Key Points
Question
Can new causes of schizophrenia be identified within the Indian population, given its unique genetic makeup and environment?
Findings
In this genome-wide association study that included 3092 individuals from southern India, a genome-wide significant association with schizophrenia was observed on chromosome 8q24.3. Bioinformatic, cellular, and animal model evidence points to NAPRT1, a gene that encodes a key niacin metabolism enzyme, as the top gene within this locus.
Meaning
These findings suggest that the genotype of the top association signal and niacin status may be relevant in schizophrenia susceptibility and treatment.
This genome-wide association study evaluates genetic loci for schizophrenia in a unique Indian population.
Abstract
Importance
Genome-wide association studies (GWASs) in European populations have identified more than 100 schizophrenia-associated loci. A schizophrenia GWAS in a unique Indian population offers novel findings.
Objective
To discover and functionally evaluate genetic loci for schizophrenia in a GWAS of a unique Indian population.
Design, Setting, and Participants
This GWAS included a sample of affected individuals, family members, and unrelated cases and controls. Three thousand ninety-two individuals were recruited and diagnostically ascertained via medical records, hospitals, clinics, and clinical networks in Chennai and surrounding regions. Affected participants fulfilled DSM-IV diagnostic criteria for schizophrenia. Unrelated control participants had no personal or family history of psychotic disorder. Recruitment, genotyping, and analysis occurred in consecutive phases beginning January 1, 2001. Recruitment was completed on February 28, 2018, and genotyping and analysis are ongoing.
Main Outcomes and Measures
Associations of single-nucleotide polymorphisms and gene expression with schizophrenia.
Results
The study population included 1321 participants with schizophrenia, 885 family controls, and 886 unrelated controls. Among participants with schizophrenia, mean (SD) age was 39.1 (11.4) years, and 52.7% were male. This sample demonstrated uniform ethnicity, a degree of inbreeding, and negligible rates of substance abuse. A novel genome-wide significant association was observed between schizophrenia and a chromosome 8q24.3 locus (rs10866912, allele A; odds ratio [OR], 1.27 [95% CI, 1.17-1.38]; P = 4.35 × 10−8) that attracted support in the schizophrenia Psychiatric Genomics Consortium 2 data (rs10866912, allele A; OR, 1.04 [95% CI, 1.02-1.06]; P = 7.56 × 10−4). This locus has undergone natural selection, with the risk allele A declining in frequency from India (approximately 72%) to Europe (approximately 43%). rs10866912 directly modifies the abundance of the nicotinate phosphoribosyltransferase gene (NAPRT1) transcript in brain cortex (normalized effect size, 0.79; 95% CI, 0.6-1.0; P = 5.8 × 10−13). NAPRT1 encodes a key enzyme for niacin metabolism. In Indian lymphoblastoid cell lines, (risk) allele A of rs10866912 was associated with NAPRT1 downregulation (AA: 0.74, n = 21; CC: 1.56, n = 17; P = .004). Preliminary zebrafish data further suggest that partial loss of function of NAPRT1 leads to abnormal brain development.
Conclusions and Relevance
Bioinformatic analyses and cellular and zebrafish gene expression studies implicate NAPRT1 as a novel susceptibility gene. Given this gene’s role in niacin metabolism and the evidence for niacin deficiency provoking schizophrenialike symptoms in neuropsychiatric diseases such as pellagra and Hartnup disease, these results suggest that the rs10866912 genotype and niacin status may have implications for schizophrenia susceptibility and treatment.
Introduction
Schizophrenia is a severe mental illness characterized by delusional beliefs, hallucinations, disordered speech, and deficits in cognitive, emotional, and social behavior. This cross-cultural disorder has a lifetime prevalence of approximately 1%1 and significant mortality. The disease imposes a substantial burden on individuals, families, and societies, ranking twelfth globally in years lived with disability.2 Onset is typically in early adulthood with a frequently chronic trajectory. Schizophrenia’s defining pathophysiology is poorly understood, and current treatments have limited efficacy. High heritability (approximately 0.8)3 has driven the search for genetic variants, the identification of which will contribute to unravelling disease mechanisms and provide essential biological understanding needed for improved evidence-based therapeutics and personalized treatments.
Most schizophrenia genome-wide association studies (GWASs) have been conducted in Europeans, with a minority in African American and East Asian populations. Common genetic variants are ancient and shared across ethnicities, with evidence of shared common genetic variation for schizophrenia between major global populations.4 The Psychiatric Genomics Consortium 2 (PGC2),5 using predominantly European ancestry samples, has provided major insights into the genetic basis of the disorder by identifying 108 genome-wide significant loci from a study of 36 989 cases and 113 075 controls. These loci explain approximately 7% of the liability to disease, but whole-genome analysis methods6 suggest that variants tagged by common single-nucleotide polymorphisms (SNPs) (minor allele frequency, > .01) on GWAS arrays collectively explain approximately 50% of the genetic liability.7 The remaining heritability will likely be accounted for by additional common SNPs of small effect size identified as GWAS sample sizes increase7 and by rare variants that are largely population specific8 and poorly correlated with common SNPs in GWASs.6 We report herein, to our knowledge, the first schizophrenia GWAS in an Indian population, recruited from ethnically homogeneous schizophrenia pedigrees and unrelated cases and controls.9
Methods
Study Participants
All participants gave written informed consent, and the study was approved by relevant institutional ethics committees at each participating institution. This study followed the Strengthening the Reporting of Genetic Association Studies (STREGA) reporting guideline. Our sample was recruited at The Schizophrenia Research Foundation, Chennai, India, and consists almost exclusively of individuals of Tamil ethnicity (>97%) (eTables 1-3 in the Supplement). Within Tamil ethnicity, we initially focused on Tamil Brahmin and other Brahmin castes to maximize sample homogeneity; we subsequently recruited other castes. We used standardized instruments (administered in Tamil where necessary), including the Diagnostic Interview for Genetic Studies,10 the Family Interview for Genetic Studies,11 DSM-IV diagnostic criteria, and the consensus diagnostic procedure (eMethods in the Supplement). The GWAS sample included (1) a family data set ascertained for multiple affected family members (of 1376 individuals, 505 with schizophrenia [36.7%]) and (2) a case-control data set (of 1716 individuals, 816 with schizophrenia [47.6%]). The total sample included 3092 individuals, 1321 (42.7%) of whom had schizophrenia. For details of genotyping and preimputation, imputation, and postimputation quality control, see the eMethods and eFigures 1-3 in the Supplement. Recruitment, genotyping, and analysis occurred in consecutive phases beginning January 1, 2001. Recruitment was completed on February 28, 2018, and genotyping and analysis are ongoing.
