The arterivirus nsp4-encoded 3C-like protease (3CLpro) plays an important role in virus replication and immune evasion, making it an attractive target for antiviral therapeutics. Previous work suggested that PRRSV nsp4 suppresses type I IFN production by cleaving NEMO at a single site. In contrast, the present study demonstrates that both EAV and PRRSV nsp4 cleave NEMO at multiple sites and that this strategy is essential for disruption of type I IFN production. Moreover, we reveal that EAV nsp4 also cleaves NEMO at glutamine 205 (Q205), which is not targeted by PRRSV nsp4. Notably, targeting a glutamine in NEMO for cleavage has been observed only with picornavirus 3C proteases (3Cpro) and coronavirus 3CLpro. In aggregate, our work expands knowledge of the innate immune evasion mechanisms associated with NEMO cleavage by arterivirus nsp4 and describes a novel substrate recognition characteristic of EAV nsp4.
KEYWORDS: 3C-like protease, equine arteritis virus, NF-κB essential modulator, interferon beta, porcine reproductive and respiratory syndrome virus
ABSTRACT
Equine arteritis virus (EAV) and porcine reproductive and respiratory syndrome virus (PRRSV) represent two members of the family Arteriviridae and pose major threats for the horse- and swine-breeding industries worldwide. A previous study suggested that PRRSV nsp4, a 3C-like protease, antagonizes interferon beta (IFN-β) production by cleaving the NF-κB essential modulator (NEMO) at a single site, glutamate 349 (E349). Here, we demonstrated that EAV nsp4 also inhibited virus-induced IFN-β production by targeting NEMO for proteolytic cleavage and that the scission occurred at four sites: E166, E171, glutamine 205 (Q205), and E349. Additionally, we found that, besides the previously reported cleavage site E349 in NEMO, scission by PRRSV nsp4 took place at two additional sites, E166 and E171. These results imply that while cleaving NEMO is a common strategy utilized by EAV and PRRSV nsp4 to antagonize IFN induction, EAV nsp4 adopts a more complex substrate recognition mechanism to target NEMO. By analyzing the abilities of the eight different NEMO fragments resulting from EAV or PRRSV nsp4 scission to induce IFN-β production, we serendipitously found that a NEMO fragment (residues 1 to 349) could activate IFN-β transcription more robustly than full-length NEMO, whereas all other NEMO cleavage products were abrogated for the IFN-β-inducing capacity. Thus, NEMO cleavage at E349 alone may not be sufficient to completely inactivate the IFN response via this signaling adaptor. Altogether, our findings suggest that EAV and PRRSV nsp4 cleave NEMO at multiple sites and that this strategy is critical for disarming the innate immune response for viral survival.
IMPORTANCE The arterivirus nsp4-encoded 3C-like protease (3CLpro) plays an important role in virus replication and immune evasion, making it an attractive target for antiviral therapeutics. Previous work suggested that PRRSV nsp4 suppresses type I IFN production by cleaving NEMO at a single site. In contrast, the present study demonstrates that both EAV and PRRSV nsp4 cleave NEMO at multiple sites and that this strategy is essential for disruption of type I IFN production. Moreover, we reveal that EAV nsp4 also cleaves NEMO at glutamine 205 (Q205), which is not targeted by PRRSV nsp4. Notably, targeting a glutamine in NEMO for cleavage has been observed only with picornavirus 3C proteases (3Cpro) and coronavirus 3CLpro. In aggregate, our work expands knowledge of the innate immune evasion mechanisms associated with NEMO cleavage by arterivirus nsp4 and describes a novel substrate recognition characteristic of EAV nsp4.
INTRODUCTION
The family Arteriviridae, which belongs to the order Nidovirales, consists of porcine reproductive and respiratory syndrome virus (PRRSV), equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV) of mice, simian hemorrhagic fever virus (SHFV), and the recently identified wobbly possum disease virus (WPDV) (1, 2). Of these arteriviruses, EAV and PRRSV are considered to be significant etiological agents in the field of veterinary research due to the economic losses that they cause in the equine- and swine-breeding industries worldwide (3). Stallions are susceptible to EAV infection; persistently infected stallions can infect and cause abortion in pregnant mares via their semen (4–6), and foals infected with EAV often suffer from fatal bronchointerstitial pneumonia or pneumoenteric syndrome (7). EAV is capable of persisting in the reproductive tract despite the presence of local mucosal antibodies and inflammatory responses (8). PRRSV infection presents as severe reproductive failure in sows and respiratory distress in piglets and growing pigs (9). As arteriviruses, EAV and PRRSV share some similar molecular properties, such as their replication strategy, capacity for establishing persistent infection, and ease of transmission from infected animals to naive animals (10, 11). These viral characteristics cause the corresponding diseases to be difficult to control. Currently available drugs and vaccines confer insufficient protection against these diseases (12, 13), so it is important to find a suitable strategy to prevent arterivirus proliferation in the host.
Arteriviruses are enveloped, single-stranded, positive-sense RNA viruses with genomes of 12 to 16 kb in size (14). The arterivirus genome consists of 10 to 12 known functional open reading frames (ORFs) (ORF1a, ORF1b, ORF2a, ORF2b, ORF3 to ORF7, ORF5a, and a transframe fusion [TF] ORF) (15). ORFs 1a and 1b, located in the 5′ three-quarters of the arterivirus genome, encode two long nonstructural precursor polyproteins, pp1a and pp1ab (16). These polyproteins are subsequently cleaved into mature nonstructural proteins (nsp’s) by ORF1a-encoded proteases (17). In EAV, ORF1a contains three proteases, two cysteine proteases located in nsp1 and nsp2 and a chymotrypsin-like serine protease in nsp4, which are involved in releasing the nsp1 to nsp3 and nsp3 to nsp12 proteins from pp1a/ab (18–20). nsp1 and nsp2 are involved in cleavage at the nsp1/2 and nsp2/3 sites, whereas nsp4 is responsible for cleavage of the remaining nsp’s (21). The nsp1α, nsp1β, nsp2, and nsp4 proteins of PRRSV are responsible for its self-cleavage processing (22, 23). Arterivirus nsp4 plays a significant role in the processing of pp1a and pp1ab for nonstructural proteins that direct genome replication and subgenomic mRNA (sg-mRNA) synthesis. Due to its indispensable role in viral replication, arterivirus nsp4 is an appealing target for antiviral drugs.
Interferons (IFNs) are crucial antiviral cytokines in innate immune responses (24, 25). Retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and Laboratory of Genetics and Physiology 2 (LGP2), which belong to the RIG-I-like receptor (RLR) family, serve as the main pattern recognition receptors (PRRs) for recognizing viral RNA (26). After sensing cytoplasmic viral RNA, pathogen-associated molecular patterns (PAMPs) are recognized by host PRRs. Subsequently, the PRRs interact with the adaptor mitochondrial antiviral signaling protein (MAVS) (also known as IPS-1/VISA/Cardif) and further recruit a bridging adaptor molecule, NF-κB essential modulator (NEMO), which is responsible for the recruitment of TRAF family member-associated NF-κB activator (TANK)-binding kinase 1 (TBK1) and IκB kinase ε (IKKε), to further activate the IKK complex. As a result, the activated IKK complex leads to the phosphorylation and activation of NF-κB and IFN regulatory factor 3 (IRF3), which directly induce IFN production (27, 28).
Given the pivotal role of the RIG-I/MDA5 pathway in IFN signaling, it is not surprising that viruses evolve strategies that target crucial molecular signaling by the viral protease to suppress IFN production. Previous work demonstrated that coronavirus (CoV) 3CL protease (3CLpro) and picornavirus-encoded 3C proteases (3Cpro) possess characteristics that suppress IFN-β production (29–32). Recent studies showed that PRRSV nsp4 could block NF-κB signaling to inhibit IFN production (33). Because infection with EAV could suppress IFN production, and viral 3CLpro and 3Cpro have been identified as IFN antagonists, here, we investigated the role of EAV 3CLpro (encoded by nsp4) in regulating the IFN response. Our results clearly demonstrated that EAV nsp4 impairs RIG-I/MDA5 signaling by cleaving the crucial adaptor molecule NEMO at multiple sites. We identified EAV nsp4 cleavage sites within NEMO and revealed that the final cleavage products were severely impaired for IFN signaling. Unlike PRRSV nsp4, EAV nsp4 can also cleave NEMO by identifying glutamine (Q) at the P1 position, the first amino acid on the left side of the recognition site. The above-described results reveal a comprehensive mechanism by which EAV nsp4 antagonizes type I IFN signaling and identify a novel substrate recognition characteristic of EAV nsp4.
RESULTS
EAV nsp4-mediated inhibition of IFN-β production requires its protease activity.
