Abstract
Purpose: The discovery of the plasmid-mediated colistin resistance genes, mcr, revealed a mechanism of transmission of colistin resistance, which is a major, global public health concern especially among individuals infected with carbapenem-resistant Gram-negative bacteria. To monitor the spread and epidemiology of mcr genes, a convenient and reliable method to detect mcr genes in clinical isolates is needed, especially in the primary care institutions. This study aimed to establish a restriction endonuclease-based multiplex loop-mediated isothermal amplification (multi-LAMP) assay to detect mcr genes (mcr-1 to mcr-5) harbored by colistin-resistant bacteria.
Methods: A triple-LAMP assay for mcr-1, mcr-3, and mcr-4 and a double-LAMP assay for mcr-2 and mcr-5 were established. The sensitivity and specificity of the LAMP reactions were determined via electrophoresis and visual detection.
Results: The sensitivity of the LAMP assay was 10-fold greater than that of PCR, with high specificity among the screened primers. Specific mcr genes were distinguished in accordance with band numbers and the fragment length of the digested LAMP amplification products. Furthermore, the LAMP assay was confirmed as a rapid and reliable diagnostic technique upon application for clinical samples, and the results were consistent with those of conventional PCR assay.
Conclusion: The multi-LAMP assay is a potentially promising method to detect mcr genes and will, if implemented, help prevent infections by drug-resistant bacteria in primary-care hospitals due to rapid and reliable surveillance. To our knowledge, this is the first study to report the application of LAMP to detect mcr-2 to mcr-5 genes and the first time that multi-LAMP has been applied to detect mcr genes.
Keywords: mcr genes, colistin resistance, multi-LAMP, rapid detection, enzyme digestion
Introduction
The misuse of antibiotics is the primary factor selecting for antibiotic resistance, followed by the unnecessary supplementation of animal feed with antibiotics.1 Among the various types of antibiotics, colistin, a last resort chemotherapeutic option against carbapenem-resistant bacteria, is faced with a growing clinical challenge of antibiotic resistance.2 Until recently, colistin resistance was believed to be chromosomally mediated; however, the discovery of plasmid-mediated colistin resistance via mcr-1 in 2015 revealed colistin resistance was capable of horizontal transmission.3,4 Subsequent studies reported that mcr-1 is expressed in many bacteria worldwide.3,5–7 Moreover, new mcr genotypes were reported, and eight more mcr genes (mcr-2 to mcr-9 )8 have been reported since the discovery of mcr-1.9–15 Currently, PCR-based methods are the most widely adopted to detect mcr genes,16 however, owing to the presence of several genotypes of mcr genes and their ability to undergo horizontal gene transfer; PCR assays are limited in clinical practice, including the primary-care hospitals and the basic quarantine stations. Therefore, a rapid, efficient, reliable, and economical method to detect mcr genes is urgently required.
Loop-mediated isothermal amplification (LAMP) is a nucleic acid amplification method conducted at a constant temperature, based on auto-cycling strand displacement of DNA synthesis by Bst DNA polymerase.17 It is a promising method to detect nucleic acids, with advantages including the non-requirement of thermo-cycling instruments, higher sensitivity, and less time-consumption. With the rapid development of the LAMP method, studies have reported increasing applications of LAMP, including nucleic acid detection from bacteria,18,19 viruses,20 and parasites.21–23 Zou et al first reported the application of LAMP to detect mcr-1, achieving higher sensitivity than traditional PCR.24 Imirzalioglu et al used the eazyplex SuperBug mcr-1 kit, developed to rapidly detect mcr-1.25 However, owing to the multiple mcr genotypes, a single LAMP cannot detect all potential target genes, thereby yielding incomplete information for nucleic acid detection.
In this study, we aimed to develop a restriction endonuclease-based multi-LAMP method to detect multiple mcr genes. We established a triple-LAMP system for the most extensively propagated mcr genes (mcr-1, mcr-3, and mcr-4) in People’s Republic of China. A double-LAMP system for mcr-2 and mcr-5 has also been established to detect all mcr genes (mcr-6 to mcr-9 had not been discovered when this study started). We designed and screened 5 sets of primers for each mcr-1 to mcr-5 genes and assessed the sensitivity and specificity of the assay through electrophoresis and visual detection, using clinical samples from hospitals in Guangzhou, People’s Republic of China. Primers for this assay were modified with restriction endonucleases and were used combinatorially for the multi-LAMP assay, and specific mcr genes were detected from the band numbers and fragment lengths of the digested LAMP-amplified products.
Materials and methods
Primers design
Sequences of mcr-1 to mcr-5 genes were downloaded from GenBank database: mcr-1 (accession number: KX886345.1); mcr-2 (accession number: NG_051171.1); mcr-3 (accession number: MG026622.1); mcr-4 (accession number: MG026621.1); and mcr-5 (accession number: MG241339.1). To determine the optimum primers, 3 primer sets for each gene were designed by using Primer Explorer (version 5, http://primerexplorer.jp/lampv5e/index.html) (Table S1). Each primer set comprises 6 primers: a forward inner primer (FIP), backward inner primer (BIP), outer forward primer (F3), outer backward primer (B3), and two loop primers (LF and LB) to accelerate the LAMP reaction.17 All primers were synthesized by Tianyi Biotech Co. (Dongguan, People’s Republic of China).
