Although human adenovirus (HAdV) can cause severe diseases that can be fatal in some populations, there are no effective treatments to combat HAdV infection. In this study, we determined that the pan-histone deacetylase (HDAC) inhibitor SAHA has inhibitory activity against several clinically relevant serotypes of HAdV. This U.S. Food and Drug Administration-approved compound affects various stages of the virus lifecycle and reduces virus yield even at low concentrations. We further report that class I HDAC activity, particularly HDAC2, is required for efficient expression of viral genes during lytic infection. Investigation of the mechanism underlying SAHA-mediated suppression of HAdV gene expression and replication will enhance current knowledge of virus-cell interaction and may aid in the development of more effective antivirals with lower toxicity for the treatment of HAdV infections.
KEYWORDS: SAHA, adenoviruses, histone deacetylase, histone deacetylase inhibitors, vorinostat
ABSTRACT
Human adenovirus (HAdV) causes minor illnesses in most patients but can lead to severe disease and death in pediatric, geriatric, and immunocompromised individuals. No approved antiviral therapy currently exists for the treatment of these severe HAdV-induced diseases. In this study, we show that the pan-histone deacetylase (HDAC) inhibitor SAHA reduces HAdV-5 gene expression and DNA replication in tissue culture, ultimately decreasing virus yield from infected cells. Importantly, SAHA also reduced gene expression from more virulent and clinically relevant serotypes, including HAdV-4 and HAdV-7. In addition to SAHA, several other HDAC inhibitors (e.g., trichostatin A, apicidin, and panobinostat) also affected HAdV gene expression. We determined that loss of class I HDAC activity, mainly HDAC2, impairs efficient expression of viral genes, and that E1A physically interacts with HDAC2. Our results suggest that HDAC activity is necessary for HAdV replication, which may represent a novel pharmacological target in HAdV-induced disease.
IMPORTANCE Although human adenovirus (HAdV) can cause severe diseases that can be fatal in some populations, there are no effective treatments to combat HAdV infection. In this study, we determined that the pan-histone deacetylase (HDAC) inhibitor SAHA has inhibitory activity against several clinically relevant serotypes of HAdV. This U.S. Food and Drug Administration-approved compound affects various stages of the virus lifecycle and reduces virus yield even at low concentrations. We further report that class I HDAC activity, particularly HDAC2, is required for efficient expression of viral genes during lytic infection. Investigation of the mechanism underlying SAHA-mediated suppression of HAdV gene expression and replication will enhance current knowledge of virus-cell interaction and may aid in the development of more effective antivirals with lower toxicity for the treatment of HAdV infections.
INTRODUCTION
Human adenovirus (HAdV) mainly causes mild, self-limiting illnesses in most immunocompetent patients. HAdV-induced diseases vary based on the virus serotype involved, but respiratory tract infections are common and can often lead to bronchitis or pneumonia (1, 2). These minor infections are thought to be underreported, leading to an underestimation of HAdV infection and prevalence (3). HAdV can also cause severe disease, including respiratory failure, disseminated infection, neurologic complications, and even death in pediatric, geriatric, and immunocompromised patients (e.g., stem cell or solid organ transplant recipients) (1, 4–6). Recently, several cases of immunocompetent patients succumbing to HAdV-induced respiratory failure was documented (7, 8). No approved antiviral therapy currently exists for the treatment of these severe HAdV infections (9, 10). Although DNA synthesis inhibitors such as cidofovir and ganciclovir are considered to be standard treatments, these drugs are often associated with serious adverse effects (11) and have limited efficacy in systemic HAdV infections (9). Brincidofovir, a phospholipid conjugate of cidofovir, is currently in clinical trials for the treatment of HAdV-induced diseases (12, 13). This drug has several advantages over cidofovir (e.g., oral delivery and increased cellular uptake), but it is still associated with gastrointestinal toxicity in some patients. Thus, novel antiviral therapies are necessary to combat severe HAdV infections.
HAdV, a nonenveloped, double-stranded DNA virus, was first isolated in the 1950s from adenoid tissue (14, 15). Since then, over 60 serotypes of HAdV (categorized into seven species) have been identified (1), of which species B, C, and E are most frequently associated with symptomatic respiratory infections (2). While species C (serotypes 1, 2, and 5) mainly causes mild infections in young children, species B (serotypes 3, 7, 14, and 21) and E (serotype 4) cause more severe infections in adults (2). A live oral vaccine against serotypes 4 and 7 was used by the U.S. military from 1971 to 1997 to limit HAdV disease in new recruits (1). The vaccine was later reintroduced in 2011 due to a substantial increase in HAdV infection within military personnel during the intervening period, but it is not available to the general public.
HAdV serotypes 2 and 5 (species C) have been the most extensively studied to gain insights on virus biology, cellular processes, and host-pathogen interactions. The 36-kbp HAdV-5 genome consists of “early” and “late” genes which are expressed before and after viral DNA replication, respectively (16, 17). Of the early genes (E1A, E1B, E2, E3, and E4), the E1A gene product is absolutely necessary for efficient virus replication, and it is the first protein expressed from the infecting virus. E1A transactivates the remaining viral coding regions, interacts with many cellular proteins, and has various other functions that promote a productive infection (18–20). The E2 transcription unit consists of two regions, E2A and E2B. The E2A region encodes the HAdV DNA-binding protein (DBP), while the E2B region codes for the precursor terminal protein (pTP) and the viral DNA polymerase. All three E2 proteins play crucial roles during HAdV DNA replication (17, 21, 22). The E3 and E4 gene products modulate the host immune system and the cellular environment to make it more conducive to virus replication (23, 24). Finally, the late transcription units, L1 to L5, are produced from alternative splicing of a common major late transcript containing five different polyadenylation sites which are all used during late infection (25). The major late promoter (MLP) is only strongly active following viral DNA replication (26), and the late regions code for structural and capsid proteins (e.g., fiber, penton, and hexon) required for progeny virion formation.
Inside the virion, the virus-encoded, highly basic protein VII (pVII) tightly condenses the HAdV genome into a structure that is refractory to transcription and replication (27). During infection, the pVII-wrapped viral DNA enters the host cell nuclei through the nuclear pores (28). In the early phase of infection, the HAdV DNA dissociates from pVII, associates with cellular proteins, including histones, and adopts a nucleoprotein structure similar to the host DNA (29, 30). The nucleosome density on the viral DNA is significantly reduced during late infection, and at least some of the newly synthesized viral DNA must associate with pre-pVII for packaging into newly formed capsids (30–32). The cellular proteins involved in mediating these processes and the role of chromatin-modifying enzymes in the epigenetic regulation of the HAdV genome has not yet been fully elucidated (33). A recent study found that the nucleosomes associated with several of the HAdV-2 early promoters and transcription start sites are acetylated at various histone 3 (H3) lysine residues, including H3K9, H3K18, and H3K27 (34). The level of acetylation was dynamic throughout infection and required interaction between HAdV-2 E1A and p300/CBP (35). In addition, valproic acid-mediated inhibition of histone deacetylase (HDAC) activity has been shown to inhibit HAdV-5 replication and spread in vitro (36), indicating the importance of HDAC activity for optimal virus replication.
In this study, we determined that the pan-HDAC inhibitor SAHA (suberoylanilide hydroxamic acid, also known as vorinostat) suppresses HAdV replication. SAHA treatment reduced viral gene transcription, protein expression, and DNA replication, leading to a significant decrease in virus yield from infected cells. Of note, SAHA reduced HAdV yield at nanomolar concentrations and was effective in decreasing gene expression from virulent serotypes 4 and 7. Trichostatin A (TSA) and several other HDAC inhibitors also exhibited a similar inhibitory activity against HAdV gene expression. We further determined that class I HDACs, specifically HDAC2, appear to play an important role in the HAdV infection process.
RESULTS
RFP expression from a novel HAdV construct serves as an efficient tool to monitor virus replication.
We generated an E1+ wild-type-like HAdV construct containing the monomeric red fluorescent protein (RFP) gene with an upstream splice acceptor site replacing the viral E3 region, thus placing RFP expression under the control of the MLP, which is only active following initiation of viral DNA replication (16, 37). This replication-competent construct was designated Ad-late/RFP (Fig. 1A). An E1-deleted, replication-deficient version of this virus, Ad(E1–)-late/RFP, and a control virus with RFP under the regulation of the ubiquitously active cytomegalovirus (CMV) enhancer/promoter, Ad(E1–)-CMV/RFP, were also created. The absence of the E1 region renders both of these latter viruses replication defective, allowing them to only replicate in E1-complementing cells, such as 293 cells.
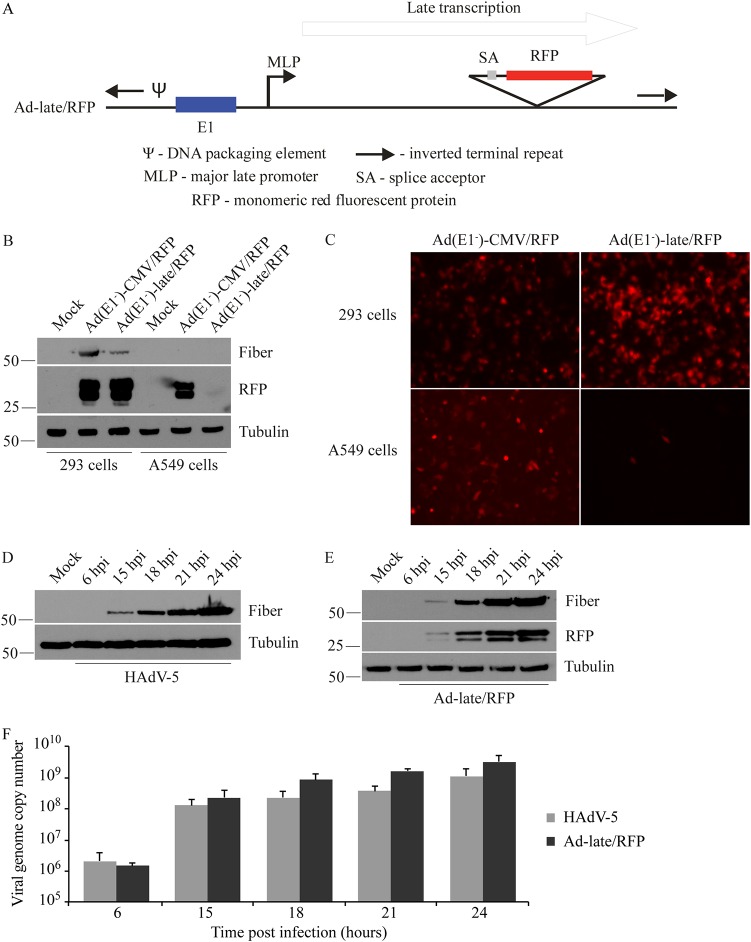
FIG 1.
