Abstract
Purpose
The purpose of this study was to investigate the relationship between epidermal growth factor receptor (EGFR) gene mutation and clinicopathological features of lung adenocarcinoma, and the prognostic and therapeutic value of EGFR.
Methods
EGFR gene mutations were detected in 424 patients with lung adenocarcinoma by amplification refractory mutation system (ARMS).
Results
The total EGFR gene mutation rate was 55.2% (234/424) and EGFR gene mutation rates were statistically different in gender, smoking status, and pathological degree (P<0.05). The overall survival (OS) time of lung adenocarcinoma patients with mutation of exon 18 was lower than those with mutation of exon 19 and exon 21 (both P<0.05), but no significant difference was seen between those with mutation of exon 19 and exon 21 (P>0.05). Among 424 cases of lung adenocarcinoma, multivariate analysis showed that EGFR gene mutation, age, gender, clinical stages, and pathological degree (P<0.05) were statistically significant prognostic factors. In multivariate analysis, prognostic factors of patients with EGFR gene mutation were associated with EGFR-TKI treatment, surgery treatment, pathological degree, clinical stages, and age (P<0.05), whereas in patients without EGFR gene mutation, prognostic factors were related to surgery treatment, pathological degree, clinical stages, gender, age, and smoking status (P<0.05).
Conclusion
The OS time of patients with mutation of exon 18 was lower than those of exon 19 and exon 21. EGFR-TKI treatment was an independent positive predictor in patients with EGFR gene mutation. Surgery treatment, age, clinical stages, and pathological degree were independent prognostic factors in Chinese patients with lung adenocarcinoma no matter whether with EGFR gene mutation or not.
Keywords: EGFR gene mutation, lung adenocarcinoma, clinicopathological features, prognosis
Introduction
Lung cancer is one of the major malignant tumors that cause cancer death,1 which has been proved to be associated with smoking,2 indicating that other genetic and environmental factors can also contribute to occurrence and development of lung cancer.3,4 EGFR is a glycoprotein that plays a vital role in cell proliferation and apoptosis and has been shown to be important in tumorigenesis.5,6 Precision treatment targeting epidermal growth factor receptor (EGFR) gene has made great progress. However, not all patients with lung cancer are sensitive to EGFR-tyrosine kinase inhibitors (EGFR-TKIs). It has been proved that EGFR gene mutation is a predictive factor of response to EGFR-TKIs treatment that leads to longer progression-free survival (PFS) in high mutation abundance patients.7,8 Some studies have examined the relationship between EGFR gene mutation and clinicopathological features.9–12 However, it is not clear whether the prognostic factors of patients with EGFR gene mutation are consistent with those of wild-type EGFR patients. Thus, our purpose was to investigate the relationship between EGFR gene mutation and clinicopathological features and to further analyze whether EGFR gene mutation has the prognostic and therapeutic implications in a large number of Chinese lung adenocarcinoma patients, which provides evidence for precision treatment of patients with lung adenocarcinoma. Finally, we further explored the prognostic factors of patients with and without EGFR gene mutation, respectively.
Materials and methods
Patients and clinical data
A total of 424 patients with pathologically confirmed lung adenocarcinoma from The Second Xiangya Hospital of Central South University between August 2012 and July 2017 were enrolled for this study, and their paraffin sections were selected to detect EGFR gene mutation. All patients in this study were approved by the Ethics Committee of The Second Xiangya Hospital of Central South University (Scientific and Research Ethics Committee, no. s02/2000) and any related procedures were performed in accordance with relevant guidelines and regulations. Written informed consent was obtained from all patients, and the written informed consent was obtained from the next of kin, caretakers, or guardians on the behalf of the minors/children participants involved in our study. TNM staging of lung adenocarcinoma was performed according to the Eighth Edition Lung Cancer Stage Classification.13 Pathological diagnosis was based on the 2011 IASLC/ATS/ERS Lung Adenocarcinoma Classification system.14 The research samples included 217 males and 207 females with an average age of 58.0±10.9 years. One hundred and thirty patients received platinum-based chemotherapy, including Cisplatin+Paclitaxel/Docetaxel/Gemcitabine/Pemetrexed and Carboplatin+Paclitaxel/Pemetrexed. One hundred and fifty-seven patients were treated with EGFR-TKIs which included Gefitinib, Erlotinib, and Icotinib. The basic information of these patients was shown in Supplementary Table S1.
Table S1.
