Abstract
Introduction:
Alzheimer apolipoprotein E (APOE) ε4/ε4 carriers have earlier disease onset and more protein aggregates than patients with other APOE genotypes. Autophagy opposes aggregation, and important autophagy genes are coordinately regulated by transcription factor EB (TFEB) binding to “coordinated lysosomal expression and regulation” (CLEAR) DNA motifs.
Methods:
Autophagic gene expression was assessed in brains of controls and Alzheimer’s disease (AD) patients parsed by APOE genotype and in a glioblastoma cell line expressing either apoE3 or apoE4. Computational modeling assessed interactions between apoE and mutated apoE with CLEAR or modified DNA.
Results:
Three TFEB-regulated mRNA transcripts—SQSTM, MAP1LC3B, and LAMP2—were lower in AD ε4/ε4 than in AD ε3/ε3 brains. Computational modeling predicted avid specific binding of apoE4 to CLEAR motifs. ApoE was found in cellular nuclei, and in vitro binding assays suggest competition between apoE4 and TFEB at CLEAR sites.
Conclusion:
ApoE4-CLEAR interactions may account for suppressed autophagy in APOE ε4/ε4 carriers and, in this way, contribute to earlier AD onset.
Keywords: Alzheimer’s disease, APOE genotype, ApoE protein, autophagy, DNA binding, TFEB, Transcription, Protein aggregation, Molecular-dynamic simulation, EMSA, PLA
1. Introduction
Inheritance of two apolipoprotein E (APOE) ε4 alleles (APOE ε4/ε4) is the single greatest genetic risk factor for development of Alzheimer’s disease (AD) [1–3], the world’s most common neurodegenerative disease [4]. The importance of the risk of having the APOE ε4 gene product, that is, the apoE4 protein, rather than either of the other two possible gene products—apoE2 or apoE3—is underscored by the fact that the odds of development of AD in APOE ε4/ε4 carriers increased 12- to 15-fold relative to carriers of either APOE ε3/ε3 or APOE ε2/ε4, and 3-fold relative to APOE ε3/ε4 carriers [3]. Sixty percent of all AD patients carry at least one APOE ε4 allele [5]. Moreover, relative to their counterparts who carry one of the five other allelic combinations of the APOE gene, Alzheimer patient carriers of APOE ε4/ε4 have conspicuous increases in the defining neuropathological changes of AD, viz., extracellular plaques of amyloid β (Aβ) [6,7] and intraneuronal paired helical filaments of hyperphosphorylated tau (P-tau) in neurofibrillary tangles [8,9]. This association suggests that the apoE4 protein, itself, may interfere with autophagic processes so as to favor proteostatic failure and aggregate buildup over clearance of unwanted proteins. Failures in proteostasis such as autophagic insufficiency are known to be early and persistent features of Alzheimer pathogenesis [10,11] and appear to be particularly accentuated in the presence of APOE ε4 [12]. Studies of autophagy-related failures have demonstrated that defects in retrograde transport [9,13] and lysosomal acidification [14,15] lead to elevations in Alzheimer-like pathology, while transcription factor EB (TFEB)-mediated activation of autophagy in various models ameliorated both Aβ [16,17] and tau pathology [18].
Despite the clear importance of the APOE ε4/ε4 genotype in both AD risk and aggregate density, at present there is no consensus as to how the presence of apoE4 proteins may directly or indirectly influence either disease risk or the genesis of early, excessive accumulations of AD-defining aggregates. Therefore, based on current knowledge regarding the importance of apoE4 in Alzheimer neuropathogenesis, we undertook a more direct approach and investigated the potential of apoE4 to interfere with autophagy by altering the expression of three essential protein elements of autophagy: sequestosome 1 (p62), LC3B, and LAMP2. These proteins are the products of the binding of TFEB to the coordinated lysosomal expression and regulation (CLEAR) DNA motif [19] for transcription of the genes SQSTM1, MAP1LC3B [20], and LAMP2 [21]. The demonstration by Theendakara and his colleagues [22] of possible productive interactions between apoE and DNA, together with recognition of the importance of TFEB/CLEAR binding in regulation of autophagy in general [20], and of Aβ [16,17] and tau [18] in particular, led us to investigate a new hypothesis, viz., that apoE4 interferes with TFEB/DNA interactions at point(s) before translation of LCB3, p62, and LAMP2 and in this way accounts, at least in part, for the observed early and persistent elevation of the numbers of plaques and neurofibrillary tangles in brains of APOE ε4/ε4 patient carriers.
2. Methods
2.1. Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized. The investigators were blinded as to the sex and genotype of the patient sources of the samples used.
2.2. Cell lines and culture
T98G cells were obtained from American Type Culture Collection. Stable transformants were generated as described previously by Wang et al. [23]. Recombinant apoE was prepared under native conditions with the intention of retaining any lipid content present. This precludes harsh approaches necessary to isolate the protein. Concerns about purity, however, are relieved by the comparative difference between the apoE3 and apoE4 preparations and by sensitivity of the binding to an antibody recognizing apoE.
2.3. Cell culture conditions
T98G cells, expressing either apoE3 or apoE4, were grown in Dulbecco’s Modified Eagle’s medium (DMEM) (Cat. No: 11995040, Thermo Fisher Scientific, Waltham, MA, USA) supplemented to 10% with fetal bovine serum (FBS) (Cat. No: 16000044, Thermo Fisher Scientific). For amino-acid starvation, cultures were washed twice with serum and amino-acid free Earle’s Balanced Salt Solution (EBSS) and incubated in EBSS at 37°C for 3 hours, whereas control (“fed”) cells were washed twice with DMEM/FBS and incubated at 37°C for 3 hours in DMEM/FBS.
2.4. Antibodies and reagents
The following commercially available antibodies were used: anti-pan 14-3-3 (sc-629, Santa Cruz Biotechnology), anti-TFEB (ab2636, Abcam), anti-p62 (BD610832, BD Biosciences), anti-actin (ab6276, Abcam), anti-LC3B (NB600–1384, Novus Biologicals), and mouse monoclonal anti-apoE (1484 273; Boehringer-Mannheim).
2.5. Immunofluorescence
Human hippocampal immunohistochemistry: Samples were acquired from human brain specimens either with pathologically diagnosed Alzheimer’s disease (without Parkinson’s disease) or age-matched controls (AMCs) from the UAMS brain bank, where they were stored as formalin-fixed, paraffin-embedded tissue. Our tissues were from patients who did not qualify as human subject research according to U.S. Department of Health and Human Services Exemption 4. Patients with Alzheimer’s disease with APOE ε3/ε3 genotype are termed “AD 3,3”; while patients with Alzheimer’s disease with APOE ε4/ε4 genotype are termed “AD 4,4”. AMCs with APOE ε3/ε3 are termed “AMC 3,3”. A total of eight Alzheimer’s disease patients and four age-matched control patients were used in TFEB nuclear localization immunofluorescence, using ab2636 at 1:50 dilution. The average age of the patients was 76 years old, with postmortem intervals between 3 and 13 hours, with an average postmortem interval of 5.3 h. Hippocampal tissue blocks were sectioned at 7-μm thickness and mounted on slides and subsequently deparaffinized in xylene, rehydrated in serial dilutions of ethanol to water, and washed with PBS + 0.1% Tween 20. Antigen retrieval was performed in boiling citrate buffer for 30 minutes; slides were blocked in Dako Animal-Free Protein Blocker, then incubated in primary antibody overnight at 4°C, washed, and then incubated in secondary antibody for 1 hour. After washing in PBS, slides were quenched for autofluorescence with 0.1% Sudan Black B in 70% EtOH, washed with water, then treated with DAPI and coverslipped in Prolong Gold Mounting Medium. A total of six images per case were analyzed, with images taken from nonadjacent locations in pyramidal cell layers in CA1. Nuclear localization was assessed by ImageJ, with DAPI used to create a mask for the TFEB channel, and TFEB intensity was divided by nuclear area to control for cell density.
