Abstract
Rationale
Increasing evidence has demonstrated that changes in the gut microbiome, including those associated with dietary influences, are associated with alterations in many physiological processes. Alcohol consumption is common across human cultures and is likely to have a major effect on the gut microbiome, but there remains a paucity of information on its effects in primates.
Objectives
The effects of chronic alcohol consumption on the primate gut microbiome and metabolome was studied in rhesus macaques that were freely drinking alcohol. The objectives of the study were to determine what changes occurred in the gut microbiome following long-term exposure to alcohol and if these changes were reversible following a period of abstinence.
Methods
Animals consuming alcohol were compared to age matched controls without access to alcohol and were studied before and after a period of abstinence. Fecal samples from rhesus macaques were used for 16S rRNA sequencing to profile the gut microbiome and for metabolomic profiling using mass spectrometry.
Results
Alcohol consumption resulted in a loss of alpha-diversity in rhesus macaques, though this was partially ameliorated by a period of abstinence. Higher levels of Firmicutes were observed in alcohol drinking animals at the expense of a number of other microbial taxa, again normalizing in part with a period of abstinence. Metabolomic changes were primarily associated with differences in glycolysis when animals were consuming alcohol and differences in fatty acids when alcohol-drinking animals became abstinent.
Conclusions
The consumption of alcohol has specific effects on the microbiome and metabolome of rhesus macaques independent of secondary influences. Many of these changes are reversed by a relatively short period of abstinence.
Keywords: alcohol, microbiome, metabolome, rhesus macaque
Introduction
Alcohol consumption is common across cultures and history, but only in the last two hundred years has society begun to associate it with negative consequences (Levine 1984). While the effects of consumption in moderate amounts remains muddled, over-consumption has been associated with a number of health-related issues (Rehm et al. 2009; Shield et al. 2013). Physiologically, it has been associated with various cancers (Corrao et al. 2004), heart disease (Taylor et al. 2009), diabetes (Baliunas et al. 2009), and more. Alcohol consumption and alcohol use disorders are also associated with major depressive disorder and anxiety disorders (Boschloo et al. 2011; Conner et al. 2009) and comorbid with a number of other psychiatric diseases although order of causation is unclear. In the past decade, the effect of alcohol on the gut microbiome has begun to garner attention as a target of both pathology and treatment (Cresci 2015; Hillemacher et al. 2018; Leclercq et al. 2018).
There have been a number of studies that have attempted to associate alcohol consumption with changes to the microbiome, the vast majority of which are correlative in nature. In humans, there has been evidence of microbiome dysbiosis associated with patients with chronic alcoholic dependence (Leclercq et al. 2014; Mutlu et al. 2012) but only with a subset of individuals and correlated neither with physiological liver disease nor period of abstinence. The individuals with greater microbiome dysbiosis also demonstrated a “leaky gut”, increased intestinal permeability and transfer of lumenal biochemicals, and had more severe alcohol use disorders, depression, and anxiety (Leclercq et al. 2014). Patients with alcohol-derived liver pathologies demonstrate distinct microbiome community profiles from controls including a significantly decreased microbial diversity (Chen et al. 2011; Sarangi et al. 2017). This work has also been extended to include other alcohol-derived liver and pancreas pathologies and to compare not only these patients to healthy controls, but also to patients with alcohol use disorders absent these additional pathologies (Ciocan et al. 2018; Dubinkina et al. 2017). These latter studies demonstrated that additional distinct changes in the microbiome occur associated with alcohol-derived diseases of the liver and pancreas beyond the changes occurring from alcohol consumption alone.
Human studies of the microbiome, however, have many challenges (Kelly et al. 2016). Inter-individual variability in human populations is high and unsurprising given the vast differences in diet and environment that can be particularly pronounced on the microbiome (Lozupone et al. 2012). In studies of alcohol, this is compounded by differences in quantity, duration, and co-variable environmental exposures. These differences in human studies are highlighted by the differences seen both in groups with comorbid liver or pancreas pathologies (Ciocan et al. 2018; Dubinkina et al. 2017) or severity of alcohol use disorder (Leclercq et al. 2014). However, while study specifics varied, a commonality was the observation of a decrease in microbiome diversity with alcohol exposure. Illuminatingly, the one exception to this decrease in diversity was observed in the citizen science American Gut Project (Kosnicki et al. 2018). It is tempting to impute sociological meaning to these differences in results, but it can suffice to demonstrate that in humans confounds in populations under study remain an important consideration.
As for much of biomedical science, the majority of foundational studies on the relationship between alcohol and the gut microbiome, particularly the gut-brain axis and behavior, are derived from rodents (Cryan and Dinan 2012; Sampson and Mazmanian 2015; Vuong et al. 2017). Some of the first studies to demonstrate gut dysbiosis resulting from alcohol were done in rats (Mutlu et al. 2009) and mice (Bull-Otterson et al. 2013; Yan et al. 2011). Since then, there have been a number of follow-up studies using rodents with varying models of alcohol administration and showing generally, but not always, similar decreases in diversity and specific shifts in microbial abundance, particularly in Firmicutes and Bacteroidetes (Jadhav et al. 2018; Kosnicki et al. 2018; Lowe et al. 2017; Peterson et al. 2017; Wang et al. 2018). It is unclear, however, how the gut microbiome of rodents can be translated to humans. The colonic contents are processed differently in rodents and the native composition of their microbiome is dissimilar (Kostic et al. 2013). Rodents also metabolize alcohol differently than humans (Cederbaum 2012) and models of alcohol consumption in rodents are often of short duration and high concentration (Bertola et al. 2013), a pattern not commonly seen in human populations.
