Abstract
Grape-derived products contain a wide array of bioactive phenolic compounds which are of significant interest to consumers and researchers for their multiple health benefits. The majority of bioavailable grape polyphenols, including the most abundant flavan-3-ols,i.e.(+)-catechin and (−)-epicatechin, undergo extensive microbial metabolism in the gut, forming metabolites that can be highly bioavailable and bioactive. To gain a better understanding in microbial metabolism of grape polyphenols and to identify bioactive metabolites, advanced analytical methods are needed to accurately quantitate microbial-derived metabolites, particularly at trace levels, in addition to their precursors. This work describes the development and validation of a high-throughput, sensitive and reproducible GC-QqQ/MS method operated under MRM mode that allowed the identification and quantification of 16 phenolic acid metabolites, along with (+)-catechin and (−)-epicatechin, in flavanol-enriched broth samples anaerobically fermented with human intestinal bacteria. Excellent sensitivity was achieved with low limits of detection and low limits of quantification in the range of 0.24–6.18 ng/mL and 0.480–12.37 ng/mL, respectively. With the exception of hippuric acid, recoveries of most analytes were greater than 85%. The percent accuracies for almost all analytes were within ±23% and precision results were all below 18%. Application of the developed method toin vitrosamples fermented with different human gut microbiota revealed distinct variations in the extent of flavanol catabolism, as well as production of bioactive phenolic acid metabolites. These results support that intestinal microbiota have a significant impact on the production of flavanol metabolites. The successful application of the established method demonstrates its applicability and robustness for analysis of grape flavanols and their microbial metabolites in biological samples.
Keywords: GC-QqQ/MS, Grape polyphenol, Phenolic acid metabolite, Gut microbiota in vitro fermentation
1. Introduction
Accumulating evidence demonstrates that the polyphenols abundant in grape-derived products have great potential in preventing or attenuating a number of chronic diseases including neurodegenerative diseases, cardiovascular diseases, inflammatory diseases and cancers [1–4]. Due to their multiple health benefits, grape polyphenol-rich foods and nutraceuticals are increasingly popular worldwide [5] and as such, there is an imperative need in understanding the bioavailability and metabolism of grape polyphenols [6,7]. The primary bioactive constituents of grape-derived products include flavan-3-ols/proanthocyanidins, anthocyanins, flavonols, resveratrol and phenolic acids [1]. However, a factor limiting the use of flavanols, including the most abundant monomers, (+)-catechin (C) and (−)-epicatechin (EC), is the poor bioavailability. Flavanols in grape-derived products exist in monomeric, oligomeric and polymeric forms, with an estimated 40% of the monomers and oligomers absorbed in the upper intestine [9]. The substantial proportion of non-absorbed flavanols then go to the colon where they undergo extensive microbial catabolism before being absorbed as low-molecular-weight phenolic acids [8–10]. Mounting evidence demonstrates that microbial-generated phenolic acid metabolites are more bioavailable and exhibit diverse bioactivities, often to a greater extent than their precursors, considering their markedly higher abundancesin vivo[9,10]. This adds complexity to the identification of the actual active agents from botanicals that exert specific health impacts. The variations in systemic absorption, metabolism, distribution and excretion of grape flavanols may also explain their varied bioefficacies in humans. One hypothesis is that the type and quantity of low-molecular-weight metabolites produced largely depend on gut microbiota, while the compositions of microbial community vary greatly among individuals [11]. This opens up a new avenue of research involving the identification of human-originated probiotic strains that are capable of producing wide varieties and high quantities of bioactive metabolites.
Analytical methods for the analysis of phenolic acid metabolites need to have high sensitivity and selectivity in view of the low concentrations of certain microbial-derived metabolites and the complex endogenous nature of biological matrices [12]. Polyphenol metabolites produced by microbial metabolism consist of mainly low molecular weight phenolic acids, generally comprised of hydroxybenzoic and hydroxycinnamic acids and their derivatives [6]. The most commonly used methods for the analysis of small molecules with moderate to high polarity include gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS). Although LC-MS has been the most widely applied technique, problems with LC-based methods include difficulties in chromatographic resolution of peaks and detection with MS due to in-source fragmentation, ion clustering and matrix effects [13,14]. With the advent of tandem mass spectrometry (MS/MS), sensitivity and selectivity of both LC and GC methods are greatly improved. Compared with LC-MS, advantages of GC-MS-based methods include better chromatographic separation with capillary columns, lower maintenance costs, fewer matrix effects, and reduced ionization suppression and adduct formation, leading to less instrumental variability [15–17]. In addition, interfacing with a triple quadrupole (QqQ) analyzer operated under multiple reaction monitoring (MRM) or selective reaction monitoring (SRM) mode provides additional analytical selectivity, sensitivity and accuracy for quantitative analysis of targeted compounds [18].
Although a number of LC-MS/MS methods have been reported [18–20], there are limited GC-MS/MS methods for the analysis of diet-derived microbial phenolic metabolites [12]. In the present work, we report a highly sensitive and reproducible GC-QqQ/MS method for the identification and quantification of 16 microbial-derived phenolic acid metabolites, along with two grape polyphenol precursors, C and EC. This method was then validated and successfully applied to an in vitro fermentation study.
2. Materials and methods
2.1. Standards and reagents
All phenolic standards (19 in total including an internal standard) used to develop and validate the method were ≥98% pure. Gallic acid (GA), caffeic acid (CA),trans-p-coumaric acid (p-CA), dihydrocoumaric acid (diHCA), 3-(3,4-dihydroxyphenyl) propionic acid (3, 4-diHPPA), 3, 4-dihydroxybenzoic acid (3, 4-diHBA), hip-puric acid (HA), homovanillic acid (HVA), 3-hydroxybenzoic acid (3-HBA), 4-hydroxybenzoic acid (4-HBA), 3-hydroxyphenylacetic acid (3-HPAA), 3-(3-hydroxyphenyl)propionic acid (3-HPPA), vanillic acid (VA),trans-cinnamic acid-d7(internal standard, IS), catechin (C) hydrate and epicatechin (EC) were purchased from Sigma-Aldrich (St Louis, MO, USA). 3, 4-Dihydroxyphenylacetic acid (3, 4-diHPAA) and ferulic acid (FA) were purchased from ChromaDex Inc. (Irvine, CA), and 5-(4-hydroxyphenyl)valeric acid (4-HPVA) from Alfa Aeser (Lancashire, UK). Formic acid, methanol (MeOH), ethyl acetate, hexane and isopropanol were purchased from Fischer Scientific (Pittsburgh, PA, USA). The derivatizing cocktail Sylon™ HTP, hexamethyldisilazane, trimethylchlorosilane and pyridine (HMDS+ TMCS +Pyridine, 3:1:9), was purchased from Sigma-Aldrich (St Louis, MO).