2.6. Duolink proximity ligation assay
Proximity ligation assay (PLA) was performed according to the manufacturer’s protocols on deparaffinized sections prepared as mentioned previously. In brief, slides were incubated overnight at 4°C with primary antibodies: anti-TFEB (1:50) and anti-pan-14-3-3, (1:200). Slides were then washed with PBS + 0.1% Tween 20, blocked with Duolink Antibody Block solution for 1 hour at 37°C, incubated in Duolink secondary antibody for 1 hour at 37°C, washed in PLA wash buffer A twice for 10 minutes, and incubated in Duolink PLA ligation solution for 30 minutes at 37°C. After another two washes in PLA wash buffer A, slides were incubated in Duolink amplification solution for 100 minutes at 37°C, after which slides were washed in PLA wash buffer B, incubated in Sudan Black B in 70% EtOH, and coverslipped with Duolink mounting medium containing DAPI.
2.7. Insoluble protein isolation and protein identification
Protein pellets isolated from T98G cell cultures were suspended in buffer containing 20-mM Hepes pH 7.4, 0.3-M NaCl, 2-mM MgCl2, 1% NP40 (w/v) with phosphatase/protease inhibitors (EMD Millipore, Billerica, MA, USA). Pellets were lyzed on ice using a Kontes homogenizer three times for 15 seconds each and sonicated thrice for 10 seconds each with an intervening 10-second incubation on ice. After centrifugation (5 minutes, 2000× g) to remove debris, protein concentrations of supernatants were determined by Bradford protein assay (Bio-Rad). Aggregate proteins were collected from equal amounts of total protein by centrifugation for 15 minutes at 14,000× g. The pellets were resuspended in buffer containing 1% sarcosyl (v/v); 0.1-M Hepes, pH 7.4; 5-mM EDTA, with protease inhibitors. Detergent-insoluble aggregate proteins were then recovered by ultracentrifugation (30 minutes, 100,000× g) and suspended in Laemmli buffer. Proteins were separated by gradient gel electrophoresis (1% sodium dodecyl sulfate [SDS], 4%−12% polyacrylamide). The separated proteins were visualized by staining with SYPRO Ruby (Invitrogen), and fluorescence intensities were quantified using ImageJ software. Protein intensities were normalized to E3 control, and the significance of difference between E3 and E4 with and without starvation was calculated by the two-tailed t-test. Proteins in lysates used for proteomic analyses were separated on 1% SDS-acrylamide gels, eluted from 1-mm slices, and incubated with trypsin. Peptides were then analyzed by LC-MS at the UAMS Proteomics Core facility and were identified by MASCOT search.
2.8. Wes Simple Western System
Protein quantification was done with the Wes Simple Western system, as per manufacturer’s specified protocols. Wes Simple Western uses an automated process of capillary gel electrophoresis. In brief, proteins were extracted from tissue with a lysis buffer comprising 50-mM Tris-HCl (pH 7.5), 150-mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate, and 0.1% SDS; lysates were quantified using a micro BCA assay reagent kit (Pierce, Rockford, IL, USA). Proteins were loaded onto Wes 25-well plates with Simple Western Sample Buffer and ProteinSimple MasterMix, containing dithiothreitol and fluorescent standards. Primary antibodies, horseradish-peroxidase-conjugated secondary antibodies, separation gel matrix, stacking gel matrix, and luminol-peroxide mixture were placed into the appropriate wells of the Simple Western plate. Chemiluminescent signal was quantified as peak area in the electropherogram (see Supplementary Fig. 2A) and also represented by the “gel view” function of the ProteinSimple software (Supplementary Fig. 2B).
2.9. RT-PCR amplification
Total RNA was extracted from cells using TriReagent RNA (Molecular Research Center, Cincinnati, OH, USA), according to the manufacturer’s instructions. Quality and quantity of RNA was determined by Agilent bioanalyzer. RT reactions were performed on equilibrated amounts of RNA using an ImProm II kit (Promega, Madison, WI, USA). PCR reactions used SybrGreen MasterMix reagents (Life Technologies; Grand Island, NY, USA). The sequences of primers were as follows: LC3B-forward (F): 5′-GTT ACG GAA AGC AGC AGT GTA-3′; LC3B-reverse (R): 5′-CAG AAG GGA GTG TGT CTG AAT G-3′; SQSTM1-F: 5′-CTG TCC CTC CTA ACA AGT GTA TC-3′; SQSTM1-R: 5′-CAC ACG ACT ATG TGA CCT CTT T-3′; LAMP2-F: 5′-GAA ATG CCA GTG TGT CCT AGA-3′; LAMP2-R: 5′-TCC CAAA GTG CTG GGA TTA C-3′; TFEB-F: 5′-CTC AAG GCC TCT GTG GAT TAC-3′; TFEB-R: 5′-AGC TGC TTG TTG GTC ATC TC-3′; PPP3-F: 5′-AGA GGC AAA GGG TTT GGA TAG-3′; PPP3-R: 5′-ATG TGC GGT GTT CAG AGA AT-3′ APOE-F: 5′-CCC AGG TCA CCC AGG AAC T-3′; APOE-R: 5′-AGT TCC GAT TTG TAG GCC-3′; 18S-F: 5′-TTC GGA CGT CTG CCC TAT CAA-3′; 18S-R: 5′-ATG GTA GGC ACG GCG ACT A-3′. PCR conditions were 40 cycles of 95°C for 10 minutes, 95°C 15 seconds, and 60°C for 60 seconds. All RT reactions were done using 1-μg total RNA in 25 μL, at a concentration of 40 ng/μL. After the RT reaction, an aliquot from each unknown sample is taken to make a pool from which the internal standard curve was made. Based on the stock concentration of 40 ng/μL, serial dilutions were made to produce a standard curve from which the concentrations of the experimental samples were determined; quantification was performed by interpolation to this standard curve. All mRNA levels were relative to 18S rRNA.
2.10. Structure modeling
A clean, full-length structure of apoE3 was first modeled using X-ray crystallography structure (PDB code: 2L7B) as a template in MODELLER 9.13 homology modeling [24]. Other variants of apoE were modeled from apoE3 structure by substituting Cys 112 to Arg (apoE4) or Arg 158 to Cys (apoE2) using the Mutagenesis Tool from Pymol [25] followed by energy minimization (5000 steps) with AMBER99SB-ILDN force field in GROMACS software package [26]. Using the same approach, other single–amino acid mutation models of apoE, including Arg112 to Ala, Arg61 to Ala, Glu255 to Ala, and Arg61 to Thr, were generated. The full-length structure of TFEB was modeled using fold recognition and the ab-initio approach from the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER). Structures were validated by analyzing the Ramachandran plot (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php) and Verify-3D program (http://services.mbi. ucla.edu/Verify_3D). All the nucleotide structures, including CLEAR and scrambled sequences, were modeled from make-na server (http://structure.usc.edu/make-na/server.html). Helix type was set to B, and sugar atom indicator was the default CNS with no hydrogens. Top and bottom codes were the default “RA.” Modeled nucleotides were then energy-minimized in GROMACS software with AMBER99SB-ILDN force field.
