Several OPXV species, including monkeypox virus and cowpox virus, cause zoonotic infection in humans. They are believed to use wild rodents as the reservoir or intermediate hosts, but the host or viral factors that are important for OPXV host range in rodents are unknown. Here, we showed that the abortive replication of several OPXV species in a Chinese hamster cell line was caused by a species-specific difference in the host antiviral factor SAMD9L, suggesting that SAMD9L divergence in different rodent species poses a barrier for cross-species OPXV infection. While the Chinese hamster SAMD9L could not be inhibited by two previously identified OPXV inhibitors of human and mouse SAMD9&L, it can be inhibited by cowpox virus CP77, indicating that OPXVs encode three SAMD9&L inhibitors with different specificities. Our data suggest that OPXV host range in broad rodent species depends on three SAMD9&L inhibitors with different specificities.
KEYWORDS: host range, host restriction factor, poxvirus, vaccinia virus
ABSTRACT
Orthopoxviruses (OPXVs) have a broad host range in mammalian cells, but Chinese hamster ovary (CHO) cells are nonpermissive for vaccinia virus (VACV). Here, we revealed a species-specific difference in host restriction factor SAMD9L as the cause for the restriction and identified orthopoxvirus CP77 as a unique inhibitor capable of antagonizing Chinese hamster SAMD9L (chSAMD9L). Two known VACV inhibitors of SAMD9 and SAMD9L (SAMD9&L), K1 and C7, can bind human and mouse SAMD9&L, but neither can bind chSAMD9L. Clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 knockout of chSAMD9L from CHO cells removed the restriction for VACV, while ectopic expression of chSAMD9L imposed the restriction for VACV in a human cell line, demonstrating that chSAMD9L is a potent restriction factor for VACV. In contrast to K1 and C7, cowpox virus CP77 can bind chSAMD9L and rescue VACV replication in cells expressing chSAMD9L, indicating that CP77 is yet another SAMD9L inhibitor but has a unique specificity for chSAMD9L. Binding studies showed that the N-terminal 382 amino acids of CP77 were sufficient for binding chSAMD9L and that both K1 and CP77 target a common internal region of SAMD9L. Growth studies with nearly all OPXV species showed that the ability of OPXVs to antagonize chSAMD9L correlates with CP77 gene status and that a functional CP77 ortholog was maintained in many OPXVs, including monkeypox virus. Our data suggest that a species-specific difference in rodent SAMD9L poses a barrier for cross-species OPXV infection and that OPXVs have evolved three SAMD9&L inhibitors with different specificities to overcome this barrier.
IMPORTANCE Several OPXV species, including monkeypox virus and cowpox virus, cause zoonotic infection in humans. They are believed to use wild rodents as the reservoir or intermediate hosts, but the host or viral factors that are important for OPXV host range in rodents are unknown. Here, we showed that the abortive replication of several OPXV species in a Chinese hamster cell line was caused by a species-specific difference in the host antiviral factor SAMD9L, suggesting that SAMD9L divergence in different rodent species poses a barrier for cross-species OPXV infection. While the Chinese hamster SAMD9L could not be inhibited by two previously identified OPXV inhibitors of human and mouse SAMD9&L, it can be inhibited by cowpox virus CP77, indicating that OPXVs encode three SAMD9&L inhibitors with different specificities. Our data suggest that OPXV host range in broad rodent species depends on three SAMD9&L inhibitors with different specificities.
INTRODUCTION
The Orthopoxvirus (OPXV) genus of the Poxviridae family consists of more than 10 closely related species, including the human pathogen variola virus (VARV) (the causative agent for smallpox), the smallpox vaccine vaccinia virus (VACV), and the emerging zoonotic agents monkeypox virus (MPXV) and cowpox virus (CPXV) (1). These viruses as well as ectromelia virus (ECTV), camelpox virus (CMLV), and teterapox virus (TATV) are known as the Old World OPXVs for originating from the Eurasian continent, while raccoonpox virus (RNCV), skunkpox virus (SKPV), and volepox virus (VPXV) are recognized as the North American OPXVs for being endemic in North America (2). VARV has been eradicated from nature, but the cessation of smallpox vaccination and the waning of anti-OPXV herd immunity have increased the risk of zoonotic OPXV infections. MPXV is highly virulent in humans (3), and the recent increase in human monkeypox cases across a wide geographic area is a concern for global health security (4). CPXV and VACV are responsible for zoonoses in Europe, Asia, and South America (5, 6). Novel OPXV species have also been discovered in recent human cases (7, 8), including Akhmeta virus (AKMV) isolated from the town of Akhmeta in the country Georgia.
OPXVs vary greatly in their animal host range (2); some, such as VARV, CMLV, and TATV, are known to infect only a single mammalian species, while others, such as MPXV, CPXV, and VACV, are capable of infecting a wide variety of mammalian hosts. OPXVs are often named after the host in which they were first isolated, while their reservoir hosts in nature are unknown. However, OPXV infections are often associated with contact with rodents. CPXV is carried and believed to be transmitted to humans and domestic animals by bank voles and striped field mice in western Europe (9, 10). MPXV is likely carried by rope squirrels in Africa (11). The spread of VACV in Brazil involves wild rodents as the reservoir or intermediate hosts (12). In contrast to their varied host range in nature, OPXV host ranges in tissue culture cells are almost universally broad (13). It was thus noteworthy that Chinese hamster ovary (CHO) cells were found to be nonpermissive for VACV, owing to a rapid shutoff of protein synthesis that resulted in a block of viral intermediate mRNA translation (14, 15). A CPXV gene from the Brighton Red (BR) strain, CP77, could rescue VACV replication in CHO cells (16). CPXV BR CP77 encodes a 668-amino-acid (aa), 77-kDa protein with nine predicted ankyrin repeats and a C-terminal F-box domain (17). CP77 is conserved in all CPXV strains, having greater than 91% amino acid sequence identity among different strains. A CP77 ortholog with approximately 90% amino acid identity is also present in many MPXV strains. In contrast, the CP77 ortholog is either deleted or fragmented in all sequenced VACV strains, while a large deletion occurs in the VARV CP77 ortholog and results in a protein of at most 452 aa.
Although the specific host pathway that restricts VACV replication in CHO cells and the mechanism by which CP77 overcomes the restriction are unknown, CP77 is long recognized as a host range gene that provides a function similar to that of the VACV host range genes K1L and C7L (18, 19). K1L encodes a 284-aa protein consisted entirely of ankyrin repeats (20), while C7L encodes a 150-aa protein forming a single β-sandwich (21). VACV with deletions in both K1L and C7L (vK1−C7−) replicates abortively in many mammalian cell lines (19); the replication defect in human cells could be rescued by any one of the three nonhomologous poxvirus genes, K1L, C7L, or CP77. K1L and CP77 are specific to OPXVs, whereas functional C7L homologs are present in nearly all mammalian poxviruses (22). Recent studies revealed that K1 and C7 provide an equivalent function for viral growth in human and mouse cells by inhibiting a common set of host restriction factors, SAMD9 (23, 24) and SAMD9L (25). Human SAMD9 and SAMD9L (SAMD9&L) are cytosolic proteins constitutively expressed in many tissues (26), and their expression can be further induced by interferons (25). Human SAMD9 (hSAMD9), human SAMD9L (hSAMD9L), and mouse SAMD9L (mSAMD9L) are all capable of blocking the replication of poxvirus mutants that lack K1L- and C7L-like genes (23–25). SAMD9&L are also recognized as tumor suppressors (27–29), and germ line mutations of human SAMD9 or SAMD9L are associated with multisystem disorders characterized by immunodeficiency and predisposition to hematological malignancies (30–35). In this study, we investigated whether CP77 functions similarly to K1 and C7 by directly targeting SAMD9 or SAMD9L. Our results not only identified CP77 as yet another OPXV inhibitor of SAMD9&L but also revealed a species-specific difference in SAMD9L that explains the specific requirement of CP77 for OPXV tropism in some rodent cells.
