The crystal structure of PssZ has been determined at 1.6 Å resolution. PssZ belongs to glycoside hydrolase family 8 and adopts a classical (α/α)6-barrel fold.
Keywords: Listeria monocytogenes, glycoside hydrolases, SAD phasing, PssZ, (α/α)6-barrel fold, biofilms
Abstract
Biofilms are microbial communities that are embedded in the extracellular matrix. The exopolysaccharide (EPS) is a key component of the biofilm matrix that maintains the structure of the biofilm and protects the bacteria from antimicrobials. Microbial glycoside hydrolases have been exploited to disrupt biofilms by breaking down EPSs. PssZ has recently been identified as a glycoside hydrolase that can disperse aggregates of Listeria monocytogenes. In this study, the crystal structure of PssZ has been determined at 1.6 Å resolution. PssZ belongs to glycoside hydrolase family 8 and adopts a classical (α/α)6-barrel fold. This architecture forms a deep groove which may serve as the substrate-binding pocket. The conserved catalytic residues (Glu72, Trp110, Asn119, Phe167, Tyr183 and Asp232) are localized at the centre of the groove. This crystal structure will help to improve the understanding of the hydrolytic mechanism of PssZ and its application as a biofilm disrupter.
1. Introduction
Biofilms, an important bacterial lifeform, are prevalent in nature, in industry and in clinical settings (Costerton et al., 1995 ▸; Stoodley et al., 2002 ▸). One of the most important features of biofilms is the biofilm matrix, which holds the biofilm cells together and protects the cells from antibiotics and the host immune response (Stewart & Costerton, 2001 ▸; Haussler & Parsek, 2010 ▸). The biofilm matrix is mainly composed of exopolysaccharide (EPS), proteins and extracellular DNA (eDNA) (Stoodley et al., 2002 ▸; Flemming & Wingender, 2010 ▸). In many bacteria, EPSs are indispensable for biofilm formation. Mutants with impaired EPS biosynthesis are severely compromised or are unable to form mature biofilms (Watnick & Kolter, 1999 ▸; Danese et al., 2000 ▸; Ma et al., 2009 ▸). Hence, factors that disrupt EPSs may disperse biofilms. Recently, microbial EPS glycoside hydrolases have been exploited as a method to prevent and disrupt diverse biofilms from both Gram-positive and Gram-negative bacteria. The noncytotoxic glycoside hydrolases PslGh and PelAh from Pseudomonas aeruginosa inhibit biofilm formation, rapidly disrupt established biofilms and increase the susceptibility of P. aeruginosa cells to killing by human neutrophils and antibiotics (Yu et al., 2015 ▸; Baker et al., 2016 ▸). These functions are achieved by degrading the Psl and Pel EPSs of P. aeruginosa. The fungal glycoside hydrolase Sph3h from Aspergillus fumigatus is able to degrade preformed A. fumigatus biofilms by hydrolyzing galactosaminogalactan (GAG) EPS. Interestingly, PelAh disrupts A. fumigatus fungal biofilms by the hydrolysis of GAG (Snarr et al., 2017 ▸). Thus, putative glycoside hydrolases from bacteria may be considered as antibiofilm agents.
Listeria monocytogenes is a ubiquitous human pathogen that thrives in diverse environments such as soil, water, animals and humans (Hamon et al., 2006 ▸; Ivanek et al., 2006 ▸). L. monocytogenes causes the foodborne disease listeriosis. Although this disease is rare, it has a high mortality rate of up to 30% (Buchanan et al., 2017 ▸). Food is considered to be the main transmission vehicle for listeriosis and has been estimated to account for 90% of listeriosis in the USA (Scallan et al., 2011 ▸). L. monocytogenes adheres to food-contact surfaces, such as metal (stainless steel), rubber, plastic (polystyrene) and glass (Poimenidou et al., 2016 ▸; Reis-Teixeira et al., 2017 ▸; Gilmartin et al., 2016 ▸; Di Bonaventura et al., 2008 ▸), and further establishes a biofilm. Therefore, the elimination of L. monocytogenes biofilms in food-processing plants is critical for food safety. PssZ is a glycoside hydrolase from L. monocytogenes that can degrade ManNAc-Gal EPS (Köseoğlu et al., 2015 ▸). The trisaccharide repeating unit structure of this EPS is {4)-β-ManpNAc-(1–4)-[α-Galp-(1–6)]-β-ManpNAc-(1-}, where ManpNAc is N-acetylmannosamine and Galp is galactose (Köseoğlu et al., 2015 ▸). PssZ degrades the ManNAc-Gal EPS to disperse aggregates of L. monocytogenes (Köseoğlu et al., 2015 ▸), suggesting that PssZ has the potential to inhibit or eradicate biofilms formed by L. monocytogenes.
