined a second GBM expression data set, the Repository of Molecular Brain Neoplasia Data (REMBRANDT) (20). In this data set, patients also received procarbazine, lomustine, and vincristine (PCV) as alkylating chemotherapy. In REMBRANDT, BCL3 expression was only informative of survival in patients whose treatment regimen included alkylating chemotherapy (fig. S1L). These data indicate that BCL3 expression was not an intrinsic prognostic factor in GBM or informative of response to IR but specifically modulated the response to alkylating chemotherapy.
Given the link between alkylation damage and MGMT promoter methylation (8), we divided patients based on MGMT promoter methylation, as assessed by the MGMT-STP27 model (21). BCL3 expression only identified distinct survival groups in tumors with high MGMT promoter methylation (Fig. 1F). Specifically, in GBM with high MGMT promoter methylation, the HR based on BCL3 expression was greater than in the entire GBM population (1.88 versus 1.41, respectively, univariate Cox regression; Fig. 1F). The data showed that the prognosis of patients with MGMT promoter methylated tumors that also had high BCL3 expression was similar to that in patients with an unmethylated MGMT promoter (Fig. 1F).
As public data sets primarily examine mRNA, to study the role of BCL-3 protein, we obtained specimens from a consecutive series of glioma patients. Individual slides and tissue microarrays (TMAs) were established, and BCL-3 protein was examined by immunohistochemistry (IHC) in 86 confirmed GBMs. IHC grading was performed in a blinded fashion based on a four-tier system that was converted into a binary scale (Fig. 1G). Clinical characteristics are noted (table S4); greater than 90% of patients received TMZ. Data analysis showed that GBM patients with low BCL-3 staining survived significantly longer than those with high staining [P < 0.0001; HR, 5.418; 95% confidence interval (CI), 2.747 to 9.647; Fig. 1H], a finding independent of age. These results support the mRNA data and emphasize the importance of BCL-3 protein in modulating the response to alkylation damage.
BCL3 is predictive in all diffuse gliomas
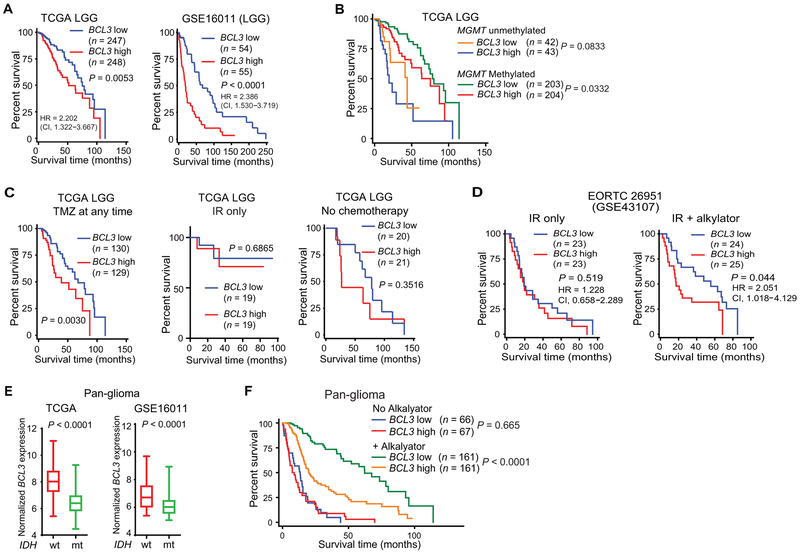
Alkylating chemotherapy is also important in the management of LGGs. Examination of multiple LGG data sets demonstrated that BCL3 expression separated these patients into distinct survival groups (Fig. 2A, fig. S2A, and table S5). In addition, our analysis showed that there was no difference in BCL3 expression between grade II and III tumors (fig. S2, B and C). Multivariate analysis that included age, isocitrate dehydrogenase 1 (IDH1) mutation, 1p/19q co-deletion, and MGMT promoter methylation showed that BCL3 expression remained associated with survival (Table 2); moreover, BCL3 separated LGG patients into different survival groups in tumors with high MGMT promoter methylation (Fig. 2B). We next partitioned patients by treatment modality. Consistent with a predictive effect, BCL3 expression was not informative in LGG patients who did not receive an alkylating agent or those treated with IR alone, whereas in patients treated with TMZ (either alone or with PCV) in combination with IR, BCL3 expression identified distinct survival groups (Fig. 2C). When data from patients treated with alkylating chemotherapy were analyzed on multivariate analysis, low numbers of patients achieving the endpoint of death precluded meaningful statistical analysis (table S6). To validate the predictive nature of BCL3, we examined LGGs from REMBRANDT. In this data set, BCL3 expression was informative of patient survival only in LGG patients who received alkylating chemotherapy, of which >95% included TMZ (fig. S2D). In addition, given the importance of randomized studies for identifying predictive factors, we looked at European Organization for Research and Treatment of Cancer (EORTC) study 26951, a randomized phase III clinical trial that examined addition of alkylating chemotherapy to IR in LGG (22). In this study, performed before TMZ was routinely used, BCL3 expression was not informative of survival time in patients treated with IR alone. However, among patients who received alkylating chemotherapy in addition to IR, those with low BCL3 expression survived significantly longer (HR, 2.051; P < 0.044; Fig. 2D). Moreover, multivariate analysis among patients that received chemotherapy showed that BCL3 expression remained significant (P = 0.006) when factoring in age and 1p/19q status (table S7; IDH1 mutation was not included due to incomplete patient data). These findings indicate that BCL3 is an independent predictor of response to alkylating chemotherapy in LGG.
Fig. 2. BCL3 is relevant in LGGs and pan-glioma patients.
(A and B) Survival curves from (A) LGG data sets separated by median BCL3 expression and (B) TCGA LGG patients separated by median BCL3 expression and median MGMT promoter methylation. (C) Survival curves in TCGA LGG patients based on median BCL3 expression and treatment (n = 338 patients total). For patients treated with TMZ, 202 received only TMZ, and 57 received TMZ and PCV. No chemotherapy group includes patients with and without IR. IR alone patients received no alkylating agents. (D) Survival curves in EORTC 26951 based on median BCL3 expression. Patients were separated by treatment modality into those treated with IR alone versus IR + alkylating chemotherapy. (E) BCL3 mRNA expression in pan-glioma patients based on IDH1 mutation status in the indicated data set. For TCGA: IDH1-wt, n = 184; IDH1-mt, n = 406. For GSE16011: IDH1-wt, n = 143; IDH1-mt, n = 83. (F) Survival curves in TCGA pan-glioma patients separated by median BCL3 expression and by alkylating chemotherapy treatment (yes/no). Kaplan-Meier curves analyzed by the log-rank test.
Table 2. Cox regression analysis in TCGA LGG patients.
BCL3 expression is also significant with inclusion of grade into analysis (P = 0.042).
| Covariate | Univariate regression | ||
|---|---|---|---|
| HR | 95% CI | P | |
| BCL3 expression | 1.431 | 1.186–1.735 | 0.000 |
| MGMTmethylation | 0.268 | 0.167–0.431 | 0.000 |
| IDH1 mutation | 0.103 | 0.062–0.173 | 0.000 |
| 1p/19q co-deletion | 0.486 | 0.281–0.837 | 0.009 |
| Grade (II or III) | 3.554 | 2.194–5.758 | 0.000 |
| Radiotherapy | 0.404 | 0.202–0.807 | 0.010 |
| Chemotherapy | 0.219 | 0.068–0.706 | 0.011 |
| Procurement method | 1.402 | 0.911–2.159 | 0.125 |
| Age | 1.060 | 1.042–1.079 | 0.000 |
| Covariate | Multivariate regression | ||
| HR | 95% CI | P | |
| BCL3 expression | 1.374 | 1.096–1.722 | 0.006 |
| MGMT methylation | 0.685 | 0.363–1.293 | 0.243 |
| IDH1 mutation | 0.401 | 0.186–0.864 | 0.020 |
| 1p/19q co-deletion | 0.646 | 0.339–1.229 | 0.183 |
| Age | 1.064 | 1.042–1.086 | 0.000 |
Given that IDH mutation status is as important a prognostic marker as tumor grade (23), we examined a pan-glioma data set (grade II, III, and IV tumors) in which patients were separated by IDH1 mutation status (24). Notably, IDH1-mutant (IDH-mt) gliomas had lower BCL3 expression than IDH–wild type (wt) (Fig. 2E). Similar results were seen with CpG island methylator phenotype (CIMP)–positive tumors compared to CIMP-negative tumors (fig. S2E). Our analysis also showed that BCL3 expression was reduced in LGG compared to GBM (P < 0.001, fig. S2F). In this pan-glioma data set, BCL3 was correlated with survival on multivariate analysis, taking IDH1 mutation, MGMT methylation, 1p/19q co-deletion, and age into consideration (Table 3). Consistent with a predictive effect, BCL3 expression separated pan-glioma patients into distinct survival groups only if they received alkylating chemotherapy in their treatment regimen (Fig. 2F). In these patients, BCL3 expression remained significant (P = 0.023) on multivariate analysis incorporating IDH1 mutation and MGMT promoter methylation (table S8). In summary, these data indicate that BCL3 is an independent predictor of response to alkylating chemotherapy in diffuse gliomas.
Table 3. Cox regression analysis in TCGA pan-glioma patients.
