Significance
Nucleosome assembly is regulated at multiple levels to impact distinct cellular processes. Defects in H3.3 incorporation into the chromatin of senescent cells lead to transcriptional reprogramming that alters gene expression. Therefore, it is important to determine how nucleosome assembly of H3.3 is controlled in senescent cells. In this paper, we provide evidence of a link between the nucleosome assembly regulator Pak2 kinase and cellular senescence and organismal aging.
Keywords: Pak2, senescence, nucleosome assembly, histone H3.3, aging
Abstract
Cellular senescence defines an irreversible cell growth arrest state linked to loss of tissue function and aging in mammals. This transition from proliferation to senescence is typically characterized by increased expression of the cell-cycle inhibitor p16INK4a and formation of senescence-associated heterochromatin foci (SAHF). SAHF formation depends on HIRA-mediated nucleosome assembly of histone H3.3, which is regulated by the serine/threonine protein kinase Pak2. However, it is unknown if Pak2 contributes to cellular senescence. Here, we show that depletion of Pak2 delayed oncogene-induced senescence in IMR90 human fibroblasts and oxidative stress–induced senescence of mouse embryonic fibroblasts (MEFs), whereas overexpression of Pak2 accelerated senescence of IMR90 cells. Importantly, depletion of Pak2 in BubR1 progeroid mice attenuated the onset of aging-associated phenotypes and extended life span. Pak2 is required for expression of genes involved in cellular senescence and regulated the deposition of newly synthesized H3.3 onto chromatin in senescent cells. Together, our results demonstrate that Pak2 is an important regulator of cellular senescence and organismal aging, in part through the regulation of gene expression and H3.3 nucleosome assembly.
Cellular senescence is a cell growth arrest state associated with loss of tissue function in mammals (1–5). Senescence can be induced prematurely by various cellular stresses. For example, excessive expression of certain oncogenes induces premature cellular senescence in a process referred to as oncogene-induced senescence (OIS) (5–7). Additionally, mouse embryonic fibroblasts (MEFs) undergo senescence under standard culture conditions with 20% oxygen due to DNA damage caused by oxidative stress (8). Cellular senescence has been directly linked to aging (9), and recent studies have demonstrated that the accumulation of senescent cells in tissues leads to organismal aging (2). BubR1 hypomorphic mice exhibit growth retardation, a shortened life span, and development of progeroid and age-related phenotypes, including cachectic dwarfism, lordokyphosis, cataracts, loss of subcutaneous fat, and impaired wound healing (10, 11). Interestingly, elimination of cells expressing p16INK4a, a senescence biomarker, significantly attenuates the progression of age-related disorders in these mice (11). Furthermore, removal of p16INK4a-expressing cells from naturally aged wild-type mice also leads to healthy life span extension (12), indicating that the accumulation of senescent cells has a negative influence on longevity and health span.
Chromatin is one principal carrier of epigenetic information in cells, and its reorganization plays a key role in cellular senescence. Generally, senescent cells exhibit extensive alterations to chromatin structures and gene expression profiles (6, 13, 14). The structure and function of chromatin is significantly affected by nucleosome occupancy during replicative aging in yeast and cellular senescence in human cells. Aged Saccharomyces cerevisiae cells exhibit decreased expression of core histones, which is coupled to decreased nucleosome density and aberrant up-regulation of associated genes (15). Notably, senescence in human cells can be accompanied by a global condensation of chromatin, wherein chromosomes are packaged into condensed structures known as senescence-associated heterochromatin foci (SAHF) (6). Functionally, SAHF silence genes that promote cell proliferation, such as E2F-regulated target genes, thereby contributing to the onset and maintenance of senescence-associated cell cycle arrest (6). Hence, studying the regulation of histone variant H3.3 and its chaperones, including the HIRA complex, in cellular senescence is important. Several lines of evidence indicate that the exchange and deposition of histones onto chromatin is implicated in cellular senescence. First, primary human cells that ectopically express HIRA, a subunit of the histone H3.3 chaperone complex composed of HIRA, ubinuclein 1 (UBN1), and calcineurin binding protein 1 (CABIN1), undergo senescence with increased SAHF formation (16, 17). Second, ectopic expression of histone variant H3.3 and its proteolytically cleaved form induces cellular senescence, most likely through stress-inducing mechanisms (18). Third, canonical histone H3.3 chaperone HIRA regulates senescent cell gene expression (14). Thus, HIRA-mediated nucleosome assembly of H3.3 promotes chromosome condensation and gene expression changes associated with cellular senescence, impacting SAHF formation.
The HIRA-mediated nucleosome assembly pathway is regulated by multiple mechanisms. We previously reported that Pak2 regulates HIRA-mediated nucleosome assembly through phosphorylation of histone H4 serine 47 (H4S47ph) to promote HIRA-mediated nucleosome assembly of histone H3.3 (19). Pak2 is an autoinhibitory kinase and a member of the p21 (Cdc42/Rac)-activated serine/threonine protein kinase (Pak) family (20, 21). Pak2 is activated by oncogenic Ras via the RAS/ERK/PAK2 pathway in rat pheochromocytoma (PC12) cells (22). Cdc42, a member of the Rho GTPase family, regulates proinflammatory gene expression in senescent human endothelial cells in vitro and promotes senescence-associated inflammation in worms and mice in vivo (23). These findings led us to postulate that Pak2 has a role in cellular senescence, in part through regulating HIRA-mediated H3.3 incorporation. To test this hypothesis, we utilize 3 senescence model systems, OIS in normal human fibroblasts, oxidative stress–induced senescence during culturing of mouse embryonic cells (MEFs), and the BubR1 hypomorphic progeroid mouse model. Results from all of these models demonstrate an important role for Pak2 in promoting gene expression and nucleosome assembly of H3.3 during cellular senescence and organismal aging.
