Abstract
The metabolome, the small molecule chemical entities involved in metabolism, has traditionally been studied with the aim of identifying biomarkers in the diagnosis and prediction of disease. However, the value of metabolomics has been redefined from a simple biomarker identification tool to a technology for the discovery of active drivers of biological processes. In this review, we describe the molecular mechanisms by which the active cell metabolome affects cellular physiology through modulation of other ‘omic’ levels, including the genome, epi-genome, transcriptome and proteome. This concept of activity screening guided by metabolomics to identify biologically active metabolites, or “activity metabolomics”, is having broad impact on biology.
Introduction
In the middle of the last century, Fritz Kahn, a German physician, had immense success with a book series on the “man machine” 1. Driven by his admiration for both technology and physiology, he drew fascinating illustrations and analogies between the human body and the recently developed industrial machines of the 20th century. Seventy years later we entered the multi-omics era, spurred by a massive advance in technology enabling the systematic quantitative characterization of the cells’ molecular “machinery” (genome, transcriptome, proteome and metabolome). The metabolome, a relatively recent entry into the omics spectrum, is represented by metabolites. These small molecular chemical entities transcend the genome and proteome, representing the most downstream stage in this dynamic system defined as metabolism. In a more visual manner, metabolism can be depicted as a machine with metabolic gears that are intertwined with the activity of genes and proteins 1. These gears are viewed as simply performing a function as an integral part of a larger system. The information flow through these different ‘omic’ levels of biochemical organization is described as the central dogma of molecular biology 2. Within this framework, the metabolome, the entity of metabolites, has become widely accepted as the dynamic and sensitive measure of the phenotype at the molecular level, placing metabolomics at the forefront of biomarker and mechanistic discoveries related to pathophysiological processes 3.
However, the perception of metabolites mainly as a downstream product - biomarkers (of gene and protein activity) - has minimized the awareness of their far-reaching regulatory activity. Metabolite activity is a fascinating aspect of metabolism given that the metabolome interacts with and actively modulates all other ‘omic’ levels (Figure 1). Through this interaction metabolites also serve as direct modulators of biological processes and phenotypes. This concept has been investigated for decades, especially through the seminal discoveries of glucose, fatty acids and other lipids as regulators of insulin secretion and sensitivity 4, the lac operon in bacteria 5, and nutrient and energy sensing by the mTOR kinase6. These findings have already shown the significant impact metabolites can have on biological systems.
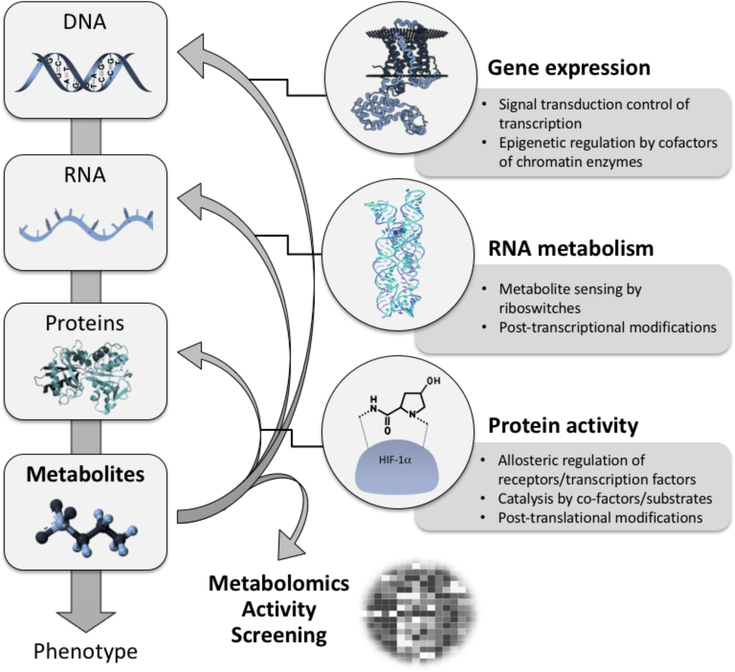
Figure 1.
Metabolites – active modulators of gene and protein activity. Metabolites actively control protein activity via allosteric regulation of transmembrane receptors and transcription factors, as enzyme co-factors and co-substrates in the catalysis of biochemical reactions, and via post-translational modifications. They also significantly influence RNA metabolism via sensing (small molecule ligands are sensed by riboswitches) and post-transcriptional modifications. Moreover, it is well known that metabolites serve as signaling molecules to control transcription factors and thus gene expression. Finally as co-factors and co-substrates of chromatin modifying enzymes they are actively involved in epigenetic regulation.
However, more recently, with the advent and evolution of metabolomics’ technologies, the discovery of active metabolites that have the capability to change cell physiology has grown rapidly. Examples of metabolite phenotype modulation include the NAD+ boosting of sirtuin activity to protect against age-associated alterations 7, lactate modulation of neuronal excitability and plasticity 8, α-ketoglutarate mediated orchestration of macrophage activation and immunity 9, malonyl-CoA controlling neurogenesis in adults 10, nitrogen metabolite balance used to enhance pertussis toxin production 11, and taurine-induced myelin basic protein expression12. Additional studies in cancer metabolism have unraveled the important and multiple roles of “oncometabolites” 13 that include fumarate, 2-hydroxyglutarate, and succinate and others. These studies highlight the intrinsic biological activity of metabolites and provide new import to the value of metabolomics as it can be harnessed to identify metabolites that act as drivers of biological processes and thus better understand their physiological role.
We describe recent developments in the field and provide an overview on metabolite discovery strategies by combining metabolomics with orthogonal experimental and computational biology approaches 14,15. This review will illustrate examples of phenotype modulation driven by processes such as macromolecule-metabolite modification and interaction. We will summarize recent conceptual advances and workflows to characterize the active metabolome in a variety of biological systems. We will also review recent technological advances that merge metabolomics with cognitive and cloud computing and further enable the activity metabolomics strategy.
Mechanisms of phenotype modulation by metabolites
Prototypes for active metabolites: Oncometabolites
How does the active metabolome drive phenotype modulation? Oncometabolites are one of the best examples of active metabolites because of their early discovery and established mechanisms of phenotype modulation in cancer cells. The accumulation of these oncometabolites in distinct types of cancer cells is a causal process in malignant transformation. Oncometabolites, including D-2 hydroxyglutarate, L-2-hydroxyglutarate, succinate and fumarate, were found in cancerous tumors that had mutations in the enzymes corresponding to the oncometabolites. Gain of function mutations were found in Isocitrate Dehydrogenase (IDH), and loss of function mutations were found in Fumarase (FH) and Succinate Dehydrogenase (SDH) (reviewed in 13). The accumulation of the oncometabolites in the tumor cells arising from these mutations resulted in a proliferative phenotype.
These oncometabolites are not only biomarkers of the diseases, depending on the activity of their respective enzymes, they can modify and interact with proteins and DNA thereby altering the proteome and the epigenome. Specifically, the activity of these oncometabolites stems from their inhibition of the α-ketoglutarate-dependent dixoxygenases, a class of enzymes that include prolyl hydroxylases 16. Inhibition of prolyl hydroxylases by the oncometabolites results in a phenotype mirroring hypoxia, known as “pseudohypoxia”, where the levels of transcription factor hypoxia inducible factor (HIF) are increased despite normal oxygen levels. In addition, oncometabolites inhibit alpha-ketoglutarate dependent TET (ten eleven translocation) enzymes and lysine demethylases. Both enzymes control removal of methyl-groups from chromatin 17, 5-methylcytosine is hydroxylated by TET in CpG dinucleotides (a step towards demethylation) and histones are demethylated by lysine demethylases 17. Thereby, histones and DNA are hypermethylated in cancer cells that have high abundances of these metabolites, illuminating the link from oncometabolites to the epigenome.
In addition to enzyme inhibition, other distinct biological activities for individual oncometabolites have been described. Fumarate can react with thiol groups of cysteine residues. This reaction results in acylation (succination) of the residue and therefore alteration of KEAP1 activity 18. Consequently, transcription factor NRF is activated, which can increase cyst size in renal cancer 18. Fumarate also mediates succination of other proteins involved in redox metabolism, a key system in cancer 19. Fumarate has also been suggested to modify other metabolites, such as glutathione 20, thereby increasing oxidative stress and senescence. Recently, D-2-hydroxyglutarate has also been shown to inhibit the activity of BCAT1, and BCAT2, two α-ketoglutarate - dependent enzymes 21. It should be noted that a number of additional metabolites have been classified as oncometabolites, including glycine, glucose, and lactate, and most of these metabolites are related to aerobic glycolysis, glutaminolysis, or one-carbon metabolism 22,23. While the full scope of the biological effects of oncometabolites is still an active area of investigation, it has been demonstrated that they modulate proteinprotein interaction, alter enzyme activity, lead to changes in the protein posttranslational modifications and modify the epigenome, all with the effect of propagating cancer.
Therefore, oncometabolites are primary examples of active metabolites. From these examples, it emerges that active metabolite crosstalk to other “omes” occurs to a largely unanticipated extent 24,25. Recent mechanistic insights have revealed that active metabolites strongly impact all layers of the omics landscape, from the genome, epigenome and transcriptome to the proteome 26–28. Within this framework the metabolome has two overarching mechanisms to control the functions of DNA, RNA and proteins: chemical modification and metabolite-macromolecule interaction that are further detailed in the following two paragraphs.