Validation of the Ad-late/RFP construct. (A) Schematic diagram of Ad-late/RFP (not drawn to scale). The RFP cDNA is under the control of the HAdV MLP and the E1 region is present. An E1-deleted, replication-defective version of the Ad-late/RFP [referred to as Ad(E1–)-late/RFP] was also generated, which is able to replicate only in E1-complementing cells. (B) 293 and A549 cells were infected with Ad(E1–)-late/RFP or Ad(E1–)-CMV/RFP at an MOI of 1. The latter is a control virus with the ubiquitously expressed cytomegalovirus enhancer/promoter driving RFP expression. Whole-cell lysates were collected at 24 hpi for the detection of RFP, fiber (a positive control for HAdV replication), and tubulin (loading control). RFP from the Ad(E1–)-CMV/RFP is present in both 293 and A549 cell lysates independent of virus replication. However, RFP from Ad(E1–)-late/RFP is only present in 293 cells. (C) Cells were infected as in panel B, and fluorescence microscopy results at 24 hpi corroborate the selective expression of RFP from Ad(E1–)-late/RFP. (D to F) A549 cells were infected with HAdV-5 or Ad-late/RFP at an MOI of 10 for 6 to 24 h. (D and E) Fiber and RFP were detected by immunoblotting cell lysates. (F) qPCR was performed on genomic DNA isolated from infected cells, using oligonucleotide primers specific for the gene encoding capsid protein hexon (error bars represent the range of three independent experiments). As indicated by viral protein and DNA levels, Ad-late/RFP is able to grow as well as wild-type HAdV.
We first verified that RFP expression from the late/RFP constructs coincided with virus replication by examining late gene expression from the E1-deleted Ad(E1–)-late/RFP and Ad(E1–)-CMV/RFP in A549 and 293 cells. The late HAdV fiber (a capsid protein) is only expressed at appreciable levels after active virus DNA replication, which can occur in the E1-complementing 293 cell line for these viruses (Fig. 1B). As expected, RFP from the Ad(E1–)-late/RFP was also only detected in 293 cells, whereas RFP production from the Ad(E1–)-CMV/RFP was independent of virus replication and occurred in both 293 and A549 cells (Fig. 1B). Similar results were observed by fluorescence microscopy of cells infected with these viruses (Fig. 1C). These observations confirm that RFP expression can be used to monitor active virus replication and can serve as an effective surrogate marker to quantify drug-induced changes in viral gene expression and replication. As such, the Ad-late/RFP construct is also ideal for conducting small-molecule screens to identify novel inhibitors of HAdV.
The E1+ replication-competent Ad-late/RFP (Fig. 1A) is used in all subsequent experiments since it is more characteristic of the wild-type HAdV-5. Comparison of protein production and DNA replication revealed that Ad-late/RFP grows similar to HAdV-5 (Fig. 1D to F). A time course of fiber expression for the two viruses show a similar extent of accumulation within the cell (Fig. 1D and E), and the kinetics of DNA replication was comparable as well (Fig. 1F). RFP expression correlates with fiber protein expression (Fig. 1E) and appears to reflect the degree of viral DNA replication. Importantly, the presence of the RFP gene in E3 does not impair Ad-late/RFP replication.
SAHA has anti-HAdV activity.
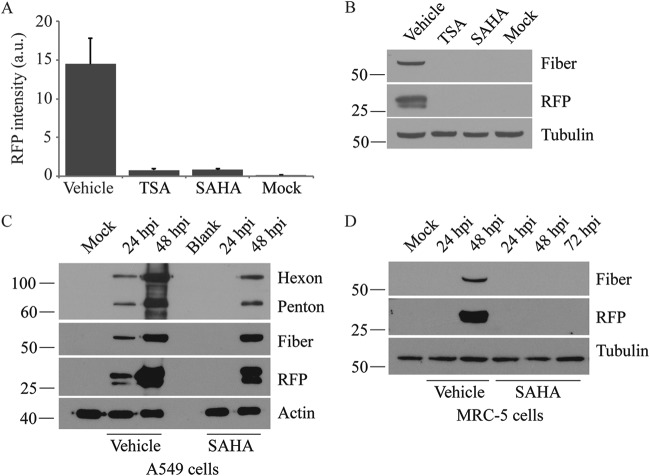
In the infected cell nucleus, HAdV associates with nucleosomes, suggesting that viral gene expression should be influenced by cellular epigenetic regulator proteins, such as histone acetyltransferases (HATs) and HDACs. Several studies have shown that HDAC inhibitors can enhance expression of reporter genes regulated by heterologous promoters within HAdV vectors (38, 39), whereas replication of the wild-type virus is inhibited (36). To test the effects of HDAC inhibitors in our system, we examined RFP expression from Ad-late/RFP in cells treated with pan-HDAC inhibitors TSA and SAHA at a concentration of 10 μM. As shown in Fig. 2A, both compounds inhibited RFP expression at 24 h postinfection (hpi). This observation was corroborated by the absence of both fiber and RFP in cell lysates following immunoblot analysis (Fig. 2B). We next examined the effects of SAHA over a longer time course of infection. As shown in Fig. 2C, in vehicle-treated cells, hexon, penton, fiber, and RFP expression were readily observed by 24 hpi. In contrast, late gene expression was undetectable at 24 hpi but could be detected at lower levels at 48 hpi in cells treated with SAHA (Fig. 2C). This is unlikely due to drug degradation over time since fresh medium containing drug was applied every 24 h in this experiment. Although we were unable to find accurate published data on the half-life of SAHA in A549 cell culture, we obtained the same results with MS-275, also known as entinostat (data not shown), which we expect to have a longer half-life than SAHA in vitro based on its significantly longer half-life (>30 h) in vivo (40, 41). We conducted a similar experiment in the MRC-5 cell line, which is derived from normal lung tissue, to confirm that these results were not specific to the transformed A549 cells. The virus life cycle is prolonged in MRC-5 cells, and we detected fiber and RFP expression from Ad-late/RFP at 48 hpi with vehicle treatment (Fig. 2D). However, neither fiber nor RFP was observed in SAHA-treated MRC-5 cells at 48 or 72 hpi. Taken together, these data illustrate that HDAC inhibitors repress the expression of HAdV late genes.
FIG 2.
Pan-HDAC inhibitor SAHA suppresses HAdV late gene expression. A549 cells were infected with Ad-late/RFP (MOI of 10) and treated with vehicle, 10 μM TSA, or 10 μM SAHA. (A) Cells were fixed at 24 hpi, and the RFP fluorescence intensities were quantified using the Cellomics HCS Platform. a.u., arbitrary units. Error bars represent the standard deviations (SD) of analytical replicates (n = 16). (B) Immunoblot analysis of cell lysates collected at 24 hpi confirmed the results in panel A. Both TSA and SAHA inhibited RFP production at 24 hpi. (C and D) A549 cells (C) and MRC-5 cells (D) were infected with Ad-late/RFP and treated with 10 μM SAHA. The expression of late proteins was analyzed by immunoblotting cell lysates collected at the indicated times. SAHA significantly reduces late gene expression in both cell lines.
SAHA reduces late gene expression from clinically relevant HAdV serotypes and at low concentrations.
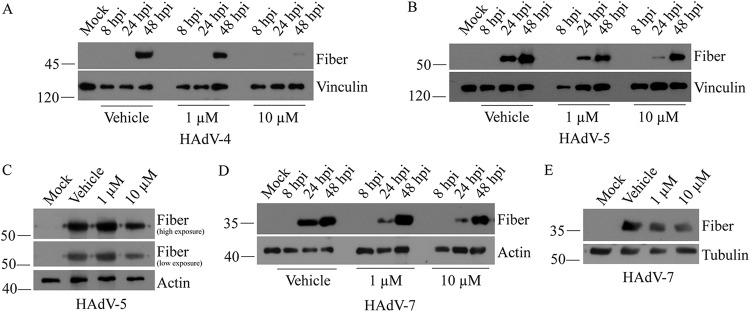
Ad-late/RFP was constructed based on HAdV-5, but it does not encode all of the wild-type HAdV genes (i.e., RFP replaces the viral E3 region). Therefore, we evaluated the efficacy of SAHA against more virulent and clinically relevant HAdV serotypes 4, 5, and 7 in A549 cells (1, 2). For HAdV-4, fiber gene expression was only detectable at 48 hpi and was significantly reduced in cells treated with 10 μM SAHA compared to vehicle (Fig. 3A). Similar results were obtained with HAdV-5 at 24 hpi: the fiber protein decreased in a dose-dependent manner (Fig. 3B). Reduced fiber was also observed at 48 hpi in the presence of 10 μM SAHA (Fig. 3C). Lastly, a modest dose-dependent decrease in fiber protein was observed for HAdV-7 at both 24 and 48 hpi (Fig. 3D and E). As with Ad-late/RFP in MRC-5 cells in Fig. 2D, late gene expression from wild-type HAdV-5 was suppressed for at least 24 h in primary mouse embryonic fibroblasts (data not shown). Thus, SAHA is inhibitory toward multiple HAdV serotypes. Our findings are consistent with those of Hoti et al. (36), who showed that HAdV-5 replication and spread were substantially reduced when treated with a different HDAC inhibitor, valproic acid.
FIG 3.
SAHA decreases late gene expression from clinically relevant HAdV serotypes. A549 cells were infected with HAdV-4 (A), HAdV-5 (B), or HAdV-7 (D) at an MOI of 10 and treated with the indicated concentrations of SAHA for 8 to 48 h. Fiber protein was detected in cell lysates by immunoblotting. (C and E) The 48-hpi samples from panels B and D were reanalyzed using shorter exposures in panels C and E, respectively, to illustrate the decrease in late gene expression more clearly.
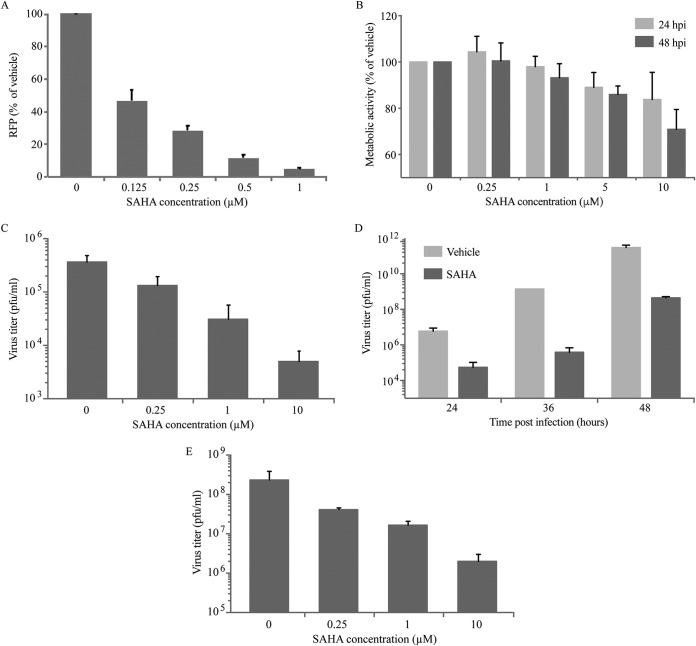
To examine the efficacy of SAHA at low concentrations, we conducted a dose-response assay. SAHA reduced RFP expression considerably at concentrations below 1 μM (Fig. 4A). For example, compared to vehicle treatment, RFP was reduced by more than 50% with 0.25 μM SAHA. Importantly, this effect was not due to compromised cell viability since the cells retained high metabolic activity (>80% at 24 hpi and >70% at 48 hpi) at all concentrations (Fig. 4B). Furthermore, the cells did not exhibit any irregular cell morphology or significant detachment from culture dishes (assessed by light microscopy and visual inspection), and minimal cell death (less than 3%) was detected with 0.25 to 10 μM SAHA via a trypan blue exclusion assay (data not shown). These observations are consistent with previous reports showing that treatment with SAHA for 24 h has a minor effect on A549 cell density and viability (42). The decrease in RFP expression was concomitant with a similar decrease in virus yield (∼100-fold or more) for Ad-late/RFP (Fig. 4C and D) and HAdV-5 (Fig. 4E) in the presence of 10 μM SAHA. In addition to valproic acid, SAHA, TSA, and MS-275, we tested 17 additional HDAC inhibitors, 11 of which were found to reduce RFP expression from the Ad-late/RFP by at least 50% at a concentration of 1 μM (Table 1). Overall, these results highlight the potential of SAHA and other pan-HDAC inhibitors as anti-HAdV agents.