In this study, we describe the cloning, expression, purification and high-resolution crystal structure of PssZ from L. monocytogenes 1.9136 (CVCC 1597). The structure adopts a classic glycoside hydrolase family 8 (GH-8) (α/α)6-barrel fold which forms a conserved groove. Structure alignment shows that the conserved residues (Glu72, Trp110, Asn119, Phe167, Tyr183 and Asp232) are located in this groove and are believed to participate in the catalytic reaction.
2. Materials and methods
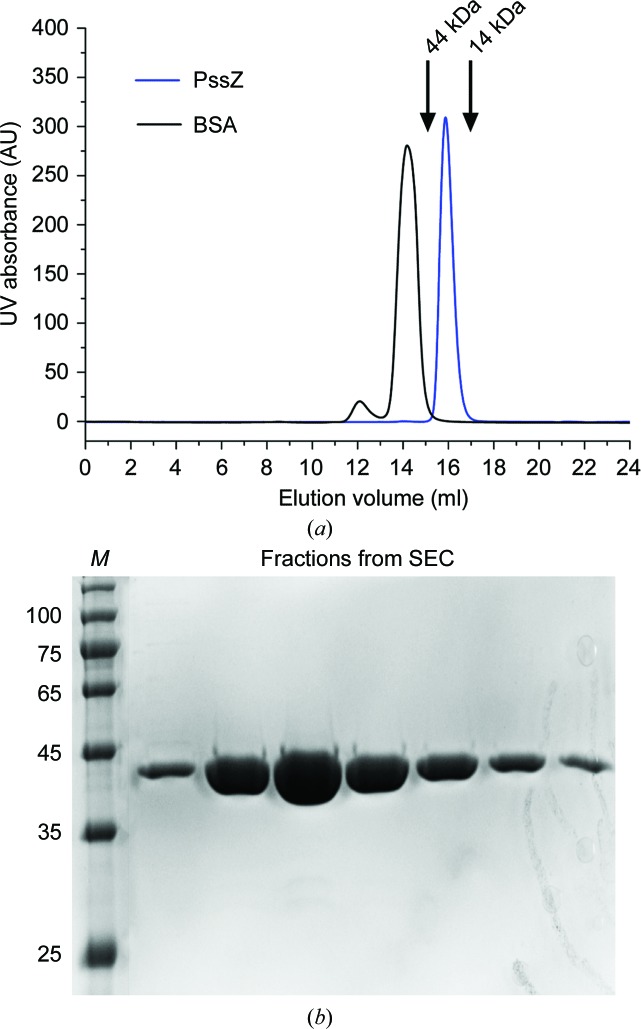
2.1. Macromolecule production
The coding sequence for PssZ (residues 23–334) was cloned into the pET-29a vector with a C-terminal hexahistidine tag. Expression of PssZ in Escherichia coli BL21 (DE3) cells was induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at an OD600 of 0.8 for 3 h at 37°C. The cells were harvested by centrifugation and resuspended in lysis buffer (20 mM Tris–HCl pH 7.4, 0.5 M NaCl, 30 mM imidazole). The cells were lysed by sonication and centrifuged at 15 000g for 30 min. The supernatant was loaded onto pre-equilibrated Ni–NTA resin (GE Healthcare) and washed with lysis buffer. PssZ was eluted with elution buffer consisting of 20 mM Tris–HCl pH 7.4, 0.5 M NaCl, 250 mM imidazole. The target protein was further purified by size-exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with 20 mM Tris–HCl pH 7.4, 0.5 M NaCl, 5%(v/v) glycerol [Fig. 1 ▸(a)]. After analysis by SDS–PAGE [Fig. 1 ▸(b)], the purified protein was concentrated to 10 mg ml−1 for crystallization. Selenomethionine (SeMet)-derivatized PssZ was produced in E. coli BL21 (DE3) cells grown in M9 medium supplemented with 1 mg ml−1 lysine, threonine and phenylalanine and 0.5 mg ml−1 leucine, isoleucine and valine, as well as 0.6 mg ml−1 l-selenomethionine. The purification of SeMet-derivatized PssZ was as described for wild-type PssZ. Macromolecule-production information is summarized in Table 1 ▸.
