Abstract
Rationale:
The immature presentation of human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) is currently a challenge for their application in disease modeling, drug screening, and regenerative medicine. Long-term culture is known to achieve partial maturation of iPSC-CMs. However, little is known about the molecular signaling circuitries that govern functional changes, metabolic output, and cellular homeostasis during long-term culture of iPSC-CMs.
Objective:
We aimed to identify and characterize critical signaling events that control functional and metabolic transitions of cardiac cells during developmental progression, as recapitulated by long-term culture of iPSC-CMs.
Methods and Results:
We combined transcriptomic sequencing with pathway network mapping in iPSC-CMs that were cultured until a late time point, day 200 (D200), in comparison to a medium time point, day 90 (D90), and an early time point, day 30 (D30). Transcriptomic landscapes of long-term cultured iPSC-CMs allowed mapping of distinct metabolic stages during development of maturing iPSC-CMs. Temporally divergent control of mitochondrial metabolism was found to be regulated by cAMP/protein kinase A (PKA)- and proteasome-dependent signaling events. The PKA/proteasome-dependent signaling cascade was mediated downstream by heat shock protein 90 (Hsp90), which in turn modulated mitochondrial respiratory chain proteins and their metabolic output. During long-term culture, this circuitry was found to initiate upregulation of iPSC-CM metabolism, resulting in increased cell contractility that reached a maximum at the D200 time point.
Conclusions:
Our results reveal a PKA/proteasome- and Hsp90-dependent signaling pathway that regulates mitochondrial respiratory chain proteins and determines cardiomyocyte energy production and functional output. These findings provide deeper insight into signaling circuitries governing metabolic homeostasis in iPSC-CMs during developmental progression.
Keywords: Human induced pluripotent stem cells, cardiomyocytes, proteasome, metabolism, mitochondria, cardiac stem cells, metabolism, mitochondria
Subject Terms: Basic Science Research, Cell Signaling/Signal Transduction
INTRODUCTION
Human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) have shown great potential in cardiovascular disease (CVD) modeling,1–7 drug screening,8 and potential therapies including transplantation of iPSC-CMs into injured myocardium.9–12 Rapid progress in the field of iPSCs and their differentiation into specific lineages such as cardiomyocytes have enabled their use for disease modeling, drug screening, and regenerative medicine.13–16 However, their immature phenotypic presentation,17, 18 which resembles early human embryonic cardiomyocytes, presents a challenge for realizing the full potential of iPSC-CMs. Although long-term culture is recognized as a valid approach for maturation of both human iPSC-CMs19 and human embryonic stem cell-derived cardiomyocytes (ESC-CMs),20, 21 little is known about the signaling circuitries that govern metabolic output and cellular homeostasis during long-term culture of iPSC-CMs.
To understand these critical signaling events, we combined transcriptomic sequencing with pathway network mapping of iPSC-CMs that were cultured until a late time point, day 200 (D200), in comparison to a medium time point, day 90 (D90), and an early time point, day 30 (D30). We found an upregulation of metabolic functions at later time points, particularly at D200, which is regulated in a cAMP/PKA-dependent and proteasome-dependent manner. The ubiquitin-proteasome system is the major cellular protein degradation pathway. After tagging with ubiquitin, proteins are bound and hydrolyzed by the 26S proteasome in an ATP-dependent manner that may be modulated by cAMP/PKA regulation.22, 23 Here we report a novel contribution of cAMP/PKA- and proteasome-dependent signaling to mitochondrial metabolic viability and functionality. This signaling pathway was found to operate differently at D30, D90, and D200 time points, and was chaperoned by Hsp90, a proteasome-interacting heat shock protein24 that is mainly localized to the cytoplasm as well as some other subcellular sites such as mitochondria.25 We propose a proteasome-concerted, Hsp90-dependent modulation of the mitochondrial respiratory chain output that feeds back into cardiomyocyte function.
METHODS
Data availability.
In order to minimize the possibility of unintentionally sharing information that can be used to re-identify private information, a subset of the data generated for this study are available at GEO database and can be accessed at GSE131236.
Culture of human iPSCs.
Human induced pluripotent stem cells (iPSCs) were grown as described previously7 on Matrigel-coated plates (ES qualified, BD Biosciences, San Diego) using chemically defined E8 medium.26 The culture medium was changed every day, and iPSCs were passaged every four days using Accutase (Global Cell Solutions).
Differentiation and culture of iPSCs into cardiomyocytes.
The derivation of human iPSC control 1 (C1) and control 2 (C2) lines from skin biopsies of healthy individuals, as well as the characterization of these cell lines, were described earlier.5 C1 and C2 iPSCs were grown to ~90% confluence. For cardiac differentiation of iPSCs, a small molecule-based monolayer protocol based on previous reports was utilized.7, 27 Afterwards, iPSC-CMs were cultured prior to experimental analysis for 30 ± 2 days (D30), 90 ± 10 days (D90), or 200 ± 20 days (D200). For culture until D90 and D200, a maintenance passage was performed between D25 and D30. Human iPSC-CMs were washed with PBS and incubated with 0.5 ml TrypleE (Life Technologies) per 1 well of a 6-well plate for 10 min at 37 °C. Subsequently, 0.5 ml RPMI with B27 supplement (Life Technologies) per well was added, followed by gentle pipetting and immediate pelleting of the detached iPSC-CMs for 5 min, 200g at room temperature (RT). Next, the cell pellet was carefully resuspended in RPMI with B27 supplement and 2% FBS. Following plating of ~ 1×10e6 iPSC-CMs per 1 well of a 6-well plate, cells were allowed to attach for 24–48 hr. Then, the culture medium was changed to RPMI with B27 supplement. The medium was changed every 4 days during long-term culture. Before experimental analysis, iPSC-CMs were passaged into culture plates of the appropriate format (6-well plates or 96-well plates) after dissociation of iPSC-CMs with Accutase or TripleE (Life Technologies) for 10 min at 37 °C. For monolayer analysis, generally ~2×10e6 iPSC-CMs were plated per 1x well of a 6-well plate or ~500,000 iPSC-CMs per 1x well of a 24-well plate. For single cell analysis, ~300,000 iPSC-CMs were plated per 1x well of a 6-well plate or ~70,000 iPSC-CMs per 1x well of a 24-well plate.
Transcriptomic sequencing and data analysis.
Total RNA was extracted from two different human iPSC-CM lines (C1 and C2) via a miRNeasy kit (Qiagen). AmpliSeq transcriptomic sequencing was performed by Macrogen Clinical Laboratory Inc. (Rockville, MD) using an Ion AmpliSeq Transcriptome Sequencing platform (3M reads). Analysis for AmpliSeq sequencing data was performed using the AmpliSeq plugin available for Ion Torrent™ sequencing platforms. The hg19 AmpliSeq transcriptome ERCC v1 was used as a reference genome for alignment of raw sequencing reads. Read count normalization was performed using the regularized logarithm (rlog) method provided in DESeq2 (PMID: 25516281). Differentially expressed genes were identified using DESeq2, and p-values were adjusted for multiple testing using the procedure of Benjamini and Hochberg. The cutoff threshold for differentially expressed gene (DEG) calling was set to log2-fold change and a p-value < 0.05.
Statistical analysis.
Statistical significance was determined using an unpaired Studentś t-test, a two-way ANOVA test or a right-tailed Fisher’s exact test. For non-normally distributed data, Mann-Whitney testing and Dunńs post-hoc test were performed. P-values <0.05 were considered statistically significant. Data are presented as mean ± standard error of mean (s.e.m.). GraphPad Prism, R, and Excel were used for statistical analysis. A minimum of n=2 independent experiments was performed for n=2 biological cell lines and per each cell line; in each independent experiment, either a duplicate or a triplicate technical replicate was performed.
An expanded Methods section is available in the online Data Supplement.
RESULTS
Phenotypic analysis of morphological transformations during long-term iPSC-CM culture.