Table S1.
The primer sequences of LAMP for mcr-1 to mcr-5 genes
| Target gene | Primers | Sequences (5ʹ-3ʹ) | Reference |
|---|---|---|---|
| mcr-1 (1) | F3 | TGATGCAGCATACTTCTGTG | This study |
| B3 | GACCGTGCCATAAGTGTC | ||
| FIP | GCGATGGGATAGGTTTGGCTGTGTTGCCGTTTTCTTGAC | ||
| BIP | TGCTGACGATCGCTGTCGGCACATAGCGATACGATGAT | ||
| LF | AAGGTAAGATTGGCGGTCG | ||
| LB | CTACTGATCACCACGCTGTT | ||
| mcr-1 (2) | F3 | TGCTACCAAGTTTGCTTGT | This study |
| B3 | CGTCTTTGGCGTGATAAATG | ||
| FIP | AGACTTGCCACGATCAAGCCTTAAGGTGGATTATCCGACTTG | ||
| BIP | GCGTTCAGCAGTCATTATGCCGTAGATTGGCATGATCGGAT | ||
| LF | CAATCGGCGCATCAAACC | ||
| LB | CGCTGCGTAGCTATGTCA | ||
| mcr-1 (3) | F3 | ATACCGCCAAATACCAAGAAA | This study |
| B3 | CATCTAAGCCAACGAGCAT | ||
| FIP | GCAGCTTATCCATCACGCCTTGATACGCTGGATCGCTTG | ||
| BIP | GCCGATTATAAATCCGCGACCAGCGGCATTCGTTATAAGGA | ||
| LF | GTCCGAATTATTATCACGCCAC | ||
| LB | CAACGCCATCTGCAACAC | ||
| mcr-2 (1) | F3 | ACGCCTAGTGGTGTTCGT | This study |
| B3 | GCGGTCAAGCGTATCTAGC | ||
| FIP | AACTTTGGCAAGCTGCGGGAATGCTGACCATGTGCAGTTC | ||
| BIP | TCGACGGCGTATTCTGTGCCGGTATTTGGCGGTATCGACA | ||
| LF | AGTCTCACGGCCATAGCC | ||
| LB | GTGTATGTTCAGCTATTTGGGTCA | ||
| mcr-2 (2) | F3 | GGGTAAGCTTGCCAGTATCG | This study |
| B3 | AGCTGCGGGAAAGTCTCA | ||
| FIP | CTCGGCTTGGTGGTCTGCACCACTGCGCCAACAGACAC | ||
| BIP | GAGCGTAAGCCACGCCTAGTGCGGCCATAGCCATTGAACTG | ||
| LF | GGCGTCTTTGGCATGATAGATG | ||
| LB | GTGTTCGTCGTCGGTGAGA | ||
| mcr-2 (3) | F3 | CAAAGACGCCGTGCAGAC | This study |
| B3 | CAGAATACGCCGTCGATGT | ||
| FIP | ACTGCACATGGTCAGCACGCGAGCGTAAGCCACGCCTA | ||
| BIP | GGCTATGGCCGTGAGACTTTCCGCCACACGATGTCACTTGG | ||
| LF | ACCGACGACGAACACCAC | ||
| LB | CTTGCCAAAGTTGATGGCTTG | ||
| mcr-3 (1) | F3 | CATTACCAATATTGCTTGTTGC | This study |
| B3 | TTGGCTGGAACAATCTCAC | ||
| FIP | GCTAACGCCTCATTTTGATTGGTTGCACTTCTTATCGCACTTA | ||
| BIP | TCAAAGGGATTCTAACTCGTGCGCAATAACCGCAATCACTAT | ||
| LF | TCATTGTGTAACTAACGATTGC | ||
| LB | CCTATCGATGTTTGCATCACTT | ||
| mcr-3 (2) | F3 | CATTACCAATATTGCTTGTTGC | This study |
| B3 | TTGGCTGGAACAATCTCAC | ||
| FIP | GCTAACGCCTCATTTTGATTGGTGCACTTCTTATCGCACTTAG | ||
| BIP | TCAAAGGGATTCTAACTCGTGCGCAATAACCGCAATCACTAT | ||
| LF | TCATTGTGTAACTAACGATTGC | ||
| LB | CCTATCGATGTTTGCATCACTT | ||
| mcr-3 (3) | F3 | CATTACCAATATTGCTTGTTGC | This study |
| B3 | TTGGCTGGAACAATCTCAC | ||
| FIP | GCTAACGCCTCATTTTGATTGGGCACTTCTTATCGCACTTAGT | ||
| BIP | TCAAAGGGATTCTAACTCGTGCGCAATAACCGCAATCACTAT | ||
| LF | TCATTGTGTAACTAACGATTGC | ||
| LB | CCTATCGATGTTTGCATCACTT | ||
| mcr-4 (1) | F3 | TGAGTTAAGGCGTTACATTGT | This study |
| B3 | CGCATGAGCTAGTATCGTTAA | ||
| FIP | TTACGACTGGCATTCTTCGCACTATTTGCAGACGCCCAT | ||
| BIP | AGTGGTTGTTGTGGGTGAAACTAGCATTGGTTGGCTTGTTA | ||
| LF | TCTAGGCCAAGTTGTTGGTATT | ||
| LB | CGCGCTCAATGAGCTATCA | ||
| mcr-4 (2) | F3 | TGTTCGAAACAACAGTGAGT | This study |
| B3 | CGCATGAGCTAGTATCGTTAA | ||
| FIP | TTACGACTGGCATTCTTCGCATTGAGCACTATTTGCAGACG | ||
| BIP | AGTGGTTGTTGTGGGTGAAACTAGCATTGGTTGGCTTGTTA | ||
| LF | GCCAAGTTGTTGGTATTCCATG | ||
| LB | CGCGCTCAATGAGCTATCA | ||
| mcr-4 (3) | F3 | CATGGAATACCAACAACTTGG | This study |