Results
Pak2 Depletion Attenuates OIS.
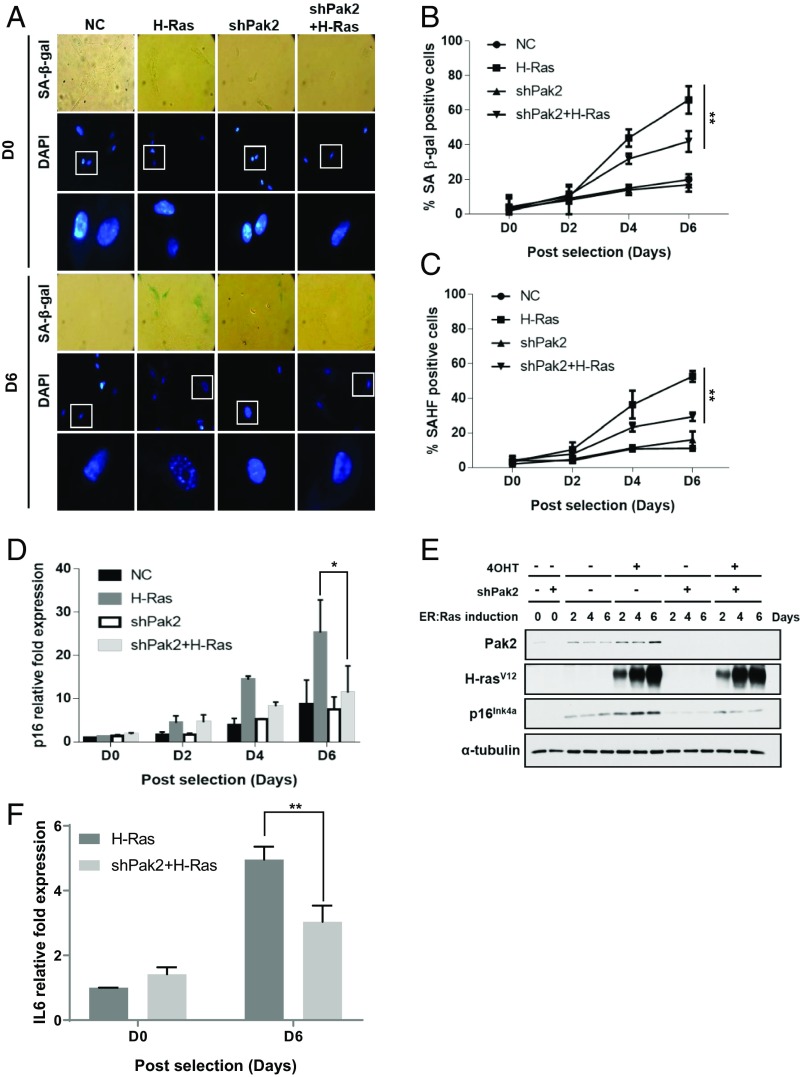
Since Pak2 regulates HIRA-mediated nucleosome assembly of H3.3, a process promoting cellular changes associated with cellular senescence, we hypothesized that Pak2 may participate in the process of cellular senescence. To test this hypothesis, we first evaluated if Pak2 regulated OIS. Exogenous expression of oncogenic Ras in human fibroblasts promotes OIS, with cells exhibiting increased p16INK4a expression and characteristically displaying one large nucleolus with heterochromatic foci at the periphery (called SAHF) as visualized by DAPI staining (5, 6). We transduced H-RasG12V-inducible IMR90 cells with short-hairpin RNA (shRNA) targeting Pak2 (shPak2). After transduction and selection, we treated cells with 4-hydroxytamoxifen (4OHT) to induce H-RasG12V expression (SI Appendix, Fig. S1A) and monitored senescence-associated β-galactosidase (SA-β-gal) activity, DAPI-positive SAHF, and p16INK4a expression, all well-known senescence markers (6, 13, 24, 25). In control cells transduced with nontargeting control (NC) shRNA, the percentage of SA-β-gal-positive cells increased beginning at day 4 following H-RasG12V induction and reached ∼60% at day 6, whereas ∼40% of Pak2-depleted cells exhibited positive SA-β-gal staining on day 6, indicating that entry into OIS was attenuated with Pak2 depletion (Fig. 1 A and B). Consistent with this idea, we observed that Pak2-depleted cells had significantly fewer SAHF-positive cells than control cells on day 6 (Fig. 1 A and C). Furthermore, Pak2 depletion using 2 independent shRNAs targeting Pak2 compromised H-RasG12V-induced expression of p16INK4a (Fig. 1 D and E and SI Appendix, Fig. S1 B and C). We also observed that Pak2 depletion significantly reduced the expression of interleukin 6 (IL-6), but not IL-8 (Fig. 1F and SI Appendix, Fig. S1D), 2 proinflammatory cytokines associated with the senescence-associated secretory phenotype (SASP), on day 6 following H-RasG12V induction. Altogether, these results indicate that Pak2 contributes to OIS, likely through regulating a subset of genes involved in senescence.
Fig. 1.
