HIV-1 subtype A is the second most prevalent subtype worldwide, with a high prevalence in Eastern Africa and Eastern Europe. However, an increase of non-B infections, including subtype A infections, has been observed in Germany and other European countries. There has simultaneously been an increased flow of refugees into Europe and especially into Germany, raising the question of whether the surge in non-B infections resulted from this increased immigration or whether German transmission chains are mainly involved. This study is the first comprehensive subtype A study from a western European country analyzing in detail its phylogenetic origin, the impact of various transmission routes, and its current spread. The results and conclusions presented provide new and substantial insights for virologists, epidemiologists, and the general public health sector. In this regard, they should be useful to those authorities responsible for developing public health intervention strategies to combat the further spread of HIV/AIDS.
KEYWORDS: Germany, HIV-1, phylogeography, reproduction numbers, risk groups, spread, subtype A, transmission cluster
ABSTRACT
HIV-1 non-B infections have been increasing in Europe for several years. In Germany, subtype A belongs to the most abundant non-B subtypes showing an increasing prevalence of 8.3% among new infections in 2016. Here we trace the origin and examine the current spread of the German HIV-1 subtype A epidemic. Bayesian coalescence and birth-death analyses were performed with 180 German HIV-1 pol sequences and 528 related and publicly available sequences to reconstruct the population dynamics and fluctuations for each of the transmission groups. Our reconstructions indicate two distinct sources of the German subtype A epidemic, with an Eastern European and an Eastern African lineage both cocirculating in the country. A total of 13 German-origin clusters were identified; among these, 6 clusters showed recent activity. Introductions leading to further countrywide spread originated predominantly from Eastern Africa when introduced before 2005. Since 2005, however, spreading introductions have occurred exclusively within the Eastern European clade. Moreover, we observed changes in the main route of subtype A transmission. The beginning of the German epidemic (1985 to 1995) was dominated by heterosexual transmission of the Eastern African lineage. Since 2005, transmissions among German men who have sex with men (MSM) have been increasing and have been associated with the Eastern European lineage. Infections among people who inject drugs dominated between 1998 and 2005. Our findings on HIV-1 subtype A infections provide new insights into the spread of this virus and extend the understanding of the HIV epidemic in Germany.
IMPORTANCE HIV-1 subtype A is the second most prevalent subtype worldwide, with a high prevalence in Eastern Africa and Eastern Europe. However, an increase of non-B infections, including subtype A infections, has been observed in Germany and other European countries. There has simultaneously been an increased flow of refugees into Europe and especially into Germany, raising the question of whether the surge in non-B infections resulted from this increased immigration or whether German transmission chains are mainly involved. This study is the first comprehensive subtype A study from a western European country analyzing in detail its phylogenetic origin, the impact of various transmission routes, and its current spread. The results and conclusions presented provide new and substantial insights for virologists, epidemiologists, and the general public health sector. In this regard, they should be useful to those authorities responsible for developing public health intervention strategies to combat the further spread of HIV/AIDS.
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) epidemic remains a major public health burden. Approximately 36.9 million people worldwide and 2.2 million people in Europe and North America are estimated to have been living with HIV in 2017, 25% of whom were undiagnosed (1–3). During 2017, 1.8 million new HIV infections occurred globally, of which about 25,300 occurred in the European Union and European Economic Area (EU/EEA) (2). Around 3,200 new infections are reported each year in Germany (2–4), making the identification of transmission networks paramount for targeted public health interventions, particularly with regard to preexposure prophylaxis (PrEP) of persons at high risk (5, 6).
Multiple subtypes and subsubtypes (A1, A2, B to D, F1, F2, H, J, and K) and numerous recombinant forms of the pandemic HIV-1 group M circulate in different geographic areas of the world. Group M is estimated to have originated around 1920 in Kinshasa, capital city of the Democratic Republic of Congo (DRC), remaining in a preepidemic state until its exponential spread around 1960 (7). The differential spread of subtypes in Africa (and subsequently worldwide) appears to reflect founder effects and seems to be linked with the geographic distribution of subtypes in different geographic regions of the DRC (8). Subtype A—the second most abundant type in the northeastern city of Kinsangani (DRC)—appears to have spread from there to Eastern Africa after 1950, growing exponentially during the 1970s and now accounting for up to 68% of infections in distinct regions of neighboring Eastern African countries such as Kenya, Uganda, and Tanzania (8–11); it is referred to here as the Eastern African lineage. Although the HIV-1 epidemic in Western African countries (Cameroon and Senegal) is clearly more divergent, subtype A (or its recombinants) is also one of the most abundant subtypes in many of these countries (10). A large epidemic of an HIV-1 subtype A variant is also seen in Eastern Europe, particularly in the Former Soviet Union (FSU) area (12–15). This subepidemic strain, here termed the Eastern European lineage or AFormer Soviet Union (AFSU), is estimated to have arrived in the town of Odessa (Ukraine) around 1984 from the DRC, and since 1993, it has spread explosively among people who inject drugs (PWID) all over the Former Soviet Union (13). The geographic range of the Eastern European clade is increasing, and this lineage has now been found in several other European countries, including Great Britain, Poland, Italy, and Germany (14, 16–20). Analysis of HIV-1 sequences from recently infected individuals in Germany revealed HIV-1 subtype A, with an increasing frequency from 5.1% in 2013/2014 to 8.3% in 2016, to be the most abundant non-B subtype among newly infected individuals in Germany (19, 20).
To better understand the origins of the HIV-1 subtype A German epidemic and to monitor the dynamics of circulating HIV variants in near real time with respect to transmission groups, country of infection, and transmitted drug resistance mutation patterns, we applied coalescence, birth-death, and multitype birth-death models of genetic analysis to new sequence data acquired in Germany between 1993 and 2015. Finally, we investigated trends for transmitted drug resistance mutations of subtype A across clusters of German origin.
RESULTS
HIV-1 subtype A in Germany results from at least two independent introductions of different geographic origins.
To elucidate the sources and dissemination routes of endemic subtype A both geographically and within transmission groups, we analyzed viral sequences collected throughout the country between 1993 and 2015. To identify subtype A variants circulating endemically in Germany, we added the 10 most closely related sequences from the Los Alamos HIV Database (n = 528; redundant entries were removed) to the set of 180 German subtype A sequences, resulting in a total of 708 subtype A sequences, here termed data set A. Of these closely related sequences, 44.7% were sampled from Africa, 30.1% from Eastern Europe, 7.8% from Germany, and 6.3% from Southern Europe and Western Asia, including Cyprus and Turkey (Table 1).
TABLE 1.
Sampling regions of the reference sequencesa
| Parameter | All samples (n = 708) | References (n = 528) | RKI samples only (n = 180) |
|---|---|---|---|
| Sampling dates | 1986–2015 | 1986–2013 | 1993–2015 |
| Sampling region, no. of samples (%) | |||
| Germany | 221 (31.2) | 41 (7.8) | 180 |
| Eastern Europe | 159 (22.5) | 159 (30.1) | |
| Northern Europe | 19 (2.7) | 19 (3.6) | |
| Western Europe | 6 (0.8) | 6 (1.1) | |
| Southern Europe | 16 (2.3) | 16 (3) | |
| Eastern Africa (KE, RW, TZ, UG, and ZM) | 166 (23.4) | 166 (31.4) | |
| Western Africa (ET, GH, ML, MR, NG, SN, TG, and BF) | 26 (3.7) | 26 (4.9) | |
| Middle Africa (CM and GA) | 41 (5.8) | 41 (7.8) | |
| South Africa (ZA) | 3 (0.4) | 3 (0.6) | |
| Western Asia (including CY and TR) | 17 (2.4) | 17 (3.2) | |
| Other area of Asia | 26 (3.7) | 26 (4.9) | |
| North America | 5 (0.7) | 5 (0.9) | |
| Unknown | 2 (0.3) | 3 (0.6) |
Definition of regions according to the M49 Standard of the United Nations (https://unstats.un.org/unsd/methodology/m49). Countries are coded according to the 2-letter codes by the International Organization for Standardization (ISO): KE, Kenya; RW, Rwanda; TZ, Tanzania; UG, Uganda; ZM, Zambia; ET, Ethiopia; GH, Ghana; ML, Mali; MR, Mauritania; NG, Nigeria; SN, Senegal; TG, Togo; BF, Burkina Faso; CM, Cameroon; GA, Gabon; ZA, South Africa; CY, Cyprus; TR, Turkey. Numbers in parentheses represent the proportion of each region to the overall (second column) and reference data set (third column).
While the ancestor of subtype A is estimated to originate most likely from Middle Africa (Fig. 1, node a; geographic probability, 0.88), our phylogeographic analysis shows that subtype A infections in Germany can be roughly attributed to two independent lineages with different origins (Fig. 1). One of these lineages originated most likely in Eastern Africa (Fig. 1, green subclade, node b; location posterior support, 0.98). The other originated from Middle and Western Africa (Fig. 1, light blue subclade, node c; location posterior support, 0.97) and has spread rapidly in Eastern European countries, including the Russian Federation (branches colored in red). The main subtype A lineages diverged early, in or around 1948 (95% highest posterior density interval [HPD], 1940 to 1955), and we estimate the introduction of subtype A into Eastern Europe to have occurred around 1990 (95% HPD, 1984 to 1991 [Fig. 1, node j]). The first introductions in Germany that led to onward countrywide spread are attributed to the Eastern African lineage and were dated back to 1983 (95% HPD, 1979 to 1989). Subsequent introductions of the Eastern European lineage have occurred frequently since the mid-1990s (Fig. 1 and 2). To ensure that the inferred geographic tree is robust and not biased by the composition of the reference data set, we tested the influence of the origin sample sizes on the migration inference data. In a second data set (data set B), we randomly downsampled the number of sequences from the four most abundant regions (e.g., Eastern Africa and Eastern Europe), obtaining comparable frequencies (13% to 14%) for each region. The inferred geographic probability of the root for each region was comparable to the full data set A (data not shown). In a second analysis, we randomized the geographic origin for each sample across the tree, resulting in equal geographic probabilities for each possible sampling location (data not shown).