| B3 | CGGCGAGGATCATAGTCT | ||
| FIP | AGTTTCACCCACAACAACCACTAGATGCGAAGAATGCCAG | ||
| BIP | CAAGCCAACCAATGCTCATACCTGCCGCATGAGCTAGTAT | ||
| LF | TAGGTTTAGTGTTCGGGTTACG | ||
| LB | GGGCTGATTGCGTTTAACG | ||
| mcr-5 (1) | F3 | CAATGGAGAATGCTGCCCTA | This study |
| B3 | GCGTGGGTATCAGCACATC | ||
| FIP | AGCCCGTTCGTAAAACCCTGACCTTGTTGGTTGCAGCCGT | ||
| BIP | AGCGGTAATGATGCGCAGCGCATGACTGGCCACAGACC | ||
| LF | CTCGCAATCCACCACACGGAT | ||
| LB | TGGCGCTCTCGCCATGA | ||
| mcr-5 (2) | F3 | ATGGATGTGCTGATACCCAC | This study |
| B3 | AGTACGAGAGCACGAGGAC | ||
| FIP | CGAATGCCCGAGATGACGTAGTCTTCGTGAAAACAAGCCGC | ||
| BIP | GACTGAACAGGCGTCATCGTCACCTTGTTCTTGAGGCCCTC | ||
| LF | GCAGGAGTGATCAAATAGCGAA | ||
| LB | GCAGACGAAGCAAGGGAAG | ||
| mcr-5 (3) | F3 | GCTGCCCTACTTGTTGGTTG | This study |
| B3 | CGAAGCGGCTTGTTTTCAC | ||
| FIP | TACCGCTTGTTTCCAGCCCGCAGCCGTATCCGTGTGGT | ||
| BIP | CTGGCGCTCTCGCCATGATTGAAGCGTGGGTATCAGCAC | ||
| LF | ACCCTGACTCTCGCAATCC | ||
| LB | TCCATGGGTCTGTGGCC |
Samples preparation
In our study, five positive controls harboring mcr-1 to mcr-5 genes were used, wherein mcr-1, mcr-3, and mcr-4 were screened in our laboratory, mcr-2 was obtained from South China Agricultural University, and mcr-5 was obtained from China Agricultural University (Table 1). Three multidrug-resistant bacterial strains (two Escherichia coli and one Klebsiella pneumonia) devoid of mcr genes, identified via PCR in our previous studies,26 were used as negative controls. All strains were stored in 30% (w/v) glycerol broth at −80°C. The strains were cultured in Luria-Bertani culture medium (OXOID, Hampshire, UK) supplemented with 2% colistin at 37°C overnight. Bacterial genomic DNA was extracted using the boiling method and recovered in 200 μL RNase-free ddH2O. pMD19-T vector containing mcr-1 to mcr-5 DNA fragments was constructed separately, as previously described.27,28 The recombinant plasmids were diluted 10-fold serially to yield 108 copies/μL to 102 copies/μL.
Table 1.
mcr genes and other resistant genes used in this study
| NO. | Genetic types | Source | Gene Role |
|---|---|---|---|
| 1 | mcr-1 | E. coli | Positive control |
| 2 | mcr-2 | Plasmid | Positive control |
| 3 | mcr-3 | E. coli | Positive control |
| 4 | mcr-4 | E. coli | Positive control |
| 5 | mcr-5 | Plasmid | Positive control |
| 6 | blaKPC-2 | K. pneumonia | Negative control |
| 7 | blaNDM-1 | E. coli | Negative control |
| 8 | blaCTX-M-9 | E. coli | Negative control |
LAMP reaction
The LAMP reaction was carried out in a 25 μL reaction mixture that contained 12.5 µL LAMP-Reaction Mix [20 mM Tris-HCl (pH 8.8), 10 mM (NH4)2SO4, 8 mM MgSO4, 10 mM KCl, 0.8 M betaine, 0.1% Tween-20, 1.4 mM deoxy-ribonucleotide triphosphates (dNTP)], 1 µL Bst 2.0 polymerase (New England Biolabs, 8,000 units/mL), 1.25 µL primer mix (2 μM each of FIP and BIP, 0.25 µM each of F3 and B3, and 1 µM each of LF and LB) (Table 2), 8.25 µL nuclease-free water, and 2 µL DNA lysate. The mixture was incubated for 60 mins at 64°C in a heated, thermostatically controlled water bath.
Table 2.
