Coronaviruses (CoVs) are emerging human and veterinary pathogens with pandemic potential. Despite the established and predicted threat these viruses pose to human health, there are currently no approved countermeasures to control infections with these viruses in humans. Viral macrodomains, enzymes that remove posttranslational ADP-ribosylation of proteins, and viral multifunctional papain-like proteases, enzymes that cleave polyproteins and remove polyubiquitin chains via deubiquitinating activity, are two important virulence factors. Here, we reveal an unanticipated interplay between the macrodomain and the PLP2 domain that is important for replication and antagonizing the host innate immune response. Targeting the interaction of these enzymes may provide new therapeutic opportunities to treat CoV disease.
KEYWORDS: coronavirus, innate immunity, interferon, macrodomain, papain-like protease, temperature sensitive, viral replication
ABSTRACT
Analysis of temperature-sensitive (ts) mutant viruses is a classic method allowing researchers to identify genetic loci involved in viral replication and pathogenesis. Here, we report genetic analysis of a ts strain of mouse hepatitis virus (MHV), tsNC11, focusing on the role of mutations in the macrodomain (MAC) and the papain-like protease 2 (PLP2) domain of nonstructural protein 3 (nsp3), a component of the viral replication complex. Using MHV reverse genetics, we generated a series of mutant viruses to define the contributions of macrodomain- and PLP2-specific mutations to the ts phenotype. Viral replication kinetics and efficiency-of-plating analysis performed at permissive and nonpermissive temperatures revealed that changes in the macrodomain alone were both necessary and sufficient for the ts phenotype. Interestingly, mutations in the PLP2 domain were not responsible for the temperature sensitivity but did reduce the frequency of reversion of macrodomain mutants. Coimmunoprecipitation studies are consistent with an interaction between the macrodomain and PLP2. Expression studies of the macrodomain-PLP2 portion of nsp3 indicate that the ts mutations enhance proteasome-mediated degradation of the protein. Furthermore, we found that during virus infection, the replicase proteins containing the MAC and PLP2 mutations were more rapidly degraded at the nonpermissive temperature than were the wild-type proteins. Importantly, we show that the macrodomain and PLP2 mutant viruses trigger production of type I interferon in vitro and are attenuated in mice, further highlighting the importance of the macrodomain-PLP2 interplay in viral pathogenesis.
IMPORTANCE Coronaviruses (CoVs) are emerging human and veterinary pathogens with pandemic potential. Despite the established and predicted threat these viruses pose to human health, there are currently no approved countermeasures to control infections with these viruses in humans. Viral macrodomains, enzymes that remove posttranslational ADP-ribosylation of proteins, and viral multifunctional papain-like proteases, enzymes that cleave polyproteins and remove polyubiquitin chains via deubiquitinating activity, are two important virulence factors. Here, we reveal an unanticipated interplay between the macrodomain and the PLP2 domain that is important for replication and antagonizing the host innate immune response. Targeting the interaction of these enzymes may provide new therapeutic opportunities to treat CoV disease.
INTRODUCTION
Coronaviruses (CoVs) are enveloped, positive-sense, single-stranded RNA viruses that primarily infect the respiratory or gastrointestinal tract. CoVs can emerge from an animal reservoir, such as bats, to infect a new species and cause epidemic or pandemic disease with high mortality. Recent emergence events exemplified by severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) in humans (1) and swine acute diarrhea syndrome coronavirus (SADS-CoV) in domestic pigs (2) have demonstrated how devastating these viruses can be within naive populations. To date, there are no approved antivirals or effective vaccines that protect humans from coronavirus diseases. Therefore, identifying viral factors that contribute to pathogenesis and characterizing novel targets for therapeutic interventions are two important approaches to facilitate the development of effective vaccines and antivirals.
The murine coronavirus, mouse hepatitis virus (MHV), is widely used as a model system to study coronavirus replication and pathogenesis in mice. Replication of the virus initiates with the engagement of the spike glycoprotein with a host cell receptor and the release of the positive-sense RNA into the cytoplasm of the cell. The large (∼32-kb) viral genomic RNA is translated to produce two long polyproteins, pp1a and pp1ab, which are processed by viral proteases, including the papain-like proteases (PLP1 and/or PLP2) and the 3C-like protease (3CLpro or Mpro), into 16 nonstructural proteins (nsp1-16, Fig. 1A). To generate the viral replication complex, the coronaviral nsp’s sequester host endoplasmic reticulum (ER) to generate convoluted membranes and double-membrane vesicles (DMVs), which are the sites of viral RNA synthesis (3, 4). The viral replication complex generates a nested set of double-stranded RNA (dsRNA) intermediates to produce copious amounts of mRNAs, which are then translated to produce the structural (spike, envelope, membrane, and nucleocapsid) and virus-specific accessory proteins. The genomic RNA and structural proteins assemble in the ER-Golgi intermediate compartment to generate infectious virus particles that are released from the cell (5, 6).
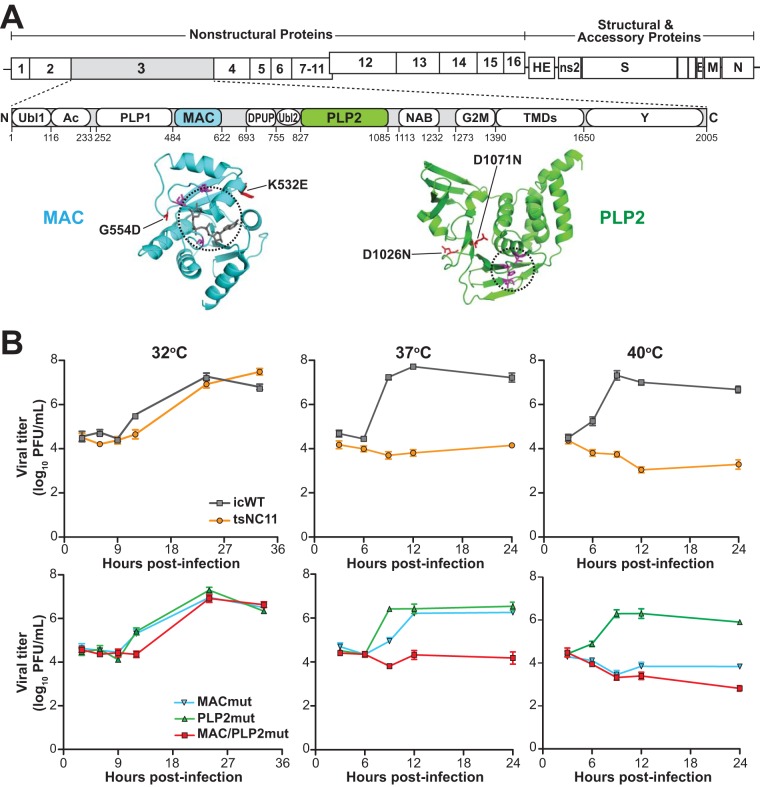
FIG 1.
Evaluation of the replication kinetics of coronavirus temperature-sensitive mutants at permissive and nonpermissive temperatures. (A) Schematic diagram of the MHV genome and the domains of nsp3. Abbreviations: Ubl1, ubiquitin-like domain 1; Ac, acidic region; PLP1, papain-like protease 1; MAC, macrodomain; DPUP, domain preceding Ubl2 and PLP2; Ubl2, ubiquitin-like domain 2; PLP2, papain-like protease 2; NAB, nucleic acid-binding domain; G2M, coronavirus group 2 marker domain; TMDs, transmembrane domains; Y, coronavirus highly conserved domain. Representative structures of the macrodomain with ribose (229E; PDB 3EWR) and PLP2 (MHV; PDB 4YPT) are shown in cyan and green with catalytic pockets circled, and the residues involved in catalysis are shown in magenta. The mutations described in this study are shown in red. (B) Growth kinetics of MHV and mutants at three temperatures. DBT cells were inoculated with the indicated virus (at a multiplicity of infection [MOI] of 5) for 1 h at 37°C and then shifted to the indicated temperatures. Culture supernatants were collected at the indicated hours postinfection and titrated in DBT cells at 32°C. The data are representative of two independent experiments. Error bars indicate ± the standard deviation (SD).