(This article was submitted to an online preprint archive [36].)
RESULTS
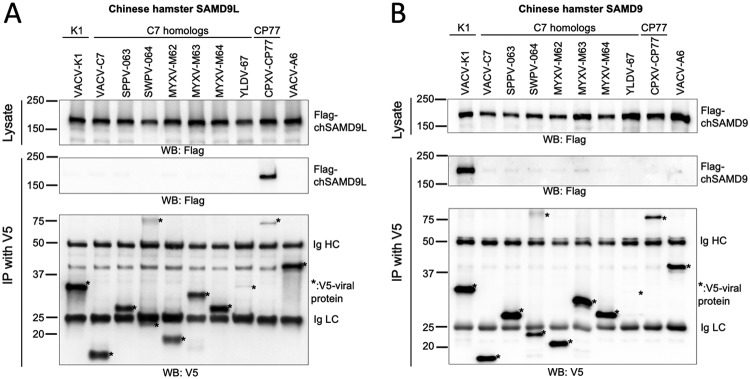
Chinese hamster SAMD9&L have unique binding specificity for poxvirus proteins.
All known poxvirus antagonists of SAMD9 and SAMD9L (SAMD9&L) were able to bind their targets in mammalian cells (21, 25). To assess whether they could also inhibit SAMD9&L from Chinese hamster, we first set out to study their binding to Chinese hamster SAMD9&L. Chinese hamster SAMD9 (chSAMD9) and SAMD9L (chSAMD9L) were cloned from CHO cells into a mammalian expression vector and transiently expressed in HEK 293FT cells. The cells were then infected with a panel of vK1−C7−-derived VACV strains expressing different V5-tagged viral proteins, including one that expressed CPXV CP77 (vCPXV-CP77). The ability of the V5-tagged viral protein to bind the Flag-tagged SAMD9 or SAMD9L was assessed by immunoprecipitation with anti-V5 antibody followed by Western blotting. All known poxvirus SAMD9&L antagonists, including K1, C7, and C7 homologs from diverse mammalian poxviruses, failed to precipitate chSAMD9L (Fig. 1A), in contrast to the binding of SAMD9&L from human and mouse by nearly all these viral proteins (21, 25). Interestingly, CP77 was able to precipitate chSAMD9L, suggesting that it is also a SAMD9L inhibitor and has a unique specificity for chSAMD9L. chSAMD9 was effectively precipitated by K1, while only a small amount of chSAMD9 was precipitated by some of the C7 homologs, and the amount was not significantly larger than that precipitated by the VACV morphogenesis protein A6 (37) (Fig. 1B). Altogether, SAMD9&L from Chinese hamster stand out among the SAMD9&Ls that have been characterized so far for their resistance to binding by many of the known poxvirus SAMD9&L inhibitors.
FIG 1.
Only CPXV CP77 can bind Chinese hamster SAMD9L, while only VACV K1 can bind Chinese hamster SAMD9. 293FT cells were transfected with a plasmid expressing Flag-tagged chSAMD9L (A) or chSAMD9 (B) and infected with a panel of vK1−C7−-derived VACVs that expressed different V5-tagged viral proteins. A virus expressing VACV-A6, a viral protein involved in viral assembly, was used as a negative control. The cell lysates were immunoprecipitated with anti-V5 antibody. Epitope-tagged proteins in the cell lysate and precipitate were detected with anti-Flag or anti-V5 antibody in Western blotting (WB). The heavy and light chains of the precipitated antibody (Ig HC and LC) served as loading controls. *, V5-tagged proteins.
chSAMD9L is required for restricting VACV replication in CHO cells, and CP77 is a SAMD9L inhibitor with a unique specificity for chSAMD9L.
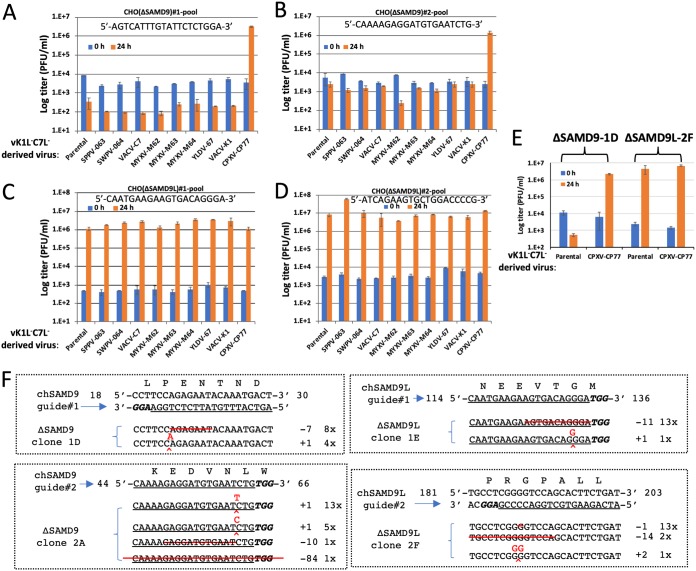
The lack of binding to chSAMD9 or chSAMD9L by K1 and C7 suggests that a failure of VACV in antagonizing chSAMD9 and/or chSAMD9L could be the reason why VACV replicates abortively in CHO cells. To test this idea, we knocked out either chSAMD9 or chSAMD9L from CHO cells by using the clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 method. For each gene knockout (KO), two independent KOs with different guide sequences (named 1 and 2) were performed, and the KO cell pools were tested for permissiveness for the panel of vK1−C7−-derived VACVs. Similar to the case for the parental CHO cells, the two chSAMD9 KO CHO cells (named ΔSAMD9) were nonpermissive for all recombinant VACVs except vCPXV-CP77, which grew more than 100-fold in titer after 24 h of infection (Fig. 2A and B). In contrast, the two chSAMD9L KO cells (named ΔSAMD9L) were permissive for vK1−C7− as well as all its derivatives (Fig. 2C and D). Several cell clones were also isolated and found to be similar to the cell pools in permissiveness for VACV (Fig. 2E). The specific KO of either chSAMD9 or chSAMD9L in the cell clones were confirmed by sequencing the region targeted by the CRISPR-Cas9 guide. Indels that result in frameshift were found (Fig. 2F). These data show that chSAMD9L but not chSAMD9 is required for restricting VACV replication in CHO cells and that CP77 has a unique capability of antagonizing chSAMD9L. The well-characterized ΔSAMD9L clone 2F was used in all subsequent experiments.
FIG 2.
The restriction of VACV by CHO cells can be abolished by knocking out chSAMD9L. (A to D) CHO cells were transduced with lentiviral vectors expressing Cas9 and a guide sequence targeting either chSAMD9 (A and B) or chSAMD9L (C and D). For each gene, two independent KOs with different guide sequences (1 [A and C] and 2 [B and D]) were performed, and the KO cell pools were infected with the panel of vK1−C7−-derived VACVs at a multiplicity of infection (MOI) of 1 PFU/cell. Viral titers at 0 and 24 h postinfection (hpi) were measured by plaque assay in Vero cells. (E) Several KO cell clones were isolated from the cell pools. Infection studies were performed on the cell clones, and the results were similar to those shown in panels A to D. Representative data with the ΔSAMD9-1D clone and the ΔSAMD9L-2F clone are shown. (F) The genotypes of the cell clones were determined by sequencing. The guide sequence is underlined, and the protospacer-adjacent motif (PAM) sequence is in bold italics. The starting and ending positions of the guide in the ORF and the encoded amino acid sequence are also shown. Shown below the target are the genomic sequences from cell clones. Red lines, deletions; ^, insertion. The numbers after the + and − denote the number of indels, and the number before the “x” denotes the number of times the sequence was detected from a total of 10 to 20 cloned PCR products.