FIG 1.
Time-scaled phylogeographic analysis reveals at least two different origins of the German HIV-1A epidemic. German samples and their 10 closest related sequences found by BLAST search (data set A) were used to construct the MCC tree. (A) Sampling locations and the inferred origin of the most recent common ancestor (MRCA) computed by discrete asymmetric trait analysis are color-coded. The node size reflects posterior probabilities. Curly braces mark the Middle and Western African and Eastern European subclade as well as the Eastern African subclade. German-origin clusters are highlighted and numbered. *, active clades; the dotted vertical line indicates the threshold for active clades (at least two introductions per cluster after 2012). The data table describes the labeled nodes. 95% HPD, highest posterior density interval. (B) Time-scaled MCC tree with discrete risk group analysis reveals distinct clusters for most transmission routes. The estimated risk group of the MRCA is color-coded. Posterior probabilities of >0.95 are marked by circles at nodes according to their value. The inset depicts the number of clusters per mean edge distance. A distance of 20 years, a posterior probability of >0.99, and a geographic probability of >0.99 were used to identify transmission clusters (TC) with German origin (highlighted in yellow and red). Roman numerals label the 14 TC fulfilling all criteria. German-origin clusters not fulfilling the distance criteria are highlighted in blue. The three TC that are parts of a bigger German-origin cluster are highlighted in red. HET, heterosexual contacts; MSM, men having sex with men; PWID, people who inject drugs; HPL, high-prevalence country.
FIG 2.
Bayesian skygrid and BDSKY analysis reveal two spatiotemporal independent subepidemics. (A) Epidemiologically unlinked samples from the Eastern African (gray curve) and Eastern European subclade (red) were analyzed separately. The logarithmic effective number of infections (Ne) × viral generation time (t) representing effective transmissions is plotted over time. 95% HPD intervals are plotted in lighter colors. (B) Estimation of Re by BDSKY analysis of the two German subepidemics using only RKI samples. Gray, samples belonging to the Eastern African variant; red, samples belonging to the Middle and Western African/Eastern European variant. (C) Time of the most recent common ancestor (tMRCA) of the German-origin clusters. Clusters within the Eastern African clade are colored in gray. Clusters within the Eastern European subclade are colored in red. 95% HPDs are depicted as bars. Small shapes represent inactive clades without recent spread. Larger shapes represent active clades with at least two infections since 2012. The shape of the cluster icon depicts the transmission route within each cluster. Squares, MSM; triangles, HET; circles, mixed clades with PWID. (D) Proportion of risk groups in all German-origin clusters (left) compared to the still active clades (right).
Recent increase in the rates of epidemic spread of the HIV-1 subtype A Eastern European lineage circulating in Germany.
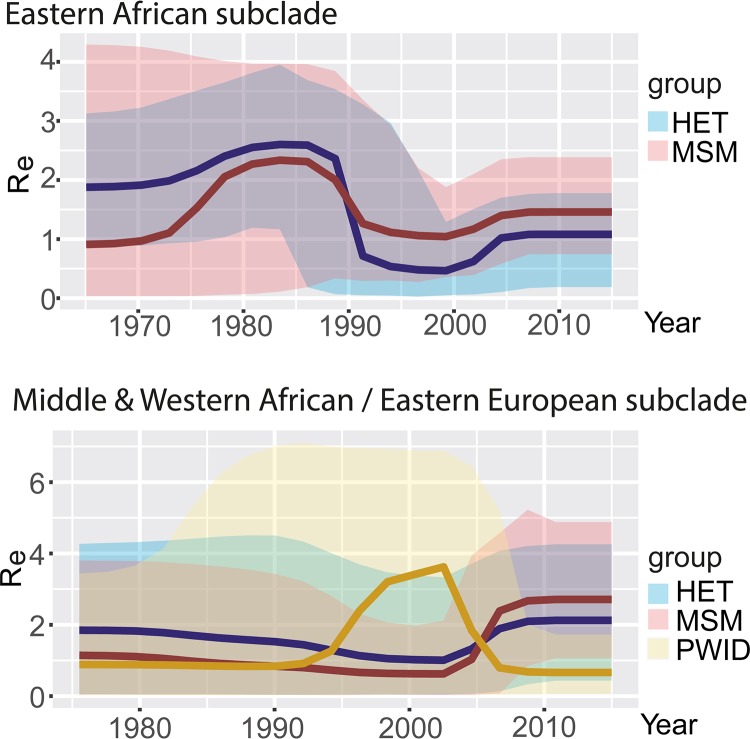
To analyze fluctuations in subtype A population dynamics in Germany, we estimated changes in the effective number of infections × generation time (Ne × t) using two data sets that included: (i) epidemiologically unlinked sequences from Eastern Africa (n = 249 [Fig. 2A, gray]) and (ii) epidemiologically unlinked samples of Middle and Western African/Eastern European origin (n = 321 [Fig. 2A, red). The reconstruction of the Eastern African dynamics (gray curve) shows an early exponential growth that reaches its plateau earlier than the Western and Middle African/Eastern European lineage (red curve). The latter shows a first phase of epidemic growth starting around 1972. While the epidemic from Eastern Africa reached a plateau in 1990, the Eastern European epidemic seems to have had a second phase of epidemic growth that started around the mid-2000s.
To estimate fluctuations in epidemic spread, we analyzed RKI sequences (see explanation in Materials and Methods; nEasternAfrica = 71; nEasternEurope = 109) using the birth-death skyline (BDSKY) model to estimate changes in the effective reproduction number (Re) over time (Fig. 2B). Re estimates the average number of secondary cases per infectious case in a population made up of both susceptible and nonsusceptible hosts. While Res for the epidemic originating in Eastern Africa are highest in the late 1980s (Re = 2.3, median) and in a second peak between 2003 and 2006 (Re = 1.6), the median estimate has been below 1 since 2008. The epidemic caused by the Eastern European lineage seems to have been slow until the mid-1990s (median Re < 1). However, it then reached a plateau at an Re of 4.5 between 1998 and 2002 before decreasing to below 1 by 2012. Notably, for the periods since 2012 we observed a strong increase of Re values (Re = 4.9) for the Eastern European lineage. Such a pattern is in line with the increase in growth rates obtained by the coalescence analysis of unlinked RKI sequences (Fig. 2A). Together, these data underline the past contribution of the Eastern African lineage to the early German epidemic, and most importantly, the increasing contribution of the Eastern European variant in recent times.
Identification and characterization of German-origin clusters and putative transmission clusters.
To characterize German clusters establishing the German epidemic in depth, we performed a clustering analysis based on a Bayesian maximum clade credibility (MCC) time tree and an equivalent maximum likelihood (ML) tree.
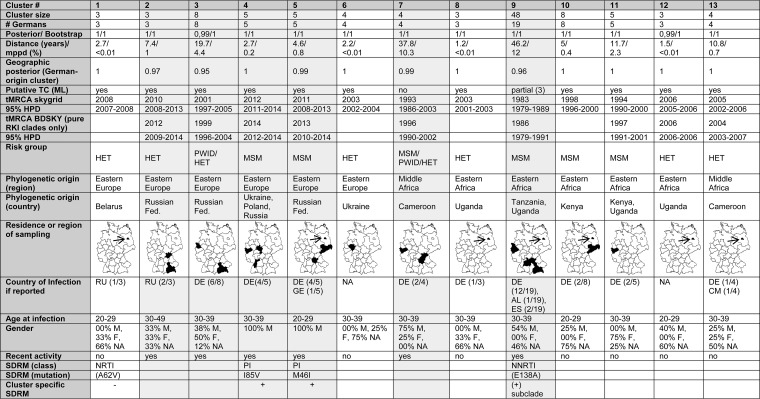
Analysis of the MCC tree revealed 13 German-origin clusters (mean size, n = 4) establishing an endemic spread in Germany. These clusters are characterized by a posterior value of 0.99 and a most recent common ancestor (MRCA) estimated to have existed in Germany (Fig. 1A and Table 2). We further analyzed whether these clusters are additionally putative transmission clusters (Fig. 1B). Using a distance cutoff of 20 years, which resembles most closely the mean pairwise patristic distance (MPPD) threshold of 4.5 in the ML tree, we found 260 sequences (36.7%) in 85 clusters, including 90 RKI sequences. Seventy of 180 RKI sequences were found in 14 clusters harboring more than 3 German sequences (Fig. 1B, highlighted in yellow and red). Of those, three clusters (namely, clusters VIII, IX, and X, highlighted red) formed a well-supported larger clade (posterior probability = 1) comprising 48 sequences, with an MRCA estimated to have existed in Germany (cluster 9* in Fig. 1; location posterior probability = 0.96). This means that 11/13 German-origin clusters fulfilled all criteria for putative transmission clusters (clusters 1 to 6, 8, and 10 to 13, highlighted in yellow in Fig. 1B). Only cluster 7 (and the combined cluster 9*, both marked in blue in Fig. 1B), failed to fulfill the distance criteria of being putative transmission clusters where individuals are linked by extremely short transmission chains. However, the sequences within these clusters are likely to be genetically linked (i.e., via longer transmission chains) and were therefore considered further in the analyses of German-origin clusters, since the main subsequent focus is on spatial spread and not on the dynamics of direct transmission.
TABLE 2.