CoV replication induces profound rearrangement of the host ER and generates viral dsRNA intermediates, processes that can be sensed by the host to activate the innate immune response. As a result, CoVs have evolved multiple strategies to counteract and delay activation of these host immune responses and establish an environment amenable to virus replication. These strategies include (i) expressing species-specific accessory proteins as modulators of innate immune responses (reviewed in reference 7), (ii) encoding highly conserved nonstructural proteins that serve as interferon (IFN) antagonists (8–10), and (iii) sequestering viral RNA in DMVs (3, 4) to prevent detection by host pattern recognition receptors. A key component in the assembly of the DMVs is nsp3 (11, 12). To date, eleven distinct nsp3 domains have been identified using either bioinformatic approaches or enzymatic studies (Fig. 1A). Here, we focus on two multifunctional components contained within nsp3, the macrodomain (MAC) and papain-like protease 2 (PLP2).
The region researchers now term the macrodomain was originally identified in the 1990s as a highly conserved domain of unknown function, termed the X domain, contained within the replicase polyprotein of rubella virus, hepatitis E virus (HEV), and coronaviruses (13–15). Structural and biochemical studies revealed that the X domain exhibited structural similarity to the cellular histone MacroH2a and catalyzed measurable ADP-ribose-1″-phosphatase (ADRP) activity (16–18), although the functional significance of this enzymatic activity was unclear. Using reverse genetics to inactivate the catalytic site of the enzyme, researchers found that ADRP activity was not essential for CoV replication in cultured cells (19). However, further studies revealed that an ADRP-catalytic mutant virus was attenuated in mice (20) and that ADRP activity in SARS-CoV and human CoV-229E mediated resistance to antiviral interferon responses (21). These findings were consistent with an essential role for enzymatic activity in vivo, although the target for the ADRP activity was still unclear. A breakthrough came in 2016 from a study revealing that the macrodomain of HEV acts as an ADP-ribose hydrolase (22). ADP-ribosylation is a known posttranslational modification that regulates cellular activities (23); therefore, viral enzymes that reverse this process could interrupt host cell signaling. For CoVs, nsp3 macrodomain activity was shown to promote MHV-induced encephalitis (24) and increase virulence during SARS-CoV infection (25).
Another highly conserved enzyme contained within nsp3 is PLP2. For MHV, PLP2 is responsible for processing the nsp3/4 junction using a highly conserved LXGG/X cleavage site (26). Studies using SARS-CoV revealed that the single papain-like protease encoded on nsp3 (termed PLpro) cleaves all three sites at the amino-terminal end of the polyprotein (27). PLpro also functions as a deubiquitinating enzyme (DUB) which is capable of removing polyubiquitin chains from substrates (28, 29). Structural studies revealed that CoV PLpro/PLP2s are similar to cellular DUBs (30). Enzymatic analysis revealed that CoV PLpro/PLP2s are multifunctional, with protease, deubiquitinating, and deISGylating activities (30–33). Viral DUB activity has been implicated as a modulator of the innate immune response to viral infection (32, 34, 35), but the target(s) of the DUB activity has not yet been identified. Thus, both the PLP2 and macrodomains of nsp3 have been independently identified as contributors to coronavirus virulence.
In this study, we characterized a temperature-sensitive (ts) MHV mutant virus containing mutations within both the macrodomain and the PLP2 domain. We investigated the contributions of these mutations to the temperature-sensitive phenotype, as well as the resulting effects on viral pathogenesis. The results presented here reveal a previously undescribed interplay between the macrodomain and the PLP2 domain that impacts replication, antagonizes the innate immune response, and contributes to viral pathogenesis. Modulating the macrodomain-PLP2 interaction may provide new opportunities for therapeutic intervention.
RESULTS
Identifying mutations associated with a temperature-sensitive phenotype.
Murine coronavirus strain tsNC11 was generated by chemical mutagenesis, plaque purified, and validated as a temperature-sensitive mutant defective in positive-sense RNA synthesis at nonpermissive temperatures (36). Complementation analysis indicated that tsNC11 harbors mutations in the open reading frame 1a (ORF1a) region of the replicase polyprotein, but the specific mutations were unknown. To identify the nucleotide changes in tsNC11, we isolated the genomic RNA from the viral supernatant, subjected it to deep sequencing, and then aligned the reads to the genomic sequence of MHV strain A59 (MHV-A59) (GenBank accession no. AY910861). In agreement with the complementation study by Schaad et al. (36), the sequence analysis revealed seven nonsynonymous substitutions in ORF1a of tsNC11. These substitutions resulted in 7 amino acid (aa) changes: two in nsp2 (I4V and T543I), four in nsp3, and one in nsp10 (P23S). The four mutations within nsp3 are distributed between the macrodomain (K532E and G554D) and the PLP2 domain (D1026N and D1071N) (Fig. 1A). As noted above, previous studies documented the importance of the macrodomain and PLP2 domain in virus replication and disease; therefore, we focused our efforts on evaluating how these substitutions contributed to the ts phenotype, the stability of the phenotype, and the pathogenesis of the virus in mice.
To evaluate the contributions of the macrodomain and PLP2 domain mutations to the ts phenotype, three mutant viruses were generated using the MHV-A59 reverse genetics system (37). The first mutant virus, designated MACmut, contains the macrodomain mutations K532E and G554D. The second virus was engineered with the D1026N and D1071N mutations within the PLP2 domain and is designated PLP2mut. The third virus, MAC/PLP2mut, combines the mutations in the macrodomain and the PLP2 domain into one virus. In addition, an isogenic wild-type MHV (icWT) was used as a control. These viruses were recovered, plaque purified, and propagated in DBT cells at a permissive temperature of 32°C. Deep-sequencing results confirmed the incorporation of the desired nucleotide changes in the RNA encoding nsp3 and revealed no additional amino acid changes within the ORF1 region.
First, we evaluated the one-step growth curves of all five viruses (tsNC11, icWT, and the three engineered mutants) at permissive (32°C) and nonpermissive (37 and 40°C) temperatures. As expected, icWT replicates to high titer at all three temperatures, whereas tsNC11 is impaired at both 37 and 40°C, as reported by Schaad et al. (36) (Fig. 1B). Analysis of the three engineered mutants revealed that the two substitutions in the PLP2 domain were not sufficient to confer a temperature-sensitive phenotype, since the kinetics of replication mirrored those of the wild-type virus. In contrast, the MACmut virus exhibited reduced virus replication at 40°C but was only slightly impaired at 37°C. The MAC/PLP2 mutant virus mirrored the kinetics of tsNC11 with impaired replication at both 37 and 40°C, implicating the mutations in both the macrodomain and the PLP2 domain as contributors to the temperature-sensitive phenotype of tsNC11.
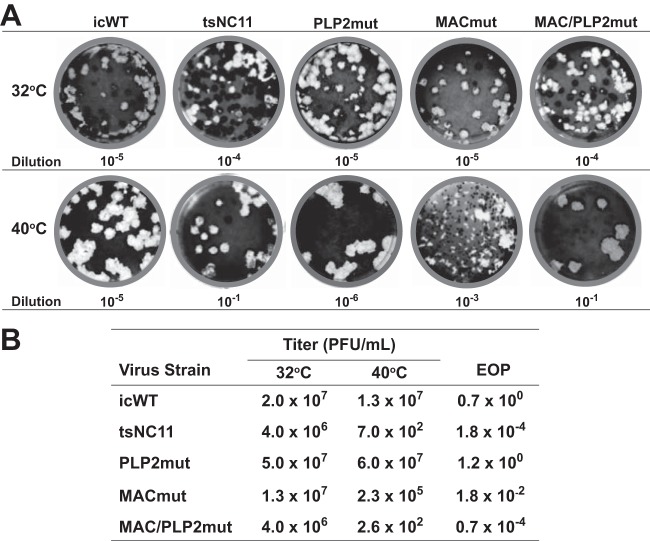
We also evaluated the plaque sizes and efficiencies of plating (EOP) of the viruses at permissive and nonpermissive temperatures. As expected, all viruses replicated to high titers and formed similarly sized plaques at 32°C (Fig. 2, upper panel). tsNC11 is profoundly temperature sensitive, with a low number of plaques detected at the 10−1 dilution on the plate incubated at the nonpermissive temperature. The tsNC11 plaques that were detected at the nonpermissive temperature exhibited a large-plaque phenotype, suggesting that these viruses may be revertants. We found that the PLP2mut virus formed large plaques at 40°C, which is consistent with the results of the kinetic analysis and indicates that the mutations in the PLP2 domain are not sufficient to cause the ts phenotype. Analysis of the MACmut virus revealed a mixed population of small and large plaques at the nonpermissive temperature, the majority of which displayed the small-plaque phenotype. The MAC/PLP2 mutant virus mirrored the plaque size and plating efficiency of tsNC11. We calculated the EOP values, which represent the ratio of viral titers obtained at 40°C and at 32°C (Fig. 2B). Again, the PLP2mut and icWT viruses had similar titers at both temperatures, resulting in an EOP of ∼1. In contrast, the MACmut virus exhibited titers that were significantly lower at 40°C than the titers obtained at 32°C (EOP of 10−2). These results indicate that the MACmut virus, but not the PLP2mut or icWT viruses, has a defect in plaque formation at the nonpermissive temperature. Taken together, these data demonstrate that the mutations in the macrodomain, but not those in the PLP2 domain, are the major determinants of the ts phenotype of tsNC11. In addition, these results are consistent with a critical role of the macrodomain in viral replication. Interestingly, we found that the MAC/PLP2 mutant virus mirrored the plaque size and low reversion frequency of tsNC11 (EOP of 10−5), supporting a role for the PLP2 domain as a genetic enhancer of the ts phenotype. A genetic enhancer, as defined by genetic studies of eukaryotic organisms, is a mutation in one gene that intensifies the phenotype caused by a mutation in another gene (38).