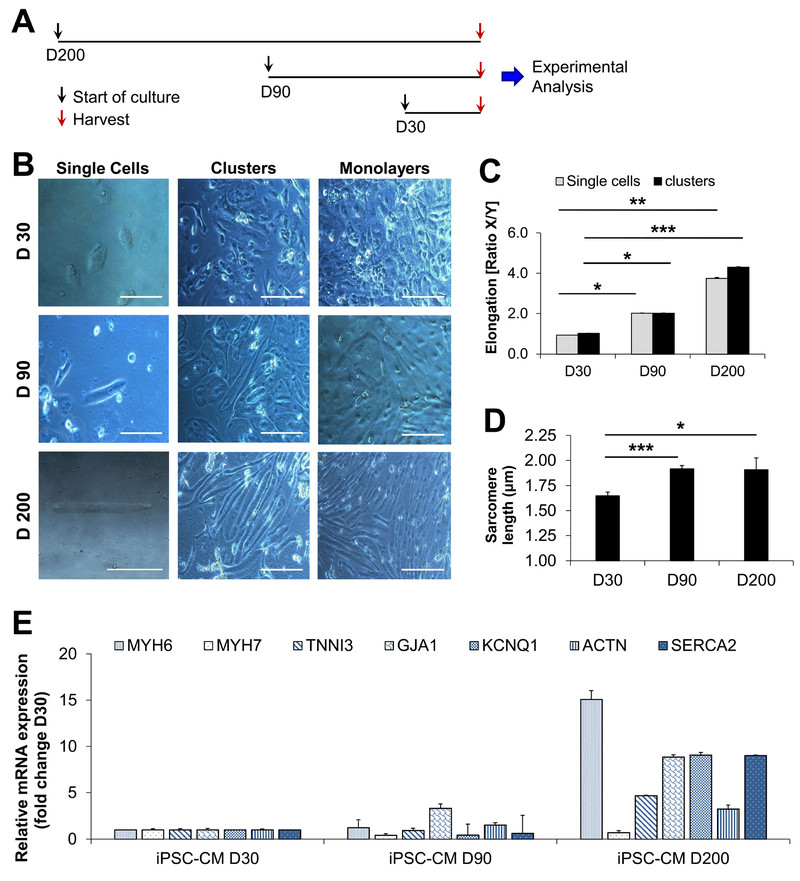
We first assessed the regulation of signal transduction in iPSC-CMs following long-term culture for more than 200 days. Characterization for iPSC-CMs at D30, D90, and D200 is shown in Fig. 1A–E. Our data demonstrate a robust morphological transformation of iPSC-CMs during long-term culture, as shown by a >4-fold increase in cell elongation (Fig. 1B–C; Online Fig. IA–B). Characterization of both iPSC and iPSC-CM populations was performed (Online Fig. II) and electrophysiological properties of iPSC-CMs were analyzed following cardiac differentiation of iPSCs (Online Fig. III). The sarcomere length of iPSC-CMs was significantly increased after long-term culture (Fig. 1D, Online Fig. IA) and increased expression of cardiac-specific transcripts was observed at D200 (Fig. 1E). We also established well-beating monolayers of D200 iPSC-CMs (Online Movies I-III). Expression of cardiac markers (e.g., TNNI3 and ACTN) was analyzed in both iPSC-CMs following long-term culture and in mouse heart tissue from postnatal day 1, day 30, and day 180 (Online Fig. IC–F), and was found to be increased at later time points. Moreover, the expression of senescence-associated markers (NFKB1 and ATGR1) was found to be increased in day 180 mouse hearts but not in human iPSC-CMs (Online Fig. IG–H).
Figure 1: Phenotypic analysis of morphological transformations during long-term culture of iPSC-CMs.
(A) Schematic outline of the experimental timeline employed for generation, long-term culture, and experimental analysis of iPSC-CMs. (B) Brightfield images of iPSC-CMs showing single cells, cell clusters, and monolayers after 30 days (D30), 90 days (D90), and 200 days (D200) of long-term culture. Images were taken with a Leica brightfield microscope at 20x magnification (clusters and monolayer, scale bar = 20 μm) and at 40x magnification (single cells, scale bar = 10 μm). (C) Quantification of the iPSC-CM diameter at D30, D90, and D200 was performed using the software Image J for both single cells and cell clusters. Vertical and horizontal region of interests (ROIs) were defined and a quotient of vertical/horizontal was calculated for each cell at D30 (n=23 cells), D90 (n=23 cells), and D200 (n=21 cells). (D) Quantification of iPSC-CM sarcomere length based on confocal images shown in Online Fig IA. (E) Relative mRNA expression levels of genes encoding for cardiac-specific transcripts were established via qRT-PCR. Data are expressed as mean ± s.e.m., n=2 independent cell lines per group, *P <0.05 as calculated by Student’s t-test. For (C), Mann-Whitney testing and Dunńs post-hoc test were performed. For single cells vs. clusters at D30, D90, D200 *P <0.05, **P <0.01, ***P <0.001.
Probing gene expression landscapes by microfluidic single-cell PCR.
To determine gene expression profiles of single iPSC-CMs, we next utilized single-cell microfluidic qRT-PCR. This platform can assess multiple cells simultaneously (e.g., 48 cells in one chip), thus enabling assessment of population heterogeneity (Fig. 2A–C, Online Fig. IV).28 Single-cell PCR (scPCR) can address the variability of gene expression levels and noise, which is challenging in single cell analysis.28–31 In unsupervised clustering, iPSC-CMs were clearly distinguished from iPSCs (Fig. 2A). Relative expression changes compared to iPSCs for all analyzed transcripts are shown in Fig. 2A–B, Online Fig. IV. Of note, scPCR analysis revealed changes in cardiac-specific, mitochondrial- and metabolism-related transcripts, as well as structure-related transcripts (Fig. 2C). Relative mRNA expression levels for transcripts such as RYR, MYL2, GJA1, and PRKAA1 were substantially increased at D90, but even more so at D200, compared to D30. To improve understanding of the transcriptional landscape associated with modulation of iPSC-CM homeostasis following long-term culture, we next employed transcriptomic profiling followed by pathway network analysis. This strategy was expected to provide insight into the dynamic regulatory networks that govern the developmental progression of iPSC-CMs during long-term culture from D30 to D200.
Figure 2: Adaptation of gene expression landscapes during long-term culture of iPSC-CMs.
(A) Heatmap of unsupervised clustering of iPSC-CM gene expression at D30, D90, and D200, as well as iPSCs, which cluster completely separately from iPSC-CMs. With the different time points, single iPSC-CMs show a trend to segregate into subsets. Relative gene expression based on Ct values is shown for iPSCs and iPSC-CMs at D30, D90, and D200. (B) Time-course analysis of iPSC-CM population heterogeneity, clustering, and segregation. Principal component analysis (PCA) is shown for iPSC-CMs following long-term culture for 30, 90, and 200 days. Expression analysis was normalized per cell for iPSCs and D30, D90, and D200 iPSC-CMs. (C) Relative mRNA expression levels of genes encoding for cardiac-specific, structural, mitochondrial, and metabolic transcripts as established via single-cell PCR. Data are shown for n=2 independent cell lines per group.
Transcriptomic profiling indicates metabolic adaptation during long-term culture.
To assess signaling mechanisms underlying mitochondrial metabolism-related changes and the molecular networks regulating them, we next subjected iPSC-CMs at D30, D90, and D200 to AmpliSeq-based transcriptomic analysis. Unsupervised clustering of significantly different expressed (SDE) genes revealed distinct transcriptional landscapes in iPSC-CMs at early (D30) versus medium (D90) and late (D200) time points (Fig. 3A). Similar to microfluidic scPCR results, D90 and D200 iPSC-CMs clustered far from D30 iPSC-CMs (Fig. 3A, Online Fig. VA). Intra-individual variation between different human iPSC-CM lines was observed and accounted for the variability between biological replicates (Fig. 3A, Online Fig. VA). Nevertheless, prominent gene expression changes were observed between D90 and D200 (Fig. 3A–C, Online Fig. VA–C). At D90, substantial changes in gene expression patterns were noted (Online Fig. VB). A Venn diagram was used to visualize SDE transcripts and related pattern changes for all possible intra-group comparisons (Fig. 3B). Transcripts in the intersections of polygons represent significantly changed transcripts between set groups (Fig. 3B). The relative directional expression changes of D200 vs D90 and D200 vs D30 were shown as a function of intensity in expression changes over time, revealing the directionality of SDE genes between these groups (Fig. 3C, Online Fig. VC). Based on these data, we expected significantly altered regulation of cellular signaling at later time points.