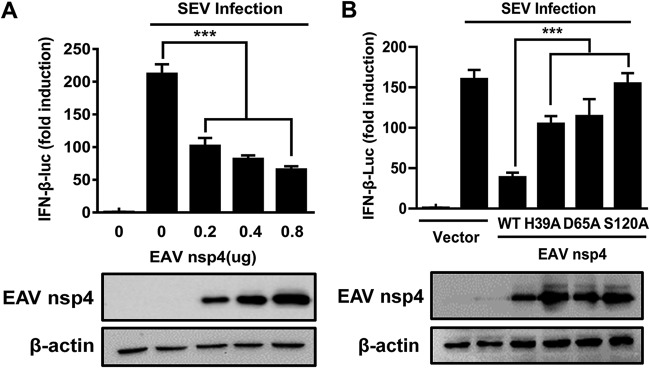
Previous studies suggested that PRRSV nsp4 inhibits IFN-β production (33). To investigate the function of EAV nsp4 in type I IFN signaling, we constructed an expression vector encoding nsp4 of EAV and determined the impact of its ectopic expression on Sendai virus (SEV)-induced IFN synthesis. Various concentrations of the EAV nsp4 expression plasmid and the IFN-β luciferase (IFN-β-Luc) reporter plasmid were cotransfected into HEK293T cells. As shown in Fig. 1A, EAV nsp4 strongly inhibited the SEV-induced IFN-β promoter activity in a dose-dependent manner. These data confirm the antagonistic property of EAV nsp4 in type I IFN signaling.
FIG 1.
EAV nsp4-mediated inhibition of IFN-β production requires its protease activity. (A) HEK293T cells were cotransfected with the IFN-β-Luc plasmid (0.1 μg), the pRL-TK plasmid (0.02 μg), and various concentrations (0.2, 0.4, or 0.8 μg) of plasmids encoding EAV nsp4. These cells were then infected with SEV at 24 h posttransfection, and their supernatants were collected 16 h after SEV infection and used in luciferase assays to analyze the expression of IFN-β. (B) HEK293T cells were transfected with plasmids encoding the EAV nsp4 wild type or mutants containing an H39A, D65A, or S120A mutation. These cells were then infected with SEV at 24 h posttransfection, and their supernatants were collected 16 h after SEV infection and used in luciferase assays to analyze the expression of IFN-β. ***, P < 0.001.
According to a previous report, the catalytic dyad of EAV nsp4 comprises the residues His39 (H39), Asp65 (D65), and Ser120 (S120), and any single mutation of these three residues could disrupt the protease activity of EAV nsp4 (3). Compared with the ectopic expression of wild-type EAV nsp4, the level of suppression of SEV-induced IFN-β promoter activity was significantly lower upon the ectopic expression of EAV nsp4-H39A, nsp4-D65A, or nsp4-S120A (Fig. 1B), revealing that the protease activity of EAV nsp4 is involved in antagonizing IFN-β production.
EAV nsp4 disrupts RIG-I/MDA5 signaling by targeting NEMO.
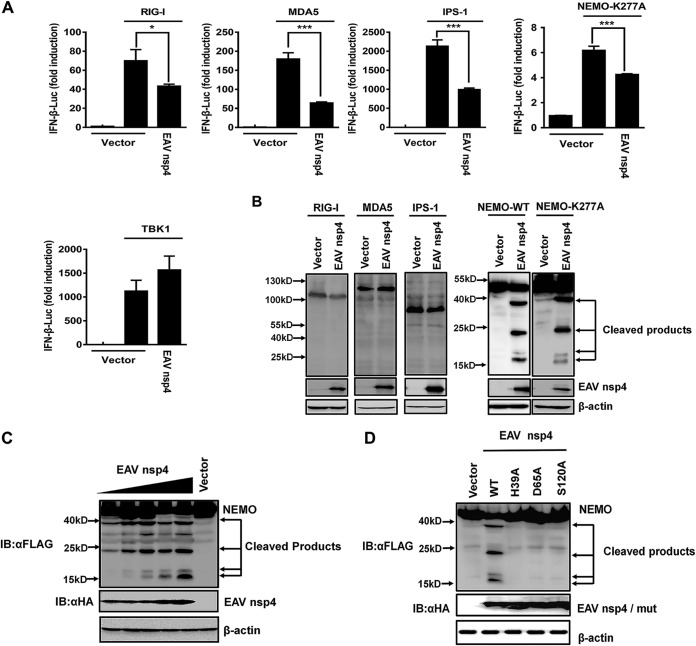
In response to infections with various RNA viruses, RIG-I/MDA5 signaling plays a pivotal role in mediating the induction of IFN-β (34). To determine the target spot at which EAV nsp4 facilitates its inhibitory role in this RIG-I/MDA5 signaling, we assessed the IFN-β induction abilities of several crucial molecules in RIG-I/MDA5 signaling, including RIG-I, MDA5, IPS-1, NEMO, and TBK1. Our results demonstrated that these five molecules significantly activated the IFN-β promoter compared with the empty vector control, and EAV nsp4 significantly inhibited the activation of the IFN-β promoter by RIG-I, MDA5, IPS-1, or NEMO-K277A, which is a constitutively active NEMO mutant (Fig. 2A). However, the TBK1-induced activation of the IFN-β promoter was not affected by EAV nsp4 (Fig. 2A), demonstrating that EAV nsp4 inhibits RIG-I/MDA5 signaling by targeting a molecule upstream of TBK1.
FIG 2.
EAV nsp4 cleaves NEMO to disrupt RIG-I/MDA5 signaling. (A) HEK293T cells were cotransfected with EAV nsp4, an IFN-β plasmid, and a pRL-TK plasmid along with the expression plasmid for RIG-I, MDA5, IPS-1, a constitutively activated form of NEMO (NEMO-K277A), or TBK1. Their supernatants were collected and used in a luciferase assay to analyze the corresponding activity of IFN-β. (B) HEK293T cells were cotransfected with Flag-appended RIG-I, MDA5, IPS-1, NEMO-WT, or NEMO-K277A expression plasmids along with EAV nsp4 or an empty vector. At 30 h posttransfection, they were harvested for Western blotting. (C) HEK293T cells were cotransfected with wild-type Flag-NEMO and different amounts (0.5, 1, 2, 3, or 4 μg) of EAV nsp4 expression plasmids or an empty vector (HA). At 30 h posttransfection, they were harvested for Western blotting. (D) Wild-type EAV nsp4 or its protease-defective mutant (H39A, D65A, or S120A) was cotransfected along with the NEMO expression plasmid and treated as described above for panel C. IB, immunoblot. *, P < 0.05; ***, P < 0.001.
Due to the participation of EAV nsp4 protease activity in IFN-β antagonism, we speculated that EAV nsp4 may target a signaling component(s) of the RIG-I/MDA5 pathway for cleavage. Thus, the signaling molecules upstream of TBK1, i.e., NEMO, RIG-I, MDA5, and IPS-1, were cotransfected into HEK293T cells along with EAV nsp4. Surprisingly, Western blotting of the resulting cell lysates revealed four new smaller fragments present under the coexpression of NEMO and EAV nsp4 compared with the control. Moreover, NEMO-K277A remained susceptible to EAV nsp4 cleavage (Fig. 2B). In contrast, no faster-migrating bands were observed for cells transfected with RIG-I, MDA5, or IPS-1 (Fig. 2B).
To further confirm the EAV nsp4-mediated cleavage of NEMO, different doses of EAV nsp4 expression plasmids were transfected into HEK293T cells along with NEMO expression plasmids. The results showed that the cleavage of NEMO gradually increased as EAV nsp4 expression increased (Fig. 2C). In contrast, no cleavage phenomenon was detected under the joint expression of NEMO with EAV nsp4-H39A, nsp4-D65A, or nsp4-S120A, suggesting that the cleavage of NEMO by EAV nsp4 depends on the protease activity of nsp4 (Fig. 2D).
Both EAV nsp4 and PRRSV nsp4 cleave multiple sites of NEMO.
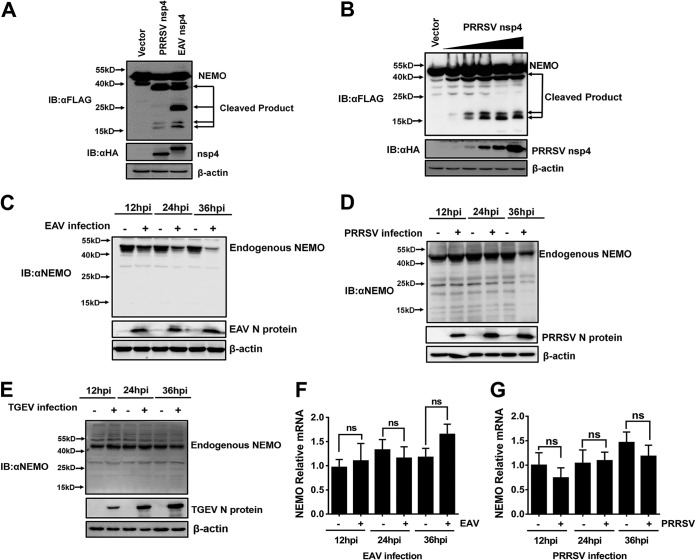
Both EAV and PRRSV, two members of the family Arteriviridae, encode nsp4 proteins with similar structures and functions (1, 3, 35). Previous research revealed that PRRSV nsp4 cleaves NEMO at glutamate 349 (E349) to produce a single faster-migrating protein band (33). To further compare the NEMO cleavage mediated by different members of the family Arteriviridae, EAV nsp4 or PRRSV nsp4 expression plasmids were transfected into HEK293T cells along with NEMO expression plasmids. Western blotting of the resulting cell lysates demonstrated that the size of the smallest-migrating protein band in EAV nsp4-mediated cleavage was similar to that in PRRSV nsp4-mediated cleavage. Interestingly, two new smaller bands (∼17 kDa in size) in the NEMO/PRRSV nsp4 coexpression samples were detected, and these two bands seemed to share a cleavage site similar to the one observed in the NEMO/EAV nsp4 coexpression samples (Fig. 3A). Only NEMO E349 has been previously reported to be cleaved by PRRSV nsp4 (28), so we further transfected different doses of PRRSV nsp4 into HEK293T cells along with NEMO. The results showed that two new smaller bands (∼17 kDa in size) were present in a PRRSV nsp4 dose-dependent manner (Fig. 3B), demonstrating that PRRSV nsp4 can cleave two residues near the N terminus of NEMO in addition to the reported cleavage residue E349. Thus, our results indicate that both the EAV and PRRSV nsp4 proteins possess the ability to cleave multiple sites of NEMO.