The primer sequences of multi-LAMP and PCR for mcr-1 to mcr-5 genes
| Target genes | multi-LAMP | Reference | PCR | Reference | ||
|---|---|---|---|---|---|---|
| Primers | Sequences(5ʹ-3ʹ)a | Primers | Sequences(5ʹ-3ʹ) | |||
| mcr-1 | F3 | TGATGCAGCATACTTCTGTG | This study | F | AGTCCGTTTGTTCTTGTGGC | 16 |
| B3 | GACCGTGCCATAAGTGTC | |||||
| FIP | GCGATGGGATAGGTTTGGCTAAGCTTGTGTTGCCGTTTTCTTGAC | |||||
| BIP | TGCTGACGATCGCTGTCGAAGCTTGCACATAGCGATACGATGAT | R | AGATCCTTGGTCTCGGCTTG | 16 | ||
| LF | AAGGTAAGATTGGCGGTCG | |||||
| LB | CTACTGATCACCACGCTGTT | |||||
| mcr-2 | F3 | CAAAGACGCCGTGCAGAC | This study | F | CAAGTGTGTTGGTCGCAGTT | 16 |
| B3 | CAGAATACGCCGTCGATGT | |||||
| FIP | ACTGCACATGGTCAGCACGCAAGCTTGAGCGTAAGCCACGCCTA | |||||
| BIP | GGCTATGGCCGTGAGACTTTCCAAGCTTGCCACACGATGTCACTTGG | R | TCTAGCCCGACAAGCATACC | 16 | ||
| LF | ACCGACGACGAACACCAC | |||||
| LB | CTTGCCAAAGTTGATGGCTTG | |||||
| mcr-3 | F3 | CATTACCAATATTGCTTGTTGC | This study | F | AAATAAAAATTGTTCCGCTTATG | 16 |
| B3 | TTGGCTGGAACAATCTCAC | |||||
| FIP | GCTAACGCCTCATTTTGATTGGAAGCTTGCACTTCTTATCGCACTTAG | |||||
| BIP | TCAAAGGGATTCTAACTCGTGCAAGCTTGCAATAACCGCAATCACTAT | R | AATGGAGATCCCCGTTTTT | 16 | ||
| LF | TCATTGTGTAACTAACGATTGC | |||||
| LB | CCTATCGATGTTTGCATCACTT | |||||
| mcr-4 | F3 | TGAGTTAAGGCGTTACATTGT | This study | F | TCACTTTCATCACTGCGTTG | 16 |
| B3 | CGCATGAGCTAGTATCGTTAA | |||||
| FIP | TTACGACTGGCATTCTTCGCAAAGCTTCTATTTGCAGACGCCCAT | |||||
| BIP | AGTGGTTGTTGTGGGTGAAACTAAGCTTAGCATTGGTTGGCTTGTTA | R | TTGGTCCATGACTACCAATG | 16 | ||
| LF | TCTAGGCCAAGTTGTTGGTATT | |||||
| LB | CGCGCTCAATGAGCTATCA | |||||
| mcr-5 | F3 | CAATGGAGAATGCTGCCCTA | This study | F | ATGCGGTTGTCTGCATTTATC | 16 |
| B3 | GCGTGGGTATCAGCACATC | |||||
| FIP | AGCCCGTTCGTAAAACCCTGACAAGCTTCTTGTTGGTTGCAGCCGT | |||||
| BIP | AGCGGTAATGATGCGCAGCGAAGCTTCATGACTGGCCACAGACC | R | TCATTGTGGTTGTCCTTTTCTG | 16 | ||
| LF | CTCGCAATCCACCACACGGAT | |||||
| LB | TGGCGCTCTCGCCATGA | |||||
Note: aUnderlining indicates the restriction enzyme sites of Hind.
Abbreviations: F3, Forward Outer Primer; B3, Backward Outer Primer; FIP, Forward Internal Primer; BIP, Backward Internal Primer; LF, Loop Forward Primer; LB, Loop Backward Primer; F, Forward Primer; R, Reverse Primer.
PCR assay
A PCR assay was performed to compare its sensitivity and the clinical detection rates with those of the LAMP assay. Each plasmid sample was amplified in 20 µL reaction mixtures containing 10 µL PCR Master Mix (Tiangen Biotech Co., Ltd., Beijing, People’s Republic of China), 400 pM primers (Table 2), and 1 µL DNA template. The cycling conditions were as follows: 3 mins at 95°C; 30 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C; 5 mins at 72°C. The PCR products were analyzed electrophoretically on a 2% agarose gel, followed by ethidium bromide staining. Images were obtained using the (Bio-Rad, Hercules, CA, USA).
Detection of LAMP products
LAMP products were detected using two methods: visual detection29 and electrophoresis. For visual detection, SYBR Green I was added into the LAMP products, where the positive reactions yielded green coloration; while the negative ones gave yellow. For electrophoresis, the LAMP products were stained with GoldView TM, analyzed electrophoretically on a 2% agarose gel, and photographed.
Multi-LAMP detection
The triple-LAMP assay was performed using a set of three primer pairs each, for mcr-1, mcr-3, and mcr-4. The double-LAMP assay was performed using a set of three primer pairs each, for mcr-2 and mcr-5. The amplification was carried out in a 25 μL reaction mixture as the LAMP reaction. Both of the amplification products of the triple-LAMP and double-LAMP were diluted by twofold, digested using Thermo Scientific Fast Digest Hind at 37°C for 15 mins and then analyzed electrophoretically on a 2% agarose gel. Through the electrophoresis results, different types of mcr genes were distinguished based on the band numbers and band locations on electropherograms.