Pak2 expression level increases in cells undergoing senescence, and its depletion attenuates OIS. (A–C) IMR90 cells containing NC, H-Ras, shPak2, and H-Ras+shPak2 were stained for SA-β-gal activity followed by DAPI staining for cell counting and SAHF detection at 0, 2, 4, and 6 d postselection. The percentage of cells containing DNA foci (SAHF positive) is reported at the indicated postselection days. Results from 3 independent experiments are shown (n = 3, mean ± SEM, **P < 0.01). (D and E) Pak2 expression level increases with senescence, and Pak2 depletion decreases p16INK4a accumulation during OIS. (D) The p16INK4a mRNA level was determined by qRT-PCR (n = 3, mean ± SEM, *P < 0.05). Values were normalized to GAPDH. Relative fold expression was calculated by normalizing to D0 NC values. (E) H-rasV12, Pak2, and p16INK4a protein expression level was tested by Western blot using lysates from IMR90 cells containing empty vector, H-Ras, shPak2, and H-Ras+shPak2. α-Tubulin was used as a loading control. (F) IL-6 mRNA expression level was determined by qRT-PCR (n = 3, mean ± SEM, **P < 0.01). Relative fold expression was calculated by normalizing to D0 NC values.
Pak2 Expression Level Increases During OIS, and Its Overexpression Promotes Cellular Senescence.
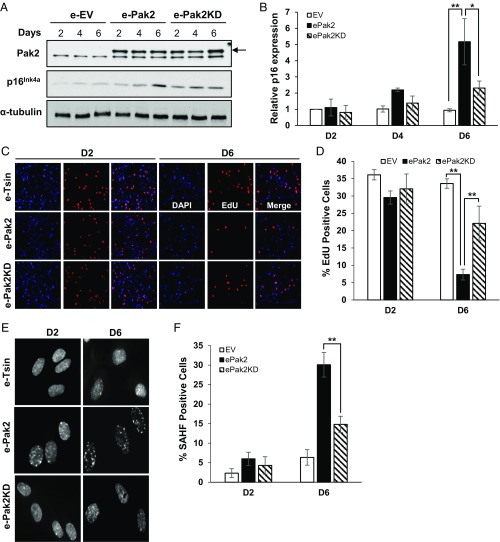
We found that Pak2 expression increased during oncogenic Ras-induced senescence (Fig. 1E and SI Appendix, Fig. S1 E and F). Therefore, we tested whether Pak2 overexpression promoted cellular senescence in normal human fibroblasts using an established senescence protocol (26). We transduced cells with a lentivirus encoding an empty vector, a vector expressing wild-type Pak2, and a vector expressing Pak2 (T402A) kinase dead mutant. Wide-type Pak2 overexpression (e-Pak2) resulted in increased p16INK4a expression as detected by immunoblotting and mRNA quantification compared with empty vector (e-EV) (Fig. 2 A and B). Furthermore, we observed a decreased incorporation of EdU (5-ethynyl-2′-deoxyuridine) in S phase cells on day 6 after overexpression of Pak2 (Fig. 2 C and D), suggesting decreased proliferation and cell cycle arrest. Additionally, the percentage of SAHF-positive cells was dramatically increased in e-Pak2 cells (Fig. 2 E and F). Interestingly, cells with ectopic expression of the Pak2 kinase dead mutant (e-Pak2 KD) exhibited a less profound effect on p16INK4a expression, the percentage of EdU-positive cells, and SAHF formation than overexpression of wild-type (WT) Pak2 kinase. Thus, overexpression of Pak2 is sufficient to promote cellular senescence in normal human fibroblasts in a manner dependent on the kinase activity of Pak2.
Fig. 2.
Overexpression of Pak2 promotes cellular senescence and reduces cell proliferation. (A) The levels of Pak2 and p16INK4a were analyzed by Western blotting using lysates from IMR90 cells without (empty vector, e-Tsin) and with overexpression of Pak2 (e-Pak2) or Pak2 kinase dead mutant (e-Pak2KD) (arrow indicates FLAG-tagged exogenous Pak2; the low band is endogenous Pak2). α-Tubulin was used as a loading control. (B) The p16INK4a mRNA levels were determined by qRT-PCR (n = 3, mean ± SEM, *P < 0.05, **P < 0.01). Values were normalized to GAPDH and then to D2 empty vector control (EV) to obtain relative fold expression. (C) Fluorescence microscopy imaging of DAPI and EdU (red) staining of IMR90 cells. EdU staining was performed at the indicated time points in IMR90 cells expressing empty vector (e-Tsin), Pak2 (e-Pak2), or Pak2 kinase dead mutant (e-Pak2KD). (D) The percentage of cells with EdU staining was calculated (mean ± SEM, **P < 0.01, n = 3 biological replicates). (E) Fluorescence microscopy imaging of DAPI staining of IMR90 cells. (F) The percentage of SAHF-positive cells (DAPI-dense foci) was calculated (mean ± SEM, **P < 0.01, n = 3 biological replicates).
Pak2 Is Required for de Novo H3.3 Deposition in Senescence Cells.