2.2. Preparation of standards
The stock solutions of all 19 standards, including an IS, were prepared in 70% aqueous MeOH with 0.1% formic acid to make a solution ofca.500 μg/mL (the actual concentration of individual standards varied from 440 μg/mL to 739 μg/mL). For compound identification and determination of retention times, each phenolic standard stock solution (including the IS) was diluted with MeOH containing 0.1% formic acid to ca.1 μg/mL for GC-MS/MS analysis as described below using full scan mode. For preparing standard working solutions used for calibration and method validation, all 18 standards excluding the IS were mixed and diluted with MeOH containing 0.1% formic acid to establish 16 concentration levels ranging from ca.0.25 ng/mL to 7000 ng/mL. The IS stock was diluted with 100% MeOH containing 0.1% formic acid to a final concentration of 2 μg/mL. Each of the calibration standard mixture (200 μL) was spiked with the 2 μg/mL IS solution (40 μL). The mixture was then transferred to a HPLC vial and dried under a gentle stream of N2. Once dried, ethyl acetate (200 μL) was added to the vial and re-dried under N2stream to ensure complete removal of water. The derivatizing cocktail (200 μL) was then added to each vial and incubated at 70 °C for 4 h prior to analysis.
2.3. GC-QqQ/MS method development and parameter optimization
2.3.1. Instrumentation
Phenolic compounds were analyzed using a Shimadzu GCMS-TQ8040 gas chromatography triple quadrupole mass spectrometer interfaced with a Shimadzu AOC6000autosampler (Shimadzu Corporation, Kyoto, Japan). Data were collected and analyzed using the GCMS Solution Software (Version 4.30, Shimadzu). Chromatographic separations were performed using a Shimadzu RP-5SilMS capillary column (30.0 m × 0.25 μm × 0.25 mm) (Shimadzu, Kyoto, Japan). Helium was used as the carrier gas and argon as the collision-induced dissociation (CID) gas.
2.3.2. Optimization of GC-QqQ/MS parameters
To ensure optimal performance of the GC-MS system for targeted analytes, column oven temperature, temperature gradient, split ratio and MRM parameters were carefully optimized. Prior to optimization, all phenolic standards were prepared at a concentration of ca.1μg/mL and derivatized individually as aforementioned. Each derivatized standard was individually injected into the GC/MS and analyzed under full scan mode to confirm the analyte identity and retention time. A standard mixture containing all 19 compounds was then analyzed under full scan mode to assess peak resolution among analytes. The column oven temperature was optimized by first exploring a range of oven programs with different temperature gradients. The temperature gradient with a specific column temperature that collectively delivered the best peak resolution, peak shape, and the highest signal to noise ratio was then selected. The final program was: initial column oven temperature set at 80 °C, held for 1 min then raised to 220 °C at a rate of 10 °C/min and held for 3.5 min, then raised to 310 °C at a rate of 20 °C/min and held for 15 min for a total run time of 38 min. The injection temperature was set at 280 °C, the ion source temperature at 200 °C and the interface temperature at 280 °C. The detector voltage was set at 0.04 kV with a signal threshold at 1000. Helium was used as the carrier gas with a column flow rate at 0.95 mL/min and a purge flow rate at 1.0 mL/min.
To enhance analytical accuracy and sensitivity, a MRM method for the QqQ analyzer was created and optimized utilizing the Shimadzu Smart MRM program. Data collected from the analysis of individual compounds with aid of the software, optimal ion transitions were selected to include in the MRM method. Two of the most intense precursor-to-product ion transitions were registered for each analyte. For the two selected product ions of each analyte, the more intense one was used as the target ion for quantification and the second one as the reference ion for structural confirmation. Preference was given for higher molecular weight product ions since they can be more easily distinguished from other small fragment ions invariably generated from endogenous compounds in the matrix. Using the Smart MRM program, collision energy for each analyte was automatically tested at 15 different levels, escalating stepwise from 3 to 45 eV to select the optimal collision energy for each transition. Following this, a MRM method with seven different time frames was set up based on the optimal collision energies for each analyte and the retention time upon acquisition of the retention times and optimal parent-to-target/reference ion transitions.
Following MRM optimization, the effects of injection split ratios on detection sensitivity, peak resolution and sharpness were examined. The analysis was performed using the optimized MRM method with injection of 1.0 μLof a standard mixture (ca.1μg/mL) at various split ratios including splitless injection and split ratios of 2, 10 and 100. For analyzing standard mixtures and samples, an injection volume of 1.0 μL with a split ratio of 2 was employed.
2.4. Method validation
2.4.1. Linearity and sensitivity
To establish linearity and sensitivity, standards were analyzed at 16 different concentration levels ranging from ca.0.25–7000 ng/mL. The standard mixtures were all spiked with IS at a concentration of 400 ng/mL. The calibration set was then analyzed in duplicate with the optimized MRM method. Calibration curves were established by plotting the analyte-to-IS peak area ratios against the exact concentration of individual analytes in the 16-point calibration set, using linear regression and the origins were not forced through zero. Linearity was determined by evaluating the coefficient of determination (R2) of each calibration curve. Analytical sensitivity for each analyte was determined by measuring the lower limit of detection (LLOD) and lower limit of quantification (LLOQ),i.e.the lowest concentration of an analyte that reaches a signal-to-noise ratio ≥3 and ≥10, respectively.
2.4.2. Preparation and extraction of quality control samples
Blank nutrient broth samples (detailed composition provided in Supplementary Material) were previously stored at −80 °C and transferred to −20 °C 24 h prior to analysis. The broth was conditioned to room temperature before processing. All spiked and blank control samples were prepared in replicates of six. The blank broth (200 μL) and 2 μg/mL IS (40 μL) were mixed in an Eppendorf tube and spiked with a standard mixture (50 μL) (diluted from a stock solution in 100% MeOH with 0.1% formic acid) to a final concentration at ca.200 ng/mL as the low quality control (LQC) level, or ca.2000 ng/mL as the high quality control (HQC) level. Control samples were only mixed with IS. Six replicates of LQC, HQC and Control samples were prepared for analysis. All samples were dried under N2and then reconstituted with 500 μLdeionized water. A reconstituted sample was transferred into an Eppendorf tube followed by addition of 4 M HC1 (100 μL). Ethyl acetate (500 μL) was added and the mixture was vigorously vortexed for1min followed by centrifugation for 5 min at 3000 × g.The upper organic layer was transferred into a HPLC vial. The extraction process was repeated two more times and the combined organic fractions were dried under a gentle stream of N2. The derivatizing cocktail (200 μL) was then added to each vial and incubated for 4 h at 70 °C prior to analysis. Original blank broth without IS nor standards were similarly extracted and derivatized to reveal the endogenous phenolic composition.
2.4.3. Recovery
Recovery of each analyte was determined in samples spiked at both LQC (ca. 200 ng/mL) and HQC (ca. 2000 ng/mL) levels with an IS concentration at ca.400 ng/mL. Blank broth spiked with IS only was also analyzed in replicates of 6 to measure endogenous phenolic compounds in the broth. All samples were analyzed in duplicate. Recovery was calculated by evaluating the analyte-to-IS response ratio of the analyte in the spiked samples versus that in standards prepared in pure solvent at corresponding concentrations. The formula below was used to calculate recovery:
where SR = IS ratio in spiked sample, BR=IS ratio in blank broth samples, and CR = IS ratio in the corresponding standard used for calibration.
