Bromodomain inhibitors have been reported to activate HIV-1 transcription in vitro, but their effect on activation of HIV-1 reservoirs during cART in vivo is unclear. We found that BETi (I-BET151) treatment reactivated HIV-1 gene expression in humanized mice during suppressive cART. Interestingly, I-BET151 preferentially reactivated HIV-1 gene expression in monocytic cells, but not in CD4 T cells, in cART-treated mice. Furthermore, I-BET151 significantly increased HIV-1 transcription in monocytic cells, but not in HIV-1-infected CD4 T cells, via CDK2-dependent mechanisms. Our findings suggest that BETi can preferentially activate monocytic HIV-1 reservoir cells and that a combination of reservoir activation agents targeting different cell types and pathways is needed to achieve reactivation of different HIV-1 reservoir cells during cART.
KEYWORDS: BET inhibitor, CDK2, HIV-1 reservoirs, I-BET151, humanized mice
ABSTRACT
Combination antiretroviral therapy (cART) effectively suppresses HIV-1 replication and enables HIV‑infected individuals to live long, productive lives. However, the persistence of HIV-1 reservoirs of both T and myeloid cells with latent or low-replicating HIV-1 in patients under cART makes HIV-1 infection an incurable disease. Recent studies have focused on the development of strategies to activate and purge these reservoirs. Bromodomain and extraterminal domain proteins (BETs) are epigenetic readers involved in modulating gene expression. Several bromodomain inhibitors (BETi) are reported to activate viral transcription in vitro in HIV-1 latency cell lines in a P-TEFb (CDK9/cyclin T1)-dependent manner. Little is known about BETi efficacy in activating HIV-1 reservoir cells under cART in vivo. Here we report that a BETi (I-BET151) efficiently activated HIV-1 reservoirs under effective cART in humanized mice in vivo. Interestingly, I-BET151 during suppressive cART in vivo activated HIV-1 gene expression only in monocytic cells and not in CD4+ T cells. We further demonstrate that BETi preferentially enhanced HIV-1 gene expression in monocytic cells rather than in T cells and that whereas CDK9 was involved in activating HIV-1 by I-BET151 in both monocytic and T cells, CDK2 enhanced HIV-1 transcription in monocytic cells but inhibited it in T cells. Our findings reveal a role for CDK2 in differential modulation of HIV-1 gene expression in myeloid cells and in T cells and provide a novel strategy to reactivate monocytic reservoirs with BETi during cART.
IMPORTANCE Bromodomain inhibitors have been reported to activate HIV-1 transcription in vitro, but their effect on activation of HIV-1 reservoirs during cART in vivo is unclear. We found that BETi (I-BET151) treatment reactivated HIV-1 gene expression in humanized mice during suppressive cART. Interestingly, I-BET151 preferentially reactivated HIV-1 gene expression in monocytic cells, but not in CD4 T cells, in cART-treated mice. Furthermore, I-BET151 significantly increased HIV-1 transcription in monocytic cells, but not in HIV-1-infected CD4 T cells, via CDK2-dependent mechanisms. Our findings suggest that BETi can preferentially activate monocytic HIV-1 reservoir cells and that a combination of reservoir activation agents targeting different cell types and pathways is needed to achieve reactivation of different HIV-1 reservoir cells during cART.
INTRODUCTION
The global implementation of combination antiretroviral therapy (cART) has transformed human immunodeficiency virus type 1 (HIV-1) infection from a fatal disease into a manageable chronic illness and has successfully blunted the HIV pandemic (1–3). HIV-1 replication can be suppressed and maintained at a level below the detection limits by current cART regimens. However, cART is still unable to cure HIV-1 infection. Drug resistance, serious non-AIDS events, and financial and societal issues associated with lifelong cART highlight the urgent need to find a cure for the disease (4–7). Recent efforts thus have focused on interventions that can yield a drug-free remission of HIV-1 replication or even eradication of replication-competent HIV-1 proviruses in patients.
The major obstacle to eradicating HIV-1 is the persistence of HIV-1 reservoirs that are able to produce infectious viral particles and result in viral relapse off cART (8–11). Although resting CD4 T cells with latent HIV-1 infection are the best-characterized source of HIV-1 reservoirs during cART in patients, myeloid cells with low replicating (low HIV-1 mRNA) have also been detected in cART-treated patients (12, 13), in simian immunodeficiency virus (SIV)-infected cART-treated nonhuman primates (NHP) (14, 15), and in HIV-infected cART-treated humanized mice (16, 17). The molecular mechanisms associated with the maintenance and reactivation of HIV latency have been extensively investigated, including transcriptional interference, deleterious mutations in the viral genome, low levels of transcriptional activators, inadequate Tat activity, and epigenetic repression (18, 19). Multiple mechanisms are most likely involved in HIV-1 reservoir persistence, and the relative importance of these mechanisms in different cell types remains to be determined.
Macrophages are susceptible to HIV-1 infection in human and animal models (20–23), and HIV-1 could be recovered from the circulating monocytes in patients treated with cART (24, 25). In addition, HIV-1 is able to infect and replicate in brain microglia cells that could persist despite cART (26, 27). Recent reports have demonstrated that humanized mice with myeloid cells only allow HIV-1 persistent infection in macrophages during cART in vivo (23), and integrated HIV-1 DNA can be detected in the bone marrow and spleen macrophages in humanized mice during cART (28). These findings highlight that monocytes or macrophages, as well as resting CD4+ T cells, are of great clinical importance in terms of HIV-1 reservoir persistence and HIV-1 cure.
It is known that bromodomain-containing protein 4 (BRD4) competes with P-TEFb and disrupts the interaction between Tat and P-TEFb, thus abrogating the ability of Tat to transactivate HIV-1 transcription (29–31). Given the important role of P-TEFb in regulating HIV gene expression, a number of bromodomain inhibitors (BETi) have been tested to activate HIV-1 gene expression in latent models of primary CD4+ T cells, lymphocytic T cell lines, and monocytic cell lines (32, 33). However, little is known about the therapeutic potential of BETi in activating viral replication in HIV-1 reservoirs during cART in vivo.
In this study, we tested how a bromodomain inhibitor (I-BET151) affected viral replication in HIV-1 reservoir cells in vivo during suppressive antiretroviral therapy in humanized mice. Our results demonstrate that I-BET151 treatment leads to reactivation of HIV-1 gene expression preferentially in monocytic cells during cART in vivo. We further demonstrate that BETi preferentially enhanced HIV-1 gene expression in monocytic cells rather than in T cells and that CDK2 enhanced HIV-1 transcription in monocytic cells but inhibited it in T cells.
(This article was submitted to an online preprint archive [34].)
RESULTS
Bromodomain inhibitor I-BET151 activates HIV-1 replication under suppressive cART in vivo.