Results
Primer selection and modification
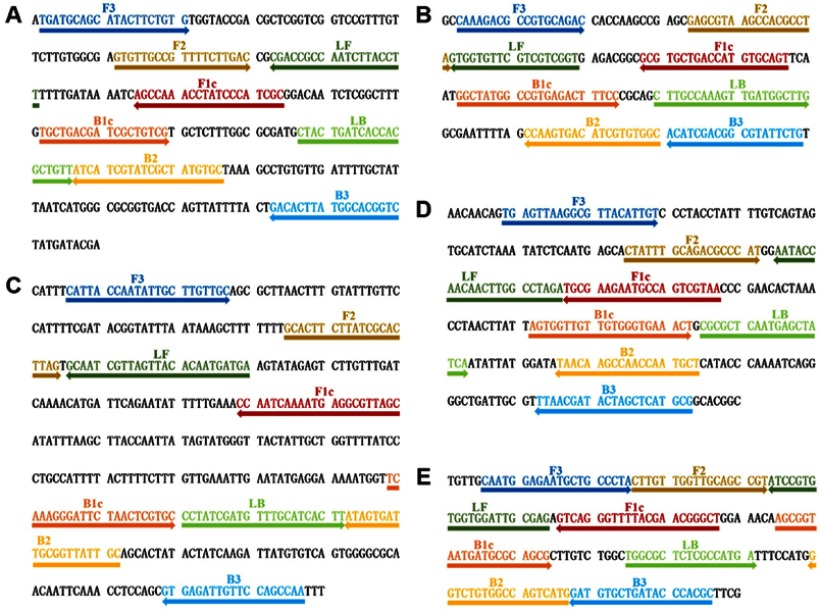
Fifteen primer sets were designed to detect mcr-1 to mcr-5 (3 sets for each gene) as shown in Table S1. To determine the optimum primers, LAMP reactions using different primer sets were conducted under the same conditions, and electropherograms of the LAMP products were compared. According to the electrophoretic analysis, the optimum primer sets were selected. The locations and sequences of selected optimal primer sets are shown in Figure 1. Additionally, for the multi-LAMP, we modified the selected optimal FIPs and BIPs by inserting the Hind restriction sites between F1c and F2 and between B1c and B2, respectively (Table 2).
Figure 1.
Locations and sequences of mcr genes used to design multi-LAMP primers. (A–E) The nucleotide sequences of the target strands of mcr-1 to mcr-5 genes. Right arrows indicate the original sequences and left arrows indicate the complementary sequences.
Sensitivity of the LAMP assay
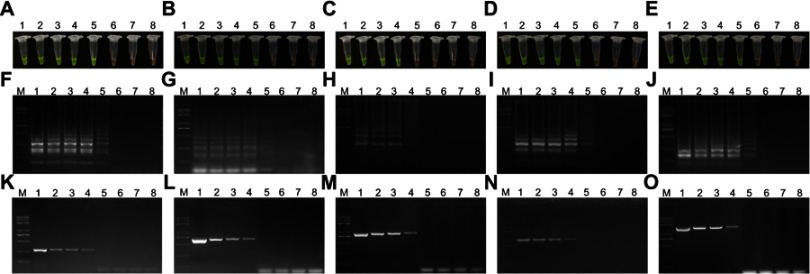
To determine the sensitivity of the LAMP assay, purified genomic DNA templates were detected via 10-fold serial dilution of known copies numbers of DNA template molecules. Comparative PCR assays were also conducted using the same DNA templates. LAMP products were further analyzed visually (Figure 2A–E) and via agarose gel electrophoresis (Figure 2F–O). These results indicated that the detection limit of the LAMP assay was 104 copies/µL for mcr-1, mcr-2, mcr-4 and mcr-5, and 105 copies/µL for mcr-3. Comparatively, agarose gel electrophoresis was conducted for the PCR products, wherein the detection limits were 105 copies/µL for mcr-1 to mcr-5 genes. All reactions were carried out at least in triplicate.
Figure 2.
Sensitivity of the loop-mediated isothermal amplification (LAMP) and polymerase chain reaction (PCR) assays. (A–E) Visual detection of the LAMP amplification products of mcr-1 to mcr-5 genes with SYBR Green I. (F–J) Agarose gel electrophoresis was conducted for the LAMP products of mcr-1 to mcr-5 genes. (K–O) Comparative agarose gel electrophoresis analysis of products of the PCR assay and the corresponding LAMP assay. Lane M, Trans 2K plus II DNA marker; Lanes 1–7, serial 10-fold dilutions of templates from 108 copies/µL to 102 copies/µL; Lane 8, negative (water).
Specificity of the LAMP assay
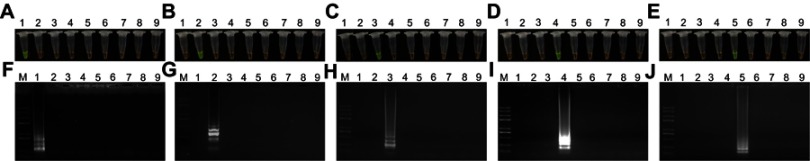
Assay specificity was assessed via direct visual detection through the addition of SYBR Green I during the reaction, followed by confirmatory evaluation via agarose gel electrophoresis. Three non-mcr genes (blaKPC-2, blaNDM-1, and blaCTX-M-9) were analyzed in addition to mcr-1 to mcr-5 genes. Our results revealed that mcr-1 was positively identified via LAMP only when amplification was performed using the mcr-1 primer set, while all non-mcr-1 genes tested negative (Figure 3A and F). The mcr-2 to mcr-5 genes were similarly analyzed, indicating that this LAMP method was specific for mcr-1 to mcr-5 genes (Figure 3B–E and G–J). All reactions were carried out at least in triplicate.