FIG 3.
Both PRRSV nsp4 and EAV nsp4 cleave multiple sites of NEMO. (A) HEK293T cells were cotransfected with Flag-tagged NEMO-WT and EAV nsp4 or PRRSV nsp4. The supernatants were harvested 30 h after transfection and analyzed by Western blotting. (B) HEK293T cells were cotransfected with Flag-tagged NEMO-WT and various concentrations (0.5, 1, 2, 3, or 4 μg) of PRRSV nsp4 expression plasmids or the empty vector. Cell lysates were collected at 30 h posttransfection and assessed by Western blotting. (C) Marc-145 cells were infected with EAV at a multiplicity of infection (MOI) of 1, and the cells were harvested for Western blotting at different time points (12, 24, or 36 h) postinfection. (D) PAM cells were infected with PRRSV strain WUH3 at an MOI of 1, after which the cells were lysed and analyzed for Western blotting at different time points (12, 24, or 36 h) postinfection. hpi, hours postinfection. (E) PK-15 cells in 6-well plates were infected with TGEV. Cells were collected at the different time points (12, 24, or 36 h) postinfection, followed by Western blotting with anti-NEMO. (F) EAV-infected Marc-145 cells were lysed at different time points (12, 24, or 36 h) postinfection, and their NEMO mRNA levels were measured by fluorescent quantitative PCR. (G) PAM cells were infected with PRRSV at an MOI of 1 and lysed at different time points (12, 24, or 36 h) postinfection. Their NEMO mRNA levels were assessed by real-time RT-PCR. ns, not significant (P > 0.05).
To further evaluate the expression of endogenous NEMO in EAV and PRRSV infection, the protein and mRNA levels of endogenous NEMO were detected in EAV- and PRRSV-infected cells. The results showed that the expression of NEMO was clearly reduced in EAV- and PRRSV-infected cells compared with mock-infected cells (Fig. 3C and D). To exclude the possibility that the decrease in the level of NEMO is a universal effect caused by any virus infection, PK-15 cells were infected with transmissible gastroenteritis virus (TGEV), a porcine intestinal coronavirus. The protein level of endogenous NEMO was detected by Western blotting. The results showed that the endogenous NEMO level was not reduced with the duration of TGEV infection (Fig. 3E), indicating that NEMO degradation is not a universal effect due to any virus infection. However, the mRNA level of endogenous NEMO in EAV- and PRRSV-infected cells was not significantly different from that in mock-infected cells (Fig. 3F and G). These findings indicate that the smaller amount of NEMO in EAV- and PRRSV-infected cells is caused by EAV and PRRSV infection and occurs in a manner that does not influence transcription.
E166, E171, and E349 are three common sites of EAV nsp4- and PRRSV nsp4-mediated NEMO cleavage.
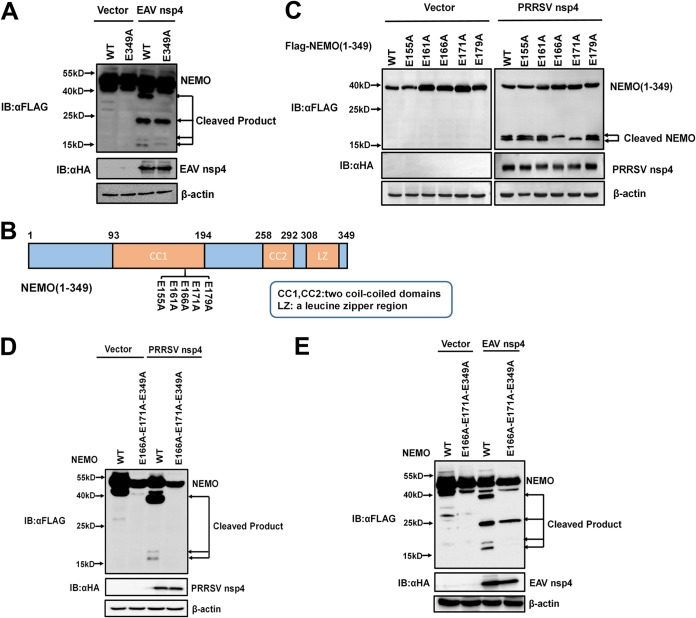
In the experiment described above, we compared cleavage characteristics of EAV nsp4 and PRRSV nsp4, and the three cleavage products of PRRSV nsp4 lie in positions similar to those of the three EAV nsp4 cleavage products (Fig. 3A). Therefore, we speculated that EAV nsp4 and PRRSV nsp4 share three common cleavage sites in NEMO. To confirm this hypothesis, NEMO with an E349A mutation (NEMO-E349A) was transfected into HEK293T cells along with EAV nsp4 (Fig. 4A). As expected, the largest cleaved band (∼40 kDa) observed in the control was absent in NEMO-E349A-transfected cells, whereas the other three cleaved bands were present in both (Fig. 4A), indicating that EAV nsp4 and PRRSV nsp4 possess the same ability to cleave NEMO E349.
FIG 4.
E166, E171, and E349 are three common sites of EAV nsp4- and PRRSV nsp4-mediated NEMO cleavage. (A) HEK293T cells were cotransfected with EAV nsp4 and Flag-tagged NEMO-WT or Flag-tagged NEMO-E349A. Cell lysates were collected at 32 h posttransfection and assessed by Western blotting. (B) Schematic representation of NEMO(1–349) and its mutants. These NEMO mutants were cloned by overlap extension PCR using NEMO(1–349) as the template and fused with an N-terminal Flag tag. (C) HEK293T cells were cotransfected with PRRSV nsp4 and wild-type NEMO or a NEMO mutant, as indicated. The cells were harvested at 30 h posttransfection and analyzed by Western blotting with anti-Flag antibody. (D and E) HEK293T cells were transfected with NEMO-WT or NEMO(E166A-E171A-E349) along with PRRSV nsp4 or EAV nsp4. Lysates of these cells were treated as described above for panel C.
After identifying NEMO E349 as one common cleavage position for EAV nsp4- and PRRSV nsp4-mediated NEMO cleavage, we next aimed to identify the two new NEMO sites cleaved by PRRSV nsp4 or EAV nsp4. Due to the ∼17-kDa molecular weight of the newly observed bands, we further analyzed the E residues in the N terminus of NEMO. A series of mutants in which the E residues among the NEMO peptides spanning amino acids (aa) 155 to 179 were targeted was constructed based on the truncation mutant NEMO(1–349) (Fig. 4B). The NEMO mutants containing E155A, E161A, E166A, E171A, or E179A mutations were each cotransfected into cells along with PRRSV nsp4. Western blotting of lysates from these cells showed that PRRSV nsp4 cleaved the NEMO mutants containing E155A, E161A, and E179A mutations into two products, whereas only a single cleavage product was present in cells transfected with NEMO mutants containing E166A or E171A (Fig. 4C). Therefore, the two potential PRRSV nsp4 cleavage positions were identified as NEMO E166 and E171.
To further confirm the sites of PRRSV nsp4-mediated NEMO cleavage, a NEMO mutant with multiple mutations, NEMO(E166A-E171A-E349A), was constructed and cotransfected into HEK293T cells along with PRRSV nsp4. The corresponding Western blot data showed that no cleaved product was present when NEMO(E166A-E171A-E349A) was overexpressed with PRRSV nsp4 (Fig. 4D), demonstrating that E166, E171, and E349 are three identified sites cleaved by PRRSV nsp4.
We next explored whether EAV nsp4 also targets E166 and E171. NEMO(E166A-E171A-E349A) was cotransfected into HEK293T cells along with EAV nsp4, and these cells were evaluated by Western blotting. The resulting blot revealed that the two smallest bands were not present in lysates from NEMO(E166A-E171A-E349A)-transfected cells (Fig. 4E). This finding indicates that EAV nsp4 and PRRSV nsp4 can target NEMO at common residues E166, E171, and E349.
EAV nsp4 cleaves NEMO by recognizing Q and E residues at the P1 position.