Figure 3: Signaling pathway mapping indicates distinct regulation of metabolic functions during long-term culture of iPSC-CMs.
(A) Heatmap showing transcriptomic profiling of iPSC-CMs at D30, D90, and D200. Cluster analysis was performed via Euclidean distance with complete linkage. Only genes that are significant in at least one pairwise comparison are shown. (B) Venn diagram of commonly expressed genes between the indicated pairwise comparison groups. Polygons show all significantly differentially expressed (SDE) genes in the pairwise group comparisons (D200 vs D90, D200 vs D30, and D90 vs D30). Overlap area between polygons highlights the indicated intra-group comparisons (D200 vs D90 and D200 vs D30; D200 vs D90 and D90 vs D30; D200 vs D30 and D90 vs D30). (C) Profiles of relative gene expression values for significantly differentially expressed transcripts between the intra-group comparisons of D200 vs D90 and D200 vs D30. Shown are directional changes of significantly differentially expressed genes as a function of time. X-axis, day 30, day 90, and day 200; y-axis, log2 (gene expression value). Blue: downregulated; red: upregulated. Directional changes are marked in blue (decrease of expression) and red (increase of expression). (D) Top networks changed for the comparison D200 vs D90 as identified by IPA analysis (Qiagen). (E) Relative alterations in IPA pathway categories (1–7) are shown for the p-value ranges of the group comparisons D30 vs. D90, D90 vs D200, and D30 vs D200. For each comparison, p-values are shown based on significantly altered transcripts in the indicated pathways and networks, as well as the p-value ranges for given functional categories generated by IPA core analysis (Qiagen, Right-Tailed Fisher’s Exact Test). 1, Cardiovascular System Development and Function; 2, Lipid Metabolism; 3, Small Molecule Biochemistry; 4, Metabolic Disease; 5, Cell-To-Cell Signaling and Interaction; 6, Cellular Assembly and Organization; and 7, Cellular Function and Maintenance. (F) Heatmap of AmpliSeq data showing the regulation of metabolism-related transcripts in iPSC-CMs at indicated time points. (G) Cellular metabolic viability following long-term culture of iPSC-CMs. Shown are measurements of absorbance at 450 nm (relative units) of the colorimetric reaction resulting from cleavage of the XTT reagent to formazan by mitochondrial respiratory chaińs succinate dehydrogenase system. Data are shown for n=2 independent cell lines per group. (H) The mitochondrial membrane potential (MMP) is analyzed via a potential sensor dye, JC-1; 590/525 nm fluorescence ratios are shown. Potential-dependent localization of JC-1 to mitochondria is causing a shift in JC-1 fluorescence emission from 525 nm to 590 nm. A lower 590/525 nm ratio indicates mitochondrial membrane depolarization. Data are expressed as mean ± s.e.m., n=2 independent cell lines as well as 2 independent experiments per group, *P <0.05, **P <0.01, ns = not significant, as calculated by Student’s t-test.
Signaling pathway analysis reveals proteasome-dependent regulation of metabolic functions.
Next, we used Ingenuity Pathway Analysis (IPA) to dissect altered signaling regulation in iPSC-CMs at late time points. IPA facilitates analysis of network components via mRNA expression data to assess transcriptional connectivity and provide insight into dynamic functions and interdependencies of physiological systems.32 Top significantly changed pathways for all intra-group comparisons are shown in Fig. 3D–E. Between D90 and D200, IPA pathway mapping analysis revealed Cardiovascular System Development and Function as the second most altered network amongst all annotated signaling networks (Fig. 3D). Concomitantly, between D30 and D90, cellular and embryonic development pathways such as hedgehog (ISL2) and Akt (ULBP1) were upregulated in iPSC-CMs. In particular, changes in cell-to-cell signaling interactions were observed (Fig. 3D–E). Relative changes for significantly altered pathways between set groups are particularly strong in the D30 and D200 comparison for Cardiovascular System Development (Fig. 3E). Intriguingly, following D200 culture, our data confirm pronounced rearrangements in signaling networks of metabolic and energy production pathways. Mitochondria are critical compartments for metabolic processes, producing the majority of cellular ATP via oxidative phosphorylation (OXPHOS).33 Amongst transcripts annotated in mitochondrial metabolic pathways, ~20% were significantly changed in at least one of the intra-group comparisons. Transcripts corresponding to signaling pathways regulating metabolic functions were drastically altered when comparing D90 and D200 (Fig. 3E–F). Correspondingly, we found a decrease in gene programs corresponding to the fetal-like, glycolysis-dependent metabolism (Online Fig. VD) from D30 to D90 and D200. Moreover, the expression of genes associated with oxidative phosphorylation and beta-oxidation was increased from D30 to D90 and D200 (Online Fig. VE). These data suggest distinct metabolic functional stages in iPSC-CMs at D30, D90, and D200, which are further corroborated by transcriptomic landscapes during long-term iPSC-CM culture (Fig. 3, Online Fig. V), particularly at D90 vs. D200 (Online Fig. VC). Molecular circuitries predicted to be involved in regulating these events are shown via an IPA-generated signaling network (Online Fig. VIA). In the most relevant intra-group comparisons, D200 vs D30 and D200 vs D90, PKA-mediated regulation of troponin I (TNNI3), as well as myosin light chain 2 (MYL2) and CASQ2 (Online Fig. VB), were predicted to be relevant signaling mechanisms (Online Fig. VIA–B). Surprisingly, IPA-based interaction network analysis suggested this to occur via contribution of the proteasome, as well as heat shock proteins (HSP) (Online Fig. VIA). We also observed upregulation of cAMP- and PKA-mediated signaling following long-term culturing (Online Fig. VIC–D). We next sought to explore the interconnected signal transduction networks governing transcriptional and metabolic regulation at the cellular level.
Upregulation of metabolic function is not dependent on increased cellular redox stress.
Validation of transcriptomic sequencing results by quantitative real-time PCR confirmed differential regulation of redox scavenging (Online Fig. VIIA, C) and metabolism-related transcripts (Online Fig. VIIB, C). However, transcriptomic sequencing also revealed increased expression of a large number of metabolism- and mitochondria-related transcripts at late time points (Fig. 3A, F), suggesting elevated metabolic activity following long-term culturing. Therefore, we next assessed whether alterations of mitochondrial function would occur in iPSC-CMs at the late time point (D200). Of note, we found significantly increased mitochondrial integrity, functional viability (Fig. 3G), and mitochondrial membrane potential (Fig. 3H) at D200. Surprisingly, increased metabolic function at the late time point was not due to an increase of mitochondrial content and mass (Online Fig. VIIIA), or to elevated levels of reactive oxygen species (ROS) (Online Fig. VIIIB). Mitochondrial energy metabolism, particularly oxidative phosphorylation, is a major source of cellular ROS. These findings suggest that at the late time point, D200, the metabolic function per mitochondrion increases, rather than higher mitochondrial numbers result in larger metabolic output. From this we conclude that a specific regulatory mechanism leading to upregulation of mitochondrial function may be active particularly in iPSC-CMs at the late time point, D200. Collectively, our data suggest that mitochondria-independent regulatory mechanisms contribute to the observed increase of metabolic function at the late time point.
Distinct cAMP-/PKA-dependent metabolic stages during long-term culture of iPSC-CMs.
