Abstract
RMEL3 is a recently identified lncRNA associated with BRAFV600E mutation and melanoma cell survival. Here, we demonstrate strong and moderate RMEL3 upregulation in BRAF and NRAS mutant melanoma cells, respectively, compared to melanocytes. High expression is also more frequent in cutaneous than in acral/mucosal melanomas, and analysis of an ICGC melanoma dataset showed that mutations in RMEL3 locus are preponderantly C > T substitutions at dipyrimidine sites including CC > TT, typical of UV signature. RMEL3 mutation does not correlate with RMEL3 levels, but does with poor patient survival, in TCGA melanoma dataset. Accordingly, RMEL3 lncRNA levels were significantly reduced in BRAFV600E melanoma cells upon treatment with BRAF or MEK inhibitors, supporting the notion that BRAFMEK- ERK pathway plays a role to activate RMEL3 gene transcription. RMEL3 overexpression, in immortalized fibroblasts and melanoma cells, increased proliferation and survival under serum starvation, clonogenic ability, and xenografted melanoma tumor growth. Although future studies will be needed to elucidate the mechanistic activities of RMEL3, our data demonstrate that its overexpression bypasses the need of mitogen activation to sustain proliferation/survival of non-transformed cells and suggest an oncogenic role for RMEL3.
Keywords: BRAFV600E, chr5:57395060-57533424 (GRCh38/hg38), CTD-2023N9.1, ENSG00000250961.1, LncGPBP1-1:1, MAPK, melanoma, mitogen, serum response
Introduction
Melanoma is a highly mutated and aggressive type of cancer originated from the malignant transformation of melanocytes. Most commonly, melanoma arises from skin melanocytes (acral and non-acral cutaneous melanoma), but it can occasionally originate from melanocytes present in other parts of the body, such as meninges, cochlea, the mucosae (mucosal melanoma), and the uvea of the eye (uveal melanoma). Acral melanoma, a relatively rare subtype, arises from non-hair-bearing skin locations, such as the palms of the hands, the soles of the feet, or the nail bed (subungual areas). The non-acral cutaneous melanoma comprises three major subtypes, superficial spreading melanoma, which is the most prevalent form and usually occurs in the trunk; nodular melanoma, the second most prevalent and highly invasive form; and lentigo maligna melanoma, associated with long-term sun-exposed skin (Scolyer, Long, & Thompson, 2011). Most non-acral cutaneous melanoma are considered sporadic in nature, and recente genomewide mutational studies show that they are associated with the ultraviolet light (UV) signature, implicating sunlight exposure as a causal factor (Hayward et al., 2017).
Whole exome sequencing (The Cancer Genome Atlas Network, 2015) led to the genomic classification of cutaneous melanoma into four subclasses according to cancer driver mutations: mutante BRAF (~52%), mutant RAS (~30%); mutant NF-1 (~14%); and triple wild-type, those with no mutations in any of the three genes. All three genes encode components of the classical mitogen-activated protein kinase (MAPK/ERK) cascade, encompassing RASRAF-MEK-ERK, the major pathway that transmits extracellular mitogen signals downstream from activated tyrosine kinase receptors to elicit diverse cellular responses, such as growth, motility, and survival. While BRAF and RAS are oncogenes, NF-1 is a tumor suppressor gene that codes for a RAS-GAP, responsible for the inactivation of RAS. Therefore, mutations in BRAF, RAS, and NF-1 genes seem to lead to equivalent constitutive activation of the MAPK/ERK pathway, and for this reason, or perhaps due to deleterious effects of coexisting mutations, they almost always are found to be mutually exclusive. Activating mutations of genes implicated in the PI3K pathway are also highly frequent in melanoma, and activation of PI3K-AKT mTOR signaling pathway cooperates with the MAPK pathway to set the scenario of sustained growth and death resistance in melanoma. Driver mutations coexist with activating mutations in the TERT gene promoter or in other genes implicated in telomere maintenance required for replicative immortality (Hayward et al., 2017).
Inhibitors of the mutated BRAF kinase (vemurafenib and dabrafenib), as well as a combination of BRAF inhibitor with MEK inhibitor (trametinib) and, more recently, immunotherapy are in clinical use to treat melanoma with unprecedented success (for review, Luke, Flaherty, Ribas, & Long, 2017; Melino et al., 2016). However, therapeutic resistance and side effects remain major challenge, and not all patients can benefit from these treatments.
The discovery of new genes with tissue-specific expression and associated with genetic events of BRAF-activating mutations may provide new insights into the field and opportunities for novel therapeutic approaches. Long non-coding RNAs (lncRNAs) have recently emerged as numerous and diversified functional RNAs with tissue, developmental stage and disease-restricted expression. Genomewide association studies have recently mapped a large proportion of disease- or trait-associated single nucleotide polymorphisms outside the protein coding regions of the genome, often in or near genes transcribed into lncRNAs (Bartonicek et al., 2017; Gong, Liu, Zhang, Miao, & Guo, 2015). There are bona fide examples of lncRNA linked to one or multiple hallmarks of cancer in a variety of cancers, including melanoma, in which many emerge as regulators of the MAPK and PI3K pathways (for review, Huang et al., 2018; Leucci, Coe, Marine, & Vance, 2016; Schmitt & Chang, 2016; Tasharrofi & Ghafouri-Fard, 2018).
RMEL3 lncRNA was first identified in a previous study of our group as a melanoma-restricted gene in a data mining analysis of ESTs exclusively originated from melanoma sources (Sousa et al., 2010). RMEL3 is not expressed in melanocytes, but it is widely expressed in nevi, primary, and metastatic melanoma. It was subsequently linked to BRAFV600E mutation, based on expression analysis in a large set of melanoma cell lines and the TCGA melanoma dataset (The Cancer Genome Atlas Network, 2015) then available (Goedert et al., 2016). The later work also demonstrates that RMEL3 knockdown leads to alterations in the expression levels of mRNA and proteins of many components of the MAPK and PI3K pathways. This result was consistent with the observed loss of clonogenic ability and increased arrest in the G1 phase of the cell cycle in a set of RMEL3-knockdown cells, with stronger effects in cells carrying BRAFV600E instead of wildtype BRAF. Here, we extended the previous studies by analyzing RMEL3 expression in a new panel of melanoma cell lines, melanocytes, and melanoma tissues, as well as examining the effects of BRAF and MEK inhibition on RMEL3 expression. We further demonstrated that enforced expression of RMEL3 in melanoma cells (Skmel-103, VM10), as well as in the immortalized murine NIH3T3 fibroblasts, promotes proliferation, clonogenic growth, cell survival, and tumor growth. Taken together, our data reinforce previous connections of RMEL3 to the MAPK pathway and demonstrate that this lncRNA plays active roles in sustaining cell survival and tumor growth.
Materials and Methods
Cell culture
Human melanoma cell lines WM278 and WM1617 were kindly provided by Dr. Meenhard Herlyn (Wistar Institute, Philadelphia, PA), and we did an HLA typing for the cells that we carry in our laboratory and confirmed that they constitute a pair of cell lines originated from the same patient, as originally described. The cells were maintained in TU medium with 2% (v/v) fetal bovine serum (FBS). All the following cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 50 U/mL penicillin, and 50 mg/ml streptomycin. The melanoma cell lines (WM1366, Skmel-147, Skmel-103, Skmel-19, Skmel-28, and UACC-62) were kindly provided by Dr. Silvya Stuchi Maria- Engler (University of São Paulo) and Dr. Marisol Soengas (CNIO, Madrid, Spain). The A375 melanoma cell line was provided by Dr. Wilson Araujo da Silva Jr. (University of São Paulo), and NIH3T3 cell line was provided by Dr. Gregg G. Gundersen (Columbia University, New York). The melanoma cell lines VM10, WM2029, MeWo, Mel-Juso, LB373, WM451LU, IPC298, COLO792, Skmel-90, 501mel, Sk-mel-119, Sk-mel-2, MDAMB435S, WM1716, COLO829, A101D, C32, G361, MALME3M, Sk-mel-5, Skmel-28 and M14, Sk-mel30, LOX-IMVI, WM115, WM88, UACC62 e UACC257 were provided by Dr. David E Fisher’s laboratory (MGH, Harvard Medical School). 501mel cells were the generous gift of Dr. Ruth Halaban (Yale University). Primary melanocytes were cultured in TIVA medium (F-10 with 10% FBS, 50 ng/ml TPA, 225 μM IBMX, 1 μM Na3VO4, and 1 mM dbcAMP; all from Sigma-Aldrich, St. Louis, MO). All cultures were kept at 37°C under a 5% CO2 humidified atmosphere.
Human melanoma tissue samples
Melanoma samples were obtained from 38 patients at the Barretos Cancer Hospital. Tumor samples were frozen in liquid nitrogen and kept in −80°C freezer until RNA extraction. Tissues samples are stored in the Barretos Cancer Hospital Biobank, which is approved by the Brazilian National Committee on Ethics in Research— CONEP. This study and the research protocols were approved by the Ethics Committee of the Barretos Cancer Hospital (HCB#548/2011).
RNA isolation
Total RNA was extracted using TRIzol® (Invitrogen, Carlsbad, CA). It was then treated with DNase I (DNA-free kit, Ambion, Austin, TX) and converted into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). All protocols were performed according to manufacturer’s instructions.
Expression analysis by RT-qPCR
For the reverse transcription reaction, 1 μg of DNase-treated total RNA was diluted to 16μL of DEPC-treated H2O and added with 4μL do Master Mix (of the High-Capacity RNA-to-cDNA Master Mix kit from Applied Biosystems). The reaction was incubated at 25°C for 5 min, 42°C for 30 min, and the reverse transcriptase was inactivated by heating at 85°C for 5 min. Equal amounts of each cDNA were assayed by qPCR, with specific primers, whose sequences are specified below, using the SYBR Green® PCR Power Mix 2x (Applied Biosystems, Foster City, CA), in an ABI PRISM 7,500 Sequence Detection System (Applied Biosystems, Foster City, CA).