Figure 1.
SDS–PAGE analysis of purified recombinant PssZ. (a) Size-exclusion chromatogram (SEC) of PssZ from the final purification column. Bovine serum albumin (BSA) was loaded as a molecular-weight control. (b) SDS–PAGE gel of PssZ from the SEC peak fraction. The protein fractions were resolved on a gradient SDS–PAGE gel (12%) and were stained with Coomassie Blue for visualization. Lane M, molecular-weight markers (labelled in kDa).
Table 1. Macromolecule-production information.
| Source organism | L. monocytogenes 1.9136 (CVCC 1597) |
| DNA source | DNA |
| Forward primer† | GGAATTCCATATGCGACCAGAGTCTAAAAAG |
| Reverse primer‡ | GGGAAGCTTTTTCTTAAGAGTTATTTCTTTATT |
| Cloning vector | pET-29a vector |
| Expression vector | pET-29a vector |
| Expression host | E. coli BL21 (DE3) |
| Complete amino-acid sequence of the construct produced§ | MRPESKKTVSAPKETTPTSTSVQTYVKENYTAKNGLIVDYKNAQEPHYLAESIGLYMEYLVEVNDSKTFQEQVSHLEKNFITEDNFIKWEATDATTTNAIVDDFRITEALYQASEKFSFPSYKKMADKILANTKKYSAEQGVPVDFYDFVHKKKADTLHLSYLNIQAMQQINYRDKAYLPIQTVNADPFFTEVFQNEQFQYADPSEVNMIDQMLIAMAYFDENGDVEPNFDNFLQTELASKGKVYARYQRETKKPSSENESTAVYAFLTQYFNKTNQAKNGKITKELLEKMDTSNPETTHFFDYINKEITLKKKHHHHHH |
The NdeI site is underlined.
The HindIII site is underlined.
The His6 tag and Lys from the pET-29a vector are shown in italics.
2.2. Crystallization
Initial screening for crystallization was performed by the sitting-drop vapour-diffusion method at 289 K using crystallization kits from Hampton Research. Crystals were observed in several conditions after three days. The crystallization condition was further optimized by altering the pH and the precipitant concentrations. Finally, the best native crystals were grown under conditions consisting of 7%(v/v) PEG 400, 24%(v/v) PEG 5000 MME, 0.1 M imidazole pH 7.0. The best SeMet-derivatized crystals were grown under conditions consisting of 0.2 M ammonium acetate pH 7.0, 26%(v/v) PEG 3350. Crystallization information is summarized in Table 2 ▸.
Table 2. Crystallization.
| Native crystal | SeMet-derivatized crystal | |
|---|---|---|
| Method | Hanging-drop vapour diffusion | Hanging-drop vapour diffusion |
| Plate type | 24-well plates | 24-well plates |
| Temperature (K) | 289 | 289 |
| Protein concentration (mg ml−1) | 15 | 10 |
| Buffer composition of protein solution | 20 mM Tris–HCl pH 7.4, 0.5 M NaCl, 5%(v/v) glycerol | 20 mM Tris–HCl pH 7.4, 0.5 M NaCl, 5%(v/v) glycerol |
| Composition of reservoir solution | 7%(v/v) PEG 400, 24% PEG 5000 MME, 0.1 M imidazole pH 7.0 | 0.2 M ammonium acetate pH 7.0, 26%(v/v) PEG 3350 |
| Volume and ratio of drop | 2 µl; 1:1 ratio | 2 µl; 1:1 ratio |
| Volume of reservoir (ml) | 0.5 | 0.5 |
2.3. Data collection and processing
Both native and SeMet-derivatized crystals were cryoprotected by soaking them in reservoir solution supplemented with 15%(v/v) glycerol for 5–10 s and were then flash-cooled in liquid nitrogen. Diffraction data were collected from native and SeMet-derivatized crystals on beamlines BL17U1 and BL19U at Shanghai Synchrotron Radiation Facility (SSRF), People’s Republic of China. All data were processed and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▸). Data-collection and processing information is summarized in Table 3 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Native | SAD | |
|---|---|---|
| Diffraction source | BL17U1, SSRF | BL19U, SSRF |
| Wavelength (Å) | 0.97853 | 0.97855 |
| Temperature (K) | 100 | 100 |
| Detector | ADSC Q315r CCD | PILATUS3 6M |
| Crystal-to-detector distance (mm) | 300 | 500 |