Metabolic chemical modification of macromolecules
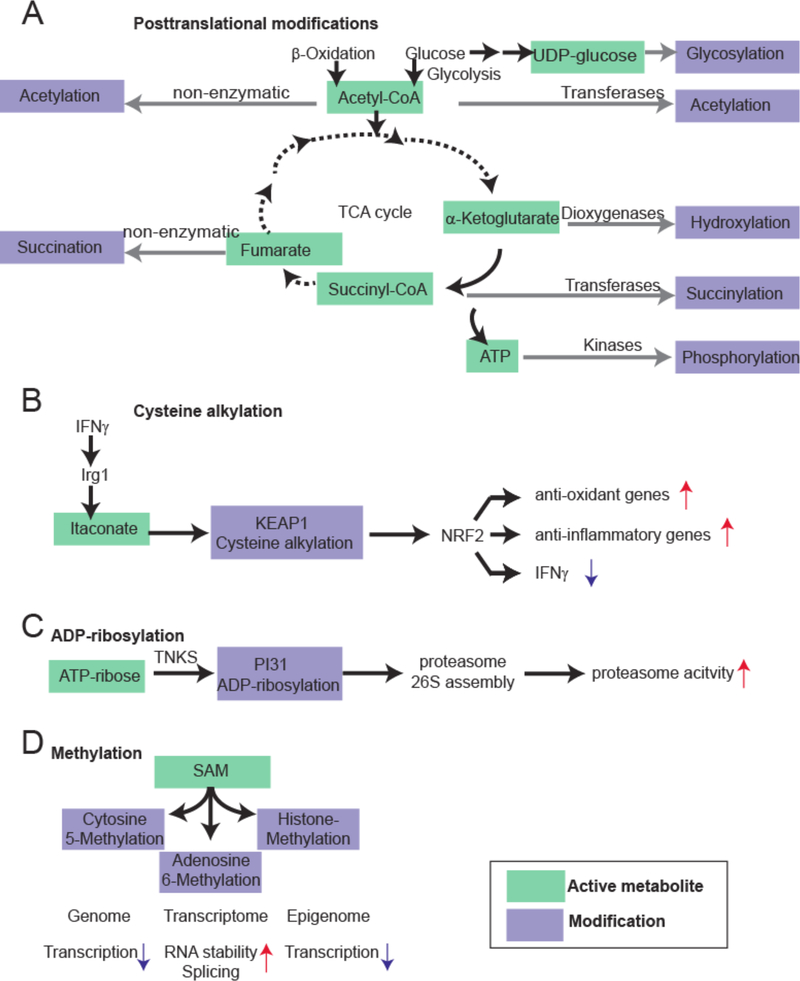
Metabolites drive pivotal covalent chemical modifications of DNA and RNA (such as methylation) and of proteins (post-translational modifications). The dynamic shape of these chemical modifications has been shown to significantly affect cellular function. Post-translational modifications of proteins involve at least dozens of different small molecules that can be covalently bound to distinct amino acids as for instance lysine acetylation (derived from Acetyl-CoA) 29 or cysteine palmitoylation 30 (derived from Acyl-CoA). It should be noted that acetylation processes also occur non-enzymatically with low overall stoichiometry and unclear functional roles 31,32. Further acylation processes with other Acyl-CoA species occur as well 31. Other metabolites responsible for posttranslational modifications include succinyl-CoA (arginine succinylation), as well as activated sugar molecules (e.g. UDP-glucose) for glycosylation, and GlcNAcylation 33,34 (Fig. 2A). Other active metabolites have shown to control anti-inflammatory responses (via alkylation of cysteine residues 35,36) (Fig. 2B), proteostasis (via proteasome ADP-ribosylation37) (Figure 2C) or enzyme activity (via lysine glutarylation 38). The active metabolites for these processes are itaconate, ATP-ribose and glutaryl-CoA, respectively. Notably, abundance of many of these metabolite-induced protein modifications are directly dependent on the metabolic state of the cell and as such represent a powerful means of phenotype modulation 29,39. DNA methylation (or a transfer of a methyl group from S-Adenosyl Methionine to cytosine) starts during embryogenesis and continues throughout the lifespan, affecting chromatin states, lineage specialization, gene expression, genome stability, and self-renewal of stem cells 40,41 (Figure 2D). Further on, various metabolites, such as S-Adenosyl Methionine, glycine, pyruvate, galactose and threonine function as co-factors for posttranscriptional RNA modifications that act as sensors and transducers of information to control metabolic rate (oxygen consumption) and protein synthesis – yet their effect is still not completely understood 42,43. Enzymatic histone modifications such as lysine acetylation (derived from Acetyl-CoA), lysine and arginine methylation (derived again from S-adenosyl-methionine), and serine phosphorylation (derived from ATP) are key controllers of the epigenome’s landscape directly influencing gene expression, chromosome packaging, DNA repair and changes in cell metabolic state 44,45.
Figure 2:
Examples of macromolecule modification by the active metabolome. A. Examples for macromolecule modifications by the tricarboxylic acid cycle (TCA) intermediates and related products. Acetyl-CoA, α-ketoglutarate, Succinyl-CoA, UDP-glucose and ATP are energy-rich molecules that can directly modify proteins, or nucleic acids. B. Cysteine alkylation by itaconate, a branched fatty acid. Recently, it was shown that itaconate is an anti-inflammatory metabolite that directly alkylates cysteine residues at KEAP1 to control expression of NRF2. KEAP1 is the primary negative regulator of NRF2. This results in increased expression of anti-oxidant and anti-inflammatory gene expression. C. ADP-ribosylation of proteasome unit PI31 to control proteostasis. By the TNKS enzyme, PI31 is ADP-ribosylated to promote proteasome 26S assembly (from the 20S subunit). This results in increased proteasome activity. D. S-Adenosylmethionine is a central methyl donor for DNA, RNA and histone methylation, thereby controlling gene expression at the level of the genome, transcriptome and epigenome.
Metabolite-macromolecule interactions
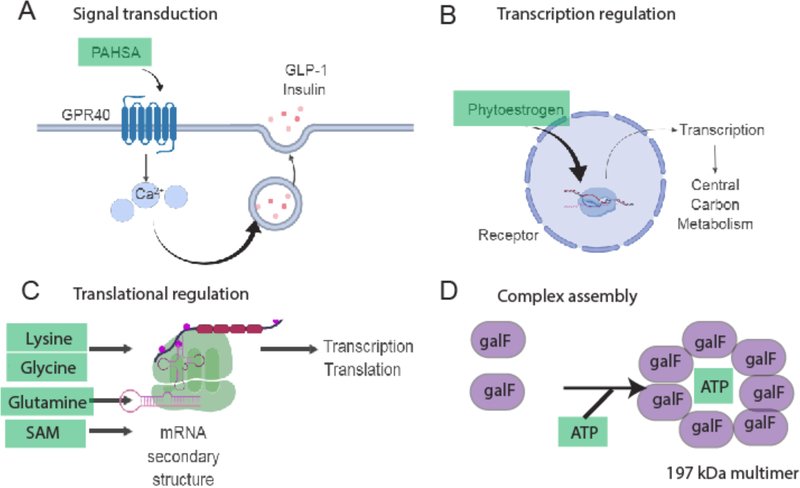
Metabolite-macromolecule noncovalent interactions represent a second mode of cellular activity regulation. A classic example is competitive binding of a metabolite to an active site of an enzyme, or allostery - the alteration of enzyme activity by binding of a small molecule at a site other than the active center. These concepts apply not only to enzymes, but also to a variety of messengerRNAs (metabolite-controlled riboswitches46), proteins such as signaling receptors, and various other molecule classes. G-protein coupled receptors (GPCRs) are the sine qua non of metabolite-activated signaling molecules and they were among the earliest identified proteins which were developed as drug targets 47. Among many other receptors, G-protein coupled receptor 91 in mice (Gpr91) is activated by succinate to control blood pressure 48 and Gpr40 is activated by palmitic acid hydroxystearic acids (PAHSA), a recently discovered class of endogenous lipids 49,50 (Fig. 3A). Binding of these receptors invokes highly specific signaling responses that lead to specialized cellular activation of signaling networks. This concept also extends to transcription factors (e.g. non-canonical activation of estrogen receptors by phytoestrogens 51), all of which crucially determine the system’s response to cues via control of expression of distinct genes (Fig. 3B). Metabolite-controlled transcription and translation is performed by riboswitches. Metabolites that control riboswitches include lysine, glutamine, cobalamin, thiamin pyrophosphate, and purines (Fig. 3C) 52. The active metabolome also controls uptake and availability of other essential nutrients: for instance, iron uptake in plants depends on the local presence of reducing agents within the roots 53. One noteworthy example is brain glucose sensing where glucose plays a pivotal signaling role controlling hormone secretion and neuronal activity as a means to regulate feeding, energy expenditure and its own homeostasis – the control of highly complex behavioral phenotypes 54. Many of these examples describe rather known biological facts of the role of active metabolites, yet they expand the active metabolome to metabolites that were typically considered common cellular building blocks: amino acids for proteins, pyrimidine and purine bases for nucleic acids biosynthesis 55 and phospholipids for cell membrane formation 14.
Figure 3.
Mechanisms for non-covalent modification of macromolecules by the active metabolome. A. Palmitic hydroxystearic acid (Pahsa) activates GPR40 to induce calcium signaling and augmentation of insulin and GLP1 release. B Phytoestrogenes activate transcription factors that control programs involved in cell metabolism and cell proliferation. C. Riboswitches are controlled by metabolites. D. Metabolites assemble higher molecular proteins, e.g. of the bacterial protein galF, a glucose-1-phosphate uridylyltransferase.