SNP Heritability and Genome-wide Association Analyses
To quantify the proportion of variance attributed to all genome-wide SNPs (SNP-based heritability) and to test for population stratification, we applied the GREML function in GCTA (genome-wide complex trait analysis), version 1.24.72212 (eMethods and eFigure 4 in the Supplement). We used GCTA’s mixed linear model–based function to conduct the analysis in the case-control and family data sets independently (eMethods in the Supplement). The final meta-analysis of both data sets did not show any inflation (λ = 1.00 [eFigure 5 in the Supplement] and λ = 0.98 [eFigure 6 in the Supplement]), confirming that our leading association signal is not a false-positive finding.
Transethnic Meta-analysis
Given the ancestral difference between our sample and the predominantly European PGC2, we conducted transethnic meta-analysis using RE2C, an extension of METASOFT,13 which accounts for heterogeneity in effect sizes of SNPs across populations. This process evaluated whether the addition of Tamil Nadu samples to PGC2 resulted in novel and/or statistically stronger, already identified genome-wide significant loci (eMethods in the Supplement).
Post-GWAS Extended Bioinformatic Analyses
We conducted the following analyses: (1) genomic profile risk scoring to capture the contribution to disease of the many SNPs that do not reach genome-wide statistical significance14; (2) statistical fine mapping analysis of Indian GWAS data with functional annotation data sets using PAINTOR, version 3.115,16 to determine which SNP at our top locus was associated with most functional annotations and reveal the SNP with the highest posterior probability of being causal and therefore the one to be prioritized for functional investigation; (3) expression quantitative trait loci (eQTL) analyses to investigate the regulatory effects of our top SNP on gene expression; (4) natural selection analyses to examine the global pattern of allele frequency distribution for the lead SNP to detect signatures of positive selection; (5) vegetarian diet analysis to test whether this environmental factor was associated with disease status in individuals homozygous for the risk allele of the lead SNP; (6) SMR (summary data–based mendelian randomization), version 0.712 analyses17 to identify genes within the top locus whose expression levels were associated with schizophrenia; (7) pathway and network connectivity enrichment analyses to identify gene sets enriched for association with disease and risk genes enriched for association with tissue types, respectively; and (8) transethnic genetic correlation for schizophrenia between India and Europe to evaluate the similarity of genetic architecture of this disease. Details are provided in eMethods in the Supplement.
Post-GWAS Functional Analyses
To assess the effect of allele-specific expression, we measured expression of genes at the rs10866912 locus in lymphoblastoid cell lines established from 60 individuals (30 cases and 30 controls) within the study population (eMethods in the Supplement). We identified the presence of 4 distinct haplotypes across the rs10866912 locus, encompassing 3054 of 3092 of the study population (98.8%) (eTable 5 in the Supplement). For naprt1 knockdown in zebrafish, we used a microRNA-mediated gene-silencing approach optimized for zebrafish as described previously18 (eMethods and eTable 4 in the Supplement). We used the 2-sided t test with significance set at P < .05 to compare findings.
Results
Clinical Sample
Of the 1321 individuals with schizophrenia (mean [SD] age, 39.1 [11.4] years), 170 (13.8%) were from consanguineous families (uncle-niece, first cousin, or second cousin); all were living with their families; 696 (52.7%) were male and 625 (47.3%) were female; 433 (32.8%) had a tertiary level of educational attainment; and 368 (27.9%) had at least part-time employment (>30% time). The clinical phenotype was homogeneous, and negative symptoms were frequently observed (660 [50.0%]). The negligible to low rates of comorbid alcohol and substance abuse further enhanced the phenotypic homogeneity of this sample (eMethods and eTables 1-3 in the Supplement).
SNP Heritability
SNP heritability on the liability scale was estimated to be 0.287 (standard error, 0.073) (eFigure 4 in the Supplement), falling within the expected range. Additional heritability analyses confirmed the lack of population stratification in our data set (eMethods in the Supplement).
Genome-wide Association Analyses
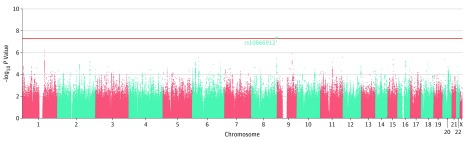
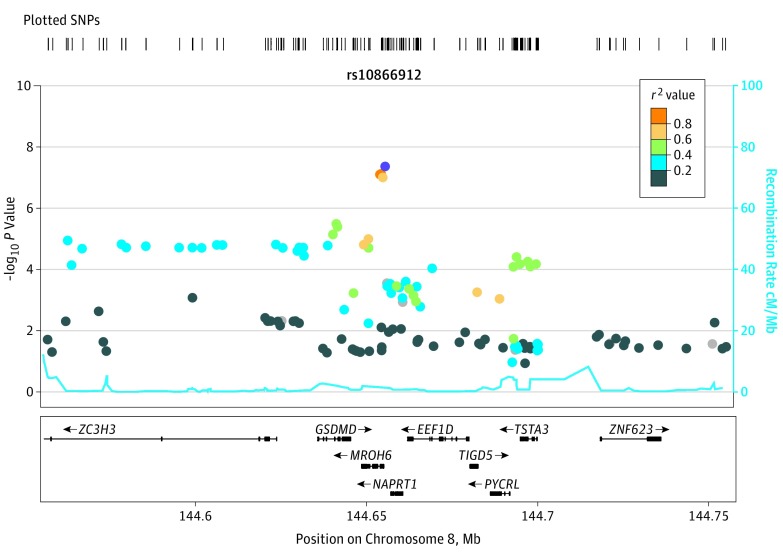
The meta-analysis genome-wide association results are summarized in Figure 1. We observed a genome-wide significant locus on chromosome 8q24.3 (rs10866912, allele A; odds ratio [OR], 1.27 [95% CI, 1.17-1.38]; P = 4.35 × 10−8; chr8:144655315; hg19). The index SNP is located approximately 7 kilobases from the 5′ end of MROH6 [GenBank NM_001100878], lying within a linkage disequilibrium (LD) block defined by r2>0.6. The 6 genes within this block are GSDMD (GenBank NM_024736), MROH6, NAPRT1 (GenBank NM_145201), EEF1D (GenBank NM_032378), TIGD5 (GenBank NM_032862), and PYCRL (GenBank NM_023078) (Figure 2). We found no signs of confounding due to caste status (666 Brahmins and 2426 non-Brahmins; rs10866912, allele C; β coefficient, 0.007; standard error [SE], 0.005; P = .22) in our data set. The top locus was replicated in PGC2 (rs10866912, allele A; OR, 1.04 [95% CI, 1.02-1.06]; P = 7.56 × 10−4) (eFigure 7 in the Supplement), with the same direction of effect in the Indian (OR, 1.27) and PGC2 (OR, 1.04) data sets.