Non-human primate models such as the rhesus macaque (Macaca mulatta) are widely used in biomedical research because of their behavioral, genetic, and physiological similarities to humans (Vallender and Miller 2013). The gastrointestinal tract of the rhesus macaque also is similar to humans (Dubois et al. 1977) and previous research has demonstrated that the compositional structure of the macaque microbiome is comparable to that of humans as well (Handley et al. 2012; McKenna et al. 2008; Yasuda et al. 2015). Rhesus macaque models of alcohol consumption have also been developed that better capture the behavioral patterns seen in humans (Platt and Rowlett 2012). Recently rhesus macaque mucosal biopsies from multiple sites in the intestinal tract were used to investigate the effects of long-term alcohol self-administration on the gut microbiome (Barr et al. 2018). In contrast to the majority of human and rodent studies, this research did not identify differences in diversity levels following alcohol consumption. It also showed decreases in Bacteroidetes and increases in Proteobacteria, again contrasting with previous work. These discrepancies are hypothesized to result from differences in duration and quantity of alcohol consumption, diet, and physiological differences. It is also likely that they are affected by differences between mucosal and lumenal microbial communities (Yasuda et al. 2015).
The reversibility, or irreversibility, of alcohol-induced changes to the gut microbiome is important in the context of treatment. Longitudinal studies can offer not only better understandings of the causes and effects of alcohol on the microbiome, but also lay the groundwork for interventional work. The animal studies, including both the human and rhesus macaque, tend to use single time points, collecting samples post-mortem or during drinking periods. Most human studies are single time point as well, but with participants varying in their last exposure to alcohol, though most of these studies are of chronic, high quantity alcohol drinkers that often have developed secondary liver diseases as a result. One study in humans included the effects of a 3-week abstinence period, finding few changes exception for an increase in total bacterial levels and a handful of specific genera (Leclercq et al. 2014).
Here, we use a rhesus macaque model to explore the effects of chronic alcohol self-administration on the gut microbiome and metabolome. We also test the effects of a short, 5-day, abstinence period. These studies are aimed at developing a rhesus macaque model of the human gut microbiome in disease that can be used to investigate translationally-relevant environmental, pharmacological, and interventional manipulations with the strong controls necessary. With this work we hope to improve our understanding of the relationship between alcohol consumption and microbiome changes, which may ultimately lead to the development of novel therapeutic tools for alcohol-related issues.
Materials and methods
Animals
Samples from 21 outbred male Indian-origin rhesus macaques (Macaca mulatta) were used in this study. At the time of study, animals were either adolescent (4–6 years; n=12) or adult (10–15 years; n=9) and weighed 7–10 kg or 10–12 kg respectively. All animals were individually housed with a 12:12 hour light/dark cycle and fed standard monkey chow (Harlan Teklad Monkey Diet, Harlan Teklad, Madison, WI). Diets were supplemented with fresh fruit but did not vary between animals. Monkeys were divided into either alcohol drinking or control groups of adolescent alcohol (n=6), adolescent control (n=6), adult alcohol (n=4) and adult control (n=5). All procedures were performed in accordance with the guidelines of the Committee on Animals of the Harvard Medical School and the National Academies Guide for Care and Use of Laboratory Animals (National Research Council 2011). All protocols were approved by the Harvard Medical School Institutional Animal Care and Use Committee.
Alcohol self-administration
For all animals, alcohol self-administration occurred in the home cage 5 days per week via a custom-designed operant drinking panel attached to one side of the cage. The panel was equipped with two response levers, two retractable sippers equipped with solenoids to minimize dripping (model #: ENV-652AM), a food pellet dispenser, and stimulus lights (Med Associates, Inc., St. Albans, VT). Sippers were attached to stainless steel liquid reservoirs fixed on the outside of the drinking panel via Tygon tubing.
Monkeys were induced to drink alcohol in a step-wise fashion similar to previous studies (Vivian et al. 2001), with each “step” lasting 30 days. Initially, only water was available from 1 spout. In this step, every lever press (fixed-ratio 1; FR1) during the 3-h session resulted in the extension of the sipper for 30-s. Depression of the sipper by the monkey resulted in fluid delivery. Within the 30-s sipper extension time, the monkey could stop drinking (i.e., release the spout) and resume drinking (i.e., displace the spout) as many times as he wanted. The actual duration and volume of intake, within the constraints of the schedule, were under the control of the subject. Between extensions of the drinking spout, all lights were off briefly, and responses had no scheduled consequences. To facilitate use of the panel and development of drinking behavior, during water training sessions, food pellets were delivered into a receptacle every 600 s (i.e., schedule-induced polydipsia; (Falk 1961; Vivian et al. 2001)).