FIG 2.
Analysis of plaque size and efficiency of plating at permissive and nonpermissive temperatures. (A) Representative plaque assays at 32 and 40°C for icWT, tsNC11, and engineered mutant viruses. The dilution of the viral stock is indicated and selected to visualize ca. 20 to 50 plaques per plate. (B) EOP data. EOP = average titer at 40°C/average titer at 32°C.
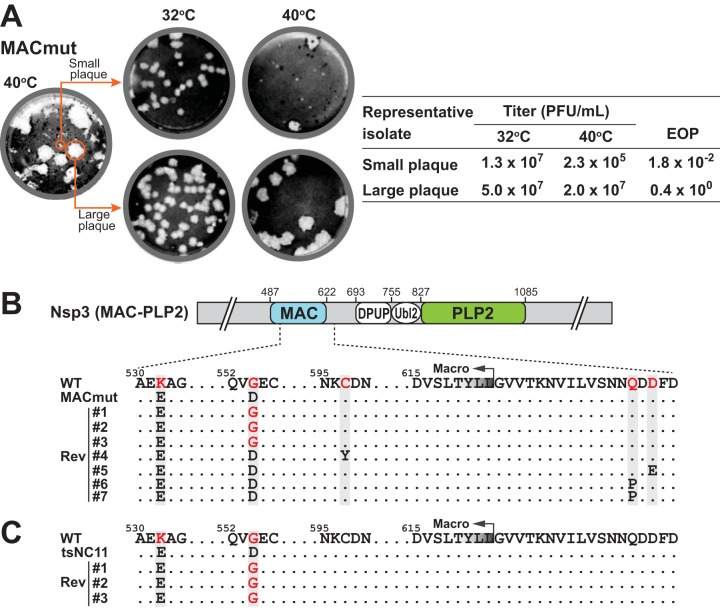
Evaluating revertants of the MACmut virus.
While generating the MACmut virus, we noticed that, in addition to the majority population having the small-plaque phenotype, a subpopulation of large plaques was also present at 40°C. The large plaques consistently appeared even after several rounds of plaque purification of the small-plaque isolates. Therefore, we sought to determine whether the small plaques were formed by temperature-sensitive viruses while the large plaques were due to revertant viruses. To address this question, we selectively isolated plaques with different sizes and propagated them at 32°C to obtain viral stocks for subsequent analysis (Fig. 3). We found that the small-plaque isolates recapitulated the phenotype of the parental MACmut virus: small plaques and similar EOP values (Fig. 3A). In contrast, the large-plaque isolates exhibited a phenotype similar to that of icWT. Sequencing results of PCR amplicons, representing the region spanning the macrodomain and the PLP2 domain, revealed that small-plaque isolates had no additional mutations in either the macrodomain or PLP2. In contrast, the large-plaque isolates had either a true reversion (D554 to G) or harbored putative suppressive mutations located within the macrodomain or the adjacent downstream sequence (Fig. 3B). Among seven large-plaque revertants, all maintained the K532E mutation, indicating it was not associated with the ts phenotype. Three isolates had the D554-to-G reversion, suggesting that it may be sufficient for the ts phenotype of the MACmut virus. We found that isolates 4 to 7 maintained the engineered mutations but had also acquired additional, potentially suppressive mutations in the downstream region. Together, these results indicate that altering the coding sequence of either the macrodomain or the downstream region is likely sufficient to revert or suppress the ts phenotype caused by the G554D mutation. We also evaluated the MAC-PLP2 region of tsNC11 large-plaque revertant viruses and found that all three isolates had the D554-to-G reversion (Fig. 3C), consistent with our findings with the MACmut revertants.
FIG 3.
Analysis of small- and large-plaque variants in the MACmut virus population. (A) MACmut isolates with distinct plaque sizes were evaluated for a ts phenotype. (B and C) Sequence analysis of individual plaque-purified revertant isolates identified mutations in the macrodomain and the adjacent downstream region in the large-plaque variants of the MACmut (B) and tsNC11 (C) viruses.
Mutations in PLP2 enhance the ts phenotype by reducing reversion frequency.
We determined that the macrodomain mutations are the major contributors to the ts phenotype; however, we noticed that the MACmut virus did not completely phenocopy tsNC11. We found that the replication of the MACmut virus was defective at 40°C but not at 37°C. In addition, the MACmut virus exhibited a higher EOP value (10−2) than that of tsNC11 (10−4) (Fig. 2B), indicating a relatively high reversion frequency. These data imply that mutations outside the macrodomain may enhance the ts phenotype by stabilizing the replication defect, thereby preventing reversion to the wild-type phenotype (38). Therefore, we sought to determine whether the addition of the PLP2 mutations observed in tsNC11 could enhance the ts phenotype of the MACmut virus and reduce reversion. We found that the MAC/PLP2mut virus exhibits a severe replication defect at both 37 and 40°C (Fig. 1B) and replicated only under permissive conditions, similar to tsNC11. The MAC/PLP2mut and tsNC11 viruses exhibited similar EOP values (∼10−4) (Fig. 2B). Of note, the low titer of the MAC/PLP2mut virus at 40°C indicates a low level of reversion to the wild-type phenotype, suggesting that the PLP2 mutations stabilize the MACmut virus. Taken together, these data demonstrate that while the PLP2 mutations are not sufficient to cause the ts phenotype, they act to enhance the ts phenotype caused by the mutation in the macrodomain. Enhancement of phenotypes has been described for other coronavirus interacting proteins (39, 40), which motivated us to determine whether the enhancement phenotype we detected here is due to an interaction between the macro and PLP2 domains.
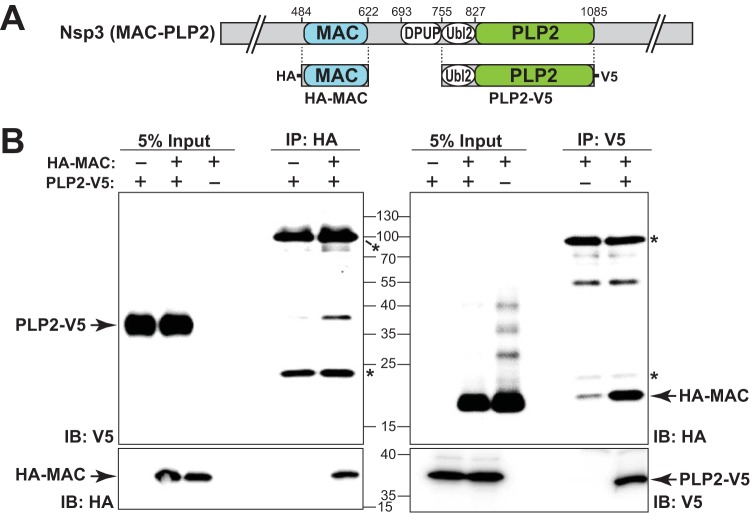
Evaluating the macrodomain interaction with the PLP2 domain.
The structures of several domains of nsp3 have been solved individually (reviewed in reference 41) or in combination (42). However, owing to the size and complexity of this protein, the complete structure of nsp3 remains unsolved. The capacity of the PLP2 mutations to enhance the ts phenotype in the presence of the macrodomain mutations raises the possibility of domain-domain interaction between the macrodomain and PLP2. To test this hypothesis, we generated plasmids that express either an epitope-tagged macrodomain (HA-MAC) or PLP2 domain (PLP2-V5) (depicted in Fig. 4A). When these plasmids were cotransfected into HEK-293T cells, expression of both the macrodomain and PLP2 was detectable by the cognate epitope antibodies (Fig. 4B). We detected HA-MAC in lysates immunoprecipitated with anti-V5, and inversely, PLP2-V5 was detected when HA-MAC was immunoprecipitated from the lysates. These results indicate that the ectopically expressed macrodomain associates with PLP2 in cell lysates, consistent with either a direct or indirect interaction.
