Viruses are master regulators of the host gene expression machinery. This is crucial to promote viral infection and to dampen host immune responses. Many viruses, including herpesviruses, express RNases that reduce host gene expression through widespread mRNA decay. However, it emerged that some mRNAs escape this fate, although it has been difficult to determine whether these escaping transcripts benefit viral infection or instead participate in an antiviral mechanism. To tackle this question, we compared the effect of the herpesviral RNases on the human transcriptome and identified a cluster of transcripts consistently escaping degradation from all tested endonucleases. Among the protected mRNAs, we identified the transcript C19ORF66 and showed that it restricts Kaposi’s sarcoma-associated herpesvirus (KSHV) infection. Collectively, these results provide a framework to explore how the control of RNA fate in the context of viral-induced widespread mRNA degradation may influence the outcome of viral infection.
KEYWORDS: Kaposi’s sarcoma-associated herpesvirus, RNA decay, endonuclease, herpesviruses, mRNA stability, C19ORF66
ABSTRACT
One striking characteristic of certain herpesviruses is their ability to induce rapid and widespread RNA decay in order to gain access to host resources. This phenotype is induced by viral endoribonucleases, including SOX in Kaposi’s sarcoma-associated herpesvirus (KSHV), muSOX in murine gammaherpesvirus 68 (MHV68), BGLF5 in Epstein-Barr virus (EBV), and vhs in herpes simplex virus 1 (HSV-1). Here, we performed comparative transcriptome sequencing (RNA-seq) upon expression of these herpesviral endonucleases in order to characterize their effect on the host transcriptome. Consistent with previous reports, we found that approximately two-thirds of transcripts were downregulated in cells expressing any of these viral endonucleases. Among the transcripts spared from degradation, we uncovered a cluster of transcripts that systematically escaped degradation from all tested endonucleases. Among these escapees, we identified C19ORF66 and reveal that this transcript is protected from degradation by its 3′ untranslated region (UTR). We then show that C19ORF66 is a potent KSHV restriction factor by impeding early viral gene expression, suggesting that its ability to escape viral cleavage may be an important component of the host response to viral infection. Collectively, our comparative approach is a powerful tool to pinpoint key regulators of the viral-host interplay and led us to uncover a novel KSHV regulator.
IMPORTANCE Viruses are master regulators of the host gene expression machinery. This is crucial to promote viral infection and to dampen host immune responses. Many viruses, including herpesviruses, express RNases that reduce host gene expression through widespread mRNA decay. However, it emerged that some mRNAs escape this fate, although it has been difficult to determine whether these escaping transcripts benefit viral infection or instead participate in an antiviral mechanism. To tackle this question, we compared the effect of the herpesviral RNases on the human transcriptome and identified a cluster of transcripts consistently escaping degradation from all tested endonucleases. Among the protected mRNAs, we identified the transcript C19ORF66 and showed that it restricts Kaposi’s sarcoma-associated herpesvirus (KSHV) infection. Collectively, these results provide a framework to explore how the control of RNA fate in the context of viral-induced widespread mRNA degradation may influence the outcome of viral infection.
INTRODUCTION
Many viruses, including alpha- and gammaherpesviruses, influenza A virus, and severe acute respiratory syndrome (SARS) coronavirus, induce widespread mRNA decay through the use of virally encoded endonucleases (1–5). This process, known as “host shutoff,” allows viruses to rapidly restrict gene expression in order to dampen immune responses and provide access to the host’s resources for viral replication (2, 6, 7). One well-studied viral endonuclease is the SOX protein encoded by Kaposi’s sarcoma-associated herpesvirus (KSHV). SOX is conserved throughout the herpesvirus family, but only gammaherpesviral SOX homologs display RNase activity in cells (8–10), and studies indicate that SOX activity is important for the in vivo viral life cycle (11, 12). Although SOX targets a degenerate RNA motif present on most mRNAs (13–15), multiple studies have shown that some transcripts robustly escape SOX-induced decay (16–21). Studying these “escapees” in aggregate is complicated, however, by the fact that multiple mechanisms can promote apparent escape. These include the lack of a targeting motif, indirect transcriptional effects, and active evasion of ribonucleolytic cleavage (16, 20–24). The latter phenotype, termed “dominant escape,” is particularly notable, as it involves a specific RNA element whose presence in the 3ʹ untranslated region (UTR) of an mRNA protects against SOX cleavage, regardless of whether the RNA contains a targeting motif (19–21). This protective RNA element was termed SRE (for SOX resistance element), but we recently showed that the SRE is also effective against a broad range of viral endonucleases. Perhaps more surprisingly, the SRE is unable to restrict endonucleolytic cleavage originating from a cellular endonuclease, making it the first identified virus-specific RNase escape element (19). We showed that this broad-acting RNA element is not characterized by a defined sequence motif (19), rendering it difficult to identify new escaping transcripts by traditional sequence search. Consequently, the host versus viral endonuclease dichotomy is the only defining characteristic of this novel type of RNA element.
Little is currently known about these types of RNA elements, how widespread they may be in the genome, and how they may contribute to the overall viral-host arms race for the control of resources. To date, only two SRE-bearing dominant escapees are known: the host interleukin-6 (IL-6) (18, 20, 21) and the growth arrest and DNA damage-inducible 45 beta (GADD45B) (19) transcripts. Both the IL-6 and GADD45B SREs were mapped to their 3ʹ UTRs and were shown to protect against an array of viral—but not host—RNases. Furthermore, while little sequence homology was detected among these SREs, we showed that they share similarity in their secondary structures; reinforcing the idea that the SRE may function as a platform to recruit a protective protein complex as previously observed (19–21). Functionally, while the beneficial role of IL-6 for KSHV during infection is well documented (25–33), the role of GADD45B is still unclear. In fact, GADD45B is repressed during KSHV latency (34), and GADD45B known proapoptotic roles may indicate that this transcript escapes to participate in an antiviral response to host shutoff.
Here, taking advantage of the ability of the SRE element to block decay from a diverse set of viral endonucleases, we sought to identify novel escaping mRNAs containing SRE or SRE-like elements in the transcriptome. Using comparative transcriptome sequencing (RNA-seq), we uncovered a cluster of 75 host mRNAs that escape degradation from four herpesviral endonucleases. Similarly to the previously identified SRE-bearing transcripts, these transcripts were spared from a range of viral—but not host—endonucleases, further supporting that our approach successfully identified novel dominant escapees. Among this list of newly identified escapees, we demonstrate that our top candidate, C19ORF66, is a negative regulator of the KSHV life cycle.
