Abstract
Background
Sepsis remains a complex medical problem and a major challenge in healthcare. Diagnostics and outcome predictions are focused on physiological parameters with less consideration given to patients’ medical background. Given the aging population, not only are diseases becoming increasingly prevalent but occur more frequently in combinations (“multimorbidity”). We hypothesized the existence of patient subgroups in critical care with distinct multimorbidity states. We further hypothesize that certain multimorbidity states associate with higher rates of organ failure, sepsis, and mortality co-occurring with these clinical problems.
Methods
We analyzed 36,390 patients from the open source Medical Information Mart for Intensive Care III (MIMIC III) dataset. Morbidities were defined based on Elixhauser categories, a well-established scheme distinguishing 30 classes of chronic diseases. We used latent class analysis to identify distinct patient subgroups based on demographics, admission type, and morbidity compositions and compared the prevalence of organ dysfunction, sepsis, and inpatient mortality for each subgroup.
Results
We identified six clinically distinct multimorbidity subgroups labeled based on their dominant Elixhauser disease classes. The “cardiopulmonary” and “cardiac” subgroups consisted of older patients with a high prevalence of cardiopulmonary conditions and constituted 6.1% and 26.4% of study cohort respectively. The “young” subgroup included 23.5% of the cohort composed of young and healthy patients. The “hepatic/addiction” subgroup, constituting 9.8% of the cohort, consisted of middle-aged patients (mean age of 52.25, 95% CI 51.85–52.65) with the high rates of depression (20.1%), alcohol abuse (47.75%), drug abuse (18.2%), and liver failure (67%). The “complicated diabetics” and “uncomplicated diabetics” subgroups constituted 9.4% and 24.8% of the study cohort respectively. The complicated diabetics subgroup demonstrated higher rates of end-organ complications (88.3% prevalence of renal failure). Rates of organ dysfunction and sepsis ranged 19.6–69% and 12.5–46.7% respectively in the six subgroups. Mortality co-occurring with organ dysfunction and sepsis ranges was 8.4–23.8% and 11.7–27.4% respectively. These adverse outcomes were most prevalent in the hepatic/addiction subgroup.
Conclusion
We identify distinct multimorbidity states that associate with relatively higher prevalence of organ dysfunction, sepsis, and co-occurring mortality. The findings promote the incorporation of multimorbidity in healthcare models and the shift away from the current single-disease paradigm in clinical practice, training, and trial design.
Electronic supplementary material
The online version of this article (10.1186/s13054-019-2486-6) contains supplementary material, which is available to authorized users.
Keywords: Sepsis, Multimorbidity, Data analytics, Machine learning, Latent class analysis
Background
Sepsis remains one of the most serious medical conditions with high mortality and poor prognosis. It is responsible for more than half of in-hospital deaths and is the most costly disease in healthcare constituting $20.3 billion or 5.2% of all hospitalization expenses [1]. Generally, risk assessment scores of mortality in critical care, such as the Simplified Acute Physiology Score [2] (SAPS II), Sepsis-related Organ Failure Score (SOFA) [3], or the Oxford Acute Severity Illness Score [4] (OASIS), focus on inpatient physiological data within 24 h of admission. Only SAPS II [4] and APACHE-IV [5] incorporate some pre-existing chronic conditions. Epidemiological studies, however, demonstrate substantial effect from underlying diseases, almost doubling mortality in sepsis [6–8]. Given these epidemiological findings, it is highly relevant to consider pre-existing morbidity states in assessing critical care mortality risk.
An increasingly aging population has led to a rise in chronic medical conditions. Multimorbidity, the state of suffering from more than one illness, has a prevalence between 13 and 54% depending on the surveyed population [9–11] and is associated with increased healthcare use, decreased quality of life, and higher mortality [9]. Nevertheless, the majority of current medical education fails to consider this; treatment protocols and clinical trial designs are based on the premise that patients have one disease [9, 12]. Further, the majority of drug candidates are identified in animal experiments under standardized conditions, homogenous treatment, and control groups. The subsequent clinical trials testing then aim to reproduce the same homogeneity in the recruited patients by excluding morbidity [13, 14]. We believe that the inability to account for the heterogeneity introduced by multimorbidity is at least in part why a large number of clinical trials fail. Previous work to address this challenge by identifying homogeneous clinical profiles led to changes in the management of heterogeneous conditions including sepsis [15], asthma [16], and acute respiratory distress syndrome [17].
A large volume of information is being collected in the critical care environment [18], yielding a broad range of public datasets that incorporate tens of thousands of patients [19]. This lends itself well to advanced analytics such as machine learning [20, 21], which allows detection of complex, clinically relevant patterns. Latent class analysis has become increasingly utilized in the discovery of clinically relevant patient subgroups from heterogenous datasets [15, 16, 22, 23]. This method assumes the existence of several unobserved groups within the data which share clinical properties that are mutually exclusive between groups. To address the outstanding issue of multimorbidity, we hypothesized the existence of clinically homogenous patient subgroups that share morbidity composition in critical care. We further hypothesized that distinct subgroups associate with greater risk of adverse health outcomes, such as organ failure, sepsis, and mortality related to these clinical problems. Awareness of such high-risk groups may help in anticipatory management, prognostics, and trial design. The findings may also help reshape our single-disease model in healthcare.
Methods
Database
We used the third edition of the Medical Information Mart in Critical Care (MIMIC3) database for our analysis. This is a single-center database containing longitudinal data on 38,597 adult patients in critical care with 53,423 distinct admissions. Further details on the database are included in (Additional file 1: Table S1). Our analysis included first-time ICU admissions for patients aged 16 or over. Readmissions to ICU were not included in the analysis.
Definition of morbidities, organ dysfunction, and sepsis
The MIMIC3 database includes more than 15,693 distinct diagnoses, which are categorized by ICD 9 and ICD 10 codes. For a more compact representation of chronic conditions, we summarized diseases using the 30 Elixhauser categories [24] based on an algorithm provided by the authors describing the MIMIC3 database [25]. The Elixhauser categories are well established to reflect chronic diseases, and they have been validated for both ICD 9 and ICD 10 [26]. Organ dysfunction was defined based on administrative criteria [6] and integrated into the sql code [25] as published previously. Criteria for sepsis were defined based on those described earlier by Angus et al. [6]. Briefly, this approach defines sepsis as a combination of organ dysfunction with concomitant bacterial or fungal infection based on ICD 9 codes and has been validated prospectively using physiological and clinical features. Mortality was defined as inpatient mortality.
