Abstract
The complex smear microbiota colonizing the surface of red-smear cheese fundamentally impacts the ripening process, appearance and shelf life of cheese. To decipher the prokaryotic composition of the cheese smear microbiome, the surface of a semi-hard surface ripened cheese was studied post-ripening by culture-based and culture-independent molecular approaches. The aim was to detect potential bacterial alterations in the composition of the cheese smear microbiota resulting from cheese storage in vacuum film-prepackaging, which is often accompanied by the development of a surface smear defect. Next-generation sequencing of amplified 16S rRNA gene fragments revealed an unexpected high diversity of a total of 132 different genera from the domains Bacteria and Archaea on the cheese surface. Beside typical smear organisms, our study revealed the presence of several microorganisms so far not associated with cheese, but related to milk, farm and cheese dairy environments. A 16S ribosomal RNA based analysis from total RNA identified the major metabolically active populations in the cheese surface smear as Actinobacteria of the genera Corynebacterium, Brevibacterium, Brachybacterium and Agrococcus. Comparison of data on a higher phylogenetic level revealed distinct differences in the composition of the cheese smear microbiome from the different samples. While the proportions of Proteobacteria and Bacteroidetes were increased in the smear of prepacked samples and in particular in defective smear, staphylococci showed an opposite trend and turned out to be strongly decreased in defective smear. In conclusion, next-generation sequencing of amplified 16S rRNA genes and 16S rRNA from total RNA extracts provided a much deeper insight into the bacterial composition of the cheese smear microbiota. The observed shifts in the microbial composition of samples from defect surface smear suggest that certain members of the Proteobacteria contribute to the observed negative organoleptic properties of the surface smear of cheese after prepacking in plastic foil.
Keywords: microbiome, next-generation sequencing, surface ripened cheese, red-smear cheese, smear defect
1. Introduction
Cheese is a fermented milk based food product evolved 8000 years ago in the Middle East. Production of cheese was originally intended for bio-preservation of a nutritious food for human nourishment and developed nowadays to a premium food product comprising more than 1000 cheese varieties [1]. The varieties of surface-ripened cheeses have a long tradition in Europe and are diversified into bacterial surface-ripened red-smear cheeses and mold surface-ripened cheeses [2]–[4].
Bacterial surface-ripened red-smear cheeses such as Comté, Gruyère, Limburger or Tilsit are characterized by a high moisture content and a high ratio of surface area to volume [5]. These physical properties favor the influence of the microbial proteolytic and lipolytic activity on the intense aroma flavor formation, characteristic for this type of cheese [4],[6]. The viscous, red-orange smear developing on the cheese surface during ripening determines not only the organoleptic properties of these cheeses, but further serves as a protection against dryness and aroma loss [4],[7]. Moreover, fast growth of the surface microbiota during ripening and the inhibition properties of the inhabiting microorganisms protect the cheese from colonization by pathogenic foodborne bacteria or mycotoxin producing molds [8]–[10].
The surface-ripening of smear cheeses is a dynamic process and starts with the deacidification of the cheese surface by yeast, followed by a successive growth of characteristic red smear bacteria [2],[11],[12]. A smear layer composed of a multi-microbial species ecosystem develops naturally on the cheese surface when the young cheese is exposed to air at a high relative humidity (>95%) and suitable temperatures (13–15 °C) [2],[13]. To enhance the establishment of a stable surface microbiota and to suppress growth of molds, the surface is smeared periodically with brine during the ripening period [2],[6]. Naturally occurring indigenous smear microorganisms originate from the cheese-making environment such as cheese brines, raw milk and from the “house microbiota” in the ripening rooms or cheese-making premises, implying wooden shelves, vats, tap water and the room air [11],[14]–[17]. At the end of the ripening process mature red-smear cheese surface is mainly composed of Gram-positive bacteria and salt-tolerant yeast [2],[4].
For quality control and food safety issues the understanding of microbial composition, dynamics and interactions is essential to improve cheese taste, flavor, texture and safety [18]. Traditionally, the composition of the complex cheese surface microbiota was studied by culture-based methods followed either by phenotypic or genotypic identification of isolates [19]–[21]. Hitherto, less time-consuming molecular methods complement or replace cultivation dependent approaches [19],[21]. State-of-the-art molecular methods do not only allow to analyze the microbial diversity, but also monitor microbial dynamics and development over space and time [18],[22]. Application of next-generation sequencing technologies for high-throughput analyses opens new dimensions in the area of food microbiology by providing a deeper and more comprehensive insight into the community structure and the metabolic activity of microbial ecosystems [18],[21],[23],[24].
The aim of this study was to analyze the cheese surface smear microbiota in order to investigate the role of the smear bacteria in the development of cheese smear quality deterioration. Today, smear-ripened cheese is stored in vacuum film-prepackaging, which promotes the occurrence of a smear defect, appearing as a very humid, sticky and unpleasant off-flavored cheese surface [25]. Previous studies ruled out a contribution of yeast and pointed to alterations in the composition or the metabolic activity of smear bacteria due to changes in physico-chemical characteristics of the cheese smear after vacuum film-prepackaging [25],[26]. Consequently, the aim of this study was to perform an in-depth analysis to compare the bacterial composition and to identify metabolically active members of the surface microbiota in unpacked and vacuum film-prepacked cheese samples of differing qualities. For this purpose, the prokaryotic composition of the surface smear microbiota was analyzed by next-generation sequencing of multiplexed 16S rDNA amplicons. Furthermore, the metabolically active members of the cheese smear microbiota were determined by next-generation sequencing of 16S rRNA from total RNA extracts. The resulting 16S rDNA and rRNA data were compared with concurrent culture-based findings of the same smear samples.
2. Materials and methods
2.1. Cheese samples
The red-smear cheese investigated in this study was a Swiss semi-hard cheese variety with protection of origin (AOP), produced from raw milk from cows fed without silage. The use of additives in the cheese production is prohibited and the produced cheeses mature on spruce boards for at least 75 days at 13–14 °C and at relative air humidity of approximately 90%.
For analysis and comparison of the surface smear microbiome of cheeses differing in their smear quality all investigated samples derived from cheeses produced on the same day from the same milk and ripened in the same cheese cellar. All cheeses were treated equally until packaging at the end of the ripening period. Then the cheese portions were vacuum prepacked in gas- and watertight standard plastic film (Csf) by the manufacturer according to the standard procedure applied and subsequently stored at 8 °C during three weeks. A temperature of 8 °C was administered to mimic the storage conditions in a display cabinet or a domestic fridge [27],[28]. As a control, one cheese portion was not film-prepacked and further stored in the cheese cellar until sampling. During storage, the unpacked and one of the vacuum film-prepacked cheese portions (vacuum film-prepacked non-defective) retained a good quality surface smear (defined as a dry, non-smeary surface with a typical cheese odor). Other vacuum film-prepacked cheese portions developed a strong smear defect during storage (vacuum film-prepacked defective), which was characterized according to a previously described defect definition scheme [26] as a humid and smeary consistency with a very intensive off-odor pronounced in an animal-like flavor.
2.2. Culture-based analysis of cheese surface smear samples
Cheese surface smear samples from cheeses with differing smear quality were collected as previously described [25]. In brief, a rectangle of 8 cm2 size and 2–3 mm thickness (corresponding to 2.3 ± 0.2 g weight) was cut out of the cheese surface by a sterile knife. The cheese smear samples were diluted in 50 ml pre-warmed (45 °C) peptone solution (pH 7) composed of 1% (w/v) peptone from casein and 8.5% (w/v) NaCl (both Merck, Darmstadt, Germany) and thereafter homogenized during 4 min in a homogenizer (Colworth Stomacher 400; Bender & Hobein, Zurich, Switzerland). Colony forming units were determined by surface plating of serial dilutions on different non-selective and selective growth media. Total aerobic mesophilic and total anaerobic bacterial colony counts were determined on Tryptic Glucose Yeast Agar (TGYA; Biolife, Milano, Italy) supplemented with 1% (w/v) peptone from casein (Merck, Darmstadt, Germany), heterotrophic marine bacteria (halotolerant bacteria) on Marine Broth Agar (MB; Becton Dickinson AG, Allschwil, Switzerland), facultative anaerobic halophilic and alcaliphilic (FAHA) bacteria on Glucose Yeast Extract Peptone Beef Extract (GYPB) Agar [10], enterococci on KF Streptococcus Agar (KFS; Becton Dickinson AG, Allschwil, Switzerland), strictly anaerobic bacteria on Differential Reinforced Clostridial Medium (DRCM; Becton Dickinson AG, Allschwil, Switzerland) supplemented with 1 mg l−1 resazurin (Sigma-Aldrich, Steinheim, Germany) and 500 mg l−1 cysteine (VWR, Dietikon, Switzerland), and Enterobacteriaceae on Violet Red Bile Glucose Agar (VRBG; Biolife, Milano, Italy). The incubation properties were as follows: TGYA aerobic respectively anaerobic (3 days 30 °C aerobic respectively anaerobic incubation, followed by 7 days 22 °C room temperature incubation under day light), MB Agar (7 days 22 °C room temperature aerobic incubation under day light), GYPB Agar (4 days 30 °C anaerobic incubation), KFS Agar (3 days 42 °C aerobic incubation), DRCM Agar (4 days 22 °C strict anaerobic incubation in anaerobic chamber), VRBG Agar (1 day 37 °C aerobic incubation), PY Agar (3 days 30 °C aerobic incubation for yeast colony counts, respectively 6 days 30 °C aerobic incubation for mold colony counts). Colony counts were determined as weighted average.