RMEL3 (f) 5’-ATGTGCTCCAAGAAAACCAGAG-3’ and (r), 5’-CTTTGTCACAGGAATACCCAAC-3’;
FOXD3 (f) 5’-TTGACGAAGCAGTCGTTGAG-3’ and (r) 5’-TCTGCGAGTTCATCAGCAAC-3’;
TBP (f) 5’- AGCTGTGATGTGAAGTTTCC-3’ and (r) 5’ TCTGGGTTTGATCATTCTGTAG-3’.
Cycle threshold (Ct) was converted to relative expression according to the 2-ΔΔCT method, using TBP (TATA-box binding protein) as endogenous control.
Pharmacological treatment and viability assay
PLX4032/vemurafenib (Sigma-Aldrich, St. Louis, MO) and PD98059 (Sigma-Aldrich, St. Louis, MO) were dissolved in DMSO to 10 mM stock solution and stored in aliquots at −20°C. Melanoma cell lines harboring BRAFV600E mutation and BRAF wild-type cell lines were seeded in 24-well plate at a density of 3×104 cells/well and treated with 1 or 10 μM PLX4032, 25 μM PD98059, or DMSO as vehicle control. Treatment with vemurafenib was carried for 6 or 48 hr and with PD98059 for 48 hr at 37°C under a 5% CO2 atmosphere. For viability assays, both adherent and floating cells were harvested, washed with PBS, and resuspended in PBS containing 10 μg/mL of propidium iodide. These cells were counted in a flow cytometer (Guava® easyCyte™ 8HT Flow Cytometry System—Millipore).
Western blot analysis
Cells were homogenized in a sample buffer containing 0.1% SDS and subjected to SDS-PAGE. Proteins were blotted onto a nitrocellulose membrane (Hybon-ECL; Amersham Biosciences, Arlington, IL). Membranes were incubated with diluted primary antibody in TBS-Tween-20 0.05% + BSA 2% overnight at 4°C. Subsequently, the membrane was incubated with corresponding secondary antibody coupled with horseradish peroxidase (Nichirei Co, Tokyo, Japan). Peroxidase reaction products were visualized using the ECL Plus detection system (Amersham Biosciences). Antibodies were used according to the manufacturer-recommended dilution. Primary antibodies anti-phosphorylated ERK1/2 and anti-total ERK1/2 were purchased from Cell Signaling Technology, Beverly, MA, USA.
RMEL3 cDNA cloning, transduction, and expression
Total RNA from WM278 melanoma cells was extracted using TRIzol (Invitrogen, Carlsbad, CA) and reverse transcribed using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). Then, 1 μg cDNA was used as template for PCR amplification of RMEL3 cDNA using a forward primer, 5’- GGATCCTGAAGGAAGCAACTGGGGA-3’, carrying a BamHI site in the 5’-end, and a reverse primer, 5’-GAATTCCCCGAGTGTGGGATCA-3’, carrying an EcoRI site in the 5’-end. Primers were designed based on the EST sequence (BQ420825) deposited in the GenBank database. PCRs were done using Elongase Enzyme Mix (Invitrogen, Carlsbad, CA), and the protocol was performed according to manufacturer’s instruction (PCR cycling conditions: 1: 94°C-3 min, 2: 94°C-45 seg, 3: 56°C-45 seg, 4: 72°C-1 min, 72°C-7 min, steps 2–4 40 cycles). The PCR produced a DNA fragment of expected size (777 bp), which was purified and cloned into the TOPO plasmid and sequenced. After confirmation of the full-length sequence (Figure S3), RMEL3 insert released with BamHI and EcoRI was subcloned into the same restriction sites of pLVX-Tight PURO plasmid, downstream of the human cytomegalovirus immediate early promoter—CMV IE promoter and a Tet-On regulatory region (Clontech Laboratories, Inc). Purified plasmidial DNA was transfected into HEK293T cells with the lentiviral expression packaging system (Lenti-X Tet-on Advanced Inducible Expression—Clontech) according to the manufacturer’s instruction. Lentiviral particles collected from HEK293T culture media were used to transduce Skmel103 melanoma cells and NIH3T3 murine fibroblast. To generate stably transduced cells expressing RMEL3 transgene, transduced cells were selected with 1 μg/mL puromycin for 3 days. All control (non-transduced cells died under this concentration of Puromycin during this period). RMEL3 expression was induced with 1 μg/mL doxycycline supplemented in the culture media. For cloning into pLJM1 lentiviral vector (Addgene-19319 plasmid), under constitutive control of the CMV promoter, RMEL3 sequence was amplified from pLVX-RMEL3 as template using a pair of primers with sequences identical to the ones described above, but with different restriction sites (NheI and EcoRI). The PCR product was digested, purified, and cloned into the NheI and EcoRI restriction sites in the pLJM1 vector. Lentiviral particles were obtained by transfecting packaging cells (HEK293T) with the packaging plasmids (pMD2.G and pSPAX2) using PEI MAX (Polysciences, Inc.) method. Lentiviral particles collected from HEK293T culture media were used to transduce VM10 melanoma cell line. Infected cells were selected with 1 μg/mL puromycin for 3 days.
Proliferation assay
Cells were seeded in 96-well culture dishes at a density of 5.000 cells/well, for Skmel103 and VM10, and 500 cells/well for NIH3T3. The melanoma cell lines Skmel103 and VM10 were cultured in FBSfree DMEM and DMEM Complete Medium, respectively, whereas NIH3T3 cells were cultured in 0.5% FBS DMEM. Culture medium was removed, and cells were fixed with 70% ethanol, at room temperature for 10 min. Ethanol was removed, and crystal violet (0.5% in water) was added and incubated for 30 min at room temperature. The fixed cells were washed five times with water, followed by addition of 10% acetic acid, and incubated for 30 min at room temperature. Absorbance at 540 nm was measured using an ELISA microplate reader.
Clonogenic assay
Cells overexpressing RMEL3 and paired controls were plated in 60-mm tissue culture dishes (600 cells/dish) in DMEM supplemented with 10% FBS. Cells were incubated at 37°C, in 5% CO2 humidified atmosphere, for 9 days (NIH3T3) and 14 days (Skmel103). After the specified period, cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet for visualization of colonies. The plates were photographed, and the colonies were quantified to determine the clonogenic ability.
Apoptosis assay
NIH3T3 cells were seeded in 24-well plates at 2 × 104 cells/well in DMEM supplemented with 10% FBS. After 24 hr, the medium was changed to FBS-free medium for 48 hr and then to 0.5% FBS medium for additional 48 hr. For detection of apoptotic and unviable
cells, we used the annexin-V, Alexa Fluor® 488 conjugate (Life Technologies), and Propidium Iodide (Life Technologies). This protocol was performed according to manufacturer’s instructions.
Tumor xenograft assay
SKMEL103 melanoma cells stably transduced with pLVX-TP (control) or pLVX-RMEL3 were cultured in complete media supplemented with 1 μg/ml doxycycline to confluence. Ten 8-week-old male nude mice were injected, subcutaneously, in the right lateral flank, with 106 pLVXTP- or pLVX-RMEL3-transduced cells. Mice were maintained with diet ad libitum and drinking water was supplemented with 1 mg/ml of doxycycline and changed every 2 days. Animals were monitored daily. Tumor size was measured on days 8, 11, 14, 17, 21, and 24 after injections, and following this period the animals were euthanized. Tumor volumes were calculated using the formula V=LxW2×0.5, where W and L are, respectively, tumor width and length. All animal protocols and experiments were performed in accordance with the National Board for the Control of Animal Experimentation in Brazil (CONCEA) and were approved by the Committee for Ethics in the Use of Animals—CEUAFMRP- USP, approval ID 177/2016.
Somatic mutation analyses
Analysis of somatic mutation in the RMEL3 locus was performed using two data sources: (1) The Cancer Genome Atlas (TCGA) data portal and (2) The International Cancer Genome Consortium (ICGC) data portal. We analyzed the RMEL3 locus for the presence of mutation in 450 melanomas of the TCGA dataset and 129 of the ICGC.
Statistical analyses
All statistical analyses were performed using GraphPad Prism 5. All numerical data were expressed as the mean ±standard deviation.