Only patients with RNA-seq data included.
| Covariate | Univariate regression | Multivariate regression | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| BCL3 expression | 2.390 | 2.065–2.765 | 0.000 | 1.467 | 1.213–1.774 | 0.000 |
| MGMT methylation | 0.227 | 0.168–0.307 | 0.000 | 0.590 | 0.416–0.835 | 0.003 |
| IDH1 mutation | 0.073 | 0.051–0.103 | 0.000 | 0.309 | 0.176–0.541 | 0.000 |
| 1p/19q co-deletion | 0.212 | 0.127–0.353 | 0.000 | 0.705 | 0.379–1.312 | 0.270 |
| Age | 1.070 | 1.058–1.080 | 0.000 | 1.052 | 1.037–1.066 | 0.000 |
BCL3 loss is a passenger event associated with survival
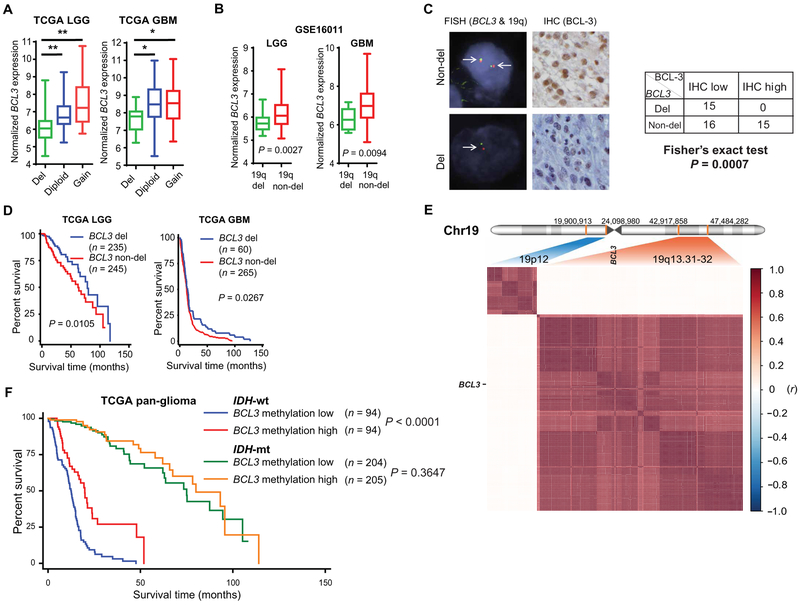
Multiple transcription factors can contribute to BCL3 mRNA expression (13); however, it is notable that BCL3 is found on 19q13, a chromosomal band deleted in 20 to 40% of all types of glioma (25-29). We therefore examined the relationship between BCL3 copy number (CN) and expression. In both LGGs and GBMs, tumors with BCL3 CN loss had significantly lower BCL3 mRNA expression (P < 0.003; Fig. 3A). Moreover, in an independent data set, LGGs and GBM with 19q deletion had significantly lower BCL3 expression than nondeleted tumors (P < 0.01; Fig. 3B). To examine the link between CN and protein expression, we performed fluorescence in situ hybridization (FISH) on TMA samples. Tumors with hemizygous BCL3 deletion had significantly lower BCL-3 than nondeleted tumors, as assessed by IHC (P = 0.0007; Fig. 3C). The association between CN and BCL3 expression raised the question of whether CN is also linked to survival. In both LGG and GBM groups, patients who had tumors with BCL3 deletion survived significantly longer than those with nondeleted tumors (P < 0.05; Fig. 3D). Moreover, BCL3 CN remained significantly associated with survival on multivariate analysis (P = 0.021; table S9). These results indicate that genetic deletion of BCL3 is associated with reduced BCL-3 expression and improved survival.
Fig. 3. Loss of BCL3 is a passenger event associated with improved survival.
(A) Relationship between BCL3 expression [on RNA sequencing (RNA-seq)] and BCL3 CN in TCGA LGG and GBM. In LGG, del: n = 169; diploid: n = 155; gain: n = 16. In GBM, del: n = 17; diploid: n = 89; gain: n = 39. (B) BCL3 expression in LGG and GBM tumors in relation to tumor 19q deletion status [determined by microsatellite polymerase chain reaction (PCR)]. LGG: del, n = 40; non-del, n = 33. GBM: del, n = 12; non-del, n = 58. (C) Representative FISH images using BCL3 and 19q specific probes (left) and BCL-3 IHC staining (high and low, right) in human gliomas. Table shows number of tumors in each category, two-sided Fisher’s exact test. (D) Kaplan-Meier curves in LGG and GBM patients based on BCL3 CN (with and without deletion). (E) Correlogram showing correlation (r, y axis) between CN of genes from the indicated regions of chromosome 19 (Chr19) in n = 514 TCGA LGG patients. For 19q13.31-32 (Chr19:42,917,858 to 47,484,282), there are 171 genes in a span of 4.5 Mbp (million base pairs); 139 of these genes have CN data available for analysis. The nearest 19p band (19p12) was used as a reference. For 19p12 (Chr19: 19,900,913 to 24,098,980), there are 59 genes in the 4.2-Mbp region, of which 36 have CN data. Location of BCL3 shown. (F) Survival in TCGA pan-glioma patients based on median BCL3 promoter methylation and IDH1 mutation status. Pearson coefficient (r) analyzed by two-sided Student’s t test. *P < 0.003 and **P < 0.0002.
The prevalence of BCL3 loss suggests either that BCL3 is a driver of glioma formation or that it is inadvertently affected by alterations targeted to a large chromosomal region. The lack of reported BCL3 mutations or translocations in glioma suggested that BCL3 loss is not a driver of these tumors (fig. S3A). Conversely, CNAs of BCL3 correlated with those of nearby genes (fig. S3B), suggesting that their CNAs are closely linked. Therefore, we examined the CN status of the genes surrounding BCL3 in 19q13.31-32. Analysis of TCGA data showed that in both GBM and LGG, loss of all the genes in this band correlated not only with BCL3 but also with the other genes in the region (Fig. 3E and fig. S3C), suggesting that BCL3 deletion occurs as a result of modifications targeted to a broad region of 19q13. This finding is consistent not only with the fact that the entire 19q arm is lost in oligodendrogliomas but also with the observation that, in astrocytic tumors and GBM, CNAs within 19q13 involve a broad region (26, 30). The above data support the hypothesis that BCL3 loss is a passenger event seen in all types of glioma.
Despite the importance of deletion, some BCL3 nondeleted gliomas also had low mRNA expression. Given the role of epigenetic modifications in regulating expression, we examined BCL3 promoter methylation. In TCGA pan-glioma data, a significant inverse correlation between BCL3 promoter methylation and expression was seen (P < 0.0001; fig. S3D). Although the correlation was evident in IDH-wt and IDH-mt tumors, this was stronger in IDH-wt tumors (fig. S3D). Analysis of the pan-glioma data set demonstrated that BCL3 promoter methylation was significantly associated with survival on multivariate analysis incorporating IDH mutation, MGMT promoter methylation, and 1p/19q co-deletion (P = 0.018; table S10). Moreover, when the data were separated by the patients’ IDH status, BCL3 promoter methylation was only informative in patients with IDH-wt tumors (Fig. 3F), a finding consistent with the stronger correlation between BCL3 methylation and expression in IDH-wt compared to IDH-mt tumors (fig. S3D). These results indicate that, in glioma, the genetic and epigenetic alterations of BCL3 that regulate expression are significantly associated with survival.
Given the correlation between BCL3 CN loss and that of other 19q13 genes, we examined whether the expression of other 19q13 genes was also associated with survival. Unlike BCL3, the mRNA expression of none of the surrounding 19q13 genes (fig. S3C) could separate TCGA GBM patients into significantly distinct survival groups, including the expression of RELB, an NF-κB subunit located on 19q13 whose CNAs are tightly linked to BCL3 and have been previously associated with GBM (fig. S1H) (2, 31) (GlioVis data portal for visualization and analysis of brain tumor expression data sets) (32). These findings indicate that the correlation between BCL3 loss and survival does not extrapolate to all genes in the 19q13 chromosomal region.
BCL-3 induces epithelial-to-mesenchymal transition in GBM
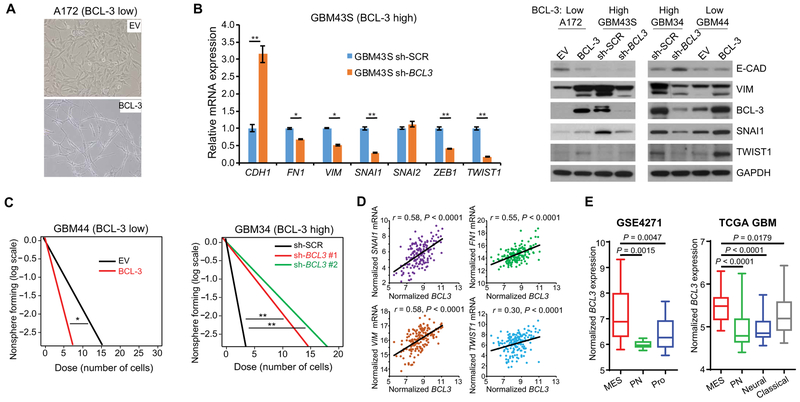
To understand how BCL-3 promotes resistance to alkylation damage, we modulated its expression in GSCs and GBM cell lines. In A172 cells that have extremely low basal BCL-3, overexpression of BCL-3 for 2 weeks changed the cell morphology to a spindle-like appearance, suggestive of epithelial-to-mesenchymal transition (EMT) (Fig. 4A). To further study this finding, we examined GSCs with low and high basal BCL-3 expression. Overexpression of BCL-3 in GBM44 GSCs and A172 cells increased the expression of several mesenchymal markers, including vimentin (VIM), SNAI1, and TWIST1, and reduced the expression of the epithelial marker E-cadherin (CDH1) (Fig. 4B and fig. S4A). Conversely, knockdown of BCL3 in GSCs expressing high basal BCL-3, GBM43S and GBM34, resulted in decreased mesenchymal marker expression (Fig. 4B). One prominent feature of mesenchymal differentiation is acquisition of a more stem cell–like phenotype (1, 33) that can be demonstrated in vitro by analysis of neurosphere formation. Loss of BCL3 reduced neurosphere formation ability in GBM34 cells, whereas overexpression of BCL-3 had the opposite effect in GBM44 GSCs, supporting the role of BCL-3 in mesenchymal differentiation (Fig. 4C). Consistent with the above, in TCGA GBM and LGG, a correlation between the expression of BCL3 and multiple EMT markers was seen (Fig. 4D and fig. S4B). Moreover, mesenchymal (MES) GBMs, as determined by the Phillips or Verhaak classification systems (1, 2), had significantly higher BCL3 expression than proneural (PN) tumors (P < 0.005; Fig. 4E and fig. S4C). Correlation of BCL3 with the Phillips MES or PN signature genes (1) demonstrated that, although there was a strong positive correlation between BCL3 and all the MES signature genes (mean correlation is 0.59, ±0.13; table S11), the correlation between BCL3 and PN signature genes was negative (mean correlation is −0.39, ±0.17; table S11) (GlioVis data portal for visualization and analysis of brain tumor expression data sets) (32). These data suggest that, in glioma, BCL-3 promotes mesenchymal differentiation.