2.3. Next-generation sequencing analysis of amplified 16S rRNA gene fragments
From each sample type (unpacked, vacuum film-prepacked non-defective and vacuum film-prepacked defective) total DNA and total RNA was extracted for subsequent next-generation sequencing analysis.
DNA was extracted from 350 µl of the previously prepared cheese smear raw extract by administering the GenEluteTM Bacterial Genomic DNA Kit (Sigma-Aldrich Chemie GmbH, Steinheim, Germany) according to the manufacturer's instruction with the following modifications: The heated incubation times were doubled. For disruption of rigid cell walls a mechanical cell treatment step was performed additionally by adding 500 µl of zirconia-silica beads (Ø 0.1 mm and 0.5 mm, mixed 1:1 (v/v)) (Carl Roth GmbH Co. KG, Karlsruhe, Germany) to the lysate followed by vigorously shaking in a bead beater (Retsch mill MM301; Retsch GMBH & CO.KG, Haan, Germany) during 2.5 min at a maximal frequency of 30 s−1 and afterwards allowed to cool on ice for 3 min. The mechanical cell disruption step was repeated before centrifugation of the assay during 1 min at 12,000 × g. The liquid supernatant, except the fat layer on top, was transferred to a column of the GenEluteTM Bacterial Genomic DNA Kit and DNA was further extracted as suggested by the manufacturer. The DNA concentration was determined using a ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, USA).
For generation of 16S rDNA amplicons different primer sets were applied, covering the whole 16S rDNA sequence corresponding to position EC1–EC1510 of the Escherichia coli 16S rRNA gene [29], and leading to size fractions of approximately 450 bp, 550 bp or 650 bp length (Table 1). PCR was administered as described before in a 50 µl total volume using 53 ng metagenomic DNA and 25 pmol primers, each in a separate PCR reaction. Properties of the amplification steps were the following: Initial denaturation at 98 °C for 2 min, followed by 25 cycles, each of 10 s 98 °C, 15 s at the according annealing temperature listed in Table 1 for the according primers applied, 30 s 72 °C and a final extension at 72 °C during 5 min. Separate PCR products were pooled for generation of a sequencing library.
Table 1. Primers used for amplification of 16S rRNA genes.
| Target | 16S rRNA gene region | Primer name | Sequence (5′–3′) | Temp (°C) | References |
| Bacteria | V1–V3 | TPU1 | AGAGTTTGATCMTGGCTCAG | 61 | [30]–[33] |
| U529R | ACCGCGGCKGCTGGC | ||||
| V3–V4 | Uni340F | CCTACGGGRBGCASCAG | 56 | [34] | |
| Uni806R | GGACTACNNGGGTATCTAAT | ||||
| V4–V6 | U515F | GTGCCAGCMGCCGCGGTAA | 70 | [30],[35]–[37] | |
| R1064 | CGACRRCCATGCANCACCT | ||||
| V5–V8 | TPU4 | GGATTAGATACCCTGGTAGTCC | 63 | [32],[38],[39] | |
| BS-R1407 | GACGGGCGGTGWGTRC | ||||
| V7–V9 | BS-F1099 | GYAACGAGCGCAACCC | 56 | [39] | |
| pB-00545 | ACGGYTACCTTGTTACG | [40] | |||
| Archaea | V1–V3 | A2Fa_minusGA | TTCCGGTTGATCCYGCCG | 59 | [41] |
| U529R | ACCGCGGCKGCTGGC | [30],[33] | |||
| V4–V6 | U515F_minus5′base | TGCCAGCMGCCGCGGTAA | 70 | [30],[35]–[37] | |
| 1048arcR-major | CGRCGGCCATGCACCWC | ||||
| 1048arcR-minor | CGRCRGCCATGYACCWC | ||||
| V7–V8 | A1040F-minusC | GAGAGGWGGTGCATGGC | 63 | [33],[42] | |
| UA1406R | ACGGGCGGTGWGTRCAA |
Amplified 16S rDNA amplicons were sequenced using GS FLX Titanium chemistry on a 454 Genome Sequencer FLX platform (Roche Diagnostics Ltd., Burgess Hill, West Sussex, United Kingdom) according to Roche 454 protocols. Sequences not passing the FLX quality controls were not considered for bioinformatics analysis, the 454-specific portions of the primers were trimmed, the raw sequences were sorted according to tag sequences and reads with low quality scores (quality score below 40) and short lengths (less than 200 bp) were removed. The resulting sequence reads were checked for chimera constructs using the program UCHIME based on the alignments to full length good quality and non-chimeric reference 16S rDNA sequences (downloaded from RDP) [43].
OTU assignment was performed using the RDP (Ribosomal Database Project) Classifier v2.7 [44] with taxonomic classification based on NCBI taxonomy (http://www.ncbi.nlm.nih.gov/taxonomy). OTUs represented by less than 5 reads were excluded from further analysis. The resulting OTUs were consolidated across different sampling points and normalized to percent counts using R statistical package.
2.4. Next generation sequencing of total RNA for taxonomic assignment of 16S rRNA transcripts
Total RNA was extracted from cheese surface smear samples applying the protocol described by Monnet and colleagues [45] and converted to cDNA. Subsequently the cDNA was sheared and the region corresponding to the 16S rRNA was selectively enriched by a set-up called vectorette [46], using the universal primer U515F as specific primer and 13 cycles of amplification. After size selection (about 600 bp) the libraries were sequenced using GS FLX Titanium chemistry on a 454 Genome Sequencer FLX platform (Roche Diagnostics Ltd., Burgess Hill, West Sussex, United Kingdom) according to Roche 454 protocols. Sequences not passing the FLX quality controls were not considered for bioinformatics analysis, the 454-specific portions of the primers were trimmed, the raw sequences were sorted according to tag sequences and reads with low quality scores (below 40) and short lengths (less than 200 bp) were removed. The resulting 16S rRNA reads were used for subsequent taxonomic classification of the metabolic active cheese surface microbiota as described previously for the sequencing of the 16S rDNA amplicons.
2.5. Nucleotide sequence accession numbers
The data from DNA and cDNA (RNA) sequencing are available through the NCBI GenBank Sequence Read Archive (SRA) database under the accession number SRP063818.
2.6. Statistical analysis
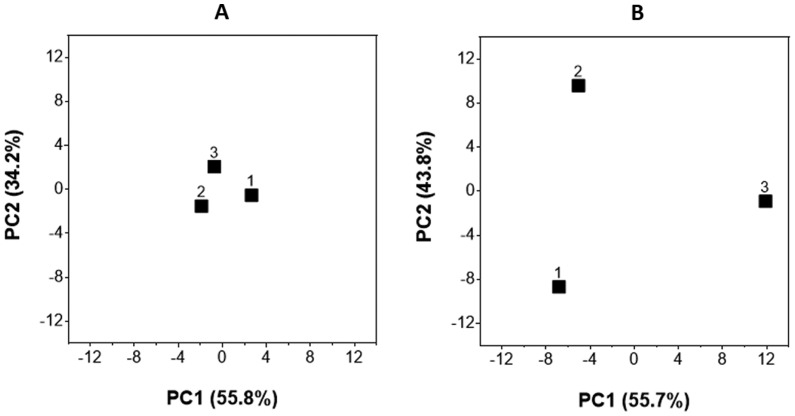
For visualization of the ordering of the cheese smear samples in reduced dimension (2D space) and for comparison between the technical approaches, principal components (PC) analysis was conducted on the correlation matrix calculated with JMP 10.0 software (SAS Institute AG, Wallisellen, Switzerland). Principal components for the first three dimensions were computed and the results for the first two principal components (PC) were visualized as PC plots. To evaluate the correlation between the applied microbial analysis techniques, results for DNA and RNA analysis were combined for each approach.