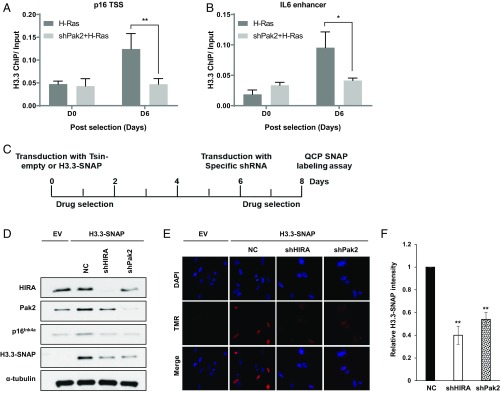
The histone H3 variant H3.3 is localized at genic regions, including promoters and enhancers, to regulate gene expression (27–29). Pak2 regulates HIRA-mediated nucleosome assembly of H3.3 (19); therefore, we tested whether Pak2 regulates H3.3 occupancy during oncogenic Ras-induced senescence. We found that H3.3 occupancy at the p16INK4a promoter and IL-6 enhancer increased on day 6 after Ras induction (Fig. 3 A and B), consistent with the increased expression of p16INK4a and IL-6 during this time frame (Fig. 1 D and F). Importantly, we observed that depletion of Pak2 resulted in reduced H3.3 occupancy at the p16INK4a promoter and IL-6 enhancer. These results support the idea that Pak2 affects gene expression, likely through its regulation of H3.3 occupancy at gene promoters and/or enhancers.
Fig. 3.
Pak2 is required for de novo H3.3 deposition in senescence cells. (A and B) H3.3 occupancy at the (A) p16INK4a transcription start site (TSS) and (B) IL-6 enhancer region was determined by ChIP-qPCR (n = 3, mean ± SEM, *P < 0.05, **P < 0.01). Values were normalized to input. (C) Experimental scheme for in vivo quench–chase–pulse (QCP)-SNAP labeling of de novo assembly of H3.3 in senescent IMR90 cells overexpressing H3.3-SNAP. (D) Western blotting analysis of lysates from IMR90 cells expressing H3.3-SNAP treated in C. Membranes were probed for the indicated antibodies. α-Tubulin was used as a loading control. (E and F) Pak2 and HIRA depletion compromised the deposition of newly synthesized H3.3 in senescent cells. Pak2 was depleted in H3.3-SNAP-tagged IMR90 cells. After old H3.3-SNAP was blocked using a nonfluorescent blocker, new H3.3-SNAP was labeled with TMR-STAR (the red fluorescent substrate for the SNAP tag) and visualized using fluorescence microscopy (E). The SNAP-TMR signal intensity was quantified and reported as mean ± SEM of 3 experiments (F, **P < 0.01). Depletion of HIRA was used as a control.
Next, we tested whether Pak2 was generally required for H3.3 nucleosome assembly in senescent cells because these cells depend on the HIRA-mediated nucleosome assembly of H3.3 at actively transcribed genes (13, 14). To monitor nucleosome assembly of H3.3 in senescent cells, we ectopically overexpressed SNAP-tagged histone H3.3 in IMR90 cells. After transduction and selection, cells were transduced with a lentivirus encoding a control shRNA or shRNA to knock down Pak2 or HIRA (as a control) (Fig. 3C). We examined the deposition of newly synthesized H3.3 in senescent cells by performing the “quench–chase–pulse” SNAP labeling assay (26, 30). We observed a dramatic loss of ectopic H3.3 expression upon HIRA depletion during senescence progression (Fig. 3D). This was not surprising as others have reported that senescent cells lacking the HIRA chaperone exhibit loss of H3.3 due to the instability of the unincorporated histone substrate (14). Interestingly, we observed that the loss of ectopic H3.3 upon Pak2 depletion is similar to that of HIRA depletion (Fig. 3D). Furthermore, analysis of the fluorescence intensity of individual cells revealed that depletion of Pak2, like HIRA, resulted in a marked reduction in H3.3 deposition (about 50%) compared with control cells (Fig. 3 E and F). Together, these results indicate that Pak2 is required for H3.3 incorporation in senescent cells.
Mouse Pak2 Is Required for the Stress-Induced Cellular Senescence in MEFs.
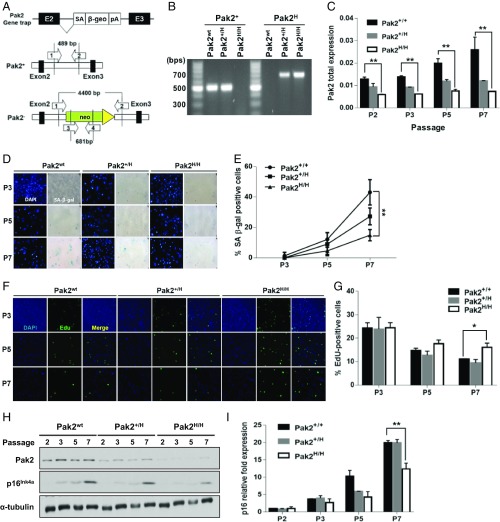
To further investigate the role of Pak2 in senescence, we tested whether Pak2 was involved in stress-induced premature senescence. Since complete knockout of Pak2 is embryonically lethal in mice (31, 32), we generated a Pak2 knockdown mouse line using a gene trap strategy (referring to H/H as homozygous for the gene trap allele) (Fig. 4 A and B). Pak2H/H MEFs expressed 30–40% of normal Pak2 levels compared with WT MEFs (Fig. 4C). To determine if these hypomorphic MEFs altered the acquisition of senescence, we passaged WT and Pak2H/H MEFs under standard culture conditions (20% oxygen), which induces MEF senescence due to oxidative stress (8), and monitored SA-β-gal activity, replicative potential (EdU staining), and p16INK4a expression. We found that the expression of Pak2 increased during MEF passaging (Fig. 4C). Moreover, the percentage of SA-β-gal-positive cells in WT Pak2 MEFs increased by passage 5 and reached about 40% by passage 7 (Fig. 4 D and E). In contrast, Pak2H/H MEF cultures contained considerably fewer SA-β-gal-positive cells (about 15% at passage 7) (Fig. 4 D and E), indicating that Pak2 depletion compromised oxidative stress–induced senescence. Consistent with this interpretation, while WT and Pak2H/H MEFs at passage 3 possessed similar percentages of EdU-positive cells, Pak2H/H MEFs had significantly more EdU-positive cells at passage 7 (Fig. 4 F and G) and exhibited reduced p16INK4a protein and mRNA levels (Fig. 4 H and I). These results indicate that Pak2 participates in oxidative stress–induced senescence.
