Abstract
Background: Esophageal squamous cell carcinoma (ESCC) is the fourth most common cause of cancer death in China. Long noncoding RNAs have emerged as critical regulators in cancer. Long intergenic noncoding RNA-p21, a kind of Long noncoding RNAs, LincRNA-p21 have been discussed dysregulated in several cancers, but its role in ESCC remains unknown. This study investigated the role of LincRNA-p21 in ESCC.
Materials and methods: The LincRNA-p21 expression level and its association with esophageal cancer was determined in 64 tumor tissues of esophageal squamous cell carcinoma patients and cells using quantitative real-time reverse transcription PCR. Fluorescence in situ hybridization of single-RNA molecular probes was used to determine subcellular localization of LincRNA-p21. CCK8 and EdU assays were used for proliferation assay, flow cytometry was performed for apoptosis and cell-cycle distribution, and 24-well Mill cell chamber was made for measuring the abilities of migration and invasion after transfected with lentivirus-expressing LincRNA-p21 in EC109 cells. Then, quantitative real-time reverse transcription PCR and Western blot detected the expression of p21. Further, UC2288, an inhibitor of p21, was used to decrease the level of p21, and flow cytometry was used to detect cell cycle. Finally, screening for differential pathways from microarray analysis and expression of p53 and cyclin D were detected by Western blot.
Results: LincRNA-p21 expression level was remarkably lower in tumor tissues versus nontumor tissues and lower in EC109 cells versus Het-1A cells. Statistical analysis found that LincRNA-p21 might enhance the risk of ESCC. We observed that LincRNA-p21 was expressed both in the nucleus and cytoplasm, and a larger proportion of LincRNA-p21 was observed in the cytoplasm. The results demonstrated that upregulating the expression of LincRNA-p21 could inhibit cell proliferation, migration, invasion, and the transition of cell cycle from G1 and promoted apoptosis of EC109. Then, we found that LincRNA-p21 promotes the expression of p21. Decreasing the level of p21 revealed that cell-cycle arrest was restored. Pathway analysis found p53 pathway was downregulated, and upregulation of LincRNA-p21 inhibited the expression of cyclin D.
Conclusion: Our study suggests that LincRNA-p21 plays as a tumor inhibitor in ESCC development and LincRNA-p21 might induce G1 arrest through p53 signal pathway.
Keywords: esophageal squamous-cell carcinoma, LincRNA-p21, G1 arrest, p53 pathway, Fluorescence in Situ Hybridization
Background
In recent years, cancer morbidity and mortality have continued to show growth trend. The 2014 China Health Statistics Yearbook reported that cancer ranked first in the death causes in which esophageal cancer ranked fourth cause of death. . Esophageal squamous cell carcinoma (ESCC) is the predominant histological type of esophageal carcinoma in the “Asian esophageal cancer belt”,1 and there is the highest incidence of ESCC in Huai'an, China. ESCC, causing serious health disorders, whose incidence rate is 62.91/10 million people and mortality is 46.75/10 million people in Huai‘an area. There is lack of typical clinical symptoms in early ESCC. Besides, patients are in advanced ESCC because of poor compliance during treatment with imaging and cytology, and 5-year survival rate is <15%. Therefore, it is particularly important to find accurately sensitive and early tumor biomarkers for diagnosing ESCC.
Long noncoding RNAs (lncRNAs) are a group of RNA with more than 200 nucleoties in length that seldom involved in protein coding but regulating gene expression in the form of RNA over various levels including epigenetic regulation, derivation regulation and post-transcriptional control.2–5 An increasing number of evidence have revealed that lncRNA involved in magnanimous cellular processes ranging from embryonic stem cell pluripotency, cell apoptosis, cell-cycle regulation, and kinds of diseases, such as cancer.6,7 lncRNA has been discerned in esophageal cancer including MALAT-1 (Metastasis-Associated in Lung Adenocarcinoma Transcript 1),8 ESCCAL-1,9 UCA1,10 LncRNA FOXCUT,11 PlncRNA1,12 HOTAIR,13–15 POU3f3,16 HNF1A-AS1,17 ANRIL,18 and taurine-upregulated gene 1 (TUG1).12
LincRNA-p21 captured our attention because it was a transcription of the p53, which is also called TP53COR1 (tumor protein P53 pathway corepressor 1). LincRNA-p21 was originally discovered in mice where it was a transcript of about 3 kb located proximal to the p21/CDKN1a gene which encodes the cell-cycle regulator.19 Since our understanding of LincRNA-p21 in esophageal, cancer biology is still in infancy. In the present study, we aimed to explore the link with LincRNA-p21 and ESCC and lay the foundation for seeking molecular biomarkers of ESCC.
Materials and methods
Chemicals
Ambion® PARIS™ system (PARIS™ Instruction Manual, Thermo Fisher Scientific, Waltham, MA, USA), BCA Protein Assay Kit (Thermo Fisher Scientific), CCK-8 Cell Proliferation/Viability Assay Kit (Sigma-Aldrich, St. Louis, MO, USA) and EdU Apollo Imaging Kit (RiboBio, Guangzhou, China), Cell Cycle (Nanjing KeyGEN, China), keyFluor 647-Annexin V/7-AAD Apoptosis Detection Kit (Nanjing KeyGEN, China), dimethyl sulfoxide (DMSO) (Sigma-Aldrich, fetal bovine serum (Gibco, Waltham, MA, USA), LincRNA-p21-lentivirus and negative control (Shandong ViGene Biosciences, Shandong, China), Matrigel (BD, Franklin Lake, NJ, USA), nuclear/cytosol fractionation kit (Biovision, San Francisco Bay, CA, USA), PBS (Gibco), PrimeScript RT Master Mix Kit (Takara, Shiga, Japan), penicillin and streptomycin (Sigma-Aldrich), RNase-Free DNase I and Purification Kit (Tiangen, Beijing, China), RiboTM lncRNA FISH Probe Mix and RiboTM Fluorescent In Situ Hybridization Kit (RiboBio, Guangzhou, China), RPMI1640 (Hyclone, Logan, UT, USA), SYBR Premix Ex Taq II Kit (Takara, Kusatsu, Japan), TRIzol reagent (Invitrogen, USA), trypsin and streptomycin (Nanjing Sunshine, Nanjing, China) were used. All primers used for this study, including LincRNA-p21, p21, β-actin, and U6, were purchased from Nanjing GenScript Company (Nanjing, China).
Clinical samples and ethics statement
Sixty-four pairs of fresh-frozen tumor samples of ESCC tumors and adjacent normal tissues were collected from Thoracic Surgery of Jiangsu Huai’an First People’s Hospital. The patients in these cases were diagnosed as primary ESCC in 2010. The eligibility criteria for the enrolled ESCC patients were: diagnosed as ESCC by pathology or endoscopy without radiotherapy or chemotherapy, Han residents have been living in Huai’an for >20 years. The study was conducted according to protocols approved by the Southeast University Affiliated Zhongda Hospital Ethics Committee, and written informed consent that was conducted in accordance with the Declaration of Helsinki was obtained from the participants.