Metabolome-driven omics alternations
From these rather reductionist approaches, an entire new layer of metabolome-driven global gene and protein level activation emerges. Foundational studies used dietary restriction in mouse models to alter metabolism and found a dramatic change in gene expression alongside amelioration of the aging phenotypes 56. It has also emerged in the last decade that control of the epigenome is determined by the metabolic status 57. Yet, only a few studies have analyzed the impact of metabolic changes or metabolite supplementation on the other omic layers in a systematic manner. One study analyzed the effect of metabolite supplementation on various yeast strains that were deficient in histidine, leucine, methionine and uracil biosynthesis 58. Subsequently, transcriptome, proteome and metabolome analysis of these strains was performed. Integrated analyses revealed that gene expression (epistasis) and protein expression is largely controlled by metabolite supplementation, suggesting a systems-wide modulation by metabolites. Among these studies, metabolic flux analysis has gained a more important role in determining time-dependent activity of the respective enzymes and thus, biochemical pathways 59. Furthermore, recent large-scale studies have revealed that metabolites mediate proteome-wide complex formations (the interactome). One recently found example is the galF complexes that are formed by the presence of ATP (galF is a UTP-glucose-1phosphate uridylyltransferase in E. coli) (Fig.3D) with yet to be determined biochemical consequences 25. The protein-metabolite interaction landscape has also been probed by protein-centric approaches in yeast and offers largely unexplored regulatory connections, including alteration of YPK, a mammalian AKT analogue by ergosterol 60.
By these means, the active metabolome empowers and mediates basic biological processes such as signal transduction, proteostasis and regulation of gene expression. This multitude of metabolite-induced and endowed omics modulations is a current focus of several investigations. Comprehensive approaches to study the metabolome, in particular the active metabolome, on a global scale have been developed only recently. Integration of metabolomics with other omics data provides a means to prioritize metabolites for functional testing and to predict metabolites with highest activity (see multi-omics integration section). Only very recently, methods have been developed to interrogate the interplay between the metabolome and the other omes, with focus on the proteome (see activity screening technology section). Further technological improvements are necessary to unravel the complexity of metabolite-induced macromolecule activity to ultimately control the phenotype. In any case, the current literature already shows that identified metabolites from metabolomic studies can modulate physiology, a concept that turns the tables on conventional omic technology thinking: metabolites are not providing a simple readout for the other omics’ layers but can act as master regulators of the biological system 26.
Discovering active metabolites
Metabolite detection, identification and quantification
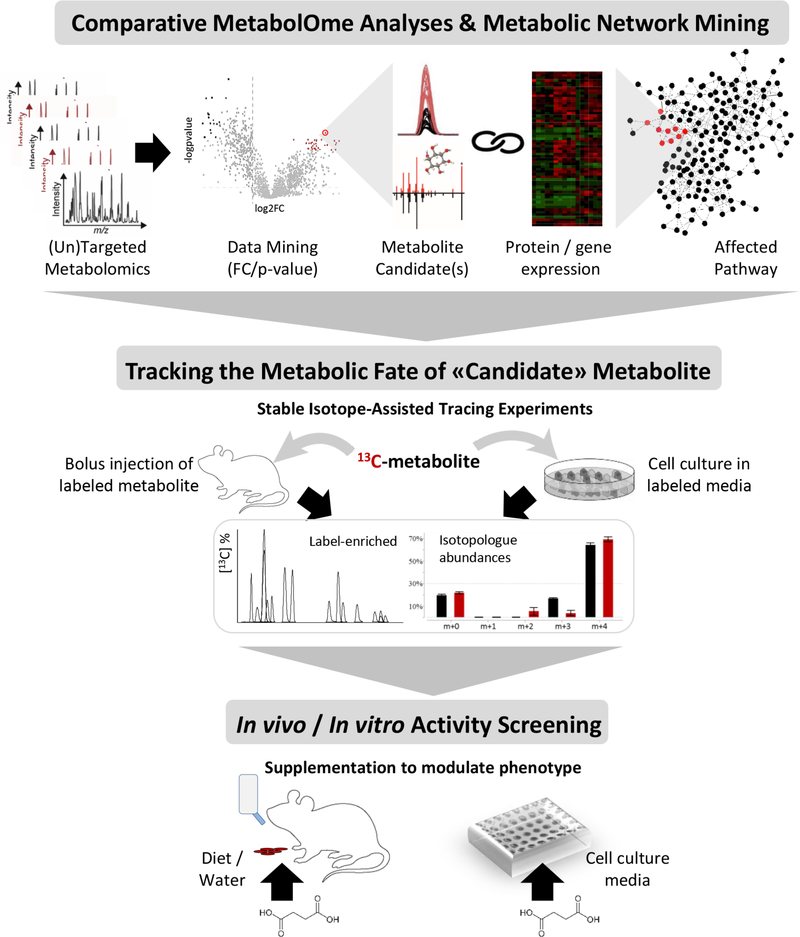
Historically, metabolite detection, identification and quantification has been accomplished using biochemical approaches, however metabolomics offers a unique framework for discovery that can, and has been, applied at multiple levels (Figure 4). The identification process is one of the most important aspects in the discovery of active metabolites. Mass spectrometry-based metabolomics data acquisition and annotation is primarily defined by identifying features [G] with a specific mass-to charge-ratio (m/z). Annotation is a multi-component and challenging aspect of feature data processing and has been extensively reviewed 61. A metabolome-based strategy for identifying candidates with biological activity would use a list of metabolites generated from statistical analysis of metabolomic datasets. Once metabolite abundance is quantified based on peak intensity, statistical filters can be adjusted depending on the experiment and data. Metabolites are typically selected for further activity screening based on specific statistical cutoff (e.g. a p-value less than 0.01 and a fold change greater than 2, as compared to control conditions). A multitude of peak detection and alignment softwares are available, including XCMS Online 62, MZmine2 63, Open-MS 64 and MS-DIAL 65. The second component is the annotation of features and identification of metabolites. This includes the use of metabolite databases and spectral libraries 66, among these the Human Metabolome Database 67,68, METLIN 69, the Birmingham Metabolite Library Nuclear Magnetic Resonance database 70, BiGG 71, MassBank 72, LipidMaps 73, mzCloud 74, the Fiehn lab GC-MS Database75, and the Golm metabolomics database76. The challenges in data analysis and sharing, especially for non-specialized labs, were recently addressed by the development of cloud-based technologies and databases (Box). Bioinformatic metabolic pathway and network analysis can be used to reduce the complexity of the data. This feature that has been recently incorporated into XCMS Online 77 and is available via MetExplorer 78 and other platforms 79–81. Aims of these approaches are to prioritize the metabolites involved in distinct modules of the metabolic network or locally enriched parts of the metabolic network, thus reflecting the true activity (while assuming that the false matches will be distributed randomly across the network). Once lists of candidates are established, their fate can be tracked using isotope based tracing through the metabolome (“flux analyses”) 82. Finally, metabolomics-guided activity screening can be performed using in vivo and in vitro phenotypic, omics and chemical biology strategies.
Figure 4.
Metabolomics-guided identification of bioactive metabolite candidate(s) followed by activity assessment. Workflow to elucidate the metabolite activity and role at the system’s level starts with the comparative metabolome analyses - a discovery-oriented untargeted or broad-scale targeted profiling analysis is followed by data mining to select the metabolite candidate(s) based on the significance (p-value), amplitude (fold change) and direction of its change (i.e. accumulation or depletion) in the studied system (e.g. cell media, mice/human plasma, etc.). To this end, statistical approaches are applied, and the metabolite is identified using spectral libraries (i.e. MS/MS matching). Protein and gene expression data (as a result of proteome and/or transcriptome analyses) are then used to support and help filter the candidate metabolites involved in the locally enriched part of the metabolic network, i.e. the same module or biochemical pathway. For example, the depletion of one a metabolite will usually be coupled by the significantly higher increased expression of its converting enzyme(s)) responsible for metabolite consumption. To gain further insights into the metabolic fate of the “metabolite-candidates” metabolite, the next step involves stable isotope-assisted (13C or 15N) tracing experiments to identify the active pathways used for metabolite catabolism. Different conditions can be tested in a longitudinal assay to gather the information about the metabolite uptake and conversions (i.e. label-enriched metabolites based on MS-based isotopologue abundance analyses, or NMR based flux analysis). Lastly, the metabolite supplementation experiments are performed using in vitro cell or organoid models, and in vivo models. These assays provide the direct information about how the phenotype is affected or modified via endogenous metabolite supplementation (ingestion through diet, drinking water or by addition to cell culture medium).
Box 1: Cloud Computing and Metabolomic Databases.
Metabolomics, a multi-disciplinary technology that is now evolving at an increasingly rapid pace, nonetheless may still be regarded as an emerging discipline. Its initial slow progression stems from the intensive resource commitment a truly comprehensive metabolomics lab can require, including high sensitivity triple quadrupole mass spectrometers [G], nuclear magnetic resonance (NMR) technologies, and high resolution mass spectrometry [G] systems for untargeted analyses. This commitment limits metabolomics accessibility to biologists due to the required expertise, instrumentation and, most importantly bioinformatics support. And while data generation occurs mainly through collaborations or fee-for-service facilities, the challenge to identify the most meaningful active metabolites and interpret its function within a biochemical context remains. To address this challenge cloud-based bioinformatic platforms represent a novel paradigm of how resources can be made available on a global scale and large data amounts can be translated into biological insights.