C19ORF66 (also annotated RyDEN, IRAV, and SVA-1) is an interferon-stimulated gene (ISG) that has been found to be upregulated upon infection by a number a viruses (35–40), including herpesviruses (41, 42), in several large-scale screens. Recently, C19ORF66 was demonstrated to repress dengue virus (DENV) replication and gene expression by interacting with the cytoplasmic poly(A) binding protein, PABPC (43), and the RNA helicase MOV10 (44), suggesting that C19ORF66 may restrict DENV infection by directly influencing the host gene expression machinery and/or directly targeting viral RNA for degradation, making it an intriguing candidate dominant escapee during KSHV infection.
Here we show that C19ORF66 is upregulated during KSHV infection and accumulates over the course of 96 h postreactivation. Knocking down C19ORF66 during KSHV infection leads to higher expression levels of early and delayed early viral genes, which results in higher yields of infectious viral particles and suggests that C19ORF66 has antiviral activity on KSHV. Taken together, these results demonstrate that SRE and SRE-like elements may be more common than anticipated in the genome and that transcripts encoding these escape elements may also function as viral restriction factors.
(This article was submitted to an online preprint archive [45].)
RESULTS
Comparative RNA-seq identifies a cluster of common escaping transcripts.
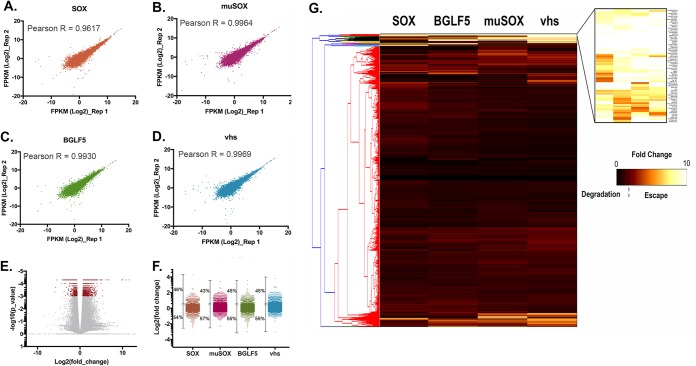
Prior analyses indicated that certain host mRNA transcripts robustly escape viral-induced RNA decay by encoding an RNA element in their 3ʹ UTRs. We demonstrated that this RNA element, herein referred to as SRE (SOX resistance element), provides protection against KSHV SOX as well as a variety of viral endonucleases. To identify mRNA transcripts containing SRE or SRE-like elements, we performed comparative RNA-seq-based analyses upon expression of the herpesviral RNA endonucleases. Pure populations of cells expressing either KSHV SOX, murine gammaherpesvirus 68 (MHV68) muSOX, Epstein-Barr virus (EBV) BGLF5, herpes simplex virus 1 (HSV-1) vhs, or an empty vector control were generated using Thy1.1-based cell sorting as described before (46). (Fig. 1 and Table S1 in the supplemental material). The reproducibility between replicate experiments was high (Fig. 1A to D), which is in line with previous reports showing that these endonucleases target transcripts in a selective/sequence-specific manner as previously observed (13). As expected, a number of transcripts were significantly affected upon expression of the various herpesviral endonucleases (Fig. 1E and F): we observed that between 55% and 60% of total mRNAs were degraded, with muSOX being the most effective of the endonucleases tested here (Fig. 1F). This rate of degradation is within the range of what was observed before (16). Gene Ontology (GO) analysis on the transcripts spared from degradation revealed that they encode proteins that have a wide array of functions, ranging from ion binding to RNA binding (see Fig. S1).
FIG 1.
Comparative RNA-seq of the herpesviral RNA endonuclease. (A to D) Scatter plots to compare gene expression expressed as log2 fragments per kilobase of transcript per million mapped reads (FPKM) among replicate experiments. The Pearson correlation coefficient, R, is shown for each plot. (E) Volcano plot of all genes differentially expressed in mock samples versus endonuclease-expressing cells. Dots represent fold change and P values as determined by CuffDiff. Significant fold changes [log10(p_value) of <0.001] are highlighted in red. (F) Distributions of fold change per endonuclease tested relative to that in the mock sample and corresponding percentages on degrading transcripts. (G) Hierarchical clustering and heatmap of RNA-seq data. Fold change in expression levels for each condition (SOX, muSOX, BGLF5, and vhs [columns]) relative to that in the mock were normalized and are represented as a heatmap. Transcripts were clustered by similarity using the complete linkage method (dendrogram on the left). A cluster representing transcripts escaping degradation by all tested endonucleases emerged and is enlarged at the top right corner.
To identify transcripts that escape degradation from all 4 endonucleases, we performed hierarchical clustering on the transcript expression data. Figure 1G shows a heatmap of the correlation matrix across all transcripts. A cluster encompassing 75 transcripts (see Table S2) represents the mRNA that escaped degradation from all 4 herpesviral endonucleases. We hypothesize that this cluster of transcripts is likely to include mRNA containing SRE or SRE-like elements.
Candidate escapees are broadly protected from cleavage by viral but not cellular endonucleases.