2.11. Protein-DNA docking
Interaction studies of apoE variants and DNA were performed in Hex 8.0. Correlation type was set to shape plus electrostatic and OPLS minimization as postprocessing. Other docking parameters were set to default. Out of 2000 possible orientations, the lowest energy complex was chosen for further analysis. DNA sequences were either generated by maintaining the same nucleotides with different positions, considered “scrambled,” or were generated with a random number generator to specify a random string of nucleotides of the same length, denoted as “random.” Hex docking energies for CLEAR sequence and scrambled sequences were plotted in Microsoft Excel.
2.12. Molecular-dynamic simulations
Atomistic simulations of protein-DNA complexes were run in the GROMACS environment with AMBER99SB-ILDN force field. Structures were first immersed in a cubical box containing SPC water molecules and counterions (NaCl). Complexes were then energy-minimized for 5000 steps with the steepest descent method and then equilibrated in two stages: stage 1 with constant temperature NVT (300 K) for 100-ps, using leap-frog integrator and modified Berendsen thermostat for temperature coupling; stage 2 with constant pressure NPT (1 bar) for 100 ps, using leap-frog algorithm as integrator and Parrinello-Rahman method for pressure coupling. Actual simulations were then performed for 10 to 50 ns with the leap-frog method as integrator. Results were then extracted from trajectories using GROMACS software package. All simulations were performed using in-house developed Linux scripts that automate the entire simulation process in GROMACS.
2.13. Binding of nuclear protein to CLEAR sequence via DNA pull-down assay
Nuclear extracts were isolated from apoE3- and apoE4-expressing T98G cells using Nuclear Extraction Kit (ab113474, Abcam), according to manufacturer’s recommendation. We designed an oligonucleotide corresponding to CLEAR sequence, biotinylated at the 5′ end (5′-Biotin-GTA GGT CAC GTG ACC GGG GTA GGT CAC GTG ACC GGG GTA GGT CAC GTG ACC GGG, IDT Technologies). Binding was performed for 8–12 hours at 4°C in binding buffer (20-mM HEPES pH 7.4, 150-mM NaCl, 1.5-mM MgC12, 1-mM dithiothreitol, 2 μg each of salmon sperm DNA and poly(dI-dC) containing 900 μg of nuclear protein and 1 μM double-stranded oligo. The protein bound oligo was isolated using streptavidin-coated beads (Invitrogen) using a magnetic rack. The bound beads were washed in 0.5X binding buffer (3X) to remove nonspecific binding. The oligo/protein-complex bound magnetic beads were suspended in Laemmli buffer and boiled for 5 minutes. The sample was then separated from magnetic beads using the magnetic rack and loaded on precast 4%–20% gradient gels (Bio-Rad) and electrophoresed under constant voltage of 90 volts for 2 hours. Gels were removed and proteins were blotted on polyvinylidene fluoride membranes and then probed with antibody against goat anti-TFEB (ab2636, Abcam) and mouse anti-apoE (Sc-13521, Santa Cruz Biotechnology). After incubating with HRP-conjugated anti-goat and anti-mouse secondary antibodies, protein bands were visualized via chemiluminescence (Clarity Western ECL, Bio-Rad).
2.14. Electrophoretic mobility shift assay
ApoE was purified from conditioned medium of T98G-apoE3-expressing or T98G-apoE4-expressing cells, as described in Wang [23]. ApoE proteins (50 ng) were mixed with BSA (5 μg) and incubated under conditions described by Mao et al. [27] with a 32P-labeled dsDNA probe containing either the CLEAR sequence (5′-GTA GGT CAC GTG ACC GGG-3′) or a scrambled sequence (5′-GTA GGT CTG CAG ACC GGG-3′). In control reactions, proteins were incubated with a 200-fold excess of nonradioactive probe before incubation with radioactive probe (cold competition) to test specificity, or with anti-apoE antibody to ensure identity of the protein in the DNA complex.
3. Results
3.1. Autophagy mRNAs in Alzheimer brain
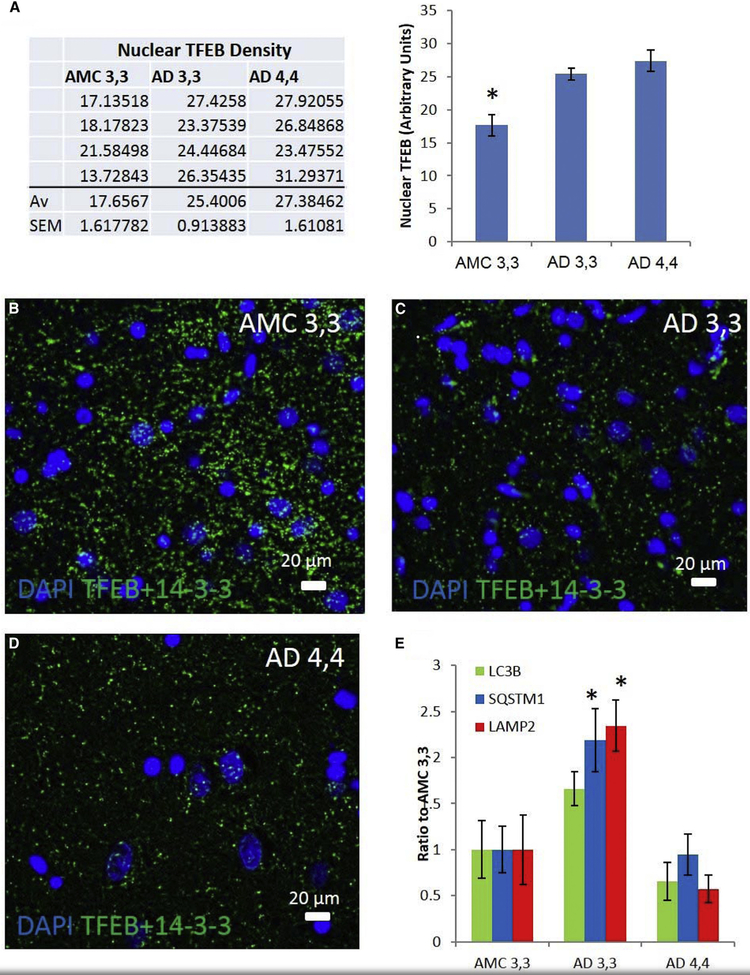
To more clearly define the role of autophagy in Alzheimer neuropathogenesis, particularly as it may be influenced by inheritance of APOE ε4/ε4, we assessed the roles played by the transcription factor TFEB, its transcriptomes, and in particular three principal autophagy proteins—LC3B, p62/SQSTM1, and LAMP2. The levels of TFEB in nuclei of hippocampal CA1 neurons in brain tissue from AMC patients were less than in analogous cells in AD patients (Fig. 1A), perhaps due to higher levels of TFEB tethered to 14-3-3, which we observed in cytoplasm of CA1 hippocampal neurons in AMC relative to that in AD patient carriers of either APOE ε3/ε3 or APOE ε4/ε4 genotype (Fig. 1B–1D). However, this tethering was probably not due to a lack of expression of the TFEB—14-3-3 untethering enzyme phosphatase PPP3 (calcineurin [28]) because the levels of PPP3 and TFEB mRNA were similar overall in AMC and AD patients and were even higher in AD 4,4 patients compared to either AMC or AD 3,3 patients (Supplementary Fig. 1A and 1B). Other autophagy-related transcripts not known to be regulated by TFEB/CLEAR, MTOR and RCAN1, are unaffected (Supplementary Fig. 1C and 1D), suggesting that this does not affect all autophagy transcripts, but may only affect those regulated by TFEB/CLEAR. Taken together, these results implicate a defect downstream of calcineurin-activated TFEB cytoplasmic-nuclear translocation in AD 4,4 patients. This might have been expected, given the elevation of calcineurin mRNA previously reported in young, healthy lymphocytes of patients with APOE ε4/ε4 [29].