Fig. 4.
Pak2 depletion attenuates oxidative stress–induced cellular senescence. (A) Schematic representation of the Pak2 gene-trapping strategy. The Pak2 gene-trap cassette was inserted into the intronic sequence between exon 2 and exon 3 of the Pak2 gene. The gene-trap cassette includes a splicing acceptor (SA), β-geo marker (β-gal and neomycin fusion), and a polyadenylation site. The arrows with numbers indicate primers used for genotyping. (B) PCR genotyping analysis of MEFs with the indicated Pak2 genotypes. The expected amplicon size of the WT allele and gene-trap allele was 500 and 700 base pairs (bp), respectively. (C) Pak2 expression in WT and Pak2+/H and Pak2H/H MEFs at various passages. Pak2 mRNA level was determined by qRT-PCR (mean ± SEM, **P < 0.01, n = 3 independent MEF lines for each genotype at each passage). Values were normalized to β-actin. (D and E) SA-β-galactosidase staining in MEF cells. The percentage of SA-β-gal-positive cells was determined for primary MEFs of the indicated genotype (Pak2 status) at the indicated passage (**P < 0.01, n = 3 independent MEF lines for each genotype at each passage). (F and G) EdU incorporation and DNA content of MEFs at different passages. The percentage of EdU-positive cells was determined for primary MEFs of each genotype at the indicated passage (n = 3 independent MEF lines for each genotype at each passage, mean ± SEM, *P < 0.05). The total number of cells was determined by DAPI staining. (H) Pak2 and p16INK4a protein expression level was tested by Western blotting using lysates from each genotype at each passage. α-Tubulin was used as a loading control. (I) The p16INK4a mRNA level was determined by qRT-PCR (n = 3 independent MEF lines for each genotype at each passage, mean ± SEM, **P < 0.01).
Depletion of Pak2 Alters Expression of Cellular Senescence-Associated Genes.
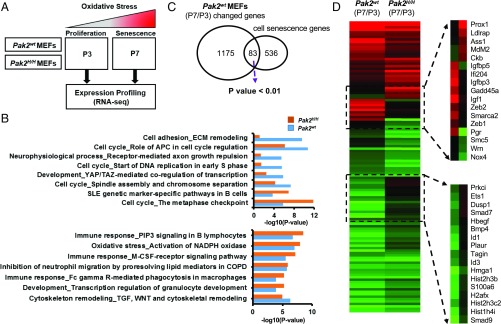
To gain further mechanistic insight into how Pak2 regulates senescence, we performed RNA-sequencing (RNA-seq) analysis on WT and Pak2H/H MEFs at 2 different passages (P3 and P7), which represent the proliferating and senescent states of WT MEFs, respectively (Fig. 5A). We identified 757 up-regulated genes and 501 down-regulated genes when WT MEF cells underwent senescence from passage 3 (P3) to passage 7 (P7). Similarly, 707 genes were up-regulated, and 486 were down-regulated in Pak2H/H MEF cells (SI Appendix, Table S1). Those genes with altered expression in both WT and Pak2H/H cells largely overlapped, suggesting that Pak2 depletion does not affect the overall gene expression program during passaging of MEFs. Of note, expression of several E2F target genes involved in the cell cycle and DNA repair and replication were down-regulated in WT MEFs at P7 and showed partial rescue in the context of Pak2 depletion (SI Appendix, Table S2). For instance, expression of several cell cycle regulators, including Prc1, Cks2, Cdc20, and Mybl2, decreased with senescence in both WT and Pak2 depletion cells. However, these genes were expressed in Pak2 depletion cells at least 2-fold higher than WT cells at P7. In addition, expression of Rpa2 and Bard1, which function in DNA synthesis and replication and DNA damage repair, respectively, was retained in Pak2-depleted cells during senescence, unlike WT cells. Moreover, pathway enrichment analysis by MetaCore from Thomson Reuters on those genes that changed expression during senescence in MEFs revealed that genes related to cell cycle, development, and cell adhesion were markedly different between WT and Pak2H/H MEFs in senescence, suggesting that Pak2 depletion affects expression of a subset of genes during senescence (Fig. 5B).
Fig. 5.