Characterization of German subtype A clustersa
#Germans, number of German sequences within a cluster. Posterior/bootstrap, the calculated posterior and bootstrap values of each German-origin cluster estimated in the MCC/ML tree. Distance, the genetic distance within a German-origin cluster either in the MCC tree (in years) or the ML tree (percent). Geographic posterior, estimated support that a cluster is of German origin. Putative TC, whether a cluster fulfills all criteria of being a putative transmission cluster (TC). tMRCA, time of the most recent common ancestor; estimated year of introduction according either to Bayesian skygrid or to BDSKY analysis. 95% HPD, 95% highest posterior density interval. Risk group, mode of transmission within a German-origin cluster. Phylogenetic origin: country of sampling of coclustering sequences. Residence or region of sampling, area of living of the patient if known. Due to its small size, the area of Berlin is indicated by an arrow. Country of infection according to the reports: RU, Russian Federation; DE, Germany; GE, Georgia; AL, Albania; ES, Spain; CM, Cameroon. Note that the country of infection was not reported for all patients. M, male; F, female; NA, no information available. Recent activity: yes, at least 2 new infections since 2012. Recently active clusters are highlighted in gray. ML tree, maximum likelihood tree. SDRM, surveillance drug resistance mutation, NRTI, nucleoside reverse transcriptase inhibitors; NNRTI, nonnucleoside reverse transcriptase inhibitors; PI, protease inhibitors. The maps were constructed using the software RegioGraph Analyse.
The clustering profile inferred using ML was highly comparable to that obtained using Bayesian phylogenetics (https://zenodo.org/record/2245808#.XKeeWtgpCpo). In the ML analysis, we found 433 sequences (61.2%) forming 88 clusters; these clusters included 117/180 (65%) of the RKI sequences. Of these, 74 sequences were found in 14 German-origin clusters (the same as in Fig. 1B and Table 2), of which 3 are again parts of the larger cluster 9*. Similar to the MCC tree analysis, cluster 7 as well as the combined cluster 9* did not fulfill the distance criteria for a putative transmission cluster in the ML analysis but were further considered as German-origin clusters.
Using molecular clock models, we next estimated the date of introduction of German-origin clusters. We found that German-origin clusters within the Eastern African clade tend to have been established for longer in Germany (Fig. 2C, colored in gray). In contrast, Middle and Western African/Eastern European lineage introductions (Fig. 2C, colored in red) tend to be more recent. Indeed, we detected four post-2008 introductions with subsequent spread in Germany within this subclade (Fig. 2C).
With one exception (cluster 9*), German clusters are well structured and do not include sequences from other countries (Table 2). Six clusters are still active, exhibiting ongoing infection events in Germany (at least two infections since 2012 [Table 2]). Four of these six active clades are of Eastern European origin and were established after 2005 (Table 2).
Clusters consisted mainly of sequences from men (74%), 24% were sequences from women, and for 2% no information on sex was available. Out of 13 German-origin clusters, 6 (46.2%) are characterized by heterosexual transmission (HET; marked blue in Fig. 1B and 2D), 5 (38.5%) are transmitted by men who have sex with men (MSM; marked red in Fig. 1B and 2D), and 2 (15.4%) are mixed clusters with sequences from people who inject drugs (PWID) coclustering with those from HET and/or MSM (green).
A comparison of the characteristics of the active clusters with all clusters revealed that heterosexual transmission plays a less pronounced role in the spread of subtype A than it did previously (Fig. 2D, right chart). Only 16.7% (−29.5%) of the active clusters are characterized by heterosexual transmission, whereas 50% of the active clusters are composed of sequences from MSM (+11.5%). Both clusters linked to PWID transmission still make up 33.3% of the active clades (+18%). Of note, the two most recently formed clusters are both MSM clusters with Eastern European origin (Fig. 2C).
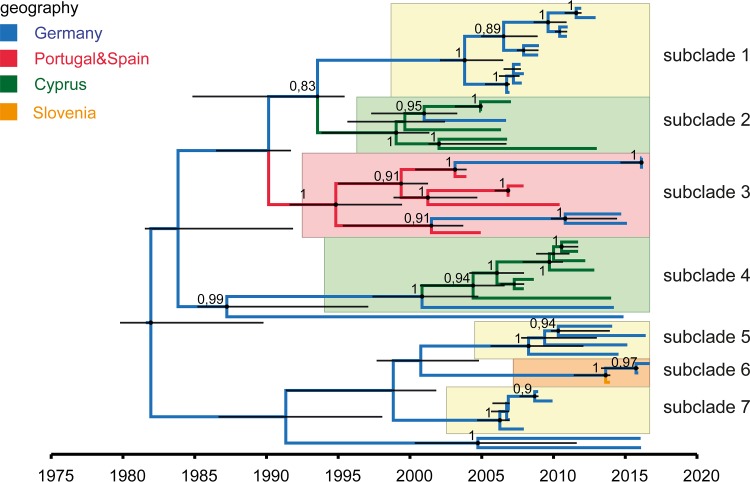
The large MSM cluster formed by 48 sequences of the Eastern African lineage (cluster 9*) has its origin around 1983 and seems to be related to Tanzania and Uganda. A detailed analysis is shown in Fig. 3. It consists of various distinct subclades: three exclusively German subclades (subclades 1, 5, and 7, highlighted in yellow), two clusters formed by Cyprian sequences (highlighted in green), one cluster consisting of sequences from Portugal, Spain, and Germany (highlighted in red), and one small clade containing sequences from Germany and Slovenia (orange). Of these 7 subclades, only two (subclades 5 and 6) suggest recent activity in Germany.
FIG 3.
MCC tree of cluster 9* within the Eastern African clade reveals seven subclades. The estimated geographic locations of the MRCA are color-coded. The various subclades are highlighted. Yellow, German clusters; green, Cyprian clades; red, cluster with Spanish, Portuguese, and German sequences; orange, mixed cluster with German and Slovenian sequences. Posterior probabilities of >0.8 are depicted at nodes. Node bars (black) indicate 95% HPDs.
Impact of transmission routes on spread.
With the exception of the two mixed PWID clades, each German cluster is characterized by a specific route of transmission (Table 2). To calculate the impact of each mode of transmission (MSM, HET, and PWID) on the spread of the epidemic, we estimated the Re for each transmission group in both major subclades using a multitype tree birth-death analysis applied to the RKI sequences (21). The effective reproduction numbers (Re) were estimated over four equidistant time intervals depending on the size of the overall tree height at each Markov chain Monte Carlo (MCMC) step (Fig. 4).
FIG 4.
Multitype birth-death (BDMM) analysis according to transmission routes and virus origin reveals a change in the impact of transmission routes over time. Changes in the Res for each transmission group in each subclade are depicted. (Upper graph) RKI sequences clustering in the Eastern African clade. (Lower graph) Res according to risk groups for RKI sequences from the Middle and Western African/Eastern European clade.
This analysis shows the changing impact of each transmission group during the German epidemic. We found that the early epidemic (1980 to 1990) was mainly driven by HET (both clades) and MSM within the Eastern African clade, strongly supporting the molecular clock discrete trait analysis of the German clusters. The BDSKY peak between 1998 and 2005 can be explained by a strong increase in the number of infections with the Middle and Western African/Eastern European virus variant among PWID (Re = 3.6). In contrast, transmissions by heterosexual contact did not play a major role in the ongoing epidemic during recent years, while infections among MSM still contribute considerably to the epidemic even though the Eastern European strain spreads much more rapidly than the Eastern African. The highest recent estimated Re value was found for the group of MSM infected with the Eastern European subtype A variant (Re = 2.9), followed by HET (Re = 2.3), whereas PWID play only a minor role (Re = 0.8).
High frequency of transmitted drug resistance mutations in German subtype A-infected individuals.
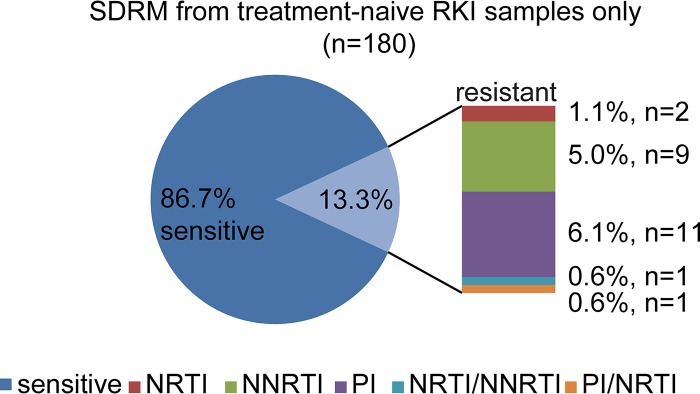
The frequency of surveillance drug resistance mutations (SDRM) in German subtype A infected individuals was 13.3% (n = 24/180) and even higher (18.9% [n = 34]) when considering the E138A RT resistance mutation (Fig. 5). Nucleoside reverse transcriptase (RT) inhibitor (NRTI) SDRM did not have a high impact on transmitted drug resistance (1.1% [n = 2]), whereas the proportions of nonnucleoside reverse transcriptase inhibitor (NNRTI) and protease (PR) inhibitor (PI) SDRM were higher, at 5% (n = 9; including E138A, 10.6% [n = 19]) and 6.1% (n = 11), respectively. Analyzing each resistance-associated position separately, the most frequent NNRTI SDRM were K103N, at 2.8% (n = 5), and Y181CIV and G190ASE, at 1.7% (n = 3) each. E138A occurred in 5.6% (n = 10) of the RKI sequences. NNRTI resistance mutations did not emerge in clusters, with one exception: a subcluster of 10 German sequences within the MSM48 clade that is no longer active (Fig. 3, subclade 1) is characterized by the E138A mutation in all infected individuals.
FIG 5.
Level of drug resistance mutations in RKI samples: transmitted surveillance drug resistance mutations (SDRM) in therapy-naive RKI samples.
Resistance mutations against protease inhibitors have also been observed. This mutation class characterizes 2.1% (n = 15) of all sequences in data set A and 6.1% (n = 11) of the RKI sequences. The most common PI SDRM in the RKI data set were M46IL (3.9% [n = 7]) and I85V (2.8% [n = 5]). Both PI SDRM occurred (with two exceptions) exclusively in the two most recent MSM clusters (namely, clusters 4* and 5*, established around 2013), accounting for 17.9% of PI mutations within clustered German sequences. This means that 3 of the 13 German-origin clusters and 3 of the 6 active clades are characterized by transmission of drug resistance mutations (MCC tree showing SDRM at https://zenodo.org/record/2245808#.XKeeWtgpCpo). However, only the M46I and I85V PI SDRM show a constant spread at this time, particularly among German MSM.