RMEL3 high levels are associated with BRAF and NRAS mutations in melanoma cell lines and to cutaneous melanoma as opposed to acral and mucosal
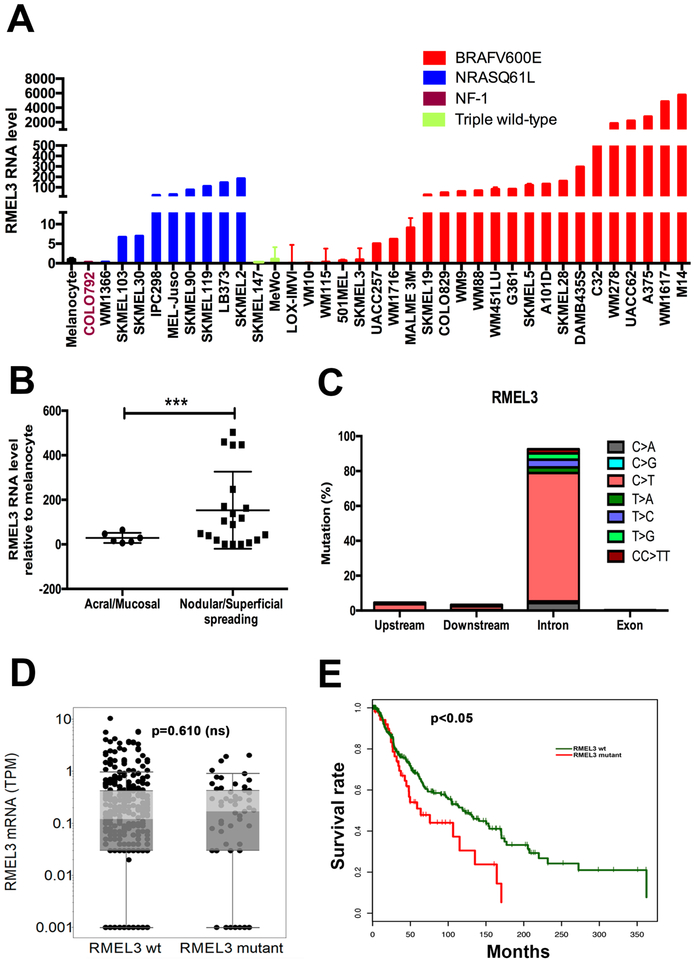
To assess the endogenous RMEL3 expression, we analyzed a panel of 36 human melanoma cell lines and 2 independent cultures of primary human melanocytes, by RT-qPCR (Figure 1a). RMEL3 showed low expression levels in two melanocyte cultures and in 9/36 (25%) of melanoma cell lines. Median levels were observed in 8/36 (22%) and high (above 40-fold higher than melanocytes) in 19/36 (53%) of cell lines. Among the lines with high expression 15/19 (~79%) harbor BRAFV600E and 4/19 (~21%) NRASQ61L. Of the lines harboring BRAFV600E 15/24 (62,5%) showed high and 9/24 (37,5%) median/low RMEL3 expression. Low levels were also observed in triple wild-type cells (Sk-mel147 and Mewo) and in one NF1 mutant (COLO792). These results extend previous studies by Sousa et al. (2010), and Goedert et al. (2016) and are in agreement with them. Analysis of RMEL3 expression across melanoma specimens from 38 patients, who were attended in the Barretos Cancer Hospital (Brazil), showed a significant association of high expression of RMEL3 to extensive superficial or nodular cutaneous (16 of 31) in contrast to acral and mucosal (0 of 7) melanomas (Figure 1b; p = 0.014—Fisher exact test). In order to clarify whether the RMEL3 locus (CTD-2023N9.1) could comprise a mutation hotspot associated with its upregulation in non-acral cutaneous melanoma, we searched for mutations in a region encompassing 20-kb upstream (including enhancer/ promoter regions) of the mapped start site of RMEL3, the entire body of the gene, and 20-kb downstream of the gene, both in a TCGA dataset of 450 melanomas (SKCM) and in an ICGC dataset of 129 melanomas (MELA-AU). We found that 108 of 450 melanomas from TCGA presented mutation in the body of the gene, including ACTBL2 which maps within the intron 1 of RMEL3 gene, with 32 mutated melanomas (Table S1). In the ICGC dataset, we found at least one mutation in 129 out of 129 melanomas, totaling 595 mutations, of which 28 mutations mapped upstream and 18 mutations downstream of the gene and the remaining majority of them were in the intronic regions, including ACTBL2 with 50 mutations (Table S2). Except for 16 insertions or deletions in ICGC and 4 in TCGA, all the others were base substitutions. A summary of the number of different types of mutations detected in the TCGA and ICGC databanks is presented in Table S3. We understand that the reduced number of mutations observed in the TCGA dataset compared to ICGC can be explained due to methodological differences, since TCGA was based on whole exome sequencing (WES) while ICGC on whole genome sequencing (WGS). This difference also explains the fact that we did not find mutations upstream nor downstream of the gene in the TCGA samples. Quantification of the base substitution mutations in the ICGC data showed high frequency (>70%) of C > T substitutions with occurrence of ~3% CC > TT double substitution, suggesting that the majority of all base substitutions in RMEL3 gene are characteristic of the UV signature mutations (Figure 1c). Analysis of the genomic context to define the neighbor nucleotides and graphical representation (as in Alexandrov et al., 2013) generated the approximate pattern of the UV mutational signature, as in COSMIC signature 7 (FigureS1). Nevertheless, none of the mutations upstream nor downstream the gene were recurrent and only three of the intronic mutations occurred twice; thus, they do not appear to characterize a hotspot. Then, we correlated the RMEL3 mutations (excluding the ACTBL2 mutations) found in TCGA samples with the RMEL3 expression levels from RNAseq data available in the TCGA, and as shown in Figure 1d, there was no statistically significant correlation between mutation and expression. Therefore, the occurrence of these mutations does not explain the differential expression observed in Figure 1a,b. Finally, using the TCGA dataset, we found that RMEL3 mutations are associated with poor patient survival rates (Figure 1e) (p < 0.05).
Figure 1. High RMEL3 expression predominates in BRAFV600E followed by NRAS mutant melanoma cell lines, as well as in nodular/ superficial spreading in comparison with acral/mucosal melanomas.
(A) RT-qPCR analysis of RMEL3 RNA levels in melanocytes and melanoma cell lines of indicated genotype regarding major câncer drivers (BRAF, NRAS, and NF1). (B) RT-qPCR analysis of RMEL3 expression in melanoma subtypes: acral/mucosal and nodular/superficial spreading cutaneous melanoma, for which patient specimens were obtained from the Barretos Cancer Hospital, São Paulo state, Brazil. In A and B, relative expression was calculated according to 2−ΔΔCT method usingTBP (Tata-box binding protein) as endogenous control, and the mean levels of RMEL3 RNA in different foreskin human melanocyte primary cultures were used as reference in both A and B, totaling the use of four different melanocyte cultures derived from different donors. Error bars represent SEM of 3 independent experiments. ***p < 0.005. Asterisks indicate statistically significant differences between groups based on Fisher’s exact test. (C) Frequency of base substitution mutations in the RMEL3 locus from a total of 579 base substitutions reported by ICGC in a set of 129 melanoma samples. (D) RMEL3 expression levels (TPM) grouped according to the occurrence (RMEL3 mutant) or not (RMEL3 wt) of somatic mutation in the RMEL3 gene detected in the TCGA melanoma dataset of 450 samples. (E) Survival curves for melanoma patients of the TCGA database carrying (RMEL3 mutant) or not (RMEL3 wt) mutations in the RMEL3 gene. Statistical analyses used chi-squared test in D and E
Acute and prolonged MAPK inhibition decreases RMEL3 RNA levels in melanoma cells
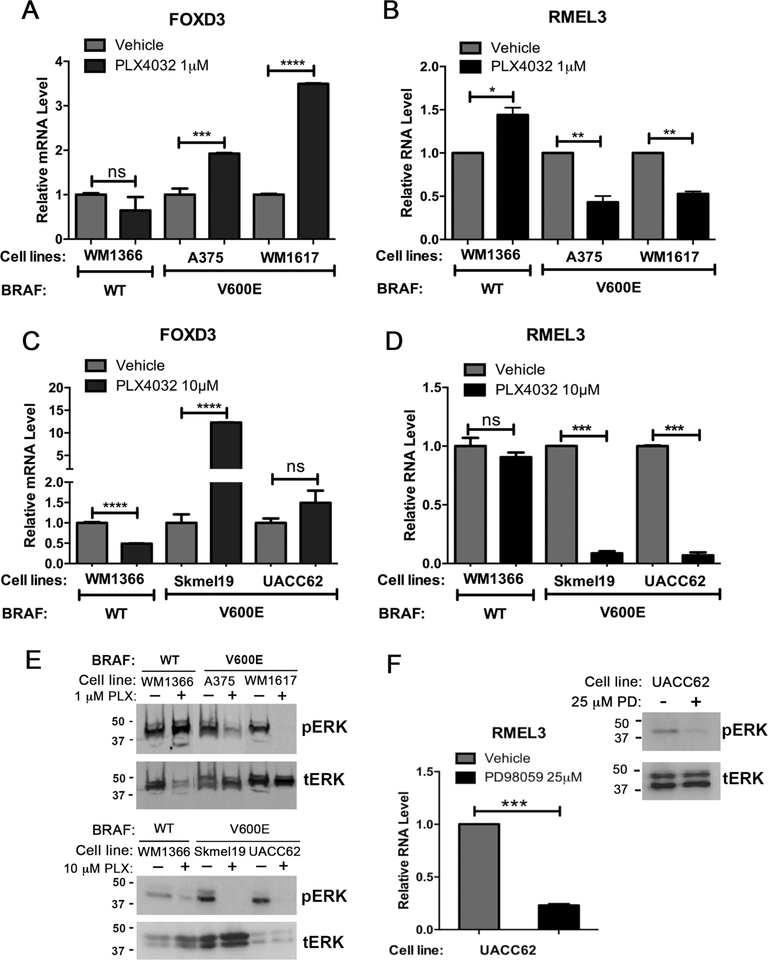
We used specific drugs against oncogenic BRAF (vemurafenib/PLX4032) and MEK1 (PD98059) in order to inhibit the MAPK pathway in a panel of five melanoma cell lines: 4 harboring BRAFV600E mutation (WM1617, A375, Skmel19, and UACC-62) and one BRAF-wt/NRAS-mutant (WM1366). As shown in Figure 2 (A–E), vemurafenib treatments were done in two conditions: low drug concentration (1.0 μM) for short exposure time (6 hr); high drug concentration (10 μM) for prolonged exposure (48 hr). RMEL3 RNA levels were reduced by approximately 50% in A375 and WM1617 BRAFV600E mutant cells after short-term, low-concentration treatment (Figure 2a) and 90% in Skmel19 and UACC62 after prolonged drug exposure (Figure 2b). Paradoxically, the NRAS mutant WM1366 cells showed an increase in RMEL3 expression following short-term treatment (Figure 2a). FOXD3 (Forkhead Box D3), which was previously shown to be upregulated after mutant BRAF inhibition (Abel & Aplin, 2010), was used here as positive control and confirmed to exhibit the expected expression pattern (Figure 2c,d). Analysis of total ERK and phosphorylated ERK (Figure 2e) demonstrates that both drug treatments effectively decreased pERK levels in the BRAFV600E mutant cells, whereas in WM1366, treatment with BRAF inhibitor increased pERK levels in the acute treatment, consistent with previously described paradoxical activation of ERK. Next, we showed that treatment of UACC62 cells with the MEK inhibitor PD98059 (25 μM for 48 hr) reduced RMEL3 RNA levels by 77%, a condition in which pERK levels were also efficiently decreased (Figure 2f). As expected, treatments for 48 hr led to growth arrest, but did not cause a significant alteration in cell death rates (Figure S2).