It was previously shown that VACV replication in CHO cells was blocked at translation of postreplicative mRNA (18). To assess the effect of SAMD9L KO on viral early and postreplicative protein synthesis, we infected the parental and ΔSAMD9L CHO cells with VACVs that expressed the luciferase gene under the control of either the synthetic early/late promoter or the late p11 promoter. In some infections, viral DNA replication was blocked to allow only early protein synthesis. The early luciferase expression measured at 8 h postinfection (hpi) was at a comparable level in BSC40 cells and ΔSAMD9L CHO cells but was reduced by 10-fold in the parental CHO cells (Fig. 3A). A similar reduction of early luciferase expression was also observed at 24 hpi (Fig. 3B). However, the greatest difference between the parental and ΔSAMD9L CHO cells was observed for postreplicative, late luciferase expression, which was reduced by ∼10,000-fold in the parental CHO cells (Fig. 3B). Altogether, the data indicate that chSAMD9L KO alleviated the block on viral protein synthesis, particularly late protein synthesis.
FIG 3.

The block in viral protein synthesis in CHO cells can be abolished by knocking out chSAMD9L. BSC40 cells and the parental and ΔSAMD9L (Δ9L, 2F clone) CHO cells were infected with VACVs that expressed a luciferase reporter under the control of either the synthetic early/late (E/L) promoter or the late p11 promoter in the presence or absence of AraC. The cells were lysed after either 8 h (A) or 24 h (B) of infection, and luciferase (LUC) activities were measured.
Expression of chSAMD9L in human cells is sufficient for recapitulating the poxvirus restriction property of CHO cells.
While many mammalian cell lines were nonpermissive for vK1−C7−, we found that some human cell lines, including breast cancer BT20 cells, expressed a low level of human SAMD9 and were thus permissive for vK1−C7−. We made a stable BT20 cell line that expressed chSAMD9L by transduction with a lentivirus encoding chSAMD9L controlled by the tetracycline-dependent promoter. The cell line (named i-chSAMD9L) expressed chSAMD9L only when induced with doxycycline and in a dose-dependent manner (Fig. 4A). When not induced to express chSAMD9L, i-chSAMD9L cells were fully permissive for vK1−C7− and its derivatives, which grew by ∼1,000-fold in titer after 24 h of infection (Fig. 4B). When induced to express chSAMD9L, the cell line became nonpermissive for vK1−C7− and its derivatives that expressed either K1 (vVACV-K1) or C7 (vVACV-C7) (Fig. 4B), recapitulating CHO cells in terms of the restriction of poxvirus. Moreover, vCPXV-CP77 was able to grow ∼100-fold in titer after 24 h of infection, again demonstrating that CP77 can antagonize chSAMD9L.
FIG 4.
Expression of chSAMD9L in human cells is sufficient for recapitulating the poxvirus restriction property of CHO cells. (A) A stable human BT20 cell line with inducible expression of chSAMD9L was established via lentiviral transduction. The cell line (i-chSAMD9L) was cultured with medium containing the indicated concentration of doxycycline (Dox). The levels of chSAMD9L and the control HSP70 protein in the cell lysates were determined by Western blotting. (B) The i-chSAMD9L cells were untreated or treated with 1 μg/ml Dox. The cells were then infected with the panel of vK1−C7−-derived VACVs. Viral titers at 0 and 24 hpi were measured by plaque assay in Vero cells.
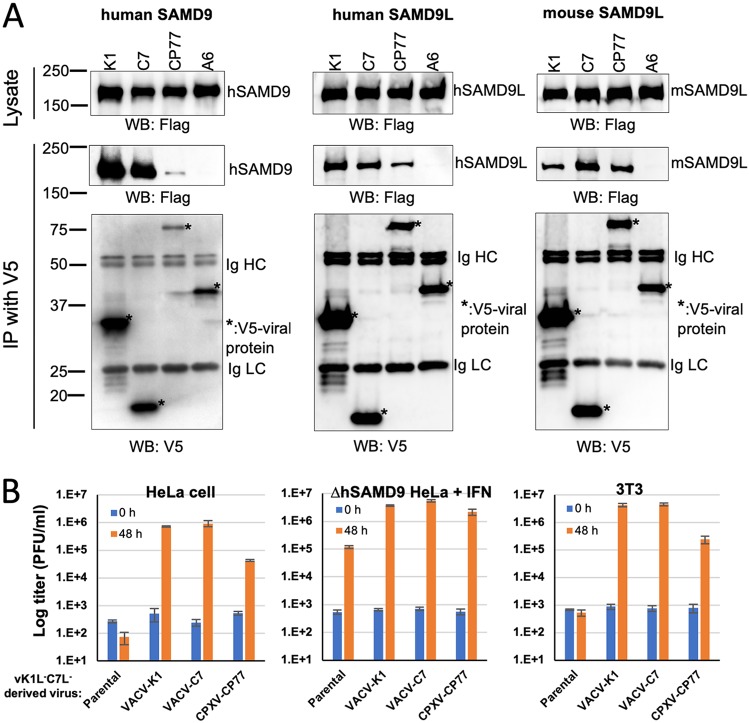
CP77 is an inhibitor of human and mouse SAMD9&L.
K1 and C7 were previously shown to inhibit human and mouse SAMD9&L. To compare the species specificities of the three OPXV SAMD9&L inhibitors, we studied the abilities of CP77 to bind and inhibit hSAMD9, hSAMD9L, and mSAMD9L. In comparison to the negative-control VACV A6 protein, CP77 could precipitate all tested SAMD9&Ls, although a smaller amount of hSAMD9 was precipitated by CP77 than by either K1 or C7 (Fig. 5A). hSAMD9 and mSAMD9L are the respective restriction factors for vK1−C7− in HeLa and 3T3 cells, while hSAMD9L could be induced by interferon (IFN) in hSAMD9 KO (ΔhSAMD9) HeLa cells to reduce the replication of vK1−C7− (25). vCPXV-CP77 could grow in HeLa and 3T3 cells, although the yield was ∼10-fold lower than that of vVACV-K1 or vVACV-C7 (Fig. 5B). In IFN-treated ΔhSAMD9 cells, the yield of vCPXV-CP77 was slightly lower than that of vVACV-K1 or vVACV-C7 and was ∼10-fold higher than that of vK1−C7−. Thus, the binding and infectivity studies showed that CP77 could inhibit hSAMD9, hSAMD9L, and mSAMD9L, albeit with a lower potency than K1 and C7.
FIG 5.
CP77 is an inhibitor of hSAMD9, hSAMD9L, and mSAMD9L. (A) 293FT cells were transfected with a plasmid expressing Flag-tagged hSAMD9, hSAMD9L, or mSAMD9L and infected with VACVs that expressed different V5-tagged viral proteins. Immunoprecipitation and detection were performed as described for Fig. 1 (B) HeLa, ΔhSAMD9 HeLa, or 3T3 cells were infected with the indicated viruses, and viral growth was determined by measuring viral titers at 0 and 48 hpi. ΔhSAMD9 HeLa cell were pretreated with IFN-β (100 U/ml) for 24 h prior to the infection.
K1 and CP77 target the same region of SAMD9&L.
A computational analysis predicated SAMD9&L to have the following domains, from the N to the C terminus (38): SAM, AlbA, SIR2, P-loop nucleoside triphosphatase (NTPase), TPR, and OB (Fig. 6A). The N-terminal 385 aa of hSAMD9, which contain the predicted SAM and AlbA domains, were reported to be sufficient for binding to a poxvirus C7 homolog (39). To find out which region of hSAMD9 is targeted by K1, we constructed hSAMD9 mutants with deletions in different domains and tested the binding of the mutants to K1. We found that aa 607 to 1172 of hSAMD9, which contain the putative NTPase and TPR domains, were sufficient for binding to K1 but not C7 (Fig. 6B). This binding was disrupted by two specific K1 substitution mutations (S2C2 or S1-mut6) that were previously shown to disrupt K1 host range function in human cells (20). Furthermore, both K1 and CP77, but not C7, can bind to the similar region of mouse SAMD9L (aa 598 to 1172), but only CP77 can bind to this region of chSAMD9L (aa 594 to 1172) (Fig. 6C). Thus, the binding to the NTPase-TPR domain by K1 and CP77 displays the same species specificity as that to the full-length protein. Altogether, the data demonstrate that K1 and CP77 target the same internal region of SAMD9&L.