Figure 1. Manhattan Plot of Observed Schizophrenia Association Signals.
Manhattan plot shows genome-wide schizophrenia associations in 3092 individuals (1321 cases and 1771 controls) from Tamil Nadu, India. The phase three 1000 Genome Project South Asian population was used to calculate linkage disequilibrium. The x-axis shows the chromosomal position, and the y-axis shows the significance of association (−log10P value). The horizontal red line represents the level of genome-wide significance (P = 5 × 10−8). Our top genome-wide significant locus is situated on chromosome 8q24.3 (rs10866912, P = 4.35 × 10−8).
Figure 2. Regional Plot of Chromosome 8q24.3 Locus (100-Kilobase Window).
The index single-nucleotide polymorphism (SNP) rs10866912 is colored purple, with other SNPs colored according to the degree of linkage disequilibrium (measured by r2 value) with the index SNP. SNPs with missing linkage disequilibrium information are shown in gray. The x-axis shows the SNP locus position on chromosome 8 (GRCh37/hg19 build). The y-axis shows the significance of association (−log10 P value) in our Indian population. Nine genes are located within the 100-kilobase window, with the direction of transcription (upstream/downstream) being annotated with arrows. cM indicates centimorgans; Mb, megabase.
We observed no significant associations between chromosome X SNPs and disease status. Further verification based on imputation batch-based meta-analysis showed consistent results (eTable 6 in the Supplement).
Transethnic Meta-analysis
Transethnic meta-analysis revealed a stronger genome-wide significance for our top locus (rs10866912) after meta-analysis with PGC2, increasing from P = 4.35 × 10−8 for India (rs10866912, allele A; OR, 1.27; 95% CI, 1.17-1.38) to P = 2.09 × 10−9 for India plus PGC2 (β coefficient, 0.05; SE, 0) (eTable 7 and eFigures 8-10 in the Supplement). This more significant P value indicates that our top SNP is in strong LD with the causal variant in the PGC2 and Indian data sets. Moreover, of the 108 PGC2 genome-wide significant loci, 23 became more significantly associated after this meta-analysis (eTable 8 in the Supplement). In addition, at our top locus, we sought to observe the difference in LD block pattern in the 2 populations that may facilitate fine mapping of the true causal SNP(s) (eFigure 9 and eFigure 10 in the Supplement).
Post-GWAS Extended Bioinformatic Analyses
For genomic profile risk scoring, PGC2 SNPs below the P value threshold of .05 had the strongest association with schizophrenia in India (eResults, eTable 9, and eFigure 11 in the Supplement). Statistical fine mapping analysis revealed that, of all the SNPs in our top locus, the top SNP rs10866912 had the highest probability of being a causal SNP (z score, 5.476; posterior probability, 1.0) (eFigure 12 in the Supplement).
To investigate possible molecular mechanisms underlying the top SNP, we interrogated eQTL databases for cis-eQTL effects of this SNP on neighboring genes. Using the Genotype-Tissue Expression project (GTEx, version 7),19 the top SNP had the strongest regulatory effect for NAPRT1 expression in brain cortex (normalized effect size, 0.79; 95% CI, 0.6-1.0; P = 5.8 × 10−13 [eFigure 13 in the Supplement]). In the Brain eQTL almanac,20 the index SNP was significantly associated with NAPRT1 expression across all brain regions (meta-analysis P = 2.9 × 10−13) and in particular with frontal cortex (133 samples; AA expression [log2 scale], 8.7; CC expression, 9.1; genotype counts: 23 for AA, 67 for AC, and 43 for CC; P = 4.7 × 10−7) and temporal cortex (133 samples; AA expression [log2 scale], 8.7; CC expression, 8.9; genotype counts: 23 for AA, 67 for AC, and 43 for CC; P = 8.7 × 10−7) (eTable 10 and eFigure 14 in the Supplement); other strongly associated SNPs in the locus (rs10866911, rs4873803, and rs4873804) also showed association with NAPRT1 expression. Importantly, the lead SNP and these other associated SNPs were not associated with expression of any other genes in the LD block (r2 > 0.6) [eTable 10 in the Supplement]). Spatiotemporal analyses using the human brain transcriptome21 indicated that the pattern of NAPRT1 expression was relatively high in most brain regions (highest in the hippocampus and neocortex) during early prenatal developmental stages, with a gradual decline in expression during postnatal development. Of the 5 other genes in the LD block, TIGD5 had a similar pattern of perinatal expression to NAPRT1 (eFigure 15A-E in the Supplement). In addition, the most active transcription factor binding to the top locus, POL2RA (NM_000937), is highly expressed during the early prenatal developmental stages in all brain regions (eFigure 15F in the Supplement), with decreasing expression during later prenatal and early postnatal stages.