Fig. 4. BCL-3 induces mesenchymal differentiation.
(A) Photomicrograph of A172 cells stably expressing BCL-3 or EV (×20 magnification). (B) Quantitative PCR (left) and representative immunoblot (right) of mesenchymal markers in GSCs and GBM cells. qPCR data show mean value relative to GAPDH normalized to sh-SCR ± SD (n = 3). (C) Limiting dilution neurosphere assays in GBM44 and GBM34 GSCs expressing the indicated constructs (n = 2 biologic replicates). (D) Correlation between mesenchymal markers and BCL3 mRNA in TCGA GBM samples (n = 166 for all genes). (E) BCL3 mRNA expression in GBM molecular subtypes. Phillips (left) and Verhaak (right) classification schemes. For GSE4271: MES, n = 22; PN, n = 9; Proliferative (Pro), n = 24. For TCGA: MES, n = 53; Classical, n = 37; Neural, n = 24; PN, n = 53. *P < 0.05 and **P < 0.01. Pearson correlation analyzed by two-sided Student’s t test.
BCL-3 induces mesenchymal differentiation via promoter-specific NF-κB dimer exchange
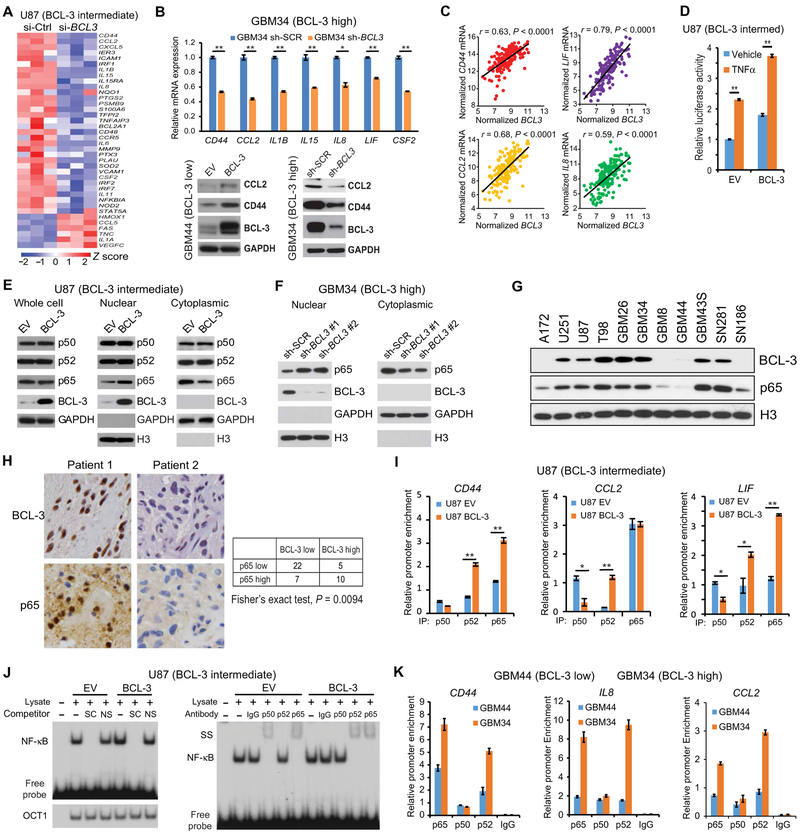
BCL-3 has no known enzymatic function and mediates its effects by regulating downstream gene expression. To understand how BCL-3 promotes mesenchymal differentiation, we examined genome-wide mRNA expression in GBM cells expressing si-BCL3 compared to si-control, a nontargeted sequence (table S12). As BCL-3 primarily acts by modulating NF-κB signaling, we looked at NF-κB–dependent factors previously identified as being specific to the MES phenotype (10). Knockdown of BCL3 caused a decrease of more than 80% of NF-κB–dependent MES genes (Fig. 5A). Among the transcripts most significantly down-regulated (>2-fold, adjusted P < 0.001) were CD44, chemokine (C-C motif) ligand 2 (CCL2), colony-stimulating factor 2 (CSF2), NF-κB–dependent factors critical for mesenchymal differentiation (10), and leukemia inhibitory factor (LIF), a primary MES signature gene (1). The BCL-3 dependence of these and several other MES genes was verified after depletion and overexpression of BCL-3 in GSCs and GBM cells (Fig. 5B and fig. S5A). In addition, in clinical GBM, a strong correlation between the expression of these factors and BCL3 was seen (Fig. 5C).
Fig. 5. BCL-3 promotes p65 nuclear translocation and NF-κB dimer exchange.
(A) Heatmap representing expression of all MES-specific NF-κB target genes [identified in (10)] in microarray analysis of U87 cells expressing si-control (Ctrl) or si-BCL3, performed using three separate biological samples. Z score normalized expression is graded by color. (B) qPCR (upper) of MES-specific genes in GBM34 GSCs. Data show mean value relative to GAPDH normalized to sh-SCR ± SD (n = 3). Bottom: Representative immunoblots in GBM44 and GBM34 GSCs expressing the indicated construct. (C) Correlation between MES-specific genes and BCL3 mRNA in TCGA GBM samples (n = 166 for all genes). (D) Relative luciferase activity in U87 cells expressing BCL-3 or EV treated with TNFα (5 ng/ml) or vehicle (12 hours). Data show mean value, normalized to vehicle-treated EV, ± SD (n = 4). (E) Representative immunoblot using the indicated cellular fractions from U87 cells expressing HA-BCL-3 or EV. (F) Immunoblot analysis with the indicated antibody using cytoplasmic and nuclear extract from GBM34 GSCs expressing two separate sh-BCL3 constructs or sh-SCR. Histone 3 (H3) antibody was used as nuclear loading control. (G) Representative immunoblot using nuclear fractions from the indicated GSCs and GBM cell lines. (H) Representative IHC images (left) of GBM samples from glioma TMAs showing nuclear p65 and BCL-3 staining in two patients. Table (right) shows numbers of tumors in each category. (I) ChIP qPCR in the indicated cells after IP. Data represent enrichment at the indicated promoter as a percentage of input ± SD (n = 2). (J) Representative electrophoretic mobility shift assay (EMSA) with CD44 κB probe using nuclear extracts from U87 cells expressing BCL-3 or EV. Supershift (SS) (right) and competition with specific (SC) or nonspecific (NS) probe (left); OCT1 binding confirms equal loading. (K) ChIP qPCR in GSCs after IP with the indicated antibodies. Data represent enrichment at the indicated promoter as a percentage of input ± SD (n = 2). *P < 0.05 and **P < 0.01. Pearson correlation analyzed by two-sided Student’s t test. Significance in IHC was calculated using two-sided Fisher’s exact test.
To study the mechanism by which BCL-3 modulates NF-κB, we first examined an NF-κB–dependent luciferase reporter. Overexpression of BCL-3 in GBM cells caused an increase in basal and tumor necrosis factor–α (TNFα)–induced NF-κB activity (Fig. 5D) and an increase in TNFα-induced CD44 expression (fig. S5B), a finding previously reported to promote mesenchymal differentiation (10). We then looked at the primary NF-κB subunits p50, p52, and p65. Although overexpression of BCL-3 did not induce a change in subunit expression, it increased p65 nuclear translocation (Fig. 5E). To examine this finding further, we depleted BCL3 in GBM34 GSCs. Knockdown of BCL3 using two distinct sh-RNA constructs decreased nuclear and increased cytoplasmic p65 (Fig. 5F). Given this finding, to study whether inherent differences in BCL-3 correlated with differences in nuclear p65, we first examined GSCs and GBM cell lines. In the cell lines examined, we found a positive correlation between BCL-3 and nuclear p65 protein (Fig. 5G). In addition, in clinical GBM specimens, we found a significant positive correlation between nuclear IHC staining of BCL-3 and p65 (P = 0.009; Fig. 5H). These results suggest that, in GBM, BCL-3 induces NF-κB activity and promotes p65 nuclear translocation.
Although BCL-3 did not alter the amount of nuclear p50 or p52, we examined whether it promoted MES differentiation by altering the chromatin recruitment of these subunits. We performed quantitative chromatin immunoprecipitation (qChIP) using primers spanning the κB sites of several BCL-3–regulated MES genes. Overexpression of BCL-3 induced recruitment of p65 to the κB sites of MES-specific genes, including CD44 and LIF (Fig. 5I and fig. S5C). Also, whereas p50 recruitment decreased at CD44, LIF, and CCL2 promoters, the recruitment of p52 increased (Fig. 5I and fig. S5C). Gel shift studies using probes corresponding to the CD44 and CCL2 κB sites showed that, in the presence of high BCL-3, p50 DNA binding was lost, whereas p52 binding was induced at both κB sites (Fig. 5J and fig. S5D). Finally, we examined endogenous chromatin enrichment of NF-κB subunits at MES promoters in patient-derived GSCs. The predominant subunits bound to MES promoters in GBM34 GSCs (BCL-3 high) were p65 and p52 (Fig. 5K). These findings indicate that high BCL-3 promotes MES gene expression by inducing promoter-specific NF-κB dimer exchange.