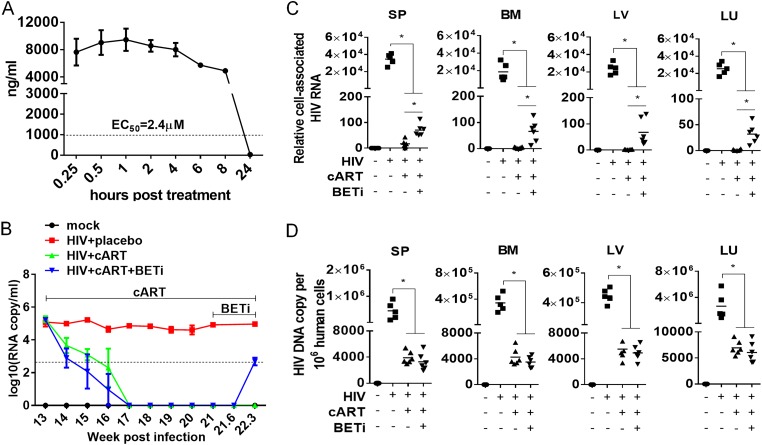
Bromodomain inhibitors have been shown to activate HIV-1 gene expression in different cell systems in vitro with latent or chronic HIV-1 infection (30–33). In order to investigate the effect of BETi on viral reservoirs in vivo, we characterized the pharmacokinetic (PK) and pharmacodynamic (PD) properties of I-BET151 in mice. We showed that at a dose of 18 mg/kg of body weight, the concentration of I-BET151 was maintained at a relative steady level (∼10 times above the 50% effective concentration [EC50] for HIV-1 activation) for at least 8 h in blood after initial administration through gavage (Fig. 1A and B). These data indicate that the dose was sufficient to activate HIV-1 gene expression in mice in vivo. Thus, we treated humanized mice with I-BET151 (18 mg/kg) once every day for activation of HIV-1 gene expression in the following experiments. To assess the activity of I-BET151 in activating HIV-1 reservoir cells, HIV-1-infected humanized mice were treated with I-BET151 for 9 days starting at 21 weeks postinfection (wpi) by daily gavage, after viremia was completely suppressed for 4 weeks by cART. Consistent with its activity in vitro, HIV-1 viremia rebound was detected on day 9 (22.3 wpi) after I-BET151 initiation (Fig. 1B). To identify the putative HIV-1 reservoir cells responding to I-BET151 treatment, animals were terminated immediately after viremia rebound was detected. Leukocytes from spleens and bone marrow, and tissues from liver and lung, were harvested and subjected to detection of cell-associated HIV-1 RNA and DNA by TaqMan real-time PCR. We demonstrated that blood viral load rebound was accompanied by a higher level of cell-associated HIV-1 RNA in human cells in different organs or tissues (Fig. 1C). However, a similar level of HIV-1 DNA in human cells was detected in tissues upon the activation of viral replication after BETi treatment (Fig. 1D), indicating no new virus spreading infection.
FIG 1.
I-BET151 treatment activates HIV-1 reservoirs under suppressive cART in vivo. (A) Pharmacokinetics/pharmacodynamics of I-BET151 in NSG mice. NSG mice were treated with 18 mg/kg of I-BET151 through gavage, and drug concentration in blood was measured by high-performance liquid chromatography (HPLC). (B to D) Humanized mice were infected with HIVJR-CSF. cART was initiated at 13 wpi. BETi treatment was initiated at 21 wpi by daily gavage (18 mg/kg) until termination at 22.3 wpi. (B) HIV plasma load. (C) Relative cell-associated viral RNA levels in human cells of lymphoid tissues (spleen [SP], bone marrow [BM], liver [LV], and lung [LU]), normalized to human CD4 mRNA level. (D) Cell-associated HIV-1 DNA copies in human cells in different tissues (spleen, bone marrow, liver, and lung) were measured by real-time PCR. Bars in dot graphs indicate mean values. *, P < 0.05.
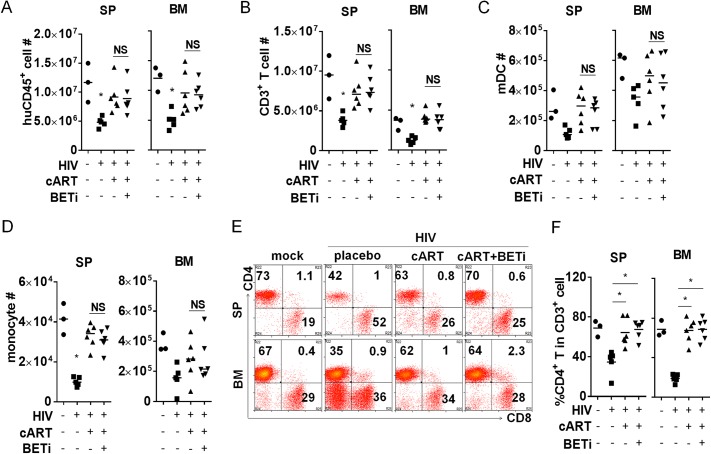
We observed no significant toxic effect of I-BET151 on the gross health of the treated mice. We found no effect on human leukocytes in treated and nontreated animals, including total numbers of human CD45+ cells (Fig. 2A), CD3+ T cells (Fig. 2B), dendritic cells (Fig. 2C), or monocytes (Fig. 2D), in the spleen or bone marrow. In addition, relative CD4+ and CD8+ T cells were not affected by I-BET151 (Fig. 2E and F). These data indicate no significant cytotoxicity of BETi treatment in vivo. We conclude that I-BET151 treatment reactivates HIV-1 gene expression during effective cART, without significant cytotoxicity to human myeloid and lymphoid cells in vivo.
FIG 2.
I-BET151 treatment shows no significant cytotoxicity in humanized mice. Animals were treated as described for Fig. 1, and cells harvested from spleen or bone marrow were counted and analyzed by flow cytometry. (A) Human CD45+ leukocyte numbers. (B) Human CD3+ T cell numbers. (C) CD3− CD14− CD11c+ dendritic cell numbers. (D) CD3− CD11c+ CD14+ monocyte numbers. (E) Plots show percentages of CD4+ T cells and CD8 T cells in the spleen and bone marrow. (F) Summary data show percentages of CD4+ T cells and CD8 T cells in CD3+ cells in spleen and bone marrow. Bars in dot graphs indicate mean values.
I-BET151 specifically activates HIV-1 replication in monocytes in vivo.