For natural selection analyses, the worldwide pattern of allele frequency distribution for the top SNP revealed a declining frequency of the risk allele (A) from African (approximately 96%) to Indian (approximately 72%) to European (approximately 43%) populations (eFigure 16 in the Supplement). Natural selection analysis using 1000 Genome Project data suggested that the top locus has undergone higher selection in Europeans than other world populations (integrated haplotype score for the European population, −2.52 [P < .01]; for the South Asian population, −1.03 [P = .30]). Similarly, the cross-population extended haplotype homozygosity test revealed more suggestive evidence of natural selection in Europeans compared with other populations (−2.16 [P < .03] [eFigure 17D in the Supplement]). The results of other tests of natural selection, corroborating these results, are shown in eFigure 17A to C in the Supplement. We observed a higher proportion of vegetarian diet in cases compared with controls (161 of 754 [21.4%] vs 154 of 783 [19.7%]; P = .67). Detailed results of other post-GWAS extended analyses, including SMR, pathway and network connectivity enrichment, transethnic genetic correlation for schizophrenia between India and Europe, and PGC2 replication in our Indian data set, are found in eResults, eFigures 18 to 21, and eTables 11 and 12 in the Supplement.
Post-GWAS Functional Analyses
Messenger RNA Expression Associated With the rs10866912 Locus
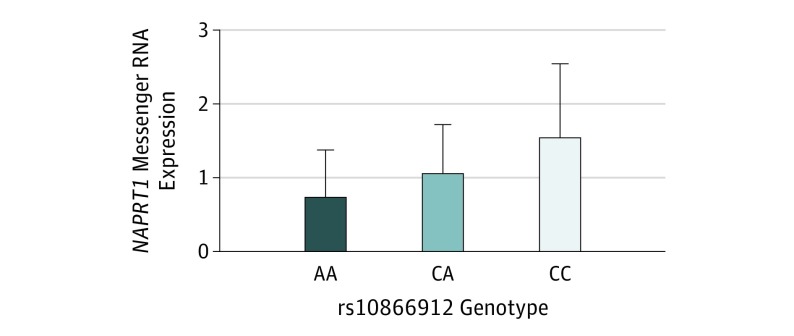
We sought to measure the effects of the index SNP on gene expression at the top locus using lymphoblastoid cell lines established from study participants. Our top locus (defined by r2>0.6) contains the following 6 genes: GSDMD, MROH6, NAPRT1, EEF1D, TIGD5, and PYCRL. All except NAPRT1 showed negligible gene expression. By contrast, NAPRT1 showed a dose-response association with the rs10866912 genotype, the A risk allele downregulating expression of NAPRT1 for AA vs CC (AA: 0.74, n = 21; CC: 1.56, n = 17; P = .004 for AA vs CC) (Figure 3).
Figure 3. Expression of NAPRT1 in Indian Lymphoblastoid Cell Lines.
Expression of NAPRT1 was analyzed in lymphoblastoid cell lines from 20 individuals (10 cases and 10 controls) of each rs10866912 genotype, including AA homozygous for risk allele, CA heterozygous, and CC homozygous for the protective allele. NAPRT1 expression shows a dose-response association with the rs10866912 genotype in these samples, with the A risk allele downregulating expression. P = .004 for AA vs CC.
This result is consistent with the CommonMind Consortium22 eQTL finding that the risk allele (A) of the index SNP, rs10866912, significantly downregulates NAPRT1 in a large postmortem collection of human brains (allele A; β coefficient, −0.45; P = 1.40×10−28). This result is also consistent with Brain eQTL almanac results showing allele-specific expression for rs10866912 (AA > AC > CC) across all measured brain regions, with the most significant result occurring in the frontal cortex (133 samples; AA expression [log2 scale], 8.7; CC expression, 9.1; genotype counts: 23 for AA, 67 for AC, and 43 for CC; P = 4.7 × 10−7) and with AA genotypes showing the least expression (eFigure 14 in the Supplement).
Zebrafish In Vivo Analysis
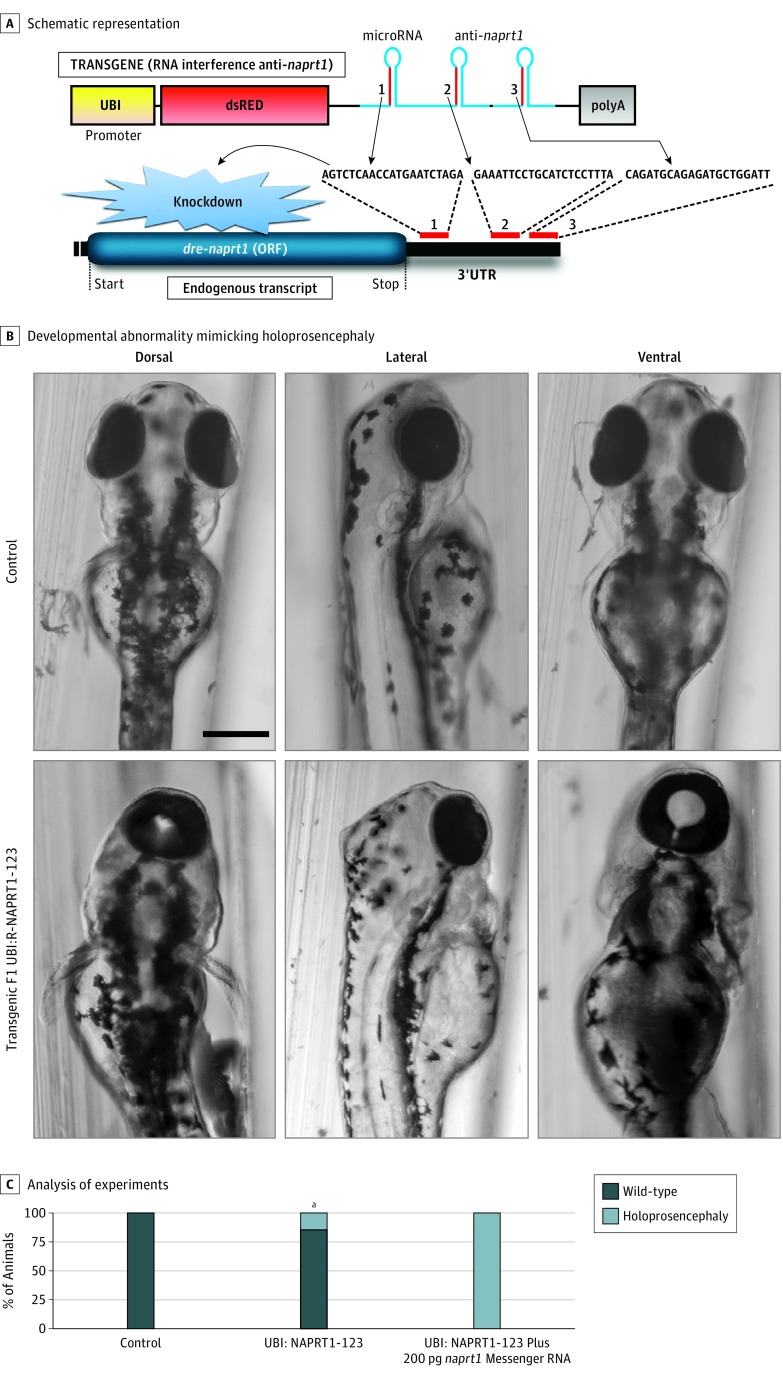
We further investigated in vivo the pathogenicity of NAPRT1 loss of function using the zebrafish model. Based on a 2016 microRNA-mediated gene-silencing technology developed for the zebrafish,23 we generated a transgene presenting a ubiquitous promoter driving expression of the dsRED marker and 3 different synthetic microRNAs targeting dre-naprt1 messenger RNA (Figure 4A). We injected this construct into the zebrafish along with transposase to promote its integration into the zebrafish genome for stable and heritable expression. Interestingly, we found that, in the injected F0 animals and in a stable F1 line (named UBI-NAPRT1-123), the expression of anti-naprt1 microRNAs trigger the developmental defect holoprosencephaly (Figure 4B), the most common congenital forebrain malformation, in which the brain fails to fully divide into 2 hemispheres. Importantly, injection of wild-type dre-naprt1 messenger RNA without microRNA target sequences into F1 UBI-NAPRT1-123 zebrafish was sufficient to fully rescue this developmental phenotype (Figure 4C), confirming both efficient endogenous naprt1 knockdown and the specificity of this defect. Further study will be required to fully understand the role of naprt1 in the development of the fish.