3. Results
3.1. Culture based analysis of microbial composition
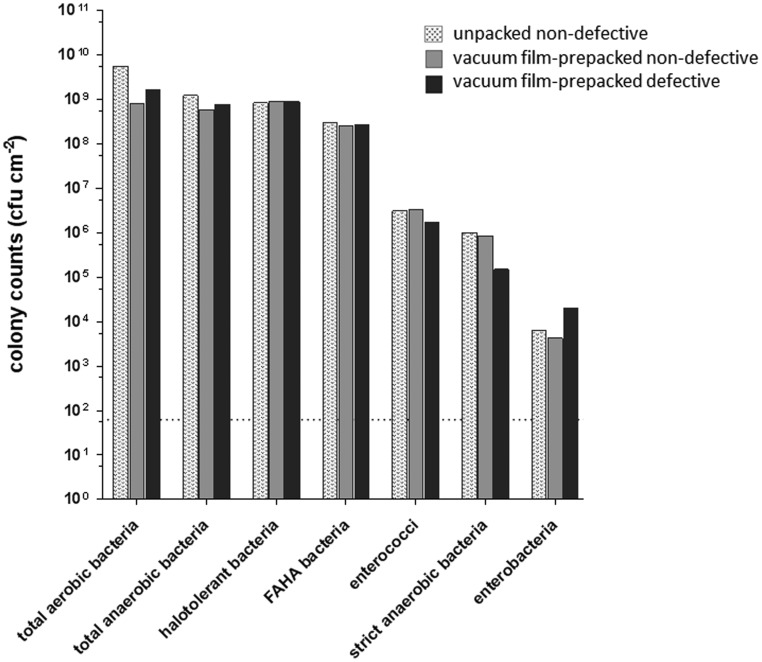
Weighted average colony counts in cfu cm−2 were determined for unpacked, vacuum film-prepacked non-defective and vacuum film-prepacked defective cheese surface smear samples. The results from the culture based investigation and comparison of the microbial composition of the differing surface smear samples are provided in Figure 1. Counts for total mesophilic aerobic bacteria, total anaerobic bacteria, halotolerant bacteria and facultative anaerobic halophilic and alcaliphilic (FAHA) bacteria were in the range of 2.5 × 108 to 5.5 × 109 cfu cm−2 for all samples analyzed. Enterococci and strictly anaerobic bacteria revealed 1.5 × 105 to 3.4 × 106 cfu cm−2, enterobacteria ranged from 1.9 × 103 to 2 × 104 cfu cm−2 in all samples.
Figure 1. Plate count results are provided as weighted average in colony forming units (cfu cm−2) for total aerobic mesophilic bacteria, total anaerobic bacteria, halotolerant bacteria, facultative anaerobic halophilic and alcaliphilic (FAHA) bacteria, and enterobacteria for unpacked non-defective, vacuum film-prepacked non-defective and vacuum film-prepacked defective cheese smear microbiota. The detection limit is indicated by the dashed horizontal line.
Overall, no substantial differences were detected that could be associated with vacuum film-prepackaging or a defective cheese surface smear. This becomes also obvious from the principal components analysis showing equidistant separation of the three sample types by the first two principal components explaining in total 90% of the data variation (Figure 2A).
Figure 2. Principal coordinates analysis plots for comparison of the microbial composition determined by culture-based analysis (A) and next-generation sequencing analysis (B) of unpacked non-defective [1], vacuum film-prepacked non-defective [2] and vacuum film-prepacked defective [3] cheese surface smear samples. The percentage variation explained by the according principal component (PC) is indicated on the according x- or y-axis.
3.2. Composition and diversity of the cheese surface microbiota analyzed by next-generation sequencing of amplified 16S rRNA gene fragments and total RNA
In order to perform the most comprehensive approach currently available for the investigation of complex microbial ecosystems, the microbial composition of the cheese surface smear was analyzed by next-generation sequencing of amplified 16S ribosomal RNA gene fragments. Furthermore, the proportion of metabolically active bacteria was investigated by high-throughput sequencing of total RNA followed by a taxonomic assignment of the obtained 16S rRNA reads.
The sequencing of amplified 16S rDNA fragments from the smear microbiota of vacuum film-prepacked cheese samples resulted in a total of 10,688 reads from defective and 10,780 reads from non-defective smear, respectively. For the sample from unpacked cheese 19,121 reads were generated. The number of reads that passed the quality check and were applied for taxonomic profiling was 14,413 reads for the sample from unpacked cheese. The sample from vacuum film-prepacked cheese with defective smear resulted in 7910 reads and 7708 reads were obtained from the sample of vacuum film-prepacked cheese with non-defective smear, respectively. With respect to total RNA sequencing 6361 to 17,952 total RNA reads were generated from vacuum film-prepacked cheese samples and 40,308 total RNA reads from the unpacked cheese sample. After quality check and restriction to reads originating from 16S rRNA the read numbers used for analysis were 2285 reads for vacuum film-prepacked cheese with defective smear, 2518 reads vacuum film-prepacked cheese with non-defective smear, and 2774 reads from unpacked cheese, respectively.
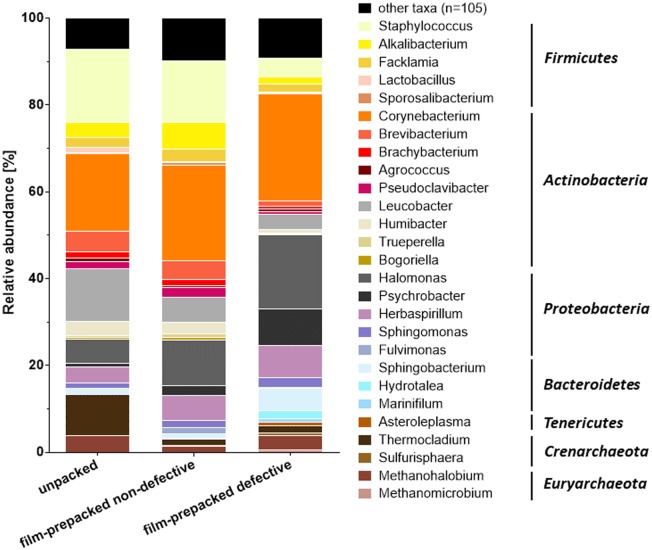
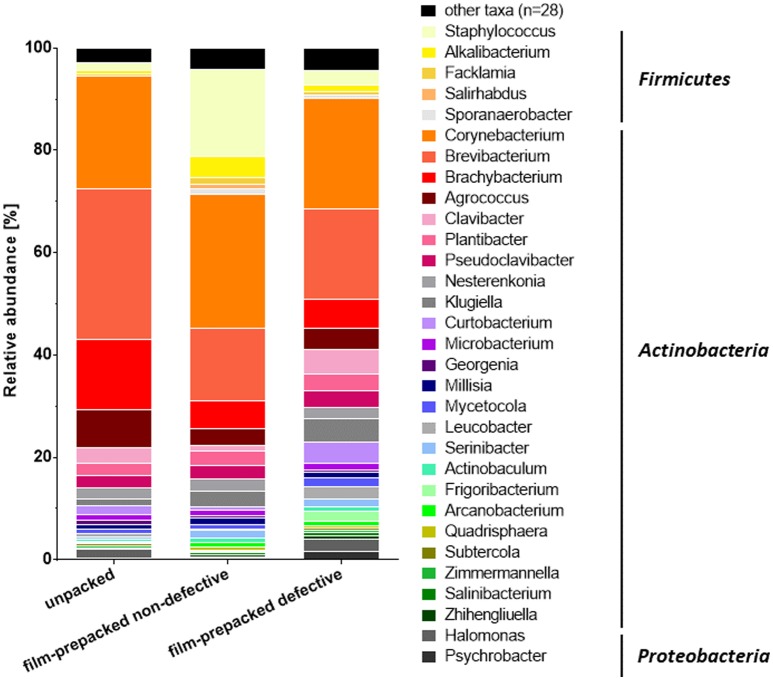
Sequencing of amplified 16S rDNA fragments revealed an extraordinarily diverse surface smear microbiota comprising 132 different OTUs (Figure 3), whereas the analysis of 16S rRNA reads resulted in only 59 different OTUs (Figure 4). However, 105 of the 132 OTUs detected by the 16S rDNA based approach revealed rather low relative abundance of less than 0.5% per sample (corresponding to less than 37 reads per sample). Therefore, they were summarized as “other taxa” in Figure 3 and Table S1. In the 16S rRNA based approach 28 of the 59 detected OTUs accounted for less than 0.5% relative abundance per sample (corresponding to less than 14 reads per sample) and were summarized as “other taxa” in Figure 4 and Table S2. Taxonomic assignment of the 16S rDNA reads on the genus level revealed that half of the reads belonged to facultative anaerobic bacteria, up to a third to strict aerobic bacteria and only 4 to 15% of the reads derived from strict anaerobic bacteria.