DISCUSSION
For decades, subtype B has been the dominating HIV-1 subtype in many European countries. However, in recent years the incidence and prevalence of other subtypes have been increasing in Western and Southern European countries, such as Great Britain, France, Italy, and Germany (2, 14, 16, 18–20, 22–25). Hence, non-B infections are becoming a more serious public health issue, which necessitates a deeper understanding of the non-B epidemics in these countries. Among HIV-1 non-B subtypes in Germany, subtype A is one of the most abundant subtypes showing increasing prevalence rates (19, 20, 26).
Analysis of 180 PR/RT sequences from HIV-1 subtype A infected individuals sampled between 1993 and 2015 revealed that subtype A has established an endemic spread in Germany which can be traced back to multiple introductions with at least two independent lineages from different geographic origins (summarized in Fig. 6).
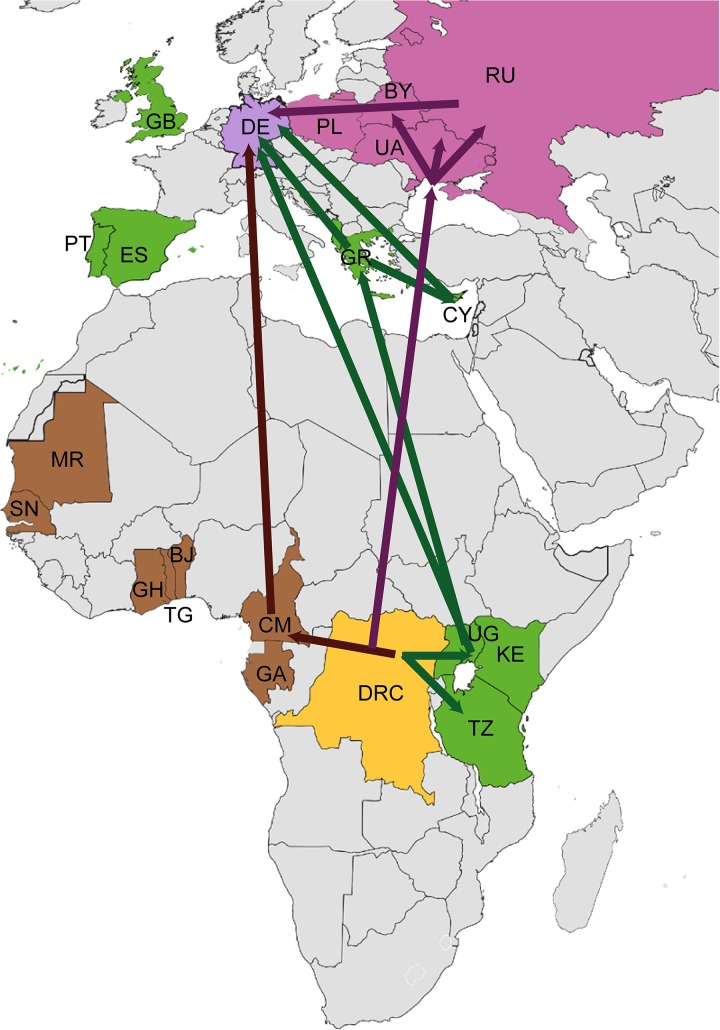
FIG 6.
Scheme of the geographic spread of HIV-1 subtype A to Germany. The subtype A origin lies in the region of today’s Democratic Republic of Congo (DRC; yellow). Subsequent spread occurred in two spatiotemporal independent routes. The first was to Eastern African countries (colored in green), such as Tanzania (TZ), Kenya (KE), and Uganda (UG), as marked by green arrows. From here spread to Germany occurred directly or indirectly (i.e., via Greece [GR] and Cyprus [CY] and GR and Albania [AL]). Other countries, such as Great Britain (GB), Portugal (PT), and Spain (ES), might also be involved but to a lesser extent. The second subtype A lineage spread from Middle and Western Africa directly (brown arrow) or indirectly via countries of the former Soviet Union (purple arrows), such as Ukraine (UA), the Russian Federation (RU), Belarus (BY), and, to a lesser extent, Poland (PL). The map was colored and modified with Adobe Illustrator and Photoshop according to our findings and published analyses by other groups (7, 8, 13, 14, 35, 74, 75) using a blank map from Wikipedia Commons (public domain).
Until now, at least 13 independent clusters could be identified which likely have a most recent common ancestor (MRCA) being infected in Germany and which show—with the exception of the MSM48 cluster 9*—an exclusively German spread. The existence of several independent clades of German origin is an indication of separate transmission networks, originating from different introductions of the virus at different times within the country. From these 13 German clades, six still show ongoing transmissions (1 HET, 2 mixed PWID, and 3 MSM clades). We observed a change between the origins of viruses entering Germany over time. Early introductions could be traced back exclusively to Eastern African variants, while later introductions originated mainly from Eastern Europe, pointing to an impact of Eastern European infections on the current HIV epidemic in Germany.
However, viruses with Eastern African origin are still circulating in Germany and are actively involved in the present epidemic. Most of the 10 closest sequences found by BLAST search of the Los Alamos database originate mainly from Uganda (n = 97), Kenya (n = 40), and Rwanda (n = 18). In Uganda, for example, subtype A is the second most abundant subtype after subtype D, with a prevalence of 25% (8, 27–29). In line with the findings of Gray and coworkers, our results from both skygrid and BDSKY analyses show an exponential growth of the subtype A epidemic, starting approximately in 1951, accelerating around 1975, and reaching a plateau in 1990 (8). According to our phylodynamic analysis, subtype A might have reached Germany in the 1980s, giving rise to the early German epidemic. However, most introductions leading to a subsequent endemic spread occurred between 1990 and 2002. Many of these are probably the result of direct migration from Eastern African countries. However, in the case of the large MSM48 clade (cluster 9*), we also observed an intra-European spread involving Cyprus, Albania, Greece, Portugal, and Spain. Subtype A spreading from Cyprus, Albania, or Greece has also been described for Italy (18) and is also most likely linked with the Spanish and Portuguese epidemic.
The epidemic described by the Western and Middle African/Eastern European clade also started in the Democratic Republic of Congo but shows a very different dynamic regarding its spread (Fig. 6, brown) (13). Most of the African sequences coclustering in this clade have been sampled in Cameroon. In addition to recombinant forms such as CRF02_AG, subtype A is the predominant subtype in this region (10, 30). Little is known about the spread of this variant across the African continent. Consistent with our Bayesian skygrid analysis, Diez-Fuertes et al. dated dispersion of this variant to 1970, finally arriving in the Ukrainian town of Odessa around 1984 (13). Odessa has emerged as the epicenter and gateway of the Eastern European HIV epidemic that started its explosive expansion among PWID in 1993. This epidemic is hallmarked by a specific subtype A variant (AFormer Soviet Union [AFSU]) which is characterized by a very low genetic diversity (12, 13, 31, 32). After the fall of the Iron Curtain, AFSU spread among PWID within the Ukraine but was also transmitted to Russia and other countries of the former Soviet Union (12, 33–44). As one of the fastest-growing epidemics in the world, the Eastern European epidemic has a big influence on the current epidemic in Europe (12). According to the European Surveillance System (TESSy [2]) and Beloukas et al. (14), 77% of all new diagnoses are made in Eastern Europe/Russia, compared to only 19.2% reported in Western Europe and 3.5% in Central Europe. This translates to a seven-times-higher incidence rate in Eastern Europe (43.2/100,000 people) than in Western Europe (6.4/100,000 people) (14). Similar to the Eastern European epidemic, the second peak of the German HIV-1 subtype A epidemic started in the mid-1990s among PWID but shows another strong increase after 2005. Interestingly, the initial spread of the subtype AFSU variant among PWID from Eastern Europe to Germany occurred closely after the fall of the Iron Curtain. However, according to the Re values we estimated for the various transmission groups, the recent reincrease is linked to MSM rather than to PWID or HET. Analysis of the most recent and fastest-growing clusters showed that these MSM are—in contrast to many HETs within the Middle and Western African/Eastern European clade—nearly exclusively of German origin and are not linked to Eastern Europe either by the known country of infection or by nationality. However, although PWID were estimated to contribute least to the current spread, this group may have made a major contribution to the transmission across risk groups. Two of the six German clades with ongoing transmission are mixed PWID clusters. This indicates that cross-risk group transmission via PWID is still relevant for subtype A and should remain a high-priority target for public health interventions.
Interestingly, alteration of the dominating risk group is not restricted to Germany and has also been observed in other Western European countries. Ragonnet-Cronin and colleagues reported a 3-fold increase in non-B infections among MSM between 2002 and 2010 in Great Britain (16). Furthermore, various groups reported similar observations for France, where the non-B strains (most likely transmitted by immigrants from sub-Saharan Africa) rose from 4% in the 1980s to 20% in just 1 decade, spreading mostly in MSM transmission networks of French origin (24, 25, 45, 46). Moreover, one-third of newly acquired HIV-1 infections in Europe occur among immigrants (including foreign-born individuals), while two-thirds are among natives (14). The predominant route is sex between men, with a considerable increase in the proportion of transmissions among MSM: 30% in 2005 to 42% in 2014 (2, 14). However, in Eastern European countries, heterosexual intercourse is still considered to be the main reason for the increased rate of infections, and transmission through PWID networks also remains high (14, 47, 48). Still, the recent relative increase in HIV subtype A infection in the group of MSM and their more frequent assignment to active clusters emphasize the fact that the spread of subtype A in Germany is also becoming increasingly associated with MSM and that subtype A is therefore the second subtype (beside subtype B) that is becoming dominated by this risk group.