Figure 4. In Vivo Functional Analysis of Zebrafish naprt1 Loss of Function.
A, Schematic representation of the F1 line UBI:NAPRT1-123 transgene used to generate stable transgenic naprt1 knockdown in zebrafish. B, Transgenic F1 UBI:NAPRT1-123 fish present a developmental abnormality mimicking holoprosencephaly. Black bar indicates 50 μm. C, Rescue experiments demonstrated that injection of dre-naprt1 (with a custom 3′UTR not recognized by the anti-naprt1 microRNAs) rescued the holoprosencephaly observed in the F1 UBI:NAPRT1-123 clutches. Analysis was performed 3 times with 50 animals per condition. ORF indicates open reading frame.
aP ≤ .02 compared with control.
Discussion
Most schizophrenia GWASs have been conducted in Europeans, with a minority in Asian and African American populations. Our results provide further evidence for a cross-ethnic polygenic contribution to schizophrenia, although caution is required when estimating disease across disparate world ancestries given differences in genetic (LD patterns, allele frequencies, and genetic architecture)24 and environmental factors.25
To our knowledge, this report describes the first Indian schizophrenia GWAS. Our sample (n = 3092) is characterized by a uniform ethnicity (>97% Tamil), a degree of inbreeding (typical F_HET coefficient range, −0.2 to 0.2), and a homogeneous schizophrenia phenotype with negligible rates of alcohol and illicit substance abuse, features that can be advantageous for genome-wide analyses.
We identified a schizophrenia locus in 8q24.3 that surpassed the threshold of genome-wide significance. It has not been possible to test replication of this finding in a separate Indian schizophrenia GWAS because, to our knowledge, none currently exist. However, our locus was replicated in the European ancestry schizophrenia PGC2 GWAS with similar direction and smaller magnitude of effect.5 Analyses suggest that this locus has undergone natural selection in Europeans, with the derived allele (C) undergoing positive selection and the ancestral or risk allele (A) declining in frequency from approximately 72% in India to approximately 43% in Europe.26 The high frequency of this Indian risk allele suggests it may have arisen recently, transforming from a neutral allele (ie, no effect on disease phenotype) to a risk allele, triggered perhaps by interaction with an environmental or lifestyle factor.24
Although the lead SNP is located closer to MROH6 than to NAPRT1 and other top SNPs lie within MROH6, cis-eQTL evidence and cellular expression studies indicate that these SNPs regulate the expression of NAPRT1. This result is consistent with analytic studies reporting that only a minority of associated genes are physically closest to the top GWAS SNP.17 To our knowledge, no prior genome-wide significant evidence exists for association between NAPRT1 and psychiatric traits, although 1 early study reports a nominal P value for a synonymous SNP, rs2290416, in exon 10 being associated with attention-deficit/hyperactivity disorder.27
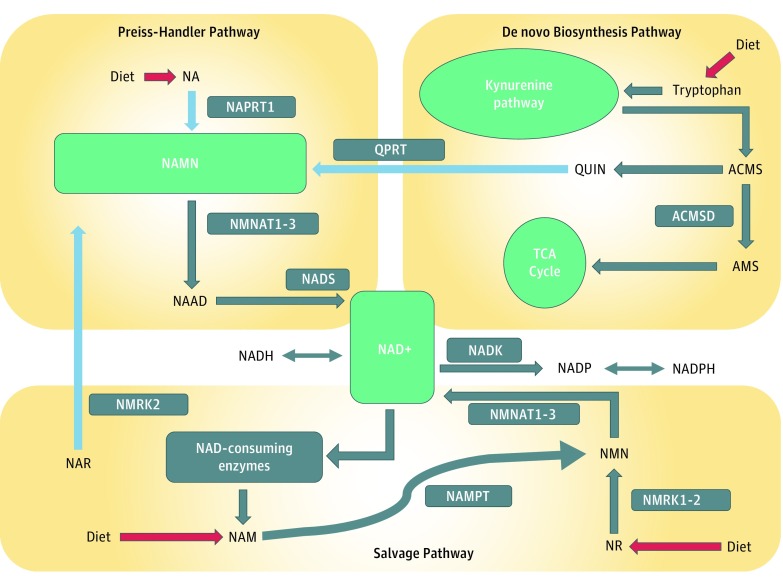
NAPRT1 is the key rate-limiting enzyme involved in metabolizing nicotinic acid (NA), the major dietary source of niacin28 (Preiss-Handler pathway, Figure 5). NAPRT1 converts NA to nicotinic acid mononucleotide (NAMN), which is then converted to nicotinamide adenine dinucleotide (NAD+), a ubiquitous coenzyme fundamental to all living cells and vital for cellular biochemistry, energy metabolism, and DNA synthesis. Levels of NAMN are maintained by 3 distinct pathways, but only the Preiss-Handler pathway operates in the brain under most conditions; thus, NAPRT1 is crucial for contributing to the production of NAMN in the brain.29 We suggest that NAPRT1 underexpression could lead to NAD+ deficiency, known to produce (1) neurodegenerative disorders such as pellagra and Hartnup disease, which can present with schizophrenialike symptoms such as auditory hallucinations, persecutory delusions, and delusional parasitosis30; and (2) oxidative stress via its negative effect on NADP levels, which have been implicated in schizophrenia.31
Figure 5. Nicotinamide Adenine Dinucleotide (NAD+) Biosynthetic Pathways.