Figure 3.
Specificity of the loop-mediated isothermal amplification (LAMP) assays. (A–E) Visual detection of the LAMP amplification products with SYBR Green I. (F–J) Agarose gel electrophoresis of the LAMP products. Lane M: Trans 2K plus II DNA marker; Lanes 1–5: mcr-1 to mcr-5 genes; Lane 6: blaKPC-2; Lane 7: blaNDM-1; Lane 8: blaCTX-M-9; Lane 9: negative (water).
Multi-LAMP detection of mcr genes
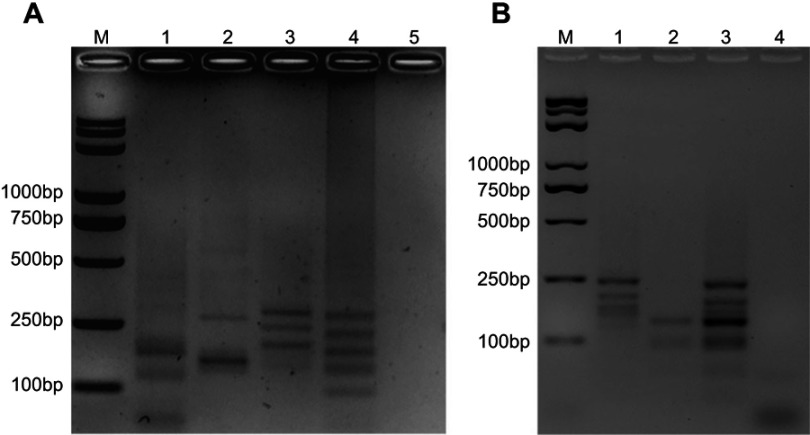
To generate the multi-LAMP system, endonuclease analysis was applied. For cleavage sites in mcr-1 to mcr-5 genes, enzyme digestion was performed using Hind restriction enzyme. For the triple-LAMP, the target sequence of mcr-1 was cleaved into 2 segments, mcr-3 into 2 segments and mcr-4 into 3 segments (Figure 4A). For the double-LAMP, the target sequence of mcr-2 was cleaved into 3 segments and mcr-5 into 2 segments (Figure 4B). The sizes of the digested products were consistent with those predicted for mcr-1 to mcr-5 genes. The DNA sequencing of the digested products confirmed the specificity of the amplification (data not shown). Through restriction endonuclease analysis of the amplification products, mcr-1 to mcr-5 genes were successfully distinguished (Figure 4A, mcr-1, mcr-3, and mcr-4; Figure 4B, mcr-2 and mcr-5).
Figure 4.
Multiplex loop-mediated isothermal amplification (multi-LAMP) detection. Agarose gel electrophoresis and enzyme digestion analysis of mcr genes was performed for the multi-LAMP products on 2% agarose gel. (A) Lane M, Trans 2K plus II DNA marker; Lane 1, restriction enzyme digestion of mcr-1 multi-LAMP products, 170 bp, 115 bp respectively; Lane 2, restriction enzyme digestion of mcr-3 multi-LAMP products, 260 bp, 155 bp respectively; Lane 3, restriction enzyme digestion of mcr-4 multi-LAMP product, 270, 230, and 185 bp respectively; Lane 4, restriction enzyme digestion of mixed mcr-1, mcr-3, and mcr-4 multi-LAMP products, 260, 225, 170, 120, and 90 bp; Lane 5, negative (water). (B) Lane M, Trans 2K plus II DNA marker; Lane 1, restriction enzyme digestion of mcr-2 multi-LAMP products, 220, 175, and 140 bp respectively; Lane 2, restriction enzyme digestion of mcr-5 multi-LAMP products, 120 bp, 90 bp respectively; Lane 3, restriction enzyme digestion of mixed mcr-2 and mcr-5 multi-LAMP products, 215, 170, 140, 120, and 90 bp, respectively; Lane 4: negative (water).
Multi-LAMP detection of clinical samples
To further evaluate the accuracy of multi-LAMP, 58 clinical bacterial samples from a previous study26 were subjected to the present assay. Among the 58 clinical samples, 12 were positive for mcr-1, five for mcr-3, and one for mcr-4, while the remaining 40 samples were mcr negative. The clinical bacterial samples were also subjected to traditional PCR analysis, which also revealed twelve mcr-1-positive samples, five mcr-3-positive samples, one mcr-4-positive sample, and forty negative samples, consistent with the results of multi-LAMP. Therefore, these findings reveal that multi-LAMP described here showed good consistency with conventional PCR analyses.