Cell culture and lentivirus transfection
The esophageal cancer cell line EC109 and human esophageal epithelial cell Het‐1A were purchased from Shanghai Tiancheng Technology Co., Ltd. The EC cell lines EC109 with wild-type p53 and a human esophageal epithelial cell line Het-1A were cultured in complete medium containing 100 U/mL each of penicillin and streptomycin and maintained at 37°C in a humidified atmosphere with 5% CO2. Lentivirus-expressing LincRNA-p21 and negative control were transfected into EC109 cells. First, cells were seeded in six-well plates, then transfected with 1×108 transducing units (TUs)/mL lentivirus (10 μL) and 5 μg/mL polybrene diluted by RPMI1640. Then, the medium was refreshed and incubation was continued for 24–96 hrs. Transfection efficiency was determined by a fluorescence microscope (FSX100 Oylmpus, Tokyo, Japan) and qRT-PCR. The infection efficiency needs to be >80% as monitored with GFP protein expression.
Total RNA extraction and qRT-PCR
Total RNA was extracted using TRIzol/chloroform according to the manufacturer’s instruction. After reverse transcription, amplification was carried out in a total volume of 20 μL containing SYBR Green real-time PCR Master Mix. The PCR reaction included an initial “hot start” at 95°C for 30 s, followed by 40 cycles of amplification. Each cycle consisted of a denaturation step at 95°C for 5 s, annealing, and fluorescence signal acquisition at 60°C for 30 s. Primer sequences used are described in Table 1. Transcription levels were normalized against β-actin. Ct (cycle threshold) values were used to calculate the relative LincRNA-p21 levels. ∆Ct = (CtLincRNA-p21− Ctβ-actin), while ∆∆Ct = (CtLincRNA-p21 in Tumor − Ctβ-actin in Tumor) − (CtLincRNA-p21 in Normal − Ctβ-actin inNormal). The fold change of LincRNA-p21 levels in ESCC is 2−∆∆Ct.
Table 1.
The primer sequences used in this study
| Gene Name | Sequence |
|---|---|
| LincRNA-p21* | 5ʹ-GGGTGGCTCACTCTTCTGGC-3ʹ (Forward) |
| 5ʹ-TGGCCTTGCCCGGGCTTGTC-3ʹ (Reverse) | |
| p21 | 5ʹ-TGGACCTGTCACTGTCTTGT-3ʹ (Forward) |
| 5ʹ-TCCTGTGGGCGGATTA-3ʹ (Reverse) | |
| β-actin | 5ʹ- CCACTGGCATCGTGATGGA-3ʹ (Forward) |
| 5ʹ- CGCTCGGTGAGGATCTTCAT-3ʹ (Reverse) | |
| U6 | 5ʹ-CTCGCTTCGGCAGCACA-3ʹ (Forward) |
| 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ (Reverse) |
Notes: *Data from Huarte19
Fluorescence in Situ Hybridization
To localize LincRNA-p21 in the EC109 cell, the FISH assay was performed using the lncRNA FISH Probe and Fluorescent in Situ Hybridization Kit according to the manufacturer’s guidelines. The probe cocktail included the U6/nuclear probe, 18S/cytoplasm probe, and the LincRNA-p21 probe (the nucleus is displayed through the DAPI,LincRNA-p21 gene appears as a red signal). Fluorescence signals were captured by the OLYMPUS laser confocal microscope FV1000 (Olympus, Tokyo, Japan).
RNA separation of nuclear and cytoplasmic fractions
The Ambion® PARIS™ system is for the isolation of RNA from nuclear and cytoplasmic of EC109 cells. Tissue or cultured cells are first homogenized in ice-cold cell disruption buffer to prepare a total cell lysate. Since the homogenization is performed quickly on ice and in the presence of detergent, both protein and RNA can be purified directly from this lysate. For RNA isolation, a part of the total cell lysate is immediately mixed with an equal volume of lysis/binding solution. Total RNA is then purified from the mixture using an RNA binding glass fiber filter. After three rapid washing steps, high-quality RNA is eluted in a concentrated form. Then, total RNA from nuclear and cytoplasm were used for qRT-PCR.
Cell proliferation assay
Cell proliferation assay was performed using the CCK-8 Cell Proliferation/Viability Assay Kit and EdU Apollo Imaging Kit as recommended in the manufacturer’s protocol.
Cell-cycle analysis
EC109 transfected with LincRNA-p21 and negative control were seeded in six-well plates and synchronized in serum-free medium for 24 hrs, then harvested and fixed in ice-cold 70% ethanol at 4°C overnight. The cells were washed with PBS and and then heated in 37°C water bath with 100 μL RNase A for 30 mins the next day. Finally, cells were stained with propidium iodide and analyzed by flow cytometry.
Apoptosis assay
LincRNA-p21-treated EC109 and negative control during logarithmic growth period were collected and detected using an Annexin V fluorescein isothiocyanate kit (APC/7-AAD) according to the protocol.
Migration and invasion assays
Cell invasion ability was measured by 24-well Mill cell chamber (corning 3422) containing a Matrigel-coated membrane, according to the protocol. After transfection, 5×105 EC109 cells in 100 μL RPMI1640 medium containing 1% fetal were seeded in the upper chamber. Then, 600 μL complete RPMI1640 medium was added to the lower chamber and incubatedfor at least 24 hrs, and noninvading cells were removed with a cotton swab and cells invading to the lower chamber were fixed with methanol, stained with 0.1% crystal violet, dried, and photographed. The number of invading cells was counted under the microscope (original magnification, ×200) in five representative fields and expressed as the average per field. The migration ability was performed in a similarity method without the Matrigel coating.
Assay for UC2288 inhibition
UC2288 were dissolved in 95% ethanol or DMSO and formulated into a 1 μmol/L stock solution, which was then diluted with RPMI1640 to the desired concentration. EC109 cells were seeded in six-well plates with complete medium after upregulating LincRNA-p21. Then, the stock solution was added to 0, 0.1, 1, 3, 10, and 30 nmol/L, respectively. Finally, cell activity, qRT, and WB were performed after incubation for 24 hrs.
Western blot
The cells were lysed in RIPA buffer that mixed with protease inhibitors after wash with PBS. Then, BCA Protein Assay Kit was used to quantify the protein concentrations. Then, a total of 20 μg protein was transferred to polyvinylidene fluoride (PVDF) membranes after separation by 10% SDS-PAGE. Next, the PVDF membrane was closed using 5% skimmed milk powder at room temperature for 2 hrs, and membranes were incubated with rabbit monoclonal anti-human β-actin (1:1,000), rabbit monoclonal anti-human p21 (1:1,000), rabbit monoclonal anti-human p53 (1:1,000), rabbit monoclonal anti-human Cyclin D (1:1,000), and rabbit monoclonal anti-human GAPDH (1:1,000) overnight at 4°C. The second day, after washing the membranes, peroxidase-conjugated goat anti-rabbit secondary antibody (1:5,000) was applied to the cells for 1 hr at room temperature. Finally, the signals were detected with the SuperSignal West Femto Trial Kit.
KEGG PATHWAY analysis
The possible pathway analysis of the identified differentially expressed proteins was elucidated by the KEGG PATHWAY database (http://www.genome.jp/kegg/pathway.html)
Statistical analysis
Data analysis was performed by SPSS 20.0. Each condition has three replicates, and data were described as the mean ± SD. Student’s t-test, paired t-test, and ANOVA were conducted to analyse quantitative variables. A Cox regression analysis was used to analyse the relationship between the risk of ESCC incidence and the level of LincRNA-p21. A P-value ≤0.05 was considered statistically significant.