FIG 6.
K1 and CP77 target a common internal region of SAMD9&L. (A) Schematics of the predicted SAMD9&L domain architecture and the SAMD9&L truncation with only the NTPase-TPR domain. (B and C) 293FT cells were transfected with a plasmid expressing the putative NTPase and TPR domains of SAMD9&L (aa 607 to 1172 for hSAMD9, aa 598 to 1172 for mSAMD9L, and aa 594 to 1172 for chSAMD9L) and infected with VACV expressing C7, K1, or CP77. Coimmunoprecipitation and Western blotting were performed as described for Fig. 1 The two K1 substitution mutations (S2C-2 or S1-mut6) were previously shown to disrupt K1 host range function in human cells (20).
The N-terminal 382 aa of CP77 are sufficient for binding to chSAMD9L.
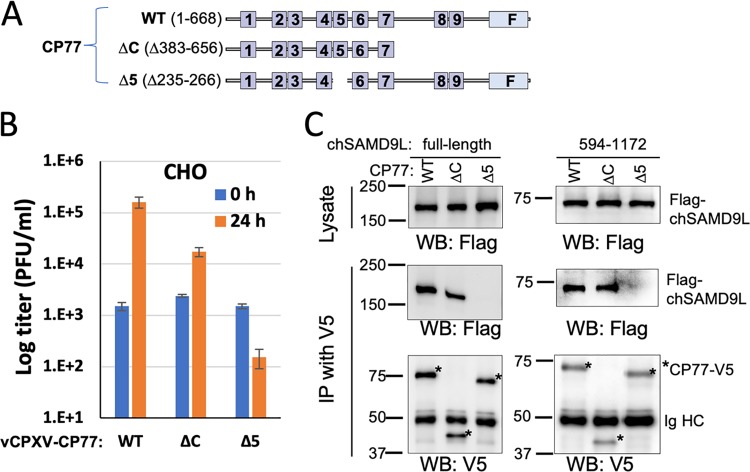
The full-length CP77 of 668 aa was predicted to contain 9 ankyrin repeats with a C-terminal F-box domain (Fig. 7A). Only the N-terminal 352 aa were reported to be necessary for the function of CP77 in CHO cells, and the deletion of ankyrin repeat 5 could disrupt CP77 function (40). To determine whether chSAMD9L binding has a similar requirement for CP77 residues, we constructed recombinant VACV from vK1−C7− by inserting truncated CP77 into the viral genome. The recombinant virus expressing the N-terminal 382 aa of CP77 (ΔC, maintaining the N-terminal seven ankyrin repeats) can grow in CHO cells, although with an ∼10-fold reduction in yield compared to that for the virus expressing the full-length CP77 (Fig. 7B). In contrast, the virus expressing a CP77 with a deletion of aa 235 to 266 (the predicted ankyrin repeat 5, Δ5) failed to grow in CHO cells. Correspondingly, CP77-ΔC can precipitate both the full length and the NTPase-TPR region of chSAMD9L, whereas CP77-Δ5 cannot precipitate either (Fig. 7C). Less full-length chSAMD9L was precipitated by CP77-Δc than by full-length CP77, correlating with the reduced growth in CHO cells of the CP77-Δc-expressing VACV. Compared to the full-length CP77, CP77-ΔC was also detected at a reduced level in the infected cells (Fig. 7C).
FIG 7.
The N-terminal 382 aa of CP77 are sufficient for binding to chSAMD9L. (A) Schematics of different CP77 constructs. Ankyrin repeats are shown as boxes and numbered. The C-terminal F-box is indicated by “F.” ΔC, deletion of aa 383 to 656 (maintaining the N-terminal seven ankyrin repeats); Δ5, deletion of aa 235 to 266 (ankyrin repeat 5). (B) CHO cells were infected with vK1−C7−-derived VACV expressing either WT or mutated CP77. Viral growth was determined as described for Fig. 2. (C) Full-length chSAMD9L and aa 594 to 1172 of chSAMD9L were subjected to coimmunoprecipitation with either WT or mutated CP77 as described for Fig. 1.
The ability of OPXVs to antagonize chSAMD9L corresponds to their CP77 gene status.
The CP77 gene is variably maintained and diversified in different OPXV species (Table 1). To assess the ability of chSAMD9L to restrict different OPXV species, we compared the growth of all available OPXV species in the parental and ΔSAMD9L CHO cells (Fig. 8). ECTV and CMLV are similar to VACV in growing in ΔSAMD9L CHO cells but failing to grow in the parental CHO cells, indicating that chSAMD9L is also a restriction factor for ECTV and CMLV. The CMLV CP77 ortholog is largely deleted, with only ∼155 nucleotides (nt) left, while the ECTV CP77 ortholog has a large deletion at the 5′ end that results in an early frameshift. On the other hand, MPXV, TATV, AKMV, SKPV, and VPXV grew in both the parental and ΔSAMD9L CHO cells with nearly the same efficiency, indicating that chSAMD9L did not pose significant restriction for their replication. These OPXV species all encode a full-length CP77 ortholog, with the two North American OPXVs (SKPV and VPXV) having the most divergent ortholog with ∼70% amino acid identity to CPXV CP77. RCNV, another North American OPXV, also grew in both the parental and ΔSAMD9L CHO cells, but the yield was more than 10-fold less in the parental cells at both 24 and 48 h postinfection (Fig. 8), indicating that chSAMD9L reduced but did not block RCNV growth in CHO cells. Interestingly, a gene fusion event in the RCNV genome resulted in an open reading frame (ORF) encoding the N-terminal 406 aa of the CP77 ortholog and the 246-aa chemokine binding protein. Altogether, the data show that the ability of OPXV species to antagonize chSAMD9L and grow in CHO cells correlates with their encoding a CP77 ortholog.
TABLE 1.
Comparison of OPXV CP77 orthologs to CPXV BR CP77
| OPXV species | CP77 |
Antagonism to chSAMD9L | |
|---|---|---|---|
| Length (aa) | Identity (%) | ||
| CPXV | 661–674 | 91–100 | Yes |
| MPXV | 659–660 | 90–91 | Yes |
| VACV | Small deletions, early frameshift | No | |
| VARV | >60-nt deletion, 355–490 | 86–88 | NDa |
| ECTV | 504-nt deletion, early frameshift | No | |
| CMLV | Large deletion (∼155 nt remaining) | No | |
| TATV | 661 | 92 | Yes |
| AKMV | 672 | 79 | Yes |
| SKPV | 633 | 69 | Yes |
| VPXV | 613 | 69 | Yes |
| RNCV | Gene fusion,b 406 | 66 | Partial |
ND, not determined.
Strain MD1964-85A. aa 1 to 406 of the CP77 ortholog is fused in frame to the N terminus of the full-length, 246-aa chemokine binding protein; the total protein length is 655 aa.
FIG 8.
The orthopoxvirus host range in CHO cells corresponds to their CP77 gene status. Parental (CHO-WT) and ΔSAMD9L (CHO-ΔSAMD9L) CHO cells were infected with the indicated orthopoxvirus species. Viral titers at 0, 24, and 48 hpi were measured by plaque assay on Vero cells.
DISCUSSION
Many infectious diseases that result in high morbidity and mortality in humans are zoonoses. VARV, an exclusively human pathogen, is believed to have evolved from an African rodent-borne virus (41) before it spread around the world and became mankind’s deadliest killer. For many viruses, divergence in viral entry receptors in different host species poses a major hurdle for cross-species transmission (42). Poxviruses, however, can enter nearly any animal cell (43). Why many poxviruses show strict host species specificity is less clear, but some host antiviral factors such as PKR and SAMD9L have shown some species-specific differences in susceptibility to poxvirus inhibitors (25, 44, 45). In this study, we revealed a species-specific difference in SAMD9L as the cause for the restriction of several OPXVs (VACV, ECTV, and CMLV) in a rodent cell, suggesting that divergence in SAMD9L (and perhaps SAMD9) in rodent species presents a major barrier for cross-species poxvirus infection. Furthermore, we identified CP77 as the third OPXV SAMD9&L inhibitor with a unique species specificity, demonstrating the sophistication brought forth by OPXVs to antagonize SAMD9&L.