Fig. 1.
AD patients demonstrate elevation of nuclear TFEB in patient carriers of both APOE ε3/ε3 (AD 3,3) and APOE ε4/ε4 (AD 4,4) compared to AMC ε3,3, while mRNAs are elevated only in AD 3,3. (A) Nuclear partitioning of TFEB was assessed by analysis of immunofluorescence histochemistry images of human hippocampal CA1 pyramidal cells in AD and AMC patients. Groups (each n = 4): AMC APOE ε4/ε4; AD 3,3; and AD 4,4 were compared by ANOVA, with Bonferroni-corrected α = 0.0167. AMC 3,3 versus AD 3,3 P = .006; AMC 3,3 versus AD 4,4 P < .01; AD 3,3 versus AD 4,4 P = .33. (B–D) Proximity ligation assay was used to illustrate the AD-related changes in cytoplasmic 14-3-3/TFEB complexes in an AMC patient, an AD 3,3, and an AD 4,4. (E) The relative levels (mRNA/18S) of MAP1LC3B, SQSTM1, and LAMP2 transcripts were determined by real-time RT-PCR in hippocampal specimens from AD and AMC patients, analyzed by disease state and separated by APOE genotype. Histogram shows means ± SEM. Significance of differences from AMC 3,3 (n = 6) was determined by two-tailed t-tests within ANOVA (Bonferroni-adjusted α < 0.02): P = .01 for AD 3,3, n = 5; and P < .01 for AD 4,4, n = 6. Abbreviations: AD, Alzheimer’s disease; AMC, age-matched control; APOE, apolipoprotein E; TFEB, transcription factor EB. *P = .05.
To determine whether TFEB-mediated transcription of autophagy mRNAs is affected by APOE genotype, we measured the mRNA levels of three TFEB-mediated autophagy genes: SQSTM1, MAP1LC3B, and LAMP2. Despite elevated nuclear localization of TFEB in AD patients relative to AMC, there was essentially no difference between AMC and AD in the levels of these three TFEB-regulated mRNA transcripts (Fig. 1E, LC3B, P = .88; SQSTM1, P = .29, and LAMP2, P = .59). However, in view of the greater P-tau and Aβ pathology characteristic of AD 4,4 patients, we separated AD cases based on their APOE genotype and, in this way, revealed a striking difference: AD 3,3 patients had levels of all three TFEB-regulated autophagy gene transcripts that were 2.5- to 4-fold greater than in AD 4,4 patients (Fig. 1E; significance by ANOVA, with Bonferroni-corrected α of 0.017: P = .013, 0.01, and 3 × 10−4 for LC3B, SQSTM1, and LAMP2, respectively), indicating that AD 3,3 patients mount a better transcriptional response than do AD 4,4 patients. Our finding of a similar pattern of expression of one of the TFEB-regulated proteins, SQSTM1/p62 (Supplementary Fig. 2), suggests that the preferential failure in autophagy in AD 4,4 patients arises more at the level of transcription rather than translation. This, along with the observation of no APOE genotype-related differences in the nuclear localization of TFEB, implies that an APOE genotype–specific dysfunction occurs at a step between TFEB translocation to the nucleus and TFEB-mediated mRNA transcription in AD brain tissue.
3.2. Aggregation and autophagy mRNAs in apoE3- or apoE4-expressing cells
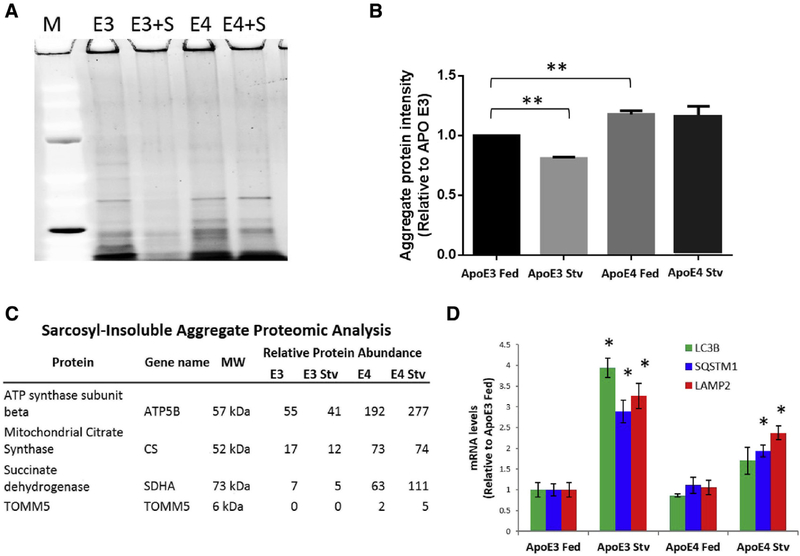
To assess the direct effects of apoE on protein aggregation, we examined clones of the T98G human astroglioma cell line stably transfected with transgenes expressing either apoE3 (T98G-apoE3) or apoE4 (T98G-apoE4). This cell line was chosen due to the fact that it produces complete, lipidated apoE molecules in vitro. In control conditions (“Fed”), T98G-apoE4 cells had ~20% higher levels of sarcosyl-insoluble intracellular aggregates than T98G-apoE3 cells (Fig. 2A and 2B, E3 vs. E4) in two different sets of clones. To test for apoE isoform-specific effects on the responsiveness of autophagy to activating conditions, we stressed cells by starvation (“Stv”) and found that under these conditions, T98G-apoE3 cells lost 22% of their aggregate content, whereas starvation had no significant effect on aggregates in T98G-apoE4 cells (Fig. 2A and 2B). These results suggested possible blunting of the apoE4-related autophagy response to the stress of starvation, which we further explored by assessing mitochondrial protein abundance through proteomics analysis (Fig. 2C). These proteins were chosen due to the fact that they would be expected to be preferentially degraded by the lysosomes, and citrate synthase in particular has been used as a marker for mitophagy [30,31]. Consistent with deficient autophagy in apoE4-expressing cells, we find that these mitochondrial proteins are enriched in the apoE4 lysates compared to apoE3 lysates. Furthermore, apoE3 cells decreased these protein concentrations under starvation stress, but apoE4 cells paradoxically elevated these concentrations. We then assessed the levels of three TFEB-mediated transcripts (Fig. 2D; protein levels, Supplementary Fig. 3). Transcript levels for SQSTM1/p62, LC3B, and LAMP2 were not different in T98G-apoE3 and T98G-apoE4 cells under fed conditions. However, under starvation stress, T98G-apoE3 cells increased these same transcripts by 3- to 4-fold, that is, to levels that averaged twice that of T98G-apoE4 cells. Similar results were obtained with two separate clones for each apoE isoform. TFEB mRNA was unchanged in all groups, but starvation did increase the TFEB-translocation activator PPP3 mRNA by at least twofold in both apoE groups (Supplementary Fig. 4), suggesting that the lack of an increase in autophagy in starved T98G-apoE4 cells is not due to inability of TFEB to translocate to the nucleus, to the available amount of TFEB, or to a failure of transcription in general, but instead is due to a failure in TFEB-mediated transcriptional upregulation.
Fig. 2.