Figure 3. Comparison of the cheese smear microbiome of unpacked, vacuum film-prepacked non-defective and vacuum film-prepacked defective cheese smear samples determined by next-generation sequencing of 16S rDNA amplicons. Columns show the relative abundance (%) of prokaryotic genera detected in each sample. The most abundant phylotypes are displayed in different colors, while taxa revealing less than 0.5% of total reads are combined and shown in black.
Figure 4. Analysis of the cheese smear microbiome of unpacked, vacuum film-prepacked non-defective and vacuum film-prepacked defective cheese smear samples performed by next-generation sequencing of total RNA extracted from smear samples. The columns display the relative abundance (%) of 16S rRNA reads for different prokaryotic genera within each cheese sample. The most abundant phylotypes are shown in different colors, taxa revealing less than 0.5% of total reads are combined and shown in black.
The 16S rRNA based analysis revealed up to 1% reads of strict anaerobic bacteria in the samples from vacuum film-prepacked cheese, whereas the unpacked cheese samples featured no strict anaerobes. The metabolically active smear community was primarily represented by aerobic Actinobacteria of the genera Corynebacterium, Brevibacterium, Brachybacterium and Agrococcus, which provided the largest numbers of 16S rRNA reads. Taken together they contributed to a relative abundance of 73% in the sample from unpacked cheese. Corynebacterium was also the most abundant taxon detected by the 16S rDNA based approach, whereas Brevibacterium, Brachybacterium and Agrococcus accounted for far less reads. Other Actinobacteria such as Clavibacter, Plantibacter, Klugiella, Microbacterium and Leucobacter constituted to a considerable amount of 19.75–37.11% relative abundance of 16S rRNA reads (Figure 4 and Table S2). Staphylococcus 16S rRNA reads represented a considerable proportion in the microbiota of the vacuum film-prepacked non-defective cheese sample, while this was not the case for the other smear samples. However, in the 16S rDNA based analysis, reads from Firmicutes were far more prominent in all three smear sample types with 8.10–23.90% relative abundance. Whereas Proteobacteria revealed up to 35.20% relative abundance and were represented by diverse genera such as Halomonas, Psychrobacter, Herbaspirillum, Sphingomonas and Fulvimonas in the rDNA based approach, they were far less represented by only few reads from Halomonas and Psychrobacter in the 16S rRNA based approach. Archaea were in principle represented by 16S rDNA reads (3–12% relative abundance) from the genera Thermocladium, Sulfurisphaera, Methanohalobium and Methanomicrobium as 16S rRNA reads from Archaea revealed less than 0.5% relative abundance. The most striking difference in the composition of the microbial community structure in the smear from unpacked, vacuum film-prepacked non-defective and vacuum film-prepacked defective cheese was the strong increase in populations of gram-negative Proteobacteria and Bacteroidetes, in particular for the sample from defective smear. The abundance of gram-positive staphylococci, which belong to the typical smear microbiota, revealed a pronounced decrease in the smear of vacuum film-prepacked defective cheese.
Overall, the differences in the microbial community structure determined by comparison of the results from the 16S rDNA and 16S rRNA based approach indicated that the majority of ribosomal RNAs derived from a few taxa which represent the metabolically active part of the cheese smear microbiota. When the results of sequencing 16S rDNA and 16S rRNA were considered separately in principal components analysis, the first principal component clearly clustered the individual samples according to the target molecules (data not shown). However, when the entire high-throughput sequencing data per smear microbiota was taken into account, the first two principal components, which explained in total 99.48% of the variation, equidistantly separated the microbiota of the unpacked, vacuum film-prepacked non-defective and vacuum film-prepacked defective cheese smear (Figure 2).
Furthermore, it shows that the different approaches applied for analysis of the surface smear microbiota provided corresponding results. The low resolution of culture-based analysis revealed a less different community structure of unpacked, vacuum film-prepacked non-defective and vacuum film-prepacked defective cheese smear samples (Figure 2A). Concurrent with the highest resolution at a global scale, high-throughput sequencing analysis resulted in greatest differences in microbial community composition of the different smear samples (Figure 2B). In summary, the applied next-generation sequencing approach allowed an in-depth analysis of the cheese smear microbiota. Whereas the detection of 16S rDNA reads demonstrated the presence of a broad hitherto unknown spectrum of specific microbial taxa, the obtained 16S rRNA reads verified the metabolic activity of a certain subset of aerobic and facultative anaerobic Actinobacteria mainly represented by typical cheese smear bacteria.
4. Discussion
Traditional culture-based analyses of complex microbiota in food systems are limited in their ability to provide a profound insight into the actual microbial composition and are strongly biased by the selected cultivation media and the cultivability of microorganisms [47]. As a faster and more accurate method providing higher specificity and sensitivity, molecular community fingerprint techniques have been state-of-the-art techniques to study the microbial diversity of complex ecosystems before they have been replaced by next-generation sequencing technologies for high-throughput purposes [20],[48]. Previous studies based on next-generation sequencing of cheese microbiota mainly focused on the analysis of individual amplified “hypervariable regions” of 16S rRNA genes [49]–[54]. In this study, the cheese surface microbiome was analyzed by a next-generation sequencing approach considering not only the different “hypervariable regions” V1–V9 of the 16S ribosomal RNA gene, but also the direct sequencing of total RNA without prior amplification. Thus, in addition to the composition and diversity, also the metabolic active proportion of the cheese surface microbiota was analyzed for the different samples from unpacked, vacuum film-prepacked non-defective and vacuum film-prepacked defective cheese. It is a preliminary study carried out on just one cheese batch produced on the same day and ripened in the same cheese cellar under identical conditions, which needs further investigation in the future to improve our knowledge on the impact of different packaging strategies on the cheese smear microbiome.
The microbial community patterns obtained from the different samples revealed an altered composition primarily caused by an increase in populations of gram-negative Proteobacteria and Bacteroidetes and a decrease in staphylococci, which are part of the typical smear microbiota. These alterations were in particular pronounced in the samples of vacuum film-prepacked cheese showing defective smear. This observation is contrary to data from culture-based investigation of the cheese surface smear defect, which did not reveal distinct differences between the different sample types. A previous study on red-smear cheese defect development [25] the stable microbial composition determined during the cheese storage period indicated a complex etiology, where more than a single factor contributes to the smear defect development. In a later study, it was shown that factors such as anaerobic conditions, an increase in water activity as well as a decrease in pH was correlated with a diminished rRNA transcriptional activity of the smear microbiota [26]. In the present study, the smear microbiota of vacuum film-prepacked cheese samples featured considerably less 16S rRNA reads and the composition of the microbial community structure appeared to be less affected compared to the 16S rDNA based next-generation sequencing approach, which revealed clear changes in the microbial population structures of the different samples.
In particular, reads from microorganisms typically for the surface of smear-ripened cheese, such as Corynebacterium, Staphylococcus, Brevibacterium, Arthrobacter, Micrococcus, Microbacterium, Alkalibacterium and Facklamia predominated the results of the 16S rDNA based analysis. With the exception of Micrococcus most of the bacteria typical for a smear cheese surface have been isolated from the surface of the same cheese variety in a previous study [25]. In this culture based study typical smear bacteria represented the major part of the smear microbiota with colony counts of 107 to 108 cfu cm−2. This was also the case in the present study were Corynebacterium, Brevibacterium and Brachybacterium accounted for the major part of identified 16S rDNA reads. This finding seems reasonable because representatives of these genera are known to contribute substantially to the typical smear-cheese characteristics during the ripening process by production of extracellular proteinases and lipases, pigments and aroma compounds [4],[8],[55],[56]. Due to the known application in starter cultures for cheese production the 16S rDNA reads from Lactobacillus detected in this study might have derived from bacteria of the cheese matrix [4],[57].