As all RKI sequences were obtained from treatment-naive patients, this allows us to get a nationwide view on SDRM in German subtype A. The analysis of these sequences revealed a higher proportion of SDRM (13.3% and 18.9% with E138A) than the nationwide level of 11% for all subtypes (19, 20). Resistance mutations against protease inhibitors as well as the E138A NNRTI mutation occur exclusively in clusters. Three out of thirteen clusters show a transmitted drug resistance mutation in all infected individuals. This is especially important for the current epidemic, as all three affected clades show ongoing activity. Furthermore, this is the most likely explanation for the higher frequency of transmitted drug resistance mutations in German subtype A. Strikingly, 6.1% of all RKI sequences harbored nonpolymorphic PI resistance mutations: M46I and I85V. These mutations were allocated to the two most recently formed German MSM clades with Eastern European origin. Both mutations might contribute to therapy failure since they confer resistance against the EACS-recommended (49) protease inhibitor-based first-line antiretroviral therapy (ART) regimen.
All analyses were performed with both birth-death and coalescence models, fortifying the accuracy of both phylogenetic estimation methods. Additionally, the use of phylogenetic birth-death modeling allowed us to estimate reproduction numbers from viral sequences separately for certain risk groups. Furthermore, we were able to extend transmission cluster detection from ML to Bayesian consensus trees, providing both a concept and a tool for comparable transmission cluster definition within both types of trees. There are some limitations to this study. Due to the relatively small sample size of sequences with German origin (especially before 2012), we observed an uncertainty of the Re and Ne estimates as indicated by the large error bars in Fig. 2 and 4, which warrants further confirmation. Moreover, the sampled German sequences strongly influence the reference data set. Missing samples in the German epidemic would therefore lead to a putative bias, particularly in the trait analyses concerned with geographic locations and transmission groups. Therefore, the dynamics of introduction might appear different or possible intra-European spread might become apparent if all German sequences had been sampled and a less biased reference data set had been used. Nevertheless, the results we obtained are both robust in several sensitivity analyses and in strong agreement with those of others using completely different data sets (discussed above [8, 18]).
In this study, we have shown that HIV-1 subtype A has been spreading in the German population for nearly 3 decades, with a current rise in incidence. While the early epidemic was established by a virus strain originating from Eastern Africa among heterosexuals and MSM, the current epidemic was established mainly among MSM and partly among PWID by the AFSU variant originating in Eastern Europe.
MATERIALS AND METHODS
Genetic data set compilation.
HIV-1 pol sequences covering the genomic regions of protease (amino acids [aa] 9 to 99) and reverse transcriptase (aa 1 to 252) were derived from (i) newly diagnosed individuals in Germany within the former diagnostic unit of the German AIDS Centre (until 1996), (ii) the German HIV-1 seroconverter cohort (1996 to 2015; sampling density, 3%) (50), and (iii) recently infected HIV newly diagnosed individuals within the German molecular surveillance (2012 to 2015; sampling density, 13%) (19, 51), here referred to as RKI sequences. Sampling densities for subtype A were roughly estimated by the estimated numbers of new infections per year according to the reports, the proportion of obtained sequences from those newly infected individuals, and the proportion of subtype A on the overall German epidemic. Sampling densities were later confirmed and partly corrected by BEAST v.2.4 analysis (see “Estimation of fluctuations in the Re” below). A preanalysis using the tool TempEst was used to ensure that the temporal signal was sufficient for subsequent molecular clock analysis and to identify sequences whose sampling dates and genetic divergence are incongruent (52). Furthermore, only sequences of pure subtype A lacking evidence of recombination according to the REGA subtyping tool v.3.0 were used for this study (53). Of 214 sequences, 180 were found to be suitable for subsequent analyses: 10 sequences from AIDS Centre diagnostic samples, 75 sequences from seroconverters, and 95 sequences from recently infected individuals (2013 to 2015). For 127 RKI sequences, additional epidemiological metadata, such as transmission route, nationality, country of infection, age, and sampling area, were available and used as trait data (Table 3).
TABLE 3.
Epidemiological data of RKI samples according to the reportsa
| Characteristics of RKI samples (n = 180) | No. (%) of samples |
|---|---|
| Risk factor | |
| Men having sex with men | 53 (29.4) |
| Heterosexuals | 54 (30) |
| People who inject drugs | 14 (7.8) |
| High-prevalence country | 8 (4.4) |
| Not reported | 51 (28.3) |
| Sex | |
| Male | 107 (59.4) |
| Female | 60 (33.3) |
| Not reported | 13 (7.2) |
| Presumed region of infection | |
| Germany | 73 (40.6) |
| Eastern Europe | 25 (13.8) |
| Africa | 18 (10) |
| Western Europe | 1 (0.6) |
| Southern Europe | 5 (2.8) |
| Asia | 5 (2.8) |
| Not reported | 53 (29.4) |
| Region of origin | |
| Germany | 73 (40.6) |
| Eastern Europe | 38 (21.1) |
| Africa | 15 (8.3) |
| Northern Europe | 1 (0.6) |
| Western Europe | 1 (0.6) |
| Southern Europe | 7 (3.9) |
| Asia | 4 (2.2) |
| Not reported | 41 (22.8) |
All data are as provided by the statutory HIV reports of new HIV diagnoses in Germany or by the HIV-1 Seroconverter study (72).
BLAST search of the Los Alamos HIV database was used to find the 10 sequences most closely related to each of the RKI sequences according to former publications (54). Duplicates and sequences shorter than 950 nucleotides (nt) were removed from the analysis. In total, the 180 subtype A pol sequences were aligned with 528 closely related subtype A sequences from the Los Alamos HIV database, resulting in 708 sequences, referred to as data set A. Sequences of German origin made up 31.6% of the entire data set (Table 1). To avoid biases due to convergent evolution introduced by drug resistance selection and deselection rather than genealogical similarity, a data set was constructed by stripping all known surveillance drug resistance mutations (SDRM) according to the methods of Bennett et al. (55) and Hofstra et al. (56), yielding a final multiple sequence alignment of 885 nt in length. To ensure that genetic distances do not differ but are comparable between the RKI sequences and the reference data set, we analyzed the frequency of the genetic distances (data not shown). The root-to-tip divergence within the final data set was additionally confirmed by TempEst (data not shown).
We adapted the size of the data set according to the requirements of the particular analyses (see below).
Maximum likelihood (ML) phylogenies were reconstructed using the Ultrafast Bootstrap approximation in IQ-TREE with 10,000 replicates (57, 58). Model selection prior to tree construction (implemented in IQ-TREE) identified the transversion nucleotide substitution model TVM+I+G as most suitable for tree construction (57, 58). The resulting ML tree was used both to confirm the topology of the Bayesian MCC tree and to verify cluster selection from the Bayesian trees based on bootstrap values and mean pairwise patristic distances [see “Identification of clusters of closely related sequences (putative transmission clusters and German-origin clusters)”]. As an outgroup for the ML tree reconstruction, we used 10 subtype C RKI sequences (GenBank accession numbers MK250696 to MK250705).
Reconstruction of time-calibrated trees.
For coalescence-based Bayesian phylogenetic analysis, a model selection implemented in BEAST v. 1.8.3 using path sampling and stepping stone was carried out for various coalescent and clock model combinations (data not shown) (59, 60). An exponential growth model and skygrid population dynamics were the best fit as coalescence models in combination with the lognormal molecular clock model (59, 61, 62). To calibrate the trees to time scale, we used the sampling dates of the tips (59). A maximum likelihood tree run by FastTree (63) was used as a starting tree. Each analysis comprised two independent Markov chain Monte Carlo (MCMC) runs of one billion generations performed with BEAST v.1.8.3 using the BEAGLE library to enhance computational speed (64). Tracer v.1.6.1 was used to inspect the convergence of each MCMC run (65), and LogCombiner (part of the BEAST package) was used to combine both runs, resulting in effective sample sizes (ESS) of >200 for all parameters. An empirical posterior tree distribution of 2,500 time-calibrated trees was generated and used for subsequent trait diffusion analyses.
To prevent biases due to variability in the density of sequence sampling, additional Bayesian skygrid analyses (61) were conducted to reconstruct the course of the German subtype A epidemic. All epidemiologically linked sequences that were identified by a transmission cluster analysis [see “Identification of clusters of closely related sequences (putative transmission clusters and German-origin clusters)”] were removed, and only one representative sequence per cluster was kept. Reduced data sets comprising (i) sequence data belonging to the Eastern African clade (n = 249) and (ii) sequence data belonging to the Middle and Western African/Eastern European clade (n = 326) were created. Each analysis was run in triplicates for one billion MCMC generations, and convergence was confirmed using Tracer v.1.6 (65).
Trait diffusion analysis.
To reconstruct the ancestral geographic locations and transmission groups throughout the evolutionary history of subtype A, we performed discrete trait diffusion analysis using symmetric and asymmetric diffusion models on an empirical tree consisting of 2500 subtrees generated by MCMC as described above (7, 54, 66, 67). As discrete traits, we considered geographic regions of sampling (for all sequences, see Table 1, “Sampling regions”), transmission groups (for RKI sequences only; see Table 3, “Risk”), and, in a separate analysis, transmitted SDRM (described below). Analyses were run for 200 million steps, and convergence was inspected using Tracer v.1.6. TreeAnnotator (part of the BEAST package) was used to generate maximum clade credibility (MCC) trees. FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) was used for visualization and annotation of the trees.
To ensure that the inferred tree and, in particular, its phylogeographic estimations were not affected by the composition of the reference data set, we additionally tested the influence of the origin sample sizes on the migration inference data. In accordance with former publications, we downsampled sequences from the most densely sampled locations (Eastern Africa and Eastern Europe, for which 170 and 159 sequences were available, respectively) in data set A, resulting in data set B, in which the numbers of sequences from the four most abundant locations (Eastern Africa, Eastern Europe, Germany, and Middle Africa) were comparable (n = 39 to 42) (7). Downsampling was conducted to avoid potential bias in spatial inference estimates that may be caused by oversampling a particular location (7, 68).