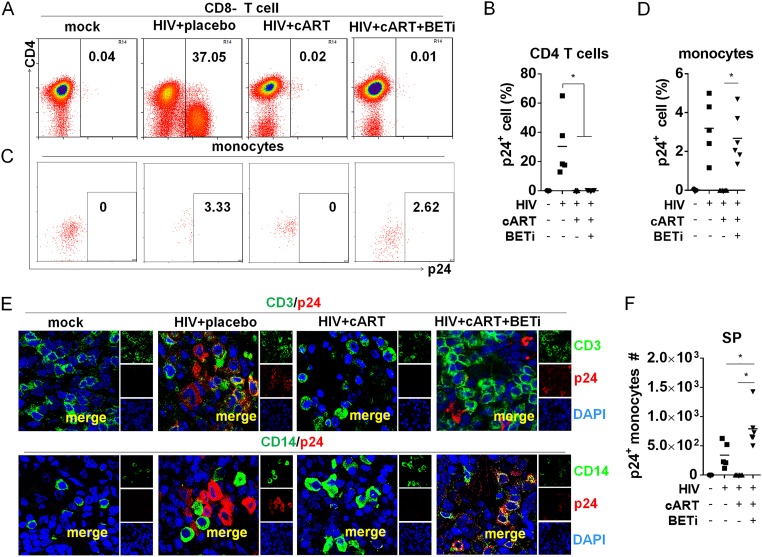
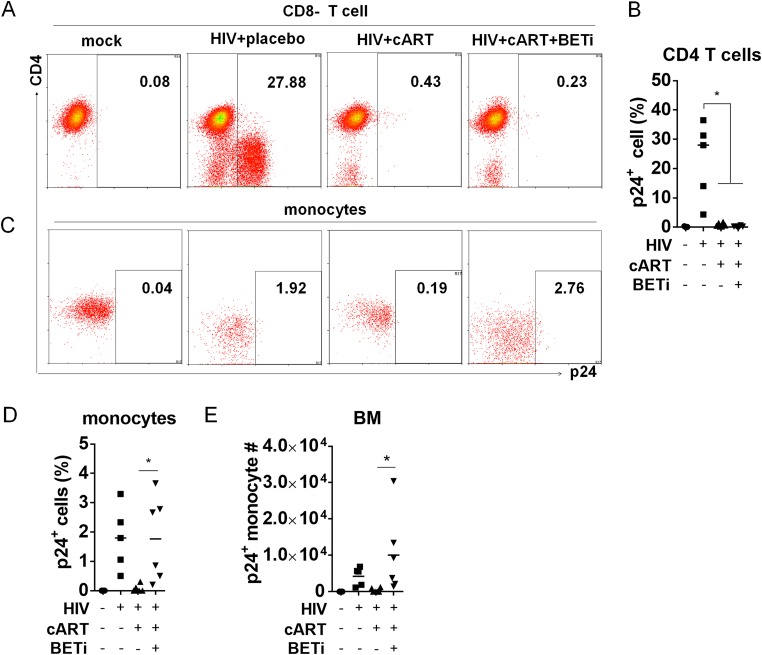
In order to define the HIV-1 reservoir cells that were activated by BETi, we investigated what cell types were activated to express HIV-1 p24 proteins by I-BET151 treatment in vivo at 22.3 wpi (Fig. 1A). Surprisingly, we did not observe any CD4 T cells with detectable p24 (Fig. 3A and B). In contrast, a significant percentage of human monocytes became p24 positive after I-BET151 treatment (Fig. 3C and D). We did not detect any significant p24 expression in other populations, including CD3− CD14− CD11c+ cells (dendritic cells) and CD3− CD11c− CD4+ CD123+ cells (pDCs) (data not shown). To confirm this finding, we performed immunofluorescence costaining of HIV-1 p24 and human CD3 or CD14 on spleen tissue sections. Consistently, we found that p24 staining was colocalized only with CD14+ cells in the I-BET151 group and not with CD3 T cells. In comparison, no p24-positive cells were detected in cART-only mice, indicating an effective suppression of HIV-1 replication by cART (Fig. 3E). Although monocyte numbers in the spleen were not altered by I-BET151 (Fig. 2D), the p24+ monocyte number was significantly increased by I-BET151 treatment (Fig. 3F). Similar findings were also obtained in the bone marrow, as I-BET151 treatment activated HIV replication only in monocytes and not CD4 T cells (Fig. 4A to D). In a separate experiment with sustained cART, the elevated viral load induced by I-BET151 treatment could be inhibited again when I-BET151 was stopped (data not shown), indicating that the observed rebound of viral RNA production was not due to the emergence of drug-resistant mutant viruses.
FIG 3.
I-BET151 treatment preferentially activates HIV replication in monocytic cells under suppressive cART in spleens. Humanized mice were treated as described for Fig. 1, and splenocytes were harvested for flow cytometry analysis. Fixed spleen tissues were also analyzed by immunofluorescent staining. (A) Histograms show percentages of HIV gag p24+ CD4+ T (CD3+ CD8−) cells in spleens. (B) Summarized percentages of p24+ CD4+ T cells in spleens. (C) Histograms show percentages of p24+ monocytic cells (CD3− CD11c+ CD14+) in spleens. (D) Summarized percentages of p24+ monocytic cells in spleens. (E) Immunofluorescence costaining of CD3 (green)/Gag p24 (red) (upper row) or CD14 (green)/Gag p24 (red) (lower row) in spleens. (F) HIV Gag p24+ monocyte numbers in spleens. Bars in dot graphs indicate mean values.
FIG 4.
I-BET151 preferentially activates HIV replication in monocytes in the bone marrow of humanized mice on cART. Humanized mice were treated as described for Fig. 1, and bone marrow cells were analyzed. (A) Histograms show percentages of HIV Gag p24+ CD4+ T cells (CD3+ CD8−). (B) Summarized percentages of p24+ CD4+ T cells. (C) Histograms show percentages of HIV Gag p24+ monocytes (CD3− CD11c+ CD14+). (D) Summarized percentages of p24+ monocytes. (E) p24+ monocyte numbers in the bone marrow (BM). Bars in dot graphs indicate mean values.
I-BET151 activates HIV-1 gene expression more efficiently in MDMs than in resting CD4 T cells.
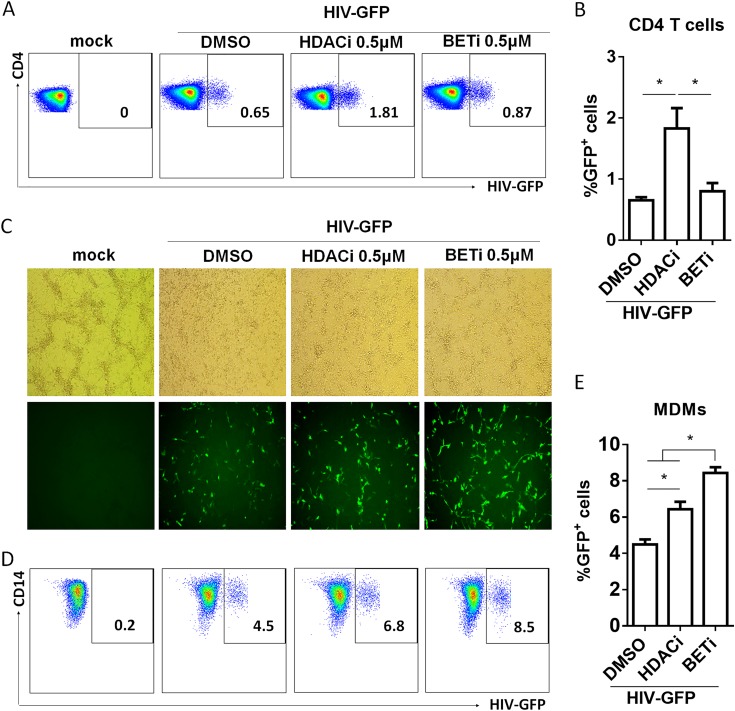
It has been reported that BETi activates HIV-1 gene transcription in latent models of both T cells and monocytic cells in vitro (32, 33). To confirm our finding, we transduced either resting CD4+ T cells or monocyte-derived macrophages (MDMs) with vesicular stomatitis virus G protein (VSV-G) HIV Duo-Fluo reporter virus. This reporter virus encodes a long terminal repeat (LTR)-driven enhanced green fluorescent protein (eGFP) marker (productive infection) and the mCherry marker driven by an EF1α promoter (35, 36). After transduction, cells were cultured for 24 h before I-BET151 treatment. GFP expression was determined at 72 h for CD4+ T cells and 36 h for MDMs after culture with I-BET151. We used suberoylanilide hydroxamic acid (SAHA), a histone deacetylase (HDAC) inhibitor reported to activate HIV LTR transcription in both cell lines and primary CD4 T cells (37–39), as a positive control. The results show that I-BET151 was unable to significantly enhance HIV-1 transcription in resting CD4+ T cells (Fig. 5A and B). In contrast, HIV-1 transcription in MDMs was significantly increased by I-BET151 (Fig. 5C to E).
FIG 5.