| Rotation range per image (°) | 1.0 | 0.5 |
| Total rotation range (°) | 360 | 360 |
| Exposure time per image (s) | 0.2 | 0.2 |
| Space group | P1211 | P1211 |
| a, b, c (Å) | 40.1, 71.6, 131.3 | 39.6, 70.7, 130.4 |
| α, β, γ (°) | 90.0, 95.8, 90.0 | 90.0, 95.9, 90.0 |
| Resolution range (Å) | 43.52–1.62 (1.64–1.62) | 43.24–2.51 (2.58–2.51) |
| Total No. of reflections | 549009 (26844) | 144732 (12294) |
| No. of unique reflections | 89287 (4350) | 42685 (3516) |
| Completeness (%) | 94.8 (93.2) | 88.7 (99.7) |
| Multiplicity | 6.1 (6.2) | 3.4 (3.5) |
| 〈I/σ(I)〉 | 11.7 (5.2) | 17.9 (7.3) |
| R merge | 0.082 (0.316) | 0.053 (0.139) |
| R p.i.m. | 0.053 (0.204) | 0.034 (0.087) |
| Overall B factor from Wilson plot (Å2) | 11.46 | 25.63 |
2.4. Structure solution and refinement
The structure of SeMet-derivatized PssZ was determined by the single-wavelength anomalous dispersion (SAD) method. The Se atoms were located by SHELXC/D (Sheldrick, 2010 ▸). Initial phasing was carried out using the PHENIX AutoSol wizard (Adams et al., 2010 ▸) and the initial models were built using the PHENIX AutoBuild wizard (Terwilliger et al., 2009 ▸). The native structure was determined with Phaser (McCoy et al., 2007 ▸) using the SeMet-derivatized structure as the search model. Refinement and model building were carried out iteratively using phenix.refine (Afonine et al., 2012 ▸) and Coot (Emsley et al., 2010 ▸). The structure was validated using MolProbity (Chen et al., 2010 ▸). Atomic coordinates and structure factors have been deposited in the Protein Data Bank (http://www.pdb.org) as entry 6r2w. All structural figures were prepared using PyMOL (DeLano, 2002 ▸). The refinement statistics are summarized in Table 4 ▸.
Table 4. Structure refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 43.54–1.62 (1.675–1.617) |
| Completeness (%) | 94.8 (93.2) |
| No. of reflections | |
| Working set | 84955 (8302) |
| Test set | 4315 (406) |
| Final R cryst | 0.1704 (0.2001) |
| Final R free | 0.1965 (0.2356) |
| No. of non-H atoms | |
| Protein | 5110 |
| Water | 1044 |
| Total | 6154 |
| R.m.s. deviations | |
| Bonds (Å) | 0.007 |
| Angles (°) | 1.14 |
| Average B factors (Å2) | |
| Overall | 16.21 |
| Protein | 14.00 |
| Water | 27.04 |
| Ramachandran plot | |
| Favoured regions (%) | 99.02 |
| Additionally allowed (%) | 0.98 |
3. Results and discussion
3.1. Overall structure of PssZ
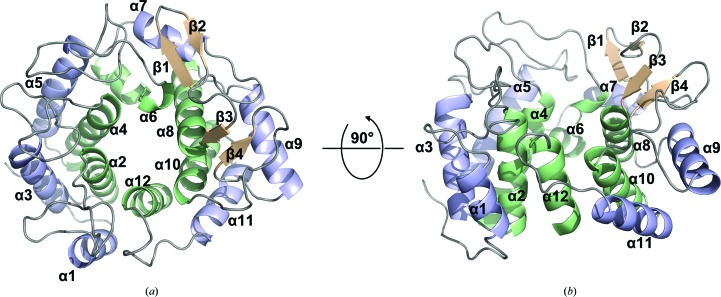
The crystal structure of native PssZ was determined by the single-wavelength anomalous dispersion (SAD) method using SeMet-derivatized PssZ. Of the total of 334 residues, the PssZ structure included residues 31–334. The final structure was refined at 1.6 Å resolution with an R work of 17.04% and an R free of 19.65% (Table 4 ▸). PssZ crystallized in space group P1211 with two molecules in the asymmetric unit. The structure of PssZ adopts a classical (α/α)6-barrel fold, in which six pairs of antiparallel α-helices (α2/α3, α4/α5, α6/α7, α8/α9, α10/α11 and α12/α1) form an inner and an outer ring [Fig. 2 ▸(a)]. This architecture forms a deep groove, which may serve as the substrate-binding pocket by comparison with other GH-8 structures (Mazur & Zimmer, 2011 ▸; Yasutake et al., 2006 ▸). Two-stranded antiparallel β-sheets are formed by the loops connecting helices α7/α8 and α9/α10, respectively [Fig. 2 ▸(a)]. The loops and two-stranded β-sheets flank the substrate-binding groove, creating a funnel on top of the hexameric rings [Fig. 2 ▸(b)].