The increased recruitment of p52 to MES promoters suggested that p52 is involved in mediating the MES differentiation induced by BCL-3. We therefore depleted p52/NFKB2 in GBM34 GSCs (BCL-3 high). Knockdown of p52/NFKB2 resulted in decreased expression of CD44, CCL2, and interleukin-8 (IL8) (fig. S5E). Moreover, in TCGA GBMs, there was a positive correlation between the expression of NFKB2 and these MES genes (fig. S5F). In addition, depletion of p52/NFKB2 not only attenuated basal NF-κB–regulated MES gene expression but also blocked the increase induced by BCL-3 overexpression (fig. S5G). These results indicate that p52 mediates BCL-3–dependent increase in MES gene expression.
CAII mediates BCL-3–dependent resistance to TMZ
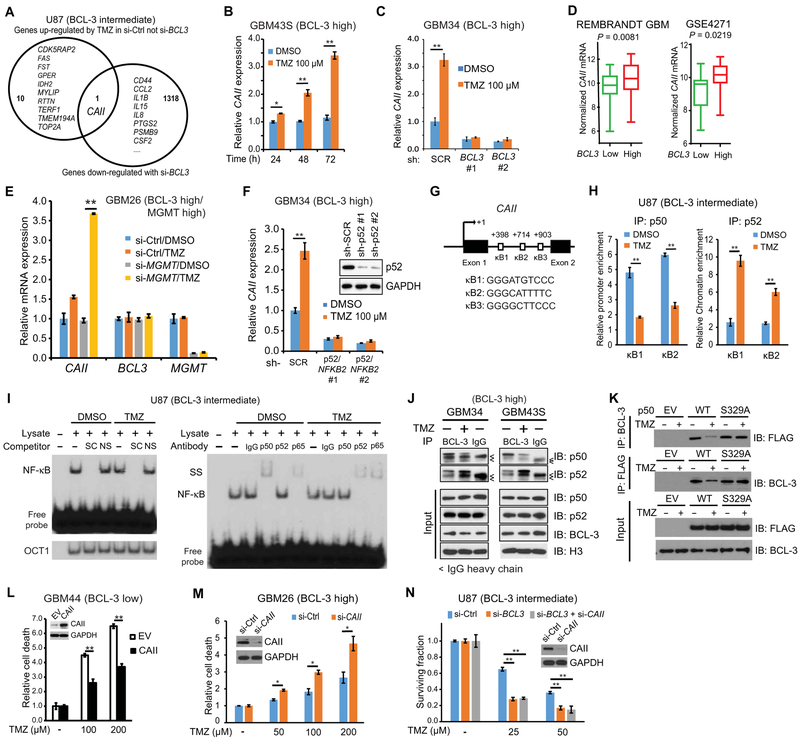
The results indicate that BCL-3 is informative specifically in patients treated with alkylating agents such as TMZ. To identify BCL-3–regulated factors that could be responsible for the resistance to TMZ and that could potentially be targeted for inhibition, we examined the gene expression response induced by TMZ, focusing on genes up-regulated by treatment. To screen for differentially expressed transcripts, we depleted BCL3 in U87 cells, a cell line with moderate BCL-3 expression that was sensitized to TMZ by BCL3 knockdown. The sole transcript induced by TMZ in the presence of BCL-3 and down-regulated with depletion of BCL3 was CAII (Fig. 6A and tables S12 and S13). CAII was induced by TMZ in GSCs (Fig. 6B and fig. S6A), and BCL-3 was required for this induction (Fig. 6C). Also, in clinical databases, tumors with low BCL3 expression had lower CAII mRNA expression than those with high BCL3 expression, as did tumors with 19q deletion compared to the nondeleted group (Fig. 6D and fig. S6, B and C). In addition, MES GBMs had higher CAII expression than PN tumors (fig. S6D). To examine the relevance of alkyl adducts for induction of CAII by TMZ, we modulated MGMT expression. Knockdown of MGMT in GBM26 GSCs (that express high MGMT and BCL-3) increased TMZ-induced CAII expression (Fig. 6E), whereas expression of MGMT in GBM cells with no MGMT blocked the effect of TMZ on CAII expression (fig. S6E). These findings suggest that TMZ induces CAII via formation of cytotoxic O6-methylguanine adducts.
Fig. 6. CAII mediates BCL-3–dependent resistance to TMZ.
(A) Significantly (P < 0.05) altered transcripts in U87 cells expressing si-BCL3 or si-control treated with TMZ (100 μM) or vehicle (24 hours). (B) CAII mRNA expression relative to GAPDH in GBM43S GSCs treated with TMZ (n = 3). (C) CAII mRNA in GBM34 GSCs expressing sh-BCL3 constructs or sh-control treated with vehicle or TMZ (48 hours) (n = 3). (D) CAII mRNA expression in patients from the indicated data set with lower and upper 33% BCL3 expression. REMBRANDT, n = 61 per group; GSE4271, n = 20 per group. (E) Expression of CAII, BCL3, and MGMT mRNA in GBM26 GSCs that express high MGMT (see fig. S7K) transfected with si-MGMT or si-control after treatment with 100 μM TMZ for 48 hours. Data show mean expression relative to GAPDH normalized to si-control, dimethyl sulfoxide (DMSO)–treated, ± SD (n = 2). (F) CAII expression relative to GAPDH in GBM34 GSCs expressing sh-control or two separate sh-p52/NFKB2 constructs after treatment with TMZ (48 hours). Data show mean value relative to GAPDH ± SD (n = 3). Inset: Immunoblot with anti-p52. (G) Location and sequence of putative human CAII κB sites. (H) ChIP qPCR in U87 cells treated with 100 μM TMZ or vehicle (24 hours). Data are normalized to immunoglobulin G (IgG) and represent enrichment as a percentage of input ± SD (n = 2). (I) Representative EMSA with κB2 probe in U87 nuclear lysate treated as in (H). Supershift (SS) (right) and competition with specific (SC) or nonspecific (NS) probe (left). OCT1 binding confirms equal loading. (J) Co-IP in GBM34 and GBM43S GSCs (both BCL-3 high) after treatment with TMZ (100 μM, 24 hours). IP and IB were performed with the indicated antibodies. Arrowheads indicate IgG heavy chain band (n = 2). (K) Reciprocal co-IP in 293T cells expressing FLAG-p50-wt, FLAG-p50-S329A, or EV after treatment with vehicle or TMZ (100 μM, 24 hours) (n = 2). (L and M) Trypan blue assays in GBM44 GSCs expressing HA-CAII or EV (L) and GBM26 GSCs expressing si-CAII or si-control (M) treated with TMZ (72 hours). Data show fold change in percent dead cells relative to vehicle ± SD (n = 3). Insets: Immunoblots with anti-CAII antibody. (N) Clonogenic assay in U87 cells expressing si-control or si-BCL3 with and without si-CAII treated with TMZ (n = 3). *P < 0.05 and **P < 0.01, two-sided Student’s t test.
No CA isoforms have yet been reported to be NF-κB target genes. Although knockdown of either p50/NFκB1 or p65 did not alter basal CAII expression, loss of p52/NFκB2 attenuated basal and TMZ-induced CAII expression in GBM34 GSCs (Fig. 6F and fig. S6F). Analysis of the CAII promoter and the upstream coding region revealed three potential κB sites (κB1, κB2, and κB3; Fig. 6G). ChIP studies demonstrated that p50, p52, and p65 are recruited to κB1 and κB2, but not κB3, and gel shift analysis showed that NF-κB binds the κB2 probe (fig. S6G). In U87 cells, TMZ decreased enrichment of p50 at κB1 and κB2, increased enrichment of p52 (Fig. 6H), and had no effect on either BCL-3 or p65 (fig. S6H). Consistent with this, gel shift studies showed that, in response to TMZ, p52 replaced p50 in the NF-κB dimer bound to the κB2 probe (Fig. 6I). The above findings suggested that TMZ modulates the interaction of BCL-3 with p50 and p52. To examine this more closely, we studied GSCs with high BCL-3 expression. Western blot analysis revealed that TMZ reduced the interaction of BCL-3 with p50 and enhanced the association of BCL-3 with p52 (Fig. 6J). Given that TMZ induces phosphorylation of p50 S329 (11) and that S329 is a residue that potentially interacts with BCL-3 (34), we examined the role of this residue in regulating the BCL-3/p50 interaction in response to TMZ. Mutation of S329 to an unphosphorylatable form blocked the dissociation of p50 and BCL-3 after TMZ treatment (Fig. 6K). Together, these findings highlight the importance of the reciprocal interaction of BCL-3 with p50 and p52 for induction of CAII by TMZ.
Finally, to examine whether CAII is involved in the cytotoxicity induced by TMZ, we altered its expression in GSCs and GBM cells. Overexpression of CAII attenuated cell death induced by TMZ (Fig. 6L and fig. S6, I and J), whereas knockdown of CAII in GBM26 GSCs led to an increase in TMZ-induced cytotoxicity (Fig. 6M). Moreover, depletion of CAII and BCL3 together did not increase the anti-glioma effect of TMZ any more than knockdown of BCL-3 alone (Fig. 6N), supporting the contention that CAII and BCL-3 act in the same pathway in response to TMZ. These findings indicate that BCL-3 promotes resistance to TMZ by induction of CAII as a result of promoter-specific NF-κB dimer exchange.