FIG 4.
Evaluation of the coimmunoprecipitation of the macrodomain and the PLP2 domain. (A) Schematic diagram of the individual constructs used to evaluate potential interactions between the macrodomain and PLP2. (B) Western blots to identify expression and coimmunoprecipitation of HA-MAC and PLP2-V5. HEK-293T cells were transfected with the indicated plasmid DNAs, lysates were prepared at 18 h posttransfection and subjected to immunoprecipitation with the indicated antibody, and the products were analyzed by SDS-PAGE and immunoblotting. The data represent the results of three independent experiments. Asterisks indicate the cross-detection of IgG chains by secondary antibody.
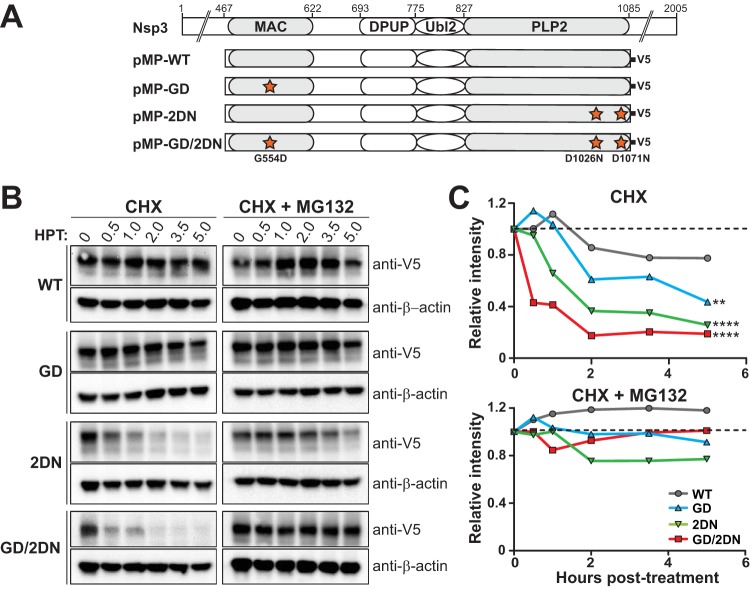
Mutations in the macrodomain and PLP2 domain affect protein stability.
Because we found that mutation in the macrodomain (G554D) is the major ts determinant and the PLP2 mutations enhance the ts phenotype, we reasoned that these mutations might alter protein folding, thereby rendering the protein unstable and susceptible to proteasome-mediated degradation. To determine whether the mutations in the macrodomain and/or the PLP2 domain alter protein stability, plasmid DNA expressing wild-type or mutant forms of MAC/PLP2 polypeptide (Fig. 5A) were transfected into HEK-293T cells. The cells were maintained at 37°C throughout the experiment. We added cycloheximide (CHX) at 16 h posttransfection to block translation and harvested cell lysates at the indicated times. The level of expressed proteins was determined by immunoblotting (Fig. 5B and C). The MAC/PLP2 (WT) protein was maintained at levels comparable to those prior to treatment up to 5 h posttreatment with CHX. In contrast, we detected rapid reductions in the levels of all of the mutant forms of the protein. Addition of the proteasome inhibitor MG132 blocked degradation of the proteins (Fig. 5B and C). These results indicate that mutations in both the MAC and PLP2 domains affect protein folding and stability, rendering the proteins more susceptible to proteasome-mediated degradation.
FIG 5.
Mutations in the macrodomain and PLP2 enhance degradation of the polypeptide. (A) Schematic diagram of constructs used to evaluate protein stability. (B and C) Western blotting was used to detect wild-type or mutant forms of MAC-PLP2 polypeptide in the presence of CHX or a combination of CHX and proteasome inhibitor MG132. HEK-293T cells were transfected with the indicated expression plasmid of wild-type (WT) or mutant forms of MAC-PLP2. At 16 h posttransfection, cells were treated with 20 μg/ml of CHX or a combination of 20 μg/ml CHX and 10 μM MG132 and then harvested at the indicated time points. Equal amounts of cell lysate were subjected to immunoblotting with anti-V5 or anti-β-actin antibodies. The intensities of the MAC/PLP2 bands relative to that of β-actin were measured and calculated with AlphaView software. The experiment was repeated two times, and representative immunoblots (B) and curves of relative intensities (C) are shown. The slope parameters of the decay curves were evaluated using nonlinear regression and two-sided t tests in comparison to WT. **, P < 0.005; ****, P < 0.0001.
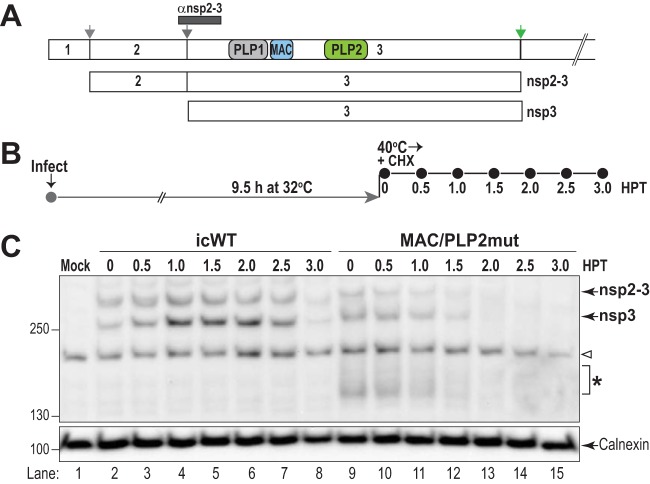
To determine whether these MAC/PLP2 mutations affect the stability of the replicase proteins during virus replication at the nonpermissive temperature, we performed temperature shift experiments as outlined in Fig. 6. We infected cells with either WT or MAC/PLP2mut virus and incubated them at the permissive temperature for 9.5 h. At this point, we added CHX to block translation and shifted the infected cells to the nonpermissive temperature. Cell lysates were collected every 30 min and evaluated using immunoblotting for the levels of nonstructural intermediate nsp2-3 and product nsp3. We found that WT nsp2-3 and nsp3 were relatively stable, with loss of detection occurring at 3 h after the temperature shift and the addition of CHX (Fig. 6C, lanes 2 to 8). In contrast, the levels of nsp2-3 and nsp3 in the MAC/PLP2mut-infected cells diminished more rapidly, with reduced levels at 1.5 h after the temperature shift and the addition of CHX (Fig. 6C, lanes 9 to 15). These results support the finding that the MAC and PLP2 mutations destabilize the replicase protein at the nonpermissive temperature.
FIG 6.
Mutations in the macrodomain and PLP2 alter the stability of replicase protein nsp3. HeLa-MHVR cells were infected with either icMHV-WT or MAC/PLP2mut virus (MOI of 5) and incubated at 32°C for 9.5 h. Then 20 μg/ml of CHX was added, and the cells were shifted to the nonpermissive temperature. Lysates were prepared every 30 min, the proteins were separated by SDS-PAGE, and nonstructural proteins nsp2-3 and nsp3 were visualized by immunoblotting. (A) Schematic diagram of MHV replicase polyprotein indicating the processing pathway and the region identified by the anti-nsp2-3 antibody. (B) Outline of the experiment. (C) Western blot evaluating the level of nsp2-3 and nsp3 proteins detected after a shift to the nonpermissive temperature. These are representative data from two independent experiments. The arrowhead indicates detections of cellular protein in all lysates. The asterisk indicates degradation products detected by anti-nsp2-3 antibody in the MAC/PLP2mut virus-infected cells.
ts mutant viruses induce interferon in macrophages and are attenuated in mice.