OPXVs have broad host range in tissue culture cells, but CHO cells, while permissive for CPXV (16), are nonpermissive for VACV (14) and ECTV (46). What host factor causes the restriction of some OPXV species in CHO cells had been enigmatic since the initial discovery more than 50 years ago (14). In this study, we solved this mystery by identifying chSAMD9L as the host restriction factor in CHO cells. We presented two complementary lines of evidence: (i) CRISPR-Cas9 KO of chSAMD9L from CHO cells completely removed the host restriction for VACV, ECTV, and CMLV, and (ii) ectopic expression of chSAMD9L in a human cell line recapitulated the poxvirus restriction property of CHO cells. We also provided a molecular explanation of why chSAMD9L could restrict some OPXV species, namely, its resistance to binding by both K1 and C7, the two SAMD9&L inhibitors in these OPXV species. Interestingly, while SAMD9 is the constitutive poxvirus restriction factor in many human cells (23–25), chSAMD9 appears not to contribute to poxvirus restriction in CHO cells. KO of chSAMD9L alone is sufficient for abolishing the restriction of a panel of VACVs, most of which did not contain K1, the only protein that was shown to bind chSAMD9. Moreover, KO of chSAMD9 had no effect on poxvirus restriction in CHO cells. CHO cells thus resemble mouse cells more than human cells in that SAMD9L is the constitutive restriction factor (25). It is unclear why chSAMD9 does not restrict poxviruses in CHO cells. We speculate that either chSAMD9 has lost the antiviral function due to some lineage-specific mutations or the chSAMD9 expression level in CHO cells is not sufficiently high. The former scenario would be analogous to the fate of mouse SAMD9, which has suffered a mouse lineage-specific gene loss (26). The latter scenario would be similar to the case for human SAMD9L, which has to be induced to a high expression level by IFN to impose restriction on vK1−C7− (25). However, our preliminary experiments in which we treated ΔSAMD9L CHO cells with mouse or universal type 1 IFN did not result in restriction of vK1−C7−. While CHO cells and a Chinese hamster peritoneal cell line are nonpermissive for VACV, there are some cell lines derived from Chinese hamster that do not restrict VACV replication (46). As ectopic expression of chSAMD9L in a human cell line alone is sufficient for restricting VACV replication, those permissive Chinese hamster cell lines may not express chSAMD9L at a sufficiently high level. Similarly, although human and mouse SAMD9&Ls are potent restriction factors for vK1−C7−, there are human and mouse cell lines that are permissive for vK1−C7− due to a low SAMD9&L expression level. It will be interesting to test whether chSAMD9L poses a restriction for OPXV species with no functional CP77 to infect Chinese hamster.
CPXV BR CP77 was found to rescue VACV replication in CHO cells more than 40 years ago (16). Since then, a number of molecular functions have been attributed to CP77, including the binding to host HMG20A, NF-κB subunit p65, and the SCF complex (17, 40). However, the molecular mechanism underlying the host range function of CP77 remained elusive. We showed in this study that CP77 coimmunoprecipitated chSAMD9L and rescued VACV replication in a human cell line that was induced to express chSAMD9L, demonstrating that CP77 is a SAMD9L inhibitor. Only the N-terminal 382 aa, containing the first seven ankyrin repeats, was essential for chSAMD9L binding, while deletion of ankyrin repeat 5 abolished the binding. Correspondingly, a CP77 mutant with only the first seven ankyrin repeats but not the mutant with deletion of ankyrin repeat 5 could rescue VACV replication in CHO cells, suggesting that the host range function of CP77 relies on its binding with chSAMD9L. This idea was further supported by comparing the growth of nearly all OPXV species in the parental and ΔSAMD9L CHO cells. OPXV species that encode a full-length CP77 ortholog (MPXV, TATV, AKMV, SKPV, and VPXV) can replicate in both the parental and ΔSAMD9L CHO cells, while OPXV species that have lost the CP77 gene (VACV, ECTV, and CMLV) can replicate only in ΔSAMD9L CHO cells. Interestingly, RNCV, which encodes a fusion protein that contains only the N-terminal 406 aa of the CP77 ortholog (47), can also replicate in CHO cells, albeit with reduced efficiency compared to that in ΔSAMD9L CHO cells. The RNCV genome is closely related to those of the other two North American OPXVs, with the largest difference being a 25-kbp deletion in the left terminal region of RCNV that removed 12 complete genes and created an in-frame gene fusion of the CP77 ortholog and the chemokine binding protein (47). The difference between the three North American OPXVs in terms of their replication in CHO cells correlates with our observation that the N-terminal seven ankyrin repeats were less effective than the full-length CP77 in inhibiting chSAMD9L. VARV encodes a CP77 ortholog varying from 355 to 490 aa in length, so VARV is predicted to be able to partially inhibit chSAMD9L.
Recently, we and others have established the importance of SAMD9&L as host restriction factors against poxviruses at the cellular and organismal levels (23–25). The identification of CP77 as yet another SAMD9&L inhibitor underscores the critical role of SAMD9&L in host defense against poxviruses and the elaborate lengths to which OPXVs have gone to evade SAMD9&L. K1 and C7 were previously shown to function equivalently in inhibiting human and mouse SAMD9&L (25). In this study, however, we uncovered differences between K1, C7, and CP77 in their targeting mechanisms and binding specificities for SAMD9&L. While a C7 ortholog was shown to target the N terminus of SAMD9 (39), both K1 and CP77 target an internal region containing the predicated NTPase and TPR domains. While any one of K1, C7, and CP77 can bind mouse SAMD9L, only K1 can bind chSAMD9, and only CP77 can bind chSAMD9L. SAMD9Ls from mouse and Chinese hamster have ∼80% amino acid sequence identity. We speculate that a need for overcoming SAMD9&L sequence divergence in different rodent species may have driven OPXVs to evolve three different inhibitors targeting different regions of SAMD9&L. The loss of K1 and/or CP77 from OPXV species with a narrow host range (VARV and CMLV) and the maintenance of all three (K1, C7, and CP77) in species presumably endemic in wild rodents (CPXV, MPXV, and North American OPXVs) also suggest that multiple SAMD9&L inhibitors are needed specifically for overcoming diverse SAMD9&Ls in rodents.
MATERIALS AND METHODS
Cells and viruses.
Vero (ATCC CCL-81), BSC40 (ATCC CRL-2761), BT20 (ATCC HTB-19), NIH/3T3 (ATCC CRL-1658), and CHO-K1 (ATCC CCL-61) cells were originally from the ATCC. HEK 293FT was from Thermo Fisher Scientific (catalog no. R70007). The HeLa cell line with an hSAMD9 knockout (ΔhSAMD9) was described before (25). The wild-type (WT) VACV WR strain, a VACV strain with deletions of K1L and C7L (vK1−C7−), and a panel of vK1−C7−-derived recombinant viruses expressing VACV K1 (vVACV-K1L) or a C7 homolog from different poxviruses (vVACV-C7L, vYLDV-67R, vMYXV-M62R, vMYXV-M63R, vMYXV-M64R, vSPPV-063, and vSWPV-064) were described before (22, 48, 49). vA6L-V5 is a VACV strain expressing VACV A6 with a V5 tag (37). Other OPXVs (50), ECTV (Moscow), CMLV (CMS), MPXV (West African clade), AKMV (51), SKPV (USA1978-WA), VPXV (USA1985-CA), TATV (Dahomey1968), and RNCV (MD1964_85A) were from the CDC and were cultured in BSC-40 cells.