ACZ enhances the anti-glioma effect of TMZ
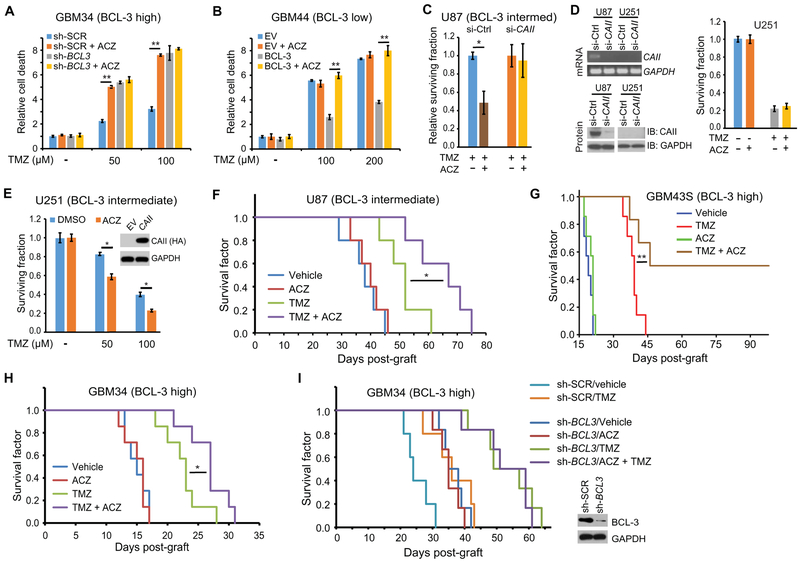
CAII is potently blocked by the CA inhibitor ACZ (35). ACZ has been reported to be effective against GBM cells (36, 37) and to enhance apoptosis by TMZ (38, 39). Consistent with this observation, we found that although ACZ alone did not alter overall clonogenic survival of GBM cells, it enhanced the ability of TMZ to reduce survival (fig. S7A). To study the role of BCL-3 in this response, patient-derived GSCs were examined. ACZ enhanced cytotoxicity by TMZ in GBM34 GSCs that express high BCL-3, and knockdown of BCL3 blocked the chemosensitizing effect of ACZ (Fig. 7A). In GBM44 GSCs that have low BCL-3 expression, ACZ did not modulate the cytotoxic effect induced by TMZ; however, overexpression of BCL-3 enabled ACZ to enhance cell death by TMZ (Fig. 7B). Similarly, depletion of BCL3 attenuated the chemosensitizing effect of ACZ on clonogenic assay (fig. S7B). These results indicate that BCL-3 is required for sensitization to TMZ by ACZ.
Fig. 7. ACZ chemosensitizes GBM xenografts to TMZ.
(A and B) Trypan blue assays at 72 hours in (A) GBM34 GSCs expressing sh-BCL3 or sh-SCR or (B) GBM44 GSCs expressing BCL-3 or EV treated with TMZ and/or 100 μM ACZ as indicated. Data show fold change in percent dead cells relative to cells transfected with EV, or sh-SCR, treated with vehicle, ± SD (n = 3). (C) Clonogenic assay in U87 cells expressing si-CAII or si-control treated with 50 μM TMZ with or without 100 μM ACZ. Data show surviving fraction relative to TMZ alone ± SD (n = 3). (D) PCR analysis of CAII mRNA and immunoblot analysis of CAII protein in U87 and U251 cells expressing si-control or si-CAII. Clonogenic assay (right) in U251 cells treated with 100 μM TMZ and/or 100 μM ACZ (n = 3). (E) Clonogenic assay in U251 cells stably expressing CAII or EV treated with TMZ and/or 100 μM ACZ (n = 3). (F) Kaplan-Meier curves of mice bearing intracranial U87 tumors (n = 5 per group) treated with TMZ on days 5, 7, and 9 (5 mg/kg per dose) and/or ACZ on days 5 to 26 (15 mg/kg per day). (G) Kaplan-Meier curves of mice bearing intracranial GBM43S PDX (n = 7 per group) treated with TMZ on days 5, 7, and 9 (5 mg/kg per dose) and/or ACZ on days 5 to 26 (15 mg/kg per day). (H) Kaplan-Meier curves of mice bearing intracranial GBM34 PDX (n = 7 per group) treated with TMZ on days 5, 7, and 9 (10 mg/kg per dose) and/or ACZ on days 5 to 26 (15 mg/kg per day). (I) Kaplan-Meier curves of mice bearing intracranial GBM34 PDX tumors stably expressing either sh-BCL3 (n = 6 per group) or sh-SCR (n = 5 per group) treated with TMZ on days 5, 7, and 9 (10 mg/kg per dose) and/or ACZ on days 5 to 26 (15 mg/kg per day). Inset: Immunoblot of cells from the indicated tumors. *P < 0.05 and **P < 0.02.
Although ACZ is possibly the most commonly used CA inhibitor (40), many other CA inhibitors are available that can potentially enhance the efficacy of TMZ (41). We examined two other clinically used general CA inhibitors, methazolamide (MZM) and topiramate (TPM). MZM enhanced cytotoxicity by TMZ in a BCL-3–dependent manner (fig. S7C), whereas the anti-epileptic agent TPM did not (fig. S7D).
Given that ACZ also acts on other CAs, albeit much less effectively (35), we also examined the requirement of CAII for the combination effect of ACZ and TMZ. Knockdown of CAII with si-RNA blocked the chemosensitizing effect of ACZ on TMZ (Fig. 7C), and overexpression of CAII in cells with low basal BCL-3 promoted chemosensitization by ACZ (fig. S7E). Moreover, in U251 GBM cells that do not express CAII, ACZ only chemosensitized these cells to TMZ when CAII was exogenously expressed (Fig. 7, D and E). The efficacy of ACZ in combination with TMZ raised the question of whether ACZ modified NF-κB activity and mesenchymal differentiation. ACZ did not affect TMZ-induced NF-κB inhibitory activity (fig. S7F), and the addition of ACZ also did not induce p65 phosphorylation or alter MES gene expression (fig. S7, G and H).
To examine combination TMZ and ACZ in vivo, we performed intracranial xenograft studies in mice. In a pilot study, U87 tumors were established and ACZ was administered for 10 days after initiation of TMZ treatment. This combination regimen did not result in a significant change in survival. However, 10 days after TMZ initiation, CAII expression was still elevated (fig. S7I), suggesting that ACZ needed to be administered for a longer period of time. We therefore altered our protocol to treat animals with daily ACZ for a total of 21 days. Using this regimen, ACZ significantly increased survival time in combination with TMZ compared to TMZ alone (P < 0.05; Fig. 7F and fig. S7J). ACZ alone did not affect survival in this model (Fig. 7F).
Next, we replicated the experiments in more clinically relevant patient-derived xenograft (PDX) models. In GBM34 and GBM43S xenografts that have high BCL-3 expression, addition of ACZ significantly increased survival compared to TMZ alone (P < 0.05; Fig. 7G and H). Addition of ACZ to TMZ in GBM43S xenografts resulted in long-term survival of several animals, a finding repeatedly seen in multiple independent experiments (Fig. 7G and fig. S7L). However, in GBM26 PDX that has high MGMT expression, addition of ACZ had no chemosensitizing effect on TMZ despite the presence of BCL-3 in this tumor (fig. S7, K and M). Finally, to specifically examine the requirement of BCL-3 for chemosensitization by ACZ in vivo, we depleted BCL3 in GBM34 tumors. Whereas expression of a control sh-RNA did not modulate the response in GBM34 tumors, knockdown of BCL3 completely blocked the ability of ACZ to increase the prosurvival effect of TMZ (Fig. 7I). Together, these data indicate that ACZ, a commonly used CA inhibitor that has no antitumor effect in GBM by itself, sensitized GBM to TMZ in a BCL-3–dependent manner.
DISCUSSION
This work identifies BCL-3 as an indicator of sensitivity to alkylating chemotherapy in GBM. Although BCL3 expression separated all GBM patients, it is primarily informative in tumors with high MGMT promoter methylation. GBM patients with high MGMT promoter methylation, who would be expected to respond well to alkylating agents, had similar survival to those with low MGMT promoter methylation if they also had high BCL3 expression. This finding is likely because cytotoxic O6-methylguanine adducts are required for BCL-3 to promote resistance to TMZ. In the presence of high MGMT expression, cytotoxic adducts are repaired before they signal to BCL-3 (fig. S8). BCL-3 was previously found to be elevated in gliomas and related to survival (42); however, in that study, patients with all grades of glioma were analyzed together, precluding determination of prognostic value. Our results indicate that only tumors with low BCL3 expression will likely benefit from adding TMZ to IR, an important clinical observation given that TMZ induces deleterious hypermutation that can cause malignant progression (43).
The data demonstrate that, in glioma, BCL3 expression is regulated by genetic, and epigenetic, modifications and that these alterations are linked to patient outcome. We see that BCL3 CN loss occurs as a result of modifications targeted to the chromosomal band 19q13. This finding, when considered with the observation that BCL-3 is a candidate oncoprotein that has never been identified as a glioma driver (4), indicates that BCL3 loss is a passenger event unrelated to glioma formation. Although the ability of passenger events to promote unintended therapeutic susceptibility has been shown in animal models (44, 45), the link between loss of BCL3 and TMZ susceptibility demonstrates the importance of passenger modification to chemosensitivity in a clinical setting. The effect of BCL3 deletion in glioma is particularly relevant given that alterations of 19q play an important role in modulating patient outcome in these tumors. Specifically, 19q, with 1p, co-deletion is predictive of response to alkylating chemotherapy in oligodendroglioma (46), whereas loss of 19q13 alone is associated with long-term survival in GBM (29, 47). The most widely accepted hypothesis as to why 19q13 loss is so prevalent in gliomas is that a glioma-specific tumor suppressor is present in the region (48). Although no 19q tumor suppressor has yet been identified in astrocytic tumors or GBM, capicua transcriptional repressor (CIC) was recently discovered in oligodendroglioma (49).