Figure 2. RMEL3 lncRNA levels decrease in BRAFV600E mutant melanoma cells after treatment with BRAF and MEK inhibitors.
(A - D) RT-qPCR expression analyses of (A, B) RMEL3 lncRNA and (C, D) FOXD3 (positive control) in melanoma cell lines treated with the BRAF inhibitor vemurafenib (PLX4032) (1 μM, 6 h or 10 μM, 48 h) and respective controls (vehicle), as indicated. (E) Western blot analysis for phosphorylated ERK (pERK) and total ERK (tERK) in total protein lysates from melanoma cells treated with BRAF inhibitor (1 μM, 6 h or 10 μM, 48 h) or vehicle. (F) RT-qPCR expression analysis of RMEL3 in UACC-62 melanoma cell line treated with the MEK inhibitor PD98059 (25 μM) or vehicle for 48 hr, and corresponding Western blot analysis for phosphorylated ERK (pERK) and total ERK (tERK) in total protein lysates of treated and control melanoma cells. For all RT-qPCR analyses, relative expression was calculated according to 2−ΔΔCT method using TBP (Tata-box binding protein) as endogenous control and the normalized Ct of the sample treated with the vehicle alone as reference. Error bars represent SEM of 3 independent experiments. *p < 0.05; **p < 0.005; ***p < 0.0005; ****p < 0.00005. Asterisks indicate statistically significant differences between groups based on unpaired parametric Student’s t test
RMEL3 expression in the NIH3T3 immortalized murine fibroblasts protects against serum withdrawal-induced growth arrest/apoptosis and promotes clonogenic ability
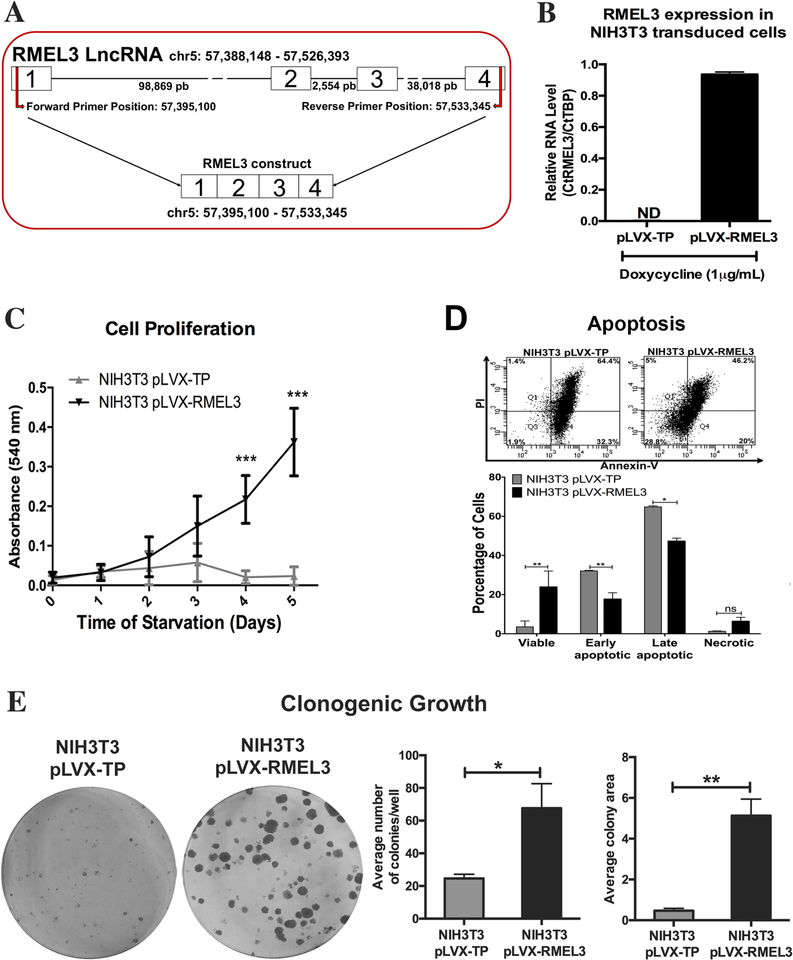
NIH3T3 murine fibroblasts constitute a well-established model in which the MAPK pathway is activated by serum stimulation to maintain cell cycle progression and survival (Meloche & Pouysségur, 2007). Thus, we used the NIH3T3 cells to investigate the potential of RMEL3 to mediate cell proliferation and survival upon serum starvation. In order to overexpress exogenous RMEL3, we cloned the human RMEL3 cDNA, as represented in Figure 3a, into lentiviral expression vectors. The full insert is 777 bp in length, and the sequence (Figure S3) was deposited in the GenBank under the accession number MH370349. Alignments of the cloned sequence with the human genomic DNA (GRCh38.p12) match four exons in the assembly NC_000005.10 (chr5: 57395100–57533345) with one gap representing a single A deletion in the position 762 of the insert, corresponding to 802 of the transcript (chromosome position 57533328 in the same assembly, near the end of exon 4). The fragment lacks 40 bp of the 5’ region of exon 1 and 79 bp of the 3’ end of exon 4. Primers matching the very end of the sequence were not efficient for amplification, what explains the option to clone a partial sequence. The full-length RMEL3 sequence is represented by CTD-2023N9.1 (LNC-GPBP1-1:1), which contains 897 bp. We achieved significant levels of RMEL3 RNA expression, in comparison with cells harboring the empty vector (below cycle threshold), upon induction with 1 μg/mL doxycycline (Figure 3b). Serum-starved NIH3T3 cells are known to exit the cell cycle and become quiescent within 48 hr, and present high death rates if serum deprivation is prolonged. However, RMEL3-overexpressing NIH3T3 cells were able to continually undergo proliferation upon serum starvation (0.5% FBS), for at least 5 days, while control cells stopped proliferation and showed high rates of death after 3 days of starvation (Figure 3c). We then evaluated the effect of RMEL3 overexpression on cell survival under serum starvation. To this end, cells were cultured in serum-free medium for 48 hr, and then, the medium was supplemented with 0.5% FBS, for a further 48-hr period, after which the cells were assayed for apoptosis (Figure 3d). By the end of this regimen, the majority (~97%) of control cells were in early or late stages of apoptotic cell death as opposed to ~65% of RMEL3-overexpressing cells (Figure 3d). Also, RMEL3-overexpressing NIH3T3 cells gained enhanced capacity of clonogenic growth as compared to control (Figure 3e), increasing both colony number and colony size.
Figure 3. Ectopic expression of human RMEL3 protects NIH3T3 murine fibroblasts from serum withdrawal-induced growth arrest/ apoptosis.