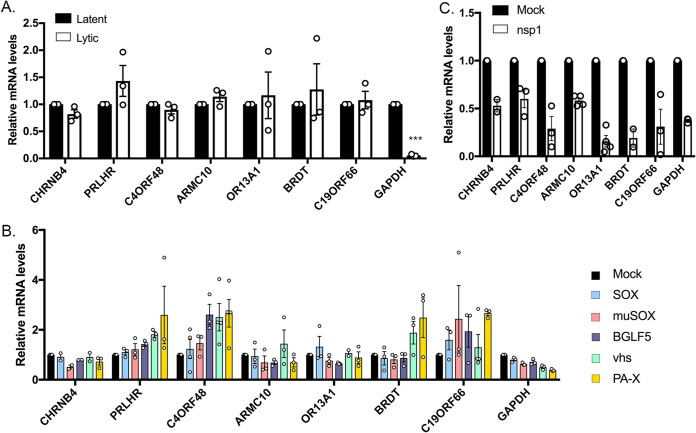
We next set out to investigate further this cluster of common escapees. The RNA-seq hits identified by hierarchical clustering were ranked by confidence (reproducibility among experimental replicates and escaped all endonucleases in all replicates). To confirm the RNA-seq data, we first examined whether the top 10% (Table S2) of this list of common escapees were resistant to host shutoff upon lytic reactivation of a KSHV-positive renal carcinoma cell line stably expressing the KSHV BAC16 (iSLK.219). iSLK.219 cells harbor a doxycycline (dox)-inducible version of the major viral lytic transactivator RTA, which promotes entry into the lytic cycle upon doxycycline treatment (47, 48). As opposed to the housekeeping gene encoding GAPDH (glyceraldehyde-3-phosphate dehydrogenase) that is naturally susceptible to host shutoff, we observed that the mRNA levels of these candidate SRE-bearing mRNAs remained unchanged in reactivated iSLK.219 cells as measured by reverse transcriptase quantitative PCR (RT-qPCR) (Fig. 2A), confirming that these transcripts are resistant to host shutoff in lytically infected cells. Additionally, we recently showed that SRE-containing transcripts are resistant to endonucleases beyond the herpesvirus family (19). We next tested the ability of these novel escapees to evade heterologous host shutoff from the influenza A virus (IAV) endonuclease (PA-X). As shown in Fig. 2B, contrary to GAPDH, the candidate transcripts were resistant to all endonucleases tested, including PA-X. Finally, one characteristic of SRE-containing mRNAs is that they are still susceptible to cleavage by cellular endonucleases (19). To test whether this was also the case for our novel candidate SRE-bearing transcripts, we monitored cleavage upon expression of the Nsp1 protein from SARS coronavirus. Nsp1 is not a nuclease but rather activates mRNA cleavage by an as yet unknown cellular endonuclease via a mechanism reminiscent of no-go decay (49, 50). Nsp1 thus allows us to induce RNA decay using a viral trigger but carried out by a cellular endonuclease. nsp1 was transfected into 293T cells, and depletion of the candidate transcripts was measured by RT-qPCR. Similar to what we observed before, the candidate escapee mRNAs were not protected in nsp1-expressing cells (Fig. 2C). Collectively, these results suggest that the escaping mRNAs identified in our comparative RNA-seq data set are broadly protected against viral but not cellular endonucleases, and we predict that these transcripts may contain an SRE or an SRE-like element that provides broad protection.
FIG 2.
Top escapees identified by RNA-seq behave like SRE-containing transcripts. (A) Total RNA was extracted from unreactivated or reactivated KSHV-positive iSLK.219 cells and subjected to RT-qPCR to measure endogenous levels of the top candidates identified by RNA-seq. (B) 293T cells were transfected with an empty vector (mock) or a plasmid expressing each of the viral endonucleases color coded on the right. After 24 h, total RNA was harvested and subjected to RT-qPCR to measure endogenous RNA levels. (C) 293T cells were transfected with an empty vector (mock) or a plasmid expressing nsp1. After 24 h, total RNA was harvested and subjected to RT-qPCR to measure endogenous RNA levels. ***, P < 0.001.
The C19ORF66 mRNA 3ʹ UTR contains an SRE.
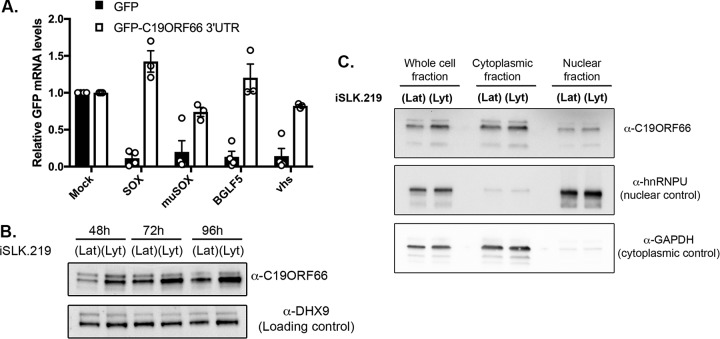
The pool of escaping transcripts did not appear to be strongly enriched for particular functions or processes when evaluated by GO term analysis. We thus proceeded to manually mine the literature to identify functions that might be important during viral infection. We were drawn to C19ORF66 (also known as RyDEN, IRAV, and SVA-1), as it was reported to be an antiviral interferon-stimulated gene (ISG) in the context of multiple viral infections (40, 43, 44). Furthermore, the transcript for C19ORF66 appeared in our comparative RNA-seq as the top escapee in all the replicates and with all the endonucleases tested. We first evaluated whether this transcript contained a putative SRE-like element in its 3ʹ UTR by testing whether it could protect green fluorescent protein (GFP) mRNA, which is normally susceptible to viral endonuclease cleavage. We fused the C19ORF66 3ʹ UTR to GFP (C19-3ʹUTR) and found that it was sufficient to confer protection from SOX and other viral endonucleases in transfected 293T cells (Fig. 3A). Thus, similar to the IL-6 and GADD45B 3ʹ UTRs, previously identified dominant escapees, C19ORF66 contains an SRE-like element in its 3ʹ UTR that is sufficient to provide protection against a range of viral endonucleases. As we previously demonstrated, there is no significant sequence conservation between the 3ʹ UTRs of these known dominant escapees. However, the highest similarities were located near the second half of C19ORF66 3ʹ UTR, and RNAfold secondary structure prediction of this UTR section revealed a long stem-loop structure with a bulge in the middle, consistent with previously found SRE structures (see Fig. S2).
FIG 3.
C19ORF66 mRNA is protected from herpesviral endonucleases by its 3ʹ UTR and accumulates in the cytoplasm of iSLK.219 cells. (A) 293T cells were transfected with a GFP reporter (GFP) or a GFP reporter containing C19ORF66 3ʹ UTR sequence along with a control empty vector (mock) or a plasmid expressing SOX, muSOX, BGLF5, or vhs. After 24 h, total RNA was harvested and subjected to RT-qPCR to measure GFP mRNA levels. (B) KSHV-positive iSLK.219 cells were reactivated for the indicated times to induce KSHV lytic cycle (lyt) or not (KSHV latent phase maintained [lat]). Cells were harvested, lysed, resolved on SDS-PAGE, and Western blotted with the indicated antibodies. (C) Unreactivated (lat) or reactivated (lyt) KSHV-positive iSLK.219 cells were fractionated into nuclear and cytoplasmic fractions and Western blotted with the indicated antibodies.
Because C19ORF66 expression was previously shown to be increased in the context of various viral infections, we next sought to investigate its expression upon KSHV lytic reactivation when host shutoff occurs. iSLK.219 cells were reactivated and total protein harvested at various time points over the course of 96 h. C19ORF66 expression was increased upon KSHV lytic reactivation and continued to accumulate over time (Fig. 3B). We previously showed that some proteins important for the resistance from SOX decay can change their subcellular localization during the KSHV lytic cycle (20), and so we proceeded to monitor C19ORF66 expression and did not find differential shuttling upon KSHV lytic reactivation (Fig. 3C). Thus, C19ORF66 escapes SOX degradation by encoding an SRE-like element on its 3ʹ UTR that allows it to escape host shutoff and accumulate in lytically infected cells.