Results
Demographic characteristics
Sixty-four cases of ESCC patients were finally included in the study. The average age of the patients was 60.47±7.47 years, ranging from 43 to 76 years. There were 13 of the 64 patients (20.30%) with lymph node metastasis (Table 2). As shown in Table 3, there is no significant difference in LincRNA-p21 expression according to age. There is no significant difference in LincRNA-p21 expression with clinical performance. However, the LincRNA-p21 expression levels were not significantly different with some risk factors such as smoking and drinking.
Table 2.
Clinical characteristics of 64 patients with ESCC
| Parameters | Class | Frequency | Proportion (%) |
|---|---|---|---|
| Age | <60 | 29 | 45.30 |
| >60 | 35 | 54.70 | |
| Sex | male | 41 | 64.10 |
| female | 23 | 35.90 | |
| Tea | never | 48 | 75.00 |
| occasionally | 8 | 12.50 | |
| Often | 8 | 12.50 | |
| Smoking | light | 25 | 39.10 |
| middle | 3 | 4.70 | |
| deep | 36 | 56.30 | |
| Drinking | yes | 20 | 31.20 |
| no | 44 | 68.80 | |
| Hot food | yes | 34 | 53.10 |
| no | 30 | 46.90 | |
| Hard food | yes | 22 | 34.40 |
| no | 42 | 65.60 | |
| Tumor site | upper | 11 | 17.20 |
| middle | 38 | 59.40 | |
| lower | 14 | 21.90 | |
| Lymphnode metastasis | yes | 13 | 20.30 |
| no | 51 | 79.70 |
Abbreviation: ESCC, esophageal squamous cell carcinoma.
Table 3.
The associations of LincRNA-p21 expression with demographic and clinical characteristics
| Parameters | Class | Expression pattern | P-value | ||
|---|---|---|---|---|---|
| Low expression | Normal | Over expression | |||
| Age | <60 | 17 | 8 | 4 | 0.561 |
| >60 | 17 | 8 | 10 | ||
| Tea | never | 27 | 11 | 10 | 0.745 |
| occasionally | 4 | 2 | 2 | ||
| often | 3 | 3 | 2 | ||
| Smoking | light | 17 | 5 | 3 | 0.265 |
| middle | 1 | 0 | 2 | ||
| deep | 16 | 11 | 9 | ||
| Drinking | no | 24 | 10 | 10 | 0.148 |
| yes | 10 | 6 | 4 | ||
| Hot food | no | 15 | 9 | 6 | 0.333 |
| yes | 19 | 7 | 8 | ||
| Hard food | no | 21 | 11 | 10 | 0.319 |
| yes | 13 | 5 | 4 | ||
| Tumor site | upper | 26 | 13 | 0 | 1.000 |
| middle | 8 | 2 | 3 | ||
| lower | 3 | 4 | 4 | ||
| Lymph node metastasis | no | 23 | 8 | 20 | 0.472 |
| yes | 9 | 4 | 0 | ||
LincRNA-p21 was down-regulating
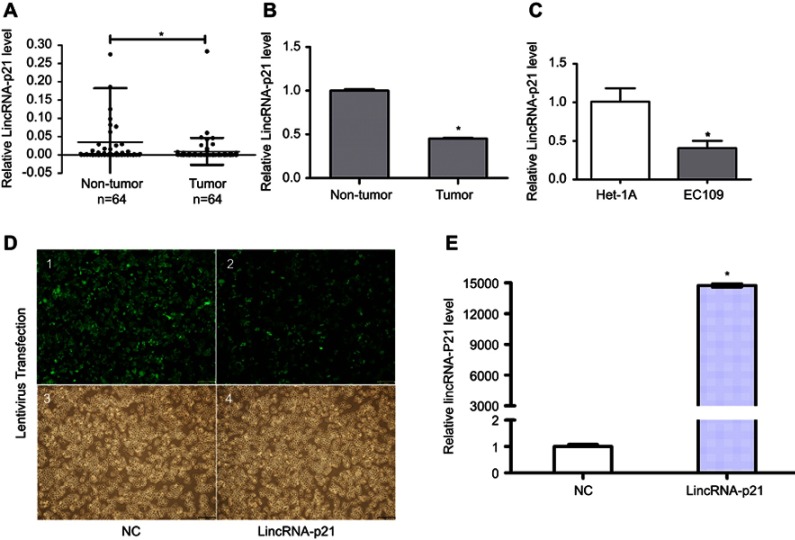
First, qRT-PCR was used to evaluate LincRNA-p21 expression levels in four esophageal cancer cell line EC109 and a normal esophageal epithelial cell line Het-1A as well as in 64 tumor tissues and paired normal tissues.
As shown in Table 4 and Figure 1, LincRNA-p21 levels in four EC cell lines were lower than those in Het-1A (Figure 1C, P<0.001). In Table 5 and Figure 1A and B, the expression level of LincRNA-p21 depressed significantly (P=0.005) in tumor tissues, as described in our previous study.20 To investigate the association between the expression of LincRNA-p21 and the risk of esophageal cancer, the LincRNA-p21 levels were analyzed by conditional logistic regression. As shown in Table 5, increased risks of developing esophageal cancer are connected with downregulation level of LincRNA-p21 (OR=3.76, P=0.014), suggesting that LincRNA-p21 may play a role as tumor suppressor gene in the development of esophageal carcinoma.
Table 4.
Relative LincRNA-p21 level in tested cells
| Group | N | ∆Ct(Mean ± SD) | Fold Change | P-value |
|---|---|---|---|---|
| Het-1A | 3 | 18.11±0.00 | 1.000 | |
| EC109 | 3 | 20.56±0.00 | 0.183 | <0.05 |
Abbreviation: CT, cycle threshold.
Figure 1.
Expression of LincRNA-p21. To evaluate the expression of LincRNA-p21 in ESCC tissues and cells, we carried out qRT-PCR test before and after application of lentivirus. Our experiments divulged that LincRNA-p21 was lower expressed in tumor tissues and EC109 cells (A–C). EC109 cells, after transfected with lentivirus, were captured by FSX100 (Olympus) at 200× magnification (D): 1 and 2, fluorescence; 3 and 4, bright field; 1 and 3, negative control; 2 and 4, virus-transfected with LincRNA-p21); LincRNA-p21 was significantly increased after transfected with LincRNA-p21-targeting lentivirus (E). Data indicate mean ± SD, n=3. *P<0.05.
Abbreviation: ESCC, esophageal squamous cell carcinoma.
Table 5.
Relative LincRNA-p21 level in tumor tissues and adjacent normal tissues
| Group | N | ∆Ct(Mean ± SD) | Fold Change | OR (95%CI) |
|---|---|---|---|---|
| Tumor | 64 | 13.54±2.94 | 0.451* | 3.76 (2.20, 9.18)# |
| Paired paracancer tissue | 64 | 12.39±3.37 | 1.000 |
Notes: *and #Significant difference compared to the Normal group (P<0.05).
Abbreviation: CT, cycle threshold.
Evaluation of transfection efficiency
qRT-PCR was used to detect the expression of LincRNA-p21 after transfection. After transfection, the expression of LincRNA-p21 was significantly higher than the control group (Figure 1D and E, P<0.05).