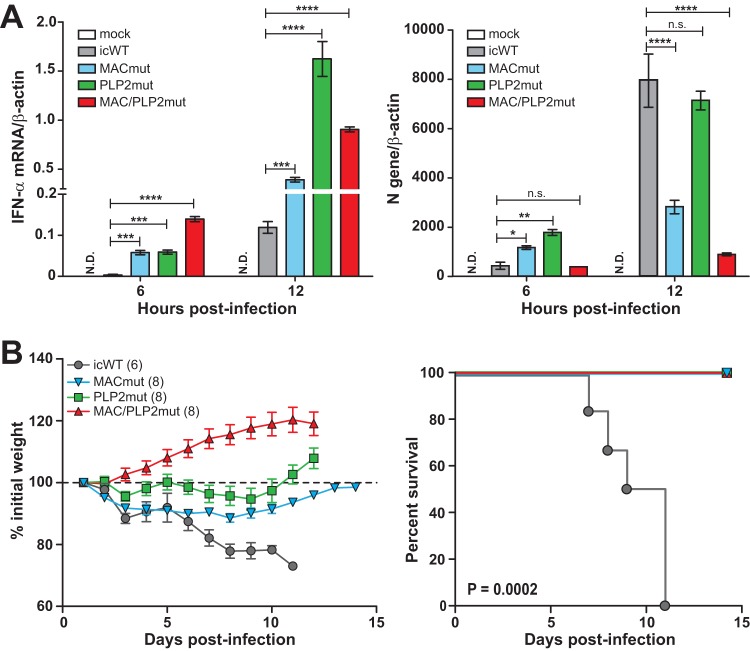
Previous studies have shown that the papain-like protease domains of MHV, SARS-CoV, and MERS-CoV antagonize the IFN response, likely through the deubiquitinating activity of these enzymes (32, 34, 35, 43–46). In addition, coronaviral macrodomains have been shown to suppress IFN production both in vitro and in vivo (20, 21, 24, 25). We sought to determine whether the mutations in the macrodomain and PLP2 modulate the type I interferon response during infection of macrophages. As shown in Fig. 7A, infection of mouse bone marrow-derived macrophages (BMDMs) with mutant viruses at the permissive temperature produced significantly more IFN-α during infection than icWT virus infection. At 12 h postinfection, the MACmut virus induced 2-fold more IFN-α than icWT virus. Furthermore, the level of N gene transcripts, which reveals the abundances of all viral mRNAs, was reduced in the MACmut-infected cells compared to that for the wild-type virus. We found that the PLP2mut virus elicited dramatically more IFN-α than WT virus, while the level of N gene expression was similar. The MAC/PLP2mut virus exhibited the most robust IFN-α response and the lowest level of N gene expression. These results show that mutations in the macrodomain and the PLP2 domain result in elevated levels of type I IFN mRNA expression during infection of macrophages, further supporting the role of the macrodomain and PLP2 in modulating host innate immunity.
FIG 7.
Macrodomain mutant viruses induce type I interferon in primary macrophages and are attenuated in mice. (A) Mouse bone marrow-derived macrophages were infected with the indicated virus (MOI of 1) at 32°C. Total RNA was extracted at the indicated time points and subjected to RT-qPCR. The mRNA levels of IFN-α (left) and the N gene (right) are presented relative to that of β-actin. The results are representative of three independent experiments and were subjected to a two-tailed, unpaired t test. Error bars indicate ± SD. ***, P < 0.001; ****, P < 0.0001; n.s., not significant; N.D., not detected. (B) Six-week-old mice were injected intracranially with either icWT or the indicated ts mutant virus (600 PFU/mouse) and monitored for weight loss. Viral pathogenicity was evaluated by body weight loss (left) and percent survival (right). The number (n) of infected mice is indicated in parentheses. Error bars indicate ± standard error of the mean. Differences in survival rates were calculated using a log-rank test.
Because the ts mutant viruses had reduced replication efficiency and elicited type I IFN production during infection of macrophages, we were interested in evaluating the pathogenicity of these viruses. To this end, C57BL/6 mice were intracranially inoculated with 600 PFU of virus and monitored for weight loss and mortality. As shown in Fig. 7B, all WT virus-infected mice lost weight rapidly and succumbed to infection by day 11 postinfection. In contrast, the mutant virus-infected mice exhibited transient or no weight loss during the infection period and all mice survived. These results demonstrate that the ts mutant viruses are attenuated in vivo, and those mutations adjacent to the catalytic sites of the macrodomains and PLP2 domains can modulate viral pathogenesis.
DISCUSSION
Identifying viral factors that modulate the immune response to viral infection provides new opportunities for developing novel antiviral interventions. Here, we described an unanticipated interplay between two previously characterized virulence factors, the macrodomain and the papain-like protease, of coronaviruses. The enzymatic activities of these domains have been implicated in removing posttranslational modifications: macrodomains remove mono- or poly-ADP-ribose from proteins (18, 22, 23), and the deubiquitinating activity of viral papain-like proteases removes mono- or polyubiquitin chains from signaling proteins (30, 32, 47). Our study stems from characterizing a temperature-sensitive mutant virus that harbored mutations within both the macrodomain and the PLP2 domain of nsp3. We found that the mutation within the macrodomain (G554D) was associated with the most significant temperature-sensitive phenotype but that this alteration of the macrodomain reverted to the wild-type phenotype at high frequency. However, viruses containing mutations in both the macrodomain and the PLP2 domain reverted less frequently, consistent with the PLP2 domain having an enhancing effect on the ts phenotype. Although these two enzymes reside within the same nsp3 polypeptide (Fig. 1A), to our knowledge this is the first suggestion of an interplay between these domains. By expressing the macrodomain and papain-like protease 2 domain on independent expression plasmids, we were able to evaluate and detect coimmunoprecipitation of the proteins, which is consistent with either a direct or an indirect interaction. Furthermore, we report that the mutations identified in the macrodomain and PLP2 domain destabilize the proteins, as revealed by proteasome-dependent degradation. Lastly, we demonstrated that these mutant viruses promote type I IFN production from macrophages in tissue culture and are attenuated in mice. This study confirms and extends previous studies that independently identified the macrodomain and the papain-like protease 2 domain as modulators of the innate immune response and virulence factors (reviewed in references 31, 35, 48, and 49).
Macrodomains have been shown to play a role in the virulence of positive-sense RNA viruses, including HEV, alphaviruses, and coronaviruses (reviewed in references 48 and 49). Studies of the alphavirus chikungunya virus (CHIKV) revealed that the macrodomain at the N terminus of nsp3 hydrolyzes ADP-ribose groups from monoribosylated proteins and that this deribosylating activity is critical for CHIKV replication in vertebrate and insect cells and for virulence in mice (50). Interestingly, viruses engineered to encode a mutation of the CHIKV macrodomain catalytic site rapidly reverted to the wild-type sequence (51), similar to the high-frequency reversion we reported for the MHV MACmut virus (Fig. 3). Studies of the role of the macrodomain during coronavirus replication indicate that catalytic activity is not required for virus replication in interferon-nonresponsive cell lines (19, 20, 24). However, catalytic activity is important for replication in primary cells and in mice, implicating the macrodomain in evading the innate immune response and promoting viral pathogenesis (20, 24, 25). Identifying the ribosylated substrates that are targeted by the viral enzymatic activity is an important future direction for this work.
Our study implicated an adjacent viral domain, the papain-like protein 2 domain, as an interacting partner with the macrodomain. Interestingly, the helicase domain adjacent to the macrodomain of HEV was found to modulate macrodomain activity. Biochemical assays revealed that the presence of the HEV helicase domain in cis enhanced the binding of the macrodomain to ADP-ribose and stimulated the hydrolase activity (22). Furthermore, we previously found that mutations in the Ubl2 domain could cause a ts phenotype and destabilize the PLP2 domain (52). Here, we found that the mutations in the macrodomain and PLP2 domain destabilized the replicase proteins, as shown by the more rapid degradation of the proteins after temperature shift. We speculate that there may be a dynamic interaction between adjacent domains within the nsp3 polyproteins.
As a multidomain protein, nsp3 must hold a sophisticated architecture to function properly and precisely. To date, four essential functions have been documented for this multidomain protein: (i) interaction of the Ubl1 domain with the nucleocapsid (N) protein, which is important for genomic RNA synthesis and encapsidation (39, 40); (ii) proteolytic processing of the N-terminal region of pp1a and pp1ab to release nsp1, nsp2, and nsp3 (26, 27); (iii) hijacking of the cellular reticular network in concert with other membrane-associated proteins (nsp4 and nsp6) to form virus-specific membrane structures for RNA synthesis (11, 12); and (iv) antagonizing of the innate immune response through the actions of the de-ADP-ribosylating activity of the macrodomain and the deubiquitinating activity of the PLP2 domain (reviewed in references 31 and 48). The removal of posttranslational modifications such as ADP-ribosylation and polyubiquitination could be directed at cellular proteins either to redirect them for use during viral replication or to subvert signaling of innate immune responses. Ultimately, structural and biochemical studies will be needed to fully investigate the multiple cis and trans interactions of nsp3 and to determine whether there is a dynamic interplay that modulates the stability, substrate specificity, and/or affinity of the enzymes and substrates.