We next sought to identify these underlying molecular functions. Unbiased pathway mapping suggested a modulatory role for proteasome and PKA functions, which serve as critical contributors to distinct regulatory stages in iPSC-CMs at D30, D90, and D200 (Fig. 3–4). The expression of genes related to PKA- and cAMP-dependent signaling pathways was significantly altered in all late time point intra-group comparisons (Online Fig. VIC–D). We therefore confirmed the known interactions of PKA with the proteasome via interactome mapping using the STRING database (Fig. 4A). This especially highlighted the chaperone Hsp90 and ubiquitin, a small regulatory protein involved in proteasome-based degradation and cellular homeostasis. PKA is known to phosphorylate troponin I (TNNI3), a component of the troponin-tropomyosin complex mediating contraction in cardiomyocytes, and also to regulate proteasome function (Fig. 4A, Online Tab. IV).34–36 Notably, we found significantly increased contraction velocity and decreased contraction duration of iPSC-CMs at the late time point (Online Fig. IXA–C). Given these observations, we set out next to assess the role of PKA in regulation of distinct metabolic stages at different time points. In addition, we sought to analyze a possible modulatory contribution of the proteasome to these effects.34, 35
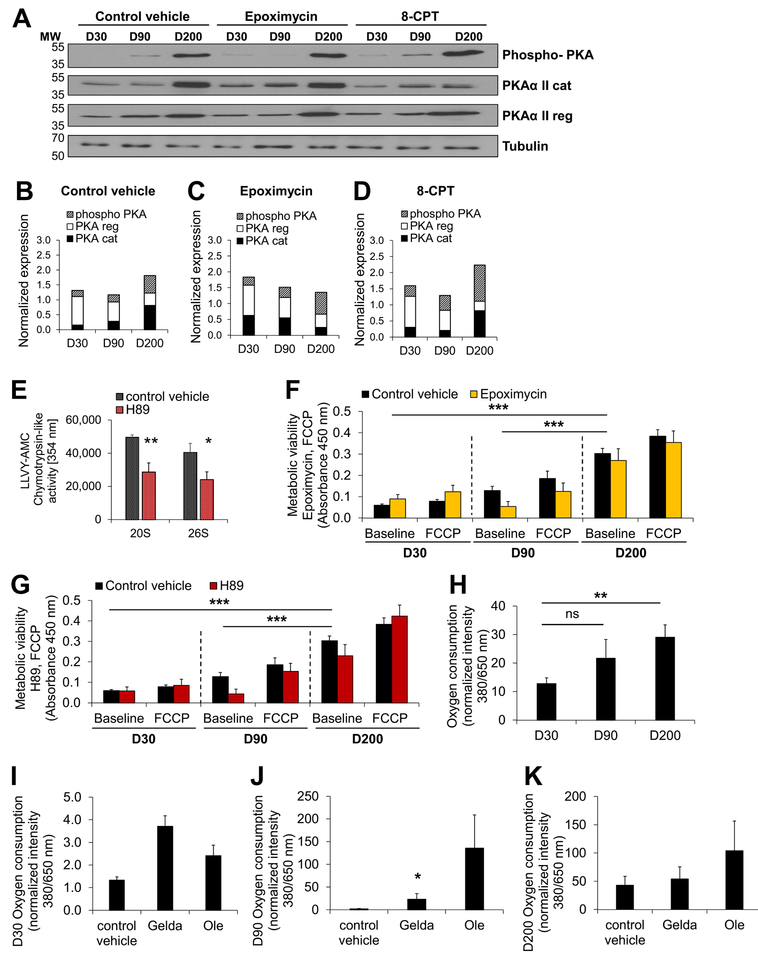
Figure 4: cAMP-/PKA-dependent regulation of distinct metabolic stages during long-term iPSC-CM culture.
(A) Interactome mapping for PKA using the STRING database. Relevant interaction partners previously identified via transcriptomic sequencing and IPA pathway mapping to be significantly changed at D90 and D200 (D90 vs D200) are highlighted in orange. Additional node colors are assigned by STRING database pre-sets for first shell of interactors. Second shell interactors are shown in grey. (B) PKA activity is measured in iPSC-CM lysates at indicated time points using a PKA activity assay kit (Promega). Data are expressed as mean ± s.e.m. Shown are n=2 independent cell lines as well as 2 independent experiments per group. *P <0.05, **P <0.01 as calculated by Student’s t-test. (C, D) Measurement of proteasome activity in iPSC-CMs following long-term culture. Fluorescence activity of the proteasome-specific molecule Suc-LLVY-AMC is detected following hydrolysis at 354 nm, indicating activity of (C) 20S and (D) 26S proteasome machinery. Data are expressed as mean ± s.e.m. Shown are n=2 independent cell lines as well as 2 independent experiments per group. *P <0.05, **P <0.01, ***P <0.001 as calculated by Student’s t-test. For (C-D), Mann-Whitney testing and Dunńs post-hoc test were performed. **P <0.01, ***P <0.001.
First, we measured PKA activity at early (D30), medium (D90), and late (D200) time points. PKA activity was highest at D200, and lowest at D90 (Fig. 4B). Following culturing for 90 days, 26S and 20S proteasome activities in iPSC-CMs were reduced (Fig. 4C–D). However, proteasomal activity was significantly increased at D200, relative to both D90 and D30 (Fig. 4C–D). The increased proteasomal activity was not due to significant expression changes of proteasome-encoding transcripts (Online Fig. XA). Treatment of iPSC-CMs with a proteasome inhibitor, epoximycin, abolished proteasomal function at all time points (Fig. 4C–D), without significantly altering mitochondrial functional viability (Online Fig. XB) or ROS levels (Online Fig. XC). Whereas the expression of proteasome- and PKA-encoding transcripts was not substantially changed (Online Fig. XA, Online Fig. XIA), heat shock proteins (HSPs) were found to be differentially expressed at different time points (Online Fig. XD). In particular, HSP90 was found to be lower at the late time point (Online Fig. XD). STRING database pathway analysis placed Hsp90 as a regulatory connection between PKA and proteasome. Likewise, ubiquitin-related transcripts were downregulated at the late time point in transcriptomic sequencing (Online Fig. XE).
Ubiquitination-dependent protein degradation by the proteasome (UDP) is a highly dynamic pathway that is important for a large variety of cellular processes, including development and differentiation, response to stress and extracellular modulators, as well as environmental adaptation. Recently, UDP was shown to aid cellular adaptation to long-term environmental changes by controlling metabolic rates.37 Therefore, downregulation of the ubiquitin-proteasome system at the late time point could contribute to upregulation of metabolic function by reducing ubiquitination and degrading metabolic protein substrates. Our data support a differential fine-tuning of regulatory mechanisms that control metabolic functions in iPSC-CMs at early (D30) versus medium (D90) and late (D200) time points. Overall, we found distinct metabolic stages in long-term cultured iPSC-CMs to correlate with PKA activity/proteasome function alterations. We next investigated if the two are linked.
Distinct metabolic stages are regulated in a PKA- and proteasome-dependent manner.
PKA- and cAMP-dependent control of the proteasome has been described previously.22, 23 To further explore a PKA proteasome-dependent regulation of differential metabolic stages in iPSC-CMs at early (D30), medium (D90), and late (D200) time points, we first assessed the levels of PKA catalytic subunits (PKAcat) and PKA regulatory subunits (PKAreg), as well as PKA phosphorylation under baseline conditions (control vehicle), after proteasome inhibition (epoximycin), and after PKA activation (8-CPT) (Fig. 5A–D). PKA expression was higher at D200 relative to D30 or D90 (Fig. 5B). In addition, iPSC-CMs at D200 displayed increased PKAcat levels, coinciding with higher PKAcat phosphorylation (phospho-PKA) at D200 (Fig. 5A–B). These findings point to a temporally divergent directional control exerted via PKA and proteasome during long-term culture (D200). To further validate these findings, we confirmed that while PKA inhibition (H89) reduced proteasome activity (Fig. 5E), PKA activity was unaffected by proteasome inhibition (Online Fig. XIB), strengthening the evidence for a PKA-dependent regulation of the proteasome.
Figure 5: Distinct metabolic stages in long-term cultured iPSC-CMs are regulated in a PKA- and proteasome-dependent manner.