LincRNA-p21 localized in nucleus and cytoplasm of EC109
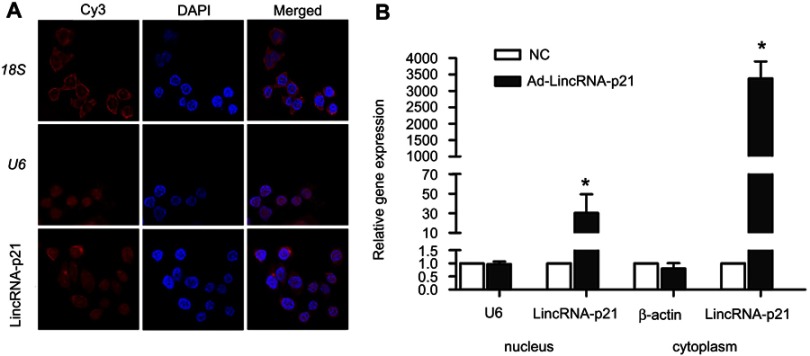
As shown in Figure 2, 18S as cytoplasmic control transcript, U6 as nuclear control transcript, and LincRNA-p21 were tagged by Cy3 (red), and the nuclei were stained with DAPI (blue). The result of fluorescence in situ hybridization revealed that LincRNA-p21 localized both in nucleus and in cytoplasm of EC109. Fluorescence intensity of LincRNA-p21 in the cytoplasm was stronger. qRT-PCR results revealed the expression of LincRNA-p21 mainly in the cytoplasm.
Figure 2.
Fluorescence in situ hybridization for LincRNA-p21 in EC109 cell. (A) Using Single Molecule lncRNA Fluorescent in Situ Hybridization (lncRNA-FISH) to fix a position on LincRNA-p21. As observed under a confocal microscope, 18S (as cytoplasmic control), U6 (as nuclear control) and LincRNA-p21 were tagged by Cy3 (red), the nuclei were stained with DAPI (blue). (B) Expression of LincRNA-p21 was detected by qRT-PCR after separation of nuclear and cytoplasmic fractions, using U6 as nuclear control transcript and actin as cytoplasmic control transcript. Data indicate mean ± SD, n=3. *P<0.05.
Function of LincRNA-p21 in EC109
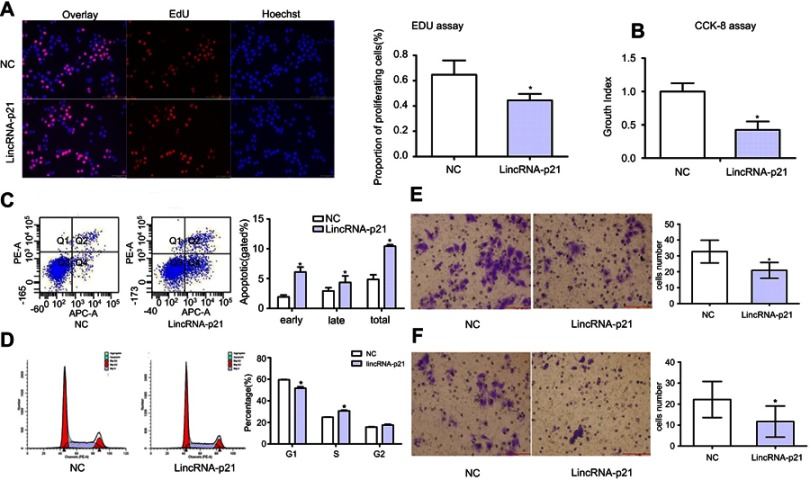
LincRNA-p21 inhibited proliferation and promotes apoptosis of EC109 As shown in Figure 3A, cells labeled in red after reaction of EdU (5-ethynyl-2′-deoxyuridine) and Apollo represent proliferative cells and cell nuclei stained with Hoechst 33342 (blue) represent the total cells. Cells were captured by FSX100 (Olympus) at 200× magnification; the percentage of proliferating cells was significantly decreased compared with the control group (*P<0.05). CCK8 cell proliferation assays also revealed that cell proliferation was inhibited (Figure 3B, *P<0.05). The result of cell apoptosis assays suggested that upregulation of LincRNA-p21 promoted apoptosis in EC109 cells (Figure 3C, *P<0.05).
Figure 3.
Function of LincRNA-p21 in EC109 cells. The cell proliferation activity was evaluated using EdU-incorporation assays and CCk-8 assay. Cells labeled in red after reaction of EdU (5-ethynyl-2′-deoxyuridine) and Apollo represent proliferative cells., cell nuclei stained with Hoechst 33,342 (blue) represent the total cells (FSX100, 200× magnification), LincRNA-p21 reduced activity of EC109 (A and B). Apoptosis rate and each phase of cell cycle were analyzed by flow cytometry (C and D). (E) The metastatic and infiltrated potential of EC09 cells was evaluated using the migration and invasion assay in vitro. The results indicated a significant decline in cells to pass through the Matrigel membrane after LincRNA-p21-knockin relative to the untreated control. Data indicate mean ± SD, n=3. *P<0.05.
LincRNA-p21 redistributes cell cycle of EC109
Flow cytometry was used to detect the number of cells in each phase of the cell cycle. The result shows that LincRNA-p21 induced G1/S cell-cycle arrest in EC109 cells. The number of cells in G1 phase was significantly increased from 51.76% to 59.63%, while cells of S and G2/M phase decreased from 30.66% to 24.79% and 17.57% to 15.56%, respectively (Figure 3D, *P<0.05).
LincRNA-p21 prohibit migration and invasion of EC109
Transwell chamber assay was employed to assess migration and invasion ability of LincRNA-p21 on EC109 cells. With LincRNA-p21 level up-regulated, the ability of EC109 cells to pass through the Matrigel membrane was weakened. The results showed that the migration and invasion ability of EC109 cells was significantly weakened than negative control group (Figure 3E and F, *P<0.05).
The effect of LincRNA-p21 on expression of p21
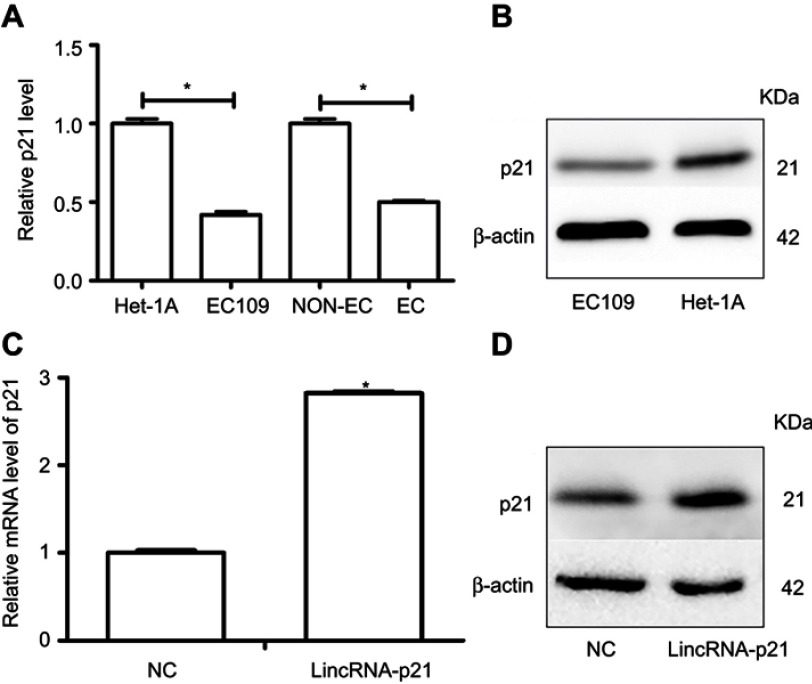
In our previous studies, we have found that mRNA of p21 downregulated both in EC cells and ESCC tissues.20 Moreover, protein of p21 was lower in EC109 cells than Het-1A cells. However, expression of p21 was upregulated both in mRNA and protein level after overexpression of LincRNA-p21 (Figure 4, Table 6).