Discussion
Colistin is a polypeptide antibiotic, belonging to the family of polymyxins. Colistin was first reported to treat infections by Gram-negative bacteria in the late 1940s. Owing to its risk of nephrotoxicity and neurotoxicity, colistin was not popularized in clinical treatment. Later, antibiotics acting on emerging carbapenem-resistant superbugs were deemed critical because of their high morbidity and mortality, framing colistin as a significant therapeutic alternative.2 Emerging colistin resistance, therefore, has confounding implications in patient care. Before 2015, the mechanisms underlying colistin were known only to involve chromosomal mutations; hence, its spread was expected to be limited to vertical transmission, which was usually stable, incapable of spreading to other bacteria and imposing a fitness cost upon the bacteria. The discovery of mcr genes signifies plasmid-meditated colistin resistance, implying a new horizontal transmission channel for the propagation of the mcr genes, increasing their rapid transmission risk and range. Moreover, colistin abuse via its supplementation in animal feed contributed to its dissemination through horizontal gene transfer.4 Therefore, analysis of mcr genes is necessary to identify colistin resistance and control its horizontal transmission.
This study established a novel multi-LAMP system to detect multiple mcr genes, with the establishment of a triple-LAMP for mcr-1, mcr-3, and mcr-4, as well as a double-LAMP for mcr-2 and mcr-5. We designed 15 primer pairs for the conserved sequences of mcr-1 to mcr-5 genes (3 for each gene). Because mcr genes share significant homology, we screened the best primer sets by considering mcr genes as positive controls. Five primer sets each for mcr-1 to mcr-5 genes were screened out and determined to be the optimal primers for the LAMP assay. Subsequent specificity analyses confirmed the optimal nature of these primer sets. We compared the sensitivity of LAMP with that of traditional PCR analysis, reporting that the detection limit of the LAMP assay was 10-fold that of conventional PCR analysis. This high sensitivity and specificity rendered LAMP suitable for early screening in clinical settings, especially in the primary medical institutions. Through restriction digestion of the LAMP products based on band numbers and fragment lengths, we successfully distinguished mcr-1, mcr-3, and mcr-4, as well as mcr-2 and mcr-5, thereby enabling multiplex detections of mcr genes. Additionally, we applied this multi-LAMP method to clinical samples to assess the reliability of our methods, with results being consistent with those of conventional PCR analyses.
Compared with traditional PCR, the present method exhibited the following advantages: 1) high specificity and sensitivity and ease of operation; 2) multiplex detections of mcr genes using the same detection system, thus reducing manual operation; 3) a total operating time of <60 mins, as opposed to 90 mins to detect mcr genes via conventional PCR analysis; 4) greater user-friendliness than conventional PCR analysis, with no requirement of specialized instruments and complicated operations. These advantages render the multi-LAMP assay promising for clinical application, especially in resource limited medical institutions.
This study has some limitations; the sensitivity and specificity of the multi-LAMP assay are relatively poorer when applied to samples containing more than one mcr genes. Nonetheless, according to other previous studies and data from our on-going experiments, it is rare for a single strain to contain multiple mcr genes in clinical samples (0.01% is according to our data; unpublished data). Therefore, our approach is applicable in most practical situations. Moreover, gene sets (mcr-1, mcr-3, and mcr-4) and gene sets (mcr-2 and mcr-5) were successfully detected, where mcr-1, mcr-3, and mcr-4 are the most prevalent mcr genes in People’s Republic of Chinaand mcr-2 and mcr-5, not yet detected in humans, complemented the whole mcr gene family. With newer mcr genes being reported (mcr-6 to mcr-9 genes during our study in 2018) in future studies, continuous efforts are needed to track newly reported mcr genes and to establish a multi-LAMP system encompassing all mcr genes.
In our future studies, we intend to investigate the LAMP-based methods for drug resistance genes of these mcr genes and to continuously track the newly reported mcr genes. Upon analyzing more mcr genes and using larger sample sizes, we intend to further assess the potential of multi-LAMP and further validate its diagnostic power. These results provide critical insights into the efficacy of this method for future clinical applications related to antibiotic resistance. Our results potentially contribute to the prevention of antibiotic resistance in health care settings by providing a system of early detection of antibiotic resistance and prescribing patterns at national, regional, and local levels.
Conclusion
In conclusion, we established a restriction digestion-based multi-LAMP method, which successfully detected mcr-1, mcr-3, and mcr-4 via a triple-LAMP and mcr-2 and mcr-5 via a double-LAMP. We anticipate that this method could rapidly help screen mcr genes and other drug-resistant genes in a variety of clinical settings. To our knowledge, this is the first study reporting the application of LAMP in the detection of mcr-2 to mcr-5 genes and the first time that multi-LAMP has been applied to detect mcr genes.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (grant numbers 81830103 and 81722030), and the Guangdong Natural Science Foundation (grant number 2017A030306012).
Disclosure
The authors report no conflicts of interest in this work.
Supplementary material
References
- 1.Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States; 2013. Available from: https://www.cdc.gov/drugresistance/pdf/ar-threats.