Pak2 regulates the expression of cellular senescence genes. (A) Experimental design for analyzing gene expression changes during oxidative stress–induced cellular senescence. RNA from Pak2 WT MEFs and Pak2H/H MEFs was collected at passages P3 and P7, representing proliferating and senescent states of WT MEF cells, respectively, for RNA-seq analysis. (B) Pathway enrichment analysis by METACORE between Pak2 WT MEFs and Pak2H/H MEFs during senescence (P7/P3 for each genotype), demonstrating enrichment of genes involved in cell cycle, cell adhesion, and development (Top). Bottom shows gene sets for which there was no enrichment. (C) Venn diagram showing the overlap between genes altered during senescence of WT MEFs and those associated with cell senescence (gene set from CSGene and Ingenuity Pathway Analysis). (D) Heat map showing gene expression changes of 120 senescence-associated genes as determined by RNA-seq in oxidative stress–induced senescence with and without Pak2 depletion; 34 genes (28%) are differentially expressed between Pak2wt MEFs and Pak2H/H MEFs during senescence.
Based on the published datasets, including the CSGene and Ingenuity Pathway Analysis databases (33), we identified 619 genes implicated in cellular senescence previously. Using this gene set, we found that the genes that were differentially expressed between P7 and P3 in both WT and Pak2H/H cells were significantly enriched for these cellular senescence genes (Fig. 5C and SI Appendix, Fig. S2A), suggesting that genes involved in senescence we identified in MEFs are likely also altered in other senescence models. As expected, most of the senescence-associated genes in WT and Pak2H/H cells were changed in a similar direction but not to the same degree. For instance, Mybl2 (B-Myb), an E2F target gene involved in cell cycle regulation, expression level decreased in both WT and Pak2H/H cells during senescence. However, its expression in Pak2H/H cells was about 3-fold greater than in WT cells at P7. We did not detect induction of any major SASP genes, including IL-6 and IL-8, during senescence, consistent with the observation that MEFs undergo senescence without activation of SASP under standard cell culture conditions, which include 20% oxygen (34). Interestingly, of all of the senescence-associated genes altered in either WT or Pak2H/H cells (120 genes), 34 genes (28%) were differentially expressed between WT and Pak2H/H MEFs during senescence. For example, the senescence inducers insulin-like growth factor 1 (IGF1) and IGF-binding protein (IGFBP5) were up-regulated in WT MEFs but not in Pak2H/H cells during senescence (Fig. 5D and SI Appendix, Fig. S2B). On the other hand, genes that were down-regulated in WT MEFs during senescence, such as Id1 and Hmga1, were up-regulated upon Pak2 depletion (Fig. 5D and SI Appendix, Fig. S2C). These results provide additional support for the idea that Pak2 depletion deregulated expression of some, but not all, genes involved in cellular senescence. Together, our results strongly support the idea that Pak2 regulates cellular senescence in murine cells and most likely in human cells by controlling the expression of a subset of genes critical for senescence.
Pak2 Depletion Delays the Onset of Aging Phenotypes and Increases Life Span in a Mouse Model of Accelerated Aging.
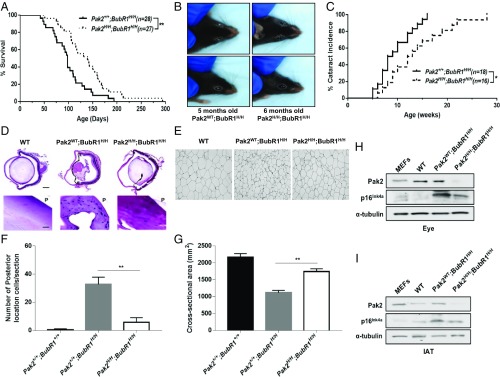
Our in vitro studies using human and mouse fibroblasts indicated that depletion of Pak2 delayed phenotypes associated with premature senescence. Since cellular senescence is causally linked to tissue aging in mammals, we hypothesized that Pak2 insufficiency slows the onset of phenotypes associated with aging in vivo. To test this idea, we utilized the BubR1 hypomorphic (BubR1H/H) premature aging (progeroid) mouse model, which has been used to demonstrate a link between accumulation of senescent cells and development of early onset aging phenotypes (10). We crossed BubR1 hypomorphic mice onto a Pak2H/H background (Pak2H/H;BubR1H/H mice) and examined the impact on progeroid phenotypes, including cataracts, body fat mass, and life span. Compared with control (Pak2+/+;BubR1H/H) mice, Pak2H/H; BubR1H/H mice had an approximate 30% life span extension (Fig. 6A). Furthermore, Pak2 insufficiency induced a modest, yet significant, delay in the latency of cataract formation in BubR1H/H mice (Fig. 6 B and C). Histological examination confirmed that Pak2H/H;BubR1H/H lenses contained fewer posteriorly located epithelial cells than Pak2+/+;BubR1H/H lenses (Fig. 6 D and F). Another early aging-associated phenotype of BubR1 hypomorphic mice is the loss of fat tissue due to accumulation of senescent cells. Quantitative NMR on 16-wk-old Pak2+/+;BubR1H/H, Pak2H/H;BubR1H/H, and WT mice revealed that while the percentage of body fat of Pak2+/+;BubR1H/H mice was reduced by about 3% compared with WT mice (SI Appendix, Fig. S3A), Pak2H/H;BubR1H/H mice retained a normal body fat content (SI Appendix, Fig. S3A). Further, histological analysis showed that the average area of individual fat cells in inguinal adipose tissue (IAT) of Pak2+/+;BubR1H/H mice declined significantly, while Pak2 insufficiency rescued this phenotype (Fig. 6 E–G). We also examined the effects of Pak2 deficiency on the expression of p16INK4a in eye and IAT tissues collected from experimental and control cohorts by RT-qPCR and Western blotting. Tissue from compound mutants exhibited a significant decrease in p16INK4a expression compared with that from BubR1 hypomorphic mice (Fig. 6 H and I and SI Appendix, Fig. S3 B and C). Interestingly, several progeroid phenotypes observed in BubR1 hypomorphic mice remained unchanged following loss of Pak2, including dwarfism, infertility, and physical function. These results indicate that Pak2 depletion rescued some, but not all, aging-associated phenotypes of the progeroid mouse model, consistent with the idea that Pak2 regulates expression of a subset of genes involved in cellular senescence.