Figure 4.
Expression of p21. Relative mRNA level of p21 was determined using qRT-PCR and protein expression of p21 was determined using Western blot. We found p21 decreased in ESCC tissues and cells (A and B). mRNA of p21 and protein of p21 were up-regulated after transfected with LincRNA-p21-targeting lentivirus (C and D). Data indicate mean ± SD, n=3. *P<0.05.
Table 6.
Seventeen significant pathways from KEGG enrichment analysis
| ID | Definition | Pathway name | Genea | DEGb | P-value |
|---|---|---|---|---|---|
| hsa05322 | Systemic lupus erythematosus | Autoimmune disease | 138 | 14 | 0.000 |
| hsa05330 | Allograft rejection | Alloimmune response | 39 | 7 | 0.000 |
| hsa04940 | Type I diabetes mellitus | Autoimmune disease | 45 | 7 | 0.000 |
| hsa04612 | Antigen processing and presentation | Immune system | 78 | 9 | 0.000 |
| hsa05332 | Graft-versus-host disease | Immune system | 43 | 6 | 0.001 |
| hsa05320 | Autoimmune thyroid disease | Autoimmune disease | 54 | 6 | 0.004 |
| hsa04650 | Natural killer cell mediated cytotoxicity | Immune system | 137 | 10 | 0.006 |
| hsa04060 | Cytokine-cytokine receptor interaction | Inflammatory defenses | 265 | 15 | 0.009 |
| hsa03050 | Proteasome | Immune responses, Stress, signaling, inflammatory | 45 | 5 | 0.009 |
| hsa04512 | ECM-receptor interaction | Tissue and organ morphogenesis | 85 | 7 | 0.011 |
| hsa04380 | Osteoclast differentiation | Immune receptors | 128 | 9 | 0.011 |
| hsa05416 | Viral myocarditis | Cardiac disease | 72 | 6 | 0.017 |
| hsa05166 | HTLV-I infection | Virus infection | 267 | 14 | 0.020 |
| hsa04145 | Phagosome | Inflammation and infectious | 155 | 9 | 0.033 |
| hsa03440 | Homologous recombination | Repair of DNA | 28 | 3 | 0.045 |
| hsa04115 | p53 signaling pathway | Signal transduction | 69 | 22 | 0.047 |
| hsa05323 | Rheumatoid arthritis | Alloimmune response | 92 | 6 | 0.048 |
Notes: aThe number of genes in the pathway. bThe number of genes differentially expressed.
LincRNA-p21 impact on EC109 cells with inhibition of p21
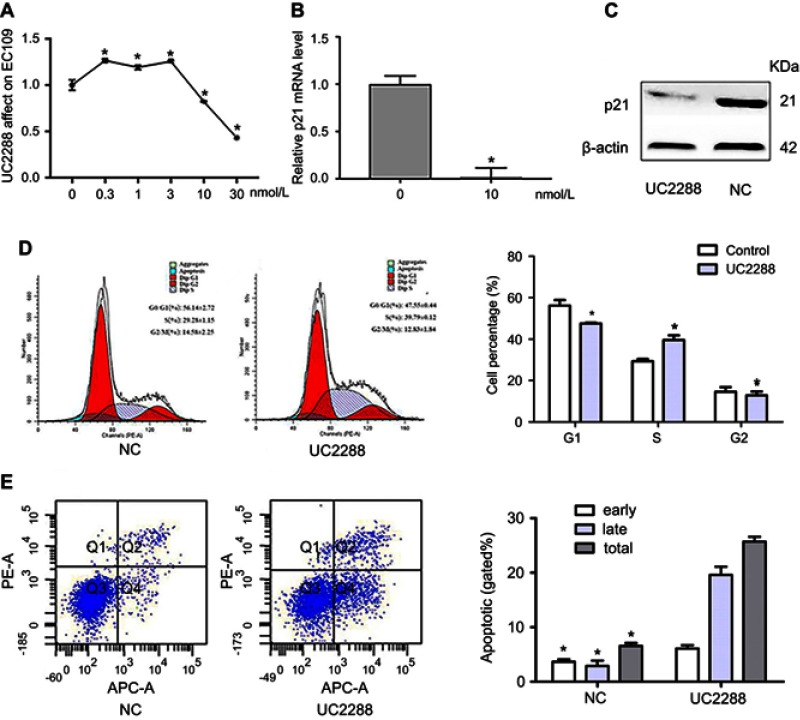
To get rid of influence of p21, we used a kind of p21 attenuator, named UC2288, which could decrease p21 mRNA expression and attenuated p21 protein levels as shown in Figure 5A–C. Then, we detected the cell apoptosis and periodic distribution in EC109 cells overexpressing LincRNA-p21. The apoptosis of EC109 cells was still high. But the periodic distribution is completely different. As shown in Figure 5, the cells of G1 phase were decreased (from 56.14% to 47.55%, P<0.01); the proportion of S phase cells were elevated (from 29.28% to 39.63%, P<0.01); the cells of G2 phase were decreased with no statistically significant. (from 14.58% to 12.83%).
Figure 5.
LinRNA-p21 affected on EC109 cell after using UC2288. (A) EC50 of UC2288, determined by cell activity, was 10 noml/L; (B and C) The result of qRT-PCR and Western blot showed that mRMA and protein of p21 were decreased; (D and E) Flow cytometry assays was performed to tested cell cycle and apoptosis, G1/S arrest was recovered with p21 decreased. (C–E) The concentration of UC2288 is 10 noml/L. Data indicate mean ± SD, n=3. *P<0.05.