To further rule out that our estimations of the location of the HIV-1 subtype A ancestor were affected by differences in the number of samples per location, we randomized tip-to-location assignments during the Bayesian inference of data set A (7). This procedure results in posterior probabilities for each location being the location of the subtype A common ancestor that are approximately equal (data not shown). This further confirms that the source location that we inferred for data set A arose from the association between phylogenetic clustering and sample location and not from the relative frequency of sampling locations (7).
Estimation of fluctuations in the Re.
To estimate changes of effective reproduction numbers (Re) during the course of the epidemic, we used the birth-death skyline (BDSKY) model (69), and to analyze the various transmission groups in more detail, we used the multitype-tree birth-death model (BDMM) (21) available in BEAST v.2.4 (21, 70). To improve computational speed, HKY+I+G was used as nucleotide substitution model. BDSKY analysis was further extended to estimate the presence of sampled ancestors according to the method of Gavryushkina et al. (71). Changes in sampling frequencies over time were also implemented (sampling proportion [SP] before 1993 = 0%, SP from 1993 to 2012 = 3%, and SP from 2012 to 2015 = 13%). To obtain reliable results, the data set was reduced to RKI samples only and split into two geographic origins (nEasternAfrica = 71; nEasternEurope = 109) as detected in the analyses using ML and BEAST v.1.8.3.
Identification of clusters of closely related sequences (putative transmission clusters and German-origin clusters).
German-origin clusters were defined as having at least 3 German sequences and a posterior value of >0.99. Furthermore, their most recent common ancestor (MRCA) was estimated to be of German origin using trait diffusion models. Clusters were defined as active when at least two recently infected patients (infected after 2013) were detected within these clusters. The tool Transmic (https://github.com/kavehyousef/code) (72) was used to identify putative transmission clusters both in ML and Bayesian MCC trees. Transmic identifies clusters of closely related sequences, possibly linked by direct transmission or very short transmission chains (putative transmission clusters). For ML trees a 99% bootstrap cutoff and 4.5% mean pairwise patristic distance (MPPD) were used as cluster thresholds as reported by other groups (16, 73). The tool was extended to allow cluster selection in MCC trees. All changes are implemented and freely available in the version published at https://github.com/kavehyousef/code. After analysis of the distribution of branch lengths within the tree, a cutoff posterior probability of >0.99 and a distance of 20 years (in a time-scaled tree) were used to identify putative transmission clusters. The distance cutoff was chosen because the number of putative transmission clusters identified was highest at this distance and strongly comparable with clusters found within the ML tree with an MPPD of 4.5% (inset in Fig. 1B and Table 2).
Analysis of transmitted SDRM.
The World Health Organization (WHO)’s list of surveillance drug resistance mutations (SDRM) was used to detect transmitted drug resistance (55), using all sequences from the RKI studies known to be treatment naive. We also considered the E138A mutation in a separate analysis because it has been recently suggested that this mutation is associated with transmitted drug resistance (56).
Ethics statement.
The studies were approved by the data protection officer of the RKI and the German Federal Commissioner for Data Protection and Freedom of Information (III-401/008#0016) as well as the ethical committee of the Charité University Medicine (Berlin, Germany; EA 2/105/05) (19, 50).
Data availability.
Reference sequences were identified and downloaded by BLAST search from the Los Alamos HIV database (https://www.hiv.lanl.gov/content/sequence/BASIC_BLAST/basic_blast.html). All unpublished German subtype A sequences used in this study were uploaded to GenBank with accession numbers MF124612 to MF124794. Subtype C sequences used as outgroup for ML analyses were also uploaded to GenBank with accession numbers MK250696 to MK250705.
The final alignment of data set A, the ML tree, the phylogeographic MCC tree as raw data, and the SDRM MCC tree can be found at https://zenodo.org/record/2245808#.XKeeWtgpCpo.
ACKNOWLEDGMENTS
We thank the laboratories participating in the InzSurv-HIV, MASTER HIV/HEP, and HIV-1 Seroconverter projects (http://www.rki.de/DE/Content/InfAZ/H/HIVAIDS/Studien/InzSurv_HIV/beteiligte_Labore.html). We also thank the sequencing facility of the RKI. Finally, we thank Stephen Norley for proofreading the manuscript and Vivi Lieu for adapting plotting scripts.
This study was part of the MASTER HIV/HEP project funded by the Bundesgesundheitsministerium/German Federal Ministry of Health (https://www.bundesgesundheitsministerium.de/en/en.html; grant number IIA5-2013-2513AUK375).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We declare that we have no competing financial, professional, or personal interests that might have influenced the performance or presentation of the work described in this article.
REFERENCES
- 1.Tebit DM, Arts EJ. 2011. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis 11:45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- 2.ECDC. 2018. European Centre for Disease Prevention and Control: HIV/AIDS surveillance in Europe 2018-2017 data, annual HIV/AIDS surveillance reports. https://ecdc.europa.eu/en/publications-data/hivaids-surveillance-europe-2018-2017-data. Accessed 18 December 2018.
- 3.UNAIDS. 2018. Fact sheet. World AIDS Day 2018. http://www.unaids.org/sites/default/files/media_asset/UNAIDS_FactSheet_en.pdf. Accessed 12 May 2018.
- 4.Robert Koch Institute. 2018. HIV-Jahresbericht 2017. Epid Bull. https://www.rki.de/DE/Content/Infekt/EpidBull/Archiv/2018/Ausgaben/47_18.pdf?__blob=publicationFile. Accessed 18 December 2018.
- 5.Chan PA, Hogan JW, Huang A, DeLong A, Salemi M, Mayer KH, Kantor R. 2015. Phylogenetic investigation of a statewide HIV-1 epidemic reveals ongoing and active transmission networks among men who have sex with men. J Acquir Immune Defic Syndr 70:428–435. doi: 10.1097/QAI.0000000000000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Wu Y, Mao L, Xia W, Zhang W, Dai L, Mehta SR, Wertheim JO, Dong X, Zhang T, Wu H, Smith DM. 2015. Targeting HIV prevention based on molecular epidemiology among deeply sampled subnetworks of men who have sex with men. Clin Infect Dis 61:1462–1468. doi: 10.1093/cid/civ526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pepin J, Posada D, Peeters M, Pybus OG, Lemey P. 2014. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science 346:56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray RR, Tatem AJ, Lamers S, Hou W, Laeyendecker O, Serwadda D, Sewankambo N, Gray RH, Wawer M, Quinn TC, Goodenow MM, Salemi M. 2009. Spatial phylodynamics of HIV-1 epidemic emergence in east Africa. AIDS 23:F9–F17. doi: 10.1097/QAD.0b013e32832faf61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hue S, Hassan AS, Nabwera H, Sanders EJ, Pillay D, Berkley JA, Cane PA. 2012. HIV type 1 in a rural coastal town in Kenya shows multiple introductions with many subtypes and much recombination. AIDS Res Hum Retroviruses 28:220–224. doi: 10.1089/aid.2011.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lihana RW, Ssemwanga D, Abimiku A, Ndembi N. 2012. Update on HIV-1 diversity in Africa: a decade in review. AIDS Rev 14:83–100. [PubMed] [Google Scholar]
- 11.Zeh C, Inzaule SC, Ondoa P, Nafisa LG, Kasembeli A, Otieno F, Vandenhoudt H, Amornkul PN, Mills LA, Nkengasong JN. 2016. Molecular epidemiology and transmission dynamics of recent and long-term HIV-1 infections in rural western Kenya. PLoS One 11:e0147436. doi: 10.1371/journal.pone.0147436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bobkova M. 2013. Current status of HIV-1 diversity and drug resistance monitoring in the former USSR. AIDS Rev 15:204–212. [PubMed] [Google Scholar]