VACVs expressing WT or mutated CPXV CP77 were constructed as follows. An intermediate virus named vK1−C7−CP77− was first constructed by deleting the CP77 gene fragments from vK1−C7− (22) by the transient dominant selection method, as described previously (52). The deletion of the CP77 gene fragment in the recombinant virus was confirmed by PCR amplification of the CP77 region of the virus. Viruses expressing V5-tagged CP77 were derived from vK1−C7−CP77− through homologous recombination with a transfer plasmid. The transfer plasmid contains (i) 500 bp of the downstream flanking region of CP77, (ii) a green fluorescent protein (GFP) gene under the control of VACV late promoter P11, (iii) CPXV BR CP77 with a C-terminal V5 tag, and (iv) ∼500 bp of the upstream flanking region of CP77 including the CP77 promoter. Specific mutations of CP77 were introduced into the plasmid through recombinant PCR as described previously (48). All constructs were confirmed by DNA sequencing. The recombinant virus construction was done according to standard protocols (53). In brief, the transfer plasmids were transfected into Vero cells that were infected with vK1−C7−CP77−. Recombinant viruses expressing GFP were picked under a fluorescence microscope and purified through four rounds of plaque isolation on Vero cells.
SAMD9 and SAMD9L expression constructs.
Expression constructs for human and mouse SAMD9L were reported before (25). The chSAMD9 ORF was PCR amplified with the primer pair 5′-GGATGACGATGACAAGGCAGAGAAACTCAACCTTCCAGAGA-3′ and 5′-GGCTCCGCGGTTAGACAATTTTAATGTCATAAGCA-3′ from cDNA synthesized from CHO cellular mRNAs. A 3×Flag tag sequence was then appended to the 5′ end of the ORF by PCR, and the final PCR product was cloned between KpnI and SacII sites of the pcDNA3.1/V5-His-Topo vector (Thermo Fisher Scientific). The chSAMD9 cDNA was completely sequenced and found to be identical to the Chinese hamster SAMD9 reference sequence in GenBank (XM_027389842.1). chSAMD9L ORF was cloned similarly but with the primer pair 5′-TGACGATGACAAGAATGAACAAGTAACTGCACCTAAATTGG-3′ and 5′-GGCTCCGCGGTTAGATTACTTTTATGCCATATGCCAGAGG-3′. The chSAMD9L cDNA was found to be identical to the Chinese hamster SAMD9L sequence (XM_027389843.1 except for a difference at codon 618 (a T-to-C substitution) that did not result in an amino acid change. Plasmids for expressing NTPase-TPR domains of SAMD9&L were constructed similarly but with PCR primers that amplify only the specific region of SAMD9&L.
Generation of CHO cells with gene knockouts.
The plasmids used for the gene knockout were constructed from lentiCRISPRv2 (Addgene plasmid 52961) according to a published protocol (54). In brief, lentiCRISPRv2 was digested with BsmBI and ligated with a pair of oligonucleotides with the specific guide sequence. For each target gene, two guide sequences were designed with the web tool CRISPOR (55). They were as follows: 5′-AGTCATTTGTATTCTCTGGA-3′ (chSAMD9 1), 5′-CAAAAGAGGATGTGAATCTG-3′ (chSAMD9 2), 5′-CAATGAAGAAGTGACAGGGA-3′ (chSAMD9L 1), and 5′-ATCAGAAGTGCTGGACCCCG-3′ (chSAMD9L 2). The lentiCRISPRv2-derived plasmids and the packaging plasmids (pMD2.G and psPAX2) were transfected into HEK 293FT cells to produce lentiviruses, which were transduced into CHO cells as described previously (25). The transduced cells were subjected to puromycin (15 μg/ml) selection for 7 days. The SAMD9 or SAMD9L genotype of the cells was identified by sequencing as described previously (25). The cellular genomic DNA was extracted using the QIAamp DNA blood minikit (Qiagen), and then ∼500 bp of DNA flanking the target site was PCR amplified and cloned to the pGEM-T vector (Promega) and the sequences of 10 to 20 clones determined by Sanger sequencing. The primer pairs were 5′-AAGAGAGCTGGGGATAATGC-3′ and 5′-GATTTCTGCAGTTCCTTGAA-3′ for chSAMD9 and 5′-AAGTAATCATATGACTACATGTAA-3′ and 5′-GTGTTCTTTTATTGAGAGCT-3′ for chSAMD9L.
Generation of a cell line that expresses chSAMD9L under control of an inducible promoter.
chSAMD9L was cloned into pCW57.1 (Addgene 41393), an “all-in-one” doxycycline-inducible lentiviral vector with rtTA-VP16-2A-puro. In an effort to increase the expression level of the inducible gene, the lentiviral vector was modified by introducing F67S and R171K substitutions into rtTA (56) and by replacing the minimal promoter downstream of the Tet response element with the EF1 alpha promoter. The plasmid was used in making lentiviruses for transduction of BT20 cells as described above. Transduced cells were selected with puromycin at 3 μg/ml.
Luciferase assay.
Cells were infected with VACV WR that expressed a luciferase reporter under the control of either the late p11 promoter (57) or the synthetic early/late (S E/L) promoter (58) in the presence or absence of cytosine arabinoside (AraC). Cells were lysed with buffer and luciferase activity measured at 8 or 24 h postinfection (hpi) according to the manufacturer’s instructions (Promega).
Viral growth analysis.
Cells in 12-well plates were incubated with 1 PFU per cell of different viruses for 2 h at room temperature. Following adsorption, the cells were washed twice with phosphate-buffered saline (PBS). One set of the cells was harvested immediately as the 0-hpi sample, while others were moved to a 37°C incubator to initiate viral entry and harvested at different times postinfection. The viral titers in the cell lysates were determined by plaque assays on Vero cells.
Immunoprecipitation and Western blot analysis.
The binding of SAMD9&L with viral proteins was determined by coimmunoprecipitation as described before (25). In brief, 293FT cells were transfected with the SAMD9 or SAMD9L expression plasmid and then infected with different VACVs. The cells were lysed on ice with lysis buffer (0.1% [wt/vol] NP-40, 50 mM Tris [pH 7.4], 150 mM NaCl), and the cleared cell lysates were mixed with V5-agarose beads (Sigma-Aldrich) for 30 min at 4°C. After washing with lysis buffer, the beads were resuspended in SDS sample buffer, and the eluted proteins were resolved by SDS-PAGE and detected by Western blotting as described previously (48). The detection antibodies were mouse monoclonal antibodies (MAb) against V5 (Sigma-Aldrich; clone V5-10) and Flag tag (Sigma-Aldrich).
ACKNOWLEDGMENTS
This work was supported by a grant from the NIAID to Y. Xiang (AI079217).