LinRNA-p21 result in G1 arrest via p53 signaling in EC109 cells
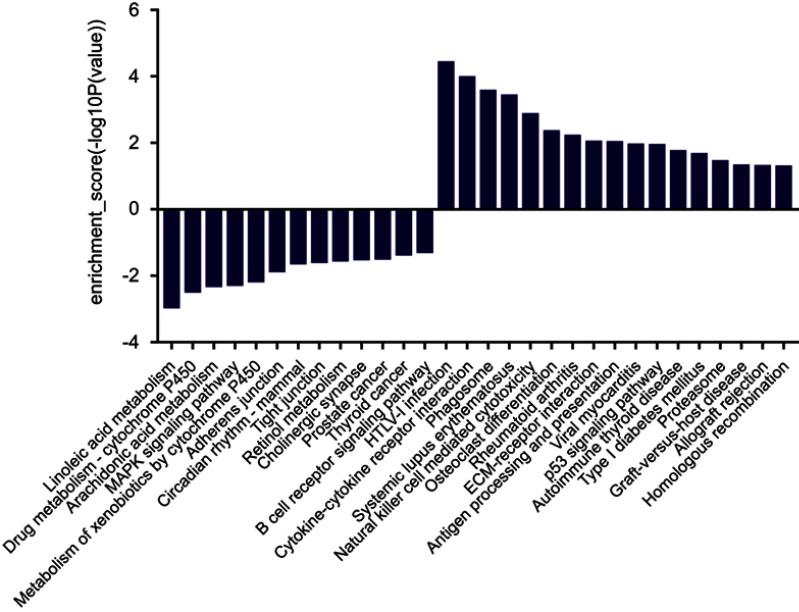
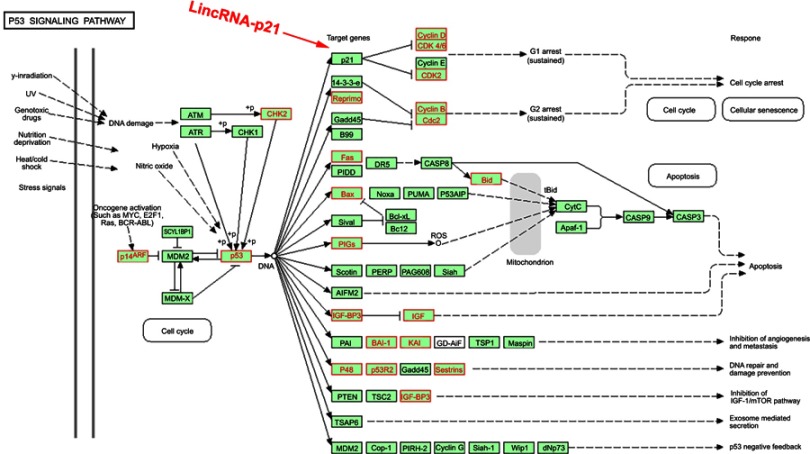
We had found that there were 17 upregulated and 13 downregulated pathways involved in differentially expressed mRNAs which may be related to the occurrence and development of ESCC.21 Upregulation pathways include protease pathways, homologous recombination pathways, and p53 signaling pathways. Downregulation pathways include MAPK signaling pathway, prostate cancer pathway, etc. (Figure 6). We concentrated on the P53 pathway is involved closely in ECSS. Particularly, p14ARF, p53, CHK2, Reprimo, cyclin D, CDK4/6, CDK2, Cyclin B, Cdc2, Fas, Bid, Bax, PIGs, IGF, BAI-1, KAI, P48, p53R2, Sestnns, and IGF-BP3 were enriched in p53 pathway (Figure 7).
Figure 6.
The histogram of KEGG pathway enrichment. The results of pathway analysis showed that there were 17 possible pathways involved in up-regulated differentially expressed mRNAs, and 13 possible pathways involved in down-regulated differentially expressed mRNAs (P<0.05).
Figure 7.
Differentially expressed genes in p53 signaling pathway. Red rectangles mean differentially expressed genes.
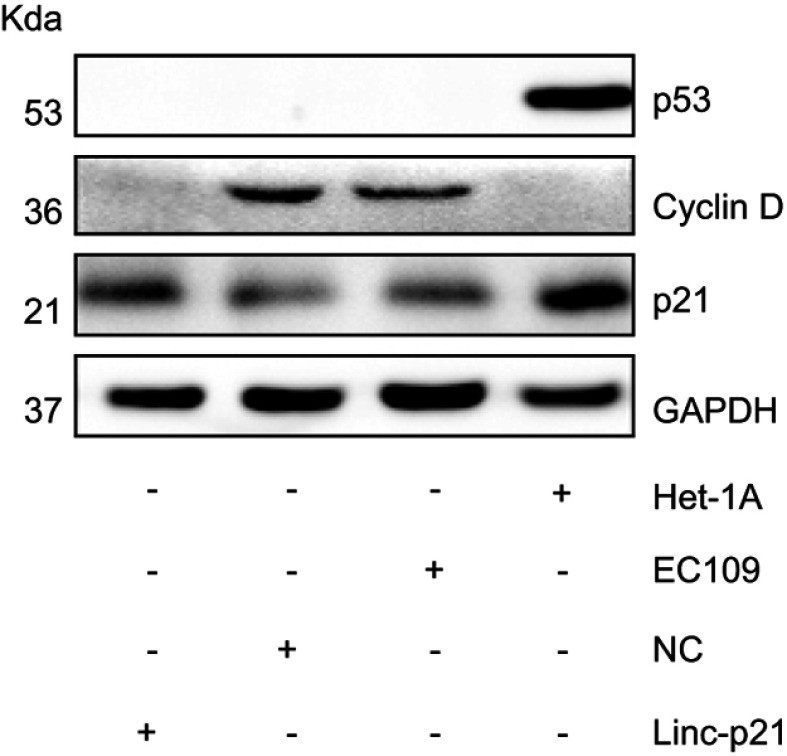
Expression proteins were then examined by Western blot. Our result showed that p21 and p53 were significantly decreased in EC109 compared to Het-1A , while the Cyclin D was significantly increased in EC109. Upexpressing LincRNA-p21 increased the expression of p21, reduced the expression of cyclin D, and had no effect on p53 protein (Figure 8). P21 and cyclin D regulate the cell cycle as the target genes for p53 signaling pathway. These results demonstrated that LincRNA-p21 was a corepressor of the p53 signaling pathway in EC109 cells.
Figure 8.
LincRNA-p21 affected on the expression of p53 and its downstream genes. Western blotting revealed a significant subsiding in Cyclin D protein expression and increasing in p21 protein expression in EC09 cells transfected with LincRNA-p21-targeting lentivirus.
Discussion
LincRNA-p21 is a regulatory factor of cell proliferation, apoptosis, and DNA damage response, which is stimulated by the p53 tumor suppressor protein.19,22,23 LincRNA-p21 is implicated in the development and progression of human diseases, particularly in cancer, which has been reported to be associated with hepatocellular carcinoma,24 colorectal cancer (CRC),25,26 non-small cell lung cancer,27,28 skin tumors, chronic lymphocytic leukemia (CLL),29 and prostate cancer.30 It has been reported that LincRNA-p21 is emerging as either oncogenes or tumor suppressor genes. For example, the expression levels of LincRNA-p21 are obviously reduced in CRC tissues and cell lines.31 Yang Ning32 also reported LincRNA-p21 is downregulated in human hepatocellular carcinoma. In some other diseases, such as atherosclerosis,33 acute respiratory distress syndrome,34 and rheumatoid arthritis (RA),35 the level of LincRNA-p21 was also decreased. Above these evidence imply LincRNA-p21 may play a role as tumor inhibitor. On the contrary, stabilization of HIF-1α by LincRNA-p21 under hypoxia conditions implies that it could also play an oncogenic role.36 In this work, we confirmed that LincRNA-p21 level was significantly downregulated in esophageal cancer cell line and tissues compared with normal tissue and cells, which was consistent with numerous previous studies performed.
Many papers mentioned that LincRNA-p21 could be a good biomarker related to human diseases.37 Wang37 provided evidence that LincRNA-p21 suppressed the tumorigenicity of cancer stem cells (CSCs) and may be potential therapeutic genes against CSCs. What’s more, in the research of Isin et al,29 it might be helpful to improve the diagnostic prediction of the malignant state for patients with prostate cancer by discriminating potential of exosomal LincRNA-p21 levels. In our study, we find LincRNA-p21 could be a risk factor of ESCC. Therefore, LincRNA-p21 levels may be an auxiliary diagnostic factor for ESCC.