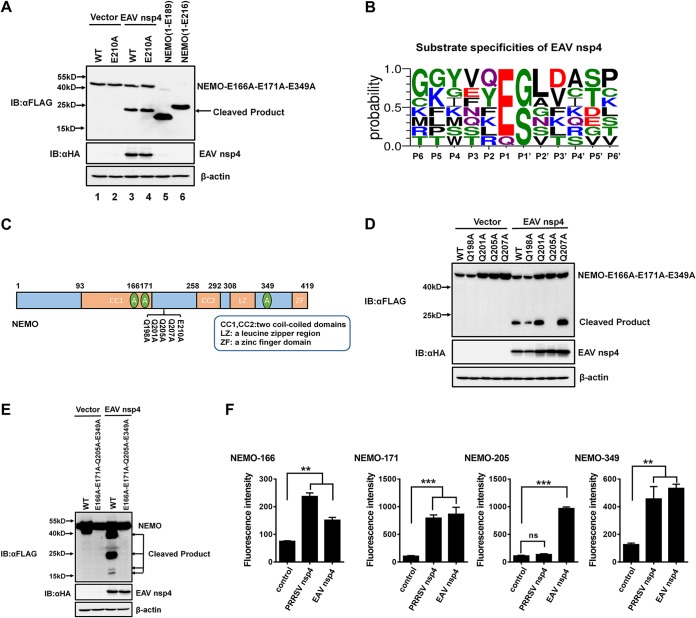
We next performed experiments to identify the unique NEMO cleavage site used by EAV nsp4. Based on the ∼20-kDa molecular weight of the fourth band and the high preference of nsp4 for E residues at the P1 position, we generated two truncation mutants, NEMO(1–189) and NEMO(1–216), to determine the cleavage position of the fourth cleavage product. The results from experiments using the two truncation mutants NEMO(1–189) and NEMO(1–216) showed that the cleavage position of the unidentified fourth cleavage product was located between residues 189 and 216 (Fig. 5A, lanes 5 and 6). However, E210 is the only E residue present among these residues, and a mutation in NEMO residue E210 did not affect the presence of the fourth cleavage product from EAV nsp4-mediated cleavage (Fig. 5A). These findings suggest that a nonclassical residue in the P1 position can be cleaved by EAV nsp4. Thus, we further analyzed the substrate specificity of EAV nsp4 in viral polyproteins from different EAV strains. The resulting data reveal that EAV nsp4 exhibits a high degree of preference for Glu (E) at the P1 position but that it also recognizes Q residues, even though the preference for them is quite low (Fig. 5B).
FIG 5.
EAV nsp4 cleaves NEMO by recognizing Q and E residues at the P1 position. (A) The estimated NEMO cleavage product produced by EAV nsp4. HEK293T cells were cotransfected with NEMO(E166A-E171A-E349A) or NEMO(E166A-E171A-E210A-E349A), along with EAV nsp4. NEMO(1–189) or NEMO(1–216) was transfected in HEK293T cells and used as a marker to indicate the location of the cleavage products. The cell lysates were treated as described in the legend of Fig. 4C. (B) Sequence logo of the polyprotein junctions cleaved by EAV nsp4 from different strains. An amino acid sequence logo of the substrate was generated by using WebLogo (http://weblogo.threeplusone.com/). (C) Schematic representation of NEMO(E166A-E171A-E349A) and its mutants. These mutants were cloned by overlap extension PCR using NEMO(E166A-E171A-E349A) as the template and fused with an N-terminal Flag tag. (D) HEK293T cells were cotransfected with Flag-tagged NEMO(E166A-E171A-E349A) or its mutants, in which the Q residues were changed to A (Q198A, Q201A, Q205A, and Q207A), along with EAV nsp4 or a vector control. The cell lysates were treated as described in the legend of Fig. 4C. (E) HEK293T cells were cotransfected with plasmids encoding wild-type NEMO or its mutant NEMO(E166A-E171A-Q205A-E349A), along with EAV nsp4. The cells were lysed at 30 h posttransfection and treated as described in the legend of Fig. 4C. (F) EAV/PRRSV nsp4 cleaved NEMO-derived substrates in vitro. Four fluorogenic peptide substrates (Dabcyl-EAATKE↓CQALEE-Edans, Dabcyl-CQALE↓GRARAAE-Edans, Dabcyl-DQLRMQ↓GQSVE-Edans, and Dabcyl-KASCQE↓SARIEDE-Edans, containing the cleavage sites of NEMO E166, E171, Q205, and E349, respectively) were introduced with EAV/PRRSV nsp4 in the FRET assays. ns, nonsignificant (P > 0.05); **, P < 0.01; ***, P < 0.001.
Using NEMO(E166A-E171A-E349A) as a template, we generated Q198A, Q201A, Q205A, and Q207A NEMO mutants by individually mutating all Q residues in the peptides spanning aa 189 to 216 of NEMO (Fig. 5C). Western blot analyses revealed that the fourth unidentified 20-kDa cleavage product was not present in cells expressing NEMO(Q205A), indicating that the 20-kDa cleavage product was caused by recognition of Q205 (Fig. 5D). To confirm this result, a NEMO mutant containing four consecutive mutations, NEMO(E166A-E171A-Q205A-E349A), was constructed. As expected, NEMO(E166A-E171A-Q205A-E349A) was resistant to cleavage (Fig. 5E). Thus, E166, E171, Q205, and E349 are the four residues in the P1 position that are recognized by EAV nsp4.
To exclude the possibility that the EAV/PRRSV nsp4-mediated NEMO cleavage observed was an artificial effect in cell culture, we further determined if the four cleavage sites of NEMO were also targeted by nsp4 in vitro. Four fluorogenic peptide substrates containing the cleavage sites (E166, E171, Q205, and E349) derived from NEMO were synthesized, and fluorescence resonance energy transfer (FRET) assays were performed to explore whether EAV or PRRSV nsp4 can cleave these peptide substrates. EAV and PRRSV nsp4 proteins were expressed as fusions to a C-terminal tandem His tag in Escherichia coli BL21(DE3) and purified by using a HisTrap HP column. When the purified proteins were incubated with four fluorogenic peptide substrates in vitro, EAV nsp4 was able to cleave all tested substrates (Fig. 5F); however, PRRSV nsp4 cleaved only three substrates, not including the substrate containing cleavage site Q205 (Fig. 5F). These results were consistent with our above-described results in vivo and confirmed that EAV/PRRSV nsp4 proteins possess the ability to cleave multiple sites of NEMO both in vivo and in vitro.
EAV and PRRSV nsp4-mediated NEMO cleavages are involved in the inhibition of type I IFN induction.
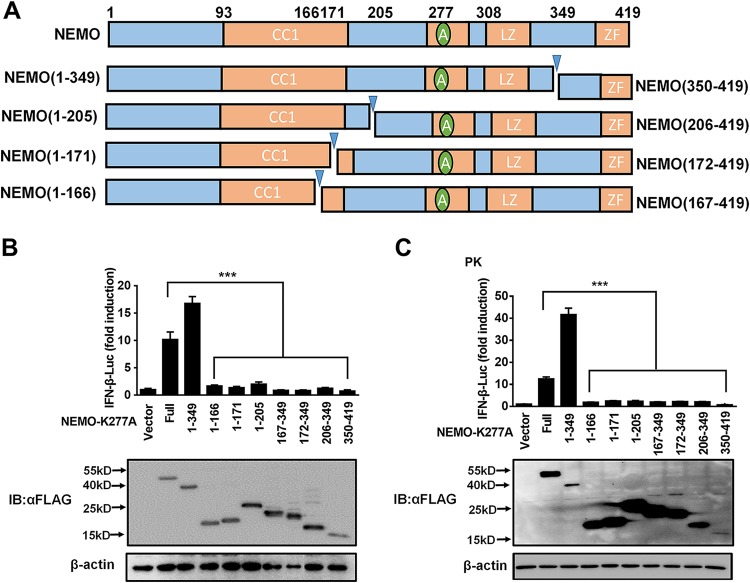
To assess whether the fragments generated from nsp4-mediatated NEMO cleavage remain active, a series of deletion mutants based on the identified nsp4 cleavage sites was constructed from the constitutively active NEMO mutant NEMO-K277A: NEMO-K277A(1–349), NEMO-K277A(1–166), NEMO-K277A(1–171), NEMO-K277A(1–205), NEMO-K277A(167–349), NEMO-K277A(172–349), NEMO-K277A(206–349), and NEMO-K277A(350–419) (Fig. 6A). Luciferase activity assays revealed that NEMO-K277A(1–349) had a relatively strong ability to activate IFN-β-dependent promoter activity in HEK293T and porcine kidney (PK-15) cells, indicating that cleavage at E349 might be not sufficient to impair NEMO function. In contrast, NEMO-K277A(1–166), NEMO-K277A(1–171), NEMO-K277A(1–205), NEMO-K277A(167–349), NEMO-K277A(172–349), NEMO-K277A(206–349), and NEMO-K277A(350–419) all failed to activate IFN-β-dependent promoter activity (Fig. 6B and C). These results demonstrate the abolished function of NEMO by all four cleavages, revealing the importance of multiple sites of NEMO cleavage by nsp4 in members of the family Arteriviridae.
FIG 6.
PRRSV and EAV nsp4-mediated NEMO cleavage is involved in the inhibition of type I IFN induction. (A) Schematic representation of NEMO-K277A and its deletion mutants. These mutants were cloned by PCR using NEMO-K277A as the template and fused with an N-terminal Flag tag. (B and C) HEK293T and PK-15 cells were separately transfected with expression plasmids for NEMO-K277A or the indicated deletion mutants along with the IFN-β-Luc plasmid and the pRL-TK plasmid. The cells were then lysed, and their IFN-β activity was assessed by luciferase assays. ns, nonsignificant (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001.
NEMO(1–349) retains the ability to activate IFN-β production.