Aggregate levels, aggregate clearance, and transcription of autophagy mRNAs are apoE isoform dependent. (A) T98G cells stably transfected with either apoE3 (E3) or apoE4 (E4) were incubated for 3 hours in control medium (“Fed”) or a medium lacking nutrients (“Stv”). Aggregates insoluble in 1% sarcosyl were collected by ultracentrifugation, resolved by sodium dodecyl sulfate - polyacrylamide gel electrophoresis, and visualized by SYPRO Ruby. (B) Aggregated proteins in such gels were quantified by densitometry, showing that apoE3 cells had less aggregate protein in the control state than apoE4 cells (**P < .01 by t-test), and that apoE3 cells respond to starvation by increasing clearance of aggregates (**P < .01 by t-test), while apoE4 cells do not. (C) Mitochondrial proteins, preferentially degraded through mitophagy (entailing lysosomal fusion), are enriched in proteomic analysis of sarcosyl-insoluble aggregates isolated from T98G cells expressing apoE4 compared to apoE3. Starvation in apoE3-expressing cells leads to a decrease in these proteins, whereas starvation in apoE4-expressing cells causes further enrichment, implying a defect in lysosomal-stress response. (D) In the fed state, the relative levels of MAP1LC3B, SQSTM1, and LAMP2 mRNAs, assessed by real-time RT-PCR, are similar in apoE3 and apoE4 cells (two-tailed t-test: LC3B, P = .48; SQSTM1, P = .69; LAMP2, P = .82). ApoE3 cells have an approximately 3-fold increased expression during starvation (one-tailed t-test: LC3B, P < .02; SQSTM1, P = .04; LAMP2, P = .04), whereas apoE4 cells are only able to increase expression by approximately 1.5-fold (one-tailed t-test: LC3B, P = .10; SQSTM1, P = .04; LAMP2, P = .02). Abbreviation: ApoE, apolipoprotein E. *P = .05.
3.3. ApoE4 modeling and simulation predict direct binding to CLEAR motifs
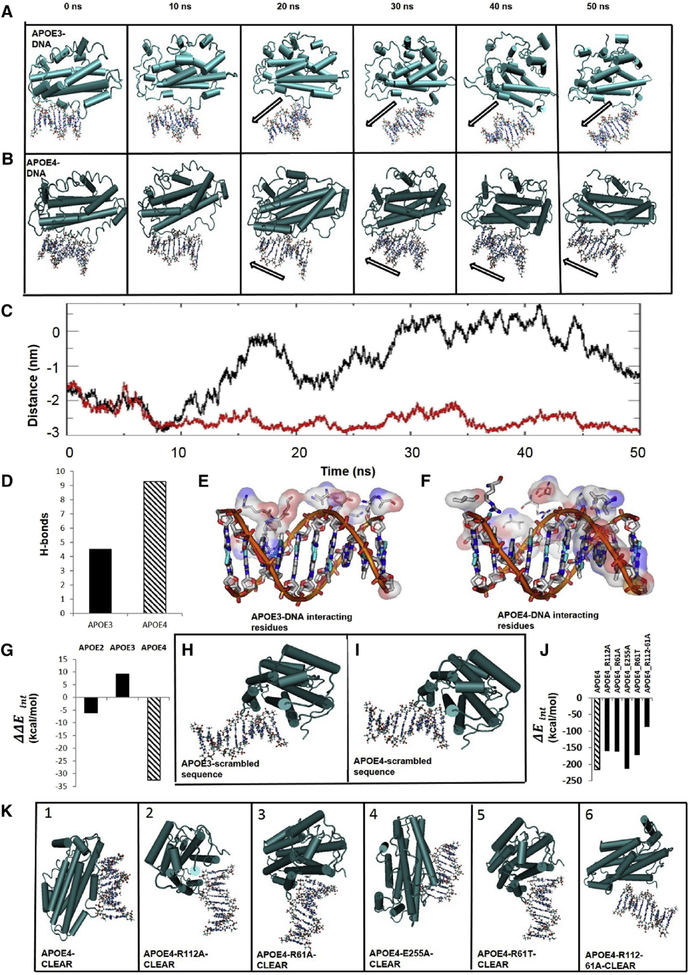
Prior studies have documented a nuclear pool of apoE that is elevated by stressors such as serum starvation [32–34], and evidence was recently provided that apoE controls transcription via direct DNA binding. Theendakara et al. [22] identified an apoE-binding region, which we noted contained a sequence (GCCACGTGAC) consistent with the consensus CLEAR motif, the enhancer bound by TFEB. To address the possibility that ApoE may bind to the CLEAR sequence of DNA, we used the HEX docking program [35] to model protein-DNA interactions in silico for apoE2, apoE3, or apoE4 binding to DNA; complexes were then simulated for 10–50 ns using the GROMACS molecular-dynamic simulation package [26]. In each case, the models predicted only weak and unstable interactions of apoE3 with CLEAR during the 50-ns simulation (Fig. 3A, Supplementary Video 1); no interactions with apoE2 (Supplementary Fig. 5); whereas apoE4 was predicted to form a stable complex with the CLEAR motif throughout the entire 50-ns simulation (Fig. 3B, and Supplementary Video 2). In addition, the center-of-mass distance between apoE3 and CLEAR increased and fluctuated over the course of the simulation (Fig. 3C, black line), whereas the distance between apoE4 and CLEAR decreased and remained relatively stable (Fig. 3C, red line). Modeling also predicted that on average, apoE4 formed twice as many hydrogen bonds with the CLEAR sequence as apoE3 (Fig. 3D). Moreover, compared to apoE3, more apoE4 amino-acid residues were predicted to interact with CLEAR DNA (Fig. 3E and 3F).
Fig. 3.
Molecular modeling predicts apoE4, but not apoE3, specifically and preferentially interacts with the CLEAR sequence on dsDNA. Simulations of interactions between apoE and CLEAR dsDNA (GTCACGTGAC), over a 50-ns period, assessed at 10-ns intervals. Interactions are visualized with apoE3 (A) or apoE4 (B) depicted in a “pipes-and-planks” format, and the CLEAR dsDNA depicted in a “ball-and-stick” format. ApoE3-CLEAR dsDNA interactions were unstable or nonexistent (A, black outline arrow; C, black trace) over 50 ns, whereas apoE4-CLEAR dsDNA interactions were early and sustained over the 50-ns period (B, black outline arrow; C, red trace). (C) Center-of-mass distance between the CLEAR sequence and either apoE3 or apoE4 over time. (D) Hydrogen bonds (H-bonds) predicted between the CLEAR sequence and either apoE3 or apoE4. ApoE3 amino-acid residues interact with the sugar-phosphate backbone but not with the base pairs (major and minor grooves) of the CLEAR dsDNA sequence (E), while there are multiple apoE4 amino-acid-residue interactions in grooves of the CLEAR dsDNA (F). (G) ApoE isoform-CLEAR interaction energies minus average interaction energies with non-CLEAR DNA sequences (Supplementary Fig. 6) predict preferential interaction of apoE4 to the CLEAR sequence over and above other DNA sequences. Visualization of interactions between a scrambled dsDNA sequence (Supplementary Fig. 6) and apoE3 (H) or apoE4 (I). Predicted interactions between CLEAR DNA and various missense mutations of apoE4 are represented quantitatively (J) and visually (K). Abbreviations: ApoE, apolipoprotein E; CLEAR, coordinated lysosomal expression and regulation.