- 2.Lim LM, Ly N, Anderson D, et al. Resurgence of colistin: a review of resistance, toxicity, pharmacodynamics, and dosing. Pharmacotherapy. 2010;30(12):1279–1291. doi: 10.1592/phco.30.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasman H, Hammerum AM, Hansen F, et al. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill. 2015;20(49). doi: 10.2807/1560-7917.ES.2015.20.49.30085 [DOI] [PubMed] [Google Scholar]
- 4.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(16)30197-9 [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Zhao XF, Che J, et al. Detection and dissemination of the colistin resistance gene, mcr-1, from isolates and faecal samples in China. J Med Microbiol. 2017;66(2):119–125. doi: 10.1099/jmm.0.000425 [DOI] [PubMed] [Google Scholar]
- 6.Yao X, Doi YH, Zeng L, Lv LC, Liu JH. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis. 2016;16(3):288–289. doi: 10.1016/S1473-3099(16)30197-9 [DOI] [PubMed] [Google Scholar]
- 7.Schwarz S, Johnson AP. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother. 2016;71(8):2066–2070. doi: 10.1093/jac/dkw274 [DOI] [PubMed] [Google Scholar]
- 8.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. mBio. 2019;10(3):e00853––19.. doi: 10.1128/mBio.00853-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xavier BB, Lammens C, Ruhal R, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21(27):8–13. doi: 10.2807/1560-7917.ES.2016.21.27.30280 [DOI] [PubMed] [Google Scholar]
- 10.Yin W, Li H, Shen Y, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio. 2017;8(3):e00543–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carattoli A, Villa L, Feudi C, et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017;22(31). doi: 10.2807/1560-7917.ES.2017.22.31.30589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. 2017;72(12):3317–3324. doi: 10.1093/jac/dkx066 [DOI] [PubMed] [Google Scholar]
- 13.AbuOun M, Stubberfield EJ, Duggett NA, et al. mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother. 2018;73(10):2904. doi: 10.1093/jac/dky272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang YQ, Li YX, Lei CW, Zhang AY, Wang HN. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother. 2018;73:1791–1795. doi: 10.1093/jac/dky111 [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Wang Y, Zhou Y, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7(1):122. doi: 10.1038/s41426-018-0124-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebelo AR, Bortolaia V, Kjeldgaard JS, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018;23(6):29–39. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan H, Chao Y, Li S, et al. Picoinjection-enabled multitarget loop-mediated isothermal amplification for detection of foodborne pathogens. Anal Chem. 2018;90(22):13173–13177. doi: 10.1021/acs.analchem.8b03233 [DOI] [PubMed] [Google Scholar]
- 19.Qian WJ, Meng YQ, Lu Y, et al. Rapid, sensitive, and carryover contamination-free loop-mediated isothermal amplification-coupled visual detection method for ‘Candidatus Liberibacter asiaticus’. J Agr Food Chem. 2017;65(38):8302–8310. doi: 10.1021/acs.jafc.7b03490 [DOI] [PubMed] [Google Scholar]
- 20.Tian B, Ma J, de la Torre TZG, et al. Rapid newcastle disease virus detection based on Loop-mediated isothermal amplification and optomagnetic readout. Acs Sensors. 2016;1(10):1228–1234. doi: 10.1021/acssensors.6b00379 [DOI] [Google Scholar]
- 21.Lodh N, Mikita K, Bosompem KM, et al. Point of care diagnosis of multiple schistosome parasites: species-specific DNA detection in urine by loop-mediated isothermal amplification (LAMP). Acta Trop. 2017;173:125–129. doi: 10.1016/j.actatropica.2017.06.015 [DOI] [PubMed] [Google Scholar]
- 22.Abbasi I, King CH, Muchiri EM, Hamburger J. Detection of Schistosoma mansoni and Schistosoma haematobium DNA by loop-mediated isothermal amplification: identification of infected snails from early prepatency. Am J Trop Med Hyg. 2010;83(2):427–432. doi: 10.4269/ajtmh.2010.09-0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbasi I, Kirstein OD, Hailu A, Warburg A. Optimization of loop-mediated isothermal amplification (LAMP) assays for the detection of Leishmania DNA in human blood samples. Acta Trop. 2016;162:20–26. doi: 10.1016/j.actatropica.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou D, Huang S, Lei H, et al. Sensitive and rapid detection of the plasmid-encoded colistin-resistance gene mcr-1 in Enterobacteriaceae isolates by loop-mediated isothermal amplification. Front Microbiol. 2017;8:2356. doi: 10.3389/fmicb.2017.02356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imirzalioglu C, Falgenhauer L, Schmiedel J, et al. Evaluation of a loop-mediated isothermal amplification-based assay for the rapid detection of plasmid-encoded colistin resistance gene mcr-1 in Enterobacteriaceae isolates. Antimicrob Agents Chemother. 2017;61(4). doi: 10.1128/AAC.02326-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong LL, Phan HTT, Shen C, et al. High rates of human fecal carriage of mcr-1-positive multidrug-resistant Enterobacteriaceae emerge in China in association with successful plasmid families. Clin Infect Dis. 2018;66(5):676–685. doi: 10.1093/cid/cix768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holton TA, Graham MW. A simple and efficient method for direct cloning of PCR products using ddT-tailed vectors. Nucleic Acids Res. 1991;19(5):1156. doi: 10.1093/nar/19.5.1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovalic D, Kwak JH, Weisblum B. General method for direct cloning of DNA fragments generated by the polymerase chain reaction. Nucleic Acids Res. 1991;19(16):4560. doi: 10.1093/nar/19.16.4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3(5):877–882. doi: 10.1038/nprot.2008.57 [DOI] [PubMed] [Google Scholar]