The most significant value of the universal access options provided by the cloud is the fast and simple sharing of resources. Among many other algorithms (see metabolite identification section), XCMS was originally developed as a metabolomics data processing algorithm to extract metabolite features out of raw mass spectrometry data and perform statistical analysis. Today it is available as a cloud-based online resource with over 27,000 users allowing them to upload mass spectrometry data files and share the processed results with an individual collaborator and/or make them publicly available. This type of cloud-based platform increases the field’s ability to compare results with those available from public repositories, and to perform metabolomic meta-analyses. Because of the conserved nature of metabolites across evolution, these analyses are transposable over a variety of phyla and species, e.g. ranging from bacteria to plants to higher organisms 137. The cloud has also addressed the issue of metabolite identification with multiple databases available, specifically databases that contain tandem mass spectrometry (MS/MS) data from defined compounds. Other cloud based approaches in metabolomics exist 138 or are emerging 139, and public online databases such as MetaboLights 105 and Metabolomics Workbench 106 enable first cloud-based analyses that will further improve in the future.
Cloud computing [G] is therefore gaining prominence in metabolomics however there are some limitations. The user of the cloud needs an efficient web connection for data uploading and processing, and may have limited flexibility and control over the resources. The provider of the cloud requires server resources that expand according to the ever-growing amount of data and, in addition, high-level security. However, even with these challenges, it appears that biology and cloud computing have already become irrevocably intertwined. This is particularly true in conjunction with the omic disciplines, that deal with data sets reaching the terabyte range and are driven by the increasing performance of high throughput methods, such as next generation sequencing or high resolution mass spectrometry-based metabolomics and proteomics. Beyond these obvious practical advantages, cloud-based computing in activity metabolomics also readily offers the ability to integrate a rich source of archived data from the multiple repositories available 79, which remains one of the key challenges of systems biology [G].
Metabolic Activity Screening Strategies
Identifying active metabolites that modulate phenotype can be achieved through multiple strategies. There is a host of examples where metabolomics in combination with orthogonal molecular biology and computational approaches has been successfully used to identify active metabolites 26. In each one of these examples the primary step in determining activity has been the use of an appropriate screening strategy. These include gene expression, protein expression and protein activity (e.g. enzyme activity), the modulation of desired cellular phenotypes, and performing in vivo phenotypic studies directly on an organism. Table 1 lists the principles behind these assays.
Table 1.
General Principles for activity screening strategies
| Technology | Mode | Principle | Application examples |
|---|---|---|---|
| Cell based assays | In vitro | Metabolite-induced cellular phenotype (e.g. cell migration) is quantified | 140 |
| Expression screening | In vitro | Metabolite-induced protein or transcript expression is quantified | 14 |
| Metabolite binding screening on protein arrays | In vitro | Metabolite-protein interactions are profiled | 83,84 |
| Chemical proteomics | In vitro | Metabolite-protein interactions are profiled | 83–86 |
| In vivo administration and phenotype screening | In vivo | Metabolite-induced molecular and patho-physiological phenotypes of model organisms are quantified | 141 |
| Mathematical modeling | In silico | Mass balance or neuronal networks are used to predict most active metabolites | 91 |
| Cognitive computing | In silico | Active metabolites are predicted based on previous studies | 142 |
In contrast to in vivo studies, cell based assays can provide high throughput and allow for the analysis of cell morphology, biophysical cell function, chemical and physiological properties, and can leverage a latitude of molecular biology tools such as fluorescent or luminescent reporters. For the fields of metabolite-epigenome and metabolite-genome interactions, these tools comprise mainly nucleotide sequencing approaches such as chromatin-immunoprecipitation and sequencing (CHIP-SEq approaches), and detection of methylated DNA using bisulfite sequencing. More recently, the field of chemical proteomics has emerged to study metabolite/small molecule-protein interactions. This can be accomplished, for instance, via protein arrays [G] that interrogate chemical binding of distinct metabolites to various proteins 83,84 or, more recently, thermal proteome profiling (TPP), a method that relies on the principle that the temperature for thermal degradation of proteins (“melting point”) is shifted upon small molecular binding. TPP involves a thermal shift to denature proteins within living cells or a cell extract in the presence of different metabolites 85. Many other chemical proteomics approaches exist 86 that are either metabolite or protein centered, including protein stability assessed from rates of oxidation (SPROX) 87 or limited proteolysis 88.
It is worth noting that some screens can be time intensive and costly, and are largely limited to specialized laboratories. In order to effectively pre-select potential candidates following metabolomics data processing, a combination of more computationally oriented activity prediction strategies with chemical approaches can be adopted (thereby accelerating the general assays delineated in Table 1). Table 2 pinpoints features of in silico and in vitro approaches, including their limitations and advantages. Beyond these technologies, more advanced prediction approaches are being adopted. Using neural networks[G] trained by large data sets of the activity, physico-chemical properties and structural information of small-molecules, which are available in the ChEMBL database89. As an example, for the enzyme cyclooxygenase 2 more than 6000 IC50 values [G] of small molecules and their physio-chemical properties are deposited in ChEMBL. Using this information to train a neural network might result in an algorithm capable of predicting the possible bioactivity of hundreds if not thousands of metabolites evolving from exploratory metabolomics. In turn, metabolomics data might be superimposed on activity constraints such as physico-chemical properties, or distinct functional groups, resulting from a neural network, thereby limiting the actual chemical space [G], which ultimately has to undergo in vitro and or in vivo bioactivity testing. Another interesting source of knowledge for such an endeavor stems from drug metabolism, where distinct molecular sites prone to metabolic activation can be predicted by specialized and highly evolved in silico approaches 90.
Table 2. Chemical biology and computational technologies to study metabolite activity.
This table delinates recently developed technologies that can be used to identify the primary target(s) and activities of a metabolite.
| Approach | Description | Advantage | Limitations | Example |
|---|---|---|---|---|
| Purification and isolation from a complex mixture | Fractionation of complex biological mixtures by chromatography (e.g. HPLC) and subsequent activity testing using biological assays | Universal, flexible regarding the biological assay | Tedious, signal overlap, minor components might be missed | Identification of PGE2 as immune modulator derived from Trichuris suis worm eggs 143, identification of bioactive drug metabolites 144,145 |
| Affinity selection mass spectrometry | Incubation of metabolite mixture and target enzymes/proteins, size exclusion separation of bound and unbound components, MS based characterization of bound fraction | Universal approach, no protein immobilization necessary | High grade of non-specific binding may be obtained. Ligand binding and not activity is assessed. | Identification of protein-metabolite and protein-protein interactions in Arabidopsis thaliana 146,147 |
| Affinity purification (chromatography) – mass spectrometry | Affinity based protein purification and MS based characterization of components. Pulldown is done from complex cellular or metabolite mixtures | Universal approach which under certain circumstances can be used in vivo (e.g. yeast cells) | Antibody-dependent. Ligand binding and not activity is assessed. | Identification of several small molecule interaction partners within ergosterol biosynthesis. 60 |
| Thermal proteome profiling | Binding of a ligand to a protein in vivo or in vitro results in increased thermal stability | Universal approach, physical stabilization of proteins | Low throughput, and potentially significant amount of non-specific binding. Requires absolute metabolite concentrations, and binding is not necessarily bioactivity. | Identification of the protein metabolite interaction between STING and 2’3’-cGAMP 148 |
| Metabolite profiling combined with orthogonal molecular biology approaches | Integration of metabolomics data with the data obtained from orthogonal molecular biology experiments (i.e. gene silencing, enzyme inhibition, etc.) | Identification of mechanism of action of a (bio)active signaling metabolite | Long-term, fastidious | Various examples 26 |
| Integrated network analysis (GAM) | Combination of transcriptional and metabolomics data for the identification of active metabolic sub-networks | Comprehensive network analysis allowing for a systems wide comparison of two biological states, e.g. control and experiment conditions | Transcriptional data mandatory, preassembled metabolic networks (species dependent) | Integration of metabolic and transcriptional data to understand macrophage immune metabolism. 24,149 |
| Flux balance analysis | Mathematical approach for the calculation of the metabolic flux through a network, in silico approach. | Easily computable. No kinetic parameters needed. |
In silico approach based on genome scale metabolic network reconstructions. Only predicts steady state. Does not predict metabolite concentrations. |
Determination of Metabolite balance to determine behaviour and composition of engineered microbial communities 150,151 |
| Metabolite set enrichment and network analysis | Computational approach based on overrepresentation and probability analysis of metabolomics data to identify the active biochemical pathways or locally enriched parts of the metabolic network associated with phenotype | Rapid, allows for direct association with biochemically relevant information | In silico approach, significant amount of false positive metabolite IDs due to high levels of redundancy and noise in metabolomics data | Multi-omics discovery of RCC6-encoded protein CSB to potentailly alter defects in DNA-repair mechanisms in Huntington’s disease -98,152,153 |
| Bioactive Natural Products Prioritization using massive multi-informational Molecular Networks | Molecular networks embedding known bioactivity and taxonomical data to highlight potentially bioactive scaffolds in crude extracts | Computational approach that facilitates the identification of potentially bioactive compounds using databases and molecular/fragmentation similarity in mass | Complete structural elucidation of active compounds remains a challenge | Isolation of new cytotoxic prenylated stilbenes of the schweinfurthin series from Macaranga tanarius 154,155 |
In addition to the use of cognitive computing (neural networks), closing mass balances of metabolic models might as well be an interesting approach. Closing mass balance means that input and output of the system are in line with the law of mass action [G]. As recently shown, closing the mass balance in metabolic models of Bordetella pertussis allowed the authors to gain significantly deeper insight into the bacteria’s biology by curating and completing their metabolic model. The metabolic model of Bordetella pertussis showed a significant nitrogen imbalance, leaving approximately 30% of nitrogen unaccounted for. Using LC-MS analysis eleven novel nitrogen containing metabolic end-products could be identified. Based on the nature of these metabolites, the authors suggested physiological roles for these alternative nitrogen sinks beyond mere nitrogen excretion. Next, the authors used their curated metabolic model to compute minimal nutrient inputs supporting the growth of Bordetella pertussis Tohama I in order to rationally design novel growth media. Most interestingly their model predicted thiosulfate as a possible sulfur source, including novel metabolic pathways for its conversion. Together with the identified nitrogen sinks, the presented approach shed light on possible host microbe interactions, as well as the metabolic flexibility of Bordetella pertussis and allowed rationally designing novel growth media, increasing pertussis toxin production 91.