To analyze the specificity of apoE4 to CLEAR, we tested scrambled CLEAR sequences (same nucleotides in a different order, scram) or random nucleotides (random, Supplementary Fig. 6) for docking with either apoE3 or apoE4. The average interaction energy (ΔE) for apoE4 binding to random DNA sequences was −188.3 kcal/mol, implying substantially less stability than for its binding to the CLEAR sequence (ΔE = −216.1). The difference between CLEAR binding and random sequences was −32.34 kcal/mol (Fig. 3G, lane 3). In contrast, apoE3 bound to altered motif sequences with an average interaction energy of −148.6 kcal/mol, indicating slightly higher stability than its complex with the CLEAR sequence at ΔE = −139.6, with a difference of +9.037 kcal/mol (Fig. 3G, lane 2; apoE2 binding was similar to apoE3, a difference of −6.110 kcal/mol, lane 1). Thus, our modeling data predicted that, relative to altered sequences of the same length, apoE4 will bind spontaneously and specifically to the CLEAR sequence (Supplementary Fig. 6), whereas neither apoE2 nor apoE3 was predicted to show a preference for the CLEAR sequence over modified sequences. Comparing the molecular orientation for the altered CLEAR sequences, apoE3 and apoE4 form tenuous complexes with a scrambled CLEAR motif (Fig. 3H and 3I); in fact, the orientation of these complexes is comparable to that of the weak interaction of apoE3 with the CLEAR DNA (Fig 3A). The predicted extensive and stable apoE4-CLEAR sequence interactions (Fig. 3B, initial panels) contrasts with those of apoE2 or apoE3, predicting that only apoE4 has the potential to bind the CLEAR motif with specificity and relatively high affinity.
The amino-acid sequences of apoE3 and apoE4 differ only at amino acid 112, which is Cys in apoE3 and Arg in apoE4 [36]. Substitution with Ala at this position compromised the apparent affinity of apoE4 for CLEAR, confirming the importance of a single amino acid in protein-DNA binding. To find other such “hotspots” that may be critical for specific apoE4 binding to CLEAR DNA, we performed a series of substitutions based on the findings of others [37]. When Arg61 was replaced with Ala or Thr (mouse apoE; Fig. 3 J, K, panels 2, 3, 5), the in silico modeling predicted a substantial shrinkage of ΔEinteraction, the energy change favoring apoE4 binding to CLEAR. Double replacement of Arg112 and Arg61 with Ala resulted in a further loss of interaction, indicating that both Arg112 and Arg61 contribute to CLEAR binding (Fig. 3 J, K, panel 6). On the other hand, replacing Glu255 with Ala did not alter the apoE4 interaction with CLEAR (Fig. 3 J, K, panel 4). ApoE4–CLEAR sequence modeling reveals close proximity of Arg112 to the phosphate backbone of the CLEAR sequence, but only in the presence of Arg61. This type of interaction is known to stabilize DNA-histone binding [38], especially in the case of two nearby Arg residues [39]. Collectively, these results predict that apoE4 will have stronger CLEAR binding than apoE3 and that this interaction greatly depends on Arg112 and Arg61, suggesting a possible therapeutic target. Together, these predictions provide further evidence for stable apoE4-CLEAR interactions and in this way, may explain at least in part, the relationship between inheritance of APOE ε4/ε4 and neuropathological evidence of greater aggregate load in patient carriers of this genotype.
3.4. ApoE4 binds CLEAR probes in vitro
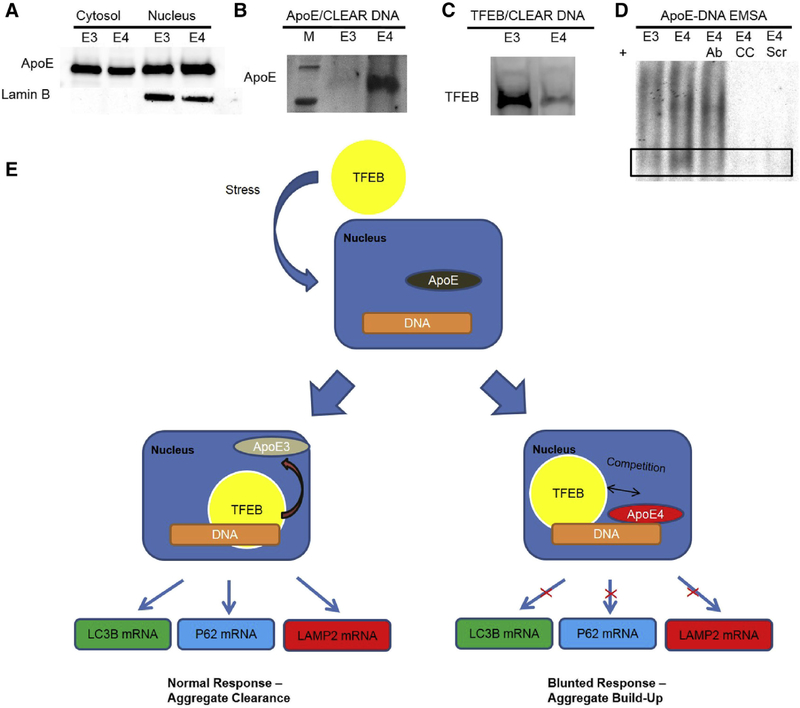
In line with reports on other cell types [32–34], we found apoE in the nucleus of both T98G-apoE3 and T98G-ApoE4 cells (Fig. 4A). To verify our predictions from in silico modeling, we performed a pull-down assay using biotinylated CLEAR DNA probes with lysate from either apoE3- or apoE4-expressing T98G cells. The protein was then run on a gel and probed with appropriate antibody. We found that more apoE protein was pulled down by CLEAR probes in the apoE4-expressing cells compared to apoE3-expressing cells (Fig. 4B). As further evidence supporting our hypothesis, TFEB binding to the CLEAR probes was diminished in apoE4 cells compared to apoE3 cells, which is consistent with apoE4 competing with TFEB for CLEAR sites (Fig. 4C). Electrophoretic mobility shift assays were also performed using a DNA probe containing the CLEAR motif. Purified apoE3 or apoE4 proteins produced in T98G cells were compared, and apoE4 produced a more intense binding reaction (Fig. 4D). Furthermore, this binding was diminished by antibody against apoE (“4 + Ab”; lane 3) and was abolished almost entirely when incubated with an excess of nonradioactive probe (cold competition, “4 + CC”; lane 4). As an additional test of specificity, we tested binding to a probe of scrambled sequence that was predicted in silico to have a poor interaction with apoE4 (“4 + scram1”; lane 5). These data demonstrate that apoE4 protein directly and specifically binds CLEAR in vitro as well as in silico.
Fig. 4.
ApoE4-expressing astroglioma cells show more apoE binding and less TFEB binding to CLEAR DNA than apoE3-expressing cells. (A) ApoE is present in the nucleus and the cytosol of apoE-expressing T98G cells. (B) Western-blot analysis of proteins pulled down by binding to CLEAR-motif dsDNA shows elevated apoE4 binding relative to apoE3. (C) ApoE4-expressing cells show less TFEB binding to CLEAR-motif dsDNA relative to cells expressing apoE3. (D) Electrophoretic mobility shift assay (EMSA), in which a 32P-labeled probe containing the CLEAR motif was incubated with 50 ng of recombinant apoE3 (lane 1, “E3”) or apoE4 (lane 2, “E4”) purified from T98G cells and resolved on a nondenaturing gel. ApoE4 was also tested in the presence of 2 μg of antibody to apoE (lane 3, “E4 Ab”), saturating levels of unlabeled CLEAR dsDNA as cold competition (lane 4, “E4 CC”), or a 32P-labeled probe of scrambled sequence (lane 5, E4 scram”), demonstrating sequence specificity for the apoE4 isoform-binding to the CLEAR sequence on dsDNA. (E) Schematic of hypothesized modulation of TFEB-CLEAR interactions by apoE. Stresses, such as starvation as we show in these cells, or other stresses that we postulate in AD, evoke nuclear translocation of TFEB. In apoE3-expressing cells, there is reduced binding of apoE to CLEAR sequences, allowing TFEB greater upregulation of transcription of autophagy genes. Conversely, apoE4 competes with TFEB for CLEAR sequences and thus blunts transcription of autophagy mRNAs. This can predispose cells to proteotoxic damage, activation of inflammatory cytokines, and ultimately to cell death; the first is of particular note in neurons as dysfunctions in autophagy result in buildup of aggregates and formation of paired helical filaments of hyperphosphorylated tau. Abbreviations: AD, Alzheimer’s disease; ApoE, apolipoprotein E; CLEAR, coordinated lysosomal expression and regulation; TFEB, transcription factor EB.