Our studies indicate that BCL-3 promotes GBM mesenchymal differentiation, an observation consistent with previous reports linking BCL-3 to EMT (50-52). We find that, in GBM cells, BCL-3 up-regulated EMT markers and that, in clinical GBM, BCL3 expression correlated strongly with MES signature gene expression. From a mechanistic standpoint, high BCL-3 led to a change in the composition of the NF-κB dimer at MES gene promoters involving replacement of p50 by p52. In addition, BCL-3 induced p65 nuclear translocation, a finding consistent with the known importance of this subunit in MES change (10). Whereas previous work demonstrates a cell-extrinsic pathway for activation of p65 in GBM cells by cytokines released from infiltrating macrophages and microglia (10), genetic and epigenetic regulation of BCL3 represents mechanisms by which cell-intrinsic pathways also contribute to promoting NF-κB–dependent mesenchymal differentiation.
An important feature of predictors like BCL-3 is that they are also informative in that they can identify pathways to improve treatment response. MGMT promoter methylation is one such predictor; however, inhibiting MGMT has not proven to be an effective chemosensitizing strategy clinically (8). Inhibition of BCL-3 is not currently feasible in patients; therefore, we searched for downstream BCL-3–regulated targets and identified CAII as a mediator of BCL-3–dependent resistance to TMZ. Although several factors have previously been reported to contribute to the antiapoptotic effects of BCL-3 (53-55), from a clinical perspective, CAII stands out because it can be effectively inhibited by ACZ. CAII mRNA expression is not prognostic in untreated GBM (GlioVis data portal) (32), yet elevated endothelial CAII has been associated with worse outcome in astrocytoma (56). Regardless of these observations, our data demonstrate that it is not basal expression but the induction of CAII by TMZ that is important in modulating response to therapy.
Although we demonstrated the predictive value of BCL3 in several independent data sets, an important limitation of our study is its retrospective nature. Ultimately, the clinical validity of using BCL-3 as a predictor in GBM will require verification in a prospective randomized trial. In addition, the question of which CA inhibitor is best for clinical use with TMZ will require further analysis. Finally, a mechanistic limitation of our study is that we have yet to determine how BCL-3 promotes p65 nuclear translocation and promoter-specific dimer exchange.
ACZ has not previously been examined in GBM xenografts; however, it was initially shown to promote chemosensitization in murine fibrosarcoma (57). More recently, ACZ was found to promote cytotoxicity of GBM cells in vitro and other cancers in vivo (37-39, 58). Although there has been significant interest in targeting other hypoxia-specific CA isoforms for GBM therapy (41, 59), ACZ is attractive because of its general clinical use and well-tolerated dosing profile (40). Using PDXs, we see that, in GBM, ACZ promoted chemosensitization specifically in the presence of high BCL-3 and low MGMT expression. Our studies suggest that, in the clinical setting, repurposing ACZ might be particularly effective in a subgroup of MGMT promoter methylated tumors that have high BCL-3 expression. Given the increase in molecular analysis of gliomas, factors such as BCL-3 might assume an important role in individualizing patient treatment, a strategy occurring with increasing frequency in cancer therapy.
MATERIALS AND METHODS
Study design
The objectives of this study were to examine the role of BCL-3 in the response of glioma to alkylating chemotherapy and to evaluate the use of ACZ as a chemosensitizer in experimental GBM. There are several design aspects relevant in this work. First, to examine the predictive role of BCL3, initially, TCGA was examined and the results were subsequently validated in other databases. Numbers of patients included or excluded in each data set are specifically noted in Materials and Methods and in each individual figure panel. To investigate survival based on BCL-3 IHC, we performed an initial power analysis based on the mRNA data from TCGA. This gave n = 74 and 32 patients per group, respectively, to have 80% power to detect a significant difference between low and high BCL-3 expression at a two-sided P < 0.05. Subsequently, after institutional review board approval, we obtained 86 consecutive GBM samples that had adequate tissue for IHC. Additional patients were not recruited. Patients were excluded from survival analysis if they died before treatment due to infection or massive hemorrhage, if no follow-up was available, or if they initially had an LGG that progressed to GBM. IHC and FISH grading was performed by two independent investigators blinded to diagnosis and to survival.
For animal studies, no statistical method was used to determine sample size. Efforts were made to achieve the scientific goals with the minimum number of animals. A sample size of five to seven animals per group was chosen on the basis of our previous experience using intracranial PDX GBMs, where we have observed 100% tumor engraftment success. After tumor implantation and before treatment, animals were randomized into the different treatment groups. Animals were excluded from the study if they were sacrificed before treatment/randomization. Determination of the survival time was performed blinded to the specific treatment group. Experiments were repeated by more than one individual to ensure reproducibility. The survival endpoint was reached when mice lost at least 20% body weight or showed symptoms of neurological deficit. Raw data are located in table S17.
Statistical analysis
Data analysis was performed using Stata/IC 13.0 statistical software (Stata Corporation, licensed to the University of Chicago, Research Computing Center). The Cox proportional hazard model was used for both univariate and multivariate analyses using the specific covariates noted in Results. For survival studies, Kaplan-Meier curves were plotted, and the log-rank test was performed for comparison of cohorts. HR and 95% CIs were calculated using the Mantel-Haenszel estimator model. For analysis of 19q CN correlation, correlation matrix analysis was used, and the R/corrplot package was used to display the correlation matrix. For box-and-whisker graphs, boxes show median and 25th and 75th percentiles, whereas whiskers show the 5th and 95th percentiles analyzed by unpaired t test. In vitro and other studies as indicated were analyzed by two-tailed Student’s t test with significance taken as P < 0.05. Pearson correlation was also analyzed by two-tailed Student’s t test.
Supplementary Material
Fig. S1. BCL3 predicts response to alkylating chemotherapy in GBM.
Fig. S2. BCL3 is relevant in LGGs and pan-glioma patients.
Fig. S3. Loss of BCL3 is a passenger event associated with survival.
Fig. S4. BCL-3 induces mesenchymal differentiation.
Fig. S5. BCL-3 promotes p65 nuclear translocation and NF-κB dimer exchange.
Fig. S6. CAII mediates BCL-3–dependent resistance to TMZ.
Fig. S7. ACZ chemosensitizes GBM xenografts to TMZ.
Fig. S8. Model demonstrating role of BCL-3 in promoting resistance to TMZ.
Table S1. Molecular profile of GBM cell lines.
Table S2. Cox regression analysis using different cutoff points for BCL3 expression in TCGA GBM patients.
Table S3. Multivariate Cox regression analysis in TCGA GBM patients treated with and without alkylating chemotherapy (TMZ).
Table S4. Clinical features of University of Chicago GBM patients.
Table S5. Cox regression analysis using different BCL3 expression cutoff points in TCGA LGG patients.
Table S6. Cox regression analysis in TCGA LGG patients with and without alkylating chemotherapy.
Table S7. Multivariate Cox regression analysis in patients from EORTC 26951 treated either with or without alkylating chemotherapy.
Table S8. Cox regression analysis of TCGA pan-glioma patients who received alkylating chemotherapy.
Table S9. Multivariate Cox regression of TCGA pan-glioma patients incorporating BCL3 CN.
Table S10. Multivariate Cox regression of TCGA pan-glioma patients incorporating BCL3 promoter methylation.
Table S11. Correlation between BCL3 mRNA expression and the expression of each MES or PN signature gene in TCGA GBM patients.
Table S14. Oligonucleotides (oligos) for EMSA.
Table S15. ChIP primers and target gene κB sites.
Table S16. Oligonucleotide sequences for qPCR.
Table S12. Genes that are up- and down-regulated (at least 1.5-fold) in U87 cells expressing si-BCL3 relative to those expressing si-control (see separate Excel file).
Table S13. Genes that are significantly (adjusted P < 0.05) up- and down-regulated in U87 cells expressing si-BCL3 or si-control after treatment with TMZ compared to vehicle (see separate Excel file).
Table S17. Raw data (see separate Excel file).
Acknowledgments:
We thank the University of Chicago Research Computing Center for support, the University of Chicago and Jonathon Hobbs for assistance with making and staining TMAs, L. Zhang for biostatistical assistance, and A. Uppal.
Funding: This work was supported by NIH grant R01CA136937 to B.Y. and the Ludwig Center for Metastasis Research; K.E.C. was a Howard Hughes Medical Institute Research Fellow.
Footnotes
SUPPLEMENTARY MATERIALS
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Expression data from microarray studies have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession GSE80729) and are openly available. In addition, data from the publicly available data sets are freely available at the locations cited for each specific database.