Fig. 6.
Pak2 depletion delays the onset of aging phenotypes and increases life span in a mouse model of accelerated aging. (A) Overall survival curves for Pak2+/+;BubR1H/H (n = 28) and Pak2H/H;BubR1H/H (n = 27) mice. The median overall survival of combined Pak2H/H;BubR1H/H mice is 159 d, about a 30% extension in life span compared with Pak2+/+;BubR1H/H mice. (**P < 0.01.) (B and C) Incidence of cataract formation in Pak2+/+;BubR1H/H (n = 18) and Pak2H/H;BubR1H/H (n = 16) mice. (*P < 0.05.) (D and F) Cross section of a cataractous lens from 4-mo-old Pak2+/+;BubR1H/H and Pak2H/H;BubR1H/H mice stained with hematoxylin and eosin (H&E). Hematoxylin stain, showing blues in bottom images, indicates posteriorly located epithelial cells (mean ± SEM, **P < 0.01, n = 4 independent biological replicates). (E and G) Cross section of IAT from 4-mo-old Pak2+/+;BubR1H/H and Pak2H/H;BubR1H/H mice stained with H&E. Cross-sectional area was obtained using ImageJ software (mean ± SEM, n = 4 independent biological replicates). (**P < 0.01.) (H and I) Western blots of eye and IAT extracts from 4-mo-old Pak2+/+;BubR1H/H, Pak2H/H;BubR1H/H, and wild-type mice probed with Pak2 and p16INK4a antibody. α-Tubulin served as loading control. MEF lysates were used as an antibody-specific band control.
Discussion
Here, we describe a function for Pak2, a kinase-regulating HIRA-mediated nucleosome assembly, in cellular senescence and organismal aging. First, depletion of Pak2 in human fibroblasts (IMR90) delayed oncogenic Ras-induced premature senescence as detected by p16INK4a expression, SAHF formation, and SA-β-gal activity. Second, overexpression of Pak2 promoted cellular senescence in IMR90 cells. Furthermore, depletion of Pak2 in MEFs delayed oxidative stress–induced senescence. Altogether, we show, using 2 different in vitro senescence models, that Pak2 is a regulator of cellular senescence. To determine the biological consequence of Pak2 depletion, we analyzed aging phenotypes in the BubR1 hypomorphic mouse model. We found that depletion of Pak2 in vivo delayed eye and body fat aging phenotypes in addition to increasing animal life span. Taken together, these results support the idea that Pak2 is an important regulator of cell senescence and organismal aging.
How does Pak2 regulate cellular senescence? Previously, we proposed that one Pak2 substrate is histone H4 serine 47. We have shown that H4S47ph increases the association of HIRA with H3.3–H4, thereby promoting nucleosome assembly of H3.3–H4S47ph by HIRA (19). HIRA is known to promote cellular senescence via deposition of H3.3 and formation of SAHF in vitro. A recent study identified that the HIRA chaperone is required for incorporation of histones H3.3–H4 into chromatin in senescent cells (14). Nonproliferating senescent cells do not engage in the S phase of the cell cycle, thus limiting histone turnover and incorporation by replication-coupled nucleosome assembly. Therefore, senescent cells have an active DNA replication-independent nucleosome assembly pathway. The effect of Pak2 depletion and overexpression on cellular senescence is similar to published results on HIRA (14, 16–18, 30). Moreover, we showed that Pak2 is required for incorporation of H3.3–H4 into chromatin during the induction of premature senescence by oncogenic Ras. Therefore, it is tempting to speculate that Pak2 functions in cellular senescence at least in part through regulating HIRA-mediated nucleosome assembly of H3.3.
Although senescence is characterized by several nonexclusive markers, there is no universal marker of senescence to date due to the transcriptional heterogeneity of senescent cells (25). Our results from 2 different model systems with different senescent stimuli, oncogenic Ras-induced human normal fibroblasts, and oxidative stress–induced MEFs indicate that Pak2 depletion likely affects expression of a subset of genes involved in cellular senescence. First, Pak2 depletion consistently altered the expression of p16INK4a during senescence at least in part through regulating H3.3 deposition at the p16INK4a promoter. In agreement with this observation, RNA-seq analysis of Pak2 hypomorphic MEFs shows that the expression of some E2F target genes, the direct downstream events of the p16INK4a/Rb pathway, is partially rescued in Pak2 depletion cells during oxidative stress–induced senescence. This would, at least in part, explain why Pak2 depletion delays stress-induced cellular senescence and why Pak2 insufficiency in vivo delayed the onset of aging phenotypes but not fully rescue. Note that although we observed altered IL-6 gene expression upon Pak2 depletion during senescence in oncogenic Ras-induced human IMR90s, under our conditions, MEFs do not likely activate SASP (34). Taken together, it would be interesting to determine how Pak2 regulates expression of a subset of genes involved in p16INK4a/Rb and E2F axis as well as the SASP pathway during cellular senescence in different settings.