It is also possible to imagine that comprehensive and diverse libraries of metabolites for high throughput could be used for testing, for example METLIN [G] currently has over 300,000 individual small molecules including metabolites and small peptides. This approach using robotic acoustic dispensing devices coupled with multiple known biological screens is already widely used in pharmaceutical and biotechnology industries and could provide a brute force approach to metabolite activity screening.
Multi-omics integration for determining activity
Beyond metabolomics, multi-omic data correlation and integration will provide an additional layer of information to improve the metabolite selection process.92 For example, metabolite candidates could be integrated with transcriptomic and proteomic data as a follow-up to pathway analysis by deducing their overlap in the selected pathways of interest. A variety of integrated data sets consisting of proteome, transcriptome and metabolome data have been generated in different research areas, ranging from cancer metabolism research 93,94, plant physiology to microbiology 95,96. The aim of the multi-omic integration is to allow for the determination of the activity of distinct metabolites through quantitative modeling, thereby allowing a targeted intervention on a specific pathway. To this end, it is very important to reduce complexity of the vast amount of data. This can be done by multiple mathematical approaches, for instance by adding metabolite, and other omics data to curated pathways, or by modeling novel pathways and fluxes by the data. It has to be stated that the area of multi-omics integration is an ongoing effort and multiple approaches are currently being pursued.
In most of the studies, the multi-omics integration is based on gene nomenclature linked to unique metabolite identifiers, and involves combining previously generated pathway information (e.g. KEGG [G], Reactome [G], Biocyc pathways [G], Recon [G] or metabolomic set enrichments (available via MetaboAnalyst platform), and the newly generated ChemRICH platform 97–101,102). The advantage of using this pathway information is the ability to reduce complexity and to filter noise. The recently introduced metabolomics guided systems biology approach integrates multi-omics data, where all other omics data layers (on gene and protein expression) are mapped onto the untargeted metabolomics-derived pathway activity information.77 Several other approaches for multi-omics pathway integration exist 94 and network-based and machine learning approaches are emerging.
Integrating the metabolome and other omic layers can be a challenging pursuit. In fact, many researchers acquire complex datasets without a preset integration strategy. Robustness of data acquisition, artefacts introduced by sample harvest (batch effects) and the distinct features of the different omic datasets must be adequately considered. The challenges in multi-omic data integration are reviewed elsewhere 103,104, yet the most important ones consist of noise removal, data prefiltering, matching of various identifiers, the selection of data dimensionality reduction methods, and finally, selecting computational approaches and mathematical models to apply to the acquired omic data, model validation, and the further integration into trans-omics network modules. The ability to streamline and efficiently advance these approaches is currently under development. Challenges also include the reality that metabolic networks cannot be sufficiently validated due to technology limitations (e.g. limited metabolite coverage as a consequence of analytical bias). Finally, as pointed out, the entirety of omics data is often not utilized in an adequate and comprehensive way 103. One limitation on the metabolomics side is the lack of appropriate archives and data sharing strategies, these concerns however are gaining more and more attention, e.g. in the MetaboLights 105 and Metabolomics Workbench 106 repositories. Finally, in a large amount of high visibility reports, a reductionist experiment is conducted as a conclusive experiment that potentially increases the impact of the study. Yet, this commonly used procedure does not take advantage of the dataset as whole, and consequently, the majority of multi-omic data is not being used 103,107. However, once acquired and integrated, the multi-layered omic strategies can be useful, especially if applied to in vivo metabolism.
It is anticipated that genome scale modeling of metabolic processes, big data analysis and machine learning strategies 108,109,110 will be integrated to further prioritize metabolite activity in a given biological system. In fact, the power of this integrative omics approach has been already shown by in vivo and in vitro modeling studies that reconciled multi-layered omic data acquisitions and metabolic and other phenotypes over a larger number of observations, and were able to accurately predict biological behavior. Yeast strains with various genetic deletions are successful model organisms to link genetic variation to metabolic phenotype, connecting the gene to its role in metabolism 111. In a machine learning approach, proteomic data was used to predict the yeast metabolome in various yeast strains that had kinome-wide knockouts 112. Multi-layered genome models, but also significantly smaller models have in fact been able to predict the entire metabolome from single datasets. One recent and successful examples was the accurately predicted growth of E. coli by metabolic and genetic interventions 113. The metabolomics-guided perspective is different: for example, mapping downstream metabolic changes onto metabolic pathways and biological networks can provide mechanistic insights, especially when associated with other ‘omic data. Within these platforms automated predictive pathway analysis enables straightforward and efficient metabolite mapping to background knowledge databases that can be either curated reference pathway databases or genome-scale networks. However, the available metabolic spectral databases only cover a fraction (up to 60%) of genome-scale metabolism, suggesting a general, yet addressable limitation of the metabolomics-guided strategy due to the “dark” metabolome 114. Flux analyses using pulsed isotope labeling of metabolites can complement traditional metabolomic studies to further decipher the numerous relationships between enzymes and metabolites, a topic reviewed elsewhere 59,82.
While the goal of these integrated approaches is the generation of accurate biological models, our ultimate goal is to use this information to identify the best candidates for activity screening. Therefore, the multidimensional integrated “omic” landscape yields unanticipated opportunities and out of the box solutions for predicting metabolites that modulate phenotype.
Perspective: Applications of activity metabolomics
The applicability of activity metabolomics is broad, for example Figure 5 illustrates how active metabolites alter phenotypes in a variety of organisms ranging from relatively simple prokaryotes to complex human physiology. For instance, activity metabolomics can enrich biotechnological applications bacteria where it has been shown to enhance the production Bordetella pertussis vaccine production or protein production in E.coli 115,11,12. The microbiome is also susceptible to modulation by modifying the mammalian metabolome 116, or the reverse, where microbiome-derived metabolites can be used to impact immune cells and satiety117. Other examples of crucial metabolite activities come from the emerging field of immunometabolism where prostaglandin E2 promotes Th2, Th17 and regulatory T cell responses, whereas it suppresses macrophage and neutrophil activity. 118,119,120. On an organ level, metabolites control pathophysiological reactions such as asthma via leukotrienes 121. In complex organisms, nutritional interventions such as supplementation of omega-3 fatty acids proved to have multiple benefits, while lacking severe side effects 122,123, and a plethora of studies has analyzed the phenotypic modulations evoked by feeding various supplemented diets to model organisms of ageing-associated diseases 124–126
Figure 5.
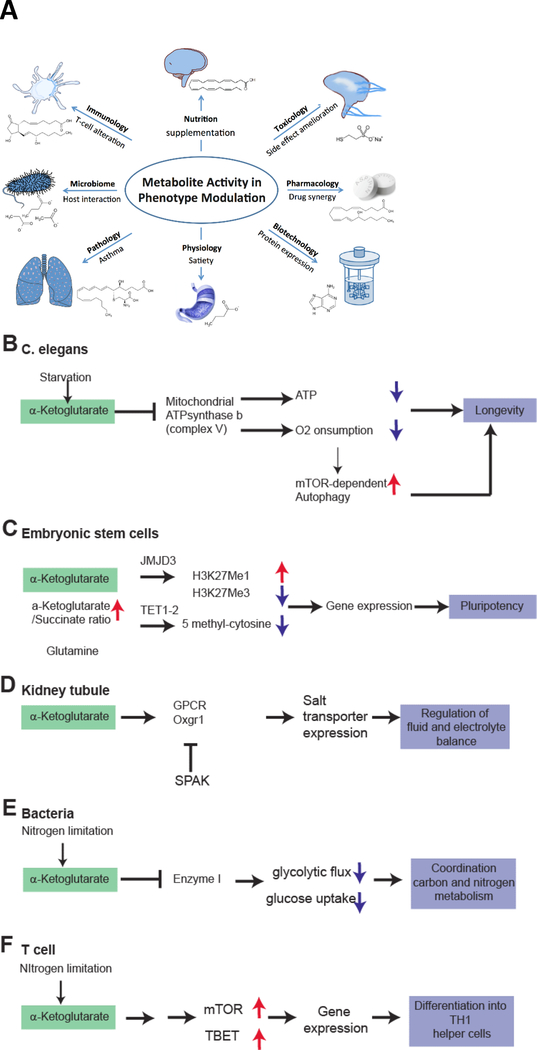
A. Metabolite Activity for Phenotype Modulation. Metabolomics has already made a significant impact in a wide variety of scientific areas through discovery of active, mainly endogenous metabolites that can regulate different biological processes and thus modulate the phenotype in health and disease. B-F. Metabolic activity of α-ketoglutarate (AKG) in a variety of prokaryote to eukaryote systems. AKG accumulates in exercise, but can also be supplemented. Direct supplementation has a wide effect on various systems via different mechanisms. Common primary AKG sensors may be ATPase b subunit (as revealed by DARTs proteomics) or PII (in plants/prokaryotes). B. In C. elegans, AKG addition leads to binding of the ATPase b subunit to inhibit ATP production, oxygen consumption, and thereby stimulating mTOR dependent autophagy. All of these mechanisms result in an extended life span of the worm. C. In embryonic stem cells, an increased AKG/Succinate ratio drives activation of JMJD3 and TET1/2 to perform epigenetic changes. These include a reduced trimethylation at H3K27Me3 and also a reduced methylation of DNA (5-methylcytosine reduced). This enables increased gene expression required for pluripotency. D. In specialized mammalian organs such as the kidney, AKG is being secreted to control organ function in a paracellular manner. Urinary AKG derived from metabolic stress results in activation of Oxgr1 and subsequent activation of salt transporters to regulate electrolyte balance and, presumably, hypertension. E. In bacteria, intracellular AKG induced by nitrogen limitation inhibits Enzyme 1 to decrease glycolytic flux and to couple nitrogen consumption to glucose consumption. F. In T-cells, AKG induces activity of mTOR and the T-cell specific transcription factor TBET via unknown mechanism to promote differentiation to TH1 cells over TH2 cells.