REFERENCES AND NOTES
- 1.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K, Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN; Cancer Genome Atlas Research Network, Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group, Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med 352, 987–996 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu C-J, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L; TCGA Research Network, The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masayesva BG, Ha P, Garrett-Mayer E, Pilkington T, Mao R, Pevsner J, Speed T, Benoit N, Moon C-S, Sidransky D, Westra WH, Califano J, Gene expression alterations over large chromosomal regions in cancers include multiple genes unrelated to malignant progression. Proc. Natl. Acad. Sci. U.S.A 101, 8715–8720 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frei III E, Gene deletion: A new target for cancer chemotherapy. Lancet 342, 662–664 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Tanaka S, Louis DN, Curry WT, Batchelor TT, Dietrich J, Diagnostic and therapeutic avenues for glioblastoma: No longer a dead end? Nat. Rev. Clin. Oncol 10, 14–26 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR, Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J. Clin. Oncol 26, 4189–4199 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou M-F, de Tribolet N, Regli L, Wick W, Kouwenhoven MCM, Hainfellner JA, Heppner FL, Dietrich P-Y, Zimmer Y, Cairncross JG, Janzer R-C, Domany E, Delorenzi M, Stupp R, Hegi ME, Stem cell–related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J. Clin. Oncol 26, 3015–3024 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, Conroy S, Long L, Lelic N, Wang S, Gumin J, Raj D, Kodama Y, Raghunathan A, Olar A, Joshi K, Pelloski CE, Heimberger A, Kim SH, Cahill DP, Rao G, Den Dunnen WFA, Boddeke HWGM, Phillips HS, Nakano I, Lang FF, Colman H, Sulman EP, Aldape K, Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell 24, 331–346 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt AM, Crawley CD, Kang S, Raleigh DR, Yu X, Wahlstrom JS, Voce DJ, Darga TE, Weichselbaum RR, Yamini B, p50 (NF-κB1) is an effector protein in the cytotoxic response to DNA methylation damage. Mol. Cell 44, 785–796 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cahill KE, Morshed RA, Yamini B, Nuclear factor-κB in glioblastoma: Insights into regulators and targeted therapy. Neuro Oncol. 18, 329–339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer S, Chen YH, Bcl-3, a multifaceted modulator of NF-κB-mediated gene transcription. Immunol. Res 42, 210–218 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS Jr., Selective activation of NF-kappa B subunits in human breast cancer: Potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene 19, 1123–1131 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Thornburg NJ, Pathmanathan R, Raab-Traub N, Activation of nuclear factor-κB p50 homodimer/Bcl-3 complexes in nasopharyngeal carcinoma. Cancer Res. 63, 8293–8301 (2003). [PubMed] [Google Scholar]
- 16.Fujita T, Nolan GP, Liou HC, Scott ML, Baltimore D, The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-kappa B p50 homodimers. Genes Dev. 7, 1354–1363 (1993). [DOI] [PubMed] [Google Scholar]
- 17.Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K, Siebenlist U, The oncoprotein Bcl-3 directly transactivates through kappa B motifs via association with DNA-binding p50B homodimers. Cell 72, 729–739 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Wang VY-F, Li Y, Kim D, Zhong X, Du Q, Ghassemian M, Ghosh G, Bcl3 phosphorylation by Akt, Erk2, and IKK is required for its transcriptional activity. Mol. Cell 67, 484–497.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, Yu ILY, Carro MS, Dai F, Tagge MJ, Ferrarese R, Bredel C, Phillips HS, Lukac PJ, Robe PA, Weyerbrock A Vogel H, Dubner S, Mobley B, He X, Scheck AC, Sikic BI, Aldape KD, Chakravarti A, Harsh GR IV, NFKBIA deletion in glioblastomas. N. Engl. J. Med 364, 627–637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madhavan S, Zenklusen J-C, Kotliarov Y, Sahni H, Fine HA, Buetow K, Rembrandt: Helping personalized medicine become a reality through integrative translational research. Mol. Cancer Res 7, 157–167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bady P, Sciuscio D, Diserens A-C, Bloch J, van den Bent MJ, Marosi C, Dietrich P-Y, Weller M, Mariani L, Heppner FL, McDonald DR, Lacombe D, Stupp R, Delorenzi M, Hegi ME, MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 124, 547–560 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erdem-Eraslan L, Gravendeel LA, de Rooi J, Eilers PHC, Idbaih A, Spliet WGM, den Dunnen WFA, Teepen JL, Wesseling P, Sillevis Smitt PAE, Kros JM, Gorlia T, van den Bent MJ, French PJ, Intrinsic molecular subtypes of glioma are prognostic and predict benefit from adjuvant procarbazine, lomustine, and vincristine chemotherapy in combination with other prognostic factors in anaplastic oligodendroglial brain tumors: A report from EORTC study 26951. J. Clin. Oncol 31, 328–336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW, The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 131, 803–820 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, Anjum S, Wang J, Manyam G, Zoppoli P, Ling S, Rao AA, Grifford M, Cherniack AD, Zhang H, Poisson L, Carlotti CG Jr., da Cunha DP, Rao A, Mikkelsen T, Lau CC, Yung WKA, Rabadan R, Huse J, Brat DJ, Lehman NL, Barnholtz-Sloan JS, Zheng S, Hess K, Rao G, Meyerson M, Beroukhim R, Cooper L, Akbani R, Wrensch M, Haussler D, Aldape KD, Laird PW, Gutmann DH; TCGA Research Network, Noushmehr H, Iavarone A, Verhaak RGW, Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164, 550–563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Deimling A, Bender B, Jahnke R, Waha A, Kraus J, Albrecht S, Wellenreuther R, Faßbender F, Nagel J, Menon AG, Louis DN, Lenartz D, Schramm J, Wiestler OD, Loci associated with malignant progression in astrocytomas: A candidate on chromosome 19q. Cancer Res. 54, 1397–1401 (1994). [PubMed] [Google Scholar]
- 26.Smith JS, Alderete B, Minn Y, Borell TJ, Perry A, Mohapatra G, Hosek SM, Kimmel D, O’Fallon J, Yates A, Feuerstein BG, Burger PC, Scheithauer BW, Jenkins RB, Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene 18, 4144–4152 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Nakamura M, Yang F, Fujisawa H, Yonekawa Y, Kleihues P, Ohgaki H, Loss of heterozygosity on chromosome 19 in secondary glioblastomas. J. Neuropathol. Exp. Neurol 59, 539–543 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J, Westphal M, Schackert G, Simon M, Tonn JC, Heese O, Krex D, Nikkhah G, Pietsch T, Wiestler O, Reifenberger G, von Deimling A, Loeffler M, Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: A prospective translational study of the German Glioma Network. J. Clin. Oncol 27, 5743–5750 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Burton EC, Lamborn KR, Feuerstein BG, Prados M, Scott J, Forsyth P, Passe S, Jenkins RB, Aldape KD, Genetic aberrations defined by comparative genomic hybridization distinguish long-term from typical survivors of glioblastoma. Cancer Res. 62, 6205–6210 (2002). [PubMed] [Google Scholar]
- 30.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR, Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc. Natl. Acad. Sci. U.S.A 104, 20007–20012 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee DW, Ramakrishnan D, Valenta J, Parney IF, Bayless KJ, Sitcheran R, The NF-κB RelB protein is an oncogenic driver of mesenchymal glioma. PLOS ONE 8, e57489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowman RL, Wang Q, Carro A, Verhaak RGW, Squatrito M, GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 19, 139–141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mani SA, Guo W, Liao M-J, Eaton E. Ng., Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang C, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA, The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manavalan B, Basith S, Choi Y-M, Lee G, Choi S, Structure-function relationship of cytoplasmic and nuclear IκB proteins: An in silico analysis. PLOS ONE 5, e15782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masereel B, Rolin S, Abbate F, Scozzafava A, Supuran CT, Carbonic anhydrase inhibitors: Anticonvulsant sulfonamides incorporating valproyl and other lipophilic moieties. J. Med. Chem 45, 312–320 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Said HM, Supuran CT, Hageman C, Staab A, Polat B, Katzer A, Scozzafava A, Anacker J, Flentje M, Vordermark D, Modulation of carbonic anhydrase 9 (CA9) in human brain cancer. Curr. Pharm. Des 16, 3288–3299 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Said HM, Hagemann C, Carta F, Katzer A, Polat B, Staab A, Scozzafava A, Anacker J, Vince GH, Flentje M, Supuran CT, Hypoxia induced CA9 inhibitory targeting by two different sulfonamide derivatives including acetazolamide in human glioblastoma. Bioorg. Med. Chem 21, 3949–3957 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Das A, Banik NL, Ray SK, Modulatory effects of acetazolomide and dexamethasone on temozolomide-mediated apoptosis in human glioblastoma T98G and U87MG cells. Cancer Invest. 