We found that the MAC/PLP2mut virus recapitulated the ts phenotype of tsNC11 (Fig. 1B). However, it is possible that some or all of the other mutations we identified by deep sequencing (I4V and T543I in nsp2 and P23S in nsp10) may contribute in a subtle way to the phenotype of tsNC11. nsp2 was shown to be dispensable for MHV and SARS-CoV replication, but deletion of the nsp2-coding sequence resulted in decreased viral replication and RNA synthesis (53). For nsp10, previous studies revealed that this protein plays critical roles in 3C-like-protease-mediated polyprotein processing and viral RNA synthesis (54, 55). The results from these studies indicate that the mutations in nsp2 and nsp10 may also contribute to a ts phenotype. While our study focuses on the contribution of the macrodomain and PLP2, further studies are needed to fully evaluate the impact of other ORF1a mutations on the replication and pathogenesis of coronaviruses.
In summary, we report what is, to our knowledge, the first indication of an interplay between the macrodomain and papain-like protease 2 domain of CoV nsp3. We found that this interplay impacts virus replication efficiency, innate immune antagonism, and virulence in mice. A detailed understanding of the relationship between the macrodomain and the PLP2 domain will require further structural and enzymatic studies. We anticipate that the genetic analysis, coimmunoprecipitation, and in vivo pathogenesis outcomes reported here will facilitate these future studies.
MATERIALS AND METHODS
Virus and cells.
Human embryonic kidney 293T (HEK-293T) cells (CRL-11268; ATCC) were cultured in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 2% l-glutamine. Delayed brain tumor (DBT) cells were grown in minimal essential medium (MEM; catalog no. 21800-0400; Gibco) supplemented with 10% tryptose phosphate broth medium, 5% heat-inactivated FBS (Atlanta Biological), 2% penicillin-streptomycin (pen/strep; HyClone), and 2% l-glutamine. The BHK-MHVR cell line was kindly provided by Mark Denison at Vanderbilt University Medical Center and cultured in DMEM (catalog no. 12100-046; Gibco) supplemented with 10% heat-inactivated FBS and G418 (0.8 mg/ml; HyClone). Differentiated BMDMs were maintained in bone marrow macrophage medium containing DMEM (catalog no. 10-017-CV; Corning) supplemented with 30% L929 cell supernatant, 20% FBS, 1% l-glutamine, 1% sodium pyruvate, and 1% pen/strep. HeLa-MHVR cells (56) were grown in DMEM (catalog no. 12100-046; Gibco) supplemented with 10% FBS, 1% l-glutamine, 0.5% HEPES, and 1% pen/strep. Temperature-sensitive MHV strain tsNC11 was propagated in DBT cells at 32°C. The infectious clone of the MHV-A59 strain (GenBank accession no. AY910861) serves as the wild-type (icWT) virus for this study.
Deep sequencing and bioinformatic analysis.
Viral RNA was extracted from the supernatant of tsNC11-infected DBT cells incubated at 32°C. Isolated RNA was sent to Genewiz, Inc., for cDNA library preparation and Illumina MiSeq high-throughput sequencing. Raw reads were subjected to pairing and trimming and aligned to the genome sequence of the synthetic construct of the MHV-A59 strain (GenBank accession no. AY910861) using Geneious software (Geneious R7). A medium-low sensitivity and an iteration of up to five times were chosen. A total of 195,824 sequences with a mean coverage of 898.8 were aligned to the MHV synthetic construct template. Polymorphisms were detected using the “find variations/SNPs” tool. Parameters included a minimum coverage of 5 with a minimum variant frequency of 25% in order for a variation to be called. The maximum variant P value was set at 10−6, and the minimum strand bias P value was set at 10−5 when exceeding 65% bias. We focused our analysis on the first 12 kb of the replicase gene, since previous studies reported the ts phenotype was associated with changes in this region (36, 57).
Generation of mutant viruses.
All infectious clones were generated using the reverse genetics system previously established for MHV-A59 (37). Mutations identified by deep sequencing within the macrodomain and PLP2 domain were introduced into plasmids A and B, respectively, and then verified by sequencing of the plasmid DNA. DNA fragments were ligated together and used for in vitro transcription of viral RNA. In vitro-transcribed genomic RNA and N gene RNA were electroporated into BHK-MHVR cells, which were overlaid onto DBT cells in a T-75 flask. These cells were incubated at the permissive temperature of 32°C to facilitate the replication of ts mutant viruses. Supernatants were collected at the time when cytopathic effect was evident, usually between 36 and 48 h postelectroporation. All infectious clone mutant viruses were plaque purified, propagated on DBT cells, and subjected to full genome sequencing to validate the genotype. These infectious clones were designated MACmut, PLP2mut, and MAC/PLP2 according to the locations of introduced mutations, as shown in Fig. 1.
Temperature-sensitive assay and one-step growth kinetics.
To determine the temperature sensitivity of mutant viruses, the efficiency of plating (EOP; titer at 40°C/titer at 32°C) of each virus was measured. DBT cells were seeded into two six-well plates at 5.0 × 105 cells/well a day prior to infection. Each viral stock supernatant was serially diluted and inoculated onto the DBT cells. After 1 h of incubation at 37°C, the inoculum was removed, and cells were subsequently overlaid with a 0.8% 2× MEM–agar mixture. One plate was incubated at 32°C for 60 h, and the second plate was incubated at 40°C for 48 h. Agarose-covered cells were fixed using 4% formaldehyde for 1 h and stained using 0.1% crystal violet solution after removal of the agarose. Plaques were counted, and titers were calculated.
To evaluate the kinetics of virus replication, a one-step growth curve was generated at each temperature. Briefly, DBT cells were infected with the designated virus at a multiplicity of infection (MOI) of 5 for 1 h at 37°C, and then the plates were incubated at the specified temperatures. The supernatants were collected at the indicated time points and titrated on DBT cells incubated at 32°C for 60 h.
Isolation and characterizations of ts revertants.
To isolate ts revertants, plaque assays were performed at 40°C. Viruses from single plaques were isolated and propagated in DBT cells at 32°C to obtain viral stocks. To determine the ts phenotype of the isolates, the isolates were titrated at both 32 and 40°C, and EOP values were calculated as described above. To identify any mutations within the region of the macrodomains and the PLP2 domains of the revertants, viral genomic RNA was extracted using TriReagent (MRC, Inc.) according to the manufacturer’s instructions and subsequently subjected to cDNA synthesis. A genomic region (nucleotides 3976 to 6101) containing the macrodomain and the PLP2 domain was amplified by PCR using specific primers (sense, 5′-CAA GAA AGG TCT TTA GGG CTG CTT-3′; antisense, 5′-GAC ACC ATC AAC CTT CTC AAA TG-3′). The PCR products were sequenced, and the sequencing results were compared to the tsNC11 sequence.
MAC and PLP2 expression plasmids.
Nucleotide sequences encoding the macrodomain (467 to 622 aa of nsp3) were amplified from a codon-optimized MHV nsp3 gene (sequence available upon request) and cloned into a pCAGGS vector with a hemagglutinin (HA) epitope tag, designated HA-MAC. The pCAGGS-PLP2 plasmid (PLP2-V5) was generated in a previous study (52). The coding sequence of the macrodomain through the PLP2 domain (467 to 1,085 aa) was inserted into pcDNA3.1 and fused with a C-terminal V5 epitope tag (pMP-WT). Mutations were introduced into these constructs using site-directed mutagenesis PCR or the Gibson Assembly technique to generate pMP-GD (G554D), pMP-2DN (D1026N/D1071N), and pMP-GD/2DN (G554D/D1026N/D1071N), which all contain a C-terminal V5 tag.
Coimmunoprecipitation.
HA-MAC and PLP2-V5 plasmids were cotransfected into HEK-293T cells in 35-mm dishes. Cells were harvested using 500 μl of lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride), and 200 μg of whole-cell lysates was used for immunoprecipitation with 1 μg of primary mouse anti-V5 (catalog no. R96025; Invitrogen) or anti-HA (MMS-101R-200; BioLegend) monoclonal antibody (Ab). Protein-Ab mixtures were rotated at 4°C overnight, and then 15-μl portions of magnetic protein G-beads (LSKMAGA02; Millipore) were added, followed by 1 h of incubation. The beads were washed three times with wash buffer (same composition as lysis buffer except for 450 mM NaCl) and eluted with 40 μl of 2× sample buffer (10% glycerol, 5% β-mercaptoethanol, 3% sodium dodecyl sulfate [SDS], 7.5 mg/ml Trizma base, bromophenol blue). Eluted products and 5% cell lysates as input were subjected to SDS-PAGE and immunoblotting with anti-V5 or anti-HA antibodies.