Figure 2.
Overall structure of PssZ. PssZ is shown in cartoon representation from top (a) and side (b) views. The inner and outer helices are shown in pale green and light blue, respectively, and the two β-sheets are shown in wheat; loops are shown in grey. Helices are labelled α1–α12 from the N-terminus to the C-terminus. Strands are labelled β1–β4 from the N-terminus to the C-terminus.
3.2. Structural comparison of PssZ with its homologues
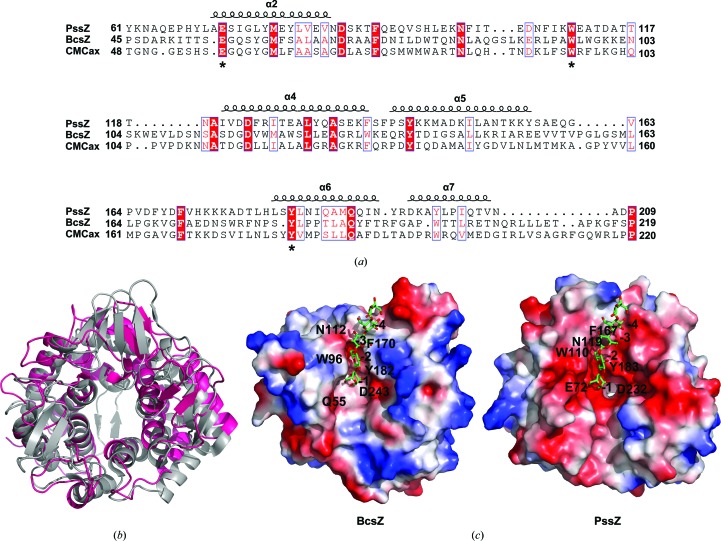
A BLAST search for similar proteins revealed that PssZ has no sequence homology to any protein in the PDB. The crystal structures of several enzymes in the GH-8 family have been determined, such as CMCax (PDB entry 1wzz; Yasutake et al., 2006 ▸) from Acetobacter xylinum and BcsZ (PDB entry 3qxq; Mazur & Zimmer, 2011 ▸) from E. coli. Pairwise structural comparison utilizing the DALI server (Holm & Rosenström, 2010 ▸) demonstrated that the r.m.s.d.s of corresponding Cα atoms between PssZ and CMCax and between PssZ and BcsZ were 3.0 Å for 268 Cα atoms (Z-score 23.2) and 3.0 Å for 280 Cα atoms (Z-score 23.7), respectively. The low r.m.s.d. values indicated overall structural similarity, whereas the sequence identities over the aligned regions were only 16% and 10%, respectively. As reported, BcsZ is one of the best characterized endo-β-1,4-glucanases, with its structure having been determined in complex with the substrate cellopentaose (Mazur & Zimmer, 2011 ▸). Structural comparison of PssZ with BcsZ showed that the main differences were the less pronounced organization of the β-sheets surrounding the substrate-binding groove of PssZ and the lack of the two-stranded β-sheet near the C-terminus of PssZ [Fig. 3 ▸(b)].