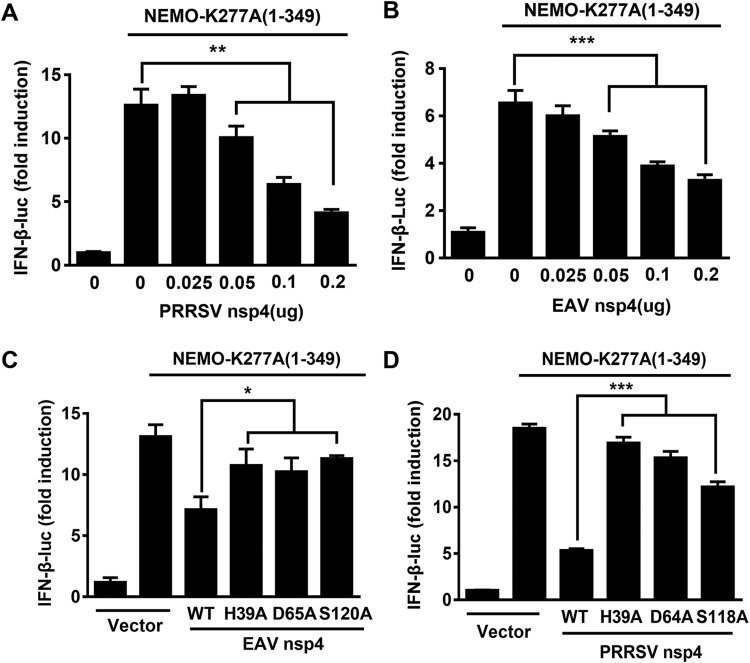
The above-described results verified that NEMO-K277A(1–349) retained the ability to activate the innate immune pathways that induce IFN-β production. To explore the detailed mechanism of this activation, NEMO-K277A(1–349) was cotransfected into HEK293T cells along with PRRSV nsp4 or EAV nsp4. The activation of the IFN-β promoter by NEMO-K277A(1–349) was significantly inhibited in the presence of PRRSV nsp4 in a dose-dependent manner (Fig. 7A), and similar results were found in the presence of EAV nsp4 (Fig. 7B). The suppression of IFN-β promoter activation by PRRSV nsp4 or EAV nsp4 was diminished upon expression of their respective enzymatic mutants (Fig. 7C and D), suggesting that the protease activity of nsp4 participates in inhibiting the NEMO-K277A(1–349)-mediated activation of IFN-β. These results indicate that both EAV nsp4 and PRRSV nsp4 target NEMO(1–349) for deep cleavage.
FIG 7.
NEMO(1–349) retains the ability to activate IFN-β production. (A and B) HEK293T cells were cotransfected with NEMO-K277A(1–349) and various concentrations of PRRSV nsp4 (A) or EAV nsp4 (B). (C) HEK293T cells were transfected with plasmids encoding EAV nsp4 or the related H39A, D65A, or S120A mutant. The cells were lysed at 28 h posttransfection and used in luciferase assays to analyze IFN-β expression. (D) PRRSV nsp4 or the related H39A, D64A, or S118A mutant was transfected into HEK293T cells for 30 h, and the cells were collected for luciferase assays. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
RIG-I-like receptor-mediated type I IFN production plays an important role in defending the host against virus invasion (34). However, during coevolution with their host, viruses always evolve a variety of strategies to escape and even inhibit host IFN responses (36, 37). Of these strategies, the cleavage of crucial signal molecules by a main viral protease is considered to be a particularly effective way for virus to escape the innate immune response (38, 39). For example, 3C proteases (3Cpro) of coxsackievirus A16, coxsackievirus A6, and enterovirus D68, which belong to the family Picornaviridae, can inhibit NF-κB activation by cleaving transforming growth factor β-activated kinase 1 (TAK1) (29). Additionally, coxsackievirus B and hepatitis C virus disrupt RIG-I/MDA5 signaling through the cleavage of MAVS by 3Cpro or NS3-4A proteases (39, 40). Here, we verified that, similarly to PRRSV nsp4, EAV nsp4 could also serve as an IFN antagonist by targeting the crucial adaptor molecule NEMO. Surprisingly, we found that catalytically inactive EAV nsp4 mutants retained slight inhibition of SEV-induced IFN production. Previous studies also reported that porcine epidemic diarrhea virus (PEDV) nsp5, porcine deltacoronavirus (PDCoV) nsp5, and foot-and-mouth disease virus (FMDV) 3Cpro mutants deprived of protease activity still retain weak inhibition of SEV-induced IFN production (31, 41, 42). It is well known that the protease-substrate interaction is the first step in the proteolytic activity of proteases. Catalytically inactive mutants of viral proteases, such as PEDV nsp5 (43) and FMDV 3Cpro (44), which lose their proteolytic activities, could form a complex with substrates. We speculate that the association between enzyme-inactive mutants and NEMO could contribute to the slight ability of catalytically inactive EAV nsp4 mutants to inhibit IFN induction. However, compared with the overexpression of wild-type EAV nsp4, the repression of SEV-mediated IFN-β promoter activation was strongly relieved with the overexpression of H39A, D65A, and S120A mutants. Thus, the proteolytic activity of EAV nsp4 is involved in suppressing IFN-β production.
A previous study on PRRSV nsp4 reported that PRRSV nsp4 blocks NF-κB signaling by targeting NEMO at a single site, E349 (33). However, we found here that the cleavage fragment NEMO(1–349) still activates IFN production (Fig. 6B). Furthermore, we additionally evaluated whether NEMO(1–349) could active the NF-κB promoter. The results showed that the cleavage fragment yielded by cleaving NEMO at E349 could also activate the NF-κB promoter compared with NEMO-K277A (data not shown). Together, the above-described data indicate that NEMO(1–349) shows a stronger ability for IFN activation than full-length NEMO. Previous work and the data shown in Fig. 2A verify that both EAV nsp4 and PRRSV nsp4 inhibit NEMO-mediated IFN activation (33), indicating that PRRSV nsp4 and EAV nsp4 each might fail to completely block only the NEMO-mediated IFN-β activation that occurs through cleavage at NEMO E349.
In recent studies, 3Cpro or 3CLpro of foot-and-mouth disease virus, hepatitis A virus, PEDV, and PDCoV were each verified to cleave NEMO at a single site to regulate IFN antagonism, despite the NEMO cleavage products not being observed in the correspondingly infected cells (30, 31, 33, 41, 42). Similar situations were also reported in work on Seneca Valley virus (SVV)-mediated MAVS, Toll/interleukin-1 (IL-1) receptor domain-containing adaptor inducing IFN-β (TRIF), and TRAF family member-associated NF-κB activator (TANK) cleavage (45). Although SVV 3Cpro could cleave MAVS, TRIF, and TANK, the corresponding cleavage products were not detected during SVV infection (45). Although we failed to detect the endogenous cleavage products of NEMO in PRRSV-infected porcine alveolar macrophages (PAM) or in EAV-infected Marc-145 cells, we observed a degradation of the NEMO protein abundance following viral infection. Thus, we speculate that the NEMO cleavage products may be short-lived during EAV or PRRSV infection. Initially, we would like to know whether arterivirus nsp4 is sufficient for endogenous NEMO degradation during EAV and PRRSV infection by reverse-genetics methods. However, the catalytically inactive mutants of EAV or PRRSV (EAV nsp4-H39A, -D65A, and -S120A and PRRSV nsp4-H39A, -D64A, and -S118A in our study) could not be rescued, and a possible cause is that the catalytically inactive nsp4 mutation also loses the ability to cleave the polyprotein of EAV or PRRSV. Future investigations to identify some mutants of nsp4 that abolish the ability to cleave NEMO but do not affect the ability to cleave the virus-encoded polyprotein will be required.
Here, we demonstrated that both PRRSV nsp4 and EAV nsp4 target NEMO at residues E166, E171, and E349, suggesting a relatively conserved identification mechanism in arterivirus nsp4-mediated NEMO cleavage. However, there is an exclusive site at Q205 for EAV-mediated NEMO cleavage, demonstrating that EAV nsp4 adopts a distinct substrate recognition characteristic for the P1 position targeting NEMO. A similar occurrence has been reported for encephalomyocarditis virus (EMCV) and SVV 3Cpro-mediated TANK cleavages. Both EMCV and SVV belong to the family Picornaviridae, and although their 3Cpro proteins target TANK at a common residue, Q291, SVV also cleaves TANK at another site, E272 (45, 46). Additional studies should be conducted to determine which part of the 3Cpro or 3C-like proteases leads to this difference in cleavage sites. Both arteriviruses and coronaviruses belong to the order Nidovirales. Previous work showed that nsp5 proteins from PEDV and PDCoV, members of the family Coronaviridae, cleave NEMO at a common residue, Q231 (31, 41). Our research group has verified that another coronavirus nsp5 could target NEMO at Q205 or Q231 (data not shown). However, EAV nsp4 can also cleave NEMO at Q205. This finding suggests that EAV nsp4 is closer to coronavirus nsp5 in evolution than PRRSV nsp4. Whether the evolutionary relationship between these viruses led to their different specific sites of NEMO cleavage by these 3C-like proteases should be studied further.