Besides microorganisms already known to predominate on the cheese surface, the surface smear microbiota was found to comprise an unexpected high microbial diversity implying many taxa hitherto not related to the cheese ecosystem. As the sampled cheeses were produced from raw milk and a high biodiversity of indigenous microorganisms is characteristic for raw milk, a major part of the cheese smear microbiota might derive from this source [57],[58]. Comparison of the genera represented by 16S rDNA and 16S rRNA sequences with scientific literature revealed manifold relations of initially unexpected taxa to the farm environment, such as the bovine microbiota, but also further relations to water, marine and salty ecosystems or soil. The genus Trueperella is known to be common in cow milk and predominates mainly bovine mastitis milk [54],[58]–[65], while Asteroleplasma is part of the cattle rumen ecosystem [66],[67]. Herbaspirillum is a genus, which is associated with roots of herbaceous plants, while Humibacter and Fulvimonas are related to soil or compost [68]–[71]. It is known that farm related microorganisms can also be transferred to cheese via raw milk used for cheese production, while salt crystals can serve as a source for microorganisms when cheese samples pass a brine bath during cheese production or are regularly smeared with brine during the ripening process. Furthermore, the 16S rDNA based approach revealed the presence of Archaea in the cheese smear microbiota. However, rDNA reads from Archaea were rare, and 16S rRNA reads accounted for less than 0.5% of all reads. The 16S rDNA based approach revealed Thermocladium, a taxon recently detected by pyrosequencing in buttermilk, and Methanomicrobium, which occurs naturally in bovine rumen. Sulfurisphaera and Methanohalobium are known to inhabit marine water systems or saline lagoons [72]–[75].
Because RNA is less stable than DNA and its expression is tightly related to the bacterial physiological status, ribosomal 16S RNA represents an appropriate marker for metabolically active and integer cells [76]–[80]. Thus, the detected 16S rRNA reads represent the composition of the metabolic active bacteria in the cheese surface smear. Consequently, the abundance of 16S rRNA reads from Actinobacteria indicates that this phylum represents the largest population of active bacteria on the cheese surface post ripening. Furthermore, Agrococcus provided a considerable number of the 16S rRNA reads in the functional communities analyzed. Although usually not numbered among the typical smear microorganisms, Agrococcus was previously detected in cheese smear and identified as a member of the “house microbiota” in dairies. Moreover, the first isolation of Agrococcus casei has been described for the surface of Gubbeen, Livarot and Tilsit cheese [11],[81]–[83].
Besides Corynebacterium, Brevibacterium, Brachybacterium and Agrococcus, which predominated the metabolic active community on the cheese surface, a variety of other genera from Actinobacteria contributed to a considerable proportion of the 16S rRNA reads. Microbacterium spp. are considered to belong to the typical smear bacteria, which frequently colonize the cheese surface in high numbers of 108 cfu cm−2 and are usually part of the “house microbiota” in cheese dairies. Representatives of Curtobacterium, Klugiella, Pseudoclavibacter, Leucobacter, and Mycetocola were occasionally described in cheese surface smear or cheese dairy and farm environment [11],[15],[56],[58],[59],[61],[63],[84]–[88]. Although Clavibacter, Plantibacter, Actinobaculum, Frigoribacterium, Zimmermannella, Arcanobacterium and Subtercola have so far not been described to inhabit cheese surface smear, recent studies indicate that members of these genera play a role in the context of milk, farm and dairy animal ecosystems [58],[61],[63],[87],[89]–[93]. Further studies indicate that raw milk may serve as a vector for the genera Georgenia, Millisia and Quadrisphaera, which usually inhabit terrestrial natural ecosystems such as soil or sludge [82],[94],[95]. The application of salt during the cheese production process is suggested as intrinsic source for microorganisms such as Salirhabdus, Nesterenkonia, Serinibacter, Salinibacterium and Zhihengliuella, which are typically isolated from marine or saline aquatic ecosystems [96]–[99]. The actual role and contribution by the various unexpected microorganisms occurring on the cheese surface remains unknown, but the previously cited literature revealed production of lipases, proteases, pigments and chemical aroma compounds or the coagulation of milk by various of these Actinobacteria.
The comparison of 16S rDNA results for samples showing varying cheese smear quality revealed a decrease of reads from staphylococci in the samples from defective smear, in particular when compared with the sample from unpacked cheese. Staphylococci represent typical cheese smear microorganisms which are known to predominate the cheese surface smear in particular at the beginning of the ripening process, providing conditions for better growth of corynebacteria [12],[100]. Thus, their decline suggests unfavorable conditions in defective smear. Another striking observation was the higher abundance of representatives from Proteobacteria and Bacteroidetes in the smear of prepacked cheese, which was most pronounced in samples from defective smear. The phylum Bacteroidetes was mainly represented by reads from Sphingobacterium and Hydrotalea, which are known to be abundant in soil and water [101],[102], but have not been associated with cheese production environment so far. The Proteobacteria were primarily represented by reads from the genera Psychrobacter and Halomonas, which have been frequently detected in high numbers in the surface smear of red-smear cheese varieties in previous studies [54],[103],[104]. Usually, high cell counts of Proteobacteria during the early stages of the ripening period decrease with time, due to the combined effects of different physical and chemical parameters [105]. For Psychrobacter celer it has been shown that the bacteria are able to successfully implant itself in cheese, regardless of its inoculation level. They contributed to the production of volatile aroma compounds such as aldehydes, ketones and sulfur compounds and had an impact on the aromatic properties of the cheese [106]. This observation is supported by whole genome sequencing of a Psychrobacter strain isolated from cheese rind, which revealed that the genome harbors enzymes that are important for cheese ripening in other bacteria, such as cystathionine/methionine beta or gamma-lyases, many proteases and peptidases, aminotransferases, and lipases [107]. Due to these properties increased levels of the bacteria may contribute to the unpleasant off-flavor that accumulates under the foil of prepacked smear cheese. For Halomonas venusta it was shown that the bacteria produce cadaverine in a cheese model for production of volatile compounds as well as biogenic amines [108]. However, Halomonas spp. are better known as potent exopolysaccharide producers [109],[110]. This property may potentially contribute to the sticky and slimy cheese surface of foil prepacked smear cheese. Thus, the combination of a decrease of typical smear organisms like staphylococci, together with an increase of members of Proteobacteria in defective smear might substantially contribute to the observed negative organoleptic properties of the surface smear of cheese after prepacking in plastic foil.
5. Conclusions
Overall, the data obtained by the high-throughput sequencing approach revealed an unexpectedly high overall diversity in the cheese smear microbiota and comprised many bacteria that have hitherto not been described for the cheese ecosystem. However, all of the detected taxa bear relation to the farm or cheese dairy environment as shown in literature. Although the next-generation sequencing approach applied in this study provides currently the most detailed microbiome analysis, a future challenge would be to trace the source of the microorganisms colonizing the surface of smear-ripened cheeses, and to monitor the microbial succession during the cheese ripening process. Therefore, a polyphasic approach by combining different molecular techniques with culture-based methods should be considered to study complex microbial ecosystems such as cheese smear. Moreover, for a better understanding of the microbial contribution to cheese ripening and for unraveling the function of certain microbial populations, a thorough analysis of the cheese smear metatranscriptome would be an indispensable prerequisite.
Acknowledgments
This study was funded by the Swiss Innovation Promotion Agency CTI, project number 10273-3. For assisting in executing experimental work, the authors like to thank most sincerely Olivia Kurtz.
Footnotes
Conflict of interest: The authors declare no conflict of interest in this paper.