Nicotinic acid mononucleotide (NAMN) levels are maintained by 3 independent pathways (see light blue arrows). First, the Preiss-Handler pathway uses dietary nicotinic acid (NA) and the enzyme nicotinic acid phosphoribosyltransferase (NAPRT1) to generate NAMN, which is then transformed into nicotinic acid adenine dinucleotide (NAAD) by NAMN transferase (NMNAT) and finally into NAD+ by NAD+ synthase (NADS). Second, the de novo synthesis pathway of NAD+ from tryptophan occurs through the kynurenine pathway to produce 2-amino-3-carboxymuconate semialdehyde (ACMS). This metabolite is converted nonenzymatically into quinolinc acid (QUIN), which is transformed into NAMN by quinolinate phosphoribosyltransferase (QPRT). Third, nicotinic acid riboside (NAR) is converted into NAMN by nicotinamide riboside kinase (NMRK2). The second and third pathways converge with the Preiss-Handler pathway via NAMN. The NAD salvage pathway recycles the nicotinamide generated as a by-product of the enzymatic activities of NAD+-consuming enzymes. Nicotinamide phosphoribosyltransferase (NAMPT) recycles nicotinamide into nicotinamide mononucleotide (NMN). NAD+ is converted to nicotinamide adenine dinucleotide phosphate (NADP) by NAD+ kinase (NADK). Pathways are described at https://reactome.org/content/detail/R-HSA-197264 and by Verdin et al.28 ACMSD indicates aminocarboxymuconate semialdehyde decarboxylase; AMS, α-aminomuconate semialdehyde; NADH, reduced form of nicotinamide adenine dinucleotide; NADPH, reduced form of nicotinamide adenine dinucleotide phosphate; NAM, nicotinamide; NR, nicotinamide riboside; and TCA, tricarboxylic acid cycle.
We further investigated whether a tryptophan-deficient (vegetarian) diet was potentially contributing to disease in the subset of 1627 samples homozygous for the rs10866912 risk allele (A). We observed a trend toward a higher proportion of vegetarian diet in cases compared with controls (21.4% vs 19.7%; P = .07), which appears to be evidence of gene-environment interaction, although further metabolic studies in this cohort would be necessary to establish the association. In the absence of this evidence and antedating clinical trials, we speculate that vegetarian diet and homozygosity of the rs10866912 risk allele (A) may additively contribute to schizophrenia susceptibility in this Indian cohort. In this context, niacin supplementation has been used to prevent congenital malformations associated with NAD+ deficiency.32
Based on post-GWAS bioinformatics analyses, cellular gene expression, and zebrafish knockdown data, NAPRT1 is the prime candidate at our locus. Our in vivo experiments showed that partial loss of function of NAPRT1 can cause abnormal brain development (holoprosencephaly) in early zebrafish (Figure 4). Holoprosencephaly is the most common human forebrain developmental defect, and many factors are known to be involved in pathogenesis.33 Evidence for such defects occurring in schizophrenia include a recent report by the ENIGMA Schizophrenia DTI Working Group34 of widespread microstructural deficits across the white matter skeleton in a large sample (1963 individuals with schizophrenia and 2359 healthy controls), with the corpus callosum being 1 of 2 regions showing greatest effects. Although only a modest effect was seen with common variants in NAPRT1 on schizophrenia risk, the fact that NAPRT1 plays an essential role in NAD+ metabolism in the brain suggests the possibility of a global effect on brain development.
Strengths and Limitations
One limitation of our study is the sample size, because large sample sizes are needed for GWAS discovery in schizophrenia. We maximized the sample size over many years according to available resources. As a consequence of this strategy, another limitation is that genotyping was conducted in 3 batches over time and used 3 different genotyping arrays. Our careful quality control steps and statistical analyses have provided strategies to overcome potential confounding introduced through these technical batch effects, but given this limitation, further replication of our results is needed before strong conclusions can be drawn. We note that this study includes one of the few Indian schizophrenia research cohorts and, to our knowledge, the only one that has undergone a GWAS. Importantly, supported by our DNA analyses, this cohort is from a single region with uniform ethnicity. Another strength of our study is the comprehensive phenotyping using the consensus diagnostic procedure (see eMethods in the Supplement) that documented a “pure” schizophrenia phenotype and negligible substance abuse.