FIG 4.
SAHA reduces RFP expression and virus yield at low concentrations. A549 cells were infected with Ad-late/RFP (MOI of 10) and treated with vehicle or the indicated concentrations of SAHA for 24 h. (A) Cells were fixed and RFP levels were quantified using the Cellomics HCS platform. (B) Cellular metabolic activity was determined by MTS assay in live cells. In both panels, changes in SAHA-treated cells were plotted relative to vehicle treatment. (C and D) Infected A549 cells were treated with various concentrations of SAHA for 24 h (C) or with 10 μM SAHA for 24 to 48 h (D). The cell lysates were subjected to plaque assay for determination of virus yield. (E) A plaque assay was conducted as in panel C with wild-type HAdV-5. Error bars represent the SD (n = 3).
TABLE 1.
HDAC inhibitors from the Cayman Epigenetics Screening Library that reduced RFP from Ad-late/RFP
| HDAC inhibitor | % RFPa |
|---|---|
| Apicidin | 12 |
| HC toxin | 13 |
| Panobinostat | 14 |
| Givinostat | 17 |
| Oxamflatin | 17 |
| Pracinostat | 20 |
| CAY10603 | 34 |
| M344 | 41 |
| 4-Iodo-SAHA | 45 |
| CBHA | 47 |
| Scriptaid | 49 |
The percentage values indicate the RFP intensities in cells treated with a 1 μM concentration of drug relative to vehicle-treated cells at 24 hpi.
SAHA treatment affects HAdV early/late gene transcription, protein expression, and DNA replication.
SAHA reduced viral late gene expression from Ad-late/RFP (Fig. 2), and also from HAdV-4, -5, and -7 (Fig. 3). However, this effect could be due to action of the drug on preceding stages of the virus life cycle, including virus attachment/internalization, viral genome chromatinization, early gene expression, DNA replication, or transcription from the MLP. Thus, we performed additional studies to determine at which stage of the virus life cycle SAHA exerted its effect.
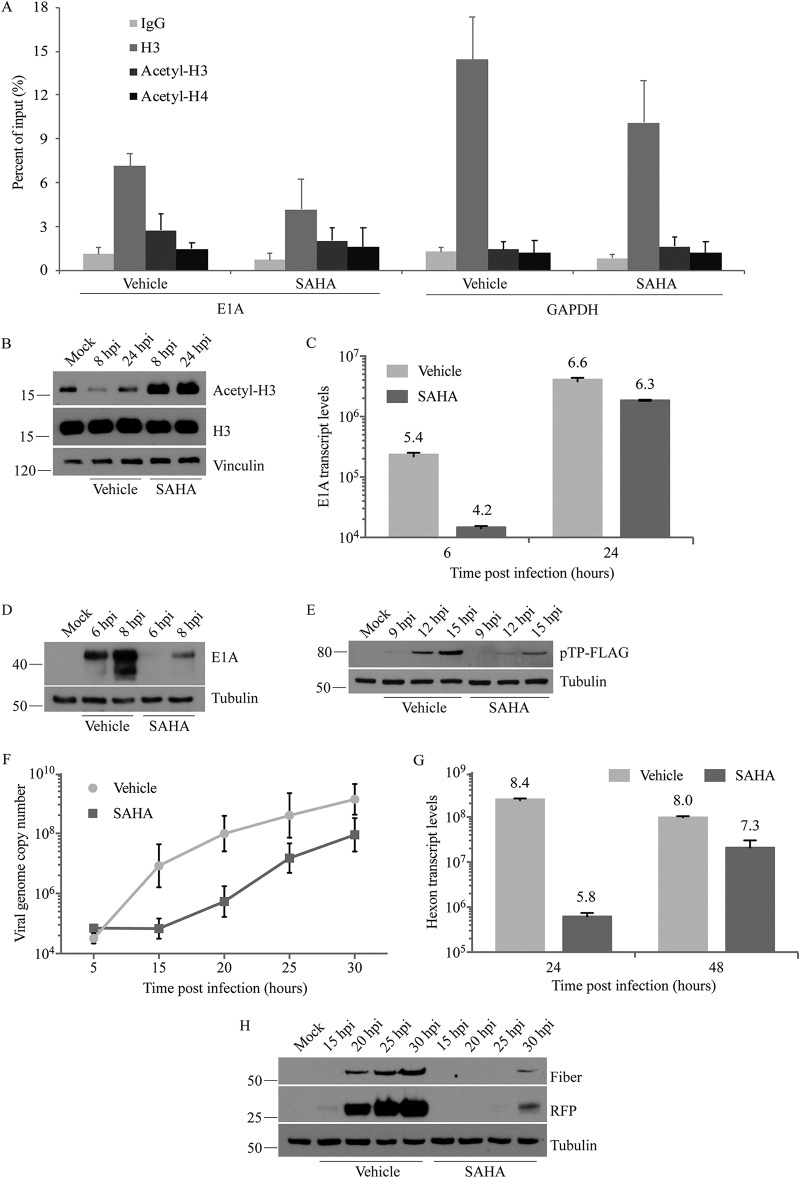
A preliminary study assessing hexon, penton, and fiber protein levels from the capsids of cell surface-attached and internalized virions indicated that the drug does not adversely affect virus attachment (data not shown), which is consistent with studies reporting that HDAC inhibitors do not downregulate cell surface expression of the coxsackie adenovirus receptor (39). HAdV DNA associates with nucleosomes soon after reaching the host cell nucleus (29, 30), so we next examined the impact of SAHA on Ad-late/RFP genome “chromatinization” by using a chromatin immunoprecipitation (ChIP) assay at 6 h following infection and drug treatment. Consistent with previous studies (29, 30), we found that the early E1A region associated with H3 (Fig. 5A). Although SAHA seems to reduce H3 association with this region (Fig. 5A), this did not reach statistical significance with four experimental replicates. HDACs are epigenetic modifiers which remove acetyl groups that neutralize the positive charge on the lysine residues of histones, thereby regulating accessibility of specific genomic regions to transcription factor binding (43). Thus, treatment of infected cells with an HDAC inhibitor could alter the posttranslational modifications present on the viral genome-associated histones. To test this possibility, we examined the acetylation levels of H3/H4 associated with the E1A region using pan-acetyl antibodies. We observed no difference in association of this region with acetylated H3 or H4 (Fig. 5A), even though overall cellular H3 acetylation was increased by SAHA as early as 8 hpi in cells infected with Ad-late/RFP (Fig. 5B). Similar results were obtained when examining SAHA-induced changes in histone acetylation and association with the late hexon region (data not shown). The cellular GAPDH (glyceraldehyde-3-phosphate dehydrogenase) region was used as a positive control for H3 association. HDAC inhibition did not affect H3 association or acetylation in this region either (Fig. 5A), which has also been observed by others (44–48). Since housekeeping genes are constitutively active, it is possible that the histones associated with these genes are already maximally acetylated and therefore not influenced by SAHA treatment.
FIG 5.
SAHA impacts multiple stages of HAdV life cycle. A549 cells and 10 μM SAHA were used in all experiments. (A) Ad-late/RFP (MOI of 10) was used in a “cold infection” to synchronize virus entry into cells. The cells were treated with vehicle or SAHA and subjected to ChIP at 6 hpi with the indicated antibodies, followed by qPCR with primers specific to HAdV E1A or cellular GAPDH regions. (B) Lysates of cells infected with Ad-late/RFP (MOI of 10) and treated with SAHA were collected at 8 or 24 hpi for immunoblot analysis. (C and G) After infection and drug treatment as in panel B, total cellular RNA was extracted at the indicated times, and cDNA was generated by reverse transcription. qPCR analysis was conducted using primers to the HAdV E1A (C) or hexon (G) regions. (D and H) Infected, SAHA-treated cell lysates were collected at the indicated times for immunoblot analysis to detect viral early (D) and late (H) proteins. (E) Replication-competent Ad(E1+)TP-F was used at an MOI of 50 for infection to assess expression from the E2 region in the presence and absence of SAHA. (F) Infection and drug treatment were carried out as in panel B, and genomic DNA was extracted at the indicated times for qPCR using primers specific to hexon. SAHA did not affect HAdV entry, DNA association with H3, or acetyl-H3/acetyl-H4 levels, but it inhibited the expression of early and late genes and viral DNA replication. All error bars represent the SD (n = 4 for ChIP and n = 2 for all other experiments).
In the HAdV life cycle, the E1 region is the first to be transcribed. We performed reverse transcription-quantitative PCR (RT-qPCR) to analyze E1A transcript levels in infected, drug-treated cells over a period of 24 h. Treatment with SAHA caused >10-fold (1.2-log) reduction in E1A transcript levels at 6 hpi (Fig. 5C), and this was concomitant with a reduction in E1A protein levels (Fig. 5D). Expression of the pTP and DBP from the E2 region was also decreased (Fig. 5E and data not shown). We next examined viral DNA replication through analysis of genome copy numbers obtained by qPCR. The genome copy numbers at 5 hpi (before the onset of viral DNA replication) was identical in cells treated with SAHA or vehicle (Fig. 5F), indicating that the compound did not impact the ability of the genomes from infecting virions to reach the nucleus. Thus, viral attachment and internalization are not affected by SAHA. Viral DNA replication, however, was decreased compared to vehicle over a 30-h time course of infection, especially at its onset at 15 to 20 hpi (Fig. 5F). Even at 48 hpi, Ad-late/RFP genome copy numbers were 3-fold lower in SAHA-treated cells (data not shown). As expected, late gene transcription was inhibited at 24 hpi with an ∼400-fold (2.6-log) decrease in hexon transcripts (Fig. 5G). Lastly, analysis of protein expression from 15 to 30 hpi revealed that SAHA treatment delays detectable late protein expression by about 10 to 15 h in A549 cells (Fig. 5H). Thus, SAHA affects multiple stages of the HAdV life cycle, ultimately leading to a reduction in virus yield.
SAHA impacts HAdV life cycle through mechanisms independent of E1A protein levels.