C19ORF66 restricts KSHV infection.
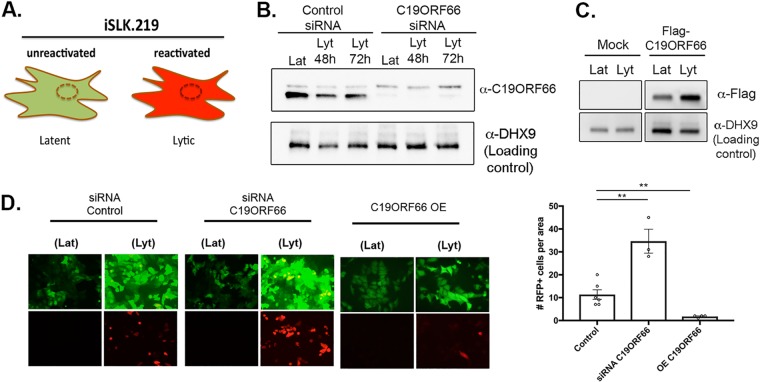
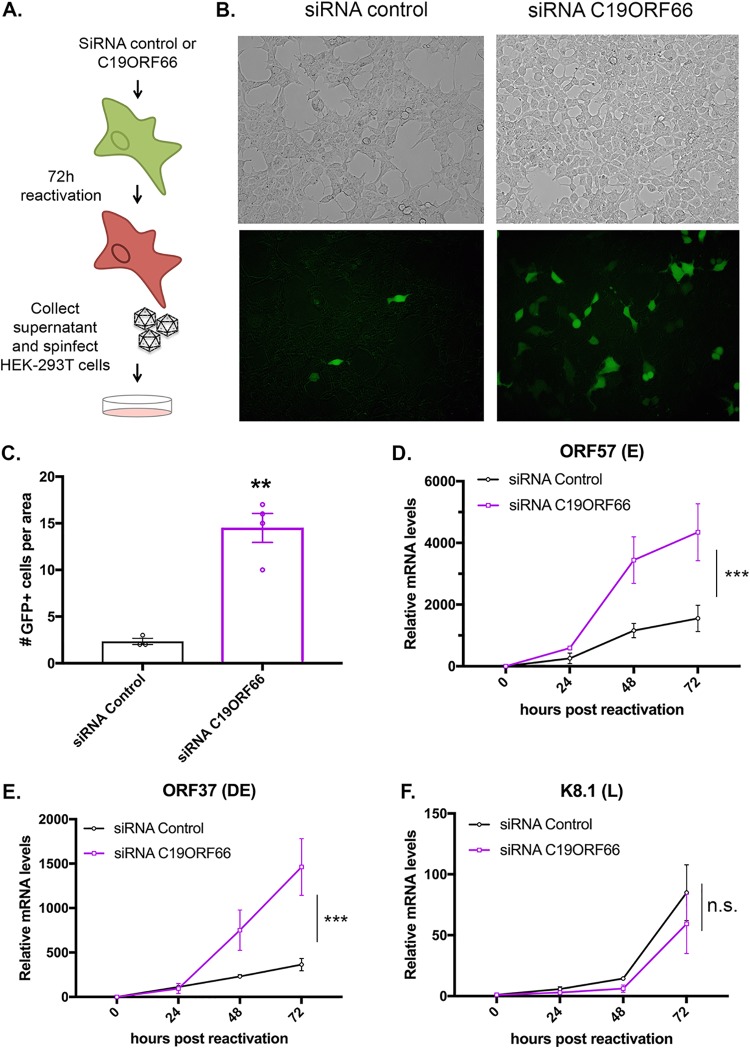
Given that C19ORF66 functions as an antiviral protein during HIV and dengue virus infection, we hypothesized that it might also play a role during KSHV infection. We thus further investigated the role of C19ORF66 in iSLK.219 cells. The recombinant KSHV.219 virus stably maintained in these cells constitutively expresses green fluorescent protein (GFP) from the EF-1 alpha promoter and can be used as a proxy for the presence of KSHV within cells. The KSHV.219 virus also encodes red fluorescent protein (RFP) under the control of the viral lytic PAN promoter (Fig. 4A). Small interfering RNA (siRNA)-mediated depletion of C19ORF66 in iSLK.219 cells during latency and at 48 h and 72 h postreactivation was efficient, reducing expression levels by 94.6%, 97%, and 97.8%, respectively (Fig. 4B). Seventy-two hours postreactivation, GFP- and RFP-positive cells were analyzed by fluorescence microscopy in siRNA C19ORF66-treated cells (or siRNA controls). C19ORF66 depletion resulted in a marked increase in the number of RFP-positive cells (Fig. 4D). Conversely, overexpression of C19ORF66 in these cells (Fig. 4C) resulted in almost no RFP detection (Fig. 4D). Taken together, these results suggest that C19ORF66 expression negatively regulates the progression of the KSHV life cycle.
FIG 4.
C19ORF66 restricts KSHV reactivation. (A) Diagram outlining the fluorescence pattern of iSLK.219 cells. iSLK.219 cells were either treated with siRNAs targeting C19ORF66 (or control nontarget siRNAs) for 48 h (B) or transfected with a Flag-tagged C19ORF66 (C). Cells were then reactivated with doxycycline and sodium butyrate and lysed, and lysates were resolved on SDS-PAGE and Western blotted with the indicated antibodies. (D) Cells were treated with the indicated siRNA or transfected with C19ORF66 (overexpression [OE]) and were checked for reactivation efficiency by monitoring the expression of GFP and RFP. Quantification of RFP-positive cells is to the right. Values represent at least four independent views of the infected cells. **, P < 0.01.