Clinicopathological features of 424 cases of lung adenocarcinoma
| Clinicopathological features | n (%) |
|---|---|
| Age (years) | |
| ≤60 | 242 (57.1%) |
| >60 | 182 (42.9%) |
| Gender | |
| Female | 207 (48.8%) |
| Male | 217 (51.2%) |
| Smoking status | |
| Smoker | 152 (35.8%) |
| Nonsmoker | 272 (64.2%) |
| Pathological degree | |
| Poor differentiation | 103 (24.3%) |
| Well and moderated differentiation | 321 (75.7%) |
| Clinical stages | |
| I+II+III | 181 (42.7%) |
| IV | 243 (57.3%) |
| Specimen types | |
| Biopsy specimen | 298 (70.3%) |
| Surgical specimen | 126 (29.7%) |
| EGFR mutation | |
| Positive | 234 (55.2%) |
| Negative | 190 (44.8%) |
| Chemotherapy | |
| Yes | 130 (30.7%) |
| No | 294 (69.3%) |
| Surgery treatment | |
| Yes | 134 (31.6%) |
| No | 290 (68.4%) |
| EGFR-TKI treatment | |
| Yes | 157 (37.0%) |
| No | 267 (63.0%) |
| Survival situation | |
| Survival | 216 (50.9%) |
| Death | 208 (49.1%) |
Clinical follow-up of 424 patients
This study collected the medical histories of admitted patients and was followed up by telephone interview to ask for survival status (survival or death), recurrence and metastasis information. Overall survival was defined as the time from the treatment initiation (diagnosis) to the date of death. If the patients died, the follow-up contents were the time and cause of death. In this study, all the dead patients died of lung cancer.
ARMS method for detecting EGFR gene mutation types
DNA extraction. Paraffin-embedded samples were cut into 5 μm, 3–8 slides were put into 1.5 mL Eppendorf tube, and an appropriate amount of xylene was added to deparaffinized, then an appropriate amount of ethanol was added to elute xylene, after that the samples dried at room temperature or 37°C until the ethanol was completely evaporated. DNA was extracted according to the manufacturer’s instructions of the QIAamp DNA FFPE Tissue kit (Qiagen, Hilden, Germany), and the DNA samples were examined for concentration and purity.
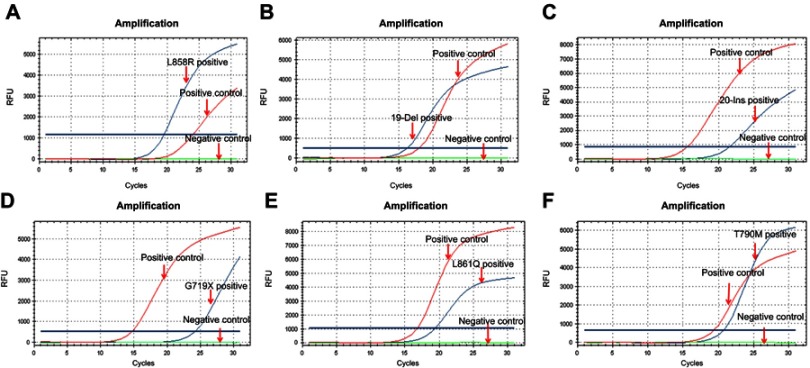
EGFR gene mutation detection. The detection of EGFR gene mutation was performed following the instructions of ADx-ARMS® EGFR Mutation Test kit (Amoydx, Xiamen, China). The representative curves of different types of EGFR gene mutation were presented in Figure 1.
Figure 1.
Representative curves of different mutation subtypes of EGFR gene.
Notes: Representative curves of L858R of exon 21 (A), 19-Del of exon 19(B), 20-Ins of exon 20(C), G719X of exon 18(D), L861Q of exon 21(E), and T790M of exon 20(F). (EGFR gene mutation was detected by ARMS method. The red curves represent positive control which presents the amplification curve, the green curves represent negative control which has no amplification curve, and the blue curves represent the lung adenocarcinoma tissue samples).
Abbreviations: EGFR, epidermal growth factor receptor; ARMS, amplification refractory mutation system.
Statistical analysis
All statistical analyses were performed using SPSS 24.0. The relationship between the rate of EGFR gene mutation and the clinicopathological features of lung adenocarcinoma was analyzed by the χ2 test or Fisher’s exact test. Logistic regression analysis was used to evaluate independent factors associated with EGFR gene status. Kaplan–Meier analysis was performed for overall survival (OS) curves, and statistical significance was assessed using the log-rank test. Additionally, Cox comparative hazards model was used to estimate the independent prognostic factors of lung adenocarcinoma. Two-sided statistical analysis was used, and the data were considered to be statistically significant when P<0.05.
Ethical approval and informed consent
All patients in this study were approved by the Ethics Committee of The Second Xiangya Hospital of Central South University (Scientific and Research Ethics Committee, no. s02/2000). Written informed consent was obtained from all patients.