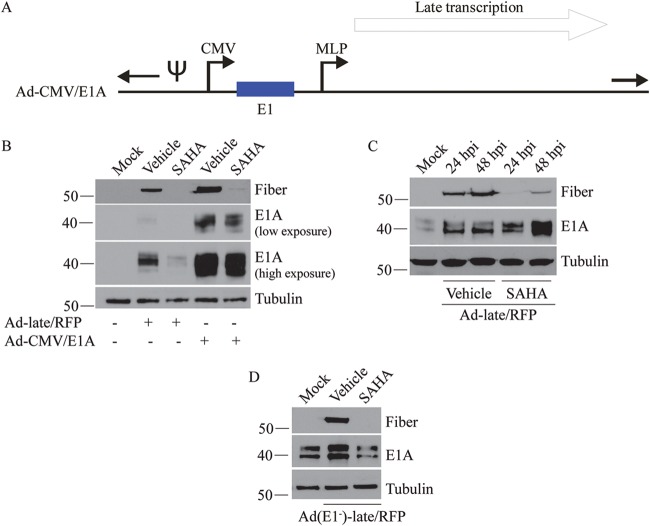
The E1A protein is required for the transcriptional activation of all other HAdV genes and for making the cellular microenvironment more conducive to virus replication (20). Since SAHA reduced E1A expression in early infection, we considered the possibility that the loss of E1A could be adversely impacting the subsequent stages of the virus life cycle (i.e., treatment with SAHA reduces E1A expression, and reduced E1A protein levels cause the decrease in expression of other viral genes). To investigate this, an Ad-CMV/E1A construct was created, where the E1 promoter was replaced with the CMV promoter (Fig. 6A). The purpose of this construct was to overexpress E1A even in the presence of SAHA. A549 cells were infected with Ad-late/RFP or Ad-CMV/E1A, treated with vehicle or SAHA, and examined for early/late gene expression at 24 hpi. Ad-CMV/E1A indeed produced very large amounts of E1A compared to Ad-late/RFP under all conditions (Fig. 6B). In vehicle-treated cells, the enhanced E1A expression from Ad-CMV/E1A was associated with a higher level of fiber expression compared to Ad-late/RFP, likely due to accelerated growth kinetics of Ad-CMV/E1A as a direct consequence of increased E1A expression. However, late gene expression was not fully rescued in SAHA-treated cells (Fig. 6B) or when 293 cells that constitutively express E1A were infected with Ad-late/RFP and Ad(E1–)-late/RFP (Fig. 6C and D). These results suggest that SAHA affects stages of the HAdV life cycle through other E1A-independent mechanisms.
FIG 6.
SAHA affects late gene expression through E1A-independent mechanisms. (A) Schematic diagram of the Ad-CMV/E1A construct (not drawn to scale). The E1 promoter was replaced with the CMV promoter to allow high level expression of the E1 proteins even in the presence of SAHA. (B) A549 cells were infected with Ad-late/RFP or Ad-CMV/E1A (MOI of 10) and treated with vehicle or 10 μM SAHA. Cell lysates were collected for immunoblot analysis of E1A and fiber at 24 h postinfection. (C) 293 cells were infected with Ad-late/RFP (MOI of 10) and treated with 10 μM SAHA for the indicated time points. (D) 293 cells were infected with Ad(E1–)-late/RFP (MOI of 10) and treated with SAHA for 24 h. Neither forced expression of E1A nor infection in the presence of existing E1A from 293 cells fully rescued fiber expression in SAHA-treated cells.
SAHA-induced increase in p21 is not responsible for reduction of HAdV replication.
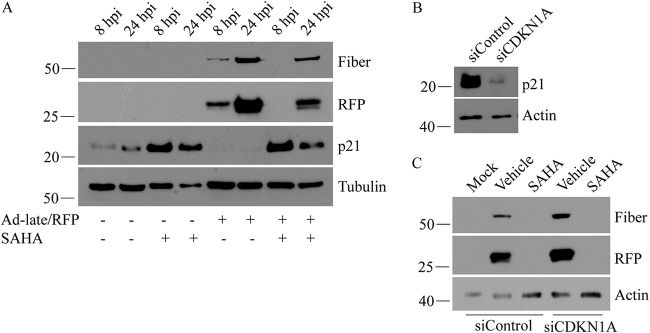
HAdV early proteins naturally induce a transition of the infected cell into S phase in order to provide an optimal environment for virus replication (49). Inactivation of cyclin-dependent kinase inhibitor p21 by the E1A protein is one of the main mechanisms by which this transition occurs (50). SAHA is a U.S. Food and Drug Administration-approved treatment for T-cell lymphoma, and its anticancer properties include the ability to induce cell cycle arrest and prevent progression to S phase through upregulation of p21 (51). Since HDACs typically repress p21 expression (52), Hoti et al. speculated that inhibition of HAdV-5 replication by valproic acid was dependent on the compound’s effects on HDAC activity, leading to p21-mediated cell cycle arrest (36). However, a direct relationship between valproic acid-induced increase in p21 and suppression of HAdV replication has not been shown. Thus, we assessed the correlation between cellular p21 levels and Ad-late/RFP replication in our system. SAHA indeed enhanced p21 protein levels in both infected and uninfected A549 cells (Fig. 7A). Consistent with literature (53), Ad-late/RFP infection caused a reduction in the cellular levels of p21 (Fig. 7A). Treatment with SAHA prevented the loss of p21 in the infected cells, but small interfering RNA (siRNA)-mediated knockdown of p21 (Fig. 7B) did not rescue Ad-late/RFP late gene expression in SAHA-treated cells (Fig. 7C). These results indicate that SAHA’s inhibitory effects on HAdV gene expression and replication was not due to increased p21 and that SAHA acts through other viral (and/or cellular) processes to impair virus replication.
FIG 7.
SAHA-induced increase in p21 is not responsible for the impairment of HAdV replication. (A) p21 levels were detected by immunoblot analysis in both infected (Ad-late/RFP, MOI of 10) and uninfected A549 cells treated with vehicle or 10 μM SAHA. SAHA treatment increases p21 expression in both infected and uninfected cells, while infection with Ad-late/RFP alone decreases expression. (B) Cells were transfected with control siRNA or siRNA specific to the CDKN1A transcript for 48 h to transiently knock down cellular p21. (C) These cells were infected with Ad-late/RFP and treated with vehicle or SAHA. Loss of p21 did not rescue late gene expression at 24 hpi following SAHA treatment.
Class I HDAC activity is necessary for efficient HAdV gene expression.
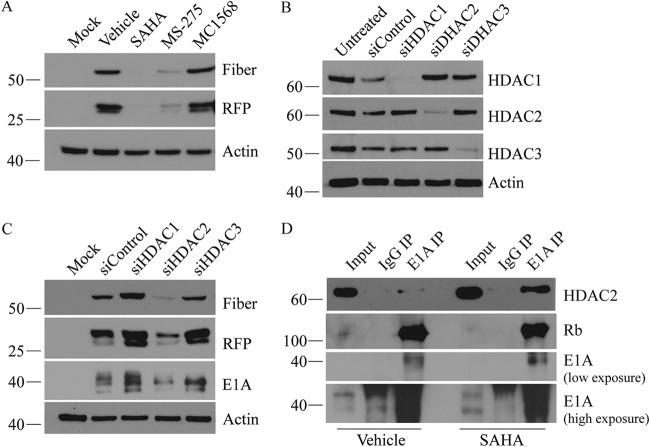
SAHA inhibits both class I and II HDACs, which are comprised of a total of ten HDAC proteins (54, 55). To determine whether suppression of HAdV was due to the inhibition of a specific class of HDAC, we first conducted class-specific HDAC inhibition studies using MS-275 (which preferentially inhibits class I HDACs 1, 2, and 3) and MC1568 (which mainly inhibits class II HDACs 4 and 5) at 10 μM (56). When Ad-late/RFP-infected cells were treated with each drug for 24 h, we observed a reduction in fiber and RFP levels with MS-275 (Fig. 8A), suggesting that inhibition of class I HDAC activity prevents efficient expression of HAdV late genes. Although treatment with MC1568 increased overall H3 acetylation (data not shown), it had no detectable effect on viral gene expression (Fig. 8A). We then knocked down class I HDACs 1, 2, and 3 with siRNAs (Fig. 8B) and found that fiber and RFP decreased only when HDAC2 activity was lost (Fig. 8C). We also investigated whether knocking down of HDAC2 alters Ad-late/RFP DNA association with histones by ChIP and DNA replication by qPCR but did not observe any discernible differences in either (data not shown). These findings suggest that HDAC2 is necessary for efficient HAdV gene expression, but the inhibition of HDAC2 activity alone does not fully account for SAHA’s anti-HAdV effects. Moreover, the HDACs we did not specifically inhibit or individually knock down may also be involved.
FIG 8.
HDAC2 activity is necessary for efficient expression of HAdV late genes. (A) A549 cells were infected with Ad-late/RFP (MOI of 10) and treated with vehicle, SAHA, MS-275, or MC1568 (10 μM each) for 24 h. Late gene expression decreased with MS-275 but not with MC1568. (B) A549 cells were transfected with 100 nM control, HDAC1, HDAC2, or HDAC3 siRNA for 48 h. (C) Cells knocked down for each HDAC were then infected with Ad-late/RFP (MOI of 10) for 24 h to analyze fiber and RFP protein expression. (D) co-IP was performed with IgG (negative control) or E1A antibody on cells infected and drug treated as in panel A. Input was 10% of the immunoprecipitation volume. HDAC2 knockdown decreased fiber and RFP expression, and the protein was found to interact with E1A.
HAdV E1A interacts with a variety of cellular proteins, including transcription factors and epigenetic regulators, such as p300/CBP and E2F1, among many others (35, 57, 58). E1A protein can alter both cellular and viral gene transcription directly by recruiting chromatin modulating proteins to specific promoters or indirectly by interacting with other components of chromatin remodeling complexes (59). We considered the possibility of a physical interaction between E1A and HDAC2 (either directly or through interaction with chromatin remodeling complexes). HDAC2 was indeed found to interact with E1A in coimmunoprecipitation experiments (Fig. 8D). Interestingly, treatment with SAHA appeared to enhance the interaction of HDAC2 with E1A protein (Fig. 8D) in a dose-dependent manner (data not shown). Rb (a positive control for interaction with E1A) was also present in the E1A coimmunoprecipitation (co-IP) samples (Fig. 8D). We did not detect an interaction between HDAC1 and E1A (data not shown).
DISCUSSION
Like several other DNA viruses and retroviruses (e.g., herpes simplex virus, Kaposi's sarcoma-associated herpesvirus, simian virus 40, and murine leukemia virus), the HAdV DNA associates with cellular histones during specific stages of the virus life cycle (60–62). Modifications of these histones has very recently been shown to modulate HAdV gene expression (34), but many questions regarding the “chromatinization” and epigenetic regulation of the HAdV genome still remain unanswered. The cellular proteins and chromatin-remodeling enzymes involved in mediating these processes have yet to be identified. In this study, we have used an E1+, RFP-expressing HAdV reporter virus (Fig. 1) to determine that the HDAC inhibitor SAHA exhibits anti-HAdV activity (Fig. 2). Given that HDACs are naturally associated with decreases in gene repression, we had expected HDAC inhibition to increase HAdV DNA-associated histone acetylation and consequently, viral gene expression. Surprisingly, SAHA reduced late gene expression from several serotypes of HAdV (Fig. 3) and was effective in reducing virus yield even at low concentrations (Fig. 4). Further investigation led us to determine that SAHA negatively impacts various steps in the HAdV life cycle throughout infection, including viral gene expression and DNA replication (Fig. 5), and that this was independent of reduced E1A protein levels (Fig. 6). However, it is unclear whether this effect is due to a complete but transient inhibition, which delayed the onset of viral gene expression and replication, or whether SAHA decreased expression throughout infection leading to a slow accumulation of transcripts/proteins/DNA over time. Furthermore, experiments conducted in MRC-5 cells (Fig. 2) indicate that SAHA’s effects last longer in primary cells. Taken together, these results highlight the potential of HDAC inhibitors as anti-HAdV compounds.