2.4.4. Precision and accuracy
To measure accuracy and precision, broth samples were spiked with standard mixture and IS same as the recovery test and analyzed in replicates of six. Precision was determined as the percent relative standard deviation (%RSD) of the measured analytes in spiked samples. Accuracy was determined by comparing measured concentration in samples with the actual spiked concentrations. Since many phenolic acids were found to exist endogenously in blank broth samples, the formula was adjusted to account for this factor as below:
where S = spiked concentration, M = measured concentration, and MB = endogenous concentration in blank broth samples.
2.5. In vitro microbial fermentation of grape flavanols and method application
To assess the applicability and efficiency of this new method, sample extracts procured from an in vitro microbial fermentation study were analyzed using the proposed GC-QqQ/MS method. Three sets of representative in vitro samples were analyzed: control samples were blank broth supplemented with C and EC and incubated without bacteria; Donor 1 and Donor 2 samples were blank broth supplemented with C and EC and fermented with gut microbiota isolated from fecal excreta of Donor 1 and Donor 2, respectively. All phenolic extracts of in vitro samples were prepared in duplicate and injected twice into the GC-MS for analysis.
2.5.1. In vitro anaerobic fermentation
Nutrient broth samples enriched with C and EC (detailed composition included in Supplementary Material) were inoculated with gut microbiota from two healthy human donors and incubated at 37 °C for 24 h. The control samples without bacteria were similarly incubated. Fermented broth samples were recovered by centrifuging at 4000 × g for 5 min. The supernatants were then acidified with 2% formic acid to a final concentration of 0.2% and stored at −80 °C.
2.5.2. Extraction of phenolic compounds from in vitro samples
Bacterial broth samples were previously stored at −80 °C and were transferred to −20 °C 24 h prior to analysis. Samples were thawed on ice and conditioned to room temperature before processing. Bacterial broth (500 μL) was mixed with 2μg/mL IS (40 μL), and then extracted and derivatized in the same manner as for quality control samples (Section 2.4.2). For each sample, 1.0 μL was injected into the GC-MS system with a split ratio of 2. Peak identity was confirmed by comparing the MS characteristics of both target and reference ions with those of authentic standards. Quantitation was achieved with calibration curves established using the analyte-to-IS peak area of target ions, and final concentrations were divided by 2.5 to compensate for the concentration change following derivatization.
3. Results and discussion
3.1. Development and optimization of GC-QqQ/MS method
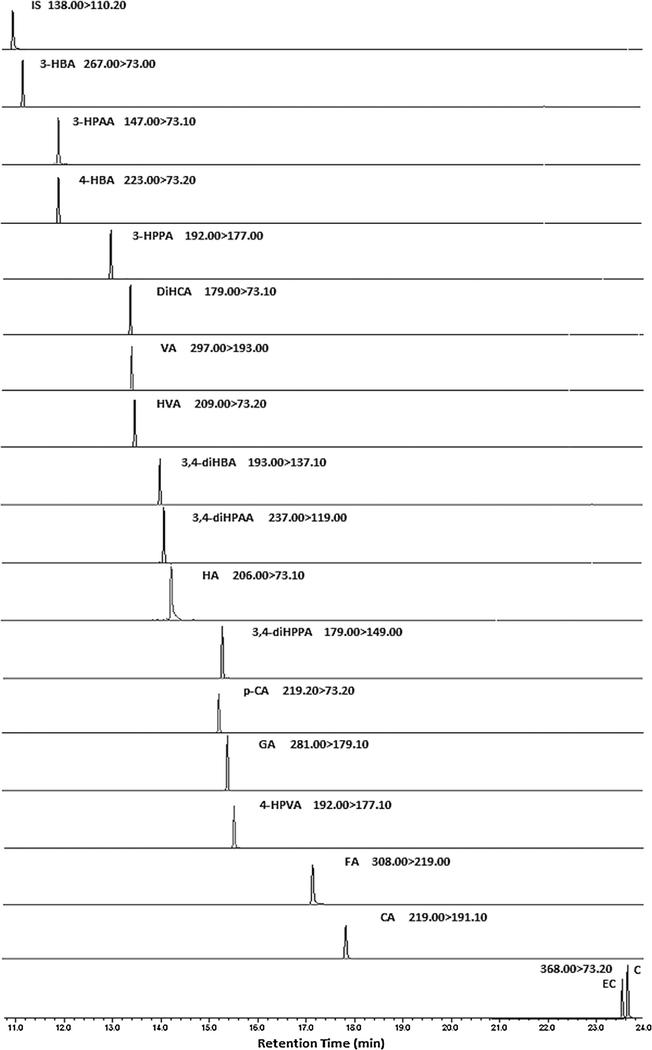
To clearly identify individual analytes and to determine retention times, individual standards at a concentration of ca.1 μg/mL were first analyzed under full scan mode. The established method demonstrated satisfactory chromatographic separation of most analytes. For two compounds that overlapped in retention time (3, 4-diHPPA and p-CA), their identification was resolved by MS operated under MRM mode. With this approach, all analytes were specifically identified and then accurately quantified. The representative chromatograms of compounds with parent-to-target ion transitions are presented in Fig. 1.
Fig.1.
Representative mass chromatograms displaying parent-to-target ion transitions for select phenolic acids, catechin, epicatechin and an internal standard. The standard mixture injected was at a concentration of ca.4000 ng/mL. Abbreviations are the same as in Table 1.
To establish the MRM method, target and reference ion transitions were selected based on the MS response of product ions, with preference taken for higher molecular weight ions with the strongest response to avoid matrix interference. Initially, the first MRM method was established with different parent to target and reference ion transitions, excluding the more prevalent m/z values that were observed for multiple compounds,i.e.73.1, 73.2, 177.0 and 149.0. However, this approach rendered reduced sensitivity, so a second MRM method was set up including these values. The comparative analysis of standard mixture using two methods revealed comparable selectivity, while the second method was more sensitive and given the study objectives was then adopted. Optimal collision energies were chosen based on those giving the greatest intensity for each transition. The use of MRM mode with multiple time frames ensures higher sensitivity and selectivity, as only the selected parent-to-target/reference ion transitions with retention times falling in the same defined time frame are scanned and allows longer dwell time for each transition. Retention times, analyte-specific parent-to-product ion transitions, dwell times and optimal collision energy for each transition are presented in Table 1. The MRM chromatograms of all 19 compounds at their selected ion transitions, obtained from a combined standard mixture are presented in Fig. 1. All peaks were well resolved, including 3,4-diHPPA and p-CA, due to the exclusive advantages of the MRM mode with defined time frames (Fig. 1). In addition, the amount of sample actually injected into the column, determined by both the inlet and outlet split ratios, can also markedly affect peak resolution and sharpness. By monitoring elution of standards with injection set to splitless, or a split ratio of2,10 or 100, we found that splitless injections led to inter-injection carryover as the transfer of the analyte vapor (diluted in carrier gas) from the inlet is much slower compared with that in split injection, while split ratios greater than 2 reduced sensitivity and trace constituents may thus be overlooked. Taking into account both peak shape and detection sensitivity, injection of 1 μL sample with a split ratio of 2 was determined to be optimal.