(A) Schematic representation of RMEL3 locus (above), its transcript (center), and the region cloned into expression vectors (below). (B) Efficiency of exogenous RMEL3 induction in cells transduced with pLVX-RMEL3 or pLVX-TP (as control) after treatment with doxycycline (1 μM, 24 h), analyzed by RT-qPCR. ND (not-detected). Relative expression was calculated according to ΔCT using TBP (Tatabox binding protein) as endogenous control. (C-E) Functional assays using NIH3T3 cells stably transduced with pLVX-RMEL3 or pLVX-TP (as control) in the presence of doxycycline (1 μM). (C) Proliferation rates. During the time course of the assay, cells were maintained under serum starvation (in medium supplemented with 0.5% FBS). After the indicated time points, cells were stained with crystal violet and cell density was quantified according to the absorbance in an ELISA microplate reader. ***p < 0.0005 (D) Apoptosis rate. Flow cytometry-based detection of annexin-V- and propidium iodide (PI)-stained cells. Dot plot from one assay representative of three replicates, and below, a summary graphics of three independent replicates. Cells were cultured in FBS-free medium for 48 h, and afterward, culture medium was supplemented with 0.5% FBS and cells were cultured for additional 48 h, when they were assayed. *p < 0.05; **p < 0.005. (E) Clonogenic ability. Cells were seeded in 60-mm-diameter plates and allowed to grow for 9 days, when they were fixed with paraformaldehyde and stained with crystal violet to reveal the colonies. *p < 0.05; **p < 0.005. Error bars represent SEM of 3 independent experiments for B-E. Asterisks indicate statistically significant differences between groups based on unpaired parametric Student’s t test
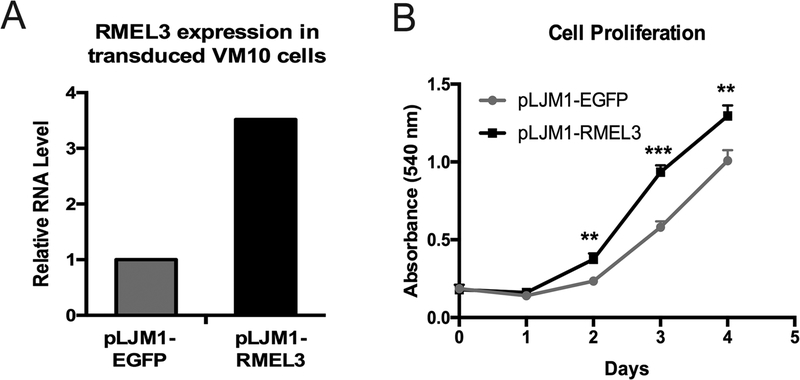
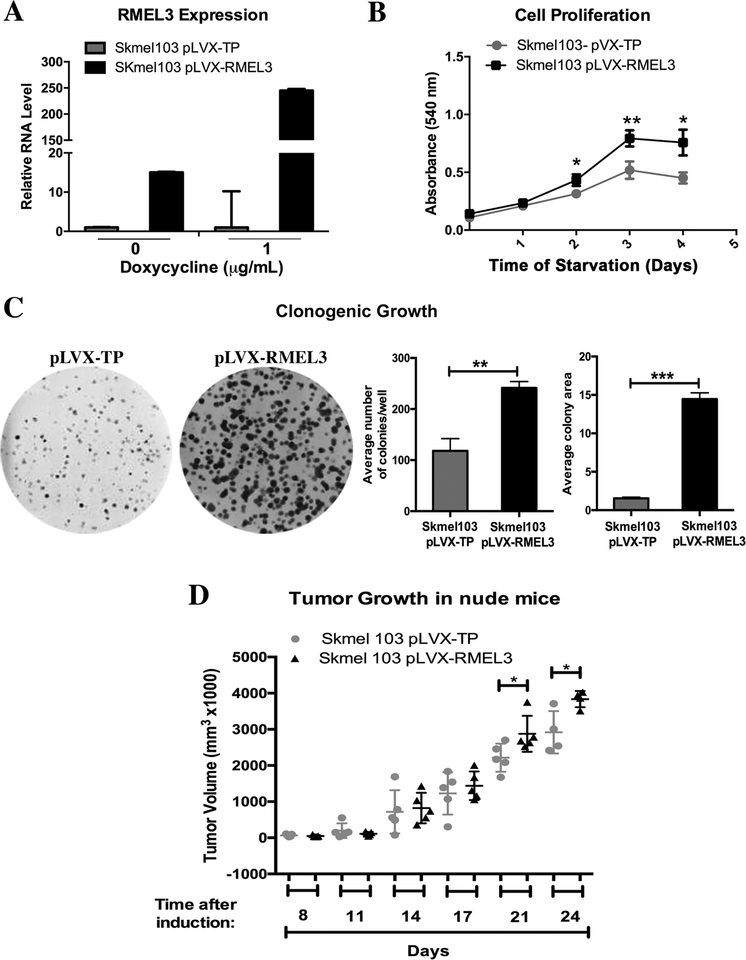
Overexpression of RMEL3 enhances proliferation, clonogenic ability, and tumorigenesis of melanoma cells
For stable expression of RMEL3 in melanoma cells, we chose VM10 line, one of the BRAFV600E mutant cells with the lowest RMEL3 expression levels (Figure 1a). Cells were transduced with lentiviral particles carrying either the construct pLJM1-RMEL3 or pLJM1-EGFP as control. The RT-qPCR data showed a gain of 3.5-fold in RMEL3 expression (Figure 4a). Proliferation assays with stably transduced cells showed a modest but significant increase in cell proliferation in VM10 pLJM1-RMEL3 cells as compared to control (Figure 4b). To further explore RMEL3 effects on melanoma cell growth and viability, we used the Tet-On inducible system. We chose SKMel103, which is a BRAF wild-type/NRAS mutant cell line with moderate to low expression levels of RMEL3. After treatment with 1 μg/mL doxycycline, cells stably transduced with pLVX-RMEL3 showed ~250-fold increase in RMEL3 RNA expression relative to pLVX-TP empty vector (Figure 5a). Variable levels of leaky expression were also observed in non-induced cultures; therefore, we chose to perform the functional analysis under doxycycline treatment only. Proliferation rates upon serum starvation were measurably higher in pLVX-RMEL3-transduced cells compared to control (Figure 5b), as well as the colony formation capacity, analyzed by clonogenic assays in DMEM Complete Medium (Figure 5c). To investigate the effect of RMEL3 overexpression in vivo, we injected nude mice on the right flank with pLVX-RMEL3-transduced SKMel103 cells and on the left flank with pLVX-TP-transduced control cells. Measurements of tumor volume showed that both RMEL3-overexpressing cells and control cells generated tumors that were comparable in size until about two weeks after injection. However, after the initial time period, tumors of the RMEL3-overexpressing cells grew more rapidly, reaching sizes significantly larger than the controls in day 21 (Figure 5d).
Figure 4. Overexpression of RMEL3 increases proliferation of a BRAFV600E mutant, RMEL3-low expresser, melanoma cell line.
(A) Efficiency of stable expression of RMEL3 RNA in VM10 melanoma cells transduced with pLJM1-RMEL3, analyzed by RT-qPCR. Relative expression was calculated according to 2−ΔΔCT method using TBP (Tata-box binding protein) as endogenous control and the normalized Ct of cells transduced with control vector (pLJM1-EGFP) as reference. (B) Proliferation of VM10 cells transduced with pLJM1-RMEL3 or pLJM1-EGFP as control. Cells were cultured in medium supplemented with 10% FBS, and after the indicated time points, they were stained with crystal violet for quantification of cell density according to the absorbance in an ELISA microplate plate reader. Error bars represent SEM of 3 independent experiments. Asterisks indicate statistically significant differences between groups based on unpaired parametric Student’s t test. Day 2 (**p < 0.0034); Day 3 (***p < 0.0004); Day 4 (**p < 0.0065)
Figure 5. Overexpression of RMEL3 in an NRAS mutant melanoma cell line increases proliferation rates, clonogenic ability, and tumor growth.
(A) Efficiency of RMEL3 transgene expression in SKMEL103 cells stably transduced with pLVX-RMEL3 or pLVX-TP (empty vector), not treated or treated with doxycycline (1 μg/mL, 24 h), as analyzed by RT-qPCR. The relative expression was calculated according to 2−ΔΔCT method using TBP (Tata-box binding protein) as endogenous control and the normalized Ct of the pLVX-TP-transduced cells as reference. (B) Proliferation of SKMEL103 cells transduced with pLVX-RMEL3 or pLVX-TP (as control), cultured in FBS-free medium in the presence of doxycycline (1 μg/mL). After the indicated time points, cells were stained with crystal violet and cell density was quantified according to the absorbance in an ELISA microplate reader. *p < 0.05; **p < 0.005. (C) Clonogenic ability of SKMEL103 cells transduced with pLVX-RMEL3 or pLVX-TP (as control). Cells were seeded in 60-mm-diameter plates with DMEM Complete Medium containing doxycycline (1 μg/mL). After 14 days, they were fixed with paraformaldehyde and stained with crystal violet. **p < 0.0014, ***p < 0.0001. Error bars represent SEM of 3 independent experiments for A-C. (D) Tumor growth. Nude mice were injected subcutaneously with 106 SKMEL103 cells, in the right flank with pLVX-RMEL3-transduced cells and in the left flank with pLVX-TP-transduced control cells. After injections, mice were maintained with diet ad libitum and drinking water was supplemented with 1 mg/ml doxycycline and changed every 2 days. *p < 0.05. This experiment was done once. Asterisks indicate statistically significant differences between groups based on unpaired parametric Student’s t test
Discussion
Previous work from our laboratory has shown that RMEL3 lncRNA is highly enriched in melanoma, particularly in tumors harboring the oncogenic BRAFV600E (Goedert et al., 2016; Sousa et al., 2010). These studies have provided first evidence of the functional relevance of this lncRNA in melanoma owning to demonstration of its restricted expression pattern and the fact that its knockdown causes cell cycle arrest and dramatic decay of melanoma cell replicative survival, especially in BRAFV600E mutant cells, while treatment with the same siRNA preserves viability of cells that do not express detectable amounts of RMEL3. These effects were consistent with many alterations, in the RMEL3-silenced cells, of central regulators of the cell cycle and apoptosis, as well as signaling transduction components of the MAPK and PI3K pathways (Goedert et al., 2016). Here, we add evidence that high RMEL3 expression is more often associated with extensive superficial/nodular cutaneous melanoma than mucosal and acral melanoma. Recently, acral and mucosal melanomas were shown not to display the UV signature mutational burden observed in cutaneous melanoma (Hayward et al, 2017). This and wealth of other evidence associating high content of UV signature mutations with cutaneous melanoma raised the possibility that the RMEL3 gene could be activated in the process of melanomagenesis due to the occurrence of a mutation hotspot, caused by UV exposure, in the promoter region of the gene itself. Analysis of TCGA and ICGC melanoma datasets, in fact, revealed a high content of UV signature mutations in the RMEL3 gene, however, lacking recurrent mutations that would characterize a hotspot. Also, RMEL3 mutation, in TCGA dataset, does not correlate with RMEL3 expression levels. Interestingly, however, RMEL3 mutation does correlate with poor patient survival raising the possibility that it could play a function as driver of tumorigenesis. Since we did not have access to expression data and survival information from ICGC, future studies should be pursued to answer whether mutations in the promoter region or additional mutations along the length of RMEL3 gene could influence its expression/function. We assume that, since mutation recurrence in the gene is rare, it is more likely that the association between RMEL3 mutation and poor patient survival rather than indicating a role of RMEL3 mutation as causative factor of tumorigenesis could just reflect an overall tumor mutational burden.
The lack of a mutation hotspot and of a correlation between mutation and expression led us to conclude that activation of the RMEL3 gene in non-acral cutaneous melanoma is more likely to result from the MAPK/ERK activation that underlies the pathogenesis of this disease. This is consistent with previous evidence that nearly 90% of cutaneous melanomas in contrast to 56% of acral and mucosal melanomas carry mutation in one of the three genes, BRAF, NRAS, or NF1 (Hayward et al, 2017). Also, because RMEL3 expression in BRAF mutant melanoma cell lines clearly reverses to the levels observed in melanocytes upon treatments with BRAF or MEK inhibitors, we favor the hypothesis that an active status of the MAPK/ERK pathway is required for RMEL3 activation in the context of melanomagenesis. Moreover, the RMEL3 upregulation observed upon vemurafenib-induced paradoxical activation of ERK, in NRASQ61R mutant WM1366 cells, reinforces the notion that RMEL3 responds to ERK pathway activation and might be involved in the development of sporadic tumors or in the relapse of BRAFi-resistant melanoma (Nazarian et al., 2010). There is precedent of lncRNAs with therapeutic potential in melanoma (Joung et al., 2017; Leucci, Vemdramin, et al., 2016).