References
- 1.Fox PF, McSweeney PLH. Cheese: An Overview. In: Fox Patrick F., McSweeney PLH, Cogan TM, et al., editors. Cheese: chemistry, physics and microbiology. Amsterdam: Elsevier; 2004. pp. 1–18. [Google Scholar]
- 2.Bockelmann W. Smear-ripened cheeses. In: Roginski H, Fuquay JW, Fox PF, editors. Encyclopaedia of Dairy Sciences. London: Academic Press; 2002. pp. 391–401. [Google Scholar]
- 3.Bockelmann W. Secondary cheese starter cultures. In: Law BA, Tamime AY, editors. Technology of cheesemaking. Oxford: Blackwell Publishing Ltd; 2010. pp. 193–230. [Google Scholar]
- 4.Brennan NM, Cogan TM, Loessner M, et al. Bacterial surface-ripened cheeses. In: Fox PF, Mcsweeney PLH, Cogan TM, et al., editors. Cheese: Chemistry, Physics and Microbiology. Amsterdam: Elsevier; 2004. pp. 199–225. [Google Scholar]
- 5.McSweeney PLH, Ottogalli G, Fox PF, et al. Cheese: Chemistry, Physics and Microbiology. Amsterdam: Elsevier; 2004. Diversity of cheese varieties: An overview; pp. 1–23. [Google Scholar]
- 6.McSweeney PLH, Fox PF. Metabolism of residual lactose and of lactate and citrate. In: Fox PF, McSweeney PLH, Cogan TM, et al., editors. Cheese: Chemistry, Physics and Microbiology. Amsterdam: Elsevier; 2004. pp. 361–371. [Google Scholar]
- 7.Reps A. Bacterial Surface-Ripened Cheeses. In: Fox PF, editor. Cheese: Chemistry, Physics and Microbiology. New York: Springer US; 1993. pp. 137–172. [Google Scholar]
- 8.Bockelmann W, Hoppe-Seyler T. The surface flora of bacterial smear-ripened cheeses from cow's and goat's milk. Int Dairy J. 2001;11:307–314. [Google Scholar]
- 9.Monnet C, Bleicher A, Neuhaus K, et al. Assessment of the anti-listerial activity of microfloras from the surface of smear-ripened cheeses. Food Microbiol. 2010;27:302–310. doi: 10.1016/j.fm.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Roth E, Schwenninger SM, Eugster-Meier E, et al. Facultative anaerobic halophilic and alkaliphilic bacteria isolated from a natural smear ecosystem inhibit Listeria growth in early ripening stages. Int J Food Microbiol. 2011;147:26–32. doi: 10.1016/j.ijfoodmicro.2011.02.032. [DOI] [PubMed] [Google Scholar]
- 11.Mounier J, Goerges S, Gelsomino R, et al. Sources of the adventitious microflora of a smear ripened cheese. J Appl Microbiol. 2006;101:668–681. doi: 10.1111/j.1365-2672.2006.02922.x. [DOI] [PubMed] [Google Scholar]
- 12.Rademaker J, Peinhopf M, Rijnen L, et al. The surface microflora dynamics of bacterial smear-ripened Tilsit cheese determined by T-RFLP DNA population fingerprint analysis. Int Dairy J. 2005;15:785–794. [Google Scholar]
- 13.Bockelmann W, Willems K, Neve H, et al. Cultures for the ripening of smear cheeses. Int Dairy J. 2005;15:719–732. [Google Scholar]
- 14.Bokulich NA, Mills DA. Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Appl Environ Microb. 2013;79:5214–5223. doi: 10.1128/AEM.00934-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Didienne R, Defargues C, Callon C, et al. Characteristics of microbial biofilm on wooden vats (“gerles”) in PDO Salers cheese. Int J Food Microbiol. 2012;156:91–101. doi: 10.1016/j.ijfoodmicro.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Mariani C, Briandet R, Chamba JF, et al. Biofilm ecology of wooden shelves used in ripening the French raw milk smear cheese Reblochon de Savoie. J Dairy Sci. 2007;90:1653–1661. doi: 10.3168/jds.2006-190. [DOI] [PubMed] [Google Scholar]
- 17.Oulahal N, Adt I, Mariani C, et al. Examination of wooden shelves used in the ripening of a raw milk smear cheese by FTIR spectroscopy. Food Control. 2009;20:658–663. [Google Scholar]
- 18.Ndoye B, Rasolofo EA, Lapointe G, et al. A review of the molecular approaches to investigate the diversity and activity of cheese microbiota. Dairy Sci Technol. 2011;91:1–30. [Google Scholar]
- 19.Jany JL, Barbier G. Culture-independent methods for identifying microbial communities in cheese. Food Microbiol. 2008;25:839–848. doi: 10.1016/j.fm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Justé A, Thomma BPHJ, Lievens B. Recent advances in molecular techniques to study microbial communities in food-associated matrices and processes. Food Microbiol. 2008;25:745–761. doi: 10.1016/j.fm.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Quigley L, O'Sullivan O, Beresford TP, et al. Molecular approaches to analysing the microbial composition of raw milk and raw milk cheese. Int J Food Microbiol. 2011;150:81–94. doi: 10.1016/j.ijfoodmicro.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Giraffa G, Carminati D. Molecular techniques in food fermentation: principles and applications. In: Cocolin L, Ercolini D, editors. Molecular Techniques in the Microbial Ecology of Fermented Foods. Berlin: Springer; 2008. pp. 1–30. [Google Scholar]
- 23.O'Sullivan DJ. Methods for analysis of the intestinal microflora. Curr Issues Intest Microbiol. 2000;1:39–50. [PubMed] [Google Scholar]
- 24.van Hijum SAFT, Vaughan EE, Vogel RF. Application of state-of-art sequencing technologies to indigenous food fermentations. Curr Opin Biotech. 2012;24:1–9. doi: 10.1016/j.copbio.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Amato L, Ritschard JS, Kurtz O, et al. Microbial composition of defect smear—a problem evolving during foil-prepacked storage of red-smear cheeses. Int Dairy J. 2012;27:77–85. [Google Scholar]
- 26.Amato L, Ritschard JS, Sprecher I, et al. Effect of packaging materials, environmental factors and rRNA transcriptional activity of surface microflora on red-smear cheese defect development. Int Dairy J. 2015;41:50–57. [Google Scholar]
- 27.Koutsoumanis K, Pavlis A, Nychas GJE, et al. Probabilistic model for Listeria monocytogenes growth during distribution, retail storage, and domestic storage of pasteurized milk. Appl Environ Microb. 2010;76:2181–2191. doi: 10.1128/AEM.02430-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laguerre O, Hoang HM, Flick D. Experimental investigation and modelling in the food cold chain: Thermal and quality evolution. Trends Food Sci Tech. 2013;29:87–97. [Google Scholar]
- 29.Brosius J, Palmer ML, Kennedy PJ, et al. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. P Natl Acad Sci. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane DJ. 16S/23S rRNA sequencing. Nucleic Acid Tech Bact Syst. 1991;1991:115–175. [Google Scholar]
- 31.Weisburg WG, Barns SM, Pelletier DA, et al. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sander A, Ruess M, Bereswill S, et al. Comparison of different DNA fingerprinting techniques for molecular typing of Bartonella henselae isolates. J Clin Microbiol. 1998;36:2973–2981. doi: 10.1128/jcm.36.10.2973-2981.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reysenbach AL, Pace NR. Reliable amplification of hyperthermophilic archaeal 16S rRNA genes by the polymerase chain reaction. In: Robb F, editor. Archaea: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. pp. 101–107. [Google Scholar]
- 34.Takai K, Horikoshi K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microb. 2000;66:5066–5072. doi: 10.1128/aem.66.11.5066-5072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hugenholtz P, Pitulle C, Hershberger KL, et al. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walters WA, Caporaso JG, Lauber CL, et al. PrimerProspector: De novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics. 2011;27:1159–1161. doi: 10.1093/bioinformatics/btr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber JA, Mark-Welch DB, Morrison HG, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Qian PY. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One. 2009;4:e7401. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Claesson MJ, Wang Q, O'Sullivan O, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu WT, Mirzabekov AD, Stahl DA. Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environ Microbiol. 2001;3:619–629. doi: 10.1046/j.1462-2920.2001.00233.x. [DOI] [PubMed] [Google Scholar]