I-BET151 treatment activates HIV-1 transcription in MDMs but not in resting CD4+ T cells in vitro. CD25− resting CD4+ T cells and MDMs were transduced with VSV Duo-Fluo I virus and cultured for 24 h before I-BET151 treatment. Cells were analyzed for GFP expression at 72 h (CD4+ T cells) or 36 h (MDMs) after I-BET151 treatment, (A) Plots show GFP+ cells in CD4+ T cells. (B) Summarized percentage of GFP+ cells in CD4+ T cells. (C) MDMs confluence (upper row) and GFP frequency (lower row) observed under a fluorescence microscope. (D) Plots show GFP expression in MDMs. (E) Summarized percentage of GFP+ cells in MDMs. Error bars indicate SD.
I-BET151 activates HIV-1 gene expression in monocytic cells via CDK2- and CDK9-dependent mechanisms.
As previously reported, inhibition of either cyclin-dependent kinase 2 (CDK2) or CDK9 suppresses HIV-1 transcription or replication (40–45), and bromodomain inhibitor-induced reactivation of HIV-1 latency is dependent on CDK9 (30, 31). In CDK2 knockdown macrophage-like cells derived from pluripotent stem cells, HIV-1 replication is significantly reduced (46). To study the mechanism of I-BET151-enhanced HIV-1 transcription in monocytic cells and CD4+ T cells, we first established cell lines transduced with a reporter HIV-1 genome expressing luciferase under the control of the HIV-1 LTR using either U937 myeloid (U937-luc) or Jurkat T (Jurkat-luc) cells. We treated cells with I-BET151 in the presence of either the highly specific and potent CDK2 inhibitor K03861 (47) or the CDK9 inhibitor LDC000067. In U937-luc cells, either the CDK2 or CDK9 inhibitor alone downregulated HIV-1 transcription relative to that in mock-treated cells. In addition, I-BET151 enhancement of HIV-1 transcription was abolished by either CDK2 (1 μM) or CDK9 (25 μM) inhibitor (Fig. 6A). Interestingly, the CDK9 inhibitor, but not the CDK2 inhibitor, suppressed I-BET151-enhanced HIV-1 gene expression in Jurkat-luc cells (Fig. 6B). In fact, the CDK2 inhibitor appeared to slightly enhance HIV-1 gene expression in the absence or presence of I-BET151. These results suggest that the different efficacies of I-BET151 in activating HIV-1 transcription in monocytic cells and CD4+ T cells may be attributed to cell-intrinsic differences in the regulation of HIV-1 transcription by CDK2.
FIG 6.
CDK2 activation by I-BET151 enhances HIV-1 transcription in monocytic cells but inhibits it in T cells. U937 cells and Jurkat cells were transduced with VSV HIV-luc reporter virus. After expansion for 6 days, cells were treated with a CDK2 or CDK9 inhibitor with or without I-BET151 treatment in triplicates. At 48 h after I-BET151 treatment, luciferase activity in cells was measured. (A) Luciferase activity in U937 cells after CDK2 or CDK9 inhibitor treatment with or without I-BET151 at 48 h after treatment. (B) Luciferase activity in Jurkat cells after CDK2 or CDK9 inhibitor treatment with or without I-BET151 at 48 h. Error bars indicate SD.
DISCUSSION
Despite the importance of HIV-1 reservoirs in resting CD4+ T cells, it is impossible to achieve a cure without considering reservoir cells like macrophages in brain tissues (48, 49). The existence of HIV-1 reservoirs in macrophages in vivo is strongly supported by Honeycutt et al. (23), who demonstrated that HIV-1 replication can be sustained in macrophages in a humanized mice model without T cells. Consistently, our study demonstrates that monocytes or macrophages are HIV-1 reservoir cells during suppressive cART in humanized mice. Importantly, bromodomain inhibitors could be used to preferentially activate this particular myeloid cell HIV-1 reservoir in vivo.
In this study, no reduction of cell-associated HIV-1 DNA in human cells solely by I-BET151 was observed after 9 days’ treatment. We speculate that the poor elimination of HIV-1 reservoir induced by I-BET151 in vivo is due to two reasons. First, macrophages are more resistant to HIV-1 infection-induced cytopathic effect than are CD4+ T cells (50, 51). As I-BET151 effectively reactivates HIV-1 replication only in monocytic cells in vivo, it is likely that the cells are simply not dying while producing viruses. Second, the immune system might be still defective or exhausted due to chronic HIV-1 infection though viral replication being suppressed by cART, as recently reported (16, 52). A recent study showed that cytotoxic T lymphocyte (CTL)-mediated cytolytic killing rate can be significantly faster than the HIV-1 lytic infection death rate in vivo (53). Given the above-mentioned possibilities, bromodomain inhibitor treatment should be combined with other therapies that can either kill HIV-1-positive cells directly (e.g., immunotoxin) or enhance the host immune response to HIV-1 infection (16) to synergistically purge HIV-1 reservoir cells.
It is known that HIV-1 reservoir cells can exist in two different statuses in vivo. One is harboring latent replication-competent proviruses expressing few to no viral genes. The long-lived resting memory CD4+ T cells are the putative latent reservoir cells most extensively studied to date (54–56). Some HIV-1 reservoir cells still support low levels of HIV-1 replication, due to either sanctuary sites that have low accessibility to cART or the cell types that are less responsive to antiretroviral drugs, such as monocytes or macrophages (57–59). A recent report has documented the existence of lymphoid tissue sanctuary sites for harboring low levels of viral replication in patients with undetectable viremia (60). In addition, brain resident macrophages or microglial cells have been implicated in maintaining HIV-1 persistent infection (48, 49). In our study, we could not distinguish if the increased level of HIV-1 replication in monocytic cells was due to activation of latent HIV provirus or further enhancement of residual low levels of viral replication. However, it is clear that BETi did not increase the number of HIV-infected cells, indicating activation of HIV-1 gene expression in preinfected target cells during cART. It is of interest to isolate these myeloid HIV-1 reservoir cells and investigate the status of their proviral DNA (latent or low replicating) and the host signaling pathways that may contribute to their persistence. Moreover, higher dosages should be tested in future studies for possible cytotoxicity effects in mice, NHP, and human patients. Our findings shed light on the development of strategies to specifically activate these myeloid HIV-1 reservoirs during cART.
In the absence of HIV-1 transactivator Tat, RNAPII pauses after transcription of the TAR sequence. P-TEFb consists of cyclin T1 and CDK9 and converts promoter-proximally paused RNA polymerase II (RNAPII) complexes into efficient elongating complexes (29). Tat recruits P-TEFb via CycT1 to TAR, allowing CDK9 to phosphorylate the C-terminal domain (CTD) of RNAPII to continue viral gene transcription (61). It has been reported that CDK2 can phosphorylate either CDK9 or Tat and thereby contributes to enhanced HIV-1 transcription (62, 63). These findings indicate both CDK9 and CDK2 are involved in the regulation of HIV-1 transcription. We indeed confirmed that I-BET151 enhanced HIV-1 gene expression in monocytic cells via mechanisms that depended on both CDK9 and CDK2. In the Jurkat T cell line, we observed that CDK9 inhibitor, but not CDK2 inhibitor, suppressed I-BET151 enhanced HIV-1 gene expression in Jurkat-luc cells. Interestingly, the CDK2 inhibitor appeared to slightly enhance HIV-1 gene expression in the absence or presence of I-BET151. Modulation of HIV-1 transcription by interactions between CDK2, CDK9, and P-TEFb is complex (62, 64) and may contribute to the different efficacies of I-BET151 in activating HIV-1 transcription in monocytic cells and CD4+ T cells. CDK2 expression is reported to be relatively low in resting CD4+ T lymphocytes (65), although other host and viral factors may be involved. It will be interesting to elucidate the exact mechanisms of the differential response of HIV-1 reactivation in myeloid cells and in T cells in future studies.