The activation of RMEL3 expression observed here in response to the status of the BRAF MEK-ERK pathway activity raises the question of how to explain the highly restricted expression observed for RMEL3 in contrast to the well-known ubiquitous and canonical roles of the MAPK pathway. Our data suggest a dependence of the degree of activation of the ERK pathway and RMEL3 expression. We observed that acute exposure to a low concentration of vemurafenib decreased RMEL3 levels by about 50%, while more intense/prolonged inhibition was sufficient to bring RMEL3 levels in BRAFV600E cells to as low as the ones detected in melanocytes. Although it is beyond the scope of this work to clarify the mechanisms of RMEL3 transcriptional regulation, we discuss some evidence from ENCODE (Dunham et al., 2012) that can be explored in future studies. The RMEL3 gene extends through a genomic region of 138,365 bp in chromosome 5p11.2 associated with remarkable features of enhancer elements. Genomewide studies of transcription factor (TF) occupancy and histone modifications show evidence of chromatin accessibility typical of a primed enhancer (DNase I sensitivity, histone H3K27Ac, and H3K4me1) in a region 11- to 13-kb upstream of the presumed transcription start site of the gene (RMEL3/CTD-2023N9.1) in several cell lines used by ENCODE, such as K562, HUVEC, NT2-D1, and others. This region was shown to be occupied by POL2A and an array of ERK-regulated TFs, such as ELK, FOS, JUN, ATF, MYC, STATs, GATA, TAL1, TEAD4, NR2F2, and CTCF. Also, high score binding was observed for at least two important co-repressors, RCOR1 and TCF7L2/TCF4. RCOR1 acts as a co-repressor of REST, whose stability is promoted by ERK inhibition (Nesti, Corson, McCleskey, Oyer, & Mandel, 2014). ERK pathway was shown to cooperate for activation of Wnt/β-catenin signaling pathway in colon cancer, leading to activation of TCF7L2-repressed genes (Lemieux, Cagnol, Beaudry, Carrier, & Rivard, 2015). Therefore, one of the above mechanisms could mediate the transcriptional regulation of RMEL3 downstream of ERK in melanoma. Notably, among the TFs occupying the RMEL3 locus, some are canonical factors for initiation of the serum response or involved in the immediate early response. Prompted by the evidence discussed above and having previously shown that RMEL3 is required for survival of BRAF mutant melanoma cells (Goedert et al., 2016), the major focus of this work was on defining the effects provoked by ectopic expression of RMEL3 in different cell lines and under a variety of culture conditions, especially serum starvation. We reasoned that if RMEL3 plays a role in, or downstream of mitogen/survival factor-stimulated signaling circuitries, under physiological conditions, its overexpression should bypass the necessity of mitogen stimulation for cell proliferation and survival. To test this hypothesis, we overexpressed RMEL3 in the immortalized, non-transformed, and widely known NIH3T3 murine fibroblasts (Meloche & Pouysségur, 2007; Wang et al., 2017). We were surprised to find that, indeed, overexpression of RMEL3 bypassed the requirement of a mitogen/survival factor stimulation to support cell proliferation and survival. These experiments also revealed that human RMEL3 lncRNA is functional in a murine cell line, and therefore, it must have conserved partners in this organism, despite low sequence conservation. Future studies will be necessary to define the underlying mechanisms of the protective role of RMEL3 against cell death caused by serum starvation.
The results discussed above led us to hypothesize that overexpression of RMEL3 would potentially enhance malignant properties of either a BRAF wild-type or a BRAF mutant melanoma cell line expressing low RMEL3 levels. Indeed, ectopic expression of RMEL3 using two different lentiviral vector systems enhanced cell proliferation in both cell lines, either cultured in complete media or under serum starvation. Most strikingly, expression mediated by a Dox-inducible vector system caused a strong increase in clonogenic ability and also promoted tumor growth in nude mice. These data are fully consistent with the phenotype shown previously for RMEL3 knockdown (Goedert et al., 2016).
In conclusion, the protective effect of RMEL3 against serum withdrawal-induced cell cycle arrest and apoptosis together with other pharmacological and functional data shown here allow us to propose that RMEL3 gene is responsive to the ERK pathway and the RMEL3 lncRNA might play a key role as an activator/effector in the mitogen/survival factor response pathways. These capabilities and the highly restricted expression pattern of RMEL3 are desirable characteristics of targets for cancer drugs.
Supplementary Material
Figure S2. Viability of melanoma cells after treatment with BRAF or MEK inhibitors. (A) Viability of WM-1366, Skmel-19 and UACC62 cells after treatment with 10 μM Vemurafenib for 48h, determined by flow cytometry using propidium iodide as indicator of dead cells. (B) Viability of UACC-62 melanoma cells after treatment with 25 μM PD98059 for 48h, assayed as in A. Error bars represent SEM of 3 independent experiments.
Figure S3. Nucleotide sequence for cloned RMEL3 cDNA. RMEL3 was PCR amplified from cDNA synthesized from total RNA extract of WM278 melanoma cell line. Reverse transcription was done using random priming. The PCR product was cloned into the TOPO vector and transferred to pLVX and pLJM1 vectors, as described in the Methods section. The sequence matches 777 pb (1–778 bp) of CTD-2023N9.1 (LncGPBP1–1:1)/ NC_000005.10 (chr5: 57395100 – 57533345) whose transcribed sequence is 897bp in length. The cloned sequence lacks 40 nt of the 5’ end and 79 nt of the 3’ plus a single A deletion (position 761 of the insert; 801 in the transcript). Cloned sequence was deposited in GenBank under the accession number MH370349.
Figure S1. The single base substitution mutations mapped in the RMEL3 locus across 129 melanoma samples of the ICGC data portal display a typical UV mutational signature. The graphic represents the rates of each base substitution calculated after distributing all single base substitutions (totalizing 558) into the 96 mutation types according to Alexandrov et al. (2013) concept. Additional feature was the presence of CC>TT double base substitutions (~3%). See Table S2 for detailed information.
Table S1. Mutations detected in the RMEL3 gene (CTD-2023N9.1), in a dataset of 450 cutaneous melanoma (SKCM) samples of the TCGA databank, with associated genomic and sample information.
Table S2. Mutations detected in the RMEL3 gene (CTD-2023N9.1), including the entire length of the gene, 20 kb upstream and 20 kb downstream of the mapped gene, in 129 cutaneous melanoma samples (MELA-AU project) of the ICGC databank, with associated genomic and sample information.
Table S3. Summary of the frequency of different types of mutations, including base substitution (C>A, C>G, C>T, T>A, T>C, T>G and CC>TT), deletions and insertions, detected in the RMEL3 locus (CTD-2023N9.1), in datasets of cutaneous melanoma samples from the TCGA (450 samples, SKCM) and ICGC (129 samples, MELA-AU project) data portals.
Significance.
Previous findings link RMEL3 with BRAFV600E and the MAPK/ERK pathway activity. Here, we show that BRAFV600E activity is required for RMEL3 upregulation and, most importantly, RMEL3 overexpression bypasses the requirement of serum supplementation for proliferation/survival of immortalized fibroblasts. Also, overexpression of RMEL3 in melanoma cells increases cell proliferation, survival, and in vivo tumor growth. Taken together, previous and present data support the hypothesis that RMEL3 lncRNA might play a key role as an activator/effector in mitogen-/survival factor-activated pathways. These capabilities and the highly restricted expression pattern of RMEL3 are desirable characteristics of targets for cancer drugs.
ACKNOWLEDGMENTS
The authors are grateful to the Barretos Cancer Hospital Biobank for the patient samples and to Dr. Sérgio Vicente Serrano and Dr. Rui Manoel Vieira Reis for their support, and especially for the patients for their permission. The authors are also thankful to Silmara Reis Banzi, Benedita Oliveira Sousa and Vivien Igras for technical assistance, and Jéssica Rodrigues Plaça for her support with bioinformatics. This work was supported by grants to EME from São Paulo Research Foundation—FAPESP (2014/18189-5; 2018/04017-9) and National Council for Scientific and Technological Development—CNPq (457603/2013-5 and 309187/2015-0). CC received fellowship from CNPq (870310/19976) and RBS from Coordination for the Improvement of Higher Education Personnel—CAPES, and CPG from FAPESP (2010/16097-5 e 2012/24056-2). VV was supported by FAPESP grants #2013/13465-1 and # 2013/08135-2. DEF gratefully acknowledges grant support from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Melanoma Research Alliance, and the NIH (5R01 AR043369; 5P01 CA163222-05; 1R01AR072304-01). EME and VV are members of Center for Cell- Therapy, CEPID/FAPESP (2013/08135-2). This study is part of the National Institute of Science and Technology in Pharmaceutical Nanotechnology: a transdisciplinary approach INCTNANOFARMA, which is supported by FAPESP (2014/50928-2) and by CNPq (465687/2014-8).