(A-D) Levels of catalytic and regulatory subunits, as well as phosphorylated PKA levels (PKAcat threonine Thr197), at indicated time points following proteasome inhibition (epoximycin) and PKA activation (8-CPT) compared to control vehicle (DMSO). (A) Immunoblot analysis and (B-D) quantification of (A), normalized to loading control (tubulin). (E) Inhibition of PKA (H89) significantly reduces proteasome activity. (F-G) Metabolic viability was measured via cleavage of the XTT reagent by mitochondrial respiratory chaińs succinate dehydrogenase system, resulting in absorbance at 450 nm (relative units) for iPSC-CMs at indicated time points. Response of iPSC-CMs to modulation of the mitochondrial respiratory chain via FCCP at indicated time points is shown. XTT-based absorbance was measured at 450 nM (relative units). (F) Proteasome inhibition (epoximycin) as well as (G) PKA inhibition (H89) decrease metabolic function of iPSC-CMs at D90. (H) Oxygen consumption measured for iPSC-CMs at indicated time points. Relative fluorescence units are shown. (I-K) Human iPSC-CMs were cultured until D30 (I), D90 (J), or D200 (K), followed by culture in presence of DMSO (control vehicle) or indicated chemical modulators. All data are expressed as mean ± s.e.m. Shown are n=2 independent cell lines as well as 2 independent experiments per group. *P <0.05, **P <0.01, ***P <0.001, ns = not significant as calculated by Student’s t-test. For (F-G), Mann-Whitney testing and Dunńs post-hoc test or Studentś t-test and Sidakś post-hoc test were performed. *P <0.05, **P <0.01, ***P <0.001. Data are expressed as mean ± s.e.m.
As PKA critically depends on cAMP, we next determined whether cellular cAMP levels [cAMP] were altered at different time points. Indeed, intracellular cAMP was the lowest in iPSC-CMs at D90, but significantly increased at D200 (Online Fig. XID). Furthermore, inhibition of the proteasome resulted in a significant rescue of cAMP levels only at D90, whereas activation of PKA (8-CPT) decreased cAMP levels as expected (Online Fig. XID). Thus, at different time points, iPSC-CMs experienced an altered regulation of PKA activity as well as of its modulator, cAMP. In addition, we found that cAMP also increased proteasome activity (Online Fig. XIE). In line with significantly elevated PKA and proteasome activity in D200 iPSC-CMs (Fig 4B–D), we found that proteasome activity at D200 was no longer dependent on cAMP (Online Fig. XIE), given the already high activity of PKA and proteasome.
To confirm that these events accounted for the observed distinct metabolic functions in D30, D90, and D200 iPSC-CMs, we next investigated if cAMP-PKA-dependent proteasomal activity could regulate metabolic function at different time points. We utilized cell viability measurements based on the activity of mitochondrial respiratory chain succinate dehydrogenase functions (see Methods), referred to in this context as “metabolic viability”. Importantly, we found that epoximycin-dependent proteasome inhibition showed no effect on metabolic viability at D30, but resulted in a substantial reduction of mitochondrial function at D90 (Fig. 5F). However, at the very late time point (D200), no significant differences in metabolic viability were observed following proteasome inhibition (Fig. 5F). In line with these results, PKA inhibition also decreased metabolic turnover at D90 but not at the other time points (D30 and D200) (Fig. 5G).
These findings indicate that modulation of mitochondrial metabolic viability by a PKA-proteasome-dependent mechanism is differentially regulated at later time points, D90 and D200. Our data support the notion that iPSC-CM homeostasis and turnover experienced a “metabolic sink” around D90 following iPSC differentiation. To explore this possibility further, we next confirmed that the PKA- and proteasome-dependent regulation of metabolism at D90 occurred also during modulation of the mitochondrial respiratory chain via uncoupling of oxidative phosphorylation from ATP synthesis, using FCCP (Fig. 5F–G). Our findings showed that both proteasome (Fig. 5F) and PKA inhibition (Fig. 5G) led to decreased mitochondrial metabolic viability of iPSC-CMs at D90, but not at D30 or D200. In line with these data, cellular respiration, measured via oxygen consumption, was increased in iPSC-CMs at D200, but not at D90 (Fig. 5H) unless chemical modulation of the PKA-proteasome pathway was applied (Fig. 5I–K).
Together, these data support the possibility that increased PKA-proteasome activity at later time points was connected to upregulation of mitochondrial metabolism, as well as a decrease in Hsp90 gene expression (Online Fig. XD) and Hsp90 protein levels, particularly at D200 (Online Fig. XIIA–B). To verify this point further and compare results to human adult cardiomyocytes, we measured Hsp90 protein levels in lysates from human heart tissue (n=4), which confirmed that Hsp90 was lowly expressed in adult human cardiac tissue (Online Fig. XIIA–B). In addition, we also validated this outcome in murine heart tissue (Online Fig. XIIC–G). In murine hearts at postnatal day 180, Hsp90 was significantly downregulated compared to postnatal days 1–2 (Online Fig. XIIC, D), and PKAcat was significantly upregulated (Online Fig. XIIC, F).
Hsp90-dependent regulation of mitochondrial metabolism via modulation of the respiratory chain.
Hsp90 is a critical player in protein folding, protein quality control, ubiquitination, and proteasome-mediated degradation, and is known to interact with the proteasome24 as well as to localize to mitochondria.25 Therefore, we next investigated whether Hsp90 could directly contribute to the proteasome-concerted hierarchical control of mitochondrial metabolic viability at different time points. Intriguingly, we found that Hsp90 inhibition using geldanamycin38, 39 upregulated metabolic viability in D30 iPSC-CMs as measured via mitochondrial succinate dehydrogenase function (Fig. 6A), thereby recapitulating the mechanism observed at the late time point, D200.
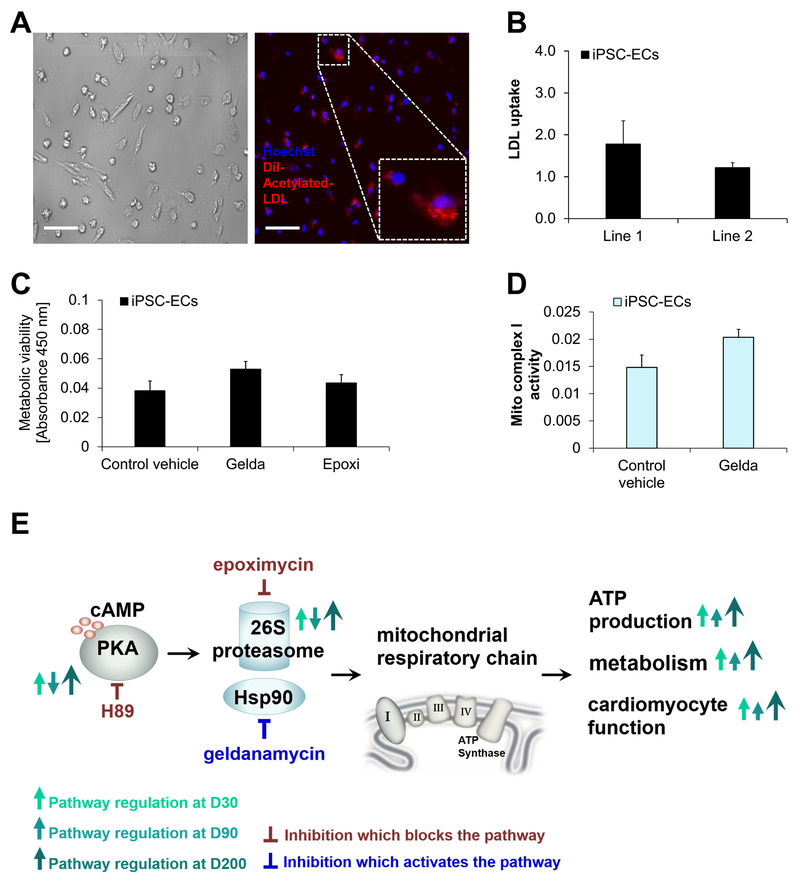
Figure 6: Hsp90 contributes to regulation of metabolism via the mitochondrial respiratory chain complex I.