Results
Subtypes analysis of EGFR gene mutation in the lung adenocarcinoma
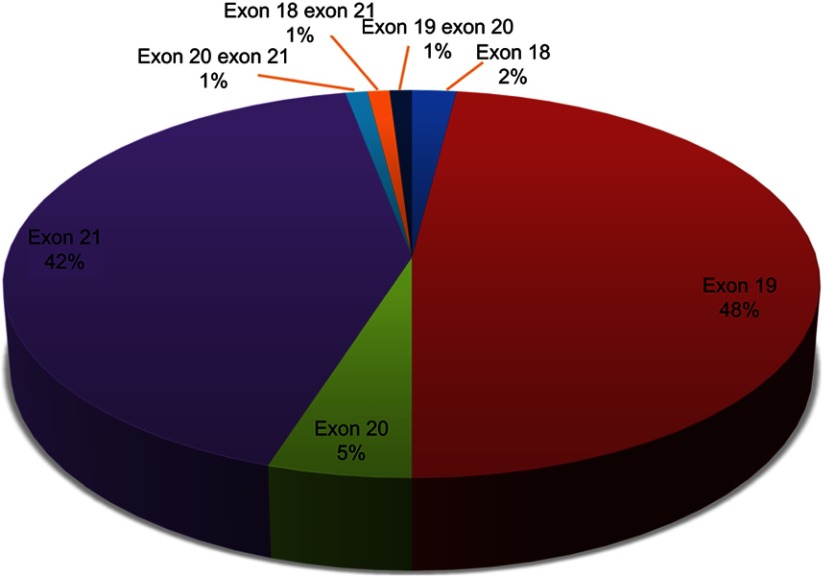
The total positive percentage of EGFR gene mutation was 55.2% (234/424) in 424 cases of lung adenocarcinoma patients (Figure 2), including exon 18 (2.0%), exon 19 (48.0%), exon 20 (5.0%), exon 21 (42.0%), and double mutations (3.0%). Six cases were identified to have EGFR gene double mutations, two of exon 18 and 21, two of exon 19 and 20, and two of exon 20 and 21. In the exon 20 mutation, 9 cases were deletion mutation and 3 cases were T790M mutation. The most common mutation was classic activating mutation L858R in exon 21 and deletion in exon 19 (19del).
Figure 2.
Proportion analysis of EGFR gene mutation subtypes in the lung adenocarcinoma patients.
Notes: Exon 18, exon 19, exon 20, and exon 21 accounted for 2%, 48%, 5%, and 42%, of mutant EGFR gene, respectively. Double mutation of exon 18 and 21, exon 19 and 20, and exon 20 and 21 accounted for 2%, respectively.
Abbreviation: EGFR, epidermal growth factor receptor.
The relationship between EGFR gene mutation and clinicopathological features of lung adenocarcinoma
The EGFR gene mutation rates were significantly higher in the female (73.9%, 153/207) than in the male patients (37.3%, 81/217) (P<0.001). Furthermore, the total EGFR gene mutation rates were evidently higher in nonsmoker patients (66.9%, 182/272) compared with smoker patients (34.2%, 52/152) (P<0.001). EGFR gene mutation rates were also statistically different in pathological degree (well and moderated differentiation vs poor differentiation: 59.8% (192/321) vs 40.8% (42/103); P=0.001). However, there was no significant association between EGFR gene mutation rates and age or clinical stages (P>0.05). Among 134 cases of lung adenocarcinoma with surgical treatment, 8 patients underwent EGFR gene mutation detection in biopsy tissue. Therefore, this analysis only included 126 cases of lung adenocarcinoma with surgical specimens, and there were significant correlations between the presence or absence of EGFR gene mutation and gender, smoking status, and pathological degree of cancer (P<0.05), but no significant differences in maximum diameter of tumor, lymph node metastasis, pleural metastasis, age, and clinical stages (P>0.05) (Table 1). EGFR gene mutation was independently associated with gender and pathological degree, but not with smoking status (Supplementary Table S2).
Table 1.
Association between the EGFR gene mutation and clinicopathological features of lung adenocarcinoma
| Clinicopathological features | Total lung adenocarcinoma (n=424) | Biopsy specimen (n=298) | Surgical specimen (n=126) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| EGFR mutation (+) | EGFR mutation (‒) | P | EGFR mutation (+) | EGFR mutation (‒) | P | EGFR mutation (+) | EGFR mutation (‒) | P | |
| Age (years) | |||||||||
| ≤60 | 132 (54.5%) | 110 (45.5%) | 0.759 | 92 (53.2%) | 81 (46.8%) | 0.835 | 40 (58.0%) | 29 (42.0%) | 0.849 |
| >60 | 102 (56.0%) | 80 (44.0%) | 68 (54.4%) | 57 (45.6%) | 34 (59.6%) | 23 (40.4%) | |||
| Gender | |||||||||
| Female | 153 (73.9%) | 54 (26.1%) | 0.000* | 103 (73.0%) | 38 (27.0%) | 0.000* | 50 (75.8%) | 16 (24.2%) | 0.000* |
| Male | 81 (37.3%) | 136 (62.7%) | 57 (36.3%) | 100 (63.7%) | 24 (40.0%) | 36 (60.0%) | |||
| Smoking status | |||||||||
| Smoker | 52 (34.2%) | 100 (65.8%) | 0.000* | 38 (34.5%) | 72 (65.5%) | 0.000* | 14 (33.3%) | 28 (66.7%) | 0.000* |