Although the mechanisms contributing to HIV-1 transcription quiescence or latency have been investigated extensively, we still do not fully understand the relative importance of each mechanism in different cell types. As we have reported previously, depletion of regulatory T cells during cART in humanized mice leads to reactivation of HIV-1 replication predominantly in memory CD4+ T cells but not in monocytic cells in humanized mice (17). In a recent report, it was shown that BET inhibitors reactivate latent HIV-1 in cells with integration sites distinct from those activated by HDAC inhibitors and PKC pathway agonists (66). It is thus possible that BET inhibitors activate HIV-1 differently in monocytic cells due to distinct signaling pathways and integration sites. This report is based on findings from the CCR5-tropic HIV-1 isolate HIVJR-CSF, which infects both myeloid cells and T cells in humanized mice in vivo (Fig. 3 and 4) (17). Other HIV-1 isolates with different coreceptor usage and macrophage tropism should be tested in order to fully understand the therapeutic effect of bromodomain inhibitors as well as other latency-reversing agents in activating latent HIV-1 in myeloid and T cells. To facilitate HIV-1 cure therapy, it is of particular importance to identify all possible HIV-1 reservoir cells in lymphoid organs and other tissues in vivo and to understand how HIV-1 maintains its low replication or latency in different cell types for future drug design and therapeutic development.
MATERIALS AND METHODS
Construction of humanized mice.
Human fetal liver tissues were obtained from elective or medically indicated termination of pregnancy through a nonprofit intermediary working with outpatient clinics (Advanced Bioscience Resources). Approval for animal work was obtained from University of North Carolina (UNC) Institutional Animal Care and Use Committee (IACUC). We constructed hu-NRG mice as previously reported (17, 67–71).
HIV-1 infection of humanized mice.
Humanized mice were infected by intravenous (i.v.) injection with HIV-1JR-CSF stocks (10 ng of p24/mouse in 50 μl) or with mock stocks in control mice.
cART regimens in hu-NRG HSC mice.
Individual tablets of tenofovir-emtricitabine (Truvada; Gilead Sciences) or raltegravir (Merck) were crushed into fine powder and manufactured as 5BXL by TestDiet based on previously published methods (16, 72).
Quantification of HIV load in plasma.
Peripheral blood was collected with EDTA as an anticoagulant at designated time points after HIV infection. Plasma was prepared by centrifugation and stored at −80°C until assay. Viral RNA was isolated from the plasma. HIV load was measured by quantitative reverse transcription-PCR (qRT-PCR) as described previously (detection limit is <400 copies/ml) (16, 17, 68).
Cell-associated HIV-1 RNA.
Total RNA was extracted from cells or tissues using an RNeasy plus minikit (Qiagen). HIV-1 RNA was detected as previously described (16, 17, 68). HIV-1 gag RNA expression was normalized to human CD4 or CD14 mRNA level, and relative HIV-1 gene expression levels were calculated according to the threshold cycle (2−ΔΔCT) (16, 73).
Cell-associated HIV-1 DNA.
Total nucleic acid was extracted from cells or tissues using a DNeasy minikit (Qiagen). HIV-1 DNA was quantified by real-time PCR. Genomic DNA of ACH2, which contains one copy of the HIV genome in each cell, was serially diluted in mouse leukocyte DNA to generate a standard curve (16).
Flow cytometry.
For HIV-1 Gag p24 staining, cells were stained with surface markers first and then permeabilized with Cytofix/Cytoperm buffer (BD Bioscience; catalog number 554714), followed by intracellular staining. Phycoerythrin (PE)-conjugated anti-human CD11c (clone 3.9, catalog number 301606), PE/Cy5-conjugated anti-human CD4 (clone RP4-T4, catalog number 300510), PE/Cy7-conjugated antihuman CD3 (clone HIT3a, catalog number 300316), Pacific blue-conjugated anti-human CD14 (clone HCD14, catalog number 325616), and allophycocyanin (APC)/Cy7-conjugated anti-human CD45 (clone H130, catalog number 304014) were purchased from Biolegend; Pacific orange-conjugated anti-mouse CD45 (clone HI30, catalog number MHCD4530), PE-TR-conjugated CD8 (clone 3B5, catalog number MHCD0817), and a LIVE/DEAD fixable aqua dead cell stain kit (catalog number L34957) were purchased from Invitrogen. Fluorescein isothiocyanate (FITC)-conjugated anti-HIV p24 (clone FH190-1-1, catalog number 6604665) was purchased from Beckman Coulter. Flow cytometry was performed using either CyAn ADP (Dako) or BD LSRFortessa (BD Biosciences), analyzed by Summit 4.3 (Beckman Coulter) or FlowJo 10 (FLOWJO, LLC) accordingly.
Immunofluorescence staining of spleens.
Mouse spleens were harvested, fixed with 10% formalin (Fisher, Fair Lawn, NJ), embedded in paraffin, and cut into 5-μm tissue sections. Antigen retrieval was performed by incubation in Diva Decloacker (Biocare Medical, Concord, CA) for 30 min at 95°C, followed by slow cooling down for 1 h. The tissue section was blocked with Background Sniper (Biocare Medical). The sections were then stained with the following primary antibodies: rabbit monoclonal anti-human CD3 (Life Span Bio Sciences, Seattle, WA; 1:100 dilution) or rabbit monoclonal anti-human CD14 (Abcam; 1:500 dilution) or mouse monoclonal anti-HIV-1 p24 (Dako; 1:5 dilution) diluted in blocking buffer (phosphate-buffered saline [PBS], 0.05% Tween 20, 5% goat serum). They were then stained with the following secondary antibodies: Alexa Fluor 594 donkey anti-mouse IgG (Life Technologies, Eugene, OR) and Alexa Fluor 488 donkey anti-rabbit IgG (Life Technologies). The sections were stained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted with an antifade mounting medium (Abcam, Cambridge, MA). Sections were analyzed by confocal microscopy (Zeiss LSM 700 confocal laser scanning microscope).
Inhibitors and treatment.
Bromodomain inhibitor I-BET151 was obtained from GlaxoSmithKline (Research Triangle Park, NC). Drug powder was dissolved in 0.5% hydroxypropyl methylcellulose (HPMC)–0.1% Tween 80 to a final concentration of 1.5 mg/ml and pH 5. Administration of the drug was through daily oral gavage at a dose of 18 mg/kg. The solvent without drug was used as a placebo. Highly potent and specific inhibitors of CDK2 (K03861) and CDK9 (LDC000067) (Selleckchem) were dissolved in DMSO and diluted to the desired concentration in medium before use.
Cell lines and culture.
Jurkat and U-937 cells were obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program, Division of AIDS. Cells were grown in RPMI 1640 (Gibco) with 10% fetal bovine serum (FBS; Invitrogen), 5% penicillin-streptomycin (Invitrogen), and 2 mM glutamine (Invitrogen). Cells were maintained at a concentration of 106/ml in T-175 flasks. Cell concentrations and cell viability were monitored throughout the experiment at all time points studied.
Primary cell isolation.