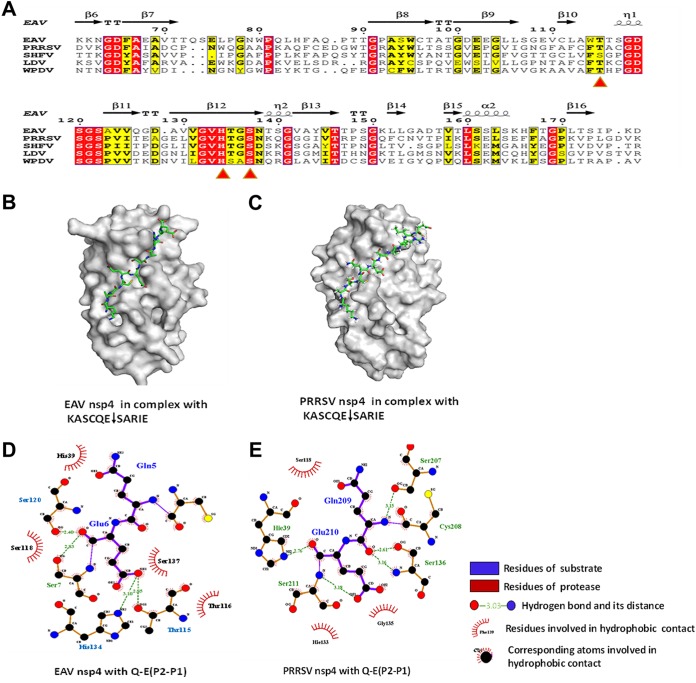
EAV nsp4 and PRRSV nsp4 are typical chymotrypsin-like serine proteases (3CLSPs) (20). A comparison between EAV nsp4 and PRRSV nsp4 revealed 36% identity in their amino acid sequences (Fig. 8A). Crystal structures revealed that the nsp4 proteins of PRRSV and EAV each contain three domains, including two chymotrypsin-like β-barrel domains and an extra C-terminal α/β-domain, mirroring a conserved structure in arterivirus nsp4 (3, 35). Additionally, recent work showed that coronavirus nsp5 proteins, which have highly conserved structures, exhibit a common P1-Q preference for substrate specificity. However, we found here that the nsp4 proteins of EAV and PRRSV, both of which belong to the family Arteriviridae, showed different preferences for substrate specificity: EAV nsp4 demonstrates a preference for Glu (E) and Gln (Q) at the P1 position to target NEMO, whereas PRRSV nsp4 prefers to recognize only Glu (E). To explore which part(s) of nsp4 causes these different recognition characteristics, we constructed two homology models for EAV and PRRSV nsp4 in complex with the same substrate, KASCQE↓SARIE (Fig. 8B and C).
FIG 8.
Homologous modeling of EAV nsp4 and PRRSV nsp4 in complex with the substrate. (A) Sequence alignment of motifs of several arterivirus 3CL proteases. The arterivirus nsp4 proteins are represented by EAV nsp4 (GenBank accession number NP_705586.1), PRRSV nsp4 (GenBank accession number AIC75887.1), WPDV nsp4 (GenBank accession number YP_009130632.2), LDV nsp4 (GenBank accession number NP_042572.1), and SHFV nsp4 (GenBank accession number YP_009109556.3). The structure-based sequence alignment was generated using the ESPript website (http://espript.ibcp.fr/ESPript/ESPript/index.php). (B and C) Homology model of EAV nsp4 (B) or PRRSV nsp4 (C) in complex with the substrate. (D and E) Distribution of hydrogen bonds and hydrophobic interactions between EAV nsp4 (D) and PRRSV nsp4 (E) with the novel substrate Q-E (P2-P1). Carbon, oxygen, and nitrogen atoms are labeled as black, red, and blue circles, respectively. Hydrophobic interactions are demonstrated by a curve with arms radiating toward the ligand residue or atoms that they contact.
The S1 pocket has been reported to play an important role in enzyme activities and substrate recognition. Thus, we analyzed the interaction between the protease and substrate motif for Q-E (P2-P1). The results showed that the S1 pocket of EAV nsp4 consisted of three residues, His134, Ser137, and Thr115, whereas only two corresponding residues, His133 and Ser136, were found in PRRSV nsp4; thus, the conformation of the S1 pocket differs between EAV nsp4 and PRRSV nsp4 (Fig. 8D and E). Additionally, Thr113 in PRRSV nsp4 is conserved with Thr115 in EAV nsp4, but Thr113 did not interact with P1 in the complex structure (Fig. 8A and E). Our data are similar to those from a previous study on the structure of arterivirus nsp4 (35). Therefore, we speculate that the different conformations of the S1 pockets of EAV nsp4 and PRRSV nsp4 may lead to P1 being able to accommodate different amino acids. However, the structure of arterivirus nsp4 in complex with substrates remains unclear. Our findings provide a suitable substrate, i.e., NEMO, for further studies on the structure of the arterivirus nsp4 complex. Whether there are any other nsp4 domains besides the S1 pocket that are responsible for different substrate recognition preferences requires further study.
NEMO, which serves as a regulatory subunit of the IKK complex, plays an indispensable role in the type I IFN production pathway (47). It has been reported that NEMO-activated IKKα/β can lead to TBK1/IKKε activation by IKKα/β-mediated TBK1/IKKε phosphorylation (47). NEMO deubiquitinylation impairs NF-κB activation (48, 49). A series of NEMO mutations (E391X, H413R, A323P, and Q157P) was reported to abrogate NF-κB activation, verifying that NEMO is a core component in regulating the NF-κB pathway (50). Different viruses can disrupt IFN signaling through interaction with NEMO or NEMO-interacting partners. According to previous studies on NEMO, the N-terminal region of NEMO is responsible for interactions with IKKs, and its C terminus interacts with upstream signaling adaptors and leads to the activation of IKKs (51). A recent study showed that p.DelQ134-R256, a truncated NEMO mutant that lacks the coiled-coil 1 (CC1) and helix-loop-helix 2 (HLX2) regions, could inhibit NF-κB signaling (52). PEDV and PDCoV 3CLpro proteins target NEMO at Q231 to divide it into two fragments, thereby disrupting NEMO downstream activity (31, 41). E166, E171, and Q205 are located within coil-coiled domains of NEMO; we verified that the cleavage fragments generated from targeting these three NEMO sites have lost the ability to induce IFN production (Fig. 6B and C). This finding indicates that the coil-coiled domains are indispensable for NEMO-induced IFN signaling.
In summary, our research revealed that arterivirus nsp4, specifically EAV nsp4 and PRRSV nsp4, cleave NEMO at multiple sites, which impairs IFN-β production. Both EAV nsp4 and PRRSV nsp4 induced NEMO cleavage at the conserved residues E166, E171, and E349, whereas only EAV nsp4 could target NEMO at site Q205. Importantly, cleavage of NEMO by PRRSV or EAV nsp4 led to suppression of the type I IFN signaling pathway. Our research provides deeper insights into strategies by which arterivirus nsp4 escapes the host immune response.
MATERIALS AND METHODS
Cells and viruses.
PK-15, HEK293T, and African green monkey kidney (Marc-145) cells were all cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS). PAM cells were cultured in RPMI 1640 (Sigma) containing 10% FBS at 37°C in a humidified 5% CO2 incubator. PRRSV strain WUH3 is a type II PRRSV that was isolated in 2006 from the brains of swine with high-fever syndrome in China (53). EAV was rescued from infectious cloning plasmid pEAV030 (54), which was a kind gift from Eric J. Snijder at Leiden University Medical Center. TGEV strain WH-1 (55), which was isolated in China in 2010, was propagated in PK-15 cells. Sendai virus (SEV) was obtained from the Centre of Virus Resource and Information at Wuhan Institute of Virology, Chinese Academy of Sciences.
Plasmids and luciferase reporter gene assay.
The DNA fragment encoding EAV nsp4 was amplified from pEAV030 and cloned into the pCAGGS-HA-C vector to generate a fusion expression plasmid with a C-terminal hemagglutinin (HA) tag. The eukaryotic expression plasmid for PRRSV nsp4 was described previously (56). EAV and PRRSV nsp4 mutants were constructed by site-directed mutagenesis to create mutant forms deprived of catalytic activity. The prokaryotic expression plasmids for EAV and PRRSV nsp4 proteins were cloned into the pET-42b vector to generate recombinant plasmids with a C-terminal His6 tag. NEMO constructs with an N-terminal Flag tag were generated by amplification of NEMO cDNA and cloned into the vector pCAGGS-Flag. A series of wild-type NEMO (NEMO-WT) or NEMO-K277A mutants was cloned by overlap extension PCR using NEMO-WT or NEMO-K277A as the template. a Flag tag was attached to the N terminus of NEMO-WT or mutants. The mutagenic primers used for this assay are available upon request. The luciferase reporter plasmids for IFN-β-Luc were described previously (41). The expression plasmids for RIG-I, IPS-1, MDA5, NEMO-K277A, TBK1, and NEMO were constructed as described previously (31, 41). All constructs were confirmed by DNA sequencing.
HEK293T or PK-15 cells were cultured in 24-well plates and transfected with IFN-β-Luc (100 ng/well) and pRL-TK (20 ng/well) plasmids, together with various amounts of the appropriate control or protein-expressing plasmid(s). The cells were infected with SEV 24 h after transfection. At 16 h postinfection, the cells were collected for the detection of luciferase and Renilla luciferase activities.
Western blot analyses.