Clustering and latent class analysis
We performed a preliminary analysis of similarities between diseases based on disease prevalence in the population using k-means clustering. We first computed disease prevalence (proportion of patients affected by the disease) in each age bracket and used Euclidean distance as similarity measure to define cluster similarities.
For the subsequent analysis of detecting subgroups of patients, we used latent class analysis (LCA) with the inputs age, admission type (elective vs non-elective), and morbidity composition (i.e., which of the 30 Elixhauser categories were present). This technique assumes the existence of unobserved (“latent”) subgroups within the study cohort and identifies them by fitting a set of mixture models to the data. In our analysis, we followed the methodological steps of determining and verifying latent subgroups as summarized by Zho et al. [27]. We chose the optimal number of subgroups based on a combination of achieving the lowest Bayesian information criteria (BIC) and Akaike information criteria (AIC) and subgroup size to be no smaller than 5% of the entire study cohort. This approach of balancing model complexity and subgroup size was adopted from previously published guidance [28], including the suggestion to remove subgroups representing significantly smaller portions of the study population. Characteristics of the latent subgroups were compared using the chi-square test for categorical variables and one-way ANOVA for continuous variables. Residual diagnostics were used to verify that the assumptions for ANOVA were not violated; expected values were calculated to verify that the assumptions for the chi-square test were not violated. We also confirmed the preferred choices between subgroups using logistic regression models assessed using the area under the receiver operator curves (AUCs) (Additional file 2: Figure S1, Table S2). Complete methodological details are described in Additional file 2. Supplementary methods.
Network visualization
The complex relationships between morbidities were visualized using network analysis, which demonstrates associations that are otherwise difficult to appreciate. In our initial approach, when analyzing the entire critical care dataset, network nodes represented variables and the co-occurrence of variables was assessed using a relative risk (RR) measure described previously [29]. This essentially represents the risk of co-occurrence for two diseases. Associations over a significance threshold of p < 0.05 were included in the network. The metrics of RR is reflected by edge width, and disease prevalence is depicted by the diameter of the nodes. In the characterization of subgroups, edges were weighted by the number of patients with the disease pair normalized to the total number of patients within the subgroup. This was used in lieu of RR, as RR may underestimate co-occurrences of highly prevalent diseases as suggested by Hidalgo et al. [29].
Results
Heterogeneous morbidity profile in the critically ill
Cohort demographics are summarized in Additional file 1: Table S1. From a population of 36,390 patients, 83.7% of admissions were due to an emergency. There was slightly greater proportion of males (57.8%) than females. Two or more medical conditions were reported for 77.3% the patients, and nearly half of the entire study population were seniors (age 65 or over). The overall prevalence of sepsis was 37.3% (95% CI 36.7–37.9%), the organ dysfunction rate was 37.5% (95% CI 37–38%), and the overall mortality was 10.9% (95% CI 10.6–11.3%). Mortality rate recorded in patients suffering from sepsis and organ dysfunction was 21.2% (95% CI 20.3–22%) and 18.4% (95% CI 17.8–19.1%) respectively, which is comparable to previous studies in critical care cohorts [6].
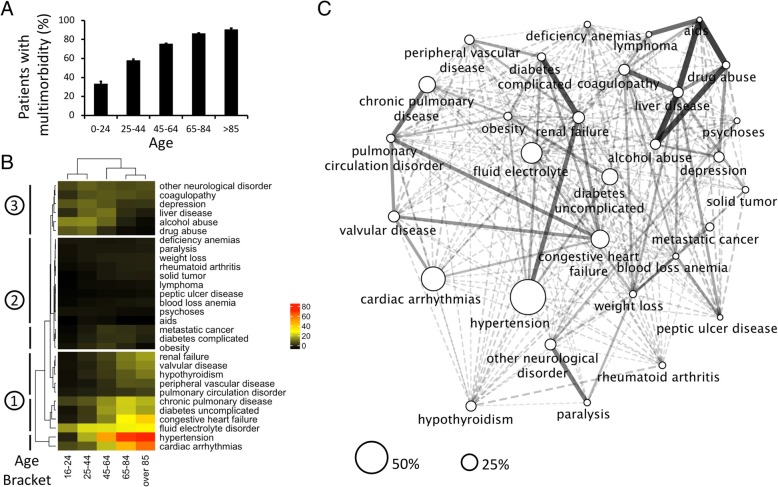
The proportion of patients with multimorbidity increased steadily with age as expected [9] (Fig. 1a). However, we found that the prevalence of individual disease groups (reflected by Elixhauser categories) had distinct patterns over age brackets (Fig. 1b). Specifically, three distinct patterns were observed: one showed increasing prevalence with age, the second had diseases with lower prevalence (range 0–3.8%), and the third had peak prevalence in the 25–44 and 45–64 age bracket but lower prevalence in older age groups. Network discovery showed a variety of disease co-occurrences over the cohort suggesting frequent associations between cardiovascular with pulmonary conditions, diabetes, renal failure, and hypertension and the co-occurrence of alcohol/drug abuse, liver failure, and coagulopathy (Fig. 1c). These findings further suggested the hypothesis that distinct subgroups existed within our cohort.
Fig. 1.