Evaluating protein stability after addition of cycloheximide.
To determine the steady-state level of protein, 0.5 μg of the specified plasmid DNA was transfected into HEK-293T cells with transfection reagent TransIT-LT1 (MIR2300; Mirus) according to the manufacturer’s recommendations. At 16 h posttransfection, the cells were treated with 20 μg/ml of cycloheximide (CHX; 5087390001; Sigma-Aldrich) or a combination of 20 μg/ml CHX and 10 μM MG132 (474790; Calbiochem), a proteasome inhibitor, and harvested at the indicated time points. Equal amounts of cell lysate were subjected to immunoblotting with anti-V5 or anti-β-actin (catalog no. A00702; GenScript) antibodies. The intensities of MAC/PLP2 bands relative to that of β-actin were measured and calculated with AlphaView software (Protein Simple). To assess the rates of decay of the protein amounts over time for the four viral protein types, we fit the two-parameter simple exponential nonlinear regression function, , using the NLIN procedure in the SAS 9.4 software package and verified the results using Minitab software, version 18. In this regression equation, θ1 is the initial viral amount parameter at time zero, and θ2 is the slope or rate of decay parameter. The slope parameters (θ2) were each compared to the WT slope using NLIN’s provided two-sided t tests, and P values of <0.05 were deemed significant.
To evaluate the steady-state level of replicase proteins nsp2-3 and nsp3, we performed a temperature shift experiment. Briefly, HeLa-MHVR cells were infected with either WT or MAC/PLP2mut virus (MOI of 5), followed by incubation at the permissive temperature for 9.5 h, when the cells were shifted to 40°C and treated with 20 μg/ml of CHX. Whole-cell lysates were prepared at 30-min intervals by the addition of lysis buffer A (4% SDS, 3% dithiothreitol, 40% glycerol, and 0.065 M Tris [pH 6.8]). The lysates were passed through a 25-gauge needle to break up aggregates, incubated at 37°C for 30 min, and loaded onto a 6% SDS-PAGE gel, followed by transfer to a nylon membrane. The membrane was incubated with a 1:2,000 dilution of rabbit polyclonal anti-nsp2-3 antibody (anti-D3) (58), followed by horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG(H+L) (SouthernBiotech), and developed with Western Lightning Plus-ECL reagents (Perkin-Elmer). The membrane was stripped and reprobed using a 1:2,000 dilution of mouse anti-calnexin antibody, followed by HRP-conjugated goat anti-mouse IgG(H+L), and then developed as described above.
RT-qPCR.
The protocol of reverse transcription quantitative PCR (RT-qPCR) was performed as described previously (9), with slight modification. Briefly, BMDMs were mock infected or infected with wild-type or mutant MHVs at an MOI of 1 and incubated at a permissive temperature of 32°C. At the indicated time points, cells were harvested for RNA extraction using an RNeasy minikit (74104; Qiagen). An equal amount of RNA was used for cDNA synthesis using an Rt2 HT first-strand kit (330401; Qiagen). To determine IFN-α11, β-actin, or MHV-A59 N gene mRNA production, qPCR was performed with specific primers for mouse IFN-α11 (PPM03050B-200; Qiagen), mouse β-actin (PPM02945B-200; Qiagen), or the MHV-A59 N gene (sense, 5′-AGC AGA CTG CAA CTA CTC AAC CCA ACT C-3′; anti-sense, 5′-GCA ATA GGC ACT CCT TGT CCT TCT GCA-3′) using RT2 SYBR green qPCR mastermix (330502; Qiagen) in the Bio-Rad CFX96 system. The thermocycler was set as follows: one step at 95°C (10 min), 40 cycles at 95°C (15 s) and 60°C (1 min), and a plate read and one step at 95°C (10 s) and a melt curve from 65 to 95°C at increments of 0.5°C/0.05 s. Samples were evaluated in triplicate, and data are representative of three independent experiments. The levels of mRNA were relative to β-actin mRNA and expressed as 2−ΔCT [ΔCT = CT(gene of interest) − CT(β-actin)], where CT is the threshold cycle.
Evaluating viral pathogenesis.
The protocol for evaluating the pathogenesis of MHV was approved by the Loyola University Chicago IACUC and previously described (59). Briefly, 6-week-old C57BL/6 female mice were purchased from the Jackson Laboratory. Mice were intracranially inoculated with 600 PFU of virus in 20 μl of phosphate-buffered saline and monitored daily for changes in body weight. Infected mice were euthanized when weight loss was >25% according to the protocol. Statistical analysis of the survival rate was evaluated using the log-rank test.
ACKNOWLEDGMENTS
We thank Ralph S. Baric, The University of North Carolina at Chapel Hill, for providing tsNC11. We thank Catherine Putonti at Loyola University Chicago for advice on the bioinformatic analysis.
This study was supported by National Institutes of Health (NIH) grant R01 AI085089 (to S.C.B.). R.C.M. was supported by an NIH T32 Training Grant for Experimental Immunology (AI007508) and an Arthur J. Schmitt Dissertation Fellowship in Leadership and Service (Arthur J. Schmitt Foundation).
X.D. and S.C.B. conceived the concept, planned the experiments, and wrote the manuscript with contributions from all authors. X.D., R.C.M., and A.O. performed specific experiments and analyzed the data. J.A.T. performed the bioinformatic analysis. T.E.O. conducted the statistical analysis.
REFERENCES
- 1.de Wit E, van Doremalen N, Falzarano D, Munster VJ. 2016. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, Fan H, Lan T, Yang X-L, Shi W-F, Zhang W, Zhu Y, Zhang Y-W, Xie Q-M, Mani S, Zheng X-S, Li B, Li J-MJ, Guo H, Pei G-Q, An X-P, Chen JJ-W, Zhou L, Mai K-J, Wu Z-X, Li D, Anderson DE, Zhang L-B, Li S-Y, Mi Z-Q, He T-T, Cong F, Guo P-J, Huang R, Luo Y, Liu X-L, Chen JJ-W, Huang Y, Sun Q, Zhang X-L-L, Wang Y-Y, Xing S-Z, Chen Y-S, Sun Y, Li J-MJ, Daszak P, Wang L-F, Shi Z-L, Tong Y-G, Ma J-Y. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol 76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knoops K, Kikkert M, Worm SHE, van den Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enjuanes L, Almazán F, Sola I, Zuñiga S. 2006. Biochemical aspects of coronavirus replication and virus-host interaction. Annu Rev Microbiol 60:211–230. doi: 10.1146/annurev.micro.60.080805.142157. [DOI] [PubMed] [Google Scholar]
- 6.Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kindler E, Thiel V, Weber F. 2016. Interaction of SARS and MERS coronaviruses with the antiviral interferon response. Adv Virus Res 96:219–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kindler E, Gil-Cruz C, Spanier J, Li Y, Wilhelm J, Rabouw HH, Züst R, Hwang M, V’kovski P, Stalder H, Marti S, Habjan M, Cervantes-Barragan L, Elliot R, Karl N, Gaughan C, van Kuppeveld FJM, Silverman RH, Keller M, Ludewig B, Bergmann CC, Ziebuhr J, Weiss SR, Kalinke U, Thiel V. 2017. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog 13:e1006195. doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng X, Hackbart M, Mettelman RC, O’Brien A, Mielech AM, Yi G, Kao CC, Baker SC. 2017. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A 114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menachery VD, Gralinski LE, Mitchell HD, Dinnon KH, Leist SR, Yount BL, McAnarney ET, Graham RL, Waters KM, Baric RS. 2018. Combination attenuation offers strategy for live-attenuated coronavirus vaccines. J Virol 92:e00710-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oudshoorn D, Rijs K, Limpens R, Groen K, Koster AJ, Snijder EJ, Kikkert M, Bárcena M. 2017. Expression and cleavage of Middle East respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. mBio 8:e01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neuman BW. 2016. Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antiviral Res 135:97–107. doi: 10.1016/j.antiviral.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez G, Wang C-Y, Frey TK. 1990. Sequence of the genome RNA of rubella virus: Evidence for genetic rearrangement during togavirus evolution. Virology 177:225–238. doi: 10.1016/0042-6822(90)90476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorbalenya AE, Koonin EV, Lai MM-C. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha-, and coronaviruses. FEBS Lett 288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin EV, Gorbalenya AE, Purdy MA, Rozanov MN, Reyes GR, Bradley DW. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci U S A 89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egloff M-P, Malet H, Putics A, Heinonen M, Dutartre H, Frangeul A, Gruez A, Campanacci V, Cambillau C, Ziebuhr J, Ahola T, Canard B. 2006. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J Virol 80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saikatendu KS, Joseph JS, Subramanian V, Clayton T, Griffith M, Moy K, Velasquez J, Neuman BW, Buchmeier MJ, Stevens RC, Kuhn P. 2005. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure 13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Putics A, Gorbalenya AE, Ziebuhr J, Putics Á́, Gorbalenya AE, Ziebuhr J. 2006. Identification of protease and ADP-ribose 1″-monophosphatase activities associated with transmissible gastroenteritis virus nonstructural protein 3. J Gen Virol 87:651–656. doi: 10.1099/vir.0.81596-0. [DOI] [PubMed] [Google Scholar]