- 41.DeLong EF. Archaea in coastal marine environments. P Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Meth. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 43.Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb. 2007;73:5261. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monnet C, Ulve V, Sarthou AS, et al. Extraction of RNA from cheese without prior separation of microbial cells. Appl Environ Microb. 2008;74:5724–5730. doi: 10.1128/AEM.00716-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riley J, Butler R, Ogilvie D, et al. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Sullivan D, Cotter P, Giblin L, et al. Nucleic acid-based approaches to investigate microbial-related cheese quality defects. Front Microbiol. 2013;4:1–15. doi: 10.3389/fmicb.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ercolini D. High-throughput sequencing and metagenomics: Moving forward in the culture-independent analysis of food microbial ecology. Appl Environ Microb. 2013;79:3148–3155. doi: 10.1128/AEM.00256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alegría Á, Szczesny P, Mayo B, et al. Biodiversity in oscypek, a traditional polish cheese, determined by culture-dependent and -independent approaches. Appl Environ Microb. 2012;78:1890–1898. doi: 10.1128/AEM.06081-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bokulich NA, Mills DA. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl Environ Microb. 2013;79:2519–2526. doi: 10.1128/AEM.03870-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lusk T, Ottesen A, White J, et al. Characterization of microflora in Latin-style cheeses by next-generation sequencing technology. BMC Microbiol. 2012;12:254. doi: 10.1186/1471-2180-12-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Masoud W, Takamiya M, Vogensen FK, et al. Characterization of bacterial populations in Danish raw milk cheeses made with different starter cultures by denaturating gradient gel electrophoresis and pyrosequencing. Int Dairy J. 2011;21:142–148. [Google Scholar]
- 53.Masoud WF, Vogensen FK, Lillevang S, et al. The fate of indigenous microbiota, starter cultures, Escherichia coli, Listeria innocua and Staphylococcus aureus in Danish raw milk and cheeses determined by pyrosequencing and quantitative real time (qRT)-PCR. Int J Food Microbiol. 2012;153:192–202. doi: 10.1016/j.ijfoodmicro.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 54.Quigley L, O'Sullivan O, Beresford TP, et al. High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl Environ Microb. 2012;78:5717–5723. doi: 10.1128/AEM.00918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bockelmann W, Krusch U, Engel G, et al. The microflora of Tilsit cheese. Part 1. Variability of the smear flora. Mol Nutr Food Res. 1997;41:208–212. [Google Scholar]
- 56.Deetae P, Bonnarme P, Spinnler H, et al. Production of volatile aroma compounds by bacterial strains isolated from different surface-ripened French cheeses. Appl Microbiol Biotech. 2007;76:1161–1171. doi: 10.1007/s00253-007-1095-5. [DOI] [PubMed] [Google Scholar]
- 57.Montel MC, Buchin S, Mallet A, et al. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int J Food Microbiol. 2014;177:136–154. doi: 10.1016/j.ijfoodmicro.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 58.Quigley L, O'Sullivan O, Stanton C, et al. The complex microbiota of raw milk. FEMS Microbiol Rev. 2013;37:664–698. doi: 10.1111/1574-6976.12030. [DOI] [PubMed] [Google Scholar]
- 59.Delbès C, Ali-Mandjee L, Montel MC. Monitoring bacterial communities in raw milk and cheese by culture-dependent and-independent 16S rRNA gene-based analyses. Appl Environ Microb. 2007;73:1882–1891. doi: 10.1128/AEM.01716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oikonomou G, Machado VS, Santisteban C, et al. Microbial diversity of bovine mastitic milk as described by pyrosequencing of metagenomic 16s rDNA. PLoS One. 2012;7:e47671. doi: 10.1371/journal.pone.0047671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schirmer BCT, Heir E, Møretrø T, et al. Microbial background flora in small-scale cheese production facilities does not inhibit growth and surface attachment of Listeria monocytogenes. J Dairy Sci. 2013;96:6161–6171. doi: 10.3168/jds.2012-6395. [DOI] [PubMed] [Google Scholar]
- 62.Schmidt VSJ, Kaufmann V, Kulozik U, et al. Microbial biodiversity, quality and shelf life of microfiltered and pasteurized extended shelf life (ESL) milk from Germany, Austria and Switzerland. Int J Food Microbiol. 2012;154:1–9. doi: 10.1016/j.ijfoodmicro.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 63.Verdier-Metz I, Gagne G, Bornes S, et al. Cow teat skin, a potential source of diverse microbial populations for cheese production. Appl Environ Microb. 2012;78:326–333. doi: 10.1128/AEM.06229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White DC, Sutton SD, Ringelberg DB. The genus Sphingomonas: Physiology and ecology. Curr Opin Biotech. 1996;7:301–306. doi: 10.1016/s0958-1669(96)80034-6. [DOI] [PubMed] [Google Scholar]
- 65.Zastempowska E, Lassa H. Genotypic characterization and evaluation of an antibiotic resistance of Trueperella pyogenes (Arcanobacterium pyogenes) isolated from milk of dairy cows with clinical mastitis. Vet Microbiol. 2012;161:153–158. doi: 10.1016/j.vetmic.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 66.Petri RM, Mapiye C, Dugan ME, et al. Subcutaneous adipose fatty acid profiles and related rumen bacterial populations of steers fed red clover or grass hay diets containing flax or sunflower-seed. PLoS One. 2014;9:e104167. doi: 10.1371/journal.pone.0104167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pitta DW, Pinchak WE, Dowd SE, et al. Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets. Microbial Ecol. 2010;59:511–522. doi: 10.1007/s00248-009-9609-6. [DOI] [PubMed] [Google Scholar]
- 68.Ahn JH, Kim SJ, Weon HY, et al. Fulvimonas yonginensis sp. nov., isolated from greenhouse soil, and emended description of the genus Fulvimonas. Int J Syst Evol Micr. 2014;64:2758–2762. doi: 10.1099/ijs.0.064642-0. [DOI] [PubMed] [Google Scholar]
- 69.Baldani JI, Baldani VLD, Seldin L, et al. Characterization of Herbaspirillum seropedicae gen. nov., sp. nov., a root-associated nitrogen-fixing bacterium. Int J Syst Bacteriol. 1986;36:86–93. [Google Scholar]
- 70.Lee SD. Humibacter antri sp. nov., an actinobacterium isolated from a natural cave, and emended description of the genus Humibacter. Int J Syst Evol Micr. 2013;63:4315–4319. doi: 10.1099/ijs.0.050708-0. [DOI] [PubMed] [Google Scholar]
- 71.Vaz-Moreira I, Nobre MF, Ferreira ACS, et al. Humibacter albus gen. nov., sp. nov., isolated from sewage sludge compost. Int J Syst Evol Micr. 2008;58:1014–1018. doi: 10.1099/ijs.0.65266-0. [DOI] [PubMed] [Google Scholar]
- 72.Chaban B, Ng SY, Jarrell KF. Archaeal habitats-from the extreme to the ordinary. Can J Microbiol. 2006;52:73–116. doi: 10.1139/w05-147. [DOI] [PubMed] [Google Scholar]
- 73.Janssen PH, Kirs M. Structure of the archaeal community of the rumen. Appl Environ Microb. 2008;74:3619–3625. doi: 10.1128/AEM.02812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jayashree S, Pushpanathan M, Rajendhran J, et al. Microbial diversity and phylogeny analysis of buttermilk, a fermented milk product, employing 16S rRNA-based pyrosequencing. Food Biotechnol. 2013;27:213–221. [Google Scholar]
- 75.Takai K, Sako Y. A molecular view of archaeal diversity in marine and terrestrial hot water environments. FEMS Microbiol Ecol. 1999;28:177–188. [Google Scholar]
- 76.Babot JD, Hidalgo M, Arganaraz-Martinez E, et al. Fluorescence in situ hybridization for detection of classical propionibacteria with specific 16S rRNA-targeted probes and its application to enumeration in Gruyère cheese. Int J Food Microbiol. 2011;145:221–228. doi: 10.1016/j.ijfoodmicro.2010.12.024. [DOI] [PubMed] [Google Scholar]
- 77.Cenciarini-Borde C, Courtois S, La SB. Nucleic acids as viability markers for bacteria detection using molecular tools. Future Microbiol. 2009;4:45–64. doi: 10.2217/17460913.4.1.45. [DOI] [PubMed] [Google Scholar]
- 78.Fey A, Eichler S, Flavier S, et al. Establishment of a Real-Time PCR-based approach for accurate quantification of bacterial RNA targets in water, using Salmonella as a model organism. Appl Environ Microb. 2004;70:3618–3623. doi: 10.1128/AEM.70.6.3618-3623.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Piir K, Paier A, Liiv A, et al. Ribosome degradation in growing bacteria. EMBO Rep. 2011;12:458–462. doi: 10.1038/embor.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vendeville A, Larivière D, Fourmentin E. An inventory of the bacterial macromolecular components and their spatial organization. FEMS Microbiol Rev. 2011;35:395–414. doi: 10.1111/j.1574-6976.2010.00254.x. [DOI] [PubMed] [Google Scholar]