4. Discussion
The evidence we provide for decreased autophagy mRNA levels in AD patient carriers of APOE ε4/ε4 (AD 4,4) relative to those with APOE ε3/ε3 (AD 3,3) helps to explain both the profound consequences of this genetic risk factor, and the well-documented autophagic insufficiency of AD, consistent with the increased levels of aggregated proteins in AD 4,4 [7,9]. Reminiscent of the increased aggregate load in AD patient carriers of APOE ε4/ε4, human astroglioma cells transformed to produce apoE4 had higher levels of protein aggregates than those expressing apoE3, with preferential enrichment of mitochondrial proteins on proteomic analysis (Fig. 2C), implying specific lysosomal dysfunction. Furthermore, the apoE4 transformants failed to increase either autophagy-related gene expression or aggregate clearance in response to starvation stress. Both in silico and in vitro evidence is consistent with apoE4 protein binding the CLEAR promoter sequence with high specificity and affinity and may imply that apoE4 competes with TFEB for the CLEAR motif. This predicted interaction between apoE4 and CLEAR was found to be dependent on Arg112 and Arg61 but not Glu255. As further evidence of sequence specificity, preferential apoE4 interaction with CLEAR sites is disrupted by mutation or scrambling of the CLEAR motif and is neither predicted nor observed for either apoE2 or apoE3. Preferential competition of apoE4 for the DNA motifs targeted by TFEB is supported by in silico dynamic modeling of interactions between CLEAR sequences and apoE2, apoE3, and apoE4 proteins. Moreover, we demonstrated that not only does apoE4 bind to CLEAR probes more effectively than apoE3, but apoE4-expressing cells also have reduced TFEB binding to these CLEAR probes.
Cataldo et al. [12] demonstrated, in a pathway that is closely related to autophagy, that early endosomes are enlarged in brains of early-stage AD patients with at least one APOE ε4 allele. Furthermore, Aβ trafficking through the endosomal system to the lysosome has been shown to be deficient in the presence of apoE4 compared to apoE3 [40]. Badia et al. showed that even in the lymphocytes of young, healthy apoE4 carriers, calcineurin is elevated both at mRNA and protein levels, and that levels of hyperphosphorylated tau are higher in those lymphocytes, consistent with reduced TFEB activation and a compensatory increase in calcineurin activity in these patients [29]. Increasing autophagy through TFEB and the CLEAR network can ameliorate several aspects of AD-like pathology in various mouse and cell culture models [16–18]. Furthermore, astrocytes from apoE4-targeted replacement mice have lower levels of autophagy and reduced ability to clear amyloid plaques [41]. A recent report noted that apoE4 expression in a P-tau mouse model caused increased neuronal death and elevations in neuroinflammation even apart from amyloidosis, further implicating the effect of apoE4 on autophagy [42]. Our cell culture and in silico models provide evidence that apoE4 directly binds to DNA at CLEAR promoter sites and in this way downregulates transcription of autophagy gene products by direct interference with TFEB-CLEAR binding (Fig. 4D), leading to increases that could account for the observed increase in apoE4 aggregate accumulation (Fig. 2A and 2B). These data provide intriguing evidence that the APOE ε4 genotype contributes to Alzheimer risk not only through various effects on amyloid clearance, as has been suggested by other investigators, but also by having a direct transcriptional effect on autophagy genes regulated by TFEB/CLEAR, thus weakening the proteostatic stress-response at the cell-autonomous level.
Competition of apoE4 with TFEB for the CLEAR motif, which we show results in reduced expression of autophagy proteins, represents a toxic gain of function that is compatible with the autosomal dominance and incomplete penetrance of the genetic association of APOE ε4 with AD. Furthermore, such apoE4-related downregulation of autophagy is remarkable for its distinction from the other functions of apoE, such as its role in lipid transport. This novel mechanistic explanation may thus afford a dramatically different therapeutic approach for the prevention or treatment of AD that would be free from side effects that may arise from strategies that could adversely impact other functions of apoE.
Here, for the first time, the negative impact of inheritance of APOE ε4 alleles may be attributed to apoE4 interactions with the CLEAR motif, which, as we show in AD 4,4 patients, lessen transcription of SQSTM1, MAP1LC3B, and LAMP2 mRNAs and thus lower levels of their three autophagy proteins, p62, LC3B, and LAMP2. This is consistent with the accrual of misfolded or damaged proteins that accumulate in aggregates, especially Aβ and P-tau in AD, and α-synuclein in Parkinson’s disease. Furthermore, any suppressive effects of apoE4 on autophagy may influence other factors such as neuroinflammation. Such suppression of autophagy, for example, could lead to increases in IL-1β [43], which is a known driver of the induction of expression of the precursor proteins of the neuropathological hallmarks of AD and Parkinson’s disease [44]. Therefore, it seems logical that our findings may be generalizable to other neurodegenerative diseases that are characterized by proteostatic failure, neuroinflammation, and aggregate formation.
Supplementary Material
RESEARCH IN CONTEXT.
Systematic review: Based on previous findings of more Aβ plaques and neurofibrillary tangles in the brains of Alzheimer patients having apolipoprotein E (APOE) ε4/ε4 genotype, we studied the role of ApoE in regulation of autophagy, through pathology studies in control (APOE ε3/ε3) and Alzheimer patients (APOE ε3/ε3 or APOE ε4/ε4), T98G cell cultures expressing either apoE3 or apoE4, computational modeling of ApoE interactions with dsDNA, DNA-protein pull-down assay, and electro-phoretic mobility shift assay. This was based on the premise that the presence of more Alzheimer-related aggregates is associated with downregulation of autophagy in APOE ε4/ε4 carriers.
Interpretation: The results reported in this article show that autophagy is downregulated at the level of transcription of three key autophagy proteins. Our results suggest that this downregulation is due to preferential and specific binding of apoE4 protein to dsDNA containing the coordinated lysosomal expression and regulation (CLEAR) site and in this way competes with transcription factor EB (TFEB), effectively preventing transcription of the mRNAs for the three key autophagy proteins. This is in contrast to both apoE2 and apoE3.
Future directions: We will continue by exploring the potential of small molecules to prevent apoE4 binding to the CLEAR site, thus allowing TFEB-mediated transcription of autophagy mRNAs for production of their protein products, as well as to further analyze the composition of apoE4-specific protein aggregates through proteomic analysis.
Acknowledgments
This work was supported by NIA AG12411 and the US Dept. of Veterans Affairs Merit Award Grant #2 I01 BX001655–05. The US National Science Foundation grants CRI CNS-0855248, EPS 0701890, EPS-0918970, and MRI CNS-0619069 supported the High Performance Computing (HPC) at the University of Arkansas at Little Rock Computational Research Center (CRC). Further funding was provided by the Windgate Foundation, the Philip R. Jonsson Foundation, the Ottenheimer Brothers Foundation, and the STOP Alzheimer’s campaign at the UAMS Foundation.
Footnotes
Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jalz.2017.07.754.