Single-molecule RNA FISH was executed to observe the location of LincRNA-p21, as we can see in Figure 2, LincRNA-p21 localized in both nucleus and cytoplasm, mainly in the cytoplasm, which is consistent with Yoon et al’s description.22
The biological function of LincRNA-p21 in many diseases has been proposed, particularly in cancers. These roles are mediated by the epigenetic regulation, and transcriptional and posttranscriptional regulation.38–40 Some lncRNA have been identified in ESCC, such as MALAT1, UCA1, Linc00152, CFLAR-AS1, and POU3F3.41–43 However, no data are available between LincRNA-p21 and ESCC so far.
There is discrepancy with previous functional studies, which have proposed LincRNA-p21 is involved in metastasis and invasion,44 metabolism and cell reprogramming,40 cell activity,45 and Warburg effect.36 Huarte et al19 revealed a role for LincRNA-p21 in a p53-dependent apoptotic response following DNA damage, and transcriptional repression by LincRNA-p21 is mediated through the physical association with hnRNP-K, which is required for proper genomic localization of hnRNP-K at repressed genes and regulation of p53-mediated apoptosis. We confirmed that the upregulating LincRNA-p21 inhibits the ability of proliferation, migration, and invasion in EC109 cell; promotes apoptosis; and contributes G1/S arrest.
To explore the role of LincRNA-p21 regulating cell-cycle block in ESCC, we had gotten inspiration from microarray results and KEGG pathway analysis which p53 signaling pathway plays a lot (Figures 6 and 7). p53 pathway is the classical cancer pathway which mediates cellular stress responses including DNA repair, cell-cycle arrest, and apoptosis.46 As shown in our paper (Figure 6), p21 and cyclin D may be essential downstream effectors of p53 pathway in the regulation of a cell process, leading to growth arrest especially in G1 arrest, which had been reported by Suk et al.47 p21 can be activated to come into play by either p53-independent48 or p53-dependent49 manner. In our study, overexpressing LincRNA-p21 increased the level of p21 while had no effect on p53, which implies that LincRNA-p21 could influence EC109 cell proliferation and cycle via enhancing p21 activity in a p53-independent manner.
However, our research was still not perfect, and we did not observe the effect of LincRNA-p21 on EC109 cells after LincRNA-p21 knockdown in LV-EC109 cells. Hence, there is a lot of work to be completed. Of course, the current work is very valuable.
To conclude, our current study validated that LincRNA-p21 expression levels were decreased in human ESCC tissues compared with adjacent normal tissues. Further, our experiments illustrated that upregulation of LincRNA-p21 led to the inhibition of ESCC functions in cell proliferation, apoptosis, cycle, migration, and invasion, suggesting that LincRNA-p21 is an important biomarker and treatment target for ESCC. In addition, LincRNA-p21 impedes the progression of EC09 cells by promoting expression of p21 and inhibiting expression of cyclin D through p53-independent pathway. However, more sufficient evidence need to be supplemented.
Conclusion
In summary, the study demonstrated that aberrantly downregulated LincRNA-p21 increases the risk of ESCC progressing. Acting as a tumor suppressor, LincRNA-p21 inhibited cell functions such as proliferation, migration, invasion and disturbing cell cycle. LincRNA-p21 plays as a tumor inhibitor in ESCC development and LincRNA-p21 might induce G1 arrest through p53 signal pathway.
Acknowledgment
This study was supported by National Natural Science Foundation of China (Nos. 81573191, 81872588, 81573108).
Abbreviation list
ESCC, esophageal squamous cell carcinoma; LincRNA-p21, large intergenic noncoding RNA-p21; qRT-PCR, quantitative real-time reverse transcription PCR; CDK, cyclin-dependent kinases; DMSO, dimethyl sulfoxide; FBS, fetal bovine serum.
Ethics approval and consent to participate
The population study was approved by the institutional review board (IRB) of the Zhongda Hospital of Southeast University, and written informed consent that conducted in accordance with the Declaration of Helsinki was obtained from the participants.
Availability of data and materials
Data and materials will be shared upon request.
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Shang L, Liu HJ, Hao JJ, et al. A panel of overexpressed proteins for prognosis in esophageal squamous cell carcinoma. PLoS One. 2014;9(10):e111045. doi: 10.1371/journal.pone.0111045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 3.Caley DP, Pink RC, Trujillano D, Carter DRF. Long noncoding RNAs, chromatin, and development. Sci World Jo. 2010;10:90–102. doi: 10.1100/tsw.2010.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):U1071–U1148. doi: 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of Histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang XY, Li M, Wang ZQ, et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290(7):3925–3935. doi: 10.1074/jbc.M114.596866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928 [DOI] [PubMed] [Google Scholar]
- 9.Hao YB, Wu W, Shi FC, et al. Prediction of long noncoding RNA functions with co-expression network in esophageal squamous cell carcinoma. BMC Cancer. 2015;15:ARTN 168. doi: 10.1186/s12885-015-1584-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Patho. 2014;7(11):7938–7944. [PMC free article] [PubMed] [Google Scholar]
- 11.Pan F, Yao J, Chen Y, et al. A novel long non-coding RNA FOXCUT and mRNA FOXC1 pair promote progression and predict poor prognosis in esophageal squamous cell carcinoma. Int J Clin Exp Patho. 2014;7(6):2838–2849. [PMC free article] [PubMed] [Google Scholar]
- 12.Wang CM, Wu QQ, Li SQ, et al. Upregulation of the long non-coding RNA PlncRNA-1 promotes esophageal squamous carcinoma cell proliferation and correlates with advanced clinical stage. Digest Dis Sci. 2014;59(3):591–597. doi: 10.1007/s10620-013-2956-7 [DOI] [PubMed] [Google Scholar]
- 13.Li X, Wu Z, Mei Q, et al. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Brit J Cancer. 2013;109(8):2266–2278. doi: 10.1038/bjc.2013.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lv XB, Lian GY, Wang HR, Song EW, Yao HR, Wang MH. Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS One. 2013;8(5):ARTN e63516. doi: 10.1371/journal.pone.0063516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen FJ, Sun M, Li SQ, et al. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinogen. 2013;52(11):908–915. doi: 10.1002/mc.21944 [DOI] [PubMed] [Google Scholar]
- 16.Li W, Zheng J, Deng JQ, et al. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146(7):1714–1726. doi: 10.1053/j.gastro.2014.03.002 [DOI] [PubMed] [Google Scholar]
- 17.Yang X, Song JH, Cheng YL, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63(6):881–890. doi: 10.1136/gutjnl-2013-305266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen DY, Zhang ZY, Mao CM, et al. ANRIL inhibits p15(INK4b) through the TGF beta 1 signaling pathway in human esophageal squamous cell carcinoma. Cell Immunol. 2014;289(1–2):91–96. doi: 10.1016/j.cellimm.2014.03.015 [DOI] [PubMed] [Google Scholar]