Heterogeneous morbidity profile in critical care patients. a Percent of patients suffering from multimorbidity progressively increases with age (average ± standard deviation). b Disease composition is non-homogenous over each age bracket but organizes into the following patterns: (1) diseases that are more prevalent increase progressively with age, (2) diseases with lower prevalence (0–3.8%), and (3) diseases with an earlier prevalence peak and lower prevalence with older population. Color code (right) represents percent prevalence. c Network discovery of disease co-occurrence shows broad range of disease associations in the critical care study cohort. Note intuitive examples such as associated cardiopulmonary conditions, diabetic nephropathy, co-occurrence of alcohol abuse, liver disease, and coagulopathy. Node size represents disease prevalence; edge width expresses relative risk for each disease pair for each disease pair; bar highlighting the multiple disease clusters in Fig 1b
Identifying distinct multimorbidity subgroups
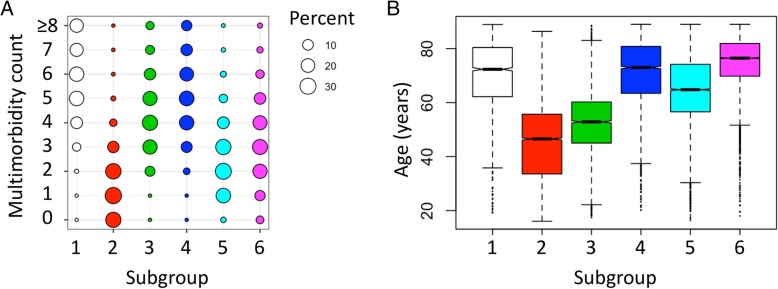
We hypothesized that with analysis of similarities at the individual patient level, we can identify subgroups of patients with distinct demographics and disease compositions. We also hypothesized that patients belonging to certain subgroups will share vulnerability to sepsis and associated death. Using latent class analysis, we identified six subgroups of patients. The number of subgroups was defined by considering metrics of model fit and subgroup size as described in the “Methods” section. Subgroups were verified using descriptive statistics as well as simulations (see the “Methods” section and Additional file 2. Supplementary methods). Disease compositions paralleled some of the patterns suggested by the previous network discovery of the entire study cohort (Fig. 1c). Subgroup characteristics are summarized in Figs. 2 and 3 and Additional file 3: Table S3.
Fig. 2.
Subgroup characteristics of the six subgroups identified in critical care cohort. a Baloonplot summary of morbidity count for each subgroup. Highest morbidity burdens are subgroups 1, 3, 4, and 6. b Boxplot of age distribution in subgroups
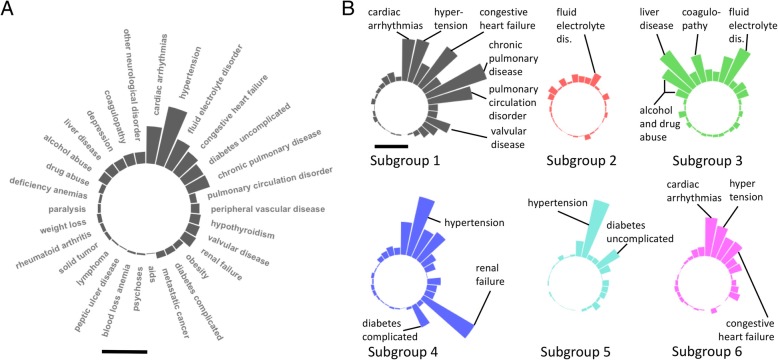
Fig. 3.
Distinct disease compositions in critical care subgroups. a Prevalence of Elixhauser’s morbidity categories for the entire cohort of 36,960 patients. Bar, 50% prevalence. b Circular barplot summary of disease composition of morbidity subgroups in critical care. Plot configuration is identical to a. Note the distinct patterns such as the complex cardiopulmonary profile in the cardiopulmonary subgroup (subgroup 1), health consequences of addiction in the hepatic/addiction subgroup (subgroup 3), and diabetic nephropathy with hypertension in the complicated diabetics subgroup (subgroup 4). Bar, 50% prevalence
In the first “cardiopulmonary” subgroup, we found a high prevalence of cardiopulmonary conditions in older patients (mean age 72.3 ± 0.27) with the highest prevalence of chronic pulmonary diseases (93.86%). The second “young” subgroup had the youngest patients with the lowest point prevalence of any morbidity compared to the rest of the subgroups. The third, “hepatic/addiction” subgroup, consisted of middle-aged patients (mean age of 52.25, 95% CI 51.85–52.65) with the high rates of depression (20.1%), alcohol abuse (47.75%), drug abuse (18.2%), liver failure (67%), and coagulopathy (41.81%). This subgroup captured 70% of patients with liver disease from the entire critical care cohort of 36,960 patients. The “complicated diabetics” and “uncomplicated diabetics” subgroups both had a high prevalence of diabetes and hypertension; a large proportion of patients in the complicated diabetics subgroup suffered from renal failure (88.3%) and complicated diabetes (35.4%). The final “cardiac” subgroup was comprised of the oldest patients with a high prevalence of cardiopulmonary problems similar to the cardiopulmonary subgroup. Statistical testing with one-way ANOVA showed significant differences between all disease prevalence between subgroups (p < 0.001) supporting the distinct disease composition of each subgroup, the basis for identification of these subgroups.
Multimorbidity subgroups vulnerable to sepsis and associated mortality
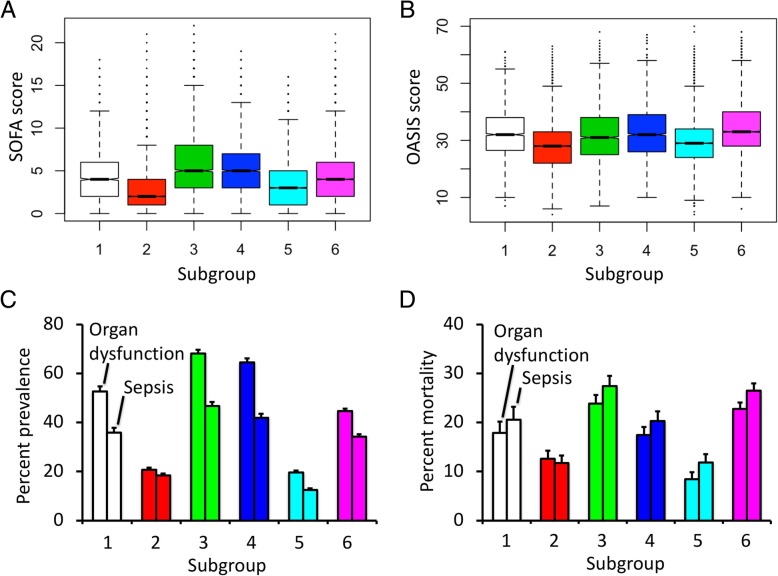
We next tested if multimorbidity subgroups were different in terms of frequencies in adverse health outcomes, such as organ dysfunction and sepsis, and whether subgroups had different mortality rate associated to these clinical problems. We first established the patterns of organ dysfunctions using Sequential Organ Failure Assessment (SOFA) score which entails a system-wise observation recorded during the first 24 h of ICU stay [3]. The system-level assessment using the SOFA subscores for respiratory, cardiovascular, renal, hepatic, coagulation, and central nervous systems paralleled the morbidity profile of each subgroup (Additional file 2: Figure S2). The highest SOFA scores were detected in the hepatic/addiction subgroup closely followed by risk scores in the complicated diabetics subgroup (Fig. 4a). OASIS risk assessment score for inpatient mortality showed higher values for the cardiopulmonary, hepatic/addiction, complicated diabetics, and cardiac subgroups (Fig. 4b). The actual rate of organ dysfunction and sepsis was highest in the hepatic/addiction subgroup followed by the complicated diabetics subgroup (Fig. 4c). High mortality subgroups were also the cardiopulmonary, hepatic/addiction, complicated diabetics, and cardiac subgroups. Mortality in patients with sepsis and organ dysfunction was highest in the hepatic/addiction subgroup, almost fourfold the mortality rates in the young and uncomplicated diabetics subgroups (Fig. 4d).