We thank Nadia Gallardo-Romero for providing the OPXVs.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Moss B. 2013. Poxviridae, p 2129–2159. In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Damon IK. 2013. Poxviruses, p 2160–2184. In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Parker S, Nuara A, Buller RM, Schultz DA. 2007. Human monkeypox: an emerging zoonotic disease. Future Microbiol 2:17–34. doi: 10.2217/17460913.2.1.17. [DOI] [PubMed] [Google Scholar]
- 4.Durski KN, McCollum AM, Nakazawa Y, Petersen BW, Reynolds MG, Briand S, Djingarey MH, Olson V, Damon IK, Khalakdina A. 2018. Emergence of Monkeypox—West and Central Africa, 1970-2017. MMWR Morb Mortal Wkly Rep 67:306–310. doi: 10.15585/mmwr.mm6710a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Megid J, Borges IA, Abrahao JS, Trindade GS, Appolinario CM, Ribeiro MG, Allendorf SD, Antunes JM, Silva-Fernandes AT, Kroon EG. 2012. Vaccinia virus zoonotic infection, Sao Paulo State, Brazil. Emerg Infect Dis 18:189–191. doi: 10.3201/eid1801.110692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivetti AV Jr, Guedes MI, Rehfeld IS, Oliveira TM, Matos AC, Abrahao JS, Kroon EG, Lobato ZI. 2013. Bovine vaccinia, a systemic infection: evidence of fecal shedding, viremia and detection in lymphoid organs. Vet Microbiol 162:103–111. doi: 10.1016/j.vetmic.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Springer YP, Hsu CH, Werle ZR, Olson LE, Cooper MP, Castrodale LJ, Fowler N, McCollum AM, Goldsmith CS, Emerson GL, Wilkins K, Doty JB, Burgado J, Gao J, Patel N, Mauldin MR, Reynolds MG, Satheshkumar PS, Davidson W, Li Y, McLaughlin JB. 2017. Novel orthopoxvirus infection in an Alaska resident. Clin Infect Dis 64:1737–1741. doi: 10.1093/cid/cix219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vora NM, Li Y, Geleishvili M, Emerson GL, Khmaladze E, Maghlakelidze G, Navdarashvili A, Zakhashvili K, Kokhreidze M, Endeladze M, Mokverashvili G, Satheshkumar PS, Gallardo-Romero N, Goldsmith CS, Metcalfe MG, Damon I, Maes EF, Reynolds MG, Morgan J, Carroll DS. 2015. Human infection with a zoonotic orthopoxvirus in the country of Georgia. N Engl J Med 372:1223–1230. doi: 10.1056/NEJMoa1407647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crouch AC, Baxby D, McCracken CM, Gaskell RM, Bennett M. 1995. Serological evidence for the reservoir hosts of cowpox virus in British wildlife. Epidemiol Infect 115:185–191. doi: 10.1017/S0950268800058258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett M, Crouch AJ, Begon M, Duffy B, Feore S, Gaskell RM, Kelly DF, McCracken CM, Vicary L, Baxby D. 1997. Cowpox in British voles and mice. J Comp Pathol 116:35–44. doi: 10.1016/S0021-9975(97)80041-2. [DOI] [PubMed] [Google Scholar]
- 11.Khodakevich L, Szczeniowski M, Manbu Ma D, Jezek Z, Marennikova S, Nakano J, Messinger D. 1987. The role of squirrels in sustaining monkeypox virus transmission. Trop Geogr Med 39:115–122. [PubMed] [Google Scholar]
- 12.Oliveira JS, Figueiredo PO, Costa GB, Assis FL, Drumond BP, da Fonseca FG, Nogueira ML, Kroon EG, Trindade GS. 2017. Vaccinia virus natural infections in Brazil: the good, the bad, and the ugly. Viruses 9:E340. doi: 10.3390/v9110340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McFadden G. 2005. Poxvirus tropism. Nat Rev Microbiol 3:201–213. doi: 10.1038/nrmicro1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drillien R, Spehner D, Kirn A. 1978. Host range restriction of vaccinia virus in Chinese hamster ovary cells: relationship to shutoff of protein synthesis. J Virol 28:843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramsey-Ewing A, Moss B. 1995. Restriction of vaccinia virus replication in CHO cells occurs at the stage of viral intermediate protein synthesis. Virology 206:984–993. doi: 10.1006/viro.1995.1021. [DOI] [PubMed] [Google Scholar]
- 16.Spehner D, Gillard S, Drillien R, Kirn A. 1988. A cowpox virus gene required for multiplication in Chinese hamster ovary cells. J Virol 62:1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang SJ, Hsiao JC, Sonnberg S, Chiang CT, Yang MH, Tzou DL, Mercer AA, Chang W. 2009. Poxvirus host range protein CP77 contains an F-box-like domain that is necessary to suppress NF-kappaB activation by tumor necrosis factor alpha but is independent of its host range function. J Virol 83:4140–4152. doi: 10.1128/JVI.01835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramsey-Ewing AL, Moss B. 1996. Complementation of a vaccinia virus host-range K1L gene deletion by the nonhomologous CP77 gene. Virology 222:75–86. doi: 10.1006/viro.1996.0399. [DOI] [PubMed] [Google Scholar]
- 19.Perkus ME, Goebel SJ, Davis SW, Johnson GP, Limbach K, Norton EK, Paoletti E. 1990. Vaccinia virus host range genes. Virology 179:276–286. doi: 10.1016/0042-6822(90)90296-4. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Meng X, Xiang Y, Deng J. 2010. Structure function studies of vaccinia virus host range protein k1 reveal a novel functional surface for ankyrin repeat proteins. J Virol 84:3331–3338. doi: 10.1128/JVI.02332-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng X, Krumm B, Li Y, Deng J, Xiang Y. 2015. Structural basis for antagonizing a host restriction factor by C7 family of poxvirus host-range proteins. Proc Natl Acad Sci U S A 112:14858–14863. doi: 10.1073/pnas.1515354112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng X, Chao J, Xiang Y. 2008. Identification from diverse mammalian poxviruses of host-range regulatory genes functioning equivalently to vaccinia virus C7L. Virology 372:372–383. doi: 10.1016/j.virol.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Wennier S, Zhang L, McFadden G. 2011. M062 is a host range factor essential for myxoma virus pathogenesis and functions as an antagonist of host SAMD9 in human cells. J Virol 85:3270–3282. doi: 10.1128/JVI.02243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sivan G, Ormanoglu P, Buehler EC, Martin SE, Moss B. 2015. Identification of restriction factors by human genome-wide RNA interference screening of viral host range mutants exemplified by discovery of SAMD9 and WDR6 as inhibitors of the vaccinia virus K1L−C7L− mutant. mBio 6:e01122-15. doi: 10.1128/mBio.01122-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng X, Zhang F, Yan B, Si C, Honda H, Nagamachi A, Sun LZ, Xiang Y. 2018. A paralogous pair of mammalian host restriction factors form a critical host barrier against poxvirus infection. PLoS Pathog 14:e1006884. doi: 10.1371/journal.ppat.1006884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li CF, MacDonald JR, Wei RY, Ray J, Lau K, Kandel C, Koffman R, Bell S, Scherer SW, Alman BA. 2007. Human sterile alpha motif domain 9, a novel gene identified as down-regulated in aggressive fibromatosis, is absent in the mouse. BMC Genomics 8:92. doi: 10.1186/1471-2164-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Topaz O, Indelman M, Chefetz I, Geiger D, Metzker A, Altschuler Y, Choder M, Bercovich D, Uitto J, Bergman R, Richard G, Sprecher E. 2006. A deleterious mutation in SAMD9 causes normophosphatemic familial tumoral calcinosis. Am J Hum Genet 79:759–764. doi: 10.1086/508069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chefetz I, Ben Amitai D, Browning S, Skorecki K, Adir N, Thomas MG, Kogleck L, Topaz O, Indelman M, Uitto J, Richard G, Bradman N, Sprecher E. 2008. Normophosphatemic familial tumoral calcinosis is caused by deleterious mutations in SAMD9, encoding a TNF-alpha responsive protein. J Invest Dermatol 128:1423–1429. doi: 10.1038/sj.jid.5701203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagamachi A, Matsui H, Asou H, Ozaki Y, Aki D, Kanai A, Takubo K, Suda T, Nakamura T, Wolff L, Honda H, Inaba T. 2013. Haploinsufficiency of SAMD9L, an endosome fusion facilitator, causes myeloid malignancies in mice mimicking human diseases with monosomy 7. Cancer Cell 24:305–317. doi: 10.1016/j.ccr.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 30.Narumi S, Amano N, Ishii T, Katsumata N, Muroya K, Adachi M, Toyoshima K, Tanaka Y, Fukuzawa R, Miyako K, Kinjo S, Ohga S, Ihara K, Inoue H, Kinjo T, Hara T, Kohno M, Yamada S, Urano H, Kitagawa Y, Tsugawa K, Higa A, Miyawaki M, Okutani T, Kizaki Z, Hamada H, Kihara M, Shiga K, Yamaguchi T, Kenmochi M, Kitajima H, Fukami M, Shimizu A, Kudoh J, Shibata S, Okano H, Miyake N, Matsumoto N, Hasegawa T. 2016. SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7. Nat Genet 48:792–797. doi: 10.1038/ng.3569. [DOI] [PubMed] [Google Scholar]