References
- Abel EV, Aplin AE (2010). FOXD3 is a mutant B-RAF-regulated inhibitor of G1-S progression in melanoma cells. Cancer Res, 70(7): 2891–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel EV, Basile KJ, Kugel CH III, Witkiewicz AK, Le K, Amaravadi RK, Karakousis GC, Xu X, Xu W, Schuchter LM, Lee JB, Ertel A, Fortina P, Aplin AE (2013). Melanoma adaptsto RAF/MEK inhibitorsthrough FOXD3-mediated upregulationof ERBB3. J Clin Invest, 123(5):2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascierto PA, Marincola FM, Atkins MB (2015). What’s new in melanoma? Combination!. J Transl Med, 13:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basile KJ, Abel EV, Aplin AE (2012). Adaptive upregulation of FOXD3 and resistance to PLX4032/4720-induced cell death in mutant B-RAF melanoma cells. Oncogene, 31: 2471–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia P, Friedlander P, Zakaria EA, Kandil E (2015). Impact of BRAF mutation status in the prognosis of cutaneous melanoma: an area of ongoing research. Ann Transl Med, 3(2):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Croyal M, Carbonnelle D, et al. IRES-dependent translation of the long non coding RNA meloe in melanoma cells produces the most immunogenic MELOE antigens. Oncotarget. 2016;7(37):59704–59713. doi: 10.18632/oncotarget.10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yang H, Xiao Y, Tang X, Li Y, Han Q, Fu J, Yang Y, Zhu Y (2016). LncRNA GAS5 is a critical regulator of metastasis phenotype of melanoma cells and inhibits tumor growth in vivo. Onco Targets Ther, 9: 4075–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yang H, Xiao Y, Tang X, Li Y, Han Q, Fu J, Yang Y, Zhu Y (2016). Lentiviral-mediated overexpression of long non- coding RNA GAS5 reduces invasion by mediating MMP2 expression and activity in human melanoma cells. Int J Oncol, 48: 1509–18. [DOI] [PubMed] [Google Scholar]
- Chen X, Dong H, Liu S, Yu L, Yan D, Yao X, Sun W, Han D, Gao G (2017). Long noncoding RNA MHENCR promotes melanoma progression via regulating miR-425/489-mediated PI3K-Akt pathway. Am. J. Transl. Res, 9(1):90–102. [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yan CC, Luo C, Ji W, Zhang Y Dai Q (2015). Constructing lncRNA functional similarity network based on lncRNA-disease associations and disease semantic similarity. Scientific Reports, 5:11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CM, Li H, Gasman S, Huang J, Schiff R, Chang EC (2011). Compartmentalized Ras Proteins Transform NIH 3T3 Cells with Different Efficiencies. Molecular and Cellular Biology, 31(5): 983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichowski K, Jänne PA (2010). Drug Discovery: Inhibitors that activate. Nature, 464, 358–359. [DOI] [PubMed] [Google Scholar]
- Cowley S, Paterson H, Kemp P, Marshall CJ (1994). Activation of MAP Kinase Kinase is Necessary and Sufficient for PC12 Differentiation and for Transformation of NIH3T3 cells. Cell, 77, 841–852. [DOI] [PubMed] [Google Scholar]
- Davies MA, Samuels Y Analysis of the genome to personalize therapy for melanoma. Oncogene, 29: 5545–5555, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Yang M, Jiang R, An N, Wang X, Liu B (2017). Long Non-Coding RNA HOTAIR Regulates the Proliferation, Self-Renewal Capacity, Tumor Formation and Migration of the Cancer Stem- Like Cell (CSC) Subpopulation Enriched from Breast Cancer Cells. PLoS ONE, 12(1): e0170860. doi:10.1371. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Engreitz JM, Ollikainen N, Guttman. (2016). Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nature Reviews, 1–15. [DOI] [PubMed] [Google Scholar]
- Esguerra JLS, Eliasson L (2014). Functional implications of long non-codinf RNAs in the pancreatic islets of Langerhans. Frontiers in Genetics, 5. doi: 10.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Fullwood MJ (2016). Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genimics Proteomics Bioinformatics, 14: 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, Khavari PA (2012). BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res, 22(6):1006–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Wei GH (2017). Genomic Insight into the Role of lncRNAs in Cancer Susceptibility. Int. J. Mol. Sci, 18, 1239. doi:10.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert L, Pereira CG, Roszik J, Placa JR, Cardoso C, Chen G, Deng W, Yennu-Nanda VG, Silva WA Jr., Davies MA, Espreafico EM (2016). RMEL3, a novel BRAFV600E-associated long noncoding RNA, is required for MAPK and PI3K signaling in melanoma. Oncotarget, 7:36711–36718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert L, Plaça JR, Fuziwara CS, Machado MCR, Plaça DR, Almeida PP, Sanches TP, Santos JFD, Corveloni AC, Pereira IEG, de Castro MM, Kimura ET, Silva WA Jr., Espreafico EM (2017). Identification of Long Noncoding RNAs Deregulated in Papillary Thyroid Cancer and Correlated with BRAFV600E Mutation by Bioinformatics Integrative Analysis. Sci Rep., 7(1):1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature, 4464, 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW, McManus MT (2013) Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLoS Genet, 9(6): e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, Patch AM, Kakavand H, Alexandrov LB et al. , (2017). Whole-genome landscapes of major melanoma subtypes. Nature, 545(7653):175–180, doi:10.1038. [DOI] [PubMed] [Google Scholar]
- Hrdlickova B, Almeida RC, Borek Z, Withoff S (2014). Genetic Variation in the non-coding genome: Involvement of micro-RNAs and long non-coding RNAs in disease. Biochimica et Biophysica Acta, 1842, 1910–1922. [DOI] [PubMed] [Google Scholar]
- Huart M (2015). The emerging role of lncRNAs in cancer. Nature Medicine, 21(11): 1253–1261, 2015. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009; 459:108–12; PMID:19295514; 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle JN, Xie F, Ellis MJ, Goss PE, Shepherd LE, Chapman JW, Chen BE, Kubo M, Furukawa Y, Momozawa Y, et al. (2016). Genetic polymorphisms in the long noncoding RNA MIR2052HG offer a pharmacogenomic basis for the response of breast cancer patients to aromatase inhibitor therapy. Cancer Res, 76, 7012–7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST, Chu LW, Zhang Z, Zhao H, Zheng SL, Isaacs WB, Xu J (2011). Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis, 32(11): 1655–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph EW, Pratilas CA, Poulikakos PI,Tadib M, Wangd W, Taylor BS, Halilovic E, Persaud Y, Xing F, Viale A, Tsai J, Chapman PB, Bollag G, Solitf DB, Rosen N (2010). The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAFselective manner. PNAS, 107(33): 14903–14908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, Gootenberg JS, Sanjana NE, Wright JB, Fulco CP, Tseng YY, Yoon CH, Boehna JS, Lander ES, Zhang F (2017). Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature, 548, 343–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NH, Lee CH, Lee AY (2010). H19 RNA downregulation stimulated melanogenesis in melasma. Pigment Cell Melanoma Res, 23: 84–92. [DOI] [PubMed] [Google Scholar]
- Koo J, Wang S, Kang N, Hur SJ, Bahk YY (2016). Induction of MAP kinase phosphatase 3 through Erk/MAP kinase activation in three oncogenic Ras (H-, K- and N-Ras)-expressing NIH/3T3 mouse embryonic fibroblast cell lines. BMB Rep, 49(7): 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacouture ME, Duvic M, Hauschild A, Prieto VG, Robert C, Schadendorf D, Kim CC, McCormack CJ, Myskowski PL, Spleiss O, Trunzer K, Su F, Nelson B, Nolop KB, Grippo JF, Lee RJ, Klimek MJ, Troy JL, Joe AK (2013). Analysis of Dermatologic Events in Vemurafenib-Treated Patients With Melanoma. The Oncoligist, 18(3): 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Sahoo A, Marchica J, Holzhauser E, Chen X, Li JL, Seki T, Govindarajan SS, Markey FB, Batish M, Lokhande SJ, Zhang S, Ray A, Perera RJ (2017). The long noncoding RNA SPRIGHTLY acts as na intranuclear organizing hub for pre-mRNA molecules. Sci. Adv, 3: e1602505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard L, Liu M, Marzese DM, Wang H, Chong K, Kawas N, Donovan NC, Kiyohara E, Hsu S, Nelson N, Izraely S, Sagi-Assif O, Witz IP, Ma XJ, Luo Y, Hoon DSB (2015). The CASC15 long intergenic non-coding RNA locus is involved in melanoma pregression and phenotype-switching. J. Invest. Dermatol, 135(10): 2464–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucci E, Vemdramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, Radaelli E, Eyckerman S, Leonelli C, Vanderheyden K, Rogiers A, Hermans E, Baatsen P, Aerts S, Amant F, Aelst SV, Oord JVD, Strooper B, Davidson I, Lafontaine DLJ, Gevaert K, Vandesompele J, Mestdagh P, Marine JC (2016). Melanoma addiction to the long non-coding RNA SAMMSON. Nature, 531, 518–522. [DOI] [PubMed] [Google Scholar]
- Li W, Notani D, Rosenfeld MG (2016). Enhancers as non-coding RNA transcription units: recente insights and future perspectives. Nature Reviews Genetics, 17: 2017–223. [DOI] [PubMed] [Google Scholar]
- Liu P, Yang H, Zhang J, Peng X, Lu Z, Tong W, Chen J (2017). The lncRNA MALAT1 acts as a competing endogenous RNA to regulate KRAS expression by sponging miR-217 in pancreatic ductal adenocarcinoma. Scientific Reports, 7: 5186 DOI:10.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Qu J, Wu DK, Lu ZL, Sun YS, Qu Q (2017). Long non-codong RNAs: a reising biotarget in colorectal cancer. Oncotarget, 8(13): 22187–22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, Pendlington Z, Welter D, Burdett T, Hindorff L, Flicek P, Cunningham F, Parkinson H (2017). The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Research, 45, D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloche S, Pouysségur J (2007). The ERK1/2 mitogen-activated protein kinase pathway as a master regulatoroftheG1-toS-phasetransition. Oncogene, 26, 3227–3239. [DOI] [PubMed] [Google Scholar]