- 13.Diez-Fuertes F, Cabello M, Thomson MM. 2015. Bayesian phylogeographic analyses clarify the origin of the HIV-1 subtype A variant circulating in former Soviet Union’s countries. Infect Genet Evol 33:197–205. doi: 10.1016/j.meegid.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Beloukas A, Psarris A, Giannelou P, Kostaki E, Hatzakis A, Paraskevis D. 2016. Molecular epidemiology of HIV-1 infection in Europe: an overview. Infect Genet Evol 46:180–189. doi: 10.1016/j.meegid.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 15.Magiorkinis G, Angelis K, Mamais I, Katzourakis A, Hatzakis A, Albert J, Lawyer G, Hamouda O, Struck D, Vercauteren J, Wensing A, Alexiev I, Asjo B, Balotta C, Gomes P, Camacho RJ, Coughlan S, Griskevicius A, Grossman Z, Horban A, Kostrikis LG, Lepej SJ, Liitsola K, Linka M, Nielsen C, Otelea D, Paredes R, Poljak M, Puchhammer-Stockl E, Schmit JC, Sonnerborg A, Stanekova D, Stanojevic M, Stylianou DC, Boucher CA, program S, Nikolopoulos G, Vasylyeva T, Friedman SR, van de Vijver D, Angarano G, Chaix ML, de Luca A, Korn K, Loveday C, Soriano V, Yerly S, Zazzi M, Vandamme AM, Paraskevis D. 2016. The global spread of HIV-1 subtype B epidemic. Infect Genet Evol 46:169–179. doi: 10.1016/j.meegid.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ragonnet-Cronin M, Lycett SJ, Hodcroft EB, Hue S, Fearnhill E, Brown AE, Delpech V, Dunn D, Leigh Brown AJ, United Kingdom HIV Drug Resistance Database . 2016. Transmission of non-B HIV subtypes in the United Kingdom is increasingly driven by large non-heterosexual transmission clusters. J Infect Dis 213:1410–1418. doi: 10.1093/infdis/jiv758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parczewski M, Urbanska A, Grzeszczuk A, Maciejewska K, Witak-Jedra M, Leszczyszyn-Pynka M. 2014. Phylogeographic analysis on the travel-related introduction of HIV-1 non-B subtypes to Northern Poland. Infect Genet Evol 27:121–130. doi: 10.1016/j.meegid.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Lai A, Bozzi G, Franzetti M, Binda F, Simonetti FR, De Luca A, Micheli V, Meraviglia P, Bagnarelli P, Di Biagio A, Monno L, Saladini F, Zazzi M, Zehender G, Ciccozzi M, Balotta C. 2016. HIV-1 A1 subtype epidemic in Italy originated from Africa and Eastern Europe and shows a high frequency of transmission chains involving intravenous drug users. PLoS One 11:e0146097. doi: 10.1371/journal.pone.0146097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauser A, Hofmann A, Hanke K, Bremer V, Bartmeyer B, Kuecherer C, Bannert N. 2017. National molecular surveillance of recently acquired HIV infections in Germany, 2013 to 2014. Euro Surveill 22:30436. doi: 10.2807/1560-7917.ES.2017.22.2.30436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauser A, Hofmann A, Meixenberger K, Altmann B, Hanke K, Bremer V, Bartmeyer B, Bannert N. 2018. Increasing proportions of HIV-1 non-B subtypes and of NNRTI resistance between 2013 and 2016 in Germany: results from the national molecular surveillance of new HIV-diagnoses. PLoS One 13:e0206234. doi: 10.1371/journal.pone.0206234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhnert D, Stadler T, Vaughan TG, Drummond AJ. 2016. Phylodynamics with migration: a computational framework to quantify population structure from genomic data. Mol Biol Evol 33:2102–2116. doi: 10.1093/molbev/msw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes GJ, Fearnhill E, Dunn D, Lycett SJ, Rambaut A, Leigh Brown AJ, UK HIV Drug Resistance Collaboration . 2009. Molecular phylodynamics of the heterosexual HIV epidemic in the United Kingdom. PLoS Pathog 5:e1000590. doi: 10.1371/journal.ppat.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox J, Castro H, Kaye S, McClure M, Weber JN, Fidler S, UK Collaborative Group on HIV Drug Resistance . 2010. Epidemiology of non-B clade forms of HIV-1 in men who have sex with men in the UK. AIDS 24:2397–2401. doi: 10.1097/QAD.0b013e32833cbb5b. [DOI] [PubMed] [Google Scholar]
- 24.Chaix ML, Seng R, Frange P, Tran L, Avettand-Fenoel V, Ghosn J, Reynes J, Yazdanpanah Y, Raffi F, Goujard C, Rouzioux C, Meyer L, ANRS PRIMO Cohort Study Group . 2013. Increasing HIV-1 non-B subtype primary infections in patients in France and effect of HIV subtypes on virological and immunological responses to combined antiretroviral therapy. Clin Infect Dis 56:880–887. doi: 10.1093/cid/cis999. [DOI] [PubMed] [Google Scholar]
- 25.Tamalet C, Ravaux I, Moreau J, Bregigeon S, Tourres C, Richet H, Abat C, Colson P. 2015. Emergence of clusters of CRF02_AG and B human immunodeficiency viral strains among men having sex with men exhibiting HIV primary infection in southeastern France. J Med Virol 87:1327–1333. doi: 10.1002/jmv.24184. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann A, Hauser A, Zimmermann R, Santos-Hovener C, Batzing-Feigenbaum J, Wildner S, Kucherer C, Bannert N, Hamouda O, Bremer V, Bartmeyer B. 2017. Surveillance of recent HIV infections among newly diagnosed HIV cases in Germany between 2008 and 2014. BMC Infect Dis 17:484. doi: 10.1186/s12879-017-2585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collinson-Streng AN, Redd AD, Sewankambo NK, Serwadda D, Rezapour M, Lamers SL, Gray RH, Wawer MJ, Quinn TC, Laeyendecker O. 2009. Geographic HIV type 1 subtype distribution in Rakai district, Uganda. AIDS Res Hum Retroviruses 25:1045–1048. doi: 10.1089/aid.2009.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baalwa J, Wang S, Parrish NF, Decker JM, Keele BF, Learn GH, Yue L, Ruzagira E, Ssemwanga D, Kamali A, Amornkul PN, Price MA, Kappes JC, Karita E, Kaleebu P, Sanders E, Gilmour J, Allen S, Hunter E, Montefiori DC, Haynes BF, Cormier E, Hahn BH, Shaw GM. 2013. Molecular identification, cloning and characterization of transmitted/founder HIV-1 subtype A, D and A/D infectious molecular clones. Virology 436:33–48. doi: 10.1016/j.virol.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ssemwanga D, Nsubuga RN, Mayanja BN, Lyagoba F, Magambo B, Yirrell D, Van der Paal L, Grosskurth H, Kaleebu P. 2013. Effect of HIV-1 subtypes on disease progression in rural Uganda: a prospective clinical cohort study. PLoS One 8:e71768. doi: 10.1371/journal.pone.0071768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mbanya D, Sama M, Tchounwou P. 2008. Current status of HIV/AIDS in Cameroon: how effective are control strategies? Int J Environ Res Public Health 5:378–383. doi: 10.3390/ijerph5050378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abecasis AB, Wensing AM, Paraskevis D, Vercauteren J, Theys K, Van de Vijver DA, Albert J, Asjo B, Balotta C, Beshkov D, Camacho RJ, Clotet B, De Gascun C, Griskevicius A, Grossman Z, Hamouda O, Horban A, Kolupajeva T, Korn K, Kostrikis LG, Kucherer C, Liitsola K, Linka M, Nielsen C, Otelea D, Paredes R, Poljak M, Puchhammer-Stockl E, Schmit JC, Sonnerborg A, Stanekova D, Stanojevic M, Struck D, Boucher CA, Vandamme AM. 2013. HIV-1 subtype distribution and its demographic determinants in newly diagnosed patients in Europe suggest highly compartmentalized epidemics. Retrovirology 10:7. doi: 10.1186/1742-4690-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hemelaar J, Gouws E, Ghys PD, Osmanov S, WHO-UNAIDS Network for HIV Isolation and Characterisation . 2011. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novitsky VA, Montano MA, Essex M. 1998. Molecular epidemiology of an HIV-1 subtype A subcluster among injection drug users in the Southern Ukraine. AIDS Res Hum Retroviruses 14:1079–1085. doi: 10.1089/aid.1998.14.1079. [DOI] [PubMed] [Google Scholar]
- 34.Saad MD, Aliev Q, Botros BA, Carr JK, Gomatos PJ, Nadai Y, Michael AA, Nasibov Z, Sanchez JL, Brix DI, Earhart KC. 2006. Genetic forms of HIV type 1 in the former Soviet Union dominate the epidemic in Azerbaijan. AIDS Res Hum Retroviruses 22:796–800. doi: 10.1089/aid.2006.22.796. [DOI] [PubMed] [Google Scholar]
- 35.Saad MD, Shcherbinskaya AM, Nadai Y, Kruglov YV, Antonenko SV, Lyullchuk MG, Kravchenko ON, Earhart KC, Sanchez JL, Birx DL, Carr JK. 2006. Molecular epidemiology of HIV type 1 in Ukraine: birthplace of an epidemic. AIDS Res Hum Retroviruses 22:709–714. doi: 10.1089/aid.2006.22.709. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez JL, Todd CS, Bautista CT, Botros BA, Khakimov MM, Giyasova GM, Yakubov SK, Abdulaeva MA, Saad MD, Graham RR, Carr JK, Earhart KC. 2006. High HIV prevalence and risk factors among injection drug users in Tashkent, Uzbekistan, 2003-2004. Drug Alcohol Depend 82(Suppl 1):S15–S22. doi: 10.1016/S0376-8716(06)80003-7. [DOI] [PubMed] [Google Scholar]
- 37.Zarandia M, Tsertsvadze T, Carr JK, Nadai Y, Sanchez JL, Nelson AK. 2006. HIV-1 genetic diversity and genotypic drug susceptibility in the Republic of Georgia. AIDS Res Hum Retroviruses 22:470–476. doi: 10.1089/aid.2006.22.470. [DOI] [PubMed] [Google Scholar]
- 38.Laga V, Vasilyev A, Lapovok I, Grigoryan S, Papoyan A, Glushchenko N, Kazennova E, Bobkova M. 2015. HIV type 1 subtype A1 dominates in Armenia. Curr HIV Res 13:219–225. doi: 10.2174/1570162X13666150407142834. [DOI] [PubMed] [Google Scholar]
- 39.Lapovok IA, Laga VY, Kazennova EV, Vasilyev AV, Dzissyuk NV, Utegenova AK, Abishev AT, Tukeev MS, Bobkova MR. 2015. Molecular epidemiological analysis of HIV-1 in Kazakhstan in 2009–2013. Vopr Virusol 60:29–37. (In Russian.) [PubMed] [Google Scholar]
- 40.Balode D, Ferdats A, Dievberna I, Viksna L, Rozentale B, Kolupajeva T, Konicheva V, Leitner T. 2004. Rapid epidemic spread of HIV type 1 subtype A1 among intravenous drug users in Latvia and slower spread of subtype B among other risk groups. AIDS Res Hum Retroviruses 20:245–249. doi: 10.1089/088922204773004978. [DOI] [PubMed] [Google Scholar]
- 41.Caplinskas S, Loukachov VV, Gasich EL, Gilyazova AV, Caplinskiene I, Lukashov VV. 2013. Distinct HIV type 1 strains in different risk groups and the absence of new infections by drug-resistant strains in Lithuania. AIDS Res Hum Retroviruses 29:732–737. doi: 10.1089/AID.2012.0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zetterberg V, Ustina V, Liitsola K, Zilmer K, Kalikova N, Sevastianova K, Brummer-Korvenkontio H, Leinikki P, Salminen MO. 2004. Two viral strains and a possible novel recombinant are responsible for the explosive injecting drug use-associated HIV type 1 epidemic in Estonia. AIDS Res Hum Retroviruses 20:1148–1156. doi: 10.1089/aid.2004.20.1148. [DOI] [PubMed] [Google Scholar]