Fig. 4.
Morbidity subgroups with higher frequencies of organ dysfunction, sepsis, and associated mortality. a SOFA score shows most severe organ dysfunction in the hepatic/addiction subgroup closely followed by the complicated diabetics. b OASIS score shows similarly high mortality risk in the cardiopulmonary, hepatic/addiction, complicated diabetics, and cardiac subgroups. c, d Prevalence of organ dysfunction, sepsis, and associated mortality is highest in the hepatic/addiction subgroup (all differences are significant with multiple testing, p < 0.001)
Disease co-occurrence in multimorbidity subgroups with higher mortality rates
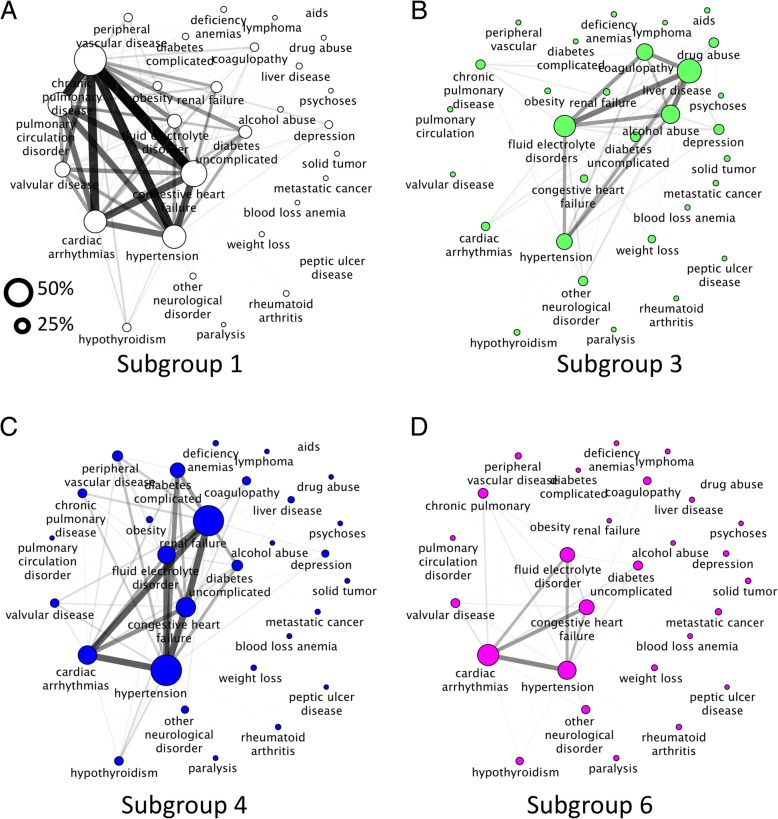
We next explored the co-occurrence of conditions for the cardiopulmonary, hepatic/addiction, complicated diabetics, and cardiac subgroups with high risk of adverse health outcomes (sepsis, organ dysfunction, and death) by visualizing prevalence and pairwise associations in a disease network (Fig. 5).
Fig. 5.
Network discovery of multimorbidity subgroups with higher rates of adverse health outcomes (organ dysfunction, sepsis, and death). Nodes represent disease prevalence, and edges express the number of patients with the disease pair normalized to the entire subgroup population. a Patients in the cardiopulmonary subgroup suffer from cardiopulmonary diseases as expected. b The hepatic/addiction subgroup has a high prevalence of health consequences of addiction. c For patients in the complicated diabetics subgroup, the dominant morbidity profile is diabetic nephropathy and hypertension. d The cardiac subgroup has a high prevalence of cardiac arrhythmias as well as other cardiac conditions
Nodes represented the Elixhauser disease categories with size defined by disease prevalence within the subgroup, and edge width was the number of patients with the disease pair normalized to the total number of patients in the subgroups (Fig. 3). Relative risk and/or Pearson’s correlation were not used in this portion of the analysis as these techniques tend to render inaccurate results for extremes of prevalence potentially resulting in under/over-estimated associations [29]. Network structure confirmed disease patterns suggested by our analysis of disease prevalence in each of the subgroups. The high mortality group consisting of younger (middle-aged) patients suggested a pattern of addiction-associated conditions (the hepatic/addiction subgroup) given the pairwise association between liver disease, coagulation disorders, alcohol excess, drug abuse, and depression. In other subgroups, the network analysis showed associations between cardiovascular-respiratory conditions (the cardiopulmonary subgroup), hypertensive-renal-diabetic with end-organ complications (the complicated diabetics subgroup), and cardiopulmonary problems (the cardiac subgroup).
Groups with lower mortality (5.31–6.02%) were either younger patients (99% below 65 years, the young subgroup) with very low disease burden or elderly with significantly higher proportion of non-emergency admissions (35.7%, the uncomplicated diabetics subgroup) than the rest of the cohort. This subgroup suffered from combination hypertensive-diabetes without end-organ complications (Additional file 2: Table S2, Figure S3).