Table 1.
GC-QqQ/MS parameters of the optimized method and retention times of targeted phenolic compounds.
| Compounda | Abbreviation | Retention Time (min) |
Dwell Time (ms) | Target Ion Transition |
Collision Energy (eV) |
Reference Ion Transition |
Collision Energ (eV) |
|---|---|---|---|---|---|---|---|
| Trans-cinnamic acid-d7 | IS | 11.00 | 25.7 | 212.0 > 110.2 | 27.0 | 110.0 > 82.1 | 15.0 |
| 3-Hydroxybenzoic acid | 3-HBA | 11.21 | 26.0 | 267.0 > 73.2 | 30.0 | 267.0 > 193.0 | 18.0 |
| 3-Hydroxyphenylacetic acid | 3-HPAA | 11.75 | 26.0 | 147.0 > 73.1 | 15.0 | 296.0 > 73.1 | 21.0 |
| 4-Hydroxybenzoic acid | 4-HBA | 11.95 | 25.7 | 223.0 > 73.1 | 27.0 | 193.0 > 73.0 | 21.0 |
| 3-(3-Hydroxyphenyl)propionic acid | 3-HPPA | 13.08 | 20.3 | 192.0 > 177.0 | 9.0 | 192.0 > 73.1 | 21.0 |
| Dihydrocoumaric acid | diHCA | 13.44 | 20.3 | 179.0 > 73.1 | 21.0 | 192.0 > 177.0 | 12.0 |
| Vanillic acid | VA | 13.47 | 20.0 | 297.0 > 193.0 | 30.0 | 297.0 > 267.0 | 15.0 |
| Homovanillic acid | HVA | 13.54 | 19.5 | 209.0 > 73.2 | 21.0 | 179.0 > 149.0 | 27.0 |
| 3,4-Dihydroxybenzoic acid | 3,4-diHBA | 14.05 | 20.0 | 193.0 > 137.0 | 21.0 | 193.0 > 165.0 | 15.0 |
| 3, 4-Dihydroxyphenylacetic acid | 3,4-diHPAA | 14.13 | 31.5 | 237.0 > 119.0 | 12.0 | 237.0 > 209.0 | 9.0 |
| Hippuric acid | HA | 14.33 | 31.5 | 206.0 > 73.1 | 21.0 | 206.0 > 73.1 | 21.0 |
| Trans-p-coumaric acid | p-CA | 15.34 | 32.0 | 219.0 > 73.1 | 27.0 | 249.0 > 73.1 | 30.0 |
| 3-(3,4-Dihydroxyphenyl)propionic acid | 3,4-diHPPA | 15.33 | 32.0 | 179.0 > 149.0 | 30.0 | 219.2 > 73.2 | 15.0 |
| Gallic acid | GA | 15.45 | 32.0 | 281.0 > 179.0 | 21.0 | 458.0 > 179.0 | 42.0 |
| 5-(4-Hydroxyphenyl)valeric acid | 4-HPVA | 15.60 | 31.5 | 192.0 > 177.0 | 15.0 | 192.0 > 73.2 | 30.0 |
| Ferulic acid | FA | 17.22 | 98.7 | 308.0 > 219.0 | 21.0 | 338.0 > 249.0 | 21.0 |
| Caffeic acid | CA | 17.90 | 98.7 | 219.0 > 191.0 | 15.0 | 396.0 > 73.0 | 27.0 |
| Epicatechin | EC | 23.62 | 73.5 | 368.0 > 73.2 | 27.0 | 179.0 > 149.0 | 21.0 |
| Catechin | C | 23.73 | 73.5 | 368.0 > 73.2 | 30.0 | 179.0 > 149.0 | 21.0 |
Listed in order of elution. IS, internal standard.
3.2. Method validation
3.2.1. Linearity and sensitivity
Dynamic linear ranges, LLODs and LLOQs were determined by analysis of the 16-point calibration set and results are presented in Table 2. All linear regression coefficient were >0.996, indicating excellent linearity. LLODs were in the range of 0.24–6.18 ng/mL and LLOQs were 0.48–12.37 ng/mL. These values demonstrate excellent sensitivity of the proposed method, especially when compared to other GC-MS methods for analyzing phenolic acid metabolites derived from dietary polyphenols [21,22]. For example, Grün et al. analyzed microbial-generated phenolic acid metabolites present in biofluids from human subjects treated with a grape polyphenol-rich extract and reported much higher LLODs, ranging from 50 to 50,000 ng/mL [21]. This study used GC-TOF/MS without targeted analysis in MRM mode, which could explain the much lower sensitivity. In a recent investigation closer to the present work, a GC-QqQ/MS method was established for the analysis of phenolic acid metabolites in plasma samples from gerbils administered with a berry (Barneris microphylla) extract [12]. Comparable LLOQs were reported, ranging from 0.5 to 16.9 ng/mL when a liquid-liquid extraction method was applied. Same as in our study, an injection volume of 1 μLwas used, but this study used splitless injection. We found that although splitless injection lead to improved sensitivity for a few analytes, it caused significant inter-sample carryover, which compromised accuracy in identification and quantitation of more compounds, and therefore splitless injection was not adopted. Even with a split ratio of 2, we achieved even lower LLOQs with complete separation of all target peaks.
Table 2.
Calibration parameters, sensitivity, recovery, accuracy and precision of the optimized GC-QqQ/MS method.