Figure 3.
Sequence and structure alignments of PssZ homologs. (a) Structure-based sequence alignment of PssZ with BcsZ (PDB entry 3qxq; Mazur & Zimmer, 2011 ▸) and CMCax (PDB entry 1wzz; Yasutake et al., 2006 ▸). Alignment was performed using ClustalX (v.1.81) and ESPript3. Identical residues are highlighted with a red background, conserved catalytic residues are boxed in blue and less conserved residues are shown as red letters. (b) Structure superimposition of PssZ (hot pink) with BcsZ (PDB entry 3qxq; grey). (c) Molecular surface-potential representation of BcsZ (PDB entry 3qxq) and PssZ. A model of the substrate in the structure with PDB code 3qxq is shown in the cleft of PssZ. The electrostatic surface potentials were generated using PyMOL in absolute mode. Areas coloured white, red and blue denote neutral, negative and positive potential, respectively.
BcsZ ultilizes three aromatic residues for sugar recognition by stacking interactions. These residues correspond to Tyr182, Trp96 and Phe170 and create three sugar-recognition subsites: −1, −2 and −3, respectively (Mazur & Zimmer, 2011 ▸). Structural comparison of PssZ with BcsZ revealed the corresponding conserved residues Tyr183, Trp110 and Phe167 in PssZ [Fig. 3 ▸(c)]. In the structure of the BcsZ–cellopentaose complex (PDB entry 3qxq; Mazur & Zimmer, 2011 ▸), the glucosyl residue at the −1 position forms a hydrogen bond to Gln55 via its OH-1 hydroxyl group and a bifurcated hydrogen bond to Asp243 via its OH-2 hydroxyl group. Superimposition of the structure of the active site revealed that two residues, Glu72 and Asp232, were also conserved in PssZ [Fig. 3 ▸(c)]. Asn119 in PssZ seems to play an identical role to Asn112 in BcsZ [Fig. 3 ▸(c)], which interacts with the glucosyl moiety at the −3 position. Sequence alignment showed that Glu72, Trp110 and Tyr183 of PssZ were highly conserved among GH-8 family members [indicated by asterisks in Fig. 3 ▸(a)], supporting the conservation of the active-site residues in the structural comparison of PssZ and BcsZ. These observations suggested that the interaction of PssZ with ManNAc-Gal EPS is similar to that of BcsZ with cellopentaose.
Here, we report the crystal structure of PssZ, a glycoside hydrolase from L. monocytogenes. The PssZ structure adopts a classical GH-8 family (α/α)6-barrel fold formed by six pairs of antiparallel helices. Structure comparisons revealed putative catalytic residues that may be associated with EPS binding in the core fold domain. The characterization of the PssZ structure provides insights into the hydrolytic mechanism, which can be explored in order to devise strategies for protection against Listeria in both healthcare and the food industry.
Supplementary Material
PDB reference: PssZ from Listeria monocytogenes, 6r2w
Acknowledgments
We thank Professor Yihua Huang at the Institute of Biophysics, Chinese Academy of Sciences for providing experimental facilities. We thank Dr Jingdong Cheng at the the Gene Center and Department of Biochemistry, Ludwig-Maximilians-Universitat München for providing technical support. We thank the staff of beamlines BL17U and BL19U at SSRF for support during data collection.
Funding Statement
This work was funded by National Natural Science Foundation of China grants 31828002 and 31570126.
References
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. [DOI] [PMC free article] [PubMed]
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Baker, P., Hill, P. J., Snarr, B. D., Alnabelseya, N., Pestrak, M. J., Lee, M. J., Jennings, L. K., Tam, J., Melnyk, R. A., Parsek, M. R., Sheppard, D. C., Wozniak, D. J. & Howell, P. L. (2016). Sci. Adv. 2, e1501632. [DOI] [PMC free article] [PubMed]
- Buchanan, R. L., Gorris, L. G. M., Hayman, M. M., Jackson, T. C. & Whiting, R. C. (2017). Food Control, 75, 1–13.
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R. & Lappin-Scott, H. M. (1995). Annu. Rev. Microbiol. 49, 711–745. [DOI] [PubMed]
- Danese, P. N., Pratt, L. A. & Kolter, R. (2000). J. Bacteriol. 182, 3593–3596. [DOI] [PMC free article] [PubMed]
- DeLano, W. (2002). PyMOL. http://www.pymol.org.