Cells were cultured in 60-mm2 flasks and treated with lysis buffer containing a protease inhibitor cocktail 30 h after transfection. The samples were detected by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) immunoblotting membrane (Millipore, USA) to determine protein expression. The membrane was then blocked with 10% milk in Tris-buffered saline–Tween (TBST) containing 0.5% Tween for 3 to 4 h, followed by incubation with anti-HA antibody (1:2,000; MBL, Japan), anti-Flag antibody (1:2,000; Macgene, China), anti-NEMO antibody (1:2,000; ABclone, China), or anti-β-actin antibody (Beyotime, China). Finally, a corresponding secondary antibody was incubated with the blots. An anti-Flag tag antibody was used for detecting RIG-I, MDA5, IPS-1, and NEMO. The expressions of PRRSV nsp4 and EAV nsp4 were each evaluated using an anti-HA tag (MBL, Japan). Endogenous NEMO was detected via an anti-NEMO tag (ABclone, China) whose antigen consists of a recombinant fusion protein containing a sequence corresponding to amino acids 1 to 419 of human NEMO. Anti-PRRSV N protein monoclonal antibody was used to detect the expression of PRRSV N protein. EAV N protein was assessed with anti-EAV N protein monoclonal antibody, which was a kind gift from XiaoJun Wang at Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
RNA extraction and quantitative real-time PCR.
To detect the mRNA expression of endogenous NEMO, cells were collected at different time points postinfection. Total RNA was extracted with TRIzol reagent (Invitrogen), and RNA (1 μg) was reverse transcribed into cDNA using avian myeloblastosis virus reverse transcriptase (TaKaRa, Japan). Quantitative real-time PCR (qPCR) experiments were each performed three times. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal reference to evaluate the mRNA expression levels. All primers for real-time PCR are listed in Table 1.
TABLE 1.
Primers used for real-time PCR
Protein expression and purification.
For nsp4 protein expression, the recombinant plasmids were transformed into E. coli BL21(DE3) and grown at 37°C in LB medium. Next, 0.8 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture medium to induce the protein until the optical density at 600 nm (OD600) reached 0.6 to 0.8. Cells were collected after incubation at 18°C for 17 h. Both PRRSV/EAV nsp4 expression and purification were carried out as described previously (43). Approximately 2.5 mg of the purified protein was obtained and frozen at −80°C for storage.
FRET-based assays for enzymatic characteristics.
Based on sites of EAV and PRRSV nsp4-mediated NEMO cleavage, we designed four fluorogenic peptide substrates, Dabcyl-EAATKE↓CQALEE-Edans, Dabcyl-CQALE↓GRARAAE-Edans, Dabcyl-DQLRMQ↓GQSVE-Edans, and Dabcyl-KASCQE↓SARIEDE-Edans (Nanjing GenScript Company), which were derived from NEMO containing four cleavage sites, E166, E171, Q205, and E349, respectively. The donor and receptor fluorophores formed a quenching pair and showed fluorescence resonance energy transfer (FRET) within the peptide (57). The increases in fluorescence due to the cleavage of the fluorogenic peptide substrates were monitored at 490 nm, with excitation at 340 nm, using a fluorescence spectrophotometer (35). His-tagged EAV or PRRSV nsp4 protein was used in the FRET assays. All reactions were performed in a buffer containing 20 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 5 mM dithiothreitol (DTT). The enzyme concentration used in the assay was 1 μM, and the substrate concentration was 10 μM.
Homology modeling.
Because the core structures of arterivirus nsp4 proteins are similar, we constructed two complex structures, EAV nsp4-substrate and PRRSV nsp4-substrate, using the coronavirus 3CLpro-substrate complex (PDB accession number 2Q6G) as a model for alignment. The distributions of hydrogen bonds and hydrophilic interactions between arterivirus nsp4 and its substrate were generated by the LigPlot+ program.
Statistical analysis.
All tests were performed in triplicate. Analyses of significant differences were performed using Student’s t test, and P values of <0.05 were considered to be statistically significant.
ACKNOWLEDGMENTS
We thank Eric J. Snijder and Xiaojun Wang for providing the reagents.
This work was supported by the National Natural Sciences Foundation of China (31490602, 31672566, and 31372467), the National Basic Research Program (973) of China (2014CB542700), the Key Technology R&D Programme of China (2015BAD12B02), and the Fundamental Research Funds for the Central Universities (2662016PY070).
REFERENCES
- 1.Kuhn JH, Lauck M, Bailey AL, Shchetinin AM, Vishnevskaya TV, Bào Y, Ng TFF, LeBreton M, Schneider BS, Gillis A, Tamoufe U, Diffo JLD, Takuo JM, Kondov NO, Coffey LL, Wolfe ND, Delwart E, Clawson AN, Postnikova E, Bollinger L, Lackemeyer MG, Radoshitzky SR, Palacios G, Wada J, Shevtsova ZV, Jahrling PB, Lapin BA, Deriabin PG, Dunowska M, Alkhovsky SV, Rogers J, Friedrich TC, O’Connor DH, Goldberg TL. 2016. Reorganization and expansion of the nidoviral family Arteriviridae. Arch Virol 161:755–768. doi: 10.1007/s00705-015-2672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, Tas A, Snijder EJ, Fang Y. 2012. Identification of porcine reproductive and respiratory syndrome virus ORF1a-encoded non-structural proteins in virus-infected cells. J Gen Virol 93:829–839. doi: 10.1099/vir.0.039289-0. [DOI] [PubMed] [Google Scholar]
- 3.Barrette-Ng IH, Ng KK-S, Mark BL, van Aken D, Cherney MM, Garen C, Kolodenko Y, Gorbalenya AE, Snijder EJ, James MNG. 2002. Structure of arterivirus nsp4. The smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole. J Biol Chem 277:39960–39966. doi: 10.1074/jbc.M206978200. [DOI] [PubMed] [Google Scholar]
- 4.Socha W, Rola J, Żmudziński JF. 2015. Variability of non-structural proteins of equine arteritis virus during persistent infection of the stallion. Pol J Vet Sci 18:255–259. doi: 10.1515/pjvs-2015-0033. [DOI] [PubMed] [Google Scholar]
- 5.Timoney PJ, McCollum WH, Roberts AW, Murphy TW. 1986. Demonstration of the carrier state in naturally acquired equine arteritis virus infection in the stallion. Res Vet Sci 41:279–280. doi: 10.1016/S0034-5288(18)30616-7. [DOI] [PubMed] [Google Scholar]
- 6.Timoney PJ, McCollum WH, Murphy TW, Roberts AW, Willard JG, Carswell GD. 1987. The carrier state in equine arteritis virus infection in the stallion with specific emphasis on the venereal mode of virus transmission. J Reprod Fertil Suppl 35:95–102. [PubMed] [Google Scholar]
- 7.Balasuriya UBR, Carossino M, Timoney PJ. 2016. Equine viral arteritis: a respiratory and reproductive disease of significant economic importance to the equine industry. Equine Vet Educ 30:497–512. [Google Scholar]
- 8.Carossino M, Wagner B, Loynachan AT, Cook RF, Canisso IF, Chelvarajan L, Edwards CL, Nam B, Timoney JF, Timoney PJ, Balasuriya UBR. 2017. Equine arteritis virus elicits a mucosal antibody response in the reproductive tract of persistently infected stallions. Clin Vaccine Immunol 24:e00215-17. doi: 10.1128/CVI.00215-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chand RJ, Trible BR, Rowland RR. 2012. Pathogenesis of porcine reproductive and respiratory syndrome virus. Curr Opin Virol 2:256–263. doi: 10.1016/j.coviro.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Balasuriya UB, Carossino M. 2017. Reproductive effects of arteriviruses: equine arteritis virus and porcine reproductive and respiratory syndrome virus infections. Curr Opin Virol 27:57–70. doi: 10.1016/j.coviro.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Balasuriya UB, Hedges JF, Smalley VL, Navarrette A, McCollum WH, Timoney PJ, Snijder EJ, MacLachlan NJ. 2004. Genetic characterization of equine arteritis virus during persistent infection of stallions. J Gen Virol 85:379–390. doi: 10.1099/vir.0.19545-0. [DOI] [PubMed] [Google Scholar]
- 12.Meng XJ. 2000. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet Microbiol 74:309–329. doi: 10.1016/S0378-1135(00)00196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowland RRR, Morrison RB. 2012. Challenges and opportunities for the control and elimination of porcine reproductive and respiratory syndrome virus. Transbound Emerg Dis 59:55–59. doi: 10.1111/j.1865-1682.2011.01306.x. [DOI] [PubMed] [Google Scholar]
- 14.Snijder EJ, Kikkert M, Fang Y. 2013. Arterivirus molecular biology and pathogenesis. J Gen Virol 94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 15.Fang Y, Treffers EE, Li Y, Tas A, Sun Z, van der Meer Y, de Ru AH, van Veelen PA, Atkins JF, Snijder EJ, Firth AE. 2012. Efficient −2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc Natl Acad Sci U S A 109:E2920–E2928. doi: 10.1073/pnas.1211145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang Y, Snijder EJ. 2010. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res 154:61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziebuhr J, Snijder EJ, Gorbalenya AE. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol 81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 18.Snijder EJ, Wassenaar AL, Spaan WJ. 1994. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J Virol 68:5755–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snijder EJ, Wassenaar AL, Spaan WJ, Gorbalenya AE. 1995. The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J Biol Chem 270:16671–16676. doi: 10.1074/jbc.270.28.16671. [DOI] [PubMed] [Google Scholar]