We next hypothesized that the reactivation defect due to C19ORF66 expression may lead to restriction of the formation of viral particles. To test this, we performed a supernatant transfer assay (Fig. 5A). iSLK.219 cells were treated with siRNA C19ORF66 (or control siRNA) and reactivated for 72 h. Supernatants containing GFP-expressing KSHV virions were collected and used to spinfect 293T cells (Fig. 5B and C). Twenty-four hours later, we observed a higher number of GFP-positive cells in the 293T cells infected with the supernatant coming from the iSLK.219 cells treated with the siRNA against C19ORF66. Since C19ORF66 seemed to affect an important step in the KSHV life cycle, we next assessed whether C19ORF66 also affects viral gene expression. Using RT-qPCR, we quantified the expression of several KSHV viral genes. Viral gene expression in KSHV unfolds as a cascade with the “early” (E) genes expressed right after lytic reactivation, followed by “delayed early” (DE) genes and finally, after viral replication, the “late” (L) genes. We harvested time points from 0 to 72 h after iSLK.219 reactivation and measured RNA levels of genes representative of each gene class upon knockdown of C19ORF66. We observed a shift in viral gene expression with early and delayed early viral genes—but not with the late gene—which were expressed earlier and at higher levels in the C19ORF66 knocked down cells as measured by RT-qPCR (Fig. 5D to F). Taken together, these results suggest that C19ORF66 may restrict the expression of certain early viral genes, which in turn results in fewer newly formed viral particles being produced by KSHV-infected cells.
FIG 5.
C19ORF66 knockdown results in higher viral production yield and higher viral gene expression levels. (A) Diagram depicting the supernatant transfer assay. (B) Supernatant transfer assay was used as a proxy for virion production and performed as described in panel A. Infection of 293T cells was monitored by imaging GFP on a fluorescence microscope. (C) Quantification of GFP-positive cells. Values represent four independent views of the infected cells. (D to F) Total RNA was extracted from iSLK.219 cells treated with siRNAs targeting C19ORF66 (or control nontarget siRNAs) for 48 h and reactivated for the indicated times. RNA was then subjected to RT-qPCR to quantify expression of the indicated viral genes. n.s., not significant; **, P < 0.01; ***, P < 0.001.
DISCUSSION
Regulation of mRNA stability has emerged as a focal point for control of the host gene expression machinery. By accelerating RNA decay, viruses can increase their access to the host translation machinery and dampen the host response to infection. RNA degradation is often driven by virally encoded endonucleases that can target a wide array of mRNA by cleaving within a specific structured element (13, 15). It is estimated that up to two-thirds of total mRNAs are degraded upon expression of these viral endonucleases (11, 13, 16). While recent studies have focused on how these viral endonucleases target mRNA, it remains unclear how and why some mRNA transcripts can escape viral-induced RNA decay. We previously demonstrated that certain transcripts escape by possessing in their 3ʹ UTR an RNA element that protects them from viral endonucleases while still allowing for normal RNA decay and cellular endonuclease cleavage (19–21). This raised a number of questions regarding how common these RNA escape elements are in the host genome and how their presence impacts the viral life cycle. Here, we reveal that a cluster of 75 host transcripts can systematically escape viral-induced endonucleolytic cleavage. We hypothesize that these may contain similar RNA escape elements as the one we previously characterized in IL-6 and GADD45B and therefore could be important regulators of the viral-host interplay. IL-6 and GADD45B escape elements (referred to as SRE and G-SRE, respectively) were shown to adopt a specific secondary structure that we hypothesized to be crucial in recruiting host proteins to the 3ʹ UTR of these escaping transcripts (19). This RNA-protein protective complex appears to be composed of core proteins as well as accessory proteins that may be transcript dependent. One future goal is thus to expand our knowledge of the known escapees by exploring the RNA-protein complexes on these newly identified escaping transcripts with the hope of understanding the protein prerequisite to forming a protective complex. More globally, the determination of whether such RNA elements impact RNA fate in uninfected cells will also be key in deciphering their role. To date, no such RNA elements have been found in viral genes, suggesting that this could be a cell-specific mechanism that has evolved in response to viral infection.
No common functions were enriched in the pool of escaping transcripts, rendering it difficult to make any definite conclusion on whether these mRNAs escape degradation to benefit the host or the virus. Instead, we hypothesize that these spared mRNAs may have both pro- and antiviral functions. Furthermore, because of the large diversity of hosts infected by members of the herpesviridae, it would be interesting to investigate whether the orthologs of the escaping transcripts in other species also contain these RNA escape elements.
Here, we also characterized the top escaping transcript in our screen, C19ORF66. Through knockdown and overexpression assays, our data indicate that C19ORF66 is restricting the expression of KSHV early and delayed early genes, resulting in lower levels of viral reactivation and reduced yield of infectious viral particles. C19ORF66 is known to be upregulated in response to type I and type II interferons (IFNs) (51, 52) and to be upregulated in response to infection by a number of unrelated viruses (35–42). Furthermore, C19ORF66 was found to interact with the NS3 protein of hepatitis C virus (53), localize to the replication complex of DENV (33), and occasionally colocalize in the cytoplasmic compartment with HIV-1 Rev and Tat proteins (40), pointing to a potential conserved role for C19ORF66 as a key player in the host-pathogen response. While it is still unclear how C19ORF66 participates in the regulation of these viruses, it was hypothesized that it may be mediated through its interaction with PABPC and LARP, two major RNA binding proteins (43). PABPC and LARP were recently shown to be relocated upon SOX-induced widespread RNA decay and to be linked to the transcription feedback loop that occurs during host shutoff (46). PABPC in particular was shown to be pivotal in triggering transcriptional repression in the nucleus after host shutoff, a process that favors expression of viral genes. It is therefore possible that C19ORF66, by interacting with PABPC, slows down PABPC relocalization to the nucleus and restricts expression of viral genes. By interacting with PABPC and LARP, C19ORF66 was also hypothesized to regulate the decay of dengue RNA by possibly influencing either translation or localization to P bodies and stress granules (54). Determining whether C19ORF66 influences the PABPC shuttling pattern is an important future goal, as well as deciphering C19ORF66 interaction pattern upon KSHV infection and lytic reactivation.
Intriguingly, the viral endonucleases tested in this study come from both related and unrelated viruses, do not share the same targeting elements on their target mRNA, and are not known to be recruited to mRNA through similar pathways. It is thus notable that within the group of common escapees was one with a conserved antiviral role. This underscores the utility of comparative approaches toward revealing broad regulators of viral infection.
Finally, none of the 3 known SREs (in IL-6, GADD45B, and now in C19ORF66) share significant sequence similarity, although they do all share similar predicted secondary structures. Thus, as predicted before, these RNA elements may function as scaffolds for recruiting a protective protein complex. Therefore, by manipulating the sequences of these RNA escape elements but maintaining the structure, these nuclease escape elements could be developed as tools to broadly inhibit viral endonucleases and open the possibility of turning these RNA elements into broad-acting antiviral RNA therapeutics.