(A-B) Human iPSC-CMs (D30) were cultured for 72 h in presence of DMSO (control vehicle) or geldanamycin (gelda) as indicated. (A) Metabolic viability was measured via the XTT reagent in iPSC-CMs at indicated time points in presence or absence of FCCP, a respiratory chain uncoupler. XTT cleavage by mitochondrial respiratory chaińs succinate dehydrogenase system results in absorbance detected at 450 nm (relative units). (B) Measurement of mitochondrial complex I activity in iPSC-CM (D30) lysates based on oxidation of NADH to NAD+ and simultaneous reduction of dye, resulting in increased absorbance (450 nm). (C-D) Oxygen consumption measured for iPSC-CMs at D30, shown as relative fluorescence units. (C) Treatment with DMSO (control vehicle) or geldanamycin (gelda). (D) siRNA knock-down of PKAreg vs. silencer control siRNA (siRNA control). (E-F) Cellular ATP production measured in D30 treated iPSC-CMs as indicated. Shown is % luminescence of control for (E) siRNA knock-down of PKAreg and Hsp90 vs. silencer control siRNA (siRNA control) and (F) PKA inhibition via H89 and control vehicle (DMSO). Data are expressed as mean ± s.e.m. Shown are n=2 independent cell lines as well as 2 independent experiments per group. *P <0.05, ***P <0.001 as calculated by Student’s t-test. For (A), Mann-Whitney testing and Dunńs post-hoc test were performed. *P <0.05, **P <0.01, ***P <0.001.
As reflected by mitochondrial dehydrogenase activity (such as succinate dehydrogenase), mitochondrial health and function are important for energy production via the mitochondrial respiratory chain. Therefore, we next tested the possibility that Hsp90 could regulate metabolic output by modulating respiratory chain function. We found that upon inhibition of Hsp90 via geldanamycin, mitochondrial complex I (MCI) activity in D30 iPSC-CMs was increased (Fig. 6B). To further probe Hsp90-dependent regulation of MCI and mitochondrial respiratory chain function, we measured cellular respiration or mitochondrial membrane potential of D30 iPSC-CMs in the presence of geldanamycin or proteasome activation via oleuropein (Fig. 6C, Online Fig. XIC, H). Oxygen consumption in D30 iPSC-CMs was increased following inhibition of Hsp90 (Fig. 6C), suggesting that Hsp90 indeed may act as a negative regulator of mitochondrial respiration via MCI. For further validation of this hypothesis, we used genetic manipulation of the proposed PKA/proteasome- and Hsp90-dependent signaling axis. We employed siRNA-based knock-down of the regulatory PKA subunit, PKAreg (PRKAR1A), which resulted in upregulation of PKA activity that is consistent with previous reports,40 as well as siRNA knock-down of Hsp90. Validation of siRNA knock-down showed a substantial reduction of PKAreg and Hsp90 protein levels following treatment with siRNAs targeting PKAreg (Online Fig. XIIIA) and Hsp90 (Online Fig. XIIIB). Of note, both upregulation of PKA activity and reduced Hsp90 protein levels were found in D200 iPSC-CMs. Concomitantly, knock-down of PKAreg increased the oxygen consumption in D30 iPSC-CMs (Fig. 6D). In addition to respiration, we measured cellular ATP production in D30 iPSC-CMs. Under aerobic conditions, most of the cellular ATP is produced by the mitochondrial respiratory chain. Importantly, we found ATP production to be significantly increased following siRNA knock-down of both PKAreg and Hsp90 (Fig. 6E). Moreover, the inhibition of PKA with H89 reduced ATP production in D30 iPSC-CMs (Fig. 6F).
We next employed genetic and chemical-based modulation to confirm that PKA/proteasome-Hsp90 signaling modulates cellular functional output. We recorded contractility parameters in D30 iPSC-CMs following either siRNA knock-down of PKAreg or Hsp90 (Online Fig. XIVA–B), versus treatment with geldanamycin or 8-CPT (Online Fig. XIVC–D). Concomitantly, the activation of the PKA/proteasome-Hsp90 cascade by PKAreg- or Hsp90 knock-down (Online Fig. XIVA, B), or PKA activation via 8-CPT as well as Hsp90 inhibition via geldanamycin (Online Fig. XIVC, D), resulted in significantly reduced contraction duration, compared to controls. These experiments indicate that positive modulation of the PKA/proteasome-Hsp90 cascade recapitulates in D30 iPSC-CMs molecular functions observed in D200 iPSC-CMs (Online Fig. IXA–C). To confirm these findings in a relevant alternative cell model, we chose another prevalent cell type of the heart, its endothelial cells. We generated iPSC-derived endothelial cells (iPSC-ECs) from the same iPSC lines used for iPSC-CM production (Fig. 7A–B). We found that inhibition of Hsp90 in iPSC-ECs resulted in upregulation of mitochondrial metabolism (Fig. 7C) as well as MCI activity (Fig. 7D). These effects were not related to alterations of mitochondrial content (Online Fig. XVA) or ROS production (Online Fig. XVB–C). Based on our findings, we propose a PKA/proteasome- and Hsp90-dependent control of mitochondrial respiratory chain that regulates cardiomyocyte function (Fig. 7E) and affects their long-term differentiation to a mature phenotype. Our data show that this mechanism experiences differential regulation at different time points.
Figure 7: Hsp90 contributes to metabolic regulation via modulation of the mitochondrial respiratory chain in iPSC-derived endothelial cells (iPSC-ECs).
(A-D) Human iPSC-ECs were derived from iPSC lines 1 and 2. (A) Uptake of low-density lipoprotein was detected using Dil-acetylated low-density lipoprotein followed by imaging with a brightfield microscope. Scale bar, 20 μm. (B) Quantification of (A) in 324 cells (line 1) and 227 cells (line 2) using Image J software. (C-D) Human iPSC-ECs were cultured for 48 hr in presence of 20 nM DMSO (control vehicle), geldanamycin (gelda) or epoximycin (epoxi) as indicated. (C) Metabolic viability was measured in iPSC-ECs was measured via XTT cleavage by mitochondrial respiratory chaińs succinate dehydrogenase system, resulting in absorbance at 450 nm (relative units). (D) Detection of mitochondrial complex I activity in iPSC-EC lysates as described above, followed by measurement of absorbance (450 nm). Data are expressed as mean ± s.e.m. Shown are n=2 independent cell lines as well as 2 independent experiments per group. (E) Schematic model of the PKA-proteasome-Hsp90 dependent axis of metabolic governance of cellular functions at D30, D90, and D200 of long-term culture. Geldanamycin treatment inhibits Hsp90 activity, which reduces its negative regulation of the mitochondrial respiratory chain, resulting in upregulation of metabolism and cardiomyocyte function.
DISCUSSION
The effects of prolonged culture on iPSC-CMs have been explored previously with respect to maturation of electrophysiological parameters such as NaV- or K-channel currents.20, 41, 42 Nevertheless, little is known so far about the molecular circuitries that regulate functional changes during prolonged iPSC-CM culture. In this study, we demonstrate the characterization of regulatory signaling cascades during long-term culture of iPSC-CMs for 200 days. Using transcriptomic sequencing at early (D30), medium (D90), and late (D200) time points, we identified major changes in mitochondrial- and metabolism-related pathways, as well as contractile elements such as TNNI3. We confirmed distinct control of cellular metabolic functions at D30, D90, and D200, which correlated with increased iPSC-CM contractile function at the late time point. Previous studies found a linkage between metabolic turnover and contractile function.33, 43–45 However, the observed upregulation of metabolic function in long-term cultured iPSC-CMs was not necessarily an intuitive result, given that long-lived mutant animal models show reduced metabolic rates46 and aging is known to reduce cellular metabolic turnover.47 We consider our findings in line with an early developmental model of human cardiac cells, as it is known that while changing from fetal to post-natal state, cardiomyocytes not only switch from glycolytic to oxidative metabolism, but also experience increased metabolic demand and upregulated metabolism.48 Our data indicate that a partial shift occurs in iPSC-CMs during long-term culture, from largely glycolytic metabolism in iPSC-CMs at D30 to increased use of oxidative phosphorylation and beta-oxidation in iPSC-CMs at D200. Subsequently, we found that cAMP-PKA-dependent upstream control of the proteasome could distinctly regulate different metabolic stages in long-term cultured iPSC-CMs. Both proteasome and PKA functions were found to be substantially decreased from D30 to D90, but were significantly increased at D200, independently of mitochondrial content and cellular ROS production.