Discussion
Our study identified multiple clinically relevant subgroups in a critical care cohort with differing frequencies of organ dysfunction, sepsis, and associated mortality rates. In addition to the expected phenotypes of multimorbid elderly with high mortality, we also found a group of younger patients suffering from health consequences of addiction with the highest rates of sepsis and organ dysfunction. Although the association of liver failure with poor outcomes in sepsis is well established, the multimorbidity configuration it occurs in is less appreciated. These findings support the shift away from the single-disease model in healthcare to a more holistic construct, putting emphasis on considering multimorbidity composition in clinical decision-making.
As expected, we found an increasing prevalence of multimorbidity with age in line with previous population studies. However, several of our identified subgroups demonstrated distinct morbidity composition and high rates of organ dysfunction, sepsis, and associated mortality. The disease combinations within these subgroups (the cardiopulmonary, hepatic/addiction, complicated diabetics, and cardiac subgroups) were in line with previous observations and can also be interpreted along the lines of shared risk factors and patho-mechanisms. For example, chronic pulmonary disease and congestive heart failure co-occur with the highest prevalence in the cardiopulmonary subgroup; this may be explained by the concept of the cardio-pulmonary continuum [30]. The complicated diabetics subgroup is associated with high rates of diabetic nephropathy and hypertension while the cardiac subgroup represents largely elderly patients with cardiovascular diseases and a higher rate of neurological conditions.
The subgroup associated with greater rates of adverse health outcomes consisted of younger/middle-aged population who suffered from a high prevalence of alcohol and/or drug addiction, associated with liver disease and coagulopathy. While liver disease is well recognized as a risk of sepsis as well as poor outcomes, our novel findings are as follows: (1) we identify complex multimorbidity phenotypes incorporating chronic liver disease and (2) we compare it with other disease configurations in terms of sepsis prevalence and mortality. The demographics and disease composition of this high-risk group in critical care paralleled the population-level data. US Center for Disease Control and Prevention showed a peak percentage of alcohol consumption in the age bracket of 25–44 and 77% of deaths from alcoholic liver disease under the age of 65 (https://www.cdc.gov/DataStatistics/). Population studies covering 175 million hospital discharges demonstrated that the 4.5 million patients suffering from liver cirrhosis were twice as likely to die while in hospital, have sepsis as the reason for admission, and die of sepsis [31, 32]. Bacterial infections are present in up to 30% of admissions with liver failure [33]. The liver has widespread functions in responding to sepsis such as synthesis of proteins for immune, coagulation, and metabolic functions as well as scavenging of endotoxins and bacteria [34–36]. These findings parallel the higher rate of organ dysfunction traceable in our own results with the highest SOFA score in the hepatic/addiction subgroup (Fig. 4a, Additional file 2: Figure S2).
Chronic conditions have long been implicated in the disparities of sepsis prevalence [8]. The presence of comorbidities that impact immune response such as chronic renal failure, diabetes, ethanol abuse, and HIV infection was found to associate with sepsis. Furthermore, cumulative conditions were associated with greater organ failure rates. Prior studies have shown that incorporating morbidity status has improved risk assessment of mortality in patients admitted to critical care [37]. The model developed by Min et al. included a multimorbidity index, a measure that incorporates individual likelihood ratio of death for over 5000 morbidities. In their model, it is assumed that each diagnosis has an independent impact on mortality. This approach carries the advantage of representing the scale of severities within disease categories not necessarily captured by Charlson’s or Elixhauser indices (for example, not all “metastatic cancers” are lethal). Our study focused on the co-occurrence of morbidities, and thereby, our approach considers the impact diseases have on clinical outcomes in combination rather than individually. In the multimorbid population, analysis of temporal trajectories towards sepsis yielded high mortality risk profiles in patients starting off with alcohol abuse, diabetes, and cardiovascular diagnosis [1] as the initial morbidity. These results parallel our findings that multimorbidity subgroups cardiopulmonary, hepatic/addiction, complicated diabetics, and cardiac associate with high sepsis-related deaths.
Conventionally, the recruitment into clinical trials in sepsis has been based on abnormal physiological parameters implying infection as cause for critical illness. Such recruitment strategies inevitably capture a heterogenous patient group where the morbidity profile is confounded by differences in the biological response to sepsis. For example, interventions often studied by clinical trials included modifiers of the inflammatory response in sepsis such as anti-TNF, anti-IL1-Ra, anti-LPS, corticosteroids, IV immunoglobulins, and activated protein C [38]. Importantly, however, conditions such as coagulopathy, chronic liver disease [39], and diabetes [40] with end-organ complications are characterized by alterations in these target mechanisms and thus may be falsely categorized as “non-responders.” Our study represents these patients in subgroups hepatic/addiction and complicated diabetics. Such subcategorization of trial patients into clinically and biologically homogeneous subgroups can help address this confounding effect without the need for stringent exclusion criteria that render the results difficult to generalize.
It is well recognized that managing sepsis patients with liver failure is challenging and requires an individualized form of goal-directed therapy [41]. Early diagnosis and treatment are essential. The early identification of the subgroups demonstrated in our study may therefore help decision-making regarding prophylactic antibiotics, consideration of early interventions, and lower threshold for goal-directed therapy to improve outcomes in sepsis.
One potential limitation of this study is that it is based on data from a single center; therefore, composition of catchment population, departmental protocols, resources, and staffing characteristics are potential limiting to the generalizability of our results [42]. Further, it is difficult to find external validation to our results due to MIMIC3 being a unique dataset with highly detailed clinical data that is unmatched in publicly available resources. Another limitation is that we analyze a cross section of the population and therefore cannot examine causality links between subgroups and vulnerability to sepsis and organ dysfunction. For the same reason, we have no information on time of diagnosis to allow us to distinguish between pre-existing medical conditions and newly incurred diagnoses. To address this issue, diagnosis-related codes assigned for the hospitalization were incorporated to filter out diseases that are the reason for the hospital stay versus those that are not.