MATERIALS AND METHODS
Cells and transfections.
293T cells (ATCC) were grown in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS). The KSHV-infected renal carcinoma human cell line iSLK.219 (kind gift from B. Glaunsinger) bearing doxycycline-inducible RTA was grown in DMEM supplemented with 10% FBS (48). KSHV lytic reactivation of the iSLK.219 cells was induced by the addition of 0.2 μg/ml doxycycline (BD Biosciences) and 110 μg/ml sodium butyrate for 72 h.
For DNA transfections, cells were plated and transfected after 24 h when 70% confluent using PolyJet (SignaGen). For small interfering RNA (siRNA) transfections, cells were reverse transfected in 6-well plates by INTERFERin (Polyplus Transfection) with 10 μM siRNAs. siRNAs were obtained from IDT as Dicer-substrate siRNA (DsiRNA; siRNA C19ORF66, hs.Ri.C19orf66.13.1).
Fractionation experiments were performed according to the REAP method (55). Briefly, cells were washed twice with ice-cold phosphate-buffered saline (PBS), and the cell pellet was lysed in 0.1% NP-40 PBS lysis buffer. The nuclei were then isolated by differential centrifugation at 10,000 × g for 10 s, and the supernatant retained as the cytoplasmic fraction. For Western blotting, the nuclei were sonicated in 0.1% NP-40 PBS lysis buffer.
Supernatant transfers were carried in iSLK.219 cells. Cells treated with siRNA were reactivated with doxycycline and sodium butyrate for 72 h, and the supernatants were collected, filtered to remove any potential whole cells, and spinfected onto 293T cells at 1,500 rpm for 1 h at 37°C. Twenty-four hours later, cells were imaged on a fluorescence microscope.
Plasmids.
The C19ORF66 3ʹ UTR was obtained as gBlocks from IDT and cloned into a pcDNA3.1 plasmid downstream of the GFP coding sequence. The C19ORF66 coding region was obtained as a gBlock from IDT and cloned in a pcDNA4 Nter-3×Flag vector. All cloning steps were performed using in-fusion cloning (Clontech TaKaRa) and were verified by sequencing.
RT-qPCR.
Total RNA was harvested using TRIzol according to the manufacture's protocol. cDNAs were synthesized from 1 μg of total RNA using AMV reverse transcriptase (Promega) and used directly for quantitative PCR (qPCR) analysis with the SYBR green qPCR kit (Bio-Rad). Signals obtained by qPCR were normalized to those for 18S.
Western blotting.
Cell lysates were prepared in lysis buffer (NaCl, 150 mM; Tris, 50 mM; NP-40, 0.5%; dithiothreitol [DTT], 1 mM; and protease inhibitor tablets) and quantified by Bradford assay. Equivalent amounts of each sample were resolved by SDS-PAGE and Western blotted with the following antibodies at 1:1,000 in TBST (Tris-buffered saline, 0.1% Tween 20): rabbit anti-C19ORF66 (Abcam), rabbit anti-DHX9/RNA helicase A (Abcam), and rabbit anti-GAPDH (Abcam). Primary antibody incubations were followed by horseradish peroxidase (HRP)-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (1:5,000; Southern Biotechnology).
RNA-seq.
Cells were transfected with constructs encoding fusion proteins between the herpesviral endonucleases (SOX, muSOX, BGLF5, and vhs) and the cell surface receptor Thy1.1 (CD90.1). Pure populations of cells expressing the endonucleases were obtained as describe before (46). Briefly, cells expressing the surface marker Thy1.1 were separated using the Miltenyi Biotec MACS cell separation system: transfected cells were incubated with anti-CD90.1 microbeads on ice for 15 min and magnetically separated according to the manufacturer’s instructions. RNA was then extracted from Thy1.1-positive cells by TRIzol and purified as described above. Purity and integrity were assessed with a bioanalyzer. After poly(A) selection, libraries were subjected to 100-base single-end sequencing on a HiSeq 4000. Using Galaxy (56), reads were then aligned to the human genome (hg38) by Bowtie2, replicates were merged using CuffCompare, and significant expression fold changes between mock and each of the endonuclease conditions were assessed by Cufflinks and CuffDiff (57). Read quality was assessed using fastqc. For graphical representation in the heatmap, fold change values were saturated by a hyperbolic tan function with a cutoff set at 10. Hierarchical clustering was generated in Python using the SciPy package with complete linkage and Euclidian distance.
Statistical analysis.
All results are expressed as means ± standard errors of the means (SEMs) of experiments independently repeated at least three times. The unpaired Student’s t test was used to evaluate the statistical difference between samples. Significance was evaluated with P values as indicated in figure legends.
Data availability.
Sequencing data from this study have been deposited in GEO under accession number GSE128866.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the Muller lab for their insights. We also thank members of the Glaunsinger lab for helpful discussions and Ella Hartenian and Sarah Gilbertson for technical help with the Thy1.1 constructs. We also thank Romain Vasseur for help with Python.
This research was supported by UMass Microbiology Startup funds to M.M. and a University Fellowship in Microbiology to W.R.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00373-19.