| Nonsmoker | 182 (66.9%) | 90 (33.1%) | 122 (64.9%) | 66 (35.1%) | 60 (71.4%) | 24 (28.6%) | |||
| Pathological degree | |||||||||
| Well/moderated | 192 (59.8%) | 129 (40.2%) | 0.001* | 121 (57.6%) | 89 (42.4%) | 0.036* | 71 (64.0%) | 40 (36.0%) | 0.001* |
| Poor | 42 (40.8%) | 61 (59.2%) | 39 (44.3%) | 49 (55.7%) | 3 (20.0%) | 12 (80.0%) | |||
| Clinical stages | |||||||||
| Stage I+II+III | 98 (54.1%) | 83 (45.9%) | 0.709 | 28 (44.4%) | 35 (55.6%) | 0.097 | 70 (59.3%) | 48 (40.7%) | 0.883 |
| Stage IV | 136 (56.0%) | 107 (44.0%) | 132 (56.2%) | 103 (43.8%) | 4 (50.0%) | 4 (50.0%) | |||
| Maximum diameter of tumor | |||||||||
| ≤3cm | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | 51(63.0%) | 30(37.0%) | 0.278 |
| >3, ≤5 cm | ‒ | ‒ | ‒ | ‒ | 18(54.5%) | 15(45.5%) | |||
| >5, ≤7cm | ‒ | ‒ | ‒ | ‒ | 4(57.1%) | 3(42.9%) | |||
| >7cm | ‒ | ‒ | ‒ | ‒ | 1(20.0%) | 4(80.0%) | |||
| Lymph node metastasis | |||||||||
| Positive | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | 28 (59.6%) | 19 (40.4%) | 0.882 |
| Negative | ‒ | ‒ | ‒ | ‒ | 46 (58.2%) | 33 (41.8%) | |||
| Pleural metastasis | |||||||||
| Positive | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | 35 (56.5%) | 27 (43.5%) | 0.609 |
| Negative | ‒ | ‒ | ‒ | ‒ | 39 (60.9%) | 25 (39.1%) | |||
Notes: For the Chi-square test,*P<0.05 is considered statistically significant, “-” missing information.
Table S2.
Summary for logistic regression analysis of factors associated with EGFR gene status in lung adenocarcinoma
| Clinicopathological features | Total lung adenocarcinoma (n=424) | Biopsy specimen (n=298) | Surgical specimen (n=126) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | |
| Gender | 0.276 | 0.159-0.479 | 0.000* | 0.240 | 0.125-0.458 | 0.000* | 0.591 | 0.180-1.936 | 0.385 |
| Smoking status | 1.583 | 0.902-2.779 | 0.109 | 0.360 | 0.706-2.605 | 0.360 | 3.323 | 0.973-11.350 | 0.055 |
| Pathological degree | 0.453 | 0.277-0.741 | 0.002* | 0.516 | 0.296-0.899 | 0.019* | 0.165 | 0.040-0.680 | 0.013* |
Note: *P<0.005.
Abbreviations: CI, confidence interval; OR, odds ratio.
Association analysis between overall survival time of lung adenocarcinoma patients and EGFR gene mutation subtypes
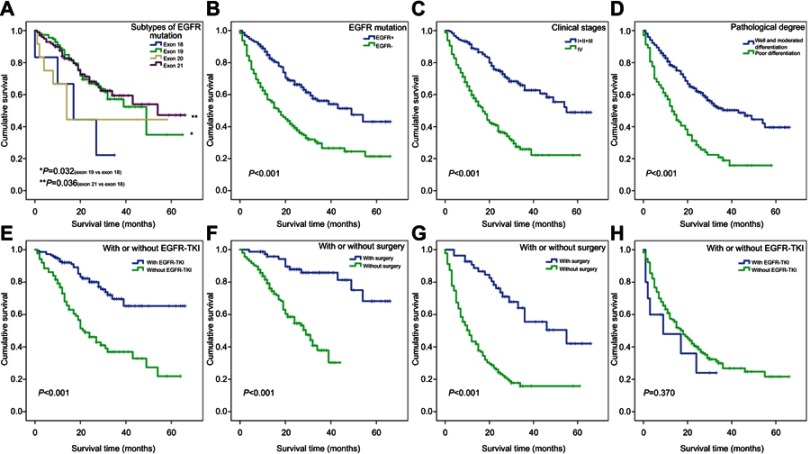
We analyzed OS time in different EGFR gene mutation subtypes based on the data of single mutation and double mutation, which was shown in Supplementary Table S3. The OS time of the exon 19 mutation group was longer than that of the exon 18 group, which had statistical significance (average survival time: 41.2 vs 19.4 months, respectively; P=0.032). The OS time of the exon 21 group was longer than exon 18 group and had statistical significance (average survival time: 43.9 vs 19.4 months, respectively; P=0.036); however, there were no significant differences between exon 18 and exon 20, exon 19 and 20, exon 19 and 21, exon 20 and exon 21 (P>0.05) (Figure 3A).
Table S3.
Survival analysis of different EGFR gene mutation types
| EGFR gene subtypes | Average survival time (SE) | 95.0% CI | |
|---|---|---|---|
| Lower | Upper | ||
| Exon 18 | 19.39 (5.02) | 9.543 | 29.234 |
| Exon 19 | 41.19 (3.34) | 34.636 | 47.741 |
| Exon 20 | 30.14 (7.82) | 14.820 | 45.457 |
| Exon 21 | 43.87 (3.03) | 37.932 | 49.807 |
| Double mutation | 26.50 (5.81) | 15.113 | 37.887 |
Abbreviations: SE, standard error; CI, confidence interval.