Senescent cells accumulate in aged tissues and contribute to age-related disease. Thus, removal of senescent cells is a potential candidate for the treatment of aging and age-related diseases. Indeed, recent animal studies have provided evidence that clearance of senescent cells delays features of aging (11, 12, 35, 36). For instance, studies using genetically engineered mice showed that removal of p16INK4a-expressing senescent cells delayed aging and protected the mice against age-related diseases (11, 12). In addition, treatment with 2 senolytic drugs (ABT-263 and ABT-737) selectively kills senescent cells in mice, rejuvenating aged tissues (35, 36). Here, we showed that deficiency of Pak2 in vivo increased life span and health span, with delayed cataract formation and increased body fat mass of the BubR1 progeroid mouse models. Therefore, we speculate that Pak2 may be a target for attenuating senescence and aging and age-related disease.
Materials and Methods
SI Appendix provides a detailed discussion of materials and methods used in this study. The RNA-seq dataset was deposited to the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE108081).
Cell Culture.
IMR90 (ATCC) cells and MEFs were maintained according to standard protocols. Detailed information on cell culture and gene transfer is described in SI Appendix.
H3-SNAP Staining and Chromatin Immunoprecipitation Assay and Real-Time PCR.
These assays were performed as described in SI Appendix. Primer sets used for chromatin immunoprecipitation (ChIP)-PCR are listed in SI Appendix, Table S3.
Immunofluorescence and Senescence Assays.
A detailed protocol is provided in SI Appendix.
Supplementary Material
Acknowledgments
We appreciate all members of the Z.Z. laboratory for helpful discussions. This work is supported by NIH Grant R35 GM118015 (to Z.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-sequencing dataset was deposited to the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE108081).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903847116/-/DCSupplemental.
References
- 1.Campisi J., d’Adda di Fagagna F., Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Collado M., Blasco M. A., Serrano M., Cellular senescence in cancer and aging. Cell 130, 223–233 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Kuilman T., Michaloglou C., Mooi W. J., Peeper D. S., The essence of senescence. Genes Dev. 24, 2463–2479 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lanigan F., Geraghty J. G., Bracken A. P., Transcriptional regulation of cellular senescence. Oncogene 30, 2901–2911 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W., Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Narita M., et al. , Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Collado M., Serrano M., The power and the promise of oncogene-induced senescence markers. Nat. Rev. Cancer 6, 472–476 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Parrinello S., et al. , Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 5, 741–747 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimauro T., David G., Chromatin modifications: The driving force of senescence and aging? Aging (Albany N.Y.) 1, 182–190 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker D. J., et al. , BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36, 744–749 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Baker D. J., et al. , Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker D. J., et al. , Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature 530, 184–189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang R., et al. , Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8, 19–30 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Rai T. S., et al. , HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev. 28, 2712–2725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Z., et al. , Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 28, 396–408 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye X., et al. , Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27, 2452–2465 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang R., Chen W., Adams P. D., Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27, 2343–2358 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duarte L. F., et al. , Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nat. Commun. 5, 5210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang B., et al. , Phosphorylation of H4 Ser 47 promotes HIRA-mediated nucleosome assembly. Genes Dev. 25, 1359–1364 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bokoch G. M., Biology of the p21-activated kinases. Annu. Rev. Biochem. 72, 743–781 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Sells M. A., et al. , Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr. Biol. 7, 202–210 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Shin E. Y., et al. , Phosphorylation of p85 beta PIX, a Rac/Cdc42-specific guanine nucleotide exchange factor, via the Ras/ERK/PAK2 pathway is required for basic fibroblast growth factor-induced neurite outgrowth. J. Biol. Chem. 277, 44417–44430 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Ito T. K., et al. , A crucial role for CDC42 in senescence-associated inflammation and atherosclerosis. PLoS One 9, e102186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakai T., et al. , Simultaneous early adenocarcinoma and mucosa-associated lymphoid tissue (MALT) lymphoma of the stomach associated with Helicobacter pylori infection. Gastric Cancer 6, 191–196 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Hernandez-Segura A., et al. , Unmasking transcriptional heterogeneity in senescent cells. Curr. Biol. 27, 2652–2660.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J. S., Zhang Z., O-linked N-acetylglucosamine transferase (OGT) interacts with the histone chaperone HIRA complex and regulates nucleosome assembly and cellular senescence. Proc. Natl. Acad. Sci. U.S.A. 113, E3213–E3220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H., et al. , RPA interacts with HIRA and regulates H3.3 deposition at gene regulatory elements in mammalian cells. Mol. Cell 65, 272–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filipescu D., Szenker E., Almouzni G., Developmental roles of histone H3 variants and their chaperones. Trends Genet. 29, 630–640 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Goldberg A. D., et al. , Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ray-Gallet D., et al. , Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44, 928–941 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Hofmann C., Shepelev M., Chernoff J., The genetics of Pak. J. Cell Sci. 117, 4343–4354 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Arias-Romero L. E., Chernoff J., A tale of two Paks. Biol. Cell 100, 97–108 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Zhao M., Chen L., Qu H., CSGene: A literature-based database for cell senescence genes and its application to identify critical cell aging pathways and associated diseases. Cell Death Dis. 7, e2053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coppé J. P., Desprez P. Y., Krtolica A., Campisi J., The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang J., et al. , Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 22, 78–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yosef R., et al. , Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat. Commun. 7, 11190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.