| Compound | LOQ (ng/mL) |
LOQ (ng/mL) |
Dynamic Linear Range | Correlation Coefficient |
%Accuracy 2000ng/mL |
%Recovery 2000ng/ml |
%Accuracy 200ng/mL |
%Recovery 200ng/ml |
% RSD 2000ng/mL | %RSD 200ng/mL |
|---|---|---|---|---|---|---|---|---|---|---|
| 3-HBA | 0.312 | 0.624 | 0.312–10222.5 | 0.9986 | −5.97 | 87.35 | −13.35 | 73.62 | 8.50 | 3.36 |
| 3-HPAA | 3.77 | 7.54 | 1.600–1638.9 | 0.9974 | 5.58 | 91.99 | −12.86 | 100.93 | 12.87 | 16.82 |
| 4-HBA | 0.240 | 0.480 | 0.970–7944.5 | 0.9982 | −7.69 | 94.49 | −18.38 | 71.75 | 5.61 | 4.83 |
| 3-HPPA | 3.28 | 6.56 | 0.448–7333.3 | 0.9967 | −0.88 | 101.13 | −13.88 | 96.65 | 6.65 | 4.49 |
| diHCA | 1.57 | 3.15 | 0.393–6444.5 | 0.9995 | −2.43 | 101.11 | −10.97 | 99.11 | 7.23 | 4.96 |
| VA | 1.00 | 3.99 | 0.498–8166.8 | 0.9988 | 0.11 | 102.04 | −6.47 | 93.83 | 9.89 | 8.19 |
| HVA | 1.17 | 2.33 | 0.583–9555.5 | 0.9999 | 2.35 | 97.83 | −10.62 | 119.16 | 7.70 | 5.12 |
| 3,4-diHBA | 1.17 | 2.35 | 0.587–9611.1 | 0.9981 | 9.07 | 97.58 | −2.79 | 89.04 | 14.27 | 8.79 |
| 3,4-diHPAA | 3.50 | 7.00 | 0.875–7166.8 | 1.0000 | 16.7 | 108.18 | −5.27 | 134.82 | 18.80 | 8.61 |
| HA | 6.18 | 12.37 | 1.546–6333.25 | 0.9984 | −41.86 | 57.9 | −15.53 | 93.72 | 18.72 | 15.70 |
| p-CA | 1.87 | 3.74 | 0.468–7666.8 | 0.9994 | 18.08 | 105.55 | −4.79 | 145.26 | 18.01 | 9.82 |
| 3,4-diHPPA | 0.94 | 3.74 | 0.468–7666.8 | 0.9995 | 9.19 | 113.37 | −22.42 | 132.14 | 18.89 | 6.58 |
| GA | 0.51 | 1.02 | 0.507–8333.25 | 0.9999 | 1.36 | 91.08 | −5.30 | 121.62 | 14.29 | 11.07 |
| 4-HPVA | 0.96 | 1.93 | 0.963–7889.0 | 0.9993 | 22.85 | 115.79 | 5.71 | 114.14 | 12.65 | 6.54 |
| FA | 3.28 | 6.56 | 1.641–1680.6 | 0.9989 | 50.5 | 137.31 | 0.68 | 93.04 | 11.22 | 11.31 |
| CA | 1.49 | 5.97 | 1.491–1527.8 | 0.9969 | 15.40 | 125.38 | −7.09 | 153.58 | 4.35 | 11.31 |
| EC | 0.98 | 3.91 | 0.488–8000.0 | 0.9976 | −42.39 | 78.17 | −29.94 | 72.16 | 6.39 | 6.82 |
| C | 1.13 | 2.25 | 0.563–9222.3 | 0.9997 | −45.11 | 73.53 | −18.99 | 79.24 | 12.84 | 7.63 |
Method was validated for in vitro analysis of phenolic acids in bacterial broth samples. Blank broth samples were spiked with concentrations of ca.200 ng/mL andca.2000 ng/mL, with accuracy (%), recovery (%) and RSD (%) determined at both concentrations. Abbreviations are the same as in Table 1.
As highlighted in this study, the use of tandem mass spectroscopy offers many advantages such as enhanced selectivity and sensitivity, for the analysis of phenolic acid metabolites which are generally low-molecular-weight and polar. High selectivity is necessary for the analysis of low quantities of phenolic acid metabolites in complex biological matrices such as plasma, urine and digesta of gut microbiota to avoid high background noise and to reduce interferences from matrix compounds that can generate same product ions as our target analytes. While methods without the use of GC-MS in MRM mode may be adequate for the analysis of changes in phenolic acid metabolites at higher concentrations, such methods are less likely to detect subtle variations in metabolite profiles across different biological samples.
3.2.2. Recovery
Percent recoveries were determined by comparing MS response ratio (peak area) of the analyte measured in spiked samples to that in standards prepared in pure solvent. For analytes existing endogenously in the broth matrix, response for components in blank broth was subtracted from that in spiked broth samples to reveal the net change in response following bacterial fermentation. Recoveries were all above 85% at HQC level (ca. 2000 ng/mL), except for C and EC, which were around 73% and HA which was 58%. At LQC level (ca. 200 ng/mL), recoveries for 3-HBA, 4-HBA, C and EC were lower than other target compounds but were all above 71% (Table 2). During our first assay, much lower recoveries of C and EC were obtained (<10%), which could be attributed to insufficient extraction from the broth into the organic solvent. During the first extraction process, we did not dry the spiked sample completely before reconstituting in deionized water. Since broth samples were spiked with standards prepared in MeOH, the methanol content introduced to the reconstituted sample mixture could have affected the extraction efficiency. Very likely, the MeOH constituent in the sample mix-tures increased the affinity of the more hydrophilic compounds, such as C and EC, to the aqueous layer and reduced their affinity for the organic layer, i.e. ethyl acetate. For this reason, we re-prepared all spiked and blank broth samples. Spiked broth samples were first dried under N2and then reconstituted in deionized water. Samples were reconstituted with 500 μL of water to closely resemble the preparation of in vitro samples, where 500 μL of broth sample was used (section 2.5.2). By using the modified extraction protocol, we obtained much higher recoveries, especially for C, EC and HA. Furthermore, a lack of salt in the aqueous layer may also reduce extraction efficacy. Viñas et al. observed that increasing salt concentrations in aqueous layer can increase the affinity of polyphenols including C and EC for organic solvent during liquid-phase extraction [23]. Future investigations into the effects of ionic strength on the extraction efficiency of polyphenols and phenolic acids during liquid-liquid extraction are warranted.
Regarding the derivatization reagent, trimethylsilyl (TMS) derived reagents including trimethylsilyl chloride (TMCS) have been successfully applied to the analysis of polyphenol-derived metabolites in plasma and other biological matrices [12,21,24]. In our study, HMDS-TMCS-pyridine (3:1:9, v /v /v) was used due to its feasibility to silyate weakly acidic polar compounds, whereas no validation study has been conducted to evaluate the efficacy of the derivatization. This is also worthy of further examination to exclude the adverse influence on recovery due to lack of efficiency or repeatability of the derivatization procedure. While the extra step of derivatization required to make phenolic compounds volatile is a drawback of GC analysis, this step can also greatly improve detection specificity, sensitivity and analyte stability [25]. In contrast to native phenolic compounds, their trimethylsilyl derivatives are more thermostable and photostable [26]. As part of an on-going project involving further investigation into microbial metabolism of grape polyphenols, we also analyzed the same phenolic acid metabolites in their native form using a LC-MS methodology. Marked degradation of phenolic acids and precursor flavanols stored at 4°C occurred on the third day in an inter-day precision study using LC-MS, while the derivatized phenolic compounds analyzed directly from the anhydrous derivatizing agent for GC-MS analysis were all well retained when stored at room temperature. (D. Zhao and E. Carry, unpublished data). This is a major advantage for analyzing a large amount of such samples as degradation can occur during a lengthy analytical sequence. Furthermore, the stability of derivatized phenolic compounds can be improved through use of different protecting groups, such astert-butyldimethylsilyl (TBDMS). TBDMS derivatization enhances the hydrolytic stability as compared to TMS derivatization [26] and has been successfully applied for GC analysis of phenolic compounds [27].