REFERENCES
- 1.Gaglia MM, Covarrubias S, Wong W, Glaunsinger BA. 2012. A common strategy for host RNA degradation by divergent viruses. J Virol 86:9527–9530. doi: 10.1128/JVI.01230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rivas HG, Schmaling SK, Gaglia MM. 2016. Shutoff of host gene expression in influenza A virus and herpesviruses: similar mechanisms and common themes. Viruses 8:102. doi: 10.3390/v8040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jagger BW, Wise HM, Kash JC, Walters K-A, Wills NM, Xiao Y-L, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong AD, Frenkel N. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc Natl Acad Sci U S A 84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol 16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abernathy E, Glaunsinger B. 2015. Emerging roles for RNA degradation in viral replication and antiviral defense. Virology 479-480:600–608. doi: 10.1016/j.virol.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaglia MM, Glaunsinger BA. 2010. Viruses and the cellular RNA decay machinery. Wiley Interdiscip Rev RNA 1:47–59. doi: 10.1002/wrna.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covarrubias S, Richner JM, Clyde K, Lee YJ, Glaunsinger BA. 2009. Host shutoff is a conserved phenotype of gammaherpesvirus infection and is orchestrated exclusively from the cytoplasm. J Virol 83:9554–9566. doi: 10.1128/JVI.01051-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowe M, Glaunsinger B, van Leeuwen D, Zuo J, Sweetman D, Ganem D, Middeldorp J, Wiertz E, Ressing ME. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc Natl Acad Sci U S A 104:3366–3371. doi: 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glaunsinger B, Ganem D. 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol Cell 13:713–723. doi: 10.1016/S1097-2765(04)00091-7. [DOI] [PubMed] [Google Scholar]
- 11.Abernathy E, Clyde K, Yeasmin R, Krug LT, Burlingame A, Coscoy L, Glaunsinger B. 2014. Gammaherpesviral gene expression and virion composition are broadly controlled by accelerated mRNA degradation. PLoS Pathog 10:e1003882. doi: 10.1371/journal.ppat.1003882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richner JM, Clyde K, Pezda AC, Cheng BYH, Wang T, Kumar GR, Covarrubias S, Coscoy L, Glaunsinger B. 2011. Global mRNA degradation during lytic gammaherpesvirus infection contributes to establishment of viral latency. PLoS Pathog 7:e1002150. doi: 10.1371/journal.ppat.1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaglia MM, Rycroft CH, Glaunsinger BA. 2015. Transcriptome-wide cleavage site mapping on cellular mRNAs reveals features underlying sequence-specific cleavage by the viral ribonuclease SOX. PLoS Pathog 11:e1005305. doi: 10.1371/journal.ppat.1005305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covarrubias S, Gaglia MM, Kumar GR, Wong W, Jackson AO, Glaunsinger BA. 2011. Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the mammalian exonuclease Xrn1. PLoS Pathog 7:e1002339. doi: 10.1371/journal.ppat.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendez AS, Vogt C, Bohne J, Glaunsinger BA. 2018. Site specific target binding controls RNA cleavage efficiency by the Kaposi's sarcoma-associated herpesvirus endonuclease SOX. Nucleic Acids Res 46:11968–11979. doi: 10.1093/nar/gky932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clyde K, Glaunsinger BA. 2011. Deep sequencing reveals direct targets of gammaherpesvirus-induced mRNA decay and suggests that multiple mechanisms govern cellular transcript escape. PLoS One 6:e19655. doi: 10.1371/journal.pone.0019655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandriani S, Ganem D. 2007. Host transcript accumulation during lytic KSHV infection reveals several classes of host responses. PLoS One 2:e811. doi: 10.1371/journal.pone.0000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glaunsinger B, Ganem D. 2004. Highly selective escape from KSHV-mediated host mRNA shutoff and its implications for viral pathogenesis. J Exp Med 200:391–398. doi: 10.1084/jem.20031881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller M, Glaunsinger BA. 2017. Nuclease escape elements protect messenger RNA against cleavage by multiple viral endonucleases. PLoS Pathog 13:e1006593. doi: 10.1371/journal.ppat.1006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller M, Hutin S, Marigold O, Li KH, Burlingame A, Glaunsinger BA. 2015. A ribonucleoprotein complex protects the interleukin-6 mRNA from degradation by distinct herpesviral endonucleases. PLoS Pathog 11:e1004899. doi: 10.1371/journal.ppat.1004899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutin S, Lee Y, Glaunsinger BA. 2013. An RNA element in human interleukin 6 confers escape from degradation by the gammaherpesvirus SOX protein. J Virol 87:4672–4682. doi: 10.1128/JVI.00159-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clyde K, Glaunsinger BA. 2010. Getting the message direct manipulation of host mRNA accumulation during gammaherpesvirus lytic infection. Adv Virus Res 78:1–42. doi: 10.1016/B978-0-12-385032-4.00001-X. [DOI] [PubMed] [Google Scholar]
- 23.Lee YJ, Glaunsinger BA. 2009. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol 7:e1000107. doi: 10.1371/journal.pbio.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abernathy E, Gilbertson S, Alla R, Glaunsinger B. 2015. Viral nucleases induce an mRNA degradation-transcription feedback loop in mammalian cells. Cell Host Microbe 18:243–253. doi: 10.1016/j.chom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sin S-H, Roy D, Wang L, Staudt MR, Fakhari FD, Patel DD, Henry D, Harrington WJ, Damania BA, Dittmer DP. 2007. Rapamycin is efficacious against primary effusion lymphoma (PEL) cell lines in vivo by inhibiting autocrine signaling. Blood 109:2165–2173. doi: 10.1182/blood-2006-06-028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miles SA, Rezai AR, Salazar-González JF, Vander Meyden M, Stevens RH, Logan DM, Mitsuyasu RT, Taga T, Hirano T, Kishimoto T. 1990. AIDS Kaposi sarcoma-derived cells produce and respond to interleukin 6. Proc Natl Acad Sci U S A 87:4068–4072. doi: 10.1073/pnas.87.11.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Screpanti I, Musiani P, Bellavia D, Cappelletti M, Aiello FB, Maroder M, Frati L, Modesti A, Gulino A, Poli V. 1996. Inactivation of the IL-6 gene prevents development of multicentric Castleman's disease in C/EBP beta-deficient mice. J Exp Med 184:1561–1566. doi: 10.1084/jem.184.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leger-Ravet MB, Peuchmaur M, Devergne O, Audouin J, Raphael M, Van Damme J, Galanaud P, Diebold J, Emilie D. 1991. Interleukin-6 gene expression in Castleman's disease. Blood 78:2923–2930. [PubMed] [Google Scholar]