Our data demonstrate that the PKA-proteasome-dependent mechanistic axis acts as a regulator of metabolic function and, consequently, of contraction rates. This cascade was enhanced during developmental progression of iPSC-CMs, and was distinctly regulated at different time points. At D90, we propose a “metabolic sink” due to threshold levels of the interdependent components assembling this intricate mechanism. Increased metabolic demands resulted in exhaustion of cellular cAMP, followed by a drop of PKA activity and resulting proteasome function, both being depleted below a level at which PKA could be still efficiently activated. Conversely, at the very late time point (D200), the significant increase in cAMP resulted in recovery of PKA-proteasome-dependent activities and concomitant regulation of metabolic functions to match cellular demands. We suggest that proteasome-dependent metabolic control was mediated via the heat shock protein, Hsp90.
Our findings provide evidence for Hsp90 as a regulator of mitochondrial respiratory chain proteins, whose function in turn is instrumental to major cellular energy production and determines the cardiomyocyte functional output. Decreased Hsp90 expression at D200 appears to signal the removal of an inhibitory function of Hsp90 for at least one complex of the mitochondrial respiratory chain, and may also prove to be critical for the proteasome-concerted upregulation of mitochondrial metabolic turnover and cardiomyocyte function at the late time point (D200). Our results provide deeper novel insight into signaling circuitries governing metabolic homeostasis in iPSC-CMs after long-term culture. Further characterization of this mechanism may uncover its possible role in the enhanced maturation of iPSC-CMs via concerted regulation of metabolic function and contractility.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
Human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) have been demonstrated to be suitable for cardiovascular disease modeling, drug screening, and potential regenerative therapies.
iPSC-CMs show an immature phenotypic presentation that resembles early human embryonic cardiomyocytes. This presents a challenge for realizing the full potential of iPSC-CMs that can be partially overcome by long-term culture of iPSC-CMs.
Little is known about signaling circuitries that govern metabolic output and cellular homeostasis during long-term culture of iPSC-CMs.
What New Information Does This Article Contribute?
Stable and reproducible generation of iPSC-CMs cultured for more than 200 days and phenotypic characterization of iPSC-CMs, including assessment of cell elongation and sarcomere length.
Gene expression landscapes during long-term culture of iPSC-CMs, as well as population heterogeneity.
Proteasome-dependent regulation of metabolic functions and contractility during long-term culture of iPSC-CMs for 200 days, which may present a model of human heart development.
Identification of distinct metabolic stages in long-term cultured iPSC-CMs that are regulated in a cAMP/PKA- and proteasome-dependent manner.
Proteasome- and Hsp90-dependent regulation of mitochondrial metabolism via modulation of the respiratory chain.
Confirmation of the PKA-proteasome-Hsp90-dependent signaling axis in a second major cell type of the heart, i.e., endothelial cells.
A novel regulatory pathway contributing to metabolic control of cardiomyocyte contractility.
Identification of new potential targets for metabolic maturation of iPSC-CMs.
This study identified a novel pathway regulating metabolic function and, consequently, contractility in human cardiac cells via PKA-proteasome and Hsp90. This pathway is upregulated particularly during developmental maturation of iPSC-CMs following long-term culture for 200 days. Modulation of this pathway in iPSC-CMs at day 30 of culture can recapitulate features of day 200 iPSC-CMs and may aid future applications of iPSC-CMs in molecular research and regenerative medicine.
ACKNOWLEDGMENTS
We are grateful for the support by the shared resource facilities at Stanford, including the Neuroscience Microscopy Service supported by National Institutes of Health (NIH) P30 NS069375, the FACS Core at the Institute for Stem Cell Biology and Regenerative Medicine, and the Stanford Shared FACS Facility.
SOURCES OF FUNDING
This work was supported by research grants from the American Heart Association (AHA) 17MERIT33610009, Burroughs Wellcome Foundation 1015009, NIH R01 HL130020, NIH R01 HL145676, NIH R01 HL146690, NIH R01 HL133272 (J.C.W.), NIH R01 HL052141 (D.M.-R.), Deutsche Forschungsgemeinschaft (German Research Foundation) Sonderforschungsbereich 1002, Projekt A12 (A.D.E.), Projekt D01 (G.H.) and Projekt D04 (K.T.), and the Clinic for Cardiology and Pneumology at the University Medical Center, Goettingen University (A.D.E.).
DISCLOSURES
D.M.-R. is the founder of ALDEA Pharmaceuticals. However, she has no role in the company, and the research in her laboratory is supported only by the NIH without being disclosed to the company. JCW is a co-founder of Khloris Biosciences but has no competing interests, as the work presented here is completely independent.
Nonstandard Abbreviations And Acronyms:
- iPSC-CMs
induced pluripotent stem cell-derived cardiomyocytes
- D30
Day 30
- D90
Day 90
- D200
Day 200
- PKA
protein kinase A
- Hsp90
heat shock protein 90
- MMP
mitochondrial membrane potential
- MCI
mitochondrial complex I
- OXPHOS
oxidative phosphorylation
- ROS
reactive oxygen species
- UDP
ubiquitination-dependent protein degradation
- PCA
principal component analysis
- FCCP
carbonyl cyanide-p-trifluoromethoxyphenylhydrazone
Footnotes
This manuscript was sent to Buddhadeb Dawn, Consulting Editor, for review by expert referees, editorial decision, and final disposition.
REFERENCES
- 1.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, Dorn T, Goedel A, Hohnke C, Hofmann F, Seyfarth M, Sinnecker D, Schomig A and Laugwitz KL. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–409. [DOI] [PubMed] [Google Scholar]
- 2.Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang YS, Schaniel C, Lee DF, Yang L, Kaplan AD, Adler ED, Rozov R, Ge Y, Cohen N, Edelmann LJ, Chang B, Waghray A, Su J, Pardo S, Lichtenbelt KD, Tartaglia M, Gelb BD and Lemischka IR. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J and Dolmetsch RE. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, Kan NG, Forcales S, Puri PL, Leone TC, Marine JE, Calkins H, Kelly DP, Judge DP and Chen HS. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494:105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N, Abilez OJ, Hu S, Ebert AD, Navarrete EG, Simmons CS, Wheeler M, Pruitt B, Lewis R, Yamaguchi Y, Ashley EA, Bers DM, Robbins RC, Longaker MT and Wu JC. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Pavlovic A, Lin S, Chen R, Hajjar RJ, Snyder MP, Dolmetsch RE, Butte MJ, Ashley EA, Longaker MT, Robbins RC and Wu JC. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4:130ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebert AD, Kodo K, Liang P, Wu H, Huber BC, Riegler J, Churko J, Lee J, de Almeida P, Lan F, Diecke S, Burridge PW, Gold JD, Mochly-Rosen D and Wu JC. Characterization of the molecular mechanisms underlying increased ischemic damage in the aldehyde dehydrogenase 2 genetic polymorphism using a human induced pluripotent stem cell model system. Sci Transl Med. 2014;6:255ra130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang P, Lan F, Lee AS, Gong T, Sanchez-Freire V, Wang Y, Diecke S, Sallam K, Knowles JW, Wang PJ, Nguyen PK, Bers DM, Robbins RC and Wu JC. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation. 2013;127:1677–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riegler J, Ebert A, Qin X, Shen Q, Wang M, Ameen M, Kodo K, Ong SG, Lee WH, Lee G, Neofytou E, Gold JD, Connolly AJ and Wu JC. Comparison of magnetic resonance imaging and serum biomarkers for detection of human pluripotent stem cell-derived teratomas. Stem Cell Reports. 2016;6:176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J and Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–24. [DOI] [PubMed] [Google Scholar]
- 11.Zimmermann WH, Melnychenko I, Wasmeier G, Didie M, Naito H, Nixdorff U, Hess A, Budinsky L, Brune K, Michaelis B, Dhein S, Schwoerer A, Ehmke H and Eschenhagen T. Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat Med. 2006;12:452–8. [DOI] [PubMed] [Google Scholar]