In our study, we obtained averaged recoveries of 98.9% and 104.7% for high and low QC levels, respectively, and all recoveries were above 71% with the only exception being HA in HQC samples. Overall, the recoveries obtained in our study were satisfactory compared to other related GC-MS studies. Previous GC-MS studies related to the analysis of phenolic acids reported averaged recoveries in the range of 70–80%, while for some compounds recoveries were as low as 20% [12,21,24]. The varied recovery values for different phenolic acids determined in our study and previous work demonstrate the challenge in simultaneous extraction and profiling of multiple phenolic analytes in biosamples.
3.2.3. Precision and accuracy
Precision and accuracy were determined by analyzing six replicates of spiked samples at both high and low concentrations. Concentration of endogenous compounds in blank broth matrix was subtracted from the measured concentration of corresponding analyte in spiked samples. Precision for analyzing each compound was evaluated by the %RSD of the measured concentrations of six replicates of QC samples. As shown in Table 2, all %RSDs were below 19%, with most being less than 10%, indicating adequate reproducibility. Percent accuracy was determined by the ratio of the measured concentration to the actual spiked concentration. For almost all phenolic compounds, percent accuracies were <23%, except for FA, C, EC and HA at HQC level, and only EC at LQC level (Table 2). While percent accuracies are adequate for in vitro analysis, possible explanations for the observed variation includes incomplete extraction from the broth sample, inefficient derivatization (especially for C and EC who have multiple hydroxyl groups, making sialylation less feasible) and the presence of considerable amount of endogenous phenolic acids. A total of nine compounds, including, 3-HBA, 4-HBA, diHCA, VA, 3, 4-diHBA, HA, p-CA, GA and FA were determined to exist endogenously at moderate concentrations. The average %RSD of these compounds in the blank broth samples was 8.4%. While this is more than acceptable, slight variations in these concentrations could have a big impact on the concentration determined in the spiked samples. Except for C and EC, other analytes with lower accuracies were among those compounds present endogenously in broth samples.
3.3. Method application to in vitro fermentation studies involving human gut microbiota
The developed and validated GC-QqQ/MS method was applied to identify and quantify 16 phenolic acids metabolites and their polyphenol precursors in bacterial fermented flavanol-enriched broth samples. The 16 phenolic acids targeted for analysis were chosen since they have been widely reported as the major microbial polyphenol catabolites and are purported with diverse bioactivities [8,20,28,29]. These 16 targeted phenolic acids have been shown to be generated from microbial metabolism of the major grape flavanols, C and EC. C and EC can be found in a variety of dietary sources, particularly in grapes, apples, tea, wines and cocoa [8]. Since the majority of C and EC are not absorbed in the small intestine, the selected phenolic acids are expected to be generated by microbiota in the colon following ingestion of flavanol-rich dietaries.
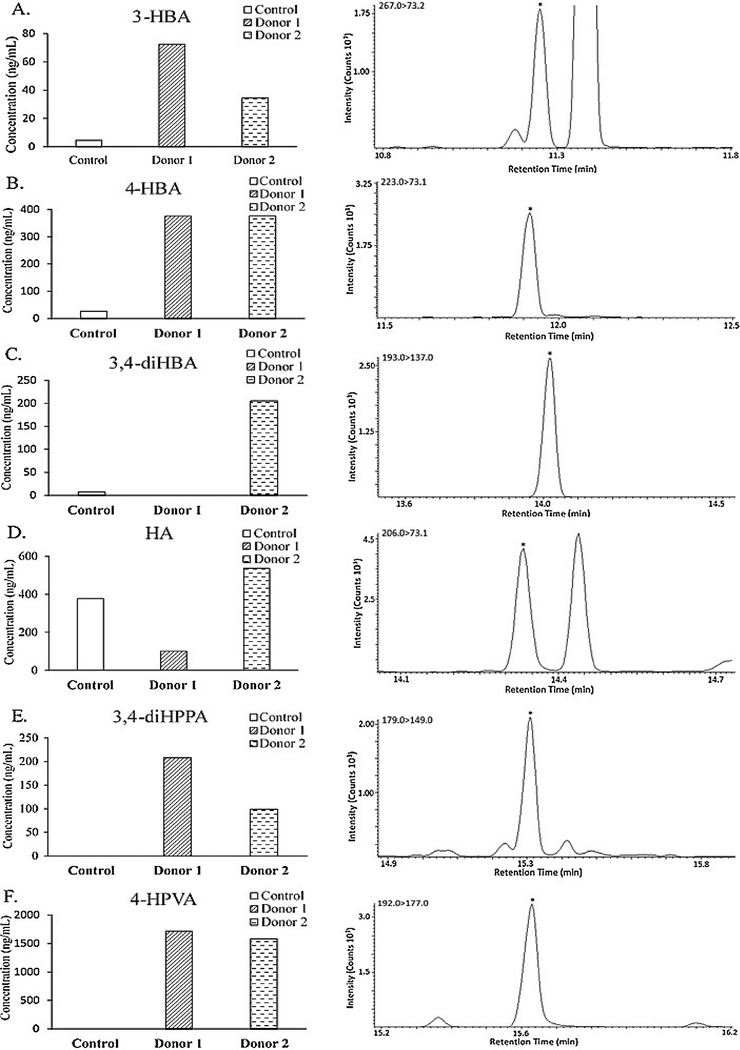
This is the first study employing GC coupled with QqQ/MS to investigate microbial-derived phenolic metabolites in in vitro fermentation studies involving human gut microbiota. In this study, C/EC-enriched nutrient broth was incubated with gut microbiota from two healthy donors for 24 h to reveal microbial activity towards grape flavanols. Clear indication of concentration variations of C, EC and their phenolic metabolites following fermentation (Table 3) verifies that the proposed method is efficient and robust. In general, all phenolic acid metabolites were detected at ng/mL level, with the lowest concentration observed for 3-HBA at 4.66 ng/mL in the control sample (Table 3). Inspection of the changes in precursor polyphenol concentrations reveals that gut microbiota originating from different individuals greatly influenced the extent of flavanol metabolism. Fermentation with Donor 1 microbiota resulted in degradation of 63% of C and 80% of EC, while with Donor 2 microbiota, over 99% of both C and EC were metabolized. This suggests that the bacterial culture from Donor 1 was less efficient at metabolizing C/EC than Donor 2. Regarding the catabolites, fermentation with bacterial culture from both donors led to significant increases in 3-HBA, 4-HBA, 4-HPVA and 3, 4-diHPPA levels (Fig. 2). Microbiota from Donor 1 were more active than Donor 2 in generating 3-HBA, 3, 4-diHPPA and 4-HPVA while decreasing HA content. In contrast, increases in 3,4-diHBA and HA were only observed in samples fermented with Donor 2 microbiota. Moreover, differences in the production of these metabolites are of particular interest due to multifarious bioactivities of selected phenolic acids. For instance, 3-HBA, 3, 4-diHBA and 4-HBA have been shown to have significant analgesic and anti-inflammatory activity [28,30], while studies have demonstrated 3,4-diHPPA to have anti-thrombolytic activity [8]. 3-HBA has been shown to accumulate in the brain and interferes with β-amyloid oligomerization which partly underlies the pathology of Alzheimer’s disease [29]. In this regard, our findings further support the well-accepted viewpoint that inter-individual differences in gut microbiota account for the varied bioefficacy of orally ingested polyphenols in the body [31]. As such, application of advanced GC-MS/MS methods for screening human bacterial culture with remarkable potentials to produce high levels and types of bioactive phenolic acid metabolites may extend a new dimension in maximizing the health benefits of grape-derived polyphenols. It is also important to note that phenolic acids are converted to phase II conjugates following enteric absorption although they are not expected to be produced by gut microorganisms in our in vitro fermentation model. In fact, GC-MS has been employed to quantitate phase II phenolic acid metabolites in urine, feces and/or plasma from human subjects and rats administered to red wine or red grape juice extracts [32,33]. However, in vivo samples in related studies were all analyzed by GC-MS following enzymatic digestion to deconjugate phase II metabolites. No GC-MS method is available for analyzing conjugated phenolic metabolites, possibly due to the difficulty in converting them into volatile compounds by derivatization. Thus, this may represent a limitation of GC-based methodology for direct analysis of conjugated phenolic acid metabolites. In addition, the advent of hydrophilic interaction liquid chromatography (HILIC), allowing better separation of polar compounds compared with conventional reversed-phase LC and GC methods [12], presents another promising option for analyzing microbial-generated polyphenol metabolites. However, as demonstrated in our study, the reproducibility, sensitivity and minimal injection volume requirement of GC-based methods warrant further investigation into their use for high-throughput analysis of phenolic acid metabolites.