In the fields of pharmacology and toxicology, metabolites are administered to reduce toxicity (e.g. to reduce methanol poisoning by administration of ethanol, or to administer scavenger metabolites of toxic metabolites, e.g. Mercaptoethansulfonat-Na for certain chemotherapeutics or acetylcysteine in the case of paracetamol poisoning) and to take advantages of drug synergies with endogenous metabolite classes 127. Even very simple and inexpensive metabolites, such as glutamine, can be utilized to treat complex diseases such as sickle cell disease as recently investigated in a phase 3 clinical trial 128,129.
These examples show the wide applicability of using active metabolites to modulate biological processes, induce shifts in cellular metabolic state and thus cell activation, differentiation or proliferation and complex tissue functions. Notably, the effect of a metabolite is determined by the context and its induced phenotype can vary largely with the biological system it is applied to. The goal of activity metabolomics is to provide the framework for this concept. α-ketoglutarate (AKG) is an example of an active metabolite that alters phenotypes in a context dependent manner: In bacteria, it regulates glucose metabolism and uptake 130. In Caenorhabditis elegans, it extends lifespan by mTOR inhibition 131. In immune cells, it supports regulatory T-cell differentiation from Th1 cells 132. In humans it is reported to increase tissue and muscle regeneration via a multitude of processes involving ERK and others 133. Once binding to a G-protein coupled receptor Oxgr1 (localized chiefly in the kidney), it translates into increased synthesis of transporters to drive hypertension via salt reabsorption134,135. A common theme in these settings may be a modulation of anabolic cell activity, and this may be the “common denominator” of AKGs role in the activity metabolome. Thus, the current challenge of activity metabolomics is to link metabolites with an organism’s phenotype in a systematic and quantitative fashion (“phenome”). This goal will require comprehensive metabolomics and phenotypic data as well as computationally integrating other omic data.
Conclusions
The concept that metabolites can act as controllers – as opposed to a set of cogs in a system – is gaining new attention as metabolomics becomes more mainstream. The term “activity metabolomics” [G] is introduced here to describe how metabolomics technologies can be employed to identify active metabolites. Central to this metabolomics-driven concept is identifying these master metabolites [G], however resolving this challenge will be achieved not by any one approach alone, but instead by computationally combining metabolomics, systems biology and bioactivity data, allowing us to identify the most active metabolites that modulate biological processes and cell physiology. And while it is easy to get lost in the details of the problems (e.g. metabolite identification, metabolite annotation, et cetera) much of these issues are functionally solved or on their way to being solved. The most intriguing ultimate goal of these technological achievements is illustrated in Figures 1 and 5: using these metabolomics-driven screening methods as the main tool to identify these master manipulators. Once accomplished, activity metabolomics has the potential to impact a multiple scientific disciplines.
Glossary
Items in this glossary are marked with a [G] in the text.
- Activity Metabolomics
Activity screening guided by metabolomics, integration of metabolomics and activity screening technologies to identify bioactive metabolites controlling the phenotype
- Biocyc
A database of organism specific metabolic and genomic pathways 97
- Chemical space
The actual physico-chemical space (degree of freedom) defined by the chemical structure in which binding/activity might occur
- Cloud computing
The use of computational resources that are not physically present but at a server at another place, via internet connection.
- High resolution mass spectrometry
An Orbitrap or a quadrupole-time of flight mass spectrometer with high mass resolution and accuracy. A commonly used instrument for untargeted metabolomics acquisition.
- Triple quadrupole mass spectrometry
A mass spectrometer consisting of three quadrupoles in a row for targeted metabolomics quantification.
- KEGG
Kyoto encyclopedia of Genes and Genomes, a collection of database for pathways, genomes, small molecules and others. 102
- Master metabolites
Key metabolites driving phenotypes in a fate determining way.
- METLIN
A database of metabolite fragmentation spectral information to identify known and unknown metabolites, current size over 100,000 metabolites.
- Neural networks
A machine learning technique. Neural networks consist of artificial neurons that translate an input into an output.
- Recon
A genome-wide integration platform for metabolism, manually curated via community efforts and automated error checking 100,136
- Systems Biology
The integration of various datasets and disciplines to understand the regulatory elements in an organism or cell as a whole.
- Metabolic flux analysis
Metabolic flux analysis is a mass spectrometry based technique that is used to examine production and consumption rates of metabolites by tracking isotopes.
- IC50 value
The half maximal inhibitory concentration. This quantitative measure indicates how much of a molecule (e.g. metabolite or drug) is needed to inhibit a distinct biological process (e.g. enzyme activity).
- Reactome
Reactome provides molecular maps of signal transduction, metabolism and other cellular processes. 99
- Law of mass action
The law of mass action defines that a reversible chemical reaction in equilibrium is directly proportional to the concentrations of the reactants.
- Protein arrays
Proteins spotted on an array to see which proteins bind a specific metabolite.
- Feature
A peak or a set of peaks across samples with a unique m/z value and retention time
Footnotes
Competing interests.
The authors declare no competing interests.
Links for a selection of metabolomics databases and data analysis resources
Metabolomics data repositories
MetaboLights https://www.ebi.ac.uk/metabolights/
MetabolomicsWorkbench http://www.metabolomicsworkbench.org/
Metabolomics repository Bordeaux http://services.cbib.u-bordeaux.fr/MERYB/home/home.php
Bioinformatic tools for multi-omics integration
MetExplore https://metexplore.toulouse.inra.fr/metexplore2/
XC-MS online https://xcmsonline.scripps.edu
Galaxy project https://usegalaxy.org
KEGG https://www.kegg.jp/
Pathview https://pathview.uncc.edu/
Reactome https://reactome.org
Biocyc https://www.biocyc.org
Recon https://www.vmh.life/
MetaboAnalyst platform https://www.metaboanalyst.ca/
ChemRICH platform www.chemrich.fiehnlab.ucdavis.edu
References
- 1.Kahn F Man in structure & function ... (Knopf AA, 1943).
- 2.Crick F Central dogma of molecular biology. Nature 227, 561–563 (1970). [DOI] [PubMed] [Google Scholar]
- 3.Johnson CH, Ivanisevic J & Siuzdak G Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol 17, 451–459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Q, Vijayakumar A & Kahn BB Metabolites as regulators of insulin sensitivity and metabolism. Nat. Rev. Mol. Cell Biol 19, 654–672 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacob F & Monod J Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol 3, 318–356 (1961). [DOI] [PubMed] [Google Scholar]
- 6.Zoncu R, Efeyan A & Sabatini DM mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol 12, 21–35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katsyuba E & Auwerx J Modulating NAD+ metabolism, from bench to bedside. EMBO J. 36, 2670–2683 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magistretti PJ & Allaman I Lactate in the brain: from metabolic end-product to signalling molecule. Nat. Rev. Neurosci 19, 235–249 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Liu P-S et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol 18, 985–994 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Knobloch M et al. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature 493, 226–230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Branco Dos Santos F et al. Probing the genome-scale metabolic landscape ofBordetella pertussis, the causative agent of whooping cough. Appl. Environ. Microbiol (2017). doi: 10.1128/AEM.01528-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giera M, Branco Dos Santos F & Siuzdak G Metabolite-Induced Protein Expression Guided by Metabolomics and Systems Biology. CellMetab. 27, 270–272 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang M, Soga T & Pollard PJ Oncometabolites: linking altered metabolism with cancer. J. Clin. Invest 123, 3652–3658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beyer BA et al. Metabolomics-based discovery of a metabolite that enhances oligodendrocyte maturation. Nat. Chem. Biol 14, 22–28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sévin DC, Fuhrer T, Zamboni N & Sauer U Nontargeted in vitro metabolomics for high-throughput identification of novel enzymes in Escherichia coli. Nat. Methods 14, 187–194 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Yang M, Su H, Soga T, Kranc KR & Pollard PJ Prolyl hydroxylase domain enzymes: important regulators of cancer metabolism. Hypoxia Auckl. NZ 2, 127–142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao M et al. Inhibition of a-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 26, 1326–1338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adam J et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 20, 524–537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang M et al. The Succinated Proteome of FH-Mutant Tumours. Metabolites 4, 640–654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng L et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat. Commun 6, 6001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McBrayer SK et al. Transaminase Inhibition by 2-Hydroxyglutarate Impairs Glutamate Biosynthesis and Redox Homeostasis in Glioma. Cell 175, 101–116.e25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wishart DS Is Cancer a Genetic Disease or a Metabolic Disease? EBioMedicine 2, 478–479 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boroughs LK & DeBerardinis RJ Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol 17, 351–359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jha AK et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Piazza I et al. A Map of Protein-Metabolite Interactions Reveals Principles of Chemical Communication. Cell 172, 358–372.e23 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Guijas C, Montenegro-Burke JR, Warth B, Spilker ME & Siuzdak G Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol 36, 316–320 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metallo CM & Vander Heiden MG Understanding metabolic regulation and its influence on cell physiology. Mol. Cell 49, 388–398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabinowitz JD & Silhavy TJ Systems biology: metabolite turns master regulator. Nature 500, 283–284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choudhary C, Weinert BT, Nishida Y, Verdin E & Mann M The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol 15, 