Figure 3.
Kaplan–Meier curves for 424 cases of lung adenocarcinoma patients with different clinicopathological features.
Notes: (A) Patients with exon 18 mutation had shorter OS than those with exon 19 and exon 21. (B–D) 424 cases of lung adenocarcinoma patients with wild-type EGFR gene, stage IV and poor differentiation had shorter survival. (E and F) 234 cases of mutant EGFR gene with EGFR-TKI treatment and surgery treatment had longer survival. (G and H) 190 cases of wild-type EGFR gene with surgery treatment had longer survival, but EGFR-TKI treatment was not significant (Kaplan–Meier analysis was used to plot the overall survival and statistical significance was assessed by log-rank test).
Abbreviations: OS, overall survival; EGFR, epidermal growth factor receptor; EGFR-TKI, EGFR-tyrosine kinase inhibitor.
Survival analysis for 424 cases of lung adenocarcinoma patients
The median survival time and average survival time of 424 patients with lung adenocarcinoma were 28.0 months and 34.9 months, respectively. Kaplan–Meier survival analysis was used to estimate the survival time of 424 cases of lung adenocarcinoma. The cumulative survival rate was 84.2% at 6 months, 72.3% at 12 months, and 42.2% at 36 months. Kaplan–Meier survival curves revealed that the average survival time of patients with EGFR gene mutation positive and EGFR gene mutation negative was 42.2 months and 26.3 months, respectively (P<0.001; Figure 3B), whereas the average survival time was 39.7 months in females and 30.6 months in males (P=0.001). There was also difference in average survival time among smoking status (smoker vs non-smoker: 29.8 vs 37.8 months, respectively; P=0.014), clinical stages (I+II+III vs IV: 46.5 vs 24.8 months, respectively; P<0.001; Figure 3C) and pathological degree of cancer (well and moderated differentiation vs poor differentiation: 39.6 vs 20.1 months, respectively; P<0.001; Figure 3D). All of these differences were statistically significant, which indicated the relationship between these prognostic factors and survival time, but there was no statistical difference in the age of patients (P=0.054). Multivariate analysis using Cox comparative hazards model analyzed these main factors determining the prognosis of patients, including EGFR gene mutation, clinical stages, pathological degree, age, and gender (P<0.05). But smoking status was not prognostic factor in multivariate analysis (P=0.144). The risk of death in patients without EGFR gene mutation was 2.4 times higher than that of patients with EGFR gene mutation, and the risk of death in patients with stage IV was 2.1 times than that of patients with stage I+II+III (Table 2).
Table 2.
Analysis between average survival time (months) and clinicopathological features in 424 cases of lung adenocarcinoma
| Clinicopathological | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Features | Survival time (SE) | 95%CI | P | Exp (B) | 95%CI | P |
| Age (years) | ||||||
| ≤60 | 38.0 (1.94) | (34.2, 41.8) | 0.054 | 0.625 | (0.501, 0.872) | 0.003* |
| >60 | 31.5 (2.10) | (27.4, 35.6) | ||||
| Gender | ||||||
| Female | 39.7 (2.11) | (35.5, 43.8) | 0.001* | 0.645 | (0.445, 0.940) | 0.022* |
| Male | 30.6 (1.95) | (26.8, 34.4) | ||||
| Smoking status | ||||||
| Smoker | 29.8 (2.30) | (25.3, 34.3) | 0.014* | 0.743 | (0.527, 1.098) | 0.144 |
| Nonsmoker | 37.8 (1.85) | (34.2, 41.4) | ||||
| Pathological degree | ||||||
| Well and moderated differentiation | 39.6 (1.69) | (36.2, 42.9) | 0.000* | 0.534 | (0.392, 0.700) | 0.000* |
| Poor differentiation | 20.1 (2.06) | (16.1, 24.1) | ||||
| Clinical stages | ||||||
| I+II+III | 46.5 (2.04) | (42.5, 50.5) | 0.000* | 0.475 | (0.192, 0.368) | 0.000* |
| IV | 24.8 (1.73) | (21.4, 28.2) | ||||
| EGFR mutation | ||||||
| Positive | 42.2 (2.04) | (38.2, 46.2) | 0.000* | 0.415 | (0.288, 0.529) | 0.000* |
| Negative | 26.3 (1.95) | (22.5, 30.1) | ||||
Note: *P<0.05.
Abbreviations: SE, standard error; 95% CI, 95% confidence interval.