- 81.Bora N, Vancanneyt M, Gelsomino R, et al. Agrococcus casei sp. nov., isolated from the surfaces of smear-ripened cheeses. Int J Syst Evol Micr. 2007;57:92–97. doi: 10.1099/ijs.0.64270-0. [DOI] [PubMed] [Google Scholar]
- 82.Li WJ, Chen HH, Kim CJ, et al. Nesterenkonia sandarakina sp. nov. and Nesterenkonia lutea sp. nov., novel actinobacteria, and emended description of the genus Nesterenkonia. Int J Syst Evol Micr. 2005;55:463–466. doi: 10.1099/ijs.0.63281-0. [DOI] [PubMed] [Google Scholar]
- 83.Mounier J, Gelsomino R, Goerges S, et al. Surface microflora of four smear-ripened cheeses. Appl Environ Microb. 2005;71:6489–6500. doi: 10.1128/AEM.71.11.6489-6500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brennan NM, Brown R, Goodfellow M, et al. Microbacterium gubbeenense sp. nov., from the surface of a smear-ripened cheese. Int J Syst Evol Micr. 2001;51:1969–1976. doi: 10.1099/00207713-51-6-1969. [DOI] [PubMed] [Google Scholar]
- 85.Mounier J, Monnet C, Jacques N, et al. Assessment of the microbial diversity at the surface of Livarot cheese using culture-dependent and independent approaches. Int J Food Microbiol. 2009;133:31–37. doi: 10.1016/j.ijfoodmicro.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 86.Rea M, Görges S, Gelsomino R, et al. Stability of the biodiversity of the surface consortia of Gubbeen, a red-smear cheese. J Dairy Sci. 2007;90:2200–2210. doi: 10.3168/jds.2006-377. [DOI] [PubMed] [Google Scholar]
- 87.Vacheyrou M, Normand AC, Guyot P, et al. Cultivable microbial communities in raw cow milk and potential transfers from stables of sixteen French farms. Int J Food Microbiol. 2011;146:253–262. doi: 10.1016/j.ijfoodmicro.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 88.Wolfe BE, Button JE, Santarelli M, et al. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell. 2014;158:422–433. doi: 10.1016/j.cell.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brandl H, Fricker-Feer C, Ziegler D, et al. Distribution and identification of culturable airborne microorganisms in a Swiss milk processing facility. J Dairy Sci. 2014;97:240–246. doi: 10.3168/jds.2013-7028. [DOI] [PubMed] [Google Scholar]
- 90.Gill JJ, Sabour PM, Gong J, et al. Characterization of bacterial populations recovered from the teat canals of lactating dairy and beef cattle by 16S rRNA gene sequence analysis. FEMS Microbiol Ecol. 2006;56:471–481. doi: 10.1111/j.1574-6941.2006.00091.x. [DOI] [PubMed] [Google Scholar]
- 91.Lawson PA, Falsen E, Åkervall E, et al. Characterization of some Actinomyces-like isolates from human clinical specimens: Reclassification of Actinomyces suis (soltys and spratling) as Actinobaculum suis comb. nov. and description of Actinobaculum schaalii sp. nov. Int J Syst Bacteriol. 1997;47:899–903. doi: 10.1099/00207713-47-3-899. [DOI] [PubMed] [Google Scholar]
- 92.Lin YC, Uemori K, de Briel DA, et al. Zimmermannella helvola gen. nov., sp. nov., Zimmermannella alba sp. nov., Zimmermannella bifida sp. nov., Zimmermannella faecalis sp. nov. and Leucobacter albus sp. nov., novel members of the family Microbacteriaceae. Int J Syst Evol Micr. 2004;54:1669–1676. doi: 10.1099/ijs.0.02741-0. [DOI] [PubMed] [Google Scholar]
- 93.Mallet A, Guéguen M, Kauffmann F, et al. Quantitative and qualitative microbial analysis of raw milk reveals substantial diversity influenced by herd management practices. Int Dairy J. 2012;27:13–21. [Google Scholar]
- 94.Maszenan AM, Tay JH, Schumann P, et al. Quadrisphaera granulorum gen. nov., sp. nov., a Gram-positive polyphosphate-accumulating coccus in tetrads or aggregates isolated from aerobic granules. Int J Syst Evol Micr. 2005;55:1771–1777. doi: 10.1099/ijs.0.63583-0. [DOI] [PubMed] [Google Scholar]
- 95.Soddell JA, Stainsby FM, Eales KL, et al. Millisia brevis gen. nov., sp. nov., an actinomycete isolated from activated sludge foam. Int J Syst Evol Micr. 2006;56:739–744. doi: 10.1099/ijs.0.63855-0. [DOI] [PubMed] [Google Scholar]
- 96.Albuquerque L, Tiago I, Rainey FA, et al. Salirhabdus euzebyi gen. nov., sp. nov., a Gram-positive, halotolerant bacterium isolated from a sea salt evaporation pond. Int J Syst Evol Micr. 2007;57:1566–1571. doi: 10.1099/ijs.0.64999-0. [DOI] [PubMed] [Google Scholar]
- 97.Hamada M, Iino T, Tamura T, et al. Serinibacter salmoneus gen. nov., sp. nov., an actinobacterium isolated from the intestinal tract of a fish, and emended descriptions of the families Beutenbergiaceae and Bogoriellaceae. Int J Syst Evol Micr. 2009;59:2809–2814. doi: 10.1099/ijs.0.011106-0. [DOI] [PubMed] [Google Scholar]
- 98.Han SK, Nedashkovskaya OI, Mikhailov VV, et al. Salinibacterium amurskyense gen. nov., sp. nov., a novel genus of the family Microbacteriaceae from the marine environment. Int J Syst Evol Micr. 2003;53:2061–2066. doi: 10.1099/ijs.0.02627-0. [DOI] [PubMed] [Google Scholar]
- 99.Zhang YQ, Schumann P, Yu LY, et al. Zhihengliuella halotolerans gen. nov., sp. nov., a novel member of the family Micrococcaceae. Int J Syst Evol Micr. 2007;57:1018–1023. doi: 10.1099/ijs.0.64528-0. [DOI] [PubMed] [Google Scholar]
- 100.Ruaro A, Andrighetto C, Torriani S, et al. Biodiversity and characterization of indigenous coagulase-negative staphylococci isolated from raw milk and cheese of North Italy. Food Microbiol. 2013;34:106–111. doi: 10.1016/j.fm.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 101.Bernardet JF, Segers P, Vanca M, et al. Cutting a gordian knot: Emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (Basonym, Cytophaga aquatilis Strohl and Tait 1978) Int J Syst Bacteriol. 1996;46:128–148. [Google Scholar]
- 102.Kämpfer P, Lodders N, Falsen E. Hydrotalea flava gen. nov., sp. nov., a new member of the phylum Bacteroidetes and allocation of the genera Chitinophaga, Sediminibacterium, Lacibacter, Flavihumibacter, Flavisolibacter, Niabella, Niastella, Segetibacter, Parasegetibacter, Terrimonas, Ferruginibacter, Filimonas and Hydrotalea to the family Chitinophagaceae fam. nov. Int J Syst Evol Micr. 2011;61:518–523. doi: 10.1099/ijs.0.023002-0. [DOI] [PubMed] [Google Scholar]
- 103.Deetae P, Mounier J, Bonnarme P, et al. Effects of Proteus vulgaris growth on the establishment of a cheese microbial community and on the production of volatile aroma compounds in a model cheese. J Appl Microbiol. 2009;107:1404–1413. doi: 10.1111/j.1365-2672.2009.04315.x. [DOI] [PubMed] [Google Scholar]
- 104.Maoz A, Mayr R, Scherer S. Temporal stability and biodiversity of two complex antilisterial cheese-ripening microbial consortia. Appl Environ Microb. 2003;69:4012–4018. doi: 10.1128/AEM.69.7.4012-4018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tornadijo M, García M, Fresno J, et al. Study of Enterobacteriaceae during the manufacture and ripening of San Simón cheese. Food Microbiol. 2001;18:499–509. [Google Scholar]
- 106.Irlinger F, Yung SA, Sarthou AS, et al. Ecological and aromatic impact of two Gram-negative bacteria (Psychrobacter celer and Hafnia alvei) inoculated as part of the whole microbial community of an experimental smear soft cheese. Int J Food Microbiol. 2012;153:332–338. doi: 10.1016/j.ijfoodmicro.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 107.Schmitz-Esser S, Dzieciol M, Nischler E, et al. Abundance and potential contribution of Gram-negative cheese rind bacteria from Austrian artisanal hard cheeses. Int J Food Microbiol. 2018;266:95–103. doi: 10.1016/j.ijfoodmicro.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 108.Delbès-Paus C, Pochet S, Helinck S, et al. Impact of Gram-negative bacteria in interaction with a complex microbial consortium on biogenic amine content and sensory characteristics of an uncooked pressed cheese. Food Microbiol. 2012;30:74–82. doi: 10.1016/j.fm.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 109.Erkorkmaz BA, Kirtel O, Ateş DÖ, et al. Development of a cost-effective production process for Halomonas levan. Bioprocess Biosyst Eng. 2018;2018:1–13. doi: 10.1007/s00449-018-1952-x. [DOI] [PubMed] [Google Scholar]
- 110.Finore I, Di Donato P, Mastascusa V, et al. Fermentation technologies for the optimization of marine microbial exopolysaccharide production. Mar Drugs. 2014;12:3005–3024. doi: 10.3390/md12053005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.