536–550 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Rana MS et al. Fatty acyl recognition and transfer by an integral membraneS-acyltransferase. Science 359, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.James AM et al. The Causes and Consequences of Nonenzymatic Protein Acylation. Trends Biochem. Sci 43, 921–932 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Weinert BT, Moustafa T, Iesmantavicius V, Zechner R & Choudhary C Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. EMBO J. 34, 2620–2632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dennis JW & Brewer CF Density-dependent lectin-glycan interactions as a paradigm for conditional regulation by posttranslational modifications. Mol. Cell. Proteomics MCP 12, 913–920 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hart GW, Slawson C, Ramirez-Correa G & Lagerlof O Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem 80, 825–858 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills EL et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature (2018). doi: 10.1038/nature25986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bambouskova M et al. Electrophilic properties of itaconate and derivatives regulate the IκBζ-ATF3 inflammatory axis. Nature (2018). doi: 10.1038/s41586-018-0052-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho-Park PF & Steller H Proteasome regulation by ADP-ribosylation. Cell 153, 614–627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan M et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 19, 605–617 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masri S & Sassone-Corsi P The circadian clock: a framework linking metabolism, epigenetics and neuronal function. Nat. Rev. Neurosci 14, 69–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petkovich DA et al. Using DNA Methylation Profiling to Evaluate Biological Age and Longevity Interventions. Cell Metab. 25, 954–960.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaefer M et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24, 1590–1595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Helm M & Alfonzo JD Posttranscriptional RNA Modifications: playing metabolic games in a cell’s chemical Legoland. Chem. Biol 21, 174–185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu X-L, Wang Y & Shen Q Epigenetic control on cell fate choice in neural stem cells. Protein Cell 3, 278–290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe A, Yamada Y & Yamanaka S Epigenetic regulation in pluripotent stem cells: a key to breaking the epigenetic barrier. Philos. Trans. R. Soc. Lond. B. Biol. Sci 368, 20120292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serganov A & Patel DJ Molecular recognition and function of riboswitches. Curr. Opin. Struct. Biol 22, 279–286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Husted AS, Trauelsen M, Rudenko O, Hjorth SA & Schwartz TW GPCR-Mediated Signaling of Metabolites. CellMetab. 25, 777–796 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Toma I et al. Succinate receptor GPR91 provides a direct link between high glucose levels and renin release in murine and rabbit kidney. J. Clin. Invest 118, 2526–2534 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Syed I et al. Palmitic Acid Hydroxystearic Acids Activate GPR40, Which Is Involved in Their Beneficial Effects on Glucose Homeostasis. Cell Metab. 27, 419–427.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yore MM et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 159, 318–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warth B et al. Metabolomics Reveals that Dietary Xenoestrogens Alter Cellular Metabolism Induced by Palbociclib/Letrozole Combination Cancer Therapy. Cell Chem. Biol 25, 291–300.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones CP & Ferré-D’Amaré AR Long-Range Interactions in Riboswitch Control of Gene Expression. Annu. Rev. Biophys 46, 455–481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rajniak J et al. Biosynthesis of redox-active metabolites in response to iron deficiency in plants. Nat. Chem. Biol. (2018). doi: 10.1038/s41589-018-0019-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinbusch L, Labouebe G & Thorens B Brain glucose sensing in homeostatic and hedonic regulation. Trends Endocrinol. Metab. TEM26, 455–466 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Cantor JR et al. Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell 169, 258–272.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee CK, Klopp RG, Weindruch R & Prolla TA Gene expression profile of aging and its retardation by caloric restriction. Science 285, 1390–1393 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Schvartzman JM, Thompson CB & Finley LWS Metabolic regulation of chromatin modifications and gene expression. J Cell Biol 217, 2247–2259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alam MT et al. The metabolic background is a global player in Saccharomyces gene expression epistasis. Nat. Microbiol 1, 15030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buescher JM et al. A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr. Opin. Biotechnol 34, 189–201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X, Gianoulis TA, Yip KY, Gerstein M & Snyder M Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses. Cell 143, 639–650 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Domingo-Almenara X, Montenegro-Burke JR, Benton HP & Siuzdak G Annotation: A Computational Solution for Streamlining Metabolomics Analysis. Anal. Chem 90, 480–489 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith CA, Want EJ, O’Maille G, Abagyan R & Siuzdak G XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem 78, 779–787 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Pluskal T, Castillo S, Villar-Briones A & Oresic M MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pfeuffer J et al. OpenMS - A platform for reproducible analysis of mass spectrometry data. J. Biotechnol 261, 142–148 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Tsugawa H et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12, 523–526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vinaixa M et al. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: State of the field and future prospects. TrAC Trends Anal. Chem 78, 23–35 (2016). [Google Scholar]
- 67.Wishart DS et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 46, D608–D617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wishart DS et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 41, D801–807 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guijas C et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem (2018). doi: 10.1021/acs.analchem.7b04424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ludwig C et al. Birmingham Metabolite Library: a publicly accessible database of 1-D 1H and 2-D 1H J-resolved NMR spectra of authentic metabolite standards (BML-NMR). Metabolomics 8, 8–18 (2012). [Google Scholar]
- 71.King ZA et al. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 44, D515–D522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Horai H et al. MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom 45, 703–714 (2010). [DOI] [PubMed] [Google Scholar]
- 73.Fahy E et al. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res 50 Suppl, S9–14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.mzCloud - Advanced Mass Spectral Database. Available at: https://www.mzcloud.org/. (Accessed: 24th October 2018)
- 75.Lai Z et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 15, 53–56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hummel J, Selbig J, Walther D & Kopka J The Golm Metabolome Database: a database for GC-MS based metabolite profiling in Metabolomics: A Powerful Tool in Systems Biology (eds. Nielsen J & Jewett MC) 75–95 (Springer Berlin Heidelberg, 2007). doi: 10.1007/4735_2007_0229 [DOI] [Google Scholar]
- 77.Huan T et al. Systems biology guided by XCMS Online metabolomics. Nat. Methods 14, 461–462 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cottret L et al. MetExplore: collaborative edition and exploration of metabolic networks. Nucleic Acids Res. 46, W495–W502 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boekel J et al. Multi-omic data analysis using Galaxy. Nat. Biotechnol 33, 137–139 (2015). [DOI] [PubMed] [Google Scholar]
- 80.Kuo T-C, Tian T-F & Tseng YJ 3Omics: a web-based systems biology tool for analysis, integration and visualization of human transcriptomic, proteomic and metabolomic data. BMC Syst. Biol 7, 64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karnovsky A et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinforma. Oxf. Engl 28, 373–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jang C, Chen L & Rabinowitz JD Metabolomics and Isotope Tracing. Cell 173, 822–837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu H et al. Global analysis of protein activities using proteome chips. Science 293, 2101–2105 (2001). [DOI] [PubMed] [Google Scholar]
- 84.Roelofs KG, Wang J, Sintim HO & Lee VT Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc. Natl. Acad. Sci. U. S. A 108, 15528–15533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Savitski MM et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346, 1255784 (2014). [DOI] [PubMed] [Google Scholar]
- 86.Diether M & Sauer U Towards detecting regulatory protein-metabolite interactions. Curr. Opin. Microbiol 39, 16–23 (2017). [DOI] [PubMed] [Google Scholar]
- 87.Tran DT, Adhikari J & Fitzgerald MC Stablelsotope Labeling with Amino Acids in Cell Culture (SILAC)-based strategy for proteome-wide thermodynamic analysis of protein-ligand binding interactions. Mol. Cell. Proteomics MCP 13, 1800–1813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Feng Y et al. Global analysis of protein structural changes in complex proteomes. Nat. Biotechnol. 32, 1036–1044 (2014). [DOI] [PubMed] [Google Scholar]
- 89.Gaulton A et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, D1100–1107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kirchmair J et al. Predicting drug metabolism: experiment and/or computation? Nat. Rev. Drug Discov 14, 387–404 (2015). [DOI] [PubMed] [Google Scholar]
- 91.Branco Dos Santos F et al. Probing the genome-scale metabolic landscape of Bordetella pertussis, the causative agent of whooping cough. Appl. Environ. Microbiol (2017). doi: 10.1128/AEM.01528-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ge H, Walhout AJM & Vidal M Integrating ‘omic’ information: a bridge between genomics and systems biology. Trends Genet. TIG 19, 551–560 (2003). [DOI] [PubMed] [Google Scholar]
- 93.Hakimi AA et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell 29, 104–116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Davidson RL, Weber RJM, Liu H, Sharma-Oates A & Viant MR Galaxy-M: a Galaxy workflow for processing and analyzing direct infusion and liquid chromatography mass spectrometry-based metabolomics data. GigaScience 5, 10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Villiers F et al. Investigating the plant response to cadmium exposure by proteomic and metabolomic approaches. Proteomics 11, 1650–1663 (2011). [DOI] [PubMed] [Google Scholar]