- Montes M, Nielsen MM, Maglieri G, Jacobsen A, Hojfeldt J, Agrawal-Singh S, Hansen K, van de Werken HJ, Pedersen JS, Lund AH (2015). The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat Commun. 6: 6967. [DOI] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee M, Attar N, Sazegar H, Chodon T, Nelson SF, McArthur G, Sosman JA, Ribas A, Lo RS (2010). Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature, 468: 973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer PA, Kee D, Dziunycz P, Sucker A, Kamsukom N, Jones R, Roden C, Chalk CJ, Ardlie K, Palescandolo E, Piris A, MacConaill LE, Robert C, Hofbauer GFL, McArthur GA, Schadendorf D, Garraway LA (2012). RAS Mutations Are Associated With the Development of Cutaneous Squamous CellTumors in Patients Treated With RAF Inhibitors. Journal of clinical oncology, 30 (3): 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmant E, Sabbagh A, Vidaud M, Bieèche I (2011). ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS, FASEB J, 25, 444–448. [DOI] [PubMed] [Google Scholar]
- Perna D, Karrethb FA, Rusta AG, Perez-Mancerab PA, Rashida M, Iorioc F, Alifrangisc C, Arendse MJ, Bosenbergf MW, Bollagg G, Tuvesonb DA, Adamsa DJ (2015). BRAF inhibitor resistance mediated by the AKT pathway in an oncogenic BRAF mouse melanoma model. PNAS, E536–E545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Haimovic A, Christos PJ, Miera ECVS, Shapiro R, Pavlick A, Berman RS, Darvishian F, Osman I (2011). Deletion of PTENP1 pseudogene in human melanoma. J Invest Dermatol. 131: 2497–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N (2010). RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature, 464: 427–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Zhao S, Erho N, Schipper M, Iyer MK, Dhanasekaran SM, Magi-Galluzzi C, Mehra R, Sahu A, Siddiqui J, Davicioni E, Den RB, Dicker AP, Karnes J, Wei JT, Klein EA, Jenkins RB, Chinnaiyan AM, Feng FY (2014). Nomination and validation of the long noncoding RNA SChLAP1 as a risk fator for metastatic prostate câncer progression: a multi-institutional high-throughput analysis. Lancelot Oncol., 15(13): 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quagliata L, Matter MS, Psicuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, Andersen JB, Hämmerle M, Tornillo L, Heim MH, Dierderichs S, Cillo C, Terracciano LM (20140. LncRNA HOTTIP/HOXA13 expression is assiciated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology, 59(3):911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rion N, Rüegg MA (2017). LncRNA-encoded peptides: More than translational noise?. Cell Research, 27: 604–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco AD, Baldassarre A, Masotti A (2012). Bioinformatics Tools and Novel Challenges in Long Non-Coding RNAs (lncRNAs) Functional Analysis. Int. J. Mol. Sci, 13, 97–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salta E, De Strooper B (2017). Noncoding RNAs in neurodegeneration. Nature Reviews, 18: 627–638. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Oghabian A, Bodiyabadu PK, Joseph WR, Leung EY, Finlay GJ, Baguley BC, Askarian-Amiri ME (2017). Multiple Isoforms of ANRIL in Melanoma Cells: Structural Complexity Suggestes Variations in Processing. Int. J. Mol. Sci, 18(7):1378. doi:10.3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann JC, Boyer LA (2013). Getting to the heart of the matter: long non-coding RNAs in cardiac development and disease. The EMBO Journal, 32: 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K, Joyce CE, Buquicchio F, Brown A, Ritz J, Distel RJ, Yoon CH, Novina CD (2016). The lncRNA SLNCR1 Mediates Melanoma Invasion through a Conserved SRA1-like Region. Cell Rep, 15:2025–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AM, Chang HY (2016). Long Noncoding RNAs in Cancer Pathways. Cancer Cell, 452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Li J, Liu Y, Ding J, Fan Y, Tian Y, Wang L, Lian Y, Wang K, Shu Y (2015). The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Molecular Cancer, 14:51 DOI 10.1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Liu Y, Wang J, Jie W, Jie D, Yun T, Li W, Yan L, Wang K, Feng J (2015). Downregulated Long Noncoding RNA BANCR Promotes the Proliferation of Colorectal Cancer Cells via Downregualtion of p21 Expression. PLoS ONE, 10(4): e0122679. doi:10.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa JF, Torrieri R, Silva RR, Pereira CG, Valente V, Torrieri E, Peronni KC, Martins W, Muto N, Francisco G, Brohem CA, Carlotti CG Jr, Maria-Engler SS, Chammas R, Espreafico EM (2010). Novel Primate-Specific Genes, RMEL 1, 2 and 3, with Highly Restricted Expression in Melanoma, Assessed by New Data Mining Tool. Plos one, 5(10): e13510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, Reis-Filho JS, Kong X, Koya RC, Flaherty KT, et al. (2012). RAS Mutations in Cutaneous Squamous-Cell Carcinomas in Patients Treated with BRAF Inhibitors. N Engl J Med, 366:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri M, Habibi M, Noroozi R, Rakhshan A, Sarrafzadeh S, Sayad A, Omrani DM, Ghafouri-Fard S (2017). HOTAIR genetic variants are associated with prostate cancer and benign prostate hyperplasia in an Iranian population. Gene, 613, 20–24. [DOI] [PubMed] [Google Scholar]
- Tao WJ, Lin H, Sun T, Samanta AK, Arlinghaus R (2011). BCR-BL oncogenic transformation of NIH 3T3 fobroblasts requires the IL-3 receptor. Oncogene, 27(22); 3194–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. (2015). Genomic Classification of Cutaneous Melanoma, In Cell, 161( 7): 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. (2012). Na integrated encyclopedia of DNA elements in the human genome. Nature, 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J, Vultur A, Herlyn M (2011). Reistance to BRAF inhibitors: Unraveling mechanisms and future treatment options. Cancer Res, 71(23): 7137–7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu H, Qiu Q, Zhang Z, Gu Y, He Z (2017). TCRP1 promotes NIH/3T3 cell transformation by over-activating PDK1 and AKT1. Oncogenesis, 6, e323; doi:10.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CF, Tan GH, Ma CC, Li L (2013). The non-coding RNA llme23 drives the malignant property of human melanoma cells. J Genet Genomics, 40: 179–188. [DOI] [PubMed] [Google Scholar]
- Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell, 38(5):662–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Liu HL, Liu ZS, Owzar K, Han YH, Su L, Wei YY, Hung JR, McLaughlin J, Brhane Y, et al. (2016). A Novel genetic variant in long non-coding RNA gene NEXN-AS1 is associated with risk of lung cancer. Sci. Rep, 6, 34234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, Zhou Y, Klibanski. (2010). Maternally Expressed Gene 3 (MEG3) Nonconding Ribonucleic Acid: Isoform Structure, Expression, and Functions. Endocrinology, 151(3): 939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Sun C, Cui Z (2017). A long noncoding RNA UCA1 promotes proliferation and predicts poor prognosis in glioma. Clin. Transl. Oncol, 19(6): 735–741. [DOI] [PubMed] [Google Scholar]
- Zheng J, Huang X, Tan W, Yu D, Du ZL, Chang J, Wei LX, Han YL, Wang CF, Che X, et al. (2015). Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat. Genet, 48, 747–759. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Yang C, Tong S, Ding Y, Deng W, Song D, Xiao K (2017). Genetic variation of long non-coding RNA TINCR contribute to the susceptibility and progression of colorectal cancer. Oncotarget, 8, 33536–33543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S2. Viability of melanoma cells after treatment with BRAF or MEK inhibitors. (A) Viability of WM-1366, Skmel-19 and UACC62 cells after treatment with 10 μM Vemurafenib for 48h, determined by flow cytometry using propidium iodide as indicator of dead cells. (B) Viability of UACC-62 melanoma cells after treatment with 25 μM PD98059 for 48h, assayed as in A. Error bars represent SEM of 3 independent experiments.
Figure S3. Nucleotide sequence for cloned RMEL3 cDNA. RMEL3 was PCR amplified from cDNA synthesized from total RNA extract of WM278 melanoma cell line. Reverse transcription was done using random priming. The PCR product was cloned into the TOPO vector and transferred to pLVX and pLJM1 vectors, as described in the Methods section. The sequence matches 777 pb (1–778 bp) of CTD-2023N9.1 (LncGPBP1–1:1)/ NC_000005.10 (chr5: 57395100 – 57533345) whose transcribed sequence is 897bp in length. The cloned sequence lacks 40 nt of the 5’ end and 79 nt of the 3’ plus a single A deletion (position 761 of the insert; 801 in the transcript). Cloned sequence was deposited in GenBank under the accession number MH370349.
Figure S1. The single base substitution mutations mapped in the RMEL3 locus across 129 melanoma samples of the ICGC data portal display a typical UV mutational signature. The graphic represents the rates of each base substitution calculated after distributing all single base substitutions (totalizing 558) into the 96 mutation types according to Alexandrov et al. (2013) concept. Additional feature was the presence of CC>TT double base substitutions (~3%). See Table S2 for detailed information.
Table S1. Mutations detected in the RMEL3 gene (CTD-2023N9.1), in a dataset of 450 cutaneous melanoma (SKCM) samples of the TCGA databank, with associated genomic and sample information.
Table S2. Mutations detected in the RMEL3 gene (CTD-2023N9.1), including the entire length of the gene, 20 kb upstream and 20 kb downstream of the mapped gene, in 129 cutaneous melanoma samples (MELA-AU project) of the ICGC databank, with associated genomic and sample information.
Table S3. Summary of the frequency of different types of mutations, including base substitution (C>A, C>G, C>T, T>A, T>C, T>G and CC>TT), deletions and insertions, detected in the RMEL3 locus (CTD-2023N9.1), in datasets of cutaneous melanoma samples from the TCGA (450 samples, SKCM) and ICGC (129 samples, MELA-AU project) data portals.