- 19.Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ying Z, Yue M, Yuhong Z, Ran L, Yuepu P, Lihong Y. Mechanism of inhibition of esophageal cancer cell proliferation by lincRNA-p21. Environ Occup Med. 2018;35(06):479–484. [Google Scholar]
- 21.Xiajun L, Miao Y, Zicheng D, Ran L, Lihong Y, Yuepu P. Preliminary study on the risk of lncRNA-UCA1 and huaian esophageal cancer. Cancer Aberration Mutat. 2014;26(06):407–411. [Google Scholar]
- 22.Yoon JH, Abdelmohsen K, Srikantan S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648–655. doi: 10.1016/j.molcel.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimitrova N, Zamudio JR, Jong RM, et al. LincRNA-p21 activates p21 in cis to promote polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54(5):777–790. doi: 10.1016/j.molcel.2014.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia M, Jiang L, Wang YD, Huang JZ, Yu M, Xue HZ. LincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through Notch signaling-induced epithelial-mesenchymal transition. Hepatol Res. 2016;46(11):1137–1144. doi: 10.1111/hepr.12659 [DOI] [PubMed] [Google Scholar]
- 25.Zhai HY, Fesler A, Schee K, Fodstad O, Flatmark K, Ju JF. Clinical significance of long intergenic noncoding RNA-p21 in colorectal cancer. Clin Colorectal Canc. 2013;12(4):261–266. doi: 10.1016/j.clcc.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 26.Wang GY, Li ZW, Zhao Q, et al. LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/beta-catenin signaling pathway. Oncol Rep. 2014;31(4):1839–1845. doi: 10.3892/or.2014.3047 [DOI] [PubMed] [Google Scholar]
- 27.Castellano JJ, Navarro A, Vinolas N, et al. LincRNA-p21 impacts prognosis in resected non-small cell lung cancer patients through angiogenesis regulation. J Thorac Oncol. 2016;11(12):2173–2182. doi: 10.1016/j.jtho.2016.07.015 [DOI] [PubMed] [Google Scholar]
- 28.Hall JR, Messenger ZJ, Tam HW, Phillips SL, Recio L, Smart RC. Long noncoding RNA lincRNA-p21 is the major mediator of UVB-induced and p53-dependent apoptosis in keratinocytes. Cell Death Dis. 2015;6:ARTN e1700. doi: 10.1038/cddis.2015.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isin M, Ozgur E, Cetin G, et al. Investigation of circulating lncRNAs in B-cell neoplasms. Clin Chim Acta. 2014;431:255–259. doi: 10.1016/j.cca.2014.02.010 [DOI] [PubMed] [Google Scholar]
- 30.Ozkan TA. Re: exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. J Urol Surg. 2015;2(4):201. doi: 10.4274/jus.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li XY, Pu JX, Jiang SY, et al. Henryin, an ent-kaurane diterpenoid, inhibits wnt signaling through interference with beta-catenin/TCF4 interaction in colorectal cancer cells. PLoS One. 2013;8(7):ARTN e68525. doi: 10.1371/journal.pone.0068525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang N, Fu Y, Zhang HB, Sima H, Zhu N, Yang GS. LincRNA-p21 activates endoplasmic reticulum stress and inhibits hepatocellular carcinoma. Oncotarget. 2015;6(29):28151–28163. doi: 10.18632/oncotarget.4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu GZ, Cai J, Han Y, et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130(17):1452–U1464. doi: 10.1161/CIRCULATIONAHA.114.011675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou WQ, Wang P, Shao QP, Wang J. Lipopolysaccharide promotes pulmonary fibrosis in acute respiratory distress syndrome (ARDS) via lincRNA-p21 induced inhibition of Thy-1 expression. Mol Cell Biochem. 2016;419(1–2):19–28. doi: 10.1007/s11010-016-2745-7 [DOI] [PubMed] [Google Scholar]
- 35.Spurlock CF 3rd, Tossberg JT, Matlock BK, Olsen NJ, Aune TM. Methotrexate inhibits NF-kappaB activity via long intergenic (noncoding) RNA-p21 induction. Arthritis Rheumatol. 2014;66(11):2947–2957. doi: 10.1002/art.38805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang F, Zhang HF, Mei YD, Wu M. Reciprocal regulation of HIF-1 alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53(1):88–100. doi: 10.1016/j.molcel.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Lei ZJ, Guo Y, et al. miRNA-regulated delivery of lincRNA-p21 suppresses beta-catenin signaling and tumorigenicity of colorectal cancer stem cells. Oncotarget. 2015;6(35):37852–37870. doi: 10.18632/oncotarget.5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Işın, M, Uysaler E, Özgür, E, et al. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front Genet. 2015;6. doi: 10.3389/fgene.2015.00360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Recio L, Phillips SL, Maynor T, Waters M, Jackson AF, Yauk CL. Differential expression of long noncoding RNAs in the livers of female B6C3F1 mice exposed to the carcinogen furan. Toxicol Sci. 2013;135(2):369–379. doi: 10.1093/toxsci/kft153 [DOI] [PubMed] [Google Scholar]
- 40.Chou SD, Murshid A, Eguchi T, Gong J, Calderwood SK. HSF1 regulation of beta-catenin in mammary cancer cells through control of HuR/elavL1 expression. Oncogene. 2015;34(17):2178–2188. doi: 10.1038/onc.2014.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bao XC, Wu HT, Zhu XH, et al. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 2015;25(1):80–92. doi: 10.1038/cr.2014.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li RQ, Ren YL, Liu WJ, Pan WT, Xu FJ, Yang M. MicroRNA-mediated silence of onco-lncRNA MALAT1 in different ESCC cells via ligand-functionalized hydroxyl-rich nanovectors. Nanoscale. 2017;9(7):2521–2530. doi: 10.1039/c6nr09668a [DOI] [PubMed] [Google Scholar]
- 43.Wang XH, Gao ZK, Liao J, et al. lncRNA UCA1 inhibits esophageal squamous-cell carcinoma growth by regulating the Wnt signaling pathway. J Toxicol Env Heal A. 2016;79(9–10):407–418. doi: 10.1080/15287394.2016.1176617 [DOI] [PubMed] [Google Scholar]
- 44.Hu HB, Jie HY, Zheng XX. Three circulating LncRNA predict early progress of esophageal squamous cell carcinoma. Cell Physiol Biochem. 2016;40(1–2):117–125. doi: 10.1159/000452529 [DOI] [PubMed] [Google Scholar]
- 45.Xia WZ, Zhuang L, Hou M. Role of lincRNA-p21 in the protective effect of macrophage inhibition factor against hypoxia/serum deprivation-induced apoptosis in mesenchymal stem cells. Int J Mol Med. 2018;42(4):2175–2184. doi: 10.3892/ijmm.2018.3767 [DOI] [PubMed] [Google Scholar]
- 46.Ye YY, He XZ, Lu FF, et al. A lincRNA-p21/miR-181 family feedback loop regulates microglial activation during systemic LPS-and MPTP-induced neuroinflammation. Cell Death Dis. 2018;9:ARTN 803. doi: 10.1038/s41419-018-1111-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7:979. doi: 10.1038/nrd2656 [DOI] [PubMed] [Google Scholar]
- 48.Suk FM, Chang CC, Lin RJ, et al. ZFP36L1 and ZFP36L2 inhibit cell proliferation in a cyclin D-dependent and p53-independent manner. Sci Rep-Uk. 2018;8:ARTN 2742. doi: 10.1038/s41598-018-21160-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Yan B. Cell cycle regulation by carboxylated multiwalled carbon nanotubes through p53-independent induction of p21 under the control of the BMP signaling pathway. Chem Res Toxicol. 2012;25(6):1212–1221. doi: 10.1021/tx300059m [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data and materials will be shared upon request.