Survival analysis for 234 cases of lung adenocarcinoma patients with EGFR gene mutation
Univariate analysis of prognostic factors using Kaplan–Meier survival curves showed that effective prognostic factors included pathological degree (well and moderated differentiation vs poor differentiation: 46.4 vs 21.3 months, respectively; P<0.001), clinical stages (I+II+III vs IV: 53.3 vs 27.9 months, respectively; P<0.001), EGFR-TKI treatment (EGFR-TKI treatment vs non-EGFR-TKI treatment: 51.0 vs 30.7 months, respectively; P<0.001; Figure 3E) and surgery treatment (non-surgical patients vs surgical patients: 27.3 vs 56.2 months, respectively; P<0.001; Figure 3F). Cox comparative hazards model revealed that the risk of death in patients with poor differentiation was 2.5 times higher than that of patients with well and moderated differentiation (P<0.001), and the risk of death in patients with stage IV was 2.3 times than that of patients with stage I+II+III (P=0.033). The risk of death in non-surgical patients was 5.2 times than that of surgical patients (P=0.001), and the risk of death in patients without receiving EGFR-TKI treatment was 7.0 times than that of patients receiving EGFR-TKI treatment (P<0.001) (Table 3).
Table 3.
Analysis of average survival time (months) in 234 cases of lung adenocarcinoma patients with EGFR mutation and 190 cases without EGFR mutation
| Prognostic factors | 234 cases with EGFR mutation | 190 cases without EGFR mutation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||||||
| Survival time (SE) | 95%CI | P | Exp (B) | 95%CI | P | Survival time (SE) | 95%CI | P | Exp (B) | 95%CI | P | |
| Age (years) | ||||||||||||
| ≤60 | 44.5 (2.58) | (39.4, 49.5) | 0.413 | 0.627 | (0.399, 0.986) | 0.043* | 29.0 (2.59) | (23.9, 34.1) | 0.043* | 0.620 | (0.423, 0.910) | 0.015* |
| >60 | 39.9 (3.05) | (33.9, 45.8) | 21.8 (2.57) | (16.8, 26.9) | ||||||||
| Gender | ||||||||||||
| Female | 41.8 (2.47) | (37.0, 46.7) | 0.001* | 1.173 | (0.521, 2.640) | 0.699 | 31.9 (3.71) | (24.7, 39.2) | 0.043* | 0.439 | (0.263, 0.734) | 0.002* |
| Male | 42.5 (3.22) | (36.2, 48.8) | 23.7 (2.17) | (19.4, 27.9) | ||||||||
| Smoking status | ||||||||||||
| Smoker | 37.0 (3.50) | (30.1, 43.8) | 0.656 | 1.393 | (0.592, 3.282) | 0.448 | 24.7 (2.59) | (19.6, 29.8) | 0.738 | 0.591 | (0.385, 0.905) | 0.016* |
| Nonsmoker | 42.9 (2.30) | (38.4, 47.4) | 26.7 (2.69) | (21.4, 32.0) | ||||||||
| Pathological degree | ||||||||||||
| Well and moderated differentiation | 46.4 (2.20) | (42.1, 50.7) | 0.000* | 0.397 | (0.243, 0.648) | 0.000* | 30.3 (2.43) | (25.6, 35.1) | 0.000* | 0.552 | (0.374, 0.813) | 0.003* |
| Poor differentiation | 21.3 (2.12) | (17.1, 25.4) | 17.0 (2.61) | (11.8, 22.1) | ||||||||
| Clinical stages | ||||||||||||
| I+II+III | 53.3 (2.55) | (48.3, 58.2) | 0.000* | 0.435 | (0.203, 0.934) | 0.033* | 39.3 (2.96) | (33.5, 45.1) | 0.000* | 0.398 | (0.244, 0.650) | 0.000* |
| IV | 27.9 (1.61) | (24.7, 31.0) | 16.0 (1.95) | (12.2, 19.8) | ||||||||
| EGFR-TKI treatment | ||||||||||||
| Yes | 51.0 (2.45) | (46.2, 55.8) | 0.000* | 0.143 | (0.088, 0.235) | 0.000* | 14.6 (4.16) | (6.5, 22.8) | 0.370 | 1.132 | (0.512, 2.501) | 0.759 |
| No | 30.7 (2.72) | (25.4, 36.0) | 26.6 (2.00) | (22.7, 30.5) | ||||||||
| Surgery treatment | ||||||||||||
| Yes | 56.2 (2.50) | (51.4, 61.1) | 0.000* | 0.194 | (0.074, 0.506) | 0.001* | 44.7 (3.52) | (37.8, 51.6) | 0.000* | 0.420 | (0.229, 0.770) | 0.005* |
| No | 27.3 (1.36) | (24.6, 30.0) | 18.6 (1.85) | (15.0, 22.3) | ||||||||
| Chemotherapy | ||||||||||||
| Yes | 40.1 (4.22) | (31.8, 48.3) | 0.939 | 1.580 | (0.875, 2.853) | 0.129 | 25.1 (2.56) | (20.1, 30.1) | 0.575 | 1.120 | (0.761, 1.648) | 0.565 |
| No | 42.6 (2.23) | (38.3, 47.0) | 27.0 (2.76) | (21.6, 32.4) | ||||||||
Note: *P<0.05.
Abbreviations: SE, standard error; 95% CI, 95% confidence interval.