- 19.Putics A, Filipowicz W, Hall J, Gorbalenya AE, Ziebuhr J. 2005. ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J Virol 79:12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eriksson KK, Cervantes-Barragán L, Ludewig B, Thiel V. 2008. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1″-phosphatase, a viral function conserved in the alpha-like supergroup. J Virol 82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuri T, Eriksson KK, Putics A, Züst R, Snijder EJ, Davidson AD, Siddell SG, Thiel V, Ziebuhr J, Weber F. 2011. The ADP-ribose-1″-monophosphatase domains of severe acute respiratory syndrome coronavirus and human coronavirus 229E mediate resistance to antiviral interferon responses. J Gen Virol 92:1899–1905. doi: 10.1099/vir.0.031856-0. [DOI] [PubMed] [Google Scholar]
- 22.Li C, Debing Y, Jankevicius G, Neyts J, Ahel I, Coutard B, Canard B. 2016. Viral macrodomains reverse protein ADP-ribosylation. J Virol 90:8478–8486. doi: 10.1128/JVI.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jankevicius G, Hassler M, Golia B, Rybin V, Zacharias M, Timinszky G, Ladurner AG. 2013. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat Struct Mol Biol 20:508–514. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fehr AR, Athmer J, Channappanavar R, Phillips JM, Meyerholz DK, Perlman S. 2015. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J Virol 89:1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fehr AR, Channappanavar R, Jankevicius G, Fett C, Zhao J, Athmer J, Meyerholz DK, Ahel I, Perlman S. 2016. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio 7:e01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanjanahaluethai A, Baker SC. 2000. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J Virol 74:7911–7921. doi: 10.1128/JVI.74.17.7911-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harcourt BH, Jukneliene D, Kanjanahaluethai A, Bechill J, Severson KM, Smith CM, Rota PA, Baker SC. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol 78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol 79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindner HA, Fotouhi-Ardakani N, Lytvyn V, Lachance P, Sulea T, Ménard R. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol 79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratia K, Saikatendu KS, Santarsiero BD, Barretto N, Baker SC, Stevens RC, Mesecar AD. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci U S A 103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mielech AM, Chen Y, Mesecar AD, Baker SC. 2014. Nidovirus papain-like proteases: Multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res 194:184–190. doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey-Elkin BA, Knaap RCM, Johnson GG, Dalebout TJ, Ninaber DK, van Kasteren PB, Bredenbeek PJ, Snijder EJ, Kikkert M, Mark BL. 2014. Crystal structure of the MERS coronavirus papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J Biol Chem 289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Báez-Santos YM, Mielech AM, Deng X, Baker S, Mesecar AD. 2014. Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the Middle East respiratory syndrome coronavirus. J Virol 88:12511–12527. doi: 10.1128/JVI.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devaraj SG, Wang N, Chen Z, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng C-T, Baker SC, Li K. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem 282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niemeyer D, Mösbauer K, Klein EM, Sieberg A, Mettelman RC, Mielech AM, Dijkman R, Baker SC, Drosten C, Müller MA. 2018. The papain-like protease determines a virulence trait that varies among members of the SARS-coronavirus species. PLoS Pathog 14:e1007296. doi: 10.1371/journal.ppat.1007296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaad MC, Stohlman SA, Egbert J, Lum K, Fu K, Wei T, Baric RS. 1990. Genetics of mouse hepatitis virus transcription: identification of cistrons which may function in positive and negative strand RNA synthesis. Virology 177:634–645. doi: 10.1016/0042-6822(90)90529-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yount B, Denison MR, Weiss SR, Ralph S, Baric RS. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J Virol 76:11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herman RK, Yochem J. 16 September 2005. Genetic enhancers, p 1–11. In The C. elegans Research Community, WormBook (ed), WormBook. doi: 10.1895/wormbook.1.27.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hurst KR, Ye R, Goebel SJ, Jayaraman P, Masters PS. 2010. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J Virol 84:10276–10288. doi: 10.1128/JVI.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hurst KR, Koetzner CA, Masters PS. 2013. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J Virol 87:9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei J, Kusov Y, Hilgenfeld R. 2018. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res 149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Savinov SN, Mielech AM, Cao T, Baker SC, Mesecar AD. 2015. X-ray structural and functional studies of the three tandemly linked domains of nonstructural protein 3 (nsp3) from murine hepatitis virus reveal conserved functions. J Biol Chem 290:25293–25306. doi: 10.1074/jbc.M115.662130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng D, Chen G, Guo B, Cheng G, Tang H. 2008. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res 18:1105–1113. doi: 10.1038/cr.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X, Chen X, Bian G, Tu J, Xing Y, Wang Y, Chen Z. 2014. Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease. J Gen Virol 95:614–626. doi: 10.1099/vir.0.059014-0. [DOI] [PubMed] [Google Scholar]
- 45.Mielech AM, Kilianski A, Baez-Santos YM, Mesecar AD, Baker SC. 2014. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology 450–451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. 2009. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-κB signaling. J Virol 83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ratia K, Kilianski A, Baez-Santos YM, Baker SC, Mesecar A. 2014. Structural basis for the ubiquitin-linkage specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog 10:e1004113. doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fehr AR, Jankevicius G, Ahel I, Perlman S. 2018. Viral macrodomains: unique mediators of viral replication and pathogenesis. Trends Microbiol 26:598–610. doi: 10.1016/j.tim.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leung AKL, McPherson RL, Griffin DE. 2018. Macrodomain ADP-ribosylhydrolase and the pathogenesis of infectious diseases. PLoS Pathog 14:e1006864. doi: 10.1371/journal.ppat.1006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McPherson RL, Abraham R, Sreekumar E, Ong S-E, Cheng S-J, Baxter VK, Kistemaker H, Filippov DV, Griffin DE, Leung A. 2017. ADP-ribosylhydrolase activity of Chikungunya virus macrodomain is critical for virus replication and virulence. Proc Natl Acad Sci U S A 114:1666–1671. doi: 10.1073/pnas.1621485114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abraham R, Hauer D, McPherson RL, Utt A, Kirby IT, Cohen MS, Merits A, Leung AKL, Griffin DE. 2018. ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc Natl Acad Sci U S A 115:E10457–E10466. doi: 10.1073/pnas.1812130115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mielech AM, Deng X, Chen Y, Kindler E, Wheeler DL, Mesecar AD, Thiel V, Perlman S, Baker SC. 2015. Murine coronavirus ubiquitin-like domain is important for papain-like protease stability and viral pathogenesis. J Virol 89:4907–4917. doi: 10.1128/JVI.00338-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graham RL, Sims AC, Brockway SM, Baric RS, Denison MR. 2005. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J Virol 79:13399–13411. doi: 10.1128/JVI.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Donaldson EF, Sims AC, Graham RL, Denison MR, Baric RS. 2007. Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis. J Virol 81:6356–6368. doi: 10.1128/JVI.02805-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donaldson EF, Graham RL, Sims AC, Denison MR, Baric RS. 2007. Analysis of murine hepatitis virus strain A59 temperature-sensitive mutant TS-LA6 suggests that nsp10 plays a critical role in polyprotein processing. J Virol 81:7086–7098. doi: 10.1128/JVI.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gallagher TM. 1996. Murine coronavirus membrane fusion is blocked by modification of thiols buried within the spike protein. J Virol 70:4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baric RS, Fu K, Schaad MC, Stohlman SA. 1990. Establishing a genetic recombination map for murine coronavirus strain A59 complementation groups. Virology 177:646–656. doi: 10.1016/0042-6822(90)90530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schiller JJ, Kanjanahaluethai A, Baker SC. 1998. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology 242:288–302. doi: 10.1006/viro.1997.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng X, StJohn SE, Osswald HL, O’Brien A, Banach BS, Sleeman K, Ghosh AK, Mesecar AD, Baker SC. 2014. Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J Virol 88:11886–11898. doi: 10.1128/JVI.01528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]