- 20.Snijder EJ, Wassenaar AL, van Dinten LC, Spaan WJ, Gorbalenya AE. 1996. The arterivirus nsp4 protease is the prototype of a novel group of chymotrypsin-like enzymes, the 3C-like serine proteases. J Biol Chem 271:4864–4871. [DOI] [PubMed] [Google Scholar]
- 21.Snijder EJ, Wassenaar AL, Spaan WJ. 1992. The 5′ end of the equine arteritis virus replicase gene encodes a papainlike cysteine protease. J Virol 66:7040–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Aken D, Zevenhovendobbe J, Gorbalenya AE, Snijder EJ. 2006. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J Gen Virol 87:3473–3482. doi: 10.1099/vir.0.82269-0. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z, Liu S, Sun W, Chen L, Yoo D, Li F, Ren S, Guo L, Cong X, Li J, Zhou S, Wu J, Du Y, Wang J. 2016. Nuclear export signal of PRRSV NSP1α is necessary for type I IFN inhibition. Virology 499:278–287. doi: 10.1016/j.virol.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 24.Ke H, Yoo D. 2017. The viral innate immune antagonism and an alternative vaccine design for PRRS virus. Vet Microbiol 209:75–89. doi: 10.1016/j.vetmic.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loo Y-M, Gale M Jr.. 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawai T, Akira S. 2006. Innate immune recognition of viral infection. Nat Immunol 7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 29.Rui Y, Su J, Wang H, Chang J, Wang S, Zheng W, Cai Y, Wei W, Gordy JT, Markham R, Kong W, Zhang W, Yu XF. 2017. Disruption of MDA5-mediated innate immune responses by the 3C proteins of coxsackievirus A16, coxsackievirus A6, and enterovirus D68. J Virol 91:e00546-17. doi: 10.1128/JVI.00546-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang D, Fang L, Wei D, Zhang H, Luo R, Chen H, Li K, Xiao S. 2014. Hepatitis A virus 3C protease cleaves NEMO to impair induction of beta interferon. J Virol 88:10252–10258. doi: 10.1128/JVI.00869-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu X, Fang L, Wang D, Yang Y, Chen J, Ye X, Foda MF, Xiao S. 2017. Porcine deltacoronavirus nsp5 inhibits interferon-β production through the cleavage of NEMO. Virology 502:33–38. doi: 10.1016/j.virol.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei X, Xiao X, Xue Q, Jin Q, He B, Wang J. 2013. Cleavage of interferon regulatory factor 7 by enterovirus 71 3C suppresses cellular responses. J Virol 87:1690–1698. doi: 10.1128/JVI.01855-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang C, Zhang Q, Guo XK, Yu ZB, Xu AT, Tang J, Feng WH. 2014. Porcine reproductive and respiratory syndrome virus nonstructural protein 4 antagonizes beta interferon expression by targeting the NF-κB essential modulator. J Virol 88:10934–10945. doi: 10.1128/JVI.01396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoneyama M, Fujita T. 2007. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev 18:545–551. doi: 10.1016/j.cytogfr.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 35.Tian X, Lu G, Gao F, Peng H, Feng Y, Ma G, Bartlam M, Tian K, Yan J, Hilgenfeld R, Gao GF. 2009. Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of porcine reproductive and respiratory syndrome virus (PRRSV). J Mol Biol 392:977–993. doi: 10.1016/j.jmb.2009.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beura LK, Sarkar SN, Kwon B, Subramaniam S, Jones C, Pattnaik AK, Osorio FA. 2010. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J Virol 84:1574–1584. doi: 10.1128/JVI.01326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol 89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 38.Bender S, Reuter A, Eberle F, Einhorn E, Binder M, Bartenschlager R. 2015. Activation of type I and III interferon response by mitochondrial and peroxisomal MAVS and inhibition by hepatitis C virus. PLoS Pathog 11:e1005264. doi: 10.1371/journal.ppat.1005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang T, Coyne CB. 2011. The coxsackievirus B 3Cpro protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog 7:e1001311. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anggakusuma, Frentzen A, Gürlevik E, Yuan Q, Steinmann E, Ott M, Staeheli P, Schmid-Burgk J, Schmidt T, Hornung V, Kuehnel F, Pietschmann T. 2015. Control of hepatitis C virus replication in mouse liver-derived cells by MAVS-dependent production of type I and type III interferons. J Virol 89:3833–3845. doi: 10.1128/JVI.03129-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang D, Fang L, Shi Y, Zhang H, Gao L, Peng G, Chen H, Li K, Xiao S. 2016. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J Virol 90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang D, Fang L, Li K, Zhong H, Fan J, Ouyang C, Zhang H, Duan E, Luo R, Zhang Z, Liu X, Chen H, Xiao S. 2012. Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J Virol 86:9311–9322. doi: 10.1128/JVI.00722-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye G, Deng F, Shen Z, Luo R, Zhao L, Xiao S, Fu ZF, Peng G. 2016. Structural basis for the dimerization and substrate recognition specificity of porcine epidemic diarrhea virus 3C-like protease. Virology 494:225–235. doi: 10.1016/j.virol.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zunszain PA, Knox SR, Sweeney TR, Yang J, Roque-Rosell N, Belsham GJ, Leatherbarrow RJ, Curry S. 2010. Insights into cleavage specificity from the crystal structure of foot-and-mouth disease virus 3C protease complexed with a peptide substrate. J Mol Biol 395:375–389. doi: 10.1016/j.jmb.2009.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qian S, Fan W, Liu T, Wu M, Zhang H, Cui X, Zhou Y, Hu J, Wei S, Chen H, Li X, Qian P. 2017. Seneca Valley virus suppresses host type I interferon production by targeting adaptor proteins MAVS, TRIF, and TANK for cleavage. J Virol 91:e00823-17. doi: 10.1128/JVI.00823-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang L, Xiong T, Yu H, Zhang Q, Zhang K, Li C, Hu L, Zhang Y, Zhang L, Liu Q, Wang S, He X, Bu Z, Cai X, Cui S, Li J, Weng C. 2017. Encephalomyocarditis virus 3C protease attenuates type I interferon production through disrupting TANK-TBK1-IKKε-IRF3 complex. Biochem J 474:2051–2065. doi: 10.1042/BCJ20161037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang R, Jiang Q, Zhou X, Wang C, Guan Y, Tao J, Xi J, Feng JM, Jiang Z. 2017. MAVS activates TBK1 and IKKε through TRAFs in NEMO dependent and independent manner. PLoS Pathog 13:e1006720. doi: 10.1371/journal.ppat.1006720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, Acquaviva R, Formisano S, Vito P, Leonardi A. 2006. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem 281:18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 49.Keusekotten K, Elliott PR, Glockner L, Fiil BK, Damgaard RB, Kulathu Y, Wauer T, Hospenthal MK, Gyrd-Hansen M, Krappmann D, Hofmann K, Komander D. 2013. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell 153:1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maubach G, Schmädicke AC, Naumann M. 2017. NEMO links nuclear factor-κB to human diseases. Trends Mol Med 23:1138–1155. doi: 10.1016/j.molmed.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 51.Godwin RC, Melvin RL, Gmeiner WH, Salsbury FR Jr.. 2017. Binding site configurations probe the structure and dynamics of the zinc finger of NEMO (NF-kappaB essential modulator). Biochemistry 56:623–633. doi: 10.1021/acs.biochem.6b00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bal E, Laplantine E, Hamel Y, Dubosclard V, Boisson B, Pesacatore A, Picard C, Hadj-Rabia S, Royer G, Steffann J, Bonnefont JP, Ursini VM, Vabres P, Munnich A, Casanova JL, Bodemer C, Weil R, Agou F, Smahi A. 2017. Lack of interaction between NEMO and SHARPIN impairs linear ubiquitination and NF-κB activation, and leads to incontinentia pigmenti. J Allergy Clin Immunol 140:1671.e2–1682.e2. doi: 10.1016/j.jaci.2016.11.056. [DOI] [PubMed] [Google Scholar]
- 53.Li B, Fang L, Liu S, Zhao F, Jiang Y, He K, Chen H, Xiao S. 2010. The genomic diversity of Chinese porcine reproductive and respiratory syndrome virus isolates from 1996 to 2009. Vet Microbiol 146:226–237. doi: 10.1016/j.vetmic.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 54.van Dinten LC, den Boon JA, Wassenaar AL, Spaan WJ, Snijder EJ. 1997. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc Natl Acad Sci U S A 94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding Z, An K, Xie L, Wu W, Zhang R, Wang D, Fang Y, Chen H, Xiao S, Fang L. 2017. Transmissible gastroenteritis virus infection induces NF-kappaB activation through RLR-mediated signaling. Virology 507:170–178. doi: 10.1016/j.virol.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jing H, Zhou Y, Fang L, Ding Z, Wang D, Ke W, Chen H, Xiao S. 2017. DExD/H-box helicase 36 signaling via myeloid differentiation primary response gene 88 contributes to NF-κB activation to type 2 porcine reproductive and respiratory syndrome virus infection. Front Immunol 8:1365. doi: 10.3389/fimmu.2017.01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matayoshi E, Wang G, Krafft G, Erickson J. 1990. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science 247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]