Part of the limitations is that ICD codes have poor reproducibility between coders which may impact the robustness of results from administrative databases [43]. However, these inaccuracies are dependent on the diagnostic category of interest, particularly how well the condition is reported and if it is covered by specific codes. For example, ICD9 codes were useful at identifying idiopathic thrombocytic purpura [44] and brain metastasis [45], both of which have specific ICD9 codes. On the other hand, ICD9 codes do not perform well for conditions without specific codes that require for example a combination of less granular code or code that is under-reported. Such examples are catheter-acquired urinary tract infection (required combination of diagnosis and procedural code) or statin-related rhabdomyolysis (lack of ICD9 code for rhabdomyolysis). The comorbidity profile in our study relies on broad disease categories of Elixhauser, which cover multiple code categories validated against ICD9 codes with good performance [24, 26]. The other labels used in our study such as organ dysfunction and sepsis were also derived based on administrative definitions. Although earlier assessments showed low coding rates (approximately 43% of acute kidney injuries for example [46]), retrospective studies from between 2005 and 2013 show improving rates of documentation in organ dysfunction [47]. There are also limitations to using the Angus criteria in our study to identify sepsis in patients. This system relies on ICD codes to establish co-occurrence of infection and organ dysfunction. Although imperfect, it has shown reasonable performance and is better compared to other administrative sepsis definitions [48, 49]. Finally, our study is limited to patients that survive long enough or are deemed appropriate for intensive care. Therefore, our mortality numbers are very likely to underestimate true figures. Nevertheless, our study uses real-world hospital data and provides a description of the population in the first 24 h in critical care, a very relevant time frame for intervention and management. This provides critical information on the risk of sepsis and associate mortality.
Conclusions
The increasing prevalence of multimorbidity creates complexity in medical diagnostics and treatment decisions. Our study focuses on the population of patients in critical care and examines the relevance of multimorbidity in ICU outcomes including the rate of sepsis, one of the highly relevant conditions. We identify several patient groups susceptible to adverse health outcomes in critical care. The patient group with the highest rate of sepsis, organ dysfunction, and mortality was a subgroup of patients suffering from health consequences of addiction. While this work is hypothesis generating, the findings support the shift away from the current single-disease model and promote a subgroup-specific approach in medical training, treatment, and trial design.
Additional files
Table S1. Demographics and outcome summary of the study cohort (DOCX 22 kb)
Supplementary methods. Figure S1. Graph summary of ROC curves depicting predictive performance of LCA input variables at differentiating a given subgroup from the remaining subgroups. Table S2. Table summary of classifier performance (assessed using area under the receiver operating curve, AUC) at predicting subgroup membership based on the input variables used in the LCA (age, sex, type of admission, and morbidities). Figure S2. Violin plot summary of organ systems impairment in the multimorbidity subgroups. Figure S3. Network summary of the multimorbidity subgroups with lower rates of sepsis and death. (ZIP 524 kb)
Table S3. Summary of morbidity composition of subgroups. (DOCX 18 kb)
Acknowledgements
The authors would like to thank Ling Chen for her help with parts of the analysis and Erik Drysdale and Lauren Erdman for their helpful discussions on the manuscript. We would also like to thank Dr. Alistair Johnson for his guidance with the MIMIC3 database.
Authors’ contributions
ZZ and NG conceived the study. ZZ carried our data analysis. ZZ, AL, MC, and NG wrote the paper. ZZ, AL, and NG revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Medical Research Council and the Engineering and Physical Sciences Research Council grant MR/N00583X/1 “Manchester Molecular Pathology Innovation Centre (MMPathIC): bridging the gap between biomarker discovery and health and wealth”; and supported by researchers at the NIHR Manchester Biomedical Research Centre. Zsolt Zador: National Institute for Health Research, Academic Clinical Lectureship, award number: CL-2014-06-004. The study does not publish individual patient data. The patient data in the MIMIC3 database has been deidentified and therefore patient consent for the study was waived.
Availability of data and materials
MIMIC 3 database is available at https://mimic.physionet.org/about/mimic/. Codes used to generate data tables are accessible at https://github.com/MIT-LCP/mimic-code
Ethics approval and consent to participate
The study is based entirely on publicly available data.
Consent for publication
All authors have read and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Michael D. Cusimano and Nophar Geifman are senior authors.
References
- 1.Beck MK, Jensen AB, Nielsen AB, Perner A, Moseley PL, Brunak S. Diagnosis trajectories of prior multi-morbidity predict sepsis mortality. Sci Rep. 2016;6:1–9. [DOI] [PMC free article] [PubMed]