- 12.Riegler J, Tiburcy M, Ebert A, Tzatzalos E, Raaz U, Abilez OJ, Shen Q, Kooreman NG, Neofytou E, Chen VC, Wang M, Meyer T, Tsao PS, Connolly AJ, Couture LA, Gold JD, Zimmermann WH and Wu JC. Human Engineered Heart Muscles Engraft and Survive Long Term in a Rodent Myocardial Infarction Model. Circ Res. 2015;117:720–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mummery C, van der Heyden MA, de Boer TP, Passier R, Ward D, van den Brink S, van Rooijen M and van de Stolpe A. Cardiomyocytes from human and mouse embryonic stem cells. Methods Mol Med. 2007;140:249–72. [DOI] [PubMed] [Google Scholar]
- 14.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J and Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–40. [DOI] [PubMed] [Google Scholar]
- 15.Paige SL, Osugi T, Afanasiev OK, Pabon L, Reinecke H and Murry CE. Endogenous Wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS One. 2010;5:e11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ and Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109:E1848–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Pabon L and Murry CE. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res. 2014;114:511–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veerman CC, Kosmidis G, Mummery CL, Casini S, Verkerk AO and Bellin M. Immaturity of human stem-cell-derived cardiomyocytes in culture: fatal flaw or soluble problem? Stem Cells Dev. 2015;24:1035–52. [DOI] [PubMed] [Google Scholar]
- 19.Kamakura T, Makiyama T, Sasaki K, Yoshida Y, Wuriyanghai Y, Chen J, Hattori T, Ohno S, Kita T, Horie M, Yamanaka S and Kimura T. Ultrastructural maturation of human-induced pluripotent stem cell-derived cardiomyocytes in a long-term culture. Circ J. 2013;77:1307–14. [DOI] [PubMed] [Google Scholar]
- 20.Lundy SD, Zhu WZ, Regnier M and Laflamme MA. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013;22:1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuppusamy KT, Jones DC, Sperber H, Madan A, Fischer KA, Rodriguez ML, Pabon L, Zhu WZ, Tulloch NL, Yang X, Sniadecki NJ, Laflamme MA, Ruzzo WL, Murry CE and Ruohola-Baker H. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc Natl Acad Sci U S A. 2015;112:E2785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lokireddy S, Kukushkin NV and Goldberg AL. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proc Natl Acad Sci U S A. 2015;112:E7176–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myeku N, Clelland CL, Emrani S, Kukushkin NV, Yu WH, Goldberg AL and Duff KE. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat Med. 2016;22:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamano T, Mizukami S, Murata S, Chiba T, Tanaka K and Udono H. Hsp90-mediated assembly of the 26 S proteasome is involved in major histocompatibility complex class I antigen processing. J Biol Chem. 2008;283:28060–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Budas GR, Churchill EN, Disatnik MH, Sun L and Mochly-Rosen D. Mitochondrial import of PKCepsilon is mediated by HSP90: a role in cardioprotection from ischaemia and reperfusion injury. Cardiovasc Res. 2010;88:83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM and Thomson JA. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ and Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Freire V, Ebert AD, Kalisky T, Quake SR and Wu JC. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nat Protoc. 2012;7:829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elowitz MB, Levine AJ, Siggia ED and Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–6. [DOI] [PubMed] [Google Scholar]
- 30.Rajan S, Djambazian H, Dang HC, Sladek R and Hudson TJ. The living microarray: a high-throughput platform for measuring transcription dynamics in single cells. BMC Genomics. 2011;12:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narsinh KH, Sun N, Sanchez-Freire V, Lee AS, Almeida P, Hu S, Jan T, Wilson KD, Leong D, Rosenberg J, Yao M, Robbins RC and Wu JC. Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J Clin Invest. 2011;121:1217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kramer A, Green J, Pollard J Jr. and Tugendreich S Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong G, Song M, Csordas G, Kelly DP, Matkovich SJ and Dorn GW, 2nd. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350:aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin JT, Chang WC, Chen HM, Lai HL, Chen CY, Tao MH and Chern Y. Regulation of feedback between protein kinase A and the proteasome system worsens Huntington’s disease. Mol Cell Biol. 2013;33:1073–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu S, Lai L, Zuo Q, Dai F, Wu L, Wang Y, Zhou Q, Liu J, Liu J, Li L, Lin Q, Creighton CJ, Costello MG, Huang S, Jia C, Liao L, Luo H, Fu J, Liu M, Yi Z, Xiao J and Li X. PKA turnover by the REGgamma-proteasome modulates FoxO1 cellular activity and VEGF-induced angiogenesis. J Mol Cell Cardiol. 2014;72:28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asai M, Tsukamoto O, Minamino T, Asanuma H, Fujita M, Asano Y, Takahama H, Sasaki H, Higo S, Asakura M, Takashima S, Hori M and Kitakaze M. PKA rapidly enhances proteasome assembly and activity in in vivo canine hearts. J Mol Cell Cardiol. 2009;46:452–62. [DOI] [PubMed] [Google Scholar]
- 37.Nakatsukasa K, Okumura F and Kamura T. Proteolytic regulation of metabolic enzymes by E3 ubiquitin ligase complexes: lessons from yeast. Crit Rev Biochem Mol Biol. 2015;50:489–502. [DOI] [PubMed] [Google Scholar]
- 38.Whitesell L, Mimnaugh EG, De Costa B, Myers CE and Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci U S A. 1994;91:8324–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matts RL, Brandt GE, Lu Y, Dixit A, Mollapour M, Wang S, Donnelly AC, Neckers L, Verkhivker G and Blagg BS. A systematic protocol for the characterization of Hsp90 modulators. Bioorg Med Chem. 2011;19:684–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JH, Han JS, Kong J, Ji Y, Lv X, Lee J, Li P and Kim JB. Protein Kinase A Subunit Balance Regulates Lipid Metabolism in Caenorhabditis elegans and Mammalian Adipocytes. J Biol Chem. 2016;291:20315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sartiani L, Bettiol E, Stillitano F, Mugelli A, Cerbai E and Jaconi ME. Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem Cells. 2007;25:1136–44. [DOI] [PubMed] [Google Scholar]
- 42.Ivashchenko CY, Pipes GC, Lozinskaya IM, Lin Z, Xiaoping X, Needle S, Grygielko ET, Hu E, Toomey JR, Lepore JJ and Willette RN. Human-induced pluripotent stem cell-derived cardiomyocytes exhibit temporal changes in phenotype. Am J Physiol Heart Circ Physiol. 2013;305:H913–22. [DOI] [PubMed] [Google Scholar]
- 43.Kiyooka T, Oshima Y, Fujinaka W, Iribe G, Mohri S and Shimizu J. Energy for myocardial Ca2+ handling per beat increases with heart rate in excised cross-circulated canine heart. Tokai J Exp Clin Med. 2014;39:51–8. [PubMed] [Google Scholar]
- 44.Willis BC, Salazar-Cantu A, Silva-Platas C, Fernandez-Sada E, Villegas CA, Rios-Argaiz E, Gonzalez-Serrano P, Sanchez LA, Guerrero-Beltran CE, Garcia N, Torre-Amione G, Garcia-Rivas GJ and Altamirano J. Impaired oxidative metabolism and calcium mishandling underlie cardiac dysfunction in a rat model of post-acute isoproterenol-induced cardiomyopathy. Am J Physiol Heart Circ Physiol. 2015;308:H467–77. [DOI] [PubMed] [Google Scholar]
- 45.Lee SR, Ko TH, Kim HK, Marquez J, Ko KS, Rhee BD and Han J. Influence of starvation on heart contractility and corticosterone level in rats. Pflugers Arch. 2015;467:2351–60. [DOI] [PubMed] [Google Scholar]
- 46.Van Voorhies WA and Ward S. Genetic and environmental conditions that increase longevity in Caenorhabditis elegans decrease metabolic rate. Proc Natl Acad Sci U S A. 1999;96:11399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunter GR, Weinsier RL, Gower BA and Wetzstein C. Age-related decrease in resting energy expenditure in sedentary white women: effects of regional differences in lean and fat mass. Am J Clin Nutr. 2001;73:333–7. [DOI] [PubMed] [Google Scholar]
- 48.de Carvalho A, Bassaneze V, Forni MF, Keusseyan AA, Kowaltowski AJ and Krieger JE. Early postnatal cardiomyocyte proliferation requires high oxidative energy metabolism. Sci Rep. 2017;7:15434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
In order to minimize the possibility of unintentionally sharing information that can be used to re-identify private information, a subset of the data generated for this study are available at GEO database and can be accessed at GSE131236.