References
- [1].Strittmatter WJ, Roses AD. Apolipoprotein E and Alzheimer disease. Proc Natl Acad Sci U S A 1995;92:4725–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Roses AD, Saunders AM, Alberts MA, Strittmatter WJ, Schmechel D, Gorder E, et al. Apolipoprotein E E4 allele and risk of dementia. Jama 1995;273:374–5. author reply 5–6. [PubMed] [Google Scholar]
- [3].Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet 2007;39:17–23. [DOI] [PubMed] [Google Scholar]
- [4].Reitz C, Brayne C, Mayeux R. Epidemiology of Alzheimer disease. Nat Rev Neurol 2011;7:137–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Riedel BC, Thompson PM, Brinton RD. Age, APOE and sex: triad of risk of Alzheimer’s disease. The J Steroid Biochem Mol Biol 2016; 160:134–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Polvikoski T, Sulkava R, Haltia M, Kainulainen K, Vuorio A, Verkkoniemi A, et al. Apolipoprotein E, dementia, and cortical deposition of beta-amyloid protein. N Engl J Med 1995;333:1242–7. [DOI] [PubMed] [Google Scholar]
- [7].Christensen DZ, Schneider-Axmann T, Lucassen PJ, Bayer TA, Wirths O. Accumulation of intraneuronal Abeta correlates with ApoE4 genotype. Acta Neuropathol 2010;119:555–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ayyadevara S, Balasubramaniam M, Parcon PA, Barger SW, Griffin WS, Alla R, et al. Proteins that mediate protein aggregation and cytotoxicity distinguish Alzheimer’s hippocampus from normal controls. Aging Cell 2016;15:924–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Aboud O, Parcon PA, DeWall KM, Liu L, Mrak RE, Griffin WS. Aging, Alzheimer’s, and APOE genotype influence the expression and neuronal distribution patterns of microtubule motor protein dynactin-P50. Front Cell Neurosci 2015;9:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med 2013;19:983–97. [DOI] [PubMed] [Google Scholar]
- [11].Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol 2005;64:113–22. [DOI] [PubMed] [Google Scholar]
- [12].Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer’s disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol 2000;157:277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lee S, Sato Y, Nixon RA. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer’s-like axonal dystrophy. J Neurosci 2011; 31:7817–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Colacurcio DJ, Nixon RA. Disorders of lysosomal acidification-The emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res Rev 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wolfe DM, Lee JH, Kumar A, Lee S, Orenstein SJ, Nixon RA. Autophagy failure in Alzheimer’s disease and the role of defective lysosomal acidification. Eur J Neurosci 2013;37:1949–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang YD, Zhao JJ. TFEB participates in the Abeta-induced pathogenesis of Alzheimer’s disease by regulating the autophagy-lysosome pathway. DNA Cel Biol 2015;34:661–8. [DOI] [PubMed] [Google Scholar]
- [17].Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, et al. Neuronal-targeted TFEB accelerates lysosomal degradation of APP, reducing Abeta generation and amyloid plaque pathogenesis. J Neurosci 2015; 35:12137–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Polito VA, Li H, Martini-Stoica H, Wang B, Yang L, Xu Y, et al. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol Med 2014;6:1142–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science 2011;332:1429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 2011; 20:3852–66. [DOI] [PubMed] [Google Scholar]
- [21].Unuma K, Aki T, Funakoshi T, Yoshida K, Uemura K. Cobalt protoporphyrin accelerates TFEB activation and lysosome reformation during LPS-induced septic insults in the rat heart. PLoS One 2013; 8:e56526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Theendakara V, Peters-Libeu CA, Spilman P, Poksay KS, Bredesen DE, Rao RV. Direct transcriptional effects of apolipoprotein E. J Neurosci 2016;36:685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang W, Moerman-Herzog AM, Slaton A, Barger SW. Presenilin 1 mutations influence processing and trafficking of the ApoE receptor apoER2. Neurobiol Aging 2017;49:145–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics 2006. Chapter 5:Unit 5 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.8. 2015.
- [26].Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem 2005;26:1701–18. [DOI] [PubMed] [Google Scholar]
- [27].Mao X, Phanavanh B, Hamdan H, Moerman-Herzog AM, Barger SW. NFkappaB-inducing kinase inhibits NFkappaB activity specifically in neurons of the CNS. J Neurochem 2016; 137:154–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cel Biol 2015;17:288–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Badia MC, Lloret A, Giraldo E, Dasi F, Olaso G, Alonso MD, et al. Lymphocytes from young healthy persons carrying the ApoE4 allele overexpress stress-related proteins involved in the pathophysiology of Alzheimer’s disease. J Alzheimers Dis 2013; 33:77–83. [DOI] [PubMed] [Google Scholar]
- [30].Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of as-says for monitoring autophagy (3rd edition). Autophagy 2016; 12:1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Amadoro G, Corsetti V, Florenzano F, Atlante A, Ciotti MT, Mongiardi MP, et al. AD-linked, toxic NH2 human tau affects the quality control of mitochondria in neurons. Neurobiol Dis 2014; 62:489–507. [DOI] [PubMed] [Google Scholar]
- [32].Panin LE, Russkikh GS, Polyakov LM. Detection of apolipoprotein A-I, B, and E immunoreactivity in the nuclei of various rat tissue cells. Biochem Biokhimiia 2000;65:1419–23. [DOI] [PubMed] [Google Scholar]
- [33].Quinn CM, Kagedal K, Terman A, Stroikin U, Brunk UT, Jessup W, et al. Induction of fibroblast apolipoprotein E expression during apoptosis, starvation-induced growth arrest and mitosis. Biochem J 2004;378:753–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kim WS, Elliott DA, Kockx M, Kritharides L, Rye KA, Jans DA, et al. Analysis of apolipoprotein E nuclear localization using green fluorescent protein and biotinylation approaches. Biochem J 2008; 409:701–9. [DOI] [PubMed] [Google Scholar]
- [35].Macindoe G, Mavridis L, Venkatraman V, Devignes MD, Ritchie DW. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res 2010;38:W445–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Huang Y, Mahley RW. Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol Dis 2014;72 Pt A:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chen HK, Ji ZS, Dodson SE, Miranda RD, Rosenblum CI, Reynolds IJ, et al. Apolipoprotein E4 domain interaction mediates detrimental effects on mitochondria and is a potential therapeutic target for Alzheimer disease. J Biol Chem 2011;286:5215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yusufaly TI, Li Y, Singh G, Olson WK. Arginine-phosphate salt bridges between histones and DNA: intermolecular actuators that control nucleosome architecture. The J Chem Phys 2014;141:165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kono H, Shirayama K, Arimura Y, Tachiwana H, Kurumizaka H. Two arginine residues suppress the flexibility of nucleosomal DNA in the canonical nucleosome core. PLoS One 2015;10:e0120635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Li J, Kanekiyo T, Shinohara M, Zhang Y, LaDu MJ, Xu H, et al. Differential regulation of amyloid-beta endocytic trafficking and lysosomal degradation by apolipoprotein E isoforms. J Biol Chem 2012; 287:44593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Simonovitch S, Schmukler E, Bespalko A, Iram T, Frenkel D, Holtzman DM, et al. Impaired autophagy in APOE4 astrocytes. J Alzheimers Dis 2016;51:915–27. [DOI] [PubMed] [Google Scholar]
- [42].Shi Y, Yamada K, Liddelow SA, Smith ST, Zhao L, Luo W, et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 2017;549:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Autophagy Harris J. and IL-1 family cytokines. Front Immunol 2013; 4:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Griffin WS, Liu L, Li Y, Mrak RE, Barger SW. Interleukin-1 mediates Alzheimer and Lewy body pathologies. J Neuroinflammation 2006; 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.