26, 352–358 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Amiri A, Uyen Le P, Moquin A, Machkalyan G, Petrecca K, Gillard JW, Yoganathan N, Maysinger D, Inhibition of carbonic anhydrase IX in glioblastoma multiforme. Eur. J. Pharm. Biopharm 109, 81–92 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Supuran CT, Acetazolamide for the treatment of idiopathic intracranial hypertension. Expert Rev. Neurother 15, 851–856 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Boyd NH, Walker K, Fried J, Hackney JR, McDonald PC, Benavides GA, Spina R, Audia A, Scott SE, Libby CJ, Tran AN, Bevensee MO, Griguer C, Nozell S, Gillespie GY, Nabors B, Bhat KP, Bar EE, Darley-Usmar V, Xu B, Gordon E, Cooper SJ, Dedhar S, Hjelmeland AB, Addition of carbonic anhydrase 9 inhibitor SLC-0111 to temozolomide treatment delays glioblastoma growth in vivo. JCI Insight 2, e92928 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J, Li L, Jiang G, Zhan H, Wang N, B-cell CLL/lymphoma 3 promotes glioma cell proliferation and inhibits apoptosis through the oncogenic STAT3 pathway. Int. J. Oncol 49, 2471–2479 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, Asthana S, Jalbert LE, Nelson SJ, Bollen AW, Gustafson WC, Charron E, Weiss WA, Smirnov IV, Song JS, Olshen AB, Cha S, Zhao Y, Moore RA, Mungall AJ, Jones SJ, Hirst M, Marra MA, Saito N, Aburatani H, Mukasa A, Berger MS, Chang SM, Taylor BS, Costello JF, Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343, 189–193 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller FL, Colla S, Aquilanti E, Manzo VE, Genovese G, Lee J, Eisenson D, Narurkar R, Deng P, Nezi L, Lee MA, Hu B, Hu J, Sahin E, Ong D, Fletcher-Sananikone E, Ho D, Kwong L, Brennan C, Wang YA, Chin L, DePinho RA, Passenger deletions generate therapeutic vulnerabilities in cancer. Nature 488, 337–342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nijhawan D, Zack TI, Ren Y, Strickland MR, Lamothe R, Schumacher SE, Tsherniak A, Besche HC, Rosenbluh J, Shehata S, Cowley GS, Weir BA, Goldberg AL, Mesirov JP, Root DE, Bhatia SN, Beroukhim R, Hahn WC, Cancer vulnerabilities unveiled by genomic loss. Cell 150, 842–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, Ramsay DA, Louis DN, Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J. Natl. Cancer Inst 90, 1473–1479 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Brat DJ, Seiferheld WF, Perry A, Hammond EH, Murray KJ, Schulsinger AR, Mehta MP, Curran WJ; Radiation Therapy Oncology Group, Analysis of 1p, 19q, 9p, and 10q as prognostic markers for high-grade astrocytomas using fluorescence in situ hybridization on tissue microarrays from Radiation Therapy Oncology Group trials. Neuro Oncol. 6, 96–103 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith JS, Tachibana I, Pohl U, Lee HK, Thanarajasingam U, Portier BP, Ueki K, Ramaswamy S, Billings SJ, Mohrenweiser HW, Louis DN, Jenkins RB, A transcript map of the chromosome 19q-arm glioma tumor suppressor region. Genomics 64, 44–50 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Bettegowda C, Agrawal N, Jiao Y, Sausen M, Wood LD, Hruban RH, Rodriguez FJ, Cahill DP, McLendon R, Riggins G, Velculescu VE, Oba-Shinjo SM, Marie SKN, Vogelstein B, Bigner D, Yan H, Papadopoulos N, Kinzler KW, Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 333, 1453–1455 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shostak K, Zhang X, Hubert P, Göktuna SI, Jiang Z, Klevernic I, Hildebrand J, Roncarati P, Hennuy B, Ladang A, Somja J, Gothot A, Close P, Delvenne P, Chariot A, NF-κB-induced KIAA1199 promotes survival through EGFR signalling. Nat. Commun 5, 5232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Massoumi R, Kuphal S, Hellerbrand C, Haas B, Wild P, Spruss T, Pfeifer A, Fässler R, Bosserhoff AK, Down-regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J. Exp. Med 206, 221–232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wakefield AM, Soukupova J, Montagne A, Ranger JJ, French R, Muller WJ, Clarkson RWE, Bcl3 selectively promotes metastasis of ERBB2-driven mammary tumors. Cancer Res. 15, 745–755 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Viatour P, Bentires-Alj M, Chariot A, Deregowski V, de Leval L, Merville M-P, Bours V, NF-κB2/p100 induces Bcl-2 expression. Leukemia 17, 1349–1356 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Kashatus D, Cogswell P, Baldwin AS, Expression of the Bcl-3 proto-oncogene suppresses p53 activation. Genes Dev. 20, 225–235 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mansour NM, Bernal GM, Wu L, Crawley CD, Cahill KE, Voce DJ, Balyasnikova IV, Zhang W, Spretz R, Nunez L, Larsen GF, Weichselbaum RR, Yamini B, Decoy receptor DcR1 is induced in a p50/Bcl3–dependent manner and attenuates the efficacy of temozolomide. Cancer Res. 75, 2039–2048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haapasalo J, Nordfors K, Järvelä S, Bragge H, Rantala I, Parkkila A-K, Haapasalo H, Parkkila S, Carbonic anhydrase II in the endothelium of glial tumors: A potential target for therapy. Neuro Oncol. 9, 308–313 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teicher BA, Liu SD, Liu JT, Holden SA, Herman TS, A carbonic anhydrase inhibitor as a potential modulator of cancer therapies. Anticancer Res. 13, 1549–1556 (1993). [PubMed] [Google Scholar]
- 58.Bayat Mokhtari R, Baluch N, Ka Hon Tsui M, Kumar S, Homayouni TS, Aitken K, Das B, Baruchel S, Yeger H, Acetazolamide potentiates the anti-tumor potential of HDACi, MS-275, in neuroblastoma. BMC Cancer 17, 156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neri D, Supuran CT, Interfering with pH regulation in tumours as a therapeutic strategy. Nat. Rev. Drug Discov 10, 767–777 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Yamini B, Yu X, Dolan ME, Wu MH, Darga TE, Kufe DW, Weichselbaum RR, Inhibition of nuclear factor-κB activity by temozolomide involves O6-methylguanine–induced inhibition of p65 DNA binding. Cancer Res. 67, 6889–6898 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Tsen AR, Long PM, Driscoll HE, Davies MT, Teasdale BA, Penar PL, Pendlebury WW, Spees JL, Lawler SE, Viapiano MS, Jaworski DM, Triacetin-based acetate supplementation as a chemotherapeutic adjuvant therapy in glioma. Int. J. Cancer 134, 1300–1310 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nandhu MS, Hu B, Cole SE, Erdreich-Epstein A, Rodriguez-Gil DJ, Viapiano MS, Novel paracrine modulation of Notch–DLL4 signaling by fibulin-3 promotes angiogenesis in high-grade gliomas. Cancer Res. 74, 5435–5448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA, Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9, 391–403 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Lan X, Jörg DJ, Cavalli FMG, Richards LM, Nguyen LV, Vanner RJ, Guilhamon P, Lee L, Kushida MM, Pellacani D, Park NI, Coutinho FJ, Whetstone H, Selvadurai HJ, Che C, Luu B, Carles A, Moksa M, Rastegar N, Head R, Dolma S, Prinos P, Cusimano MD, Das S, Bernstein M, Arrowsmith CH, Mungall AJ, Moore RA, Ma Y, Gallo M, Lupien M, Pugh TJ, Taylor MD, Hirst M, Eaves CJ, Simons BD, Dirks PB, Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature 549, 227–232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu Y, Smyth GK, ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Gravendeel LAM, Kouwenhoven MCM, Gevaert O, de Rooi JJ, Stubbs AP, Duijm JE, Daemen A, Bleeker FE, Bralten LBC, Kloosterhof NK, De Moor B, Eilers PHC, van der Spek PJ, Kros JM, Sillevis Smitt PAE, van den Bent MJ, French PJ, Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 69, 9065–9072 (2009). [DOI] [PubMed] [Google Scholar]
- 67.Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, Mischel PS, Nelson SF, Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 64, 6503–6510 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Joy A, Ramesh A, Smirnov I, Reiser M, Misra A, Shapiro WR, Mills GB, Kim S, Feuerstein AG, AKT pathway genes define 5 prognostic subgroups in glioblastoma. PLOS ONE 9, e100827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee Y, Scheck AC, Cloughesy TF, Lai A, Dong J, Farooqi HK, Liau LM, Horvath S, Mischel PS, Nelson SF, Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med. Genomics 1 52 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rich JN, Hans C, Jones B, Iversen ES, McLendon RE, Ahmed Rasheed BK, Dobra A, Dressman HK, Bigner DD, Nevins JR, West M, Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 65, 4051–4058 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Petalidis LP, Oulas A, Backlund M, Wayland MT, Liu L, Plant K, Happerfield L, Freeman TC, Poirazi P, Collins VP, Improved grading and survival prediction of human astrocytic brain tumors by artificial neural network analysis of gene expression microarray data. Mol. Cancer Ther 7, 1013–1024 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AWM, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LGT, Huse JT, Mellinghoff IK, Chan TA, IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483, 479–483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. BCL3 predicts response to alkylating chemotherapy in GBM.
Fig. S2. BCL3 is relevant in LGGs and pan-glioma patients.
Fig. S3. Loss of BCL3 is a passenger event associated with survival.
Fig. S4. BCL-3 induces mesenchymal differentiation.
Fig. S5. BCL-3 promotes p65 nuclear translocation and NF-κB dimer exchange.
Fig. S6. CAII mediates BCL-3–dependent resistance to TMZ.
Fig. S7. ACZ chemosensitizes GBM xenografts to TMZ.
Fig. S8. Model demonstrating role of BCL-3 in promoting resistance to TMZ.
Table S1. Molecular profile of GBM cell lines.
Table S2. Cox regression analysis using different cutoff points for BCL3 expression in TCGA GBM patients.
Table S3. Multivariate Cox regression analysis in TCGA GBM patients treated with and without alkylating chemotherapy (TMZ).
Table S4. Clinical features of University of Chicago GBM patients.
Table S5. Cox regression analysis using different BCL3 expression cutoff points in TCGA LGG patients.
Table S6. Cox regression analysis in TCGA LGG patients with and without alkylating chemotherapy.
Table S7. Multivariate Cox regression analysis in patients from EORTC 26951 treated either with or without alkylating chemotherapy.
Table S8. Cox regression analysis of TCGA pan-glioma patients who received alkylating chemotherapy.
Table S9. Multivariate Cox regression of TCGA pan-glioma patients incorporating BCL3 CN.
Table S10. Multivariate Cox regression of TCGA pan-glioma patients incorporating BCL3 promoter methylation.
Table S11. Correlation between BCL3 mRNA expression and the expression of each MES or PN signature gene in TCGA GBM patients.
Table S14. Oligonucleotides (oligos) for EMSA.
Table S15. ChIP primers and target gene κB sites.
Table S16. Oligonucleotide sequences for qPCR.
Table S12. Genes that are up- and down-regulated (at least 1.5-fold) in U87 cells expressing si-BCL3 relative to those expressing si-control (see separate Excel file).
Table S13. Genes that are significantly (adjusted P < 0.05) up- and down-regulated in U87 cells expressing si-BCL3 or si-control after treatment with TMZ compared to vehicle (see separate Excel file).
Table S17. Raw data (see separate Excel file).