Survival analysis for 190 cases of lung adenocarcinoma patients without EGFR gene mutation
Kaplan–Meier survival curves revealed that the survival time of patients without EGFR gene mutation was associated with surgery treatment (Figure 3G), age, gender, clinical stages, and pathological degree (P<0.05), which were considered as statistically significant difference, but there was no significance on EGFR-TKI treatment (Figure 3H), smoking status, and chemotherapy (P>0.05). Cox comparative hazards model revealed that age, gender, smoking status, pathological degree, clinical stages, and surgery treatment (P<0.05) were independent prognostic factors for lung adenocarcinoma patients without EGFR gene mutation, but no significance on chemotherapy and EGFR-TKI treatment (P>0.05). The risk of death in non-surgical patients was 2.4 times higher than that of surgical patients (P=0.005), the risk of death in patients with stage IV was 2.5 times than that of patients with stage I+II+III (P<0.001), and the risk of death in patients more than 60 years was 1.6 times than that of patients younger than 60 years (P=0.015). The risk of death among male, smoker, and poor differentiation patients was 2.3 times, 1.7 times, and 1.8 times than that of female, non-smoker, and well and moderated differentiation patients, respectively (P<0.05; Table 3).
Discussion
EGFR is a part of the ErbB family of transmembrane receptor tyrosine kinases, which overexpresses and/or mutates in some lung cancer. EGFR gene mutation has been reported as a predictive factor for EGFR-TKI treatment.5,15 In this study, the total positive percentage of EGFR gene mutation was 55.2% (234/424) in 424 cases of lung adenocarcinoma patients, including exon 18 (2.0%), exon 19 (48.0%), exon 20 (5.0%) and exon 21 (42.0%). We found that exon 19 deletion (Del19) and exon 21 Leu858Arg substitution (L858R) were the most common mutations subtypes, which was consistent with previous reports.16 In addition, 6 cases of double mutations were also observed, including double mutations of exon 18 and 21, double mutations of exon 19 and 20, and double mutations of exon 20 and 21, suggesting a co-mutation of EGFR gene in patients with lung adenocarcinoma. In the exon 20 mutation, we found 3 cases of T790M mutation, accounting for acquired resistance to EGFR-TKI treatment.17
In our study, the prevalence of EGFR gene mutation was higher in female, nonsmoker, and well and moderated differentiation, which was similar to previous reports from the East Asian countries.9,10,18 Furthermore, we also found that the rate of EGFR gene mutation was not associated with specimen types, and there were no significant differences in maximum diameter of tumor, lymph node metastasis, and pleural metastasis of surgical specimen. Of note, logistic regression analysis demonstrated that EGFR gene mutation was independently associated with gender and pathological degree, but not with smoking status, which needed to be further investigated in large samples. The most likely reason for the result was that smoking status was related to gender, which was considered as a confounding factor, and there was no uniform standard for assessing smoking status. In this study, some patients may accept second-hand smoke but were still identified as non-smokers. In further studies, we will pay more attention to the standard definition of smoking status. Our findings provided further evidence that EGFR gene mutation varied by clinicopathological features which showed some predictors for EGFR gene mutation. So far, the results of survival difference between Del19 and L858R subtypes were still inconsistent.19 In our study, the survival time of exon 18 group was statistically lower than that of exon 19 group and exon 21 group, but the survival time between exon 19 group and exon 21 group was not statistically different, suggesting there were no differences in the survival time of Del19 and L858R subtypes in the patients with lung adenocarcinoma, which needed to be further investigated.
Multivariate analysis showed that clinicopathological features, following as age, gender, clinical stages, pathological degree, and EGFR gene mutation were independent prognostic factors in the 424 cases of lung adenocarcinoma patients. Patients with EGFR gene mutation tended to have better survival in univariate and multivariate analysis. Smoking status was significant in univariate analysis, but not in multivariate analysis. These findings suggested that EGFR gene mutation, clinical stages, and pathological degree were more important prognostic factors than smoking status. Multivariate analysis identified that the following clinicopathological features, such as age, pathological degree, clinical stages, surgery treatment, and EGFR-TKI treatment, were significantly associated with survival time in patients with EGFR gene mutation. In multivariate analysis of patients without EGFR gene mutation, surgery treatment, younger patients, non-smoker female, early clinical stage, and well and moderated differentiation showed better survival, thus, suggesting both in patients with or without EGFR gene mutation could benefit from the early clinical stage, younger, well and moderated differentiation, and surgery treatment. The results of the present study also showed that EGFR-TKI treatment was a prognostic factor in patients with EGFR gene mutation rather than in patients without EGFR mutation, further indicating EGFR-TKI treatment has favorable efficacy in patients with EGFR mutation.8,20
In conclusion, clinicopathological features, such as gender, smoking status, and pathological degree of cancer were predictors of EGFR gene mutation. EGFR gene mutation had important prognostic and therapeutic implications in lung adenocarcinoma patients, and EGFR-TKI treatment was a more important prognostic factor in patients with EGFR gene mutation than in patients without EGFR gene mutation. Surgery treatment, age, clinical stages, and pathological degree were independent factors in Chinese lung adenocarcinoma patients no matter with or without EGFR gene mutation.