Conclusions
We have identified NAPRT1 as a novel susceptibility gene for schizophrenia in an Indian population. Further studies in larger Indian samples are needed to replicate this novel genome-wide significant association.
eMethods. Data Collection and Analysis
eResults. Findings
eFigure 1. MDS Analysis Using 1000g SAS Population
eFigure 2. MDS Analysis Using 1000g SAS Populations and Our Indian Data Set
eFigure 3. MDS Analysis Using 1000g Super Populations
eFigure 4. SNP Heritability
eFigure 5. Q-Q Plot for Indian Meta-analysis
eFigure 6. Q-Q Plot for Indian Batch–Based Meta-analysis
eFigure 7. Regional Plot of Indian Chromosome 8q24.3 Locus (100-Kb Window) in PGC2 Data Set
eFigure 8. Manhattan Plot for PGC2-Indian Meta-analysis Using Metasoft-RE2C
eFigure 9. Regional Plot of Chromosome 8q24.3 Locus (100-Kb Window) in Cross Population Meta-analysis Results With SAS LD
eFigure 10. Regional Plot of Chromosome 8q24.3 Locus (100Kb window) in Cross Population Meta-analysis Results With EUR LD
eFigure 11. Genomic Profile Risk (PRS) Scoring
eFigure 12. Fine Mapping of Indian Top Locus
eFigure 13. Summary of the eQTL Results of NAPRT1in GTEx Heat Map
eFigure 14. Lead SNP, rs10866912, Is Associated (eQTL) With Brain Expression of NAPRT1
eFigure 15. Spatiotemporal Brain Expression Profiling of Genes in Top Locus
eFigure 16. Worldwide Pattern of Allele Frequencies for Indian Top SNP (rs10866912)
eFigure 17. Natural Selection Analyses Between South Asian and European Populations
eFigure 18. Manhattan Plot for Gene-Based Association Using MAGMA Version 1.06
eFigure 19. Network Connectivity Results for Indian Data Set Using MAGNUM
eFigure 20. Regional Plot of Indian SNP Data Set at PGC2 Top Locus (Chromosome 6p22:28303247-287712247; GRCh37/hg19 Build)
eFigure 21. Regional Plot of Top Replicated PGC2 Locus (Chromosome 2q32.3:193848340-194028340; GRCh37/hg19 Build) in Indian SNP Data Set
eTable 1. Indian Clinical Data
eTable 2. Clinical Characteristics
eTable 3. Symptoms and Treatment
eTable 4. Forward and Reverse Primer Sequences for DRE-NAPRT1 mRNA
eTable 5. Study Population Haplotypes Present in Our Top Locus
eTable 6. Imputation Batch-Based Meta-analysis for Top Indian Locus
eTable 7. Transethnic Meta-analysis Results for Indian Top Locus With PGC2 Using Metasoft-RE2C
eTable 8. Transethnic Meta-analysis Results for 108 PGC2 Significant Regions Using Metasoft-RE2C
eTable 9. Risk Profile Scoring–Sign Test P-values for SNPs (PT<.05) Sharing the Same Direction of Effects
eTable 10. BRAINEAC cis-eQTL Results for the Top SNP and Genes in Indian Top Locus (100-Kb Window)
eTable 11. SMR Results Using the Brain-eMeta eQTL Data Set for Our Top SNP, rs10866912
eTable 12. Indian Replication Results for PGC2 Top Loci (96/108)
eReferences
References
- 1.Saha S, Chant D, Welham J, McGrath J. A systematic review of the prevalence of schizophrenia. PLoS Med. 2005;2(5):e141. doi: 10.1371/journal.pmed.0020141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GBD 2015 Disease and Injury Incidence and Prevalence Collaborators Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388(10053):1545-1602. doi: 10.1016/S0140-6736(16)31678-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60(12):1187-1192. doi: 10.1001/archpsyc.60.12.1187 [DOI] [PubMed] [Google Scholar]
- 4.de Candia TR, Lee SH, Yang J, et al. ; International Schizophrenia Consortium; Molecular Genetics of Schizophrenia Collaboration . Additive genetic variation in schizophrenia risk is shared by populations of African and European descent. Am J Hum Genet. 2013;93(3):463-470. doi: 10.1016/j.ajhg.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421-427. doi: 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SH, DeCandia TR, Ripke S, et al. ; Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ); International Schizophrenia Consortium (ISC); Molecular Genetics of Schizophrenia Collaboration (MGS) . Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44(3):247-250. doi: 10.1038/ng.1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visscher PM, Wray NR, Zhang Q, et al. . 10 Years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101(1):5-22. doi: 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson MR, Wegmann D, Ehm MG, et al. . An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337(6090):100-104. doi: 10.1126/science.1217876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thara R, Srinivasan T, John S, et al. . Design and clinical characteristics of a homogeneous schizophrenia pedigree sample from Tamil Nadu, India. Aust N Z J Psychiatry. 2009;43(6):561-570. doi: 10.1080/00048670902873631 [DOI] [PubMed] [Google Scholar]
- 10.Nurnberger JI Jr, Blehar MC, Kaufmann CA, et al. ; NIMH Genetics Initiative . Diagnostic Interview for Genetic Studies: rationale, unique features, and training. Arch Gen Psychiatry. 1994;51(11):849-859. doi: 10.1001/archpsyc.1994.03950110009002 [DOI] [PubMed] [Google Scholar]
- 11.Maxwell ME. Family Interview for Genetic Studies (FIGS): A Manual for FIGS. Bethesda, MD: Clinical Neurogenetics Branch, Intramural Research Program, NIMH; 1992. [Google Scholar]
- 12.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76-82. doi: 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee CH, Eskin E, Han B. Increasing the power of meta-analysis of genome-wide association studies to detect heterogeneous effects. Bioinformatics. 2017;33(14):i379-i388. doi: 10.1093/bioinformatics/btx242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell SM, Wray NR, Stone JL, et al. ; International Schizophrenia Consortium . Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748-752. doi: 10.1038/nature08185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kichaev G, Roytman M, Johnson R, et al. . Improved methods for multi-trait fine mapping of pleiotropic risk loci. Bioinformatics. 2017;33(2):248-255. doi: 10.1093/bioinformatics/btw615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kichaev G, Yang WY, Lindstrom S, et al. . Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10(10):e1004722. doi: 10.1371/journal.pgen.1004722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Z, Zhang F, Hu H, et al. . Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481-487. doi: 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