Human blood buffy coats were ordered from Gulf Coast Regional Blood Center, and peripheral blood mononuclear cells (PBMCs) were separated with Ficoll (Ficoll-Paque Plus; GE Healthcare). Resting CD4+ T cells were purified by negative selection using a CD4 separation kit (Miltenyi Biotec) according to the manufacturer’s instructions. CD25− CD4+ T cells were subsequently isolated from CD4+ T cells by negative selection using CD25 beads (Miltenyi Biotec). CD14+ CD16− monocytes were isolated using the EasySep human monocyte isolation kit (STEMCELL). All cell purifications were performed according to the manufacturers’ instructions. The purity of CD25− CD4+ T cells was checked by staining with anti-CD4-FITC and anti-CD25-PE (Biolegend). The purity of monocytes was checked by staining with anti-CD3-FITC and anti-CD14-PE (Biolegend). Cell purity was analyzed by Guava Easycyte 8HT (EMD Millipore). The purity of each isolated population was over 90% (data not shown).
Primary cell transduction and culture.
CD4+ T cells were transduced with VSV-G Duo-Fluo reporter virus at a multiplicity of infection (MOI) of 0.1 and cultured in RPMI 1640 complemented with 10% FBS and interleukin 2 (IL-2) (10 U/ml) for 24 h before I-BET151 treatment. Monocyte-derived macrophages (MDMs) were obtained by culturing monocytes with macrophage colony-stimulating factor (M-CSF; 50 ng/ml) and granulocyte-macrophage colony-stimulating factor (GM-CSF; 50 ng/ml) in RPMI 1640 with 10% FBS for 7 days. MDMs were harvested and transduced with VSV-G Duo-Fluo reporter virus at an MOI of 0.1 and cultured for 24 h before I-BET151 treatment.
In vitro activation of HIV in cell lines.
Cells were transduced with VSV-G HIV-luc, in which luciferase gene expression is under the control of the HIV LTR, at an MOI of 1. After culture for 6 days, cells were treated with the CDK2 inhibitor (K03861, 1 μM) (1) or CDK9 inhibitor (LDC000067, 25 μM) (Selleck Chemicals) alone or in the presence of I-BET151. At 48 h after treatment, cells were washed with PBS and lysed in 100 μl of passive lysis buffer (Promega). Cell lysates were transferred into 96-well plates, and luciferase activity was detected by adding 50 μl of substrate into each well and read with a GloMax 96 microplate luminometer.
Statistical analysis.
Unpaired 2-tailed t test was applied for in vivo experiment with three samples in each group or in vitro assay with triplicates in each group. Mann-Whitney test was applied for all other analyses. All comparisons were performed using GraphPad Prism (GraphPad Software, San Diego, CA). A P value of <0.05 was considered statistically significant. All data were reported as means ± standard deviations (SD).
ACKNOWLEDGMENTS
We thank Qiong He and Adrain Kingsberry for technical assistance. We thank the University of North Carolina (UNC) Division of Laboratory Medicine for animal care and the UNC CFAR, Flow Cytometry Core Facility, and UNC Animal Histopathology Core Facility. We thank all members in the Su laboratory for critical reading and/or discussion of the paper and for their input and assistance.
This study was supported in part by the U.S. National Institutes of Health (AI080432, AI077454, and AI095097 to L.S.) and by GlaxoSmithKline.
REFERENCES
- 1.Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 2.Palella FJ Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N Engl J Med 338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 3.Broder S. 2010. The development of antiretroviral therapy and its impact on the HIV-1/AIDS pandemic. Antiviral Res 85:1–18. doi: 10.1016/j.antiviral.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu DC, Sereti I. 2016. Serious non-AIDS events: therapeutic targets of immune activation and chronic inflammation in HIV infection. Drugs 76:533–549. doi: 10.1007/s40265-016-0546-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cihlar T, Fordyce M. 2016. Current status and prospects of HIV treatment. Curr Opin Virol 18:50–56. doi: 10.1016/j.coviro.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Suphanchaimat R, Sommanustweechai A, Khitdee C, Thaichinda C, Kantamaturapoj K, Leelahavarong P, Jumriangrit P, Topothai T, Wisaijohn T, Putthasri W. 2014. HIV/AIDS health care challenges for cross-country migrants in low- and middle-income countries: a scoping review. HIV AIDS Auckl 6:19–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnighausen T, Salomon JA, Sangrujee N. 2012. HIV treatment as prevention: issues in economic evaluation. PLoS Med 9:e1001263. doi: 10.1371/journal.pmed.1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katlama C, Deeks SG, Autran B, Martinez-Picado J, van Lunzen J, Rouzioux C, Miller M, Vella S, Schmitz JE, Ahlers J, Richman DD, Sekaly RP. 2013. Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs. Lancet 381:2109–2117. doi: 10.1016/S0140-6736(13)60104-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. 2009. The challenge of finding a cure for HIV infection. Science 323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 11.Margolis DM. 2014. How might we cure HIV? Curr Infect Dis Rep 16:392. doi: 10.1007/s11908-014-0392-2. [DOI] [PubMed] [Google Scholar]
- 12.Natarajan V, Bosche M, Metcalf JA, Ward DJ, Lane HC, Kovacs JA. 1999. HIV-1 replication in patients with undetectable plasma virus receiving HAART. Lancet 353:119–120. doi: 10.1016/S0140-6736(05)76156-0. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Picado J, Deeks SG. 2016. Persistent HIV-1 replication during antiretroviral therapy. Curr Opin HIV AIDS 11:417–423. doi: 10.1097/COH.0000000000000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mavigner M, Habib J, Deleage C, Rosen E, Mattingly C, Bricker K, Kashuba A, Amblard F, Schinazi RF, Jean S, Cohen J, McGary C, Paiardini M, Wood MP, Sodora DL, Silvestri G, Estes J, Chahroudi A. 2018. Simian immunodeficiency virus persistence in cellular and anatomic reservoirs in antiretroviral therapy-suppressed infant rhesus macaques. J Virol 92:e00562-18. doi: 10.1128/JVI.00562-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malzahn J, Shen C, Caruso L, Ghosh P, Sankapal SR, Barratt-Boyes S, Gupta P, Chen Y. 2012. Effect of early anti-retroviral therapy on the pathogenic changes in mucosal tissues of SIV infected rhesus macaques. Virol J 9:269. doi: 10.1186/1743-422X-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng L, Ma J, Li J, Li D, Li G, Li F, Zhang Q, Yu H, Yasui F, Ye C, Tsao LC, Hu Z, Su L, Zhang L. 2017. Blocking type I interferon signaling enhances T cell recovery and reduces HIV-1 reservoirs. J Clin Invest 127:269–279. doi: 10.1172/JCI90745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li G, Nunoya JI, Cheng L, Reszka-Blanco N, Tsao LC, Jeffrey J, Su L. 2017. Regulatory T cells contribute to HIV-1 reservoir persistence in CD4+ T cells through cyclic adenosine monophosphate-dependent mechanisms in humanized mice in vivo. J Infect Dis 216:1579–1591. doi: 10.1093/infdis/jix547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cary DC, Fujinaga K, Peterlin BM. 2016. Molecular mechanisms of HIV latency. J Clin Invest 126:448–454. doi: 10.1172/JCI80565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donahue DA, Wainberg MA. 2013. Cellular and molecular mechanisms involved in the establishment of HIV-1 latency. Retrovirology 10:11. doi: 10.1186/1742-4690-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igarashi T, Brown CR, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin MA. 2001. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc Natl Acad Sci U S A 98:658–663. doi: 10.1073/pnas.021551798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M. 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 22.Koenig S, Gendelman HE, Orenstein JM, Dal Canto MC, Pezeshkpour GH, Yungbluth M, Janotta F, Aksamit A, Martin MA, Fauci AS. 1986. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science 233:1089–1093. doi: 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- 23.Honeycutt JB, Wahl A, Baker C, Spagnuolo RA, Foster J, Zakharova O, Wietgrefe S, Caro-Vegas C, Madden V, Sharpe G, Haase AT, Eron JJ, Garcia JV. 2016. Macrophages sustain HIV replication in vivo independently of T cells. J Clin Invest 126:1353–1366. doi: 10.1172/JCI84456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambotte O, Taoufik Y, de Goer MG, Wallon C, Goujard C, Delfraissy JF. 2000. Detection of infectious HIV in circulating monocytes from patients on prolonged highly active antiretroviral therapy. J Acquir Immune Defic Syndr 23:114–119. doi: 10.1097/00126334-200002010-00002. [DOI] [PubMed] [Google Scholar]
- 25.Crowe SM, Sonza S. 2000. HIV-1 can be recovered from a variety of cells including peripheral blood monocytes of patients receiving highly active antiretroviral therapy: a further obstacle to eradication. J Leukoc Biol 68:345–350. [PubMed] [Google Scholar]
- 26.Price RW, Brew B, Sidtis J, Rosenblum M, Scheck AC, Cleary P. 1988. The brain in AIDS: central nervous system HIV-1 infection and AIDS dementia complex. Science 239:586–592. doi: 10.1126/science.3277272. [DOI] [PubMed] [Google Scholar]
- 27.Gorry PR, Howard JL, Churchill MJ, Anderson JL, Cunningham A, Adrian D, McPhee DA, Purcell DF. 1999. Diminished production of human immunodeficiency virus type 1 in astrocytes results from inefficient translation of gag, env, and nef mRNAs despite efficient expression of Tat and Rev. J Virol 73:352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arainga M, Edagwa B, Mosley RL, Poluektova LY, Gorantla S, Gendelman HE. 2017. A mature macrophage is a principal HIV-1 cellular reservoir in humanized mice after treatment with long acting antiretroviral therapy. Retrovirology 14:17. doi: 10.1186/s12977-017-0344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. 2007. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A 104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu P, Qu X, Shen Y, Jiang Z, Wang P, Zeng H, Ji H, Deng J, Yang X, Li X, Lu H, Zhu H. 2016. The BET inhibitor OTX015 reactivates latent HIV-1 through P-TEFb. Sci Rep 6:24100. doi: 10.1038/srep24100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan H, Lu P, Shen Y, Wang Y, Jiang Z, Yang X, Zhong Y, Yang H, Khan IU, Zhou M, Li B, Zhang Z, Xu J, Lu H, Zhu H. 2017. The bromodomain and extraterminal domain inhibitor bromosporine synergistically reactivates latent HIV-1 in latently infected cells. Oncotarget 8:94104–94116. doi: 10.18632/oncotarget.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darcis G, Kula A, Bouchat S, Fujinaga K, Corazza F, Ait-Ammar A, Delacourt N, Melard A, Kabeya K, Vanhulle C, Van Driessche B, Gatot JS, Cherrier T, Pianowski LF, Gama L, Schwartz C, Vila J, Burny A, Clumeck N, Moutschen M, De Wit S, Peterlin BM, Rouzioux C, Rohr O, Van Lint C. 2015. An in-depth comparison of latency-reversing agent combinations in various in vitro and ex vivo HIV-1 latency models identified bryostatin-1+JQ1 and ingenol-B+JQ1 to potently reactivate viral gene expression. PLoS Pathog 11:e1005063. doi: 10.1371/journal.ppat.1005063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee C, Archin N, Michaels D, Belkina AC, Denis GV, Bradner J, Sebastiani P, Margolis DM, Montano M. 2012. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J Leukoc Biol 92:1147–1154. doi: 10.1189/jlb.0312165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li G, Zhang Z, Reszka-Blanco N, Li F, Chi L, Ma J, Jeffrey J, Cheng L, Su L. 2019. Specific activation in vivo of HIV-1 by a bromodomain inhibitor from monocytic cells in humanized mice under antiretroviral therapy. bioRxiv 10.1101/375535. [DOI] [PMC free article] [PubMed]
- 35.Calvanese V, Chavez L, Laurent T, Ding S, Verdin E. 2013. Dual-color HIV reporters trace a population of latently infected cells and enable their purification. Virology 446:283–292. doi: 10.1016/j.virol.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chavez L, Calvanese V, Verdin E. 2015. HIV latency is established directly and early in both resting and activated primary CD4 T cells. PLoS Pathog 11:e1004955. doi: 10.1371/journal.ppat.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei DG, Chiang V, Fyne E, Balakrishnan M, Barnes T, Graupe M, Hesselgesser J, Irrinki A, Murry JP, Stepan G, Stray KM, Tsai A, Yu H, Spindler J, Kearney M, Spina CA, McMahon D, Lalezari J, Sloan D, Mellors J, Geleziunas R, Cihlar T. 2014. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog 10:e1004071. doi: 10.1371/journal.ppat.1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasmussen TA, Schmeltz Sogaard O, Brinkmann C, Wightman F, Lewin SR, Melchjorsen J, Dinarello C, Ostergaard L, Tolstrup M. 2013. Comparison of HDAC inhibitors in clinical development: effect on HIV production in latently infected cells and T-cell activation. Hum Vaccin Immunother 9:993–1001. doi: 10.4161/hv.23800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ammosova T, Berro R, Kashanchi F, Nekhai S. 2005. RNA interference directed to CDK2 inhibits HIV-1 transcription. Virology 341:171–178. doi: 10.1016/j.virol.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 41.Debebe Z, Ammosova T, Breuer D, Lovejoy DB, Kalinowski DS, Kumar K, Jerebtsova M, Ray P, Kashanchi F, Gordeuk VR, Richardson DR, Nekhai S. 2011. Iron chelators of the di-2-pyridylketone thiosemicarbazone and 2-benzoylpyridine thiosemicarbazone series inhibit HIV-1 transcription: identification of novel cellular targets—iron, cyclin-dependent kinase (CDK) 2, and CDK9. Mol Pharmacol 79:185–196. doi: 10.1124/mol.110.069062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agbottah E, de La Fuente C, Nekhai S, Barnett A, Gianella-Borradori A, Pumfery A, Kashanchi F. 2005. Antiviral activity of CYC202 in HIV-1-infected cells. J Biol Chem 280:3029–3042. doi: 10.1074/jbc.M406435200. [DOI] [PubMed] [Google Scholar]