Our data suggest a role of HDACs, specifically class I HDACs, in modulating HAdV gene expression (Fig. 8). Knocking down HDAC2 reduced HAdV late gene expression, and we further showed that HDAC2 interacts with the early protein E1A (Fig. 8). E1A could be involved in redirecting or recruiting HDAC2 to protein substrates that are important for viral replication. HDAC2 is well known for its ability to modify histones, and has specific targets on H3 and H4 (e.g., H3K9 and H4K16). Although SAHA treatment did not appear to cause a change in acetylation status of H3 or H4 in the E1A/hexon coding regions we examined (Fig. 5), loss of HDAC2 activity could affect acetylation at the promoter regions or transcriptional start sites of the early and late genes, since H3 acetylation was observed at HAdV-2 early promoters (34). Interestingly, knockdown of the histone acetyltransferase Tip60 and reduced H4 acetylation actually resulted in increased HAdV-5 E1A promoter activity (63). Thus, HAdV may have a complex or temporal requirement for histone acetylation for proper promoter function. Alternatively, HDAC2 may even target specific lysine acetyl groups on the E1A protein itself to regulate its function (64, 65). However, HDAC2-E1A association is likely not restricted to the enzyme’s active site as association increases in the presence of SAHA, a competitive inhibitor which occupies the active site of HDAC2 and prevents substrate binding (Fig. 8). It is also possible that HDAC2 binding interferes with other E1A functions (e.g., interaction with other proteins) or that the interaction opposes the beneficial functions of HDAC2, contributing to the impairment of virus replication observed with SAHA.
Our studies have uncovered a specific role for HDAC2 in activating HAdV gene expression, which cannot be compensated for by HDAC1 or HDAC3. Knockdown of HDAC1 and HDAC3 slightly enhanced expression of HAdV genes (Fig. 8). This observation is similar to that of Wang et al. for the yeast HDACs Rpd3 and Hos2 (66). Although both are class I HDACs, Rpd3 represses the yeast GAL1 gene, while Hos2 activates GAL1. It is also intriguing that HDAC1 cannot fully compensate for the loss of HDAC2 activity in our system. The two enzymes coexist in several corepressor complexes, and while they are generally known to be functionally redundant and compensatory, some studies report that HDAC2 has distinct targets and plays distinct roles in certain cellular processes (67, 68). For example, Feng et al. found that the respiratory syncytial virus decreases histone acetylation by upregulating HDAC2 expression in primary airway epithelial cells (69). However, we did not observe any such changes in HDAC2 levels in immunoblot analyses over a 48-h time course of HAdV-5 infection in A549 cells (data not shown).
It is important to note that HDAC2 knockdown alone did not recapitulate the full extent of SAHA-mediated inhibition (compare Fig. 8A and C). While this may reflect the relative efficiency of drug treatment versus siRNA-mediated knockdown, it may also suggest that other cellular pathways or processes affected by SAHA, besides those regulated through HDAC2, are important for HAdV function during a productive infection. In addition to modifying histone acetylation status, HDACs have numerous other nonhistone targets, including proteins involved in transcription regulation, cell proliferation, metabolism, apoptosis, and immune response (70). HDAC inhibitors have been shown to alter the expression level and function of cellular proteins p21, E2F1, and SP1 (71–74), all of which can impact upon optimal viral gene expression and growth. The transcription factors E2F1 and SP1 both play a role in transcriptional control of E1A (75, 76). E2F1 and SP1 also bind to the E2 promoter and the MLP, respectively, and binding is required for the maximum expression of proteins from those regions (17, 58, 77–79). Although we were unable to rescue HAdV gene expression via knockdown of p21 (Fig. 7) or overexpression of E2F1 or SP1 (data not shown), a more sensitive experimental design may be required to conclusively exclude involvement of these proteins. SAHA’s effects on HAdV may also be a cumulative consequence of several factors, as opposed to changes in a single pathway or protein. While further investigation is required to fully understand the underlying molecular mechanism, our findings enhance the current knowledge of the HAdV infection process and elucidate the need of HDAC2 activity for optimal viral gene expression and replication. Future studies should also focus on determining the in vivo efficacy of SAHA against HAdV in the Syrian hamster or the cotton rat model, both of which are permissive for HAdV infection.
The costs associated with HAdV-induced disease are significant in terms of medical expenses, lost work hours, and loss of life in some populations. Thus, development of novel and effective antiviral therapies is necessary to reduce disease pathogenesis and increase survival rates in severe infections. Our results show that SAHA led to a significant reduction in virus yield at low concentrations and was effective in reducing gene expression from virulent serotypes 4 and 7. Understanding the molecular mechanism underlying SAHA’s inhibitory effects on HAdV may make it possible to design better compounds with longer-lasting effects and lower toxicity for the treatment of HAdV infections. As HDAC inhibitors have been reported to exhibit antiviral activity against other viruses, including respiratory syncytial virus and hepatitis C virus (69, 80), these novel compounds may be useful in treating a variety of viral infections.
MATERIALS AND METHODS
Cell lines and reagents.
The 293 cell line was kindly provided by Frank Graham (McMaster University) (81). The A549-based cell line used in our experiments constitutively express the green fluorescent protein from an integrated lentivirus vector and has been previously described (82). These cells grow and support HAdV replication identical to the parental A549 cell line (CCL-185; American Type Culture Collection [ATCC]) and, for simplicity, will be referred to as A549 cells in this study. Both 293 and A549 cells were grown in minimum essential medium (MEM; Sigma-Aldrich) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Sigma-Aldrich), 2 mM GlutaMAX (Invitrogen), and 1× antibiotic-antimycotic (Invitrogen). MRC-5 cells (CCL-171; ATCC) were grown similarly in Dulbecco minimum essential medium (Sigma-Aldrich). The following seeding densities were used unless otherwise indicated: 2 × 104 cells per well for a 96-well plate, 4 × 105 cells per well for a 12-well plate, 1.2 × 106 cells per well for a 6-well plate, and 8.8 × 106 cells for a 10-cm dish.
SAHA and TSA were purchased from Sigma (SML0061 and T8552) and MC1568 was purchased from Cayman Chemicals (catalog no. 16265). MS-275 was kindly provided by Jean-Simon Diallo (Ottawa Hospital Research Institute). All compounds were dissolved in dimethyl sulfoxide.
Production of HAdV constructs.
All HAdV constructs used this study are based on HAdV-5 and were constructed using a combination of conventional cloning and bacterial recombination (83). To generate the Ad-late/RFP virus, the monomeric red fluorescent protein (RFP) (84) was excised from pRP2483 (85) with BamHI and cloned into BamHI-digested pDC5 (85). As such, the RFP gene was placed behind a splice acceptor derived from the HAdV-40 long fiber gene, generating pRP3085. A PvuI fragment from pRP3085 was cloned into PacI-digested pDC9 (85), placing the RFP cassette within the E3 deletion in an HAdV right-end shuttle plasmid. This construct was designated pRP3087 and then recombined into an HAdV E1/E3-deleted genomic plasmid pRP2468, generating pRP3088. pRP3088 was recombined with pXC1 (86), producing pRP3089, an HAdV genomic plasmid containing an intact E1 region, with the E3 region replaced by splice acceptor-RFP cassette. pRP3089 was recovered as a virus and designated Ad-late/RFP for this study. pRP3088 was also recombined with pAVH5 (85) to generate pRP3090, which is an HAdV genomic plasmid containing an enhanced green fluorescent protein under regulation of the human CMV enhancer/promoter and the bovine hormone polyadenylation sequence replacing the E1 region, as well as the splice acceptor-RFP cassette replacing the E3 region. pRP3090 was recovered as a virus and designated Ad(E1–)-late/RFP in this study.
Ad-CMV/E1A (AdRP3169) contains a small murine CMV promoter from pMH4 (87) inserted at 350 bp of the conventional HAdV-5 genome, followed by the E1 region beginning at 420 bp of the HAdV-5 genome. It is deleted of the E3 region. Ad(E1–)-CMV/RFP (AdRP2619) and Ad(E1+)TP-F (AdRP3000) constructs have been described previously (17, 88, 89). All viruses were propagated in 293 cells and purified by cesium chloride buoyant density centrifugation using standard procedures (90). Viral titers were determined by plaque assay on 293 cells as previously reported (17).
Infections and drug treatment.
Unless otherwise indicated, medium was removed from confluent monolayers of A549 cells before infecting with the appropriate HAdV constructs at the indicated multiplicity of infection (MOI). MOIs were calculated as the PFU per cell, and an MOI of 10 was used for most experiments. Virus inoculums were diluted in phosphate-buffered saline (PBS; without CaCl2 and MgCl2; Sigma-Aldrich) and added to the cell monolayer for 1 h at 37°C with periodic rocking. Medium containing either vehicle or drug was then added to the cells and incubated in a humidified CO2 incubator at 37°C until the indicated time points. This medium (with vehicle or drug) was replaced every 24 h of infection.
For synchronized or cold infections, the cells were incubated with the virus inoculum for 30 min on ice, allowing the viruses to attach but not enter the cells. Infection started when warm media (containing vehicle or drug) was added, and the cells were placed at 37°C.
Immunoblot analysis.
Protein samples were prepared in 2× Laemmli buffer, subjected to SDS-PAGE and transferred on a polyvinylidene difluoride membrane (Millipore) as previously described (17). The membrane was blocked in 5% milk prepared in Tris-buffered saline containing 0.2% Tween 20 (Thermo Fisher Scientific) and probed with antibodies diluted in 5% milk solution. The following primary antibodies were used: HAdV-5 fiber (1/10,000 dilution; MS-1027-P0; Neomarkers), anti-HAdV-5 (1/10,000; ab6982; Abcam), HAdV-5 E1A (1/5,000; ab31686 [Abcam] or MA5-13643 [Invitrogen]), HDAC1 (1/2,000; 5356P; Cell Signaling), HDAC2 (1/5,000; ab16032; Abcam), HDAC3 (1/2,000; 3949S; Cell Signaling), acetyl-H3 (1/2,000; 06-599; Millipore), vinculin (1/10,000; ab129002; Abcam), tubulin (1/5,000; CP06; Calbiochem), actin (1/10,000; A1978; Sigma-Aldrich), Rb (1/5,000; 554136; BD Pharmingen), FLAG (M2, 1/5,000; 200-301-383S; Rockland), and RFP (1/5,000; ab62341; Abcam). The p21 antibody (1/1,000; catalog no. 2947; New England Biolabs) was kindly provided by Lynn Megeney (Ottawa Hospital Research Institute). The membranes were then washed twice in Tris-buffered saline–Tween and incubated with the appropriate secondary antibodies conjugated to horseradish peroxidase (HRP). Blots were developed using Pierce ECL Western blotting substrate (Thermo Fisher Scientific) or Immobilon Classico Western HRP substrate (Millipore). All immunoblot data are representative of two or more independent experiments.