- 43.Vinogradova A, Gafurova E, Munoz-Nieto M, Rakhmanova A, Osmanov S, Thomson MM. 2010. Short communication: molecular epidemiology of HIV type 1 in the Republic of Dagestan, Russian Federation: virtually uniform circulation of subtype A, former Soviet Union variant, with predominance of the V77I(PR) subvariant. AIDS Res Hum Retroviruses 26:395–400. doi: 10.1089/aid.2009.0205. [DOI] [PubMed] [Google Scholar]
- 44.Beyrer C, Patel Z, Stachowiak JA, Tishkova FK, Stibich MA, Eyzaguirre LM, Carr JK, Mogilnii V, Peryshkina A, Latypov A, Strathdee SA. 2009. Characterization of the emerging HIV type 1 and HCV epidemics among injecting drug users in Dushanbe, Tajikistan. AIDS Res Hum Retroviruses 25:853–860. doi: 10.1089/aid.2008.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barin F, Courouce AM, Pillonel J, Buzelay L. 1997. Increasing diversity of HIV-1M serotypes in French blood donors over a 10-year period (1985–1995). Retrovirus Study Group of the French Society of Blood Transfusion. AIDS 11:1503–1508. doi: 10.1097/00002030-199712000-00015. [DOI] [PubMed] [Google Scholar]
- 46.Brand D, Moreau A, Cazein F, Lot F, Pillonel J, Brunet S, Thierry D, Le Vu S, Plantier JC, Semaille C, Barin F. 2014. Characteristics of patients recently infected with HIV-1 non-B subtypes in France: a nested study within the mandatory notification system for new HIV diagnoses. J Clin Microbiol 52:4010–4016. doi: 10.1128/JCM.01141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bobkov A, Cheingsong-Popov R, Selimova L, Ladnaya N, Kazennova E, Kravchenko A, Fedotov E, Saukhat S, Zverev S, Pokrovsky V, Weber J. 1997. An HIV type 1 epidemic among injecting drug users in the former Soviet Union caused by a homogeneous subtype A strain. AIDS Res Hum Retroviruses 13:1195–1201. doi: 10.1089/aid.1997.13.1195. [DOI] [PubMed] [Google Scholar]
- 48.Bobkov AF, Kazennova EV, Selimova LM, Khanina TA, Ryabov GS, Bobkova MR, Sukhanova AL, Kravchenko AV, Ladnaya NN, Weber JN, Pokrovsky VV. 2004. Temporal trends in the HIV-1 epidemic in Russia: predominance of subtype A. J Med Virol 74:191–196. doi: 10.1002/jmv.20177. [DOI] [PubMed] [Google Scholar]
- 49.Ryom L, Boesecke C, Gisler V, Manzardo C, Rockstroh JK, Puoti M, Furrer H, Miro JM, Gatell JM, Pozniak A, Behrens G, Battegay M, Lundgren JD, EACS Governing Board . 2016. Essentials from the 2015 European AIDS Clinical Society (EACS) guidelines for the treatment of adult HIV-positive persons. HIV Med 17:83–88. doi: 10.1111/hiv.12322. [DOI] [PubMed] [Google Scholar]
- 50.Meixenberger K, Hauser A, Jansen K, Yousef KP, Fiedler S, von Kleist M, Norley S, Somogyi S, Hamouda O, Bannert N, Bartmeyer B, Kücherer C. 2014. Assessment of ambiguous base calls in HIV-1 pol population sequences as a biomarker for identification of recent infections in HIV-1 incidence studies. J Clin Microbiol 52:2977–2983. doi: 10.1128/JCM.03289-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hauser A, Meixenberger K, Machnowska P, Fiedler S, Hanke K, Hofmann A, Bartmeyer B, Bremer V, Bannert N, Kuecherer C. 2018. Robust and sensitive subtype-generic HIV-1 pol genotyping for use with dried serum spots in epidemiological studies. J Virol Methods 259:32–38. doi: 10.1016/j.jviromet.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 52.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pineda-Pena AC, Faria NR, Imbrechts S, Libin P, Abecasis AB, Deforche K, Gomez-Lopez A, Camacho RJ, de Oliveira T, Vandamme AM. 2013. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: performance evaluation of the new REGA version 3 and seven other tools. Infect Genet Evol 19:337–348. doi: 10.1016/j.meegid.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 54.Bezemer D, Faria NR, Hassan A, Hamers RL, Mutua G, Anzala O, Mandaliya K, Cane P, Berkley JA, Rinke de Wit TF, Wallis C, Graham SM, Price MA, Coutinho RA, Sanders EJ. 2014. HIV type 1 transmission networks among men having sex with men and heterosexuals in Kenya. AIDS Res Hum Retroviruses 30:118–126. doi: 10.1089/aid.2013.0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, Heneine W, Kantor R, Jordan MR, Schapiro JM, Vandamme AM, Sandstrom P, Boucher CA, van de Vijver D, Rhee SY, Liu TF, Pillay D, Shafer RW. 2009. Drug resistance mutations for surveillance of transmitted HIV-1 drug resistance: 2009 update. PLoS One 4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hofstra LM, Sauvageot N, Albert J, Alexiev I, Garcia F, Struck D, Van de Vijver DA, Asjo B, Beshkov D, Coughlan S, Descamps D, Griskevicius A, Hamouda O, Horban A, Van Kasteren M, Kolupajeva T, Kostrikis LG, Liitsola K, Linka M, Mor O, Nielsen C, Otelea D, Paraskevis D, Paredes R, Poljak M, Puchhammer-Stockl E, Sonnerborg A, Stanekova D, Stanojevic M, Van Laethem K, Zazzi M, Zidovec Lepej S, Boucher CA, Schmit JC, Wensing AM, Program S. 2016. Transmission of HIV drug resistance and the predicted effect on current first-line regimens in Europe. Clin Infect Dis 62:655–663. doi: 10.1093/cid/civ963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Minh BQ, Nguyen MA, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baele G, Li WL, Drummond AJ, Suchard MA, Lemey P. 2013. Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Mol Biol Evol 30:239–243. doi: 10.1093/molbev/mss243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, Suchard MA. 2013. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol 30:713–724. doi: 10.1093/molbev/mss265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Griffiths RC, Tavare S. 1994. Sampling theory for neutral alleles in a varying environment. Philos Trans R Soc Lond B Biol Sci 344:403–410. doi: 10.1098/rstb.1994.0079. [DOI] [PubMed] [Google Scholar]
- 63.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ayres DL, Darling A, Zwickl DJ, Beerli P, Holder MT, Lewis PO, Huelsenbeck JP, Ronquist F, Swofford DL, Cummings MP, Rambaut A, Suchard MA. 2012. BEAGLE: an application programming interface and high-performance computing library for statistical phylogenetics. Syst Biol 61:170–173. doi: 10.1093/sysbio/syr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1.6. http://beast.bio.ed.ac.uk/Tracer.
- 66.Bielejec F, Lemey P, Baele G, Rambaut A, Suchard MA. 2014. Inferring heterogeneous evolutionary processes through time: from sequence substitution to phylogeography. Syst Biol 63:493–504. doi: 10.1093/sysbio/syu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bielejec F, Rambaut A, Suchard MA, Lemey P. 2011. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 27:2910–2912. doi: 10.1093/bioinformatics/btr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Avise JC. 2000. Phylogeography: the history and formation of species. Harvard University Press, Cambridge, MA. [Google Scholar]
- 69.Stadler T, Kuhnert D, Bonhoeffer S, Drummond AJ. 2013. Birth-death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV). Proc Natl Acad Sci U S A 110:228–233. doi: 10.1073/pnas.1207965110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bouckaert R, Heled J, Kuhnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gavryushkina A, Welch D, Stadler T, Drummond AJ. 2014. Bayesian inference of sampled ancestor trees for epidemiology and fossil calibration. PLoS Comput Biol 10:e1003919. doi: 10.1371/journal.pcbi.1003919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pouran Yousef K, Meixenberger K, Smith MR, Somogyi S, Gromöller S, Schmidt D, Gunsenheimer-Bartmeyer B, Hamouda O, Kücherer C, von Kleist M. 2016. Inferring HIV-1 transmission dynamics in Germany from recently transmitted viruses. J Acquir Immune Defic Syndr 73:356–363. doi: 10.1097/QAI.0000000000001122. [DOI] [PubMed] [Google Scholar]
- 73.Ragonnet-Cronin M, Hodcroft E, Hue S, Fearnhill E, Delpech V, Brown AJ, Lycett S, Database U. 2013. Automated analysis of phylogenetic clusters. BMC Bioinformatics 14:317. doi: 10.1186/1471-2105-14-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salemi M, de Oliveira T, Ciccozzi M, Rezza G, Goodenow MM. 2008. High-resolution molecular epidemiology and evolutionary history of HIV-1 subtypes in Albania. PLoS One 3:e1390. doi: 10.1371/journal.pone.0001390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paraskevis D, Magiorkinis E, Magiorkinis G, Sypsa V, Paparizos V, Lazanas M, Gargalianos P, Antoniadou A, Panos G, Chrysos G, Sambatakou H, Karafoulidou A, Skoutelis A, Kordossis T, Koratzanis G, Theodoridou M, Daikos GL, Nikolopoulos G, Pybus OG, Hatzakis A, Multicentre Study on HIVH . 2007. Increasing prevalence of HIV-1 subtype A in Greece: estimating epidemic history and origin. J Infect Dis 196:1167–1176. doi: 10.1086/521677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Reference sequences were identified and downloaded by BLAST search from the Los Alamos HIV database (https://www.hiv.lanl.gov/content/sequence/BASIC_BLAST/basic_blast.html). All unpublished German subtype A sequences used in this study were uploaded to GenBank with accession numbers MF124612 to MF124794. Subtype C sequences used as outgroup for ML analyses were also uploaded to GenBank with accession numbers MK250696 to MK250705.
The final alignment of data set A, the ML tree, the phylogeographic MCC tree as raw data, and the SDRM MCC tree can be found at https://zenodo.org/record/2245808#.XKeeWtgpCpo.