- 43.Salerno D, Hasham MG, Marshall R, Garriga J, Tsygankov AY, Grana X. 2007. Direct inhibition of CDK9 blocks HIV-1 replication without preventing T-cell activation in primary human peripheral blood lymphocytes. Gene 405:65–78. doi: 10.1016/j.gene.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medina-Moreno S, Dowling TC, Zapata JC, Le NM, Sausville E, Bryant J, Redfield RR, Heredia A. 2017. Targeting of CDK9 with indirubin 3′-monoxime safely and durably reduces HIV viremia in chronically infected humanized mice. PLoS One 12:e0183425. doi: 10.1371/journal.pone.0183425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okamoto M, Hidaka A, Toyama M, Hosoya T, Yamamoto M, Hagiwara M, Baba M. 2015. Selective inhibition of HIV-1 replication by the CDK9 inhibitor FIT-039. Antiviral Res 123:1–4. doi: 10.1016/j.antiviral.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 46.Jerebtsova M, Kumari N, Xu M, de Melo GB, Niu X, Jeang KT, Nekhai S. 2012. HIV-1 resistant CDK2-knockdown macrophage-like cells generated from 293T cell-derived human induced pluripotent stem cells. Biology (Basel) 1:175–195. doi: 10.3390/biology1020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alexander LT, Mobitz H, Drueckes P, Savitsky P, Fedorov O, Elkins JM, Deane CM, Cowan-Jacob SW, Knapp S. 2015. Type II inhibitors targeting CDK2. ACS Chem Biol 10:2116–2125. doi: 10.1021/acschembio.5b00398. [DOI] [PubMed] [Google Scholar]
- 48.Thompson KA, Cherry CL, Bell JE, McLean CA. 2011. Brain cell reservoirs of latent virus in presymptomatic HIV-infected individuals. Am J Pathol 179:1623–1629. doi: 10.1016/j.ajpath.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hellmuth J, Valcour V, Spudich S. 2015. CNS reservoirs for HIV: implications for eradication. J Virus Erad 1:67–71. [PMC free article] [PubMed] [Google Scholar]
- 50.Gendelman HE, Orenstein JM, Martin MA, Ferrua C, Mitra R, Phipps T, Wahl LA, Lane HC, Fauci AS, Burke DS. 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J Exp Med 167:1428–1441. doi: 10.1084/jem.167.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carter CA, Ehrlich LS. 2008. Cell biology of HIV-1 infection of macrophages. Annu Rev Microbiol 62:425–443. doi: 10.1146/annurev.micro.62.081307.162758. [DOI] [PubMed] [Google Scholar]
- 52.Zhen A, Rezek V, Youn C, Lam B, Chang N, Rick J, Carrillo M, Martin H, Kasparian S, Syed P, Rice N, Brooks DG, Kitchen SG. 2017. Targeting type I interferon-mediated activation restores immune function in chronic HIV infection. J Clin Invest 127:260–268. doi: 10.1172/JCI89488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao Y, Cartwright EK, Silvestri G, Perelson AS. 2018. CD8+ lymphocyte control of SIV infection during antiretroviral therapy. PLoS Pathog 14:e1007350. doi: 10.1371/journal.ppat.1007350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 55.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 56.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jorajuria S, Dereuddre-Bosquet N, Becher F, Martin S, Porcheray F, Garrigues A, Mabondzo A, Benech H, Grassi J, Orlowski S, Dormont D, Clayette P. 2004. ATP binding cassette multidrug transporters limit the anti-HIV activity of zidovudine and indinavir in infected human macrophages. Antivir Ther 9:519–528. [PubMed] [Google Scholar]
- 58.Zha W, Wang G, Xu W, Liu X, Wang Y, Zha BS, Shi J, Zhao Q, Gerk PM, Studer E, Hylemon PB, Pandak WM Jr, Zhou H. 2013. Inhibition of P-glycoprotein by HIV protease inhibitors increases intracellular accumulation of berberine in murine and human macrophages. PLoS One 8:e54349. doi: 10.1371/journal.pone.0054349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choo EF, Leake B, Wandel C, Imamura H, Wood AJ, Wilkinson GR, Kim RB. 2000. Pharmacological inhibition of P-glycoprotein transport enhances the distribution of HIV-1 protease inhibitors into brain and testes. Drug Metab Dispos 28:655–660. [PubMed] [Google Scholar]
- 60.Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Kosakovsky Pond SL, Chung YS, Penugonda S, Chipman JG, Fletcher CV, Schacker TW, Malim MH, Rambaut A, Haase AT, McLean AR, Wolinsky SM. 2016. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 530:51–56. doi: 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451–462. doi: 10.1016/S0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 62.Breuer D, Kotelkin A, Ammosova T, Kumari N, Ivanov A, Ilatovskiy AV, Beullens M, Roane PR, Bollen M, Petukhov MG, Kashanchi F, Nekhai S. 2012. CDK2 regulates HIV-1 transcription by phosphorylation of CDK9 on serine 90. Retrovirology 9:94. doi: 10.1186/1742-4690-9-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ammosova T, Berro R, Jerebtsova M, Jackson A, Charles S, Klase Z, Southerland W, Gordeuk VR, Kashanchi F, Nekhai S. 2006. Phosphorylation of HIV-1 Tat by CDK2 in HIV-1 transcription. Retrovirology 3:78. doi: 10.1186/1742-4690-3-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garber ME, Mayall TP, Suess EM, Meisenhelder J, Thompson NE, Jones KA. 2000. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA. Mol Cell Biol 20:6958–6969. doi: 10.1128/MCB.20.18.6958-6969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pauls E, Ruiz A, Badia R, Permanyer M, Gubern A, Riveira-Muñoz E, Torres-Torronteras J, Alvarez M, Mothe B, Brander C, Crespo M, Menéndez-Arias L, Clotet B, Keppler OT, Martí R, Posas F, Ballana E, Esté JA. 2014. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J Immunol 193:1988–1997. doi: 10.4049/jimmunol.1400873. [DOI] [PubMed] [Google Scholar]
- 66.Abner E, Stoszko M, Zeng L, Chen HC, Izquierdo-Bouldstridge A, Konuma T, Zorita E, Fanunza E, Zhang Q, Mahmoudi T, Zhou MM, Filion GJ, Jordan A. 2018. A new quinoline BRD4 inhibitor targets a distinct latent HIV-1 reservoir for reactivation from other “shock” drugs. J Virol 92:e02056-17. doi: 10.1128/JVI.02056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang Q, Zhang L, Wang R, Jeffrey J, Washburn ML, Brouwer D, Barbour S, Kovalev GI, Unutmaz D, Su L. 2008. FoxP3+CD4+ regulatory T cells play an important role in acute HIV-1 infection in humanized Rag2−/−gammaC−/− mice in vivo. Blood 112:2858–2868. doi: 10.1182/blood-2008-03-145946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li G, Cheng M, Nunoya J, Cheng L, Guo H, Yu H, Liu YJ, Su L, Zhang L. 2014. Plasmacytoid dendritic cells suppress HIV-1 replication but contribute to HIV-1 induced immunopathogenesis in humanized mice. PLoS Pathog 10:e1004291. doi: 10.1371/journal.ppat.1004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oswald-Richter K, Grill SM, Shariat N, Leelawong M, Sundrud MS, Haas DW, Unutmaz D. 2004. HIV infection of naturally occurring and genetically reprogrammed human regulatory T-cells. PLoS Biol 2:E198. doi: 10.1371/journal.pbio.0020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L, Jiang Q, Li G, Jeffrey J, Kovalev GI, Su L. 2011. Efficient infection, activation, and impairment of pDCs in the BM and peripheral lymphoid organs during early HIV-1 infection in humanized rag2(−)/(−)gamma C(−)/(−) mice in vivo. Blood 117:6184–6192. doi: 10.1182/blood-2011-01-331173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang L, Kovalev GI, Su L. 2007. HIV-1 infection and pathogenesis in a novel humanized mouse model. Blood 109:2978–2981. doi: 10.1182/blood-2006-07-033159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Halper-Stromberg A, Lu CL, Klein F, Horwitz JA, Bournazos S, Nogueira L, Eisenreich TR, Liu C, Gazumyan A, Schaefer U, Furze RC, Seaman MS, Prinjha R, Tarakhovsky A, Ravetch JV, Nussenzweig MC. 2014. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell 158:989–999. doi: 10.1016/j.cell.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]