Quantitative real-time PCR.
Medium was removed from infected cells at the indicated time points, and the cells were incubated overnight at 37°C in SDS-proteinase K buffer (10 mM Tris-HCl [pH 7.4], 10 mM EDTA, 1% [wt/vol] SDS, 1 mg/ml proteinase K). DNA was extracted from the cell lysates using the standard phenol-chloroform method and ethanol precipitation and then dissolved in 1× Tris-EDTA (TE) buffer. qPCR was conducted as previously described (89) using 200 ng of genomic DNA per reaction. The following primer sets were used: 5′-CTT ACC CCC AAC GAG TTT GA and 5′-GGA GTA CAT GCG GTC CTT GT for HAdV hexon, 5′-CCA TTA AAC CAG TTG CCG TGA GAG and 5′-GGC GTT TAC AGC TCA AGT CCA AAG for HAdV E1A, and 5′-GGC CGG TGC TGA GTA TGT CG and 5′-TTC AAG TGG GCC CCG GCC TT for cellular GAPDH. Viral genome copy numbers were calculated from the CT values using a standard curve obtained from serial dilutions of the pRP3089 plasmid.
RNA extraction and RT-qPCR.
Medium was removed from infected cells and total cellular RNA was extracted using TRIzol Reagent (Thermo Fisher Scientific) and a PureLink RNA minikit (Ambion) according to the manufacturer’s protocol. To synthesize cDNA from the extracted RNA by reverse transcription (RT), a mixture of random hexamers (New England Biolabs) and the Moloney murine leukemia virus reverse transcriptase (New England Biolabs) was used according to the manufacturer’s protocol. Then, 800 ng of RNA was used per RT reaction. qPCR was then conducted as described above using 2 μl of the cDNA mixture.
Plaque assay to determine virus yield.
Monolayers of A549 cells were infected with Ad-late/RFP or HAdV-5 (MOI of 10), and the virus inoculum was removed after 1 h of infection. The cells were washed with PBS, and fresh medium containing vehicle or various concentrations of SAHA was added. After 24 to 48 h of infection, the cells were collected by scraping into the medium, sucrose (diluted in 10 mM Tris) was added to a final concentration of 4% (vol/vol), and the samples were flash-frozen in liquid nitrogen. For the plaque assay, monolayers of A549 cells were infected with serial dilutions of this cell lysate. After 1 h of infection at 37°C, the cells were overlaid with medium containing agarose (50% [vol/vol] of a 1% [wt/vol] agarose solution, 43% clear 2× MEM, 5% FBS, 1% GlutaMAX, and 1% antibiotic-antimycotic). Plaques were counted 8 to 10 days later.
Fluorescence microscopy.
Live imaging of infected A549 cells was performed 24 h after infection using a 10× objective on a Zeiss Axio Observer.Z1 microscope equipped with an Axiocam MRC camera and ZEN 2 software for image processing.
Coimmunoprecipitation.
Ten-centimeter dishes of A549 cells were infected with Ad-late/RFP (MOI of 10) and treated with vehicle or drug. After 24 h, the medium was removed, and the cells were collected in 500 μl of cold PBS. After centrifugation at 1,500 × g, the volumes of the cell pellets were estimated, and the pellets were resuspended in twice the volume of lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 2 mM MgCl2, 0.5 mM EDTA, 0.5% Triton X-100, and protease inhibitors) for 30 min on ice. Cell lysates were cleared by centrifugation and diluted to 1 ml with additional lysis buffer. After ∼1.5 h of preclearing with protein G-beads (1004D; Dynabeads), equal volumes of lysate were added to 30 μl of protein G-beads that had been preincubated for 5 h with 4 μg of E1A antibody (ab31686; Abcam) or IgG (5415S; Cell Signaling) as a negative control. Next, 10% of the immunoprecipitation volume was retained as an input sample. The antibody-bead complex was incubated with the cell lysate overnight at 4°C. The protein-bound beads were washed twice with lysis buffer and eluted with 2× Laemmli buffer for 15 min at 65°C. β-Mercaptoethanol was added to a final concentration of 0.05% prior to subjecting the samples to SDS-PAGE and immunoblotting with the indicated antibodies.
Chromatin immunoprecipitation.
Ten-centimeter dishes of A549 cells were infected with Ad-late/RFP (MOI of 10) using the synchronized infection method and treated with vehicle or drug. After 6 h of infection, the medium was removed, and the cells were washed in cold PBS. Then, 8 to 10 aliquots of roughly 500,000 cells were suspended in 1 ml of medium each, and 100 μl of formaldehyde buffer (50 mM HEPES-KOH, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 11% formaldehyde) was added to allow protein-DNA cross-linking. After 10 min of incubation on a rotating platform, the cells were treated with 50 μl of 2.5 M glycine for 5 min and washed three times in cold PBS (containing calcium and magnesium) with centrifugation at 1,350 × g for 5 min at 4°C. The PBS was discarded, and the cell pellets were flash frozen and stored at –80°C overnight.
To prepare the antibody-bead complex for immunoprecipitation, 50 μl of magnetic protein G-beads (1004D; Dynabeads) was washed in blocking solution (5% bovine serum albumin in PBS) three times, followed by incubation in 250 μl of blocking buffer containing 10 μg of IgG (SC-2027X; Santa Cruz), histone H3 (ab1791; Abcam), acetyl-H3 (ab47915; Abcam), or acetyl-H4 (06-598; Millipore) antibody overnight at 4°C to allow antibody-bead complex formation. The unbound antibody was removed from the beads prior to the addition of sheared DNA for ChIP (see below).
The frozen cell pellets were resuspended in 140 μl of lysis buffer (0.1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8], 1× protease inhibitor) and sonicated for 10 min at 4°C using a Covaris S220 sonicator to 500- to 1,000-bp fragments. The sheared DNA samples were then diluted to 0.1 μg/μl in lysis buffer. Equal amounts of DNA were added to 100 μl of the various antibody-bead complexes and incubated overnight at 4°C with rotation. The next day, the beads (now complexed with DNA) were washed for 5 min each in low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8], 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris HCl [pH 8], 500 mM NaCl), wash buffer (0.25 M LiCl, 1% GEPAL, 1% deoxycholic acid, 1 mM EDTA, 10 mM Tris [pH 8]), and 1× TE buffer. The DNA was eluted from the beads in 200 μl of elution buffer (50 mM Tris-HCl [pH 8], 10 mM, 1% SDS) using a thermomixer at 65°C for 15 min. The eluate was separated from the beads and incubated overnight at 65°C with 20 μg of proteinase K (Sigma-Aldrich) to reverse DNA-protein cross-link and digest the proteins. The next day, the DNA samples were treated with 20 μg of RNase (Sigma-Aldrich) for 30 min at 37°C. One volume of phenol-chloroform was added to each sample, which were vortexed briefly and centrifuged at 15,000 × g for 15 min at 4°C. The aqueous phase was collected, and the following were added prior to overnight incubation at –80°C: 1/10 volume of 3 M sodium acetate, 5 μg of glycogen (Roche), 5 μg of linear acrylamide (Thermo Fisher Scientific), and 2.5 volumes of 99% ethanol. The next day, all of the samples were centrifuged for 30 min at 4°C to obtain a DNA pellet, which was washed in 70% ethanol, air dried, and dissolved in 20 μl of 1× TE buffer. qPCR was performed as described above using 2 μl of this ChIP DNA.
Quantification of RFP intensity.
Next, 96-well plates of A549 cells were infected with Ad-late/RFP (MOI of 10) and treated with vehicle or HDAC inhibitors. Virus inoculum and medium containing drug were added to the existing cell medium simultaneously. After 24 h, the medium was removed, and the cells were washed with PBS and fixed with 4% paraformaldehyde (pH 7). The cell nuclei were stained with 10 μg/ml Hoechst 33342 (Life Technologies) for ∼20 min before RFP quantification using a Cellomics high content screening (HCS) platform and the Cellomics navigator version of the HCS Studio software (Thermo Fisher Scientific). A 10× objective and the following filters were used: 386excitation (ex) nm for Hoechst and 549ex nm for RFP. Roughly 1,000 cells (Hoechst-stained intact nuclei) were analyzed per well, and their total RFP intensity was measured. Further information on the quantification parameters is available upon request.
siRNA-mediated knockdown.
Pooled siRNA targeting human HDAC1 (M-003493-02), HDAC2 (M-003495-02), HDAC3 (M-003496-02), p21 (M-003471-00), and siGenome non-targeting control siRNA were obtained from Dharmacon. A total of 5 × 104 A549 cells were plated in 24-well plates and transfected with 100 nM siRNA using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions for A549 cells. Transfection medium was replaced with regular medium after 5 h of treatment. Knockdown was confirmed at 48 h posttransfection via immunoblot analysis, and the knockdown cells were infected at this point.
MTS metabolic activity assays.
Plates (96 well) of A549 cells were prepared and infected/drug treated as described above for RFP quantification. At 24 h postinfection, the relative metabolic activity was determined using a CellTiter 96 AQueous nonradioactive cell proliferation assay (Promega) according to the manufacturer’s instructions. Cells were incubated for 45 min at 37°C with 20 μl of MTS substrate, and absorbance readings were obtained at 490 nm using a SpectraMax 190 plate spectrophotometer.
ACKNOWLEDGMENTS
We thank Jean-Simon Diallo for providing the MS-275 used in this study and Michael Rudnicki for sharing the co-IP protocol. We also thank William Stanford for kindly sharing the ChIP protocol and for access to equipment such as the Covaris ultrasonicator and the Cellomics HCS platform.
This research was supported by grants to R.J.P. from the Canadian Institute of Health Research (MOP-142316) and the Natural Sciences and Engineering Research Council of Canada (RGPIN-2014-04810). B.S. was supported by the Ontario Graduate Scholarship and the Queen Elizabeth II Graduate Scholarship in Science and Technology from the Ontario Government.