Table 3.
Concentrations of phenolic acid metabolites and precursor polyphenols (cate-chin/epicatechin) in in vitro bacterial broth samples.
| Compound | Blank | Control | Donor 1 | Donor 2 |
|---|---|---|---|---|
| 3-HBA | 4.91 | 4.66 | 72.4 | 34.5 |
| 3-HPAA | N.D. | N.D. | N.D. | N.D. |
| 4-HBA | 35.5 | 25.9 | 375 | 377 |
| 3-HPPA | 0.0 | N.D. | N.D. | N.D. |
| diHCA | 23.4 | 22.5 | 41.3 | 97.2 |
| VA | 67.6 | 61.8 | 15.7 | 15.1 |
| HVA | N.D. | N.D. | N.D. | N.D. |
| 3,4-diHBA | 17.7 | 7.40 | N.D. | 205 |
| 3,4-diHPAA | N.D. | N.D. | N.D. | N.D. |
| HA | 731 | 379 | 101 | 536 |
| p-CA | 89.2 | 62.7 | 146 | 69.4 |
| 3,4-diHPPA | N.D. | N.D. | 208 | 99.0 |
| GA | 72.1 | 60.0 | 89.8 | 107 |
| 4-HPVA | N.D. | N.D. | 1717 | 1586 |
| FA | 69.0 | 62.3 | 196 | 58.0 |
| CA | N.D. | N.D. | 92.3 | N.D. |
| EC | N.D. | 4145 | 814 | 235 |
| C | N.D. | 3968 | 1478 | 177 |
Data is expressed in ng/mL. Abbreviations are the same as in Table 1. N.D. = not detected.
The Blank sample refers to non-incubated nutrient broth, used to determine endogenous levels of phenolic acids. The Control sample represents broth sample enriched with EC and C only.Donor 1 and Donor 2 represent EC and C enriched broth samples inoculated with intestinal microbiota from human donors 1 and 2, respectively. Control and donor samples were incubated for 24 h at 37 °C prior to processing. Abbreviations are the same as in Table 1.
Fig. 2.
Concentration changes and MS identification of select phenolic acid metabolites derived from catechin/epicatechin in broth samples fermented with human gut microbiota in vitro.Left column (charts A-F): average concentrations (ng/mL) of phenolic acid metabolites in control and samples fermented with microbiota from Donor 1 and 2. Right column: mass chromatograms showing the corresponding peaks of parent-to-target ion transition for identifying the target analyte, with chromatograms A, B, D, E and F selected from a representative Donor 1 sample and chromatogram C from a representative Donor 2 sample. Abbreviations are the same as in Table 1.
4. Conclusions
This study presents a highly sensitive and reproducible GC-QqQ/MS method for the analysis of 16 phenolic acid metabolites derived from two major grape flavanols, C and EC. This is the first study that developed and validated a GC-QqQ/MS method operated under MRM mode with multiple time frames for targeted analysis of phenolic acid metabolites derived from grape flavanols with implications on metabolic activity from human gut microbiota. This method proved to be highly sensitive, accurate and efficient for the analysis of phenolic compounds, and was successfully applied to a study of in vitro fermented bacterial broth supplemented with two major grape flavanols. Analysis of these samples revealed intensive metabolism of precursor polyphenols by human intestinal bacteria, resulting in depleted polyphenol precursors and elevated phenolic acid concentrations. Marked differences in the production of diverse phenolic acid metabolites were observed between flavanol-enriched broths fermented with microbiota from different human donors. Although not fully comprehensive, accurate and valuable information in relation to the complex interactions between gut microbiota and grape flavanols can be obtained by quantifying the phenolic acid metabolites using the proposed method. Given the increased interest in linking possible health benefits to the consumption of polyphenol-rich foods and critical roles of gut microbiota in modulating the bioefficacy of dietary polyphenols, the method described in this work may be widely applied to investigate microbial biotransformation of other dietary polyphenols.
Supplementary Material
Acknowledgements
This work was done as part of the Core B research program in NIH Botanical Center with funds provided by NIH ODS and NCCAM IP50AT008661 −01 to Mt. Sinai and with Supplemental Funding by the NIH to Mt. Sinai (Grant number 0254-3831-4609). We thank Shimadzu Instruments for the analytical instruments used to conduct this research. Funds were also provided by the New Jersey Agricultural Experiment Station Hatch Project Number NJ12158. The authors are very thankful to Dr. Mario Ferruzzi in North Carolina State University and Dr. Rick Dixon, University of North Texas for their constructive suggestions.
Footnotes
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jpba.2018.06.034.