- 2.Gall JR, Lemeshow S, Saulnier F. A new simplified acute physiology score (SAPS II) based on a European/North American multicenter study. JAMA. 1993;270:2957–2963. doi: 10.1001/jama.1993.03510240069035. [DOI] [PubMed] [Google Scholar]
- 3.Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996;22:707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- 4.Johnson Alistair E. W., Kramer Andrew A., Clifford Gari D. A New Severity of Illness Scale Using a Subset of Acute Physiology and Chronic Health Evaluation Data Elements Shows Comparable Predictive Accuracy*. Critical Care Medicine. 2013;41(7):1711–1718. doi: 10.1097/CCM.0b013e31828a24fe. [DOI] [PubMed] [Google Scholar]
- 5.Zimmerman Jack E., Kramer Andrew A., McNair Douglas S., Malila Fern M. Acute Physiology and Chronic Health Evaluation (APACHE) IV: Hospital mortality assessment for today’s critically ill patients*. Critical Care Medicine. 2006;34(5):1297–1310. doi: 10.1097/01.CCM.0000215112.84523.F0. [DOI] [PubMed] [Google Scholar]
- 6.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Esper AM, Martin GS. The impact of cormorbid conditions on critical illness. Crit Care Med. 2011;39:2728–2735. doi: 10.1097/CCM.0b013e318236f27e. [DOI] [PubMed] [Google Scholar]
- 8.Esper AM, Moss M, Lewis CA, Nisbet R, Mannino DM, Martin GS. The role of infection and comorbidity: factors that influence disparities in sepsis. Crit Care Med. 2006;34:2576–2582. doi: 10.1097/01.CCM.0000239114.50519.0E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts KC, Rao DP, Bennett TL, Loukine L, Jayaraman GC. Prevalence and patterns of chronic disease multimorbidity and associated determinants in Canada. Chronic Dis Inj Can. 2015;35:87–94. doi: 10.24095/hpcdp.35.6.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnett Karen, Mercer Stewart W, Norbury Michael, Watt Graham, Wyke Sally, Guthrie Bruce. Epidemiology of multimorbidity and implications for health care, research, and medical education: a cross-sectional study. The Lancet. 2012;380(9836):37–43. doi: 10.1016/S0140-6736(12)60240-2. [DOI] [PubMed] [Google Scholar]
- 11.Wolff JL, Starfield B, Anderson G. Prevalence, expenditures, and complications of multiple chronic conditions in the elderly. Arch Intern Med. 2012;162:2269–2276. doi: 10.1001/archinte.162.20.2269. [DOI] [PubMed] [Google Scholar]
- 12.Farmer C, Fenu E, O’Flynn N, Guthrie B. Clinical assessment and management of multimorbidity: summary of NICE guidance. BMJ. 2016;354:i4843. doi: 10.1136/bmj.i4843. [DOI] [PubMed] [Google Scholar]
- 13.Van Spall Harriette G. C., Toren Andrew, Kiss Alex, Fowler Robert A. Eligibility Criteria of Randomized Controlled Trials Published in High-Impact General Medical Journals. JAMA. 2007;297(11):1233. doi: 10.1001/jama.297.11.1233. [DOI] [PubMed] [Google Scholar]
- 14.Espeland MA, Crimmins EM, Grossardt BR, Crandall JP, Gelfond JAL, Harris TB, et al. Clinical trials targeting aging and age-related multimorbidity. J Gerontol A Biol Sci Med Sci. 2017;72:355-61. [DOI] [PMC free article] [PubMed]
- 15.Scicluna BP, van Vught LA, Zwinderman AH, Wiewel MA, Davenport EE, Burnham KL, et al. Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Respir Med. 2017;5:816–826. doi: 10.1016/S2213-2600(17)30294-1. [DOI] [PubMed] [Google Scholar]
- 16.Anderson Gary P. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. The Lancet. 2008;372(9643):1107–1119. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- 17.Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorani Marco D., Hemphill J. Claude, Morabito Diane, Rosenthal Guy, Manley Geoffrey T. New Approaches to Physiological Informatics in Neurocritical Care. Neurocritical Care. 2007;7(1):45–52. doi: 10.1007/s12028-007-0043-7. [DOI] [PubMed] [Google Scholar]
- 19.Johnson AE, Pollard TJ, Shen L, Lehman LH, Feng M, Ghassemi M, et al. Data descriptor: MIMIC-III, a freely accessible critical care database. Background Summary. 2016;3:1–9. Article number 160035. [DOI] [PMC free article] [PubMed]
- 20.Zador Zsolt, Huang Wendy, Sperrin Matthew, Lawton Michael T. Multivariable and Bayesian Network Analysis of Outcome Predictors in Acute Aneurysmal Subarachnoid Hemorrhage: Review of a Pure Surgical Series in the Post-International Subarachnoid Aneurysm Trial Era. Operative Neurosurgery. 2017;14(6):603–610. doi: 10.1093/ons/opx163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zador Zsolt, Sperrin Matthew, King Andrew T. Predictors of Outcome in Traumatic Brain Injury: New Insight Using Receiver Operating Curve Indices and Bayesian Network Analysis. PLOS ONE. 2016;11(7):e0158762. doi: 10.1371/journal.pone.0158762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calfee CS, Delucchi K, Parsons PE, Taylor B, Ware LB, Matthay MA. Latent class analysis of ARDS subphenotypes: analysis of data from two randomized controlled trials. Lancet Respir Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall Marlous, Dondo Tatendashe B., Yan Andrew T., Mamas Mamas A., Timmis Adam D., Deanfield John E., Jernberg Tomas, Hemingway Harry, Fox Keith A. A., Gale Chris P. Multimorbidity and survival for patients with acute myocardial infarction in England and Wales: Latent class analysis of a nationwide population-based cohort. PLOS Medicine. 2018;15(3):e1002501. doi: 10.1371/journal.pmed.1002501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elixhauser A, Steiner C, Harris DR, Coffey RM. Comorbidity measures for use with administrative data. Med Care. 1998;36:8–27. doi: 10.1097/00005650-199801000-00004. [DOI] [PubMed] [Google Scholar]
- 25.https://github.com/MIT-LCP/mimic-code (Accessed 22 Mar 2018).