- 31.Chen DH, Below JE, Shimamura A, Keel SB, Matsushita M, Wolff J, Sul Y, Bonkowski E, Castella M, Taniguchi T, Nickerson D, Papayannopoulou T, Bird TD, Raskind WH. 2016. Ataxia-pancytopenia syndrome is caused by missense mutations in SAMD9L. Am J Hum Genet 98:1146–1158. doi: 10.1016/j.ajhg.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tesi B, Davidsson J, Voss M, Rahikkala E, Holmes TD, Chiang SCC, Komulainen-Ebrahim J, Gorcenco S, Rundberg Nilsson A, Ripperger T, Kokkonen H, Bryder D, Fioretos T, Henter J-I, Möttönen M, Niinimäki R, Nilsson L, Pronk CJ, Puschmann A, Qian H, Uusimaa J, Moilanen J, Tedgård U, Cammenga J, Bryceson YT. 2017. Gain-of-function SAMD9L mutations cause a syndrome of cytopenia, immunodeficiency, MDS, and neurological symptoms. Blood 129:2266–2279. doi: 10.1182/blood-2016-10-743302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amano N, Narumi S, Hayashi M, Takagi M, Imai K, Nakamura T, Hachiya R, Sasaki G, Homma K, Ishii T, Hasegawa T. 2017. Genetic defects in pediatric-onset adrenal insufficiency in Japan. Eur J Endocrinol 177:187–194. doi: 10.1530/EJE-17-0027. [DOI] [PubMed] [Google Scholar]
- 34.Bluteau O, Sebert M, Leblanc T, Peffault de Latour R, Quentin S, Lainey E, Hernandez L, Dalle J-H, Sicre de Fontbrune F, Lengline E, Itzykson R, Clappier E, Boissel N, Vasquez N, Da Costa M, Masliah-Planchon J, Cuccuini W, Raimbault A, De Jaegere L, Adès L, Fenaux P, Maury S, Schmitt C, Muller M, Domenech C, Blin N, Bruno B, Pellier I, Hunault M, Blanche S, Petit A, Leverger G, Michel G, Bertrand Y, Baruchel A, Socié G, Soulier J. 2018. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood 131:717–732. doi: 10.1182/blood-2017-09-806489. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz JR, Ma J, Lamprecht T, Walsh M, Wang S, Bryant V, Song G, Wu G, Easton J, Kesserwan C, Nichols KE, Mullighan CG, Ribeiro RC, Klco JM. 2017. The genomic landscape of pediatric myelodysplastic syndromes. Nat Commun 8:1557. doi: 10.1038/s41467-017-01590-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F, Meng X, Townsend M, Satheshkumar P, Xiang Y. 2019. Identification of CP77 as the third orthopoxvirus SAMD9L inhibitor with a unique specificity for a rodent SAMD9L. bioRxiv 10.1101/551556. [DOI] [PMC free article] [PubMed]
- 37.Meng X, Embry A, Sochia D, Xiang Y. 2007. Vaccinia virus A6L encodes a virion core protein required for formation of mature virion. J Virol 81:1433–1443. doi: 10.1128/JVI.02206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mekhedov SL, Makarova KS, Koonin EV. 2017. The complex domain architecture of SAMD9 family proteins, predicted STAND-like NTPases, suggests new links to inflammation and apoptosis. Biol Direct 12:13. doi: 10.1186/s13062-017-0185-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nounamo B, Li Y, O’Byrne P, Kearney AM, Khan A, Liu J. 2017. An interaction domain in human SAMD9 is essential for myxoma virus host-range determinant M062 antagonism of host anti-viral function. Virology 503:94–102. doi: 10.1016/j.virol.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsiao JC, Chao CC, Young MJ, Chang YT, Cho EC, Chang W. 2006. A poxvirus host range protein, CP77, binds to a cellular protein, HMG20A, and regulates its dissociation from the vaccinia virus genome in CHO-K1 cells. J Virol 80:7714–7728. doi: 10.1128/JVI.00207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Carroll DS, Gardner SN, Walsh MC, Vitalis EA, Damon IK. 2007. On the origin of smallpox: correlating variola phylogenics with historical smallpox records. Proc Natl Acad Sci U S A 104:15787–15792. doi: 10.1073/pnas.0609268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parrish CR, Holmes EC, Morens DM, Park EC, Burke DS, Calisher CH, Laughlin CA, Saif LJ, Daszak P. 2008. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev 72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moss B. 2012. Poxvirus cell entry: how many proteins does it take? Viruses 4:688–707. doi: 10.3390/v4050688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng C, Haller SL, Rahman MM, McFadden G, Rothenburg S. 2016. Myxoma virus M156 is a specific inhibitor of rabbit PKR but contains a loss-of-function mutation in Australian virus isolates. Proc Natl Acad Sci U S A 113:3855–3860. doi: 10.1073/pnas.1515613113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elde NC, Child SJ, Geballe AP, Malik HS. 2009. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature 457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen W, Drillien R, Spehner D, Buller RM. 1992. Restricted replication of ectromelia virus in cell culture correlates with mutations in virus-encoded host range gene. Virology 187:433–442. doi: 10.1016/0042-6822(92)90445-U. [DOI] [PubMed] [Google Scholar]
- 47.Smithson C, Tang N, Sammons S, Frace M, Batra D, Li Y, Emerson GL, Carroll DS, Upton C. 2017. The genomes of three North American orthopoxviruses. Virus Genes 53:21–34. doi: 10.1007/s11262-016-1388-9. [DOI] [PubMed] [Google Scholar]
- 48.Meng X, Xiang Y. 2006. Vaccinia virus K1L protein supports viral replication in human and rabbit cells through a cell-type-specific set of its ankyrin repeat residues that are distinct from its binding site for ACAP2. Virology 353:220–233. doi: 10.1016/j.virol.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 49.Meng X, Jiang C, Arsenio J, Dick K, Cao J, Xiang Y. 2009. Vaccinia virus K1L and C7L inhibit antiviral activities induced by type I interferons. J Virol 83:10627–10636. doi: 10.1128/JVI.01260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emerson GL, Li Y, Frace MA, Olsen-Rasmussen MA, Khristova ML, Govil D, Sammons SA, Regnery RL, Karem KL, Damon IK, Carroll DS. 2009. The phylogenetics and ecology of the orthopoxviruses endemic to North America. PLoS One 4:e7666. doi: 10.1371/journal.pone.0007666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao J, Gigante C, Khmaladze E, Liu P, Tang S, Wilkins K, Zhao K, Davidson W, Nakazawa Y, Maghlakelidze G, Geleishvili M, Kokhreidze M, Carroll DS, Emerson G, Li Y. 2018. Genome sequences of Akhmeta virus, an early divergent Old World orthopoxvirus. Viruses 10:E252. doi: 10.3390/v10050252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Falkner FG, Moss B. 1990. Transient dominant selection of recombinant vaccinia viruses. J Virol 64:3108–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Earl PL, Moss B, Wyatt LS, Carroll MW. 1998. Generation of recombinant vaccinia viruses. Curr Protoc Mol Biol 43:16.17.1–16.17.19. [DOI] [PubMed] [Google Scholar]
- 54.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Concordet JP, Haeussler M. 2018. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Das AT, Tenenbaum L, Berkhout B. 2016. Tet-On systems for doxycycline-inducible gene expression. Curr Gene Ther 16:156–167. doi: 10.2174/1566523216666160524144041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bengali Z, Satheshkumar PS, Yang Z, Weisberg AS, Paran N, Moss B. 2011. Drosophila S2 cells are non-permissive for vaccinia virus DNA replication following entry via low pH-dependent endocytosis and early transcription. PLoS One 6:e17248. doi: 10.1371/journal.pone.0017248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Townsley AC, Weisberg AS, Wagenaar TR, Moss B. 2006. Vaccinia virus entry into cells via a low-pH-dependent endosomal pathway. J Virol 80:8899–8908. doi: 10.1128/JVI.01053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]