References
- [1].Pasinetti GM, et al. , Roles of resveratrol and other grape-derived polyphenols in Alzheimer’s disease prevention and treatment, Biochimica et Biophysica Acta (BBA) - Mol. Basis Dis. 1852 (6) (2015) 1202–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wang J, et al. , Grape derived polyphenols attenuate tau neuropathology in a mouse model of Alzheimer’s disease, J. Alzheimers Dis. 22 (2) (2010) 653–661. [DOI] [PubMed] [Google Scholar]
- [3].Xia E-Q, et al. , Biological activities of polyphenols from grapes, Int. J. Mol. Sei.11 (2) (2010) 622–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chen L, et al. , Intracellular signaling pathways of inflammation modulated by dietary flavonoids: the most recent evidence, Crit. Rev. Food Sei. Nutr. (2017) 1–17. [DOI] [PubMed] [Google Scholar]
- [5].Georgiev V, Ananga A, Tsolova V, Recent advances and uses of grape flavonoids as nutraceuticals, Nutrients 6 (1) (2014) 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Heleno SA, et al. , Bioactivity of phenolic acids: metabolites versus parent compounds: a review, Food Chem. 173 (2015) 501–513. [DOI] [PubMed] [Google Scholar]
- [7].Rechner AR, et al. , The metabolic fate of dietary polyphenols in humans, Free Radie. Biol. Med. 33 (2) (2002) 220–235. [DOI] [PubMed] [Google Scholar]
- [8].Monagas M, et al. , Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites, Food Funct. 1 (3) (2010) 233–253. [DOI] [PubMed] [Google Scholar]
- [9].Ou K, Gu L, Absorption and metabolism of proanthocyanidins, J. Funct. Foods 7(2014)43–53. [Google Scholar]
- [10].Teng H, Chen L, Polyphenols and bioavailability: an update, Crit. Rev. Food Sei. Nutr. (2018) (Qust-accepted): p. 00–00. [DOI] [PubMed] [Google Scholar]
- [11].Gross G, et al. , In vitro bioconversion of polyphenols from black tea and red wine/grape juice by human intestinal microbiota displays strong interindividual variability, Agric. Food. Chem. 58 (18) (2010) 10236–10246. [DOI] [PubMed] [Google Scholar]
- [12].Bustamante L, et al. , Evaluation of microextraction by packed sorbent, liquid-liquid microextraction and derivatization pretreatment of diet-derived phenolic acids in plasma by gas chromatography with triple quadrupole mass spectrometry, J. Sep. Sei. 40 (17) (2017) 3487–3496. [DOI] [PubMed] [Google Scholar]
- [13].Kalili KM, de Villiers A, Recent developments in the HPLC separation of phenolic compounds, J. Sep. Sei. 34 (8) (2011) 854–876. [DOI] [PubMed] [Google Scholar]
- [14].Ostrowski W, et al. , Mass spectrometric behavior of phenolic acids standards and their analysis in the plant samples with LC/ESI/MS system, J. Chromatogr. B 967 (2014)21–27. [DOI] [PubMed] [Google Scholar]
- [15].Kind T, et al. , FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry, Anal. Chem. 81 (24) (2009) 10038–10048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lu D, et al. , Analysis of twenty phenolic compounds in human urine: hydrochloric acid hydrolysis, solid-phase extraction based on K2C03-treated silica, and gas chromatography tandem mass spectrometry, Anal. Bioanal. Chem. 407 (14) (2015) 4131–4141. [DOI] [PubMed] [Google Scholar]
- [17].Venn RF, Principles and Practice of Bioanalysis, CRC Press, 2008. [Google Scholar]
- [18].Becerra-Herrera M, et al. , Determination of phenolic compounds in olive oil: new method based on liquid-liquid micro extraction and ultra high performance liquid chromatography-triple-quadrupole mass spectrometry, LWT-Food Sei. Technol. 57 (1) (2014) 49–57. [Google Scholar]
- [19].Chang Y.-x., et al. , Simultaneous determination of four phenolic acids and seven alkaloids in rat plasma after oral administration of traditional Chinese medicinal preparation Jinqi Jiangtang Tablet by LC-ESI-MS/MS, J. Pharm. Biomed. Anal. 117 (2016) 1–10. [DOI] [PubMed] [Google Scholar]
- [20].Sa’nchez-Pata’n F, et al. , Determination of microbial phenolic acids in human faeces by UPLC-ESI-TQ. MS, J. Agric. Food. Chem. 59 (6) (2011) 2241–2247. [DOI] [PubMed] [Google Scholar]
- [21].Grün CH, et al. , GC-MS methods for metabolic profiling of microbial fermentation products of dietary polyphenols in human and in vitro intervention studies, J. Chromatogr. B 871 (2) (2008) 212–219. [DOI] [PubMed] [Google Scholar]
- [22].Marsol-Vall A, et al. , Injection-port derivatization coupled to GC-MS/MS for the analysis of glycosylated and non-glycosylated polyphenols in fruit samples, Food Chem. 204 (2016) 210–217. [DOI] [PubMed] [Google Scholar]
- [23].Vinas P, et al. , Directly suspended droplet microextraction with in injection-port derivatization coupled to gas chromatography-mass spectrometry for the analysis of polyphenols in herbal infusions, fruits and functional foods, J. Chromatogr. A 1218 (5) (2011) 639–646. [DOI] [PubMed] [Google Scholar]
- [24].Zhang K, Zuo Y, GC-MS determination of flavonoids and phenolic and benzoic acids in human plasma after consumption of cranberry juice, J. Agric. Food. Chem. 52 (2) (2004) 222–227. [DOI] [PubMed] [Google Scholar]
- [25].Ferreira AMC, et al. , In situ aqueous derivatization as sample preparation technique for gas chromatographic determinations, Chromatogr. A 1296 (2013)70–83. [DOI] [PubMed] [Google Scholar]
- [26].Halket JM, Zaikin VG, Derivatization in mass spectrometry—1. Silylation, Eur. J. Mass Spectrom. 9 (1) (2003) 1–21. [DOI] [PubMed] [Google Scholar]
- [27].Heberer T, Stan H-J, Detection of more than 50 substituted phenols as their t-butyldimethylsilyl derivatives using gas chromatography-mass spectrometry, Anal. Chim. Acta 341 (1) (1997) 21–34. [Google Scholar]
- [28].Khan SA, Chatterjee SS, Kumar V, Low dose aspirin like analgesic and anti-inflammatory activities of mono-hydroxybenzoic acids in stressed rodents, Life Sei. 148 (2016) 53–62. [DOI] [PubMed] [Google Scholar]
- [29].Wang D, et al. , Role of intestinal microbiota in the generation of polyphenol-derived phenolic acid mediated attenuation of Alzheimer’s disease β-amyloid oligomerization, Mol. Nutr. Food Res. 59 (6) (2015) 1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lende AB, et al. , Anti-inflammatory and analgesic activity of protocatechuic acid in rats and mice, Inflammopharmacology 19 (5) (2011) 255. [DOI] [PubMed] [Google Scholar]
- [31].Manach C, et al. , Bioavailability and bioefficacy of polyphenols in humans. L Review of 97 bioavailability studies, Am. J. Clin. Nutr. 81 (1) (2005) 230S–242S. [DOI] [PubMed] [Google Scholar]
- [32].Gonthier M-P, et al. , Microbial aromatic acid metabolites formed in the gut account for a major fraction of the polyphenols excreted in urine of rats fed red wine polyphenols, J. Nutr. 133 (2) (2003) 461–467. [DOI] [PubMed] [Google Scholar]
- [33].Grün CH, et al. , GC-MS methods for metabolic profiling of microbial fermentation products of dietary polyphenols in human and in vitro intervention studies, J. Chromatogr. B 871 (2) (2008) 212–219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.