- Di Bonaventura, G., Piccolomini, R., Paludi, D., D’Orio, V., Vergara, A., Conter, M. & Ianieri, A. (2008). J. Appl. Microbiol. 104, 1552–1561. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Flemming, H. C. & Wingender, J. (2010). Nature Rev. Microbiol. 8, 623–633. [DOI] [PubMed]
- Gilmartin, N., Gião, M. S., Keevil, C. W. & O’Kennedy, R. (2016). Int. J. Food Microbiol. 219, 50–55. [DOI] [PubMed]
- Hamon, M., Bierne, H. & Cossart, P. (2006). Nature Rev. Microbiol. 4, 423–434. [DOI] [PubMed]
- Haussler, S. & Parsek, M. R. (2010). J. Bacteriol. 192, 2941–2949. [DOI] [PMC free article] [PubMed]
- Holm, L. & Rosenström, P. (2010). Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed]
- Ivanek, R., Gröhn, Y. T. & Wiedmann, M. (2006). Foodborne Pathog. Dis. 3, 319–336. [DOI] [PubMed]
- Köseoğlu, V. K., Heiss, C., Azadi, P., Topchiy, E., Güvener, Z. T., Lehmann, T. E., Miller, K. W. & Gomelsky, M. (2015). Mol. Microbiol. 96, 728–743. [DOI] [PubMed]
- Ma, L., Conover, M., Lu, H., Parsek, M. R., Bayles, K. & Wozniak, D. J. (2009). PLoS Pathog. 5, e1000354. [DOI] [PMC free article] [PubMed]
- Mazur, O. & Zimmer, J. (2011). J. Biol. Chem. 286, 17601–17606. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Poimenidou, S. V., Chrysadakou, M., Tzakoniati, A., Bikouli, V. C., Nychas, G. J. & Skandamis, P. N. (2016). Int. J. Food Microbiol. 237, 164–171. [DOI] [PubMed]
- Reis-Teixeira, F. B. D., Alves, V. F. & de Martinis, E. C. P. (2017). Braz. J. Microbiol. 48, 587–591. [DOI] [PMC free article] [PubMed]
- Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., Jones, J. L. & Griffin, P. M. (2011). Emerg. Infect. Dis. 17, 7–15. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2010). Acta Cryst. D66, 479–485. [DOI] [PMC free article] [PubMed]
- Snarr, B. D., Baker, P., Bamford, N. C., Sato, Y., Liu, H., Lehoux, M., Gravelat, F. N., Ostapska, H., Baistrocchi, S. R., Cerone, R. P., Filler, E. E., Parsek, M. R., Filler, S. G., Howell, P. L. & Sheppard, D. C. (2017). Proc. Natl Acad. Sci. USA, 114, 7124–7129. [DOI] [PMC free article] [PubMed]
- Stewart, P. S. & Costerton, J. W. (2001). Lancet, 358, 135–138. [DOI] [PubMed]
- Stoodley, P., Sauer, K., Davies, D. G. & Costerton, J. W. (2002). Annu. Rev. Microbiol. 56, 187–209. [DOI] [PubMed]
- Terwilliger, T. C., Adams, P. D., Read, R. J., McCoy, A. J., Moriarty, N. W., Grosse-Kunstleve, R. W., Afonine, P. V., Zwart, P. H. & Hung, L.-W. (2009). Acta Cryst. D65, 582–601. [DOI] [PMC free article] [PubMed]
- Watnick, P. I. & Kolter, R. (1999). Mol. Microbiol. 34, 586–595. [DOI] [PMC free article] [PubMed]
- Yasutake, Y., Kawano, S., Tajima, K., Yao, M., Satoh, Y., Munekata, M. & Tanaka, I. (2006). Proteins, 64, 1069–1077. [DOI] [PubMed]
- Yu, S., Su, T., Wu, H., Liu, S., Wang, D., Zhao, T., Jin, Z., Du, W., Zhu, M.-J., Chua, S. L., Yang, L., Zhu, D., Gu, L. & Ma, L. Z. (2015). Cell Res. 25, 1352–1367. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: PssZ from Listeria monocytogenes, 6r2w