- 29.Xie J, Pan H, Yoo S, Gao S-J. 2005. Kaposi's sarcoma-associated herpesvirus induction of AP-1 and interleukin 6 during primary infection mediated by multiple mitogen-activated protein kinase pathways. J Virol 79:15027–15037. doi: 10.1128/JVI.79.24.15027-15037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.An J, Sun Y, Sun R, Rettig MB. 2003. Kaposi's sarcoma-associated herpesvirus encoded vFLIP induces cellular IL-6 expression: the role of the NF-kappaB and JNK/AP1 pathways. Oncogene 22:3371–3385. doi: 10.1038/sj.onc.1206407. [DOI] [PubMed] [Google Scholar]
- 31.Santarelli R, Gonnella R, Di Giovenale G, Cuomo L, Capobianchi A, Granato M, Gentile G, Faggioni A, Cirone M. 2014. STAT3 activation by KSHV correlates with IL-10, IL-6 and IL-23 release and an autophagic block in dendritic cells. Sci Rep 4:4241. doi: 10.1038/srep04241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng H, Chu JT, Rettig MB, Martinez-Maza O, Sun R. 2002. Rta of the human herpesvirus 8/Kaposi sarcoma-associated herpesvirus up-regulates human interleukin-6 gene expression. Blood 100:1919–1921. doi: 10.1182/blood-2002-01-0015. [DOI] [PubMed] [Google Scholar]
- 33.McCormick C, Ganem D. 2005. The kaposin B protein of KSHV activates the p38/MK2 pathway and stabilizes cytokine mRNAs. Science 307:739–741. doi: 10.1126/science.1105779. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Happel C, Ziegelbauer JM. 2017. Kaposi's sarcoma-associated herpesvirus microRNAs target GADD45B to protect infected cells from cell cycle arrest and apoptosis. J Virol 91:e02045-16. doi: 10.1128/JVI.02045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR, Castro E, Akondy R, Rinfret A, Yassine-Diab B, Said EA, Chouikh Y, Cameron MJ, Clum R, Kelvin D, Somogyi R, Greller LD, Balderas RS, Wilkinson P, Pantaleo G, Tartaglia J, Haddad EK, Sékaly R-P. 2008. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med 205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harvey SAK, Romanowski EG, Yates KA, Gordon YJ. 2005. Adenovirus-directed ocular innate immunity: the role of conjunctival defensin-like chemokines (IP-10, I-TAC) and phagocytic human defensin-alpha. Invest Ophthalmol Vis Sci 46:3657–3665. doi: 10.1167/iovs.05-0438. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Nikrad MP, Phang T, Gao B, Alford T, Ito Y, Edeen K, Travanty EA, Kosmider B, Hartshorn K, Mason RJ. 2011. Innate immune response to influenza A virus in differentiated human alveolar type II cells. Am J Respir Cell Mol Biol 45:582–591. doi: 10.1165/rcmb.2010-0108OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zapata JC, Carrion R, Patterson JL, Crasta O, Zhang Y, Mani S, Jett M, Poonia B, Djavani M, White DM, Lukashevich IS, Salvato MS. 2013. Transcriptome analysis of human peripheral blood mononuclear cells exposed to Lassa virus and to the attenuated Mopeia/Lassa reassortant 29 (ML29), a vaccine candidate. PLoS Negl Trop Dis 7:e2406. doi: 10.1371/journal.pntd.0002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kash JC, Mühlberger E, Carter V, Grosch M, Perwitasari O, Proll SC, Thomas MJ, Weber F, Klenk H-D, Katze MG. 2006. Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J Virol 80:3009–3020. doi: 10.1128/JVI.80.6.3009-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong W, Contreras D, Ignatius Irudayam J, Ali A, Yang OO, Arumugaswami V. 2016. C19ORF66 is an interferon-stimulated gene (ISG) which Inhibits human immunodeficiency virus-1. BioRxiv. doi: 10.1101/050310. [DOI]
- 41.Bull TM, Meadows CA, Coldren CD, Moore M, Sotto-Santiago SM, Nana-Sinkam SP, Campbell TB, Geraci MW. 2008. Human herpesvirus-8 infection of primary pulmonary microvascular endothelial cells. Am J Respir Cell Mol Biol 39:706–716. doi: 10.1165/rcmb.2007-0368OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyazaki D, Haruki T, Takeda S, Sasaki S-I, Yakura K, Terasaka Y, Komatsu N, Yamagami S, Touge H, Touge C, Inoue Y. 2011. Herpes simplex virus type 1-induced transcriptional networks of corneal endothelial cells indicate antigen presentation function. InvestOphthalmol Vis Sci 52:4282–4293. doi: 10.1167/iovs.10-6911. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki Y, Chin W-X, Han Q, Ichiyama K, Lee CH, Eyo ZW, Ebina H, Takahashi H, Takahashi C, Tan BH, Hishiki T, Ohba K, Matsuyama T, Koyanagi Y, Tan Y-J, Sawasaki T, Chu JJH, Vasudevan SG, Sano K, Yamamoto N. 2016. Characterization of RyDEN (C19orf66) as an interferon-stimulated cellular inhibitor against dengue virus replication. PLoS Pathog 12:e1005357. doi: 10.1371/journal.ppat.1005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balinsky CA, Schmeisser H, Wells AI, Ganesan S, Jin T, Singh K, Zoon KC. 2017. IRAV (FLJ11286), an interferon-stimulated gene with antiviral activity against dengue virus, interacts with MOV10. J Virol 91:e01606-16. doi: 10.1128/JVI.01606-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez W, Srivastav A, Muller M. 2018. C19ORF66 broadly escapes viral-induced endonuclease cleavage and restricts Kaposi sarcoma associated herpesvirus (KSHV). bioRxiv doi: 10.1101/506410. [DOI] [PMC free article] [PubMed]
- 46.Gilbertson S, Federspiel JD, Hartenian E, Cristea IM, Glaunsinger B. 2018. Changes in mRNA abundance drive shuttling of RNA binding proteins, linking cytoplasmic RNA degradation to transcription. Elife 7:e37663. doi: 10.7554/eLife.37663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J Virol 77:4205–4220. doi: 10.1128/JVI.77.7.4205-4220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myoung J, Ganem D. 2011. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J Virol Methods 174:12–21. doi: 10.1016/j.jviromet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narayanan K, Ramirez SI, Lokugamage KG, Makino S. 2015. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res 202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. 2011. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog 7:e1002433. doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmeisser H, Mejido J, Balinsky CA, Morrow AN, Clark CR, Zhao T, Zoon KC. 2010. Identification of alpha interferon-induced genes associated with antiviral activity in Daudi cells and characterization of IFIT3 as a novel antiviral gene. J Virol 84:10671–10680. doi: 10.1128/JVI.00818-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, Agaugué S, Meiffren G, Pradezynski F, Faria BF, Chantier T, Le Breton M, Pellet J, Davoust N, Mangeot PE, Chaboud A, Penin F, Jacob Y, Vidalain PO, Vidal M, André P, Rabourdin-Combe C, Lotteau V. 2008. Hepatitis C virus infection protein network. Mol Syst Biol 4:230. doi: 10.1038/msb.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi H, Suzuki Y. 2017. Cellular control of dengue virus replication: role of interferon-inducible genes In Aparecida Sperança M. (ed), Dengue-immunopathology and control strategies. InTech, London, UK. [Google Scholar]
- 55.Nabbi A, Riabowol K. 2015. Rapid isolation of nuclei from cells in vitro. Cold Spring Harb Protoc 2015:769–772. doi: 10.1101/pdb.prot083733. [DOI] [PubMed] [Google Scholar]
- 56.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data from this study have been deposited in GEO under accession number GSE128866.