In the second step, the volume of a 4% w/v ethanol solution (95% ethanol diluted with tap water; Pharmco Products, Brookfield, CT) required to deliver a dose of 0.5 g/kg alcohol was made available to drink on the opposite sipper; in the third step, the volume of 4% w/v ethanol solution was increased to deliver a dose of 1 g/kg; and, finally, in the fourth step, the volume of 4% w/v ethanol solution was increased again to deliver a dose of 1.5 g/kg. Other aspects of the self-administration procedure (e.g., sipper extension time, fixed-ratio, session length, scheduled delivery of pellets) were the same as during water only availability (step 1). Following induction, the adult animals were given concurrent access to both water and ethanol (0.5 – 16% w/v depending on phase of study) and could press either lever to obtain the fluid of their choice. Pellet delivery also was discontinued.
All adolescent animals self-administered alcohol for a period of 3 months (i.e., step-wise induction), while the adult animals had been allowed to self-administer alcohol for 5 years with irregular breaks as necessary for standard animal husbandry. All animals reliably self-administered alcohol across the period of study (Sawyer et al. 2014). Average daily alcohol consumption in the adolescents ranged from 0.5 – 1.5 g/kg depending on the induction step; whereas the average daily consumption in adults ranged from 0.8 – 2.2 g/kg depending on the animal. These intake levels routinely produce blood alcohol levels ≥ 80 mg/dL, the legal limit in the United States for humans operating a motor vehicle (Sawyer et al. 2014). Average alcohol consumption levels and blood alcohol levels for individual animals are shown in Table 1. Control animals were age-matched, housed in the same facility and exposed to identical conditions as the alcohol self-administering animals, but did not have access to alcohol.
Table 1.
Individual alcohol consumption and blood alcohol levels (BAL) for rhesus macaques used in this study. Values for adult alcohol group reflect intakes/BALs from the year prior to sample collection. Values for the adolescent alcohol group reflect intakes from the first 3 months of alcohol availability during step-wise induction; BAL was determined on the final day of 1.5 g/kg intake.
| Monkey | Groupa | Average Alcohol g/kg (± SEM) | Average BAL mg/dl ((± SEM) |
|---|---|---|---|
| MM-33 | Adult alcohol | 2.2 (0.08) | 128.5 (1.3) |
| MM-162 | Adult alcohol | 1.8 (0.01) | 105.5 (3.7) |
| MM-247 | Adult alcohol | 1.6 (0.07) | 88.6 (2.3) |
| MM-267 | Adult alcohol | 1.8 (0.09) | 106.4 (5.0) |
| MM-107 | Adolescent alcohol | 1.0 (0.05) | 104.4 (-) |
| MM-118 | Adolescent alcohol | 0.9 (0.06) | 109.2 (-) |
| MM-119 | Adolescent alcohol | 0.9 (0.05) | 111.4 (-) |
| MM-121 | Adolescent alcohol | 1.0 (0.05) | 105.4 (-) |
| MM-128 | Adolescent alcohol | 0.9 (0.05) | 101.4 (-) |
| MM-132 | Adolescent alcohol | 1.0 (0.06) | 107.3 (-) |
Fecal sample collection
Fresh fecal samples were collected from cage trays from all animals under conditions of standard feeding and exposure, or lack thereof, to alcohol. All samples were collected in the morning between roughly 9AM and 10AM. Samples from each animal were collected serially immediately following one another and all groups were collected contemporaneously. Alcohol availability was then withheld from the self-administering animals for a period of 5 days and fecal samples collected in the same manner. There were no observable signs of withdrawal during this period. Samples were immediately flash frozen and stored at −80°C for future analysis.
DNA isolation and sequencing
DNA was extracted from 200 mg of the fecal samples from each subject using the FastDNA™ SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA) following manufacturer’s protocols. Concentration of the DNA samples was measured using a NanoDrop™ 1000 spectrophotometer (Thermo Scientific, Waltham, MA). Amplification and sequencing of the V4 region was performed as previously described (Yasuda et al. 2015; Yatsunenko et al. 2012). Briefly, 25 μg of genomic DNA from each sample was amplified using HotMaster Taq DNA Polymerase and HotMasterMix (Quantabio, Beverly, MA) and primers designed to cover the V4 region of 16S RNA incorporating Illumina adapters and a barcode sequence (515F: GTGCCAGCMGCCGCGGTAA; 806R: GGACTACHVGGGTWTCTAAT). Standard cycling conditions were used: initial denaturation at 94°C for 3 minutes; 30 cycles of denaturation at 94°C for 45 seconds, annealing at 50°C for 60 seconds, and elongation at 72°C for 5 minutes; and a final elongation step at 72°C for 10 minutes.
Amplicons were quantified (Caliper LabChipGX; PerkinElmer, Waltham, MA), pooled in equimolar concentrations, and size selected (375–