REFERENCES
- 1.Binder AM, Biggs HM, Haynes AK, Chommanard C, Lu X, Erdman DD, Watson JT, Gerber SI. 2017. Human adenovirus surveillance: United States, 2003-2016. MMWR Morb Mortal Wkly Rep 66:1039–1042. doi: 10.15585/mmwr.mm6639a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott MK, Chommanard C, Lu X, Appelgate D, Grenz L, Schneider E, Gerber SI, Erdman DD, Thomas A. 2016. Human adenovirus associated with severe respiratory infection, Oregon, USA, 2013-2014. Emerg Infect Dis 22:1044–1051. doi: 10.3201/eid2206.151898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kajon AE, Lamson DM, Bair CR, Lu X, Landry ML, Menegus M, Erdman DD, St George K. 2018. Adenovirus type 4 respiratory infections among civilian adults, northeastern United States, 2011-2015(1). Emerg Infect Dis 24:201–209. doi: 10.3201/eid2402.171407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kojaoghlanian T, Flomenberg P, Horwitz MS. 2003. The impact of adenovirus infection on the immunocompromised host. Rev Med Virol 13:155–171. doi: 10.1002/rmv.386. [DOI] [PubMed] [Google Scholar]
- 5.Ying B, Tollefson AE, Spencer JF, Balakrishnan L, Dewhurst S, Capella C, Buller RM, Toth K, Wold WS. 2014. Ganciclovir inhibits human adenovirus replication and pathogenicity in permissive immunosuppressed Syrian hamsters. Antimicrob Agents Chemother 58:7171–7181. doi: 10.1128/AAC.03860-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caravokyri C, Leppard KN. 1996. Human adenovirus type 5 variants with sequence alterations flanking the E2A gene: effects on E2 expression and DNA replication. Virus Genes 12:65–75. doi: 10.1007/BF00370002. [DOI] [PubMed] [Google Scholar]
- 7.Bhatti Z, Dhamoon A. 2017. Fatal adenovirus infection in an immunocompetent host. Am J Emerg Med 35:1034.e1–1034.e2. doi: 10.1016/j.ajem.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Adriana EK, Michael GI. 2016. Severe infections with human adenovirus 7d in 2 adults in family, Illinois, USA, 2014. Emerg Infect Dis J 22:730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ronchi A, Doern C, Brock E, Pugni L, Sanchez PJ. 2014. Neonatal adenoviral infection: a seventeen year experience and review of the literature. J Pediatr 164:529–535.e1-4. doi: 10.1016/j.jpeds.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Lenaerts L, De Clercq E, Naesens L. 2008. Clinical features and treatment of adenovirus infections. Rev Med Virol 18:357–374. doi: 10.1002/rmv.589. [DOI] [PubMed] [Google Scholar]
- 11.Keyes A, Mathias M, Boulad F, Lee YJ, Marchetti MA, Scaradavou A, Spitzer B, Papanicolaou GA, Wieczorek I, Busam KJ. 2016. Cutaneous involvement of disseminated adenovirus infection in an allogeneic stem cell transplant recipient. Br J Dermatol 174:885–888. doi: 10.1111/bjd.14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Detweiler CJ, Mueller SB, Sung AD, Saullo JL, Prasad VK, Cardona DM. 2018. Brincidofovir (CMX001) toxicity associated with epithelial apoptosis and crypt drop out in a hematopoietic cell transplant patient: challenges in distinguishing drug toxicity from GVHD. J Pediatr Hematol Oncol 40(6):e364–e368. doi: 10.1097/mph.0000000000001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramsay ID, Attwood C, Irish D, Griffiths PD, Kyriakou C, Lowe DM. 2017. Disseminated adenovirus infection after allogeneic stem cell transplant and the potential role of brincidofovir: case series and 10 year experience of management in an adult transplant cohort. J Clin Virol 96:73–79. doi: 10.1016/j.jcv.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Rowe WP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG. 1953. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc Soc Exp Biol Med 84:570–573. doi: 10.3181/00379727-84-20714. [DOI] [PubMed] [Google Scholar]
- 15.Hilleman MR, Werner JH. 1954. Recovery of new agent from patients with acute respiratory illness. Proc Soc Exp Biol Med 85:183–188. doi: 10.3181/00379727-85-20825. [DOI] [PubMed] [Google Scholar]
- 16.Thomas GP, Mathews MB. 1980. DNA replication and the early to late transition in adenovirus infection. Cell 22:523–533. doi: 10.1016/0092-8674(80)90362-1. [DOI] [PubMed] [Google Scholar]
- 17.Saha B, Parks RJ. 2017. Human adenovirus type 5 vectors deleted of early region 1 (E1) undergo limited expression of early replicative E2 proteins and DNA replication in non-permissive cells. PLoS One 12:e0181012. doi: 10.1371/journal.pone.0181012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flint J. 1999. Adenoviruses: basic biology to gene therapy, p 17–30. R. G. Landes Company, Austin, TX. [Google Scholar]
- 19.Frisch SM, Mymryk JS. 2002. Adenovirus-5 E1A: paradox and paradigm. Nat Rev Mol Cell Biol 3:441–452. doi: 10.1038/nrm827. [DOI] [PubMed] [Google Scholar]
- 20.Berk AJ. 1986. Adenovirus promoters and E1A transactivation. Annu Rev Genet 20:45–79. doi: 10.1146/annurev.ge.20.120186.000401. [DOI] [PubMed] [Google Scholar]
- 21.de Jong RN, Meijer LAT, van der Vliet PC. 2003. DNA binding properties of the adenovirus DNA replication priming protein pTP. Nucleic Acids Res 31:3274–3286. doi: 10.1093/nar/gkg405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker EJ, Botting CH, Webster A, Hay RT. 1998. Adenovirus DNA polymerase: domain organization and interaction with preterminal protein. Nucleic Acids Res 26:1240–1247. doi: 10.1093/nar/26.5.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horwitz MS. 2004. Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J Gene Med 6:S172–S183. doi: 10.1002/jgm.495. [DOI] [PubMed] [Google Scholar]
- 24.Weitzman MD. 2005. Functions of the adenovirus E4 proteins and their impact on viral vectors. Front Biosci 10:1106–1117. doi: 10.2741/1604. [DOI] [PubMed] [Google Scholar]
- 25.Prescott JC, Falck-Pedersen E. 1992. Varied poly(A) site efficiency in the adenovirus major late transcription unit. J Biol Chem 267:8175–8181. [PubMed] [Google Scholar]
- 26.Young CS. 2003. The structure and function of the adenovirus major late promoter. Curr Top Microbiol Immunol 272:213–249. [DOI] [PubMed] [Google Scholar]
- 27.Korn R, Horwitz MS. 1986. Adenovirus DNA synthesis in vitro is inhibited by the virus-coded major core protein. Virology 150:342–351. doi: 10.1016/0042-6822(86)90299-0. [DOI] [PubMed] [Google Scholar]
- 28.Strunze S, Engelke MF, Wang IH, Puntener D, Boucke K, Schleich S, Way M, Schoenenberger P, Burckhardt CJ, Greber UF. 2011. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe 10:210–223. doi: 10.1016/j.chom.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Komatsu T, Nagata K. 2012. Replication-uncoupled histone deposition during adenovirus DNA replication. J Virol 86:6701–6711. doi: 10.1128/JVI.00380-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giberson A, Saha B, Campbell K, Christou C, Poulin KL, Parks RJ. 2018. Human adenoviral DNA association with nucleosomes containing histone variant H3.3 during the early phase of infection is not dependent on viral transcription or replication. Biochem Cell Biol 96:797–807. doi: 10.1139/bcb-2018-0117. [DOI] [PubMed] [Google Scholar]
- 31.Daniell E, Groff DE, Fedor MJ. 1981. Adenovirus chromatin structure at different stages of infection. Mol Cell Biol 1:1094–1105. doi: 10.1128/MCB.1.12.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beyer AL, Bouton AH, Hodge LD, Miller OL Jr.. 1981. Visualization of the major late R strand transcription unit of adenovirus serotype 2. J Mol Biol 147:269–295. doi: 10.1016/0022-2836(81)90441-1. [DOI] [PubMed] [Google Scholar]
- 33.Giberson AN, Davidson AR, Parks RJ. 2012. Chromatin structure of adenovirus DNA throughout infection. Nucleic Acids Res 40:2369–2376. doi: 10.1093/nar/gkr1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu E, Pennella MA, Zemke NR, Eng C, Berk AJ. 2018. Adenovirus E1A activation domain regulates H3 acetylation affecting varied steps in transcription at different viral promoters. J Virol 92:e00805-18. doi: 10.1128/jvi.00805-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelka P, Ablack JN, Torchia J, Turnell AS, Grand RJ, Mymryk JS. 2009. Transcriptional control by adenovirus E1A conserved region 3 via p300/CBP. Nucleic Acids Res 37:1095–1106. doi: 10.1093/nar/gkn1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoti N, Chowdhury W, Hsieh JT, Sachs MD, Lupold SE, Rodriguez R. 2006. Valproic acid, a histone deacetylase inhibitor, is an antagonist for oncolytic adenoviral gene therapy. Mol Ther 14:768–778. doi: 10.1016/j.ymthe.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Tollefson AE, Scaria A, Hermiston TW, Ryerse JS, Wold LJ, Wold WS. 1996. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol 70:2296–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross PJ, Kennedy MA, Parks RJ. 2009. Host cell detection of noncoding stuffer DNA contained in helper-dependent adenovirus vectors leads to epigenetic repression of transgene expression. J Virol 83:8409–8417. doi: 10.1128/JVI.00796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldsmith ME, Aguila A, Steadman K, Martinez A, Steinberg SM, Alley MC, Waud WR, Bates SE, Fojo T. 2007. The histone deacetylase inhibitor FK228 given prior to adenovirus infection can boost infection in melanoma xenograft model systems. Mol Cancer Ther 6:496–505. doi: 10.1158/1535-7163.MCT-06-0431. [DOI] [PubMed] [Google Scholar]
- 40.Kelly WK, O’Connor OA, Krug LM, Chiao JH, Heaney M, Curley T, MacGregore-Cortelli B, Tong W, Secrist JP, Schwartz L, Richardson S, Chu E, Olgac S, Marks PA, Scher H, Richon VM. 2005. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J Clin Oncol 23:3923–3931. doi: 10.1200/JCO.2005.14.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryan QC, Headlee D, Acharya M, Sparreboom A, Trepel JB, Ye J, Figg WD, Hwang K, Chung EJ, Murgo A, Melillo G, Elsayed Y, Monga M, Kalnitskiy M, Zwiebel J, Sausville EA. 2005. Phase I and pharmacokinetic study of MS-275, a histone deacetylase inhibitor, in patients with advanced and refractory solid tumors or lymphoma. J Clin Oncol 23:3912–3922. doi: 10.1200/JCO.2005.02.188. [DOI] [PubMed] [Google Scholar]
- 42.Lee JH, Choy ML, Ngo L, Foster SS, Marks PA. 2010. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc Natl Acad Sci U S A 107:14639–14644. doi: 10.1073/pnas.1008522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo RX, Dean DC. 1999. Chromatin remodeling and transcriptional regulation. J Natl Cancer Inst 91:1288–1294. doi: 10.1093/jnci/91.15.1288. [DOI] [PubMed] [Google Scholar]