Acknowledgments
The work was supported by grants from The National Natural Science Foundation of China (No:81773218, 81703009, 81472773) and The Natural Science Foundations of Hunan Province (No: 2017JJ3457).
Ethical approval
The present study was approved by the Ethics Committee of The Second Xiangya Hospital of Central South University (Scientific and Research Ethics Committee, no. s02/2000).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Supplementary materials
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3(10):733–744. doi: 10.1038/nrc1190 [DOI] [PubMed] [Google Scholar]
- 3.Wogan GN, Hecht SS, Felton JS, Conney AH, Loeb LA. Environmental and chemical carcinogenesis. Semin Cancer Biol. 2004;14(6):473–486. doi: 10.1016/j.semcancer.2004.06.010 [DOI] [PubMed] [Google Scholar]
- 4.Thun MJ, Henley SJ, Calle EE. Tobacco use and cancer: an epidemiologic perspective for geneticists. Oncogene. 2002;21(48):7307–7325. doi: 10.1038/sj.onc.1205807 [DOI] [PubMed] [Google Scholar]
- 5.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088 [DOI] [PubMed] [Google Scholar]
- 6.Steuer CE, Ramalingam SS. Targeting EGFR in lung cancer: lessons learned and future perspectives. Mol Aspects Med. 2015;45:67–73. doi: 10.1016/j.mam.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Zhang M, Tang W, et al. Mutation abundance affects the therapeutic efficacy of EGFR-TKI in patients with advanced lung adenocarcinoma: a retrospective analysis. Cancer Biol Ther. 2018;19(8):687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938 [DOI] [PubMed] [Google Scholar]
- 9.Sun PL, Seol H, Lee HJ, et al. High incidence of EGFR mutations in Korean men smokers with no intratumoral heterogeneity of lung adenocarcinomas: correlation with histologic subtypes, EGFR/TTF-1 expressions, and clinical features. J Thorac Oncol. 2012;7(2):323–330. doi: 10.1097/JTO.0b013e3182381515 [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Zhang Y, Liu B, Hu M, Zhou N, Zhi X. Associations between epidermal growth factor receptor mutations and histological subtypes of lung adenocarcinoma according to the IASLC/ATS/ERS classification in Chinese patients. Thorac Cancer. 2017;8(6):600–605. doi: 10.1111/1759-7714.12489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ninomiya H, Hiramatsu M, Inamura K, et al. Correlation between morphology and EGFR mutations in lung adenocarcinomas significance of the micropapillary pattern and the hobnail cell type. Lung Cancer. 2009;63(2):235–240. doi: 10.1016/j.lungcan.2008.04.017 [DOI] [PubMed] [Google Scholar]
- 12.Clay TD, Russell PA, Do H, et al. Associations between the IASLC/ATS/ERS lung adenocarcinoma classification and EGFR and KRAS mutations. Pathology. 2016;48(1):17–24. doi: 10.1016/j.pathol.2015.11.002 [DOI] [PubMed] [Google Scholar]
- 13.Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. The Eighth edition lung cancer stage classification. Chest. 2017;151(1):193–203. doi: 10.1016/j.chest.2016.10.010 [DOI] [PubMed] [Google Scholar]
- 14.Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J Thorac Oncol. 2011;6(2):244–285. doi: 10.1097/JTO.0b013e318206a221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida T, Ishii G, Goto K, et al. Solid predominant histology predicts EGFR tyrosine kinase inhibitor response in patients with EGFR mutation-positive lung adenocarcinoma. J Cancer Res Clin Oncol. 2013;139(10):1691–1700. doi: 10.1007/s00432-013-1495-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Zhang Z, Bie Z, et al. Epidermal growth factor receptor mutation analysis in cytological specimens and responsiveness to gefitinib in advanced non-small cell lung cancer patients. Chin J Cancer Res. 2015;27:294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hata A, Katakami N, Yoshioka H, et al. Spatiotemporal T790M heterogeneity in individual patients with EGFR-Mutant non-small-cell lung cancer after acquired resistance to EGFR-TKI. J Thorac Oncol. 2015;10(11):1553–1559. doi: 10.1097/JTO.0000000000000647 [DOI] [PubMed] [Google Scholar]
- 18.Hsieh RK, Lim KH, Kuo HT, Tzen CY, Huang MJ. Female sex and bronchioloalveolar pathologic subtype predict EGFR mutations in non-small cell lung cancer. Chest. 2005;128(1):317–321. doi: 10.1378/chest.128.1.317 [DOI] [PubMed] [Google Scholar]
- 19.Zhuo M, Zheng Q, Zhao J, et al. Survival difference between EGFR Del19 and L858R mutant advanced non-small cell lung cancer patients receiving gefitinib: a propensity score matching analysis. Chin J Cancer Res. 2017;29(6):553–560. doi: 10.21147/j.issn.1000-9604.2017.06.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji080 [DOI] [PubMed] [Google Scholar]