- 96.Zhang W, Li F & Nie L Integrating multiple ‘omics’ analysis for microbial biology: application and methodologies. Microbiol. Read. Engl 156, 287–301 (2010). [DOI] [PubMed] [Google Scholar]
- 97.Caspi R et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 42, D459–471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xia J & Wishart DS MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 38, W71–77 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fabregat A et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 46, D649–D655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Swainston N et al. Recon 2.2: from reconstruction to model of human metabolism. Metabolomics Off. J. Metabolomic Soc 12, 109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barupal DK & Fiehn O Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci. Rep 7, 14567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kanehisa M, Goto S, Sato Y, Furumichi M & Tanabe M KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Haas R et al. Designing and interpreting ‘multi-omic’ experiments that may change our understanding of biology. Curr. Opin. Syst. Biol 6, 37–45 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yugi K, Kubota H, Hatano A & Kuroda S Trans-Omics: How To Reconstruct Biochemical Networks Across Multiple ‘Omic’ Layers. Trends Biotechnol. 34, 276–290 (2016). [DOI] [PubMed] [Google Scholar]
- 105.Haug K et al. MetaboLights—an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 41, D781–D786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sud M et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 44, D463–D470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Knepper MA Proteomic pearl diving versus systems biology in cell physiology. Focus on “Proteomic mapping of proteins released during necrosis and apoptosis from cultured neonatal cardiac myocytes”. Am. J. Physiol. - Cell Physiol 306, C634–C635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nielsen J & Keasling JD Engineering Cellular Metabolism. Cell 164, 1185–1197 (2016). [DOI] [PubMed] [Google Scholar]
- 109.Chassagnole C, Noisommit-Rizzi N, Schmid JW, Mauch K & Reuss M Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnol. Bioeng 79, 53–73 (2002). [DOI] [PubMed] [Google Scholar]
- 110.Covert MW, Xiao N, Chen TJ & Karr JR Integrating metabolic, transcriptional regulatory and signal transduction models in Escherichia coli. Bioinformatics 24, 2044–2050 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Breunig JS, Hackett SR, Rabinowitz JD & Kruglyak L Genetic Basis of Metabolome Variation in Yeast. PLOS Genet. 10, e1004142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zelezniak A et al. Machine Learning Predicts the Yeast Metabolome from the Quantitative Proteome of Kinase Knockouts. Cell Syst. 7, 269–283.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim M, Rai N, Zorraquino V & Tagkopoulos I Multi-omics integration accurately predicts cellular state in unexplored conditions for Escherichia coli. Nat. Commun 7, 13090 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frainay C et al. Mind the Gap: Mapping Mass Spectral Databases in Genome-Scale Metabolic Networks Reveals Poorly Covered Areas. Metabolites 8, 51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chae YK, Kim SH & Markley JL Relationship between recombinant protein expression and host metabolome as determined by two-dimensional NMR spectroscopy. PloS One 12, e0177233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Human Microbiome Project Consortium. A framework for human microbiome research. Nature 486, 215–221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li Z et al. Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit. Gut (2017). doi: 10.1136/gutjnl-2017-314050 [DOI] [PubMed] [Google Scholar]
- 118.Funk CD Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 294, 1871–1875 (2001). [DOI] [PubMed] [Google Scholar]
- 119.Kalinski P Regulation of immune responses by prostaglandin E2. J. Immunol. Baltim. Md 1950 188, 21–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kaisar MMM et al. Dectin-1/2-induced autocrine PGE2 signaling licenses dendritic cells to prime Th2 responses. PLoS Biol. 16, e2005504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lipworth BJ Leukotriene-receptor antagonists. LancetLond. Engl. 353, 57–62 (1999). [DOI] [PubMed] [Google Scholar]
- 122.Siscovick DS et al. Omega-3 Polyunsaturated Fatty Acid (Fish Oil) Supplementation and the Prevention of Clinical Cardiovascular Disease: A Science Advisory From the American Heart Association. Circulation 135, e867–e884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kris-Etherton PM, Harris WS, Appel LJ & American Heart Association. Nutrition Committee. Fish consumption, fish oil, omega-3 fatty acids, and cardiovascular disease. Circulation 106, 2747–2757 (2002). [DOI] [PubMed] [Google Scholar]
- 124.Hur J et al. Cerebrovascular P-amyloid deposition and associated microhemorrhages in a Tg2576 Alzheimer mouse model are reduced with a DHA-enriched diet. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol fj201800200R (2018). doi: 10.1096/fj.201800200R [DOI] [PubMed] [Google Scholar]
- 125.Grandison RC, Piper MDW & Partridge L Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462, 1061–1064 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Denzel MS et al. Hexosamine pathway metabolites enhance protein quality control and prolong life. Cell 156, 1167–1178 (2014). [DOI] [PubMed] [Google Scholar]
- 127.Serhan CN Treating inflammation and infection in the 21st century: new hints from decoding resolution mediators and mechanisms. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 31, 1273–1288 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Niihara Y et al. A Phase 3 Trial of l-Glutamine in Sickle Cell Disease. N. Engl. J. Med 379, 226–235 (2018). [DOI] [PubMed] [Google Scholar]
- 129.Morris CR et al. Erythrocyte glutamine depletion, altered redox environment, and pulmonary hypertension in sickle cell disease. Blood 111, 402–410 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Doucette CD, Schwab DJ, Wingreen NS & Rabinowitz JD a-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat. Chem. Biol 7, 894–901 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chin RM et al. The metabolite a-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature 510, 397–401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Klysz D et al. Glutamine-dependent a-ketoglutarate production regulates the balance between T helper 1 cell and regulatory T cell generation. Sci. Signal 8, ra97 (2015). [DOI] [PubMed] [Google Scholar]
- 133.Coudray-Lucas C, Le Bever H, Cynober L, De Bandt JP & Carsin H Ornithine alpha-ketoglutarate improves wound healing in severe burn patients: a prospective randomized double-blind trial versus isonitrogenous controls. Crit. Care Med. 28, 1772–1776 (2000). [DOI] [PubMed] [Google Scholar]
- 134.Grimm PR et al. Integrated compensatory network is activated in the absence of NCC phosphorylation. J. Clin. Invest 125, 2136–2150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Grimm PR & Welling PA a-Ketoglutarate drives electroneutral NaCl reabsorption in intercalated cells by activating a G-protein coupled receptor, Oxgrl. Curr. Opin. Nephrol. Hypertens 26, 426–433 (2017). [DOI] [PubMed] [Google Scholar]
- 136.Brunk E et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol 36, 272–281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Patti GJ et al. Meta-analysis of global metabolomic data identifies metabolites associated with life-span extension. Metabolomics Off. J. Metabolomic Soc 10, 737–743 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kamburov A, Cavill R, Ebbels TMD, Herwig R & Keun HC Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics 27, 2917–2918 (2011). [DOI] [PubMed] [Google Scholar]
- 139.PhenoMeNal: Processing and analysis of Metabolomics data in the Cloud | bioRxiv. Available at: https://www.biorxiv.org/content/early/2018/09/13/409151. (Accessed: 24th October 2018) [DOI] [PMC free article] [PubMed]
- 140.Boneschansker L, Yan J, Wong E, Briscoe DM & Irimia D Microfluidic platform for the quantitative analysis of leukocyte migration signatures. Nat. Commun 5, 4787 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cravatt BF et al. Chemical characterization of a family of brain lipids that induce sleep. Science 268, 1506–1509 (1995). [DOI] [PubMed] [Google Scholar]
- 142.Warth B et al. Exposome-Scale Investigations Guided by Global Metabolomics, Pathway Analysis, and Cognitive Computing. Anal. Chem. 89, 11505–11513 (2017). [DOI] [PubMed] [Google Scholar]
- 143.Laan LC et al. The whipworm (Trichuris suis) secretes prostaglandin E2 to suppress proinflammatory properties in human dendritic cells. FASEB J. 31, 719–731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Coupling HPLC to on-line, post-column (bio)chemical assays for high-resolution screening of bioactive compounds from complex mixtures - ScienceDirect. Available at: https://www.sciencedirect.com/science/article/abs/pii/S0165993609000715. (Accessed: 24th October 2018)
- 145.Tammela P, Wennberg T, Vuorela H & Vuorela P HPLC micro-fractionation coupled to a cell-based assay for automated on-line primary screening of calcium antagonistic components in plant extracts. Anal. Bioanal. Chem 380, 614–618 (2004). [DOI] [PubMed] [Google Scholar]
- 146.Veyel D et al. PROMIS, global analysis of PROtein-metabolite interactions using size separation in Arabidopsis thaliana. J. Biol. Chem 293, 12440–12453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Annis DA, Nickbarg E, Yang X, Ziebell MR & Whitehurst CE Affinity selection-mass spectrometry screening techniques for small molecule drug discovery. Curr. Opin. Chem. Biol 11, 518–526 (2007). [DOI] [PubMed] [Google Scholar]
- 148.Huber KVM et al. Proteome-wide drug and metabolite interaction mapping by thermal-stabilit profiling. Nat. Methods 12, 1055–1057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sergushichev AA et al. GAM: a web-service for integrated transcriptional and metabolic network analysis. Nucleic Acids Res. 44, W194–W200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Orth JD, Thiele I & Palsson B 0. What is flux balance analysis? Nat. Biotechnol. 28, 245–248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Harcombe WR et al. Metabolic Resource Allocation in Individual Microbes Determines Ecosystem Interactions and Spatial Dynamics. Cell Rep. 7, 1104–1115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li S et al. Predicting Network Activity from High Throughput Metabolomics. PLOS Comput. Biol 9, e1003123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pirhaji L et al. Revealing disease-associated pathways by network integration of untargeted metabolomics. Nat. Methods 13, 770–776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Olivon F et al. Bioactive Natural Products Prioritization Using Massive Multi-informational Molecular Networks. ACS Chem. Biol 12, 2644–2651 (2017). [DOI] [PubMed] [Google Scholar]
- 155.Cytotoxic Prenylated Stilbenes Isolated from Macaranga tanarius - Journal of Natural Products (ACS Publications). Available at: https://pubs.acs.org/doi/abs/10.1021/acs.jnatprod.7b00409. (Accessed: 24th October 2018) [DOI] [PubMed]