- 18.Giacomotto J, Rinkwitz S, Becker TS. Effective heritable gene knockdown in zebrafish using synthetic microRNAs. Nat Commun. 2015;6:7378. doi: 10.1038/ncomms8378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580-585. doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trabzuni D, Ryten M, Walker R, et al. . Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem. 2011;119(2):275-282. doi: 10.1111/j.1471-4159.2011.07432.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang HJ, Kawasawa YI, Cheng F, et al. . Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483-489. doi: 10.1038/nature10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fromer M, Roussos P, Sieberts SK, et al. . Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19(11):1442-1453. doi: 10.1038/nn.4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laird AS, Mackovski N, Rinkwitz S, Becker TS, Giacomotto J. Tissue-specific models of spinal muscular atrophy confirm a critical role of SMN in motor neurons from embryonic to adult stages. Hum Mol Genet. 2016;25(9):1728-1738. doi: 10.1093/hmg/ddw044 [DOI] [PubMed] [Google Scholar]
- 24.Martin AR, Gignoux CR, Walters RK, et al. . Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100(4):635-649. doi: 10.1016/j.ajhg.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorlov IP, Gorlova OY, Amos CI. Allelic spectra of risk SNPs are different for environment/lifestyle dependent versus independent diseases. PLoS Genet. 2015;11(7):e1005371. doi: 10.1371/journal.pgen.1005371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcus JH, Novembre J. Visualizing the geography of genetic variants. Bioinformatics. 2017;33(4):594-595. doi: 10.1093/bioinformatics/btw643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lasky-Su J, Neale BM, Franke B, et al. . Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(8):1345-1354. doi: 10.1002/ajmg.b.30867 [DOI] [PubMed] [Google Scholar]
- 28.Verdin E. NAD+ in aging, metabolism, and neurodegeneration. Science. 2015;350(6265):1208-1213. doi: 10.1126/science.aac4854 [DOI] [PubMed] [Google Scholar]
- 29.Galassi L, Di Stefano M, Brunetti L, et al. . Characterization of human nicotinate phosphoribosyltransferase: kinetic studies, structure prediction and functional analysis by site-directed mutagenesis. Biochimie. 2012;94(2):300-309. doi: 10.1016/j.biochi.2011.06.033 [DOI] [PubMed] [Google Scholar]
- 30.Prakash R, Gandotra S, Singh LK, Das B, Lakra A. Rapid resolution of delusional parasitosis in pellagra with niacin augmentation therapy. Gen Hosp Psychiatry. 2008;30(6):581-584. doi: 10.1016/j.genhosppsych.2008.04.011 [DOI] [PubMed] [Google Scholar]
- 31.Hardingham GE, Do KQ. Linking early-life NMDAR hypofunction and oxidative stress in schizophrenia pathogenesis. Nat Rev Neurosci. 2016;17(2):125-134. doi: 10.1038/nrn.2015.19 [DOI] [PubMed] [Google Scholar]
- 32.Shi H, Enriquez A, Rapadas M, et al. . NAD deficiency, congenital malformations, and niacin supplementation. N Engl J Med. 2017;377(6):544-552. doi: 10.1056/NEJMoa1616361 [DOI] [PubMed] [Google Scholar]
- 33.Petryk A, Graf D, Marcucio R. Holoprosencephaly: signaling interactions between the brain and the face, the environment and the genes, and the phenotypic variability in animal models and humans. Wiley Interdiscip Rev Dev Biol. 2015;4(1):17-32. doi: 10.1002/wdev.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly S, Jahanshad N, Zalesky A, et al. . Widespread white matter microstructural differences in schizophrenia across 4322 individuals: results from the ENIGMA Schizophrenia DTI Working Group. Mol Psychiatry. 2018;23(5):1261-1269. doi: 10.1038/mp.2017.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Data Collection and Analysis
eResults. Findings
eFigure 1. MDS Analysis Using 1000g SAS Population
eFigure 2. MDS Analysis Using 1000g SAS Populations and Our Indian Data Set
eFigure 3. MDS Analysis Using 1000g Super Populations
eFigure 4. SNP Heritability
eFigure 5. Q-Q Plot for Indian Meta-analysis
eFigure 6. Q-Q Plot for Indian Batch–Based Meta-analysis
eFigure 7. Regional Plot of Indian Chromosome 8q24.3 Locus (100-Kb Window) in PGC2 Data Set
eFigure 8. Manhattan Plot for PGC2-Indian Meta-analysis Using Metasoft-RE2C
eFigure 9. Regional Plot of Chromosome 8q24.3 Locus (100-Kb Window) in Cross Population Meta-analysis Results With SAS LD
eFigure 10. Regional Plot of Chromosome 8q24.3 Locus (100Kb window) in Cross Population Meta-analysis Results With EUR LD
eFigure 11. Genomic Profile Risk (PRS) Scoring
eFigure 12. Fine Mapping of Indian Top Locus
eFigure 13. Summary of the eQTL Results of NAPRT1in GTEx Heat Map
eFigure 14. Lead SNP, rs10866912, Is Associated (eQTL) With Brain Expression of NAPRT1
eFigure 15. Spatiotemporal Brain Expression Profiling of Genes in Top Locus
eFigure 16. Worldwide Pattern of Allele Frequencies for Indian Top SNP (rs10866912)
eFigure 17. Natural Selection Analyses Between South Asian and European Populations
eFigure 18. Manhattan Plot for Gene-Based Association Using MAGMA Version 1.06
eFigure 19. Network Connectivity Results for Indian Data Set Using MAGNUM
eFigure 20. Regional Plot of Indian SNP Data Set at PGC2 Top Locus (Chromosome 6p22:28303247-287712247; GRCh37/hg19 Build)
eFigure 21. Regional Plot of Top Replicated PGC2 Locus (Chromosome 2q32.3:193848340-194028340; GRCh37/hg19 Build) in Indian SNP Data Set
eTable 1. Indian Clinical Data
eTable 2. Clinical Characteristics
eTable 3. Symptoms and Treatment
eTable 4. Forward and Reverse Primer Sequences for DRE-NAPRT1 mRNA
eTable 5. Study Population Haplotypes Present in Our Top Locus
eTable 6. Imputation Batch-Based Meta-analysis for Top Indian Locus
eTable 7. Transethnic Meta-analysis Results for Indian Top Locus With PGC2 Using Metasoft-RE2C
eTable 8. Transethnic Meta-analysis Results for 108 PGC2 Significant Regions Using Metasoft-RE2C
eTable 9. Risk Profile Scoring–Sign Test P-values for SNPs (PT<.05) Sharing the Same Direction of Effects
eTable 10. BRAINEAC cis-eQTL Results for the Top SNP and Genes in Indian Top Locus (100-Kb Window)
eTable 11. SMR Results Using the Brain-eMeta eQTL Data Set for Our Top SNP, rs10866912
eTable 12. Indian Replication Results for PGC2 Top Loci (96/108)
eReferences