- 44.Kumari A, Cacan E, Greer SF, Garnett-Benson C. 2013. Turning T cells on: epigenetically enhanced expression of effector T-cell costimulatory molecules on irradiated human tumor cells. J Immunother Cancer 1:17. doi: 10.1186/2051-1426-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gavin DP, Kartan S, Chase K, Jayaraman S, Sharma RP. 2009. Histone deacetylase inhibitors and candidate gene expression: an in vivo and in vitro approach to studying chromatin remodeling in a clinical population. J Psychiatr Res 43:870–876. doi: 10.1016/j.jpsychires.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 46.To KK, Polgar O, Huff LM, Morisaki K, Bates SE. 2008. Histone modifications at the ABCG2 promoter following treatment with histone deacetylase inhibitor mirror those in multidrug-resistant cells. Mol Cancer Res 6:151–164. doi: 10.1158/1541-7786.MCR-07-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambucetti LC, Fischer DD, Zabludoff S, Kwon PO, Chamberlin H, Trogani N, Xu H, Cohen D. 1999. Histone deacetylase inhibition selectively alters the activity and expression of cell cycle proteins leading to specific chromatin acetylation and antiproliferative effects. J Biol Chem 274:34940–34947. doi: 10.1074/jbc.274.49.34940. [DOI] [PubMed] [Google Scholar]
- 48.Hu QP, Mao DA. 2016. Histone deacetylase inhibitor SAHA attenuates post-seizure hippocampal microglia TLR4/MYD88 signaling and inhibits TLR4 gene expression via histone acetylation. BMC Neurosci 17:22. doi: 10.1186/s12868-016-0264-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ben-Israel H, Kleinberger T. 2002. Adenovirus and cell cycle control. Front Biosci 7:d1369–d1395. doi: 10.2741/ben. [DOI] [PubMed] [Google Scholar]
- 50.Chattopadhyay D, Ghosh MK, Mal A, Harter ML. 2001. Inactivation of p21 by E1A leads to the induction of apoptosis in DNA-damaged cells. J Virol 75:9844–9856. doi: 10.1128/JVI.75.20.9844-9856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. 2000. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci U S A 97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson AJ, Byun DS, Popova N, Murray LB, L’Italien K, Sowa Y, Arango D, Velcich A, Augenlicht LH, Mariadason JM. 2006. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem 281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- 53.Flak MB, Connell CM, Chelala C, Archibald K, Salako MA, Pirlo KJ, Lockley M, Wheatley SP, Balkwill FR, McNeish IA. 2010. p21 promotes oncolytic adenoviral activity in ovarian cancer and is a potential biomarker. Mol Cancer 9:175. doi: 10.1186/1476-4598-9-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dokmanovic M, Clarke C, Marks PA. 2007. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res 5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 55.Butler LM, Zhou X, Xu WS, Scher HI, Rifkind RA, Marks PA, Richon VM. 2002. The histone deacetylase inhibitor SAHA arrests cancer cell growth, upregulates thioredoxin-binding protein-2, and downregulates thioredoxin. Proc Natl Acad Sci U S A 99:11700–11705. doi: 10.1073/pnas.182372299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scognamiglio A, Nebbioso A, Manzo F, Valente S, Mai A, Altucci L. 2008. HDAC-class II specific inhibition involves HDAC proteasome-dependent degradation mediated by RANBP2. Biochim Biophys Acta 1783:2030–2038. doi: 10.1016/j.bbamcr.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 57.Swaminathan S, Thimmapaya B. 1996. Transactivation of adenovirus E2-early promoter by E1A and E4 6/7 in the context of viral chromosome. J Mol Biol 258:736–746. doi: 10.1006/jmbi.1996.0283. [DOI] [PubMed] [Google Scholar]
- 58.O’Connor RJ, Hearing P. 2000. The E4-6/7 protein functionally compensates for the loss of E1A expression in adenovirus infection. J Virol 74:5819–5824. doi: 10.1128/JVI.74.13.5819-5824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gallimore PH, Turnell AS. 2001. Adenovirus E1A: remodelling the host cell, a life or death experience. Oncogene 20:7824–7835. doi: 10.1038/sj.onc.1204913. [DOI] [PubMed] [Google Scholar]
- 60.Wang GZ, Wang Y, Goff SP. 2016. Histones are rapidly loaded onto unintegrated retroviral DNAs soon after nuclear entry. Cell Host Microbe 20:798–809. doi: 10.1016/j.chom.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Milavetz BI, Balakrishnan L. 2015. Viral epigenetics. Methods Mol Biol 1238:569–596. doi: 10.1007/978-1-4939-1804-1_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knipe DM, Lieberman PM, Jung JU, McBride AA, Morris KV, Ott M, Margolis D, Nieto A, Nevels M, Parks RJ, Kristie TM. 2013. Snapshots: chromatin control of viral infection. Virology 435:141–156. doi: 10.1016/j.virol.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gupta A, Jha S, Engel DA, Ornelles DA, Dutta A. 2013. Tip60 degradation by adenovirus relieves transcriptional repression of viral transcriptional activator EIA. Oncogene 32:5017. doi: 10.1038/onc.2012.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Q, Yao H, Vo N, Goodman RH. 2000. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc Natl Acad Sci U S A 97:14323–14328. doi: 10.1073/pnas.011283598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Madison DL, Yaciuk P, Kwok RP, Lundblad JR. 2002. Acetylation of the adenovirus-transforming protein E1A determines nuclear localization by disrupting association with importin-alpha. J Biol Chem 277:38755–38763. doi: 10.1074/jbc.M207512200. [DOI] [PubMed] [Google Scholar]
- 66.Wang A, Kurdistani SK, Grunstein M. 2002. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- 67.Somanath P, Herndon Klein R, Knoepfler PS. 2017. CRISPR-mediated HDAC2 disruption identifies two distinct classes of target genes in human cells. PLoS One 12:e0185627. doi: 10.1371/journal.pone.0185627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lai Q, Du W, Wu J, Wang X, Li X, Qu X, Wu X, Dong F, Yao R, Fan H. 2017. H3K9ac and HDAC2 activity are involved in the expression of monocarboxylate transporter 1 in oligodendrocyte. Front Mol Neurosci 10:376. doi: 10.3389/fnmol.2017.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng Q, Su Z, Song S, Χu H, Zhang B, Yi L, Tian M, Wang H. 2016. Histone deacetylase inhibitors suppress RSV infection and alleviate virus-induced airway inflammation. Int J Mol Med 38:812–822. doi: 10.3892/ijmm.2016.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marks PA. 2007. Discovery and development of SAHA as an anticancer agent. Oncogene 26:1351–1356. doi: 10.1038/sj.onc.1210204. [DOI] [PubMed] [Google Scholar]
- 71.Wu Y, Ma S, Xia Y, Lu Y, Xiao S, Cao Y, Zhuang S, Tan X, Fu Q, Xie L, Li Z, Yuan Z. 2017. Loss of GCN5 leads to increased neuronal apoptosis by upregulating E2F1- and Egr-1-dependent BH3-only protein Bim. Cell Death Dis 8:e2570. doi: 10.1038/cddis.2016.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abramova MV, Pospelova TV, Nikulenkov FP, Hollander CM, Fornace AJ Jr, Pospelov VA. 2006. G1/S arrest induced by histone deacetylase inhibitor sodium butyrate in E1A + Ras-transformed cells is mediated through down-regulation of E2F activity and stabilization of beta-catenin. J Biol Chem 281:21040–21051. doi: 10.1074/jbc.M511059200. [DOI] [PubMed] [Google Scholar]
- 73.Jeon YJ, Ko SM, Cho JH, Chae JI, Shim JH. 2013. The HDAC inhibitor, panobinostat, induces apoptosis by suppressing the expression of specificity protein 1 in oral squamous cell carcinoma. Int J Mol Med 32:860–866. doi: 10.3892/ijmm.2013.1451. [DOI] [PubMed] [Google Scholar]
- 74.Hedrick E, Crose L, Linardic CM, Safe S. 2015. Histone deacetylase inhibitors inhibit rhabdomyosarcoma by reactive oxygen species-dependent targeting of specificity protein transcription factors. Mol Cancer Ther 14:2143. doi: 10.1158/1535-7163.MCT-15-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rojas JJ, Guedan S, Searle PF, Martinez-Quintanilla J, Gil-Hoyos R, Alcayaga-Miranda F, Cascallo M, Alemany R. 2010. Minimal RB-responsive E1A promoter modification to attain potency, selectivity, and transgene-arming capacity in oncolytic adenoviruses. Mol Ther 18:1960–1971. doi: 10.1038/mt.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hedjran F, Shantanu K, Tony R. 2011. Deletion analysis of Ad5 E1a transcriptional control region: impact on tumor-selective expression of E1a and E1b. Cancer Gene Ther 18:717–723. doi: 10.1038/cgt.2011.41. [DOI] [PubMed] [Google Scholar]
- 77.Reichel R, Neill SD, Kovesdi I, Simon MC, Raychaudhuri P, Nevins JR. 1989. The adenovirus E4 gene, in addition to the E1A gene, is important for trans-activation of E2 transcription and for E2F activation. J Virol 63:3643–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parks CL, Shenk T. 1997. Activation of the adenovirus major late promoter by transcription factors MAZ and Sp1. J Virol 71:9600–9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Swaminathan S, Thimmapaya B. 1995. Regulation of adenovirus E2 transcription unit. Curr Top Microbiol Immunol 199:177–194. [DOI] [PubMed] [Google Scholar]
- 80.Ai T, Xu Y, Qiu L, Geraghty RJ, Chen L. 2015. Hydroxamic acids block replication of hepatitis C virus. J Med Chem 58:785–800. doi: 10.1021/jm501330g. [DOI] [PubMed] [Google Scholar]
- 81.Graham FL, Smiley J, Russell WC, Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 82.Wong CM, Poulin KL, Tong G, Christou C, Kennedy MA, Falls T, Bell JC, Parks RJ. 2016. Adenovirus-mediated expression of the p14 fusion-associated small transmembrane protein promotes cancer cell fusion and apoptosis in vitro but does not provide therapeutic efficacy in a xenograft mouse model of cancer. PLoS One 11(3):e0151516. doi: 10.1371/journal.pone.0151516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol 70:4805–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. 2002. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A 99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Poulin KL, Lanthier RM, Smith AC, Christou C, Risco Quiroz M, Powell KL, O’Meara RW, Kothary R, Lorimer IA, Parks RJ. 2010. Retargeting of adenovirus vectors through genetic fusion of a single-chain or single-domain antibody to capsid protein IX. J Virol 84:10074–10086. doi: 10.1128/JVI.02665-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McKinnon RD, Bacchetti S, Graham FL. 1982. Tn5 mutagenesis of the transforming genes of human adenovirus type 5. Gene 19:33–42. doi: 10.1016/0378-1119(82)90186-X. [DOI] [PubMed] [Google Scholar]
- 87.Addison CL, Hitt M, Kunsken D, Graham FL. 1997. Comparison of the human versus murine cytomegalovirus immediate early gene promoters for transgene expression by adenoviral vectors. J Gen Virol 78:1653–1661. doi: 10.1099/0022-1317-78-7-1653. [DOI] [PubMed] [Google Scholar]
- 88.Wong CM, Nash LA, Del Papa J, Poulin KL, Falls T, Bell JC, Parks RJ. 2016. Expression of the fusogenic p14 FAST protein from a replication-defective adenovirus vector does not provide a therapeutic benefit in an immunocompetent mouse model of cancer. Cancer Gene Ther 23:355–364. doi: 10.1038/cgt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ross PJ, Kennedy MA, Christou C, Risco Quiroz M, Poulin KL, Parks RJ. 2011. Assembly of helper-dependent adenovirus DNA into chromatin promotes efficient gene expression. J Virol 85:3950–3958. doi: 10.1128/JVI.01787-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ross PJ, Parks RJ. 2009. Construction and characterization of adenovirus vectors. Cold Spring Harb Protoc 2009:pdb.prot5011. doi: 10.1101/pdb.prot5011. [DOI] [PubMed] [Google Scholar]