- 26.Quan H, Sundararajan V, Halfon P, Fong A, Burnand B, Luthi JC, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care. 2005;43:1130–1139. doi: 10.1097/01.mlr.0000182534.19832.83. [DOI] [PubMed] [Google Scholar]
- 27.Zhou M, Thayer WM, Bridges JFP. Using latent class analysis to model preference heterogeneity in health: a systematic review. Pharmacoeconomics. 2018;36:175–187. doi: 10.1007/s40273-017-0575-4. [DOI] [PubMed] [Google Scholar]
- 28.Nasserinejad Kazem, van Rosmalen Joost, de Kort Wim, Lesaffre Emmanuel. Comparison of Criteria for Choosing the Number of Classes in Bayesian Finite Mixture Models. PLOS ONE. 2017;12(1):e0168838. doi: 10.1371/journal.pone.0168838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hidalgo César A., Blumm Nicholas, Barabási Albert-László, Christakis Nicholas A. A Dynamic Network Approach for the Study of Human Phenotypes. PLoS Computational Biology. 2009;5(4):e1000353. doi: 10.1371/journal.pcbi.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ukena C, Mahfoud F, Kindermann M, Kindermann I, Bals R, Voors AA, et al. The cardiopulmonary continuum systemic inflammation as “common soil” of heart and lung disease. Int J Cardiol. 2010;145:172–176. doi: 10.1016/j.ijcard.2010.04.082. [DOI] [PubMed] [Google Scholar]
- 31.Gustot T, Durand F, Lebrec D, Vincent JL, Moreau R. Severe sepsis in cirrhosis. Hepatology. 2009;50:2022–2033. doi: 10.1002/hep.23264. [DOI] [PubMed] [Google Scholar]
- 32.Foreman MG, Mannino DM, Moss M. Cirrhosis as a risk factor for sepsis and death: analysis of the National Hospital Discharge Survey. Chest Am College Chest Physicians. 2003;124:1016–1020. doi: 10.1378/chest.124.3.1016. [DOI] [PubMed] [Google Scholar]
- 33.Fernández J, Gustot T. Management of bacterial infections in cirrhosis. J Hepatol. 2012;56:1–12. doi: 10.1016/S0168-8278(12)60002-6. [DOI] [PubMed] [Google Scholar]
- 34.Nesseler N, Launey Y, Aninat C, Morel F, Mallédant Y, Seguin P. Clinical review: the liver in sepsis. Crit Care. 2012;16:1–8. doi: 10.1186/cc11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canabal Juan M, Kramer David J. Management of sepsis in patients with liver failure. Current Opinion in Critical Care. 2008;14(2):189–197. doi: 10.1097/MCC.0b013e3282f6a435. [DOI] [PubMed] [Google Scholar]
- 36.Protzer U, Maini MK, Knolle PA. Living in the liver: hepatic infections. Nat Rev Immunol. 2012;12:201–213. doi: 10.1038/nri3169. [DOI] [PubMed] [Google Scholar]
- 37.Min Hua, Avramovic Sanja, Wojtusiak Janusz, Khosla Rahul, Fletcher Ross D., Alemi Farrokh, Elfadel Kheirbek Raya. A Comprehensive Multimorbidity Index for Predicting Mortality in Intensive Care Unit Patients. Journal of Palliative Medicine. 2017;20(1):35–41. doi: 10.1089/jpm.2015.0392. [DOI] [PubMed] [Google Scholar]
- 38.Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med. 2014;20:195–203. doi: 10.1016/j.molmed.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 39.Yan Jun, Li Song, Li Shulin. The Role of the Liver in Sepsis. International Reviews of Immunology. 2014;33(6):498–510. doi: 10.3109/08830185.2014.889129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trevelin SC, Carlos D, Beretta M, Da Silva JS, Cunha FQ. Diabetes mellitus and sepsis: a challenging association. Shock. 2017;47:276–286. doi: 10.1097/SHK.0000000000000778. [DOI] [PubMed] [Google Scholar]
- 41.Ford H, Systems H, Re- CW. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N Engl J Med. 2001;345:1368–1377. doi: 10.1056/NEJMoa010307. [DOI] [PubMed] [Google Scholar]
- 42.Bellomo R, Warrillow SJ, Reade MC. Why we should be wary of single-center trials. Crit Care Med. 2009;37:3114–3119. doi: 10.1097/CCM.0b013e3181bc7bd5. [DOI] [PubMed] [Google Scholar]
- 43.Misset Benoit, Nakache Didier, Vesin Aurelien, Darmon Mickael, Garrouste-Orgeas Maite, Mourvillier Bruno, Adrie Christophe, Pease Sebastian, Costa de Beauregard Marie-Aliette, Goldgran Toledano Dany, Metais Elisabeth, Timsit Jean Francois, Outcomerea Database Investigators The. Reliability of diagnostic coding in intensive care patients. Critical Care. 2008;12(4):R95. doi: 10.1186/cc6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segal Jodi B., Powe Neil R. Accuracy of identification of patients with immune thrombocytopenic purpura through administrative records: A data validation study. American Journal of Hematology. 2003;75(1):12–17. doi: 10.1002/ajh.10445. [DOI] [PubMed] [Google Scholar]
- 45.Eichler April F., Lamont Elizabeth B. Utility of administrative claims data for the study of brain metastases: a validation study. Journal of Neuro-Oncology. 2009;95(3):427–431. doi: 10.1007/s11060-009-9943-z. [DOI] [PubMed] [Google Scholar]
- 46.Wilson Francis Perry, Bansal Amar D., Jasti Sravan K., Lin Jennie J., Shashaty Michael G.S., Berns Jeffrey S., Feldman Harold I., Fuchs Barry D. The impact of documentation of severe acute kidney injury on mortality. Clinical Nephrology. 2013;80(12):417–425. doi: 10.5414/CN108072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhee C, Murphy MV, Li L, Platt R, Klompas M; Centers for Disease Control and Prevention Epicenters Program. Improving documentation and coding for acute organ dysfunction biases estimates of changing sepsis severity and burden: a retrospective study. Crit Care. 2015;19:338. [DOI] [PMC free article] [PubMed]
- 48.Iwashyna Theodore J., Odden Andrew, Rohde Jeffrey, Bonham Catherine, Kuhn Latoya, Malani Preeti, Chen Lena, Flanders Scott. Identifying Patients With Severe Sepsis Using Administrative Claims. Medical Care. 2014;52(6):e39–e43. doi: 10.1097/MLR.0b013e318268ac86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson Alistair E. W., Aboab Jerome, Raffa Jesse D., Pollard Tom J., Deliberato Rodrigo O., Celi Leo A., Stone David J. A Comparative Analysis of Sepsis Identification Methods in an Electronic Database*. Critical Care Medicine. 2018;46(4):494–499. doi: 10.1097/CCM.0000000000002965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Demographics and outcome summary of the study cohort (DOCX 22 kb)
Supplementary methods. Figure S1. Graph summary of ROC curves depicting predictive performance of LCA input variables at differentiating a given subgroup from the remaining subgroups. Table S2. Table summary of classifier performance (assessed using area under the receiver operating curve, AUC) at predicting subgroup membership based on the input variables used in the LCA (age, sex, type of admission, and morbidities). Figure S2. Violin plot summary of organ systems impairment in the multimorbidity subgroups. Figure S3. Network summary of the multimorbidity subgroups with lower rates of sepsis and death. (ZIP 524 kb)
Table S3. Summary of morbidity composition of subgroups. (DOCX 18 kb)
Data Availability Statement
MIMIC 3 database is available at https://mimic.physionet.org/about/mimic/. Codes used to generate data tables are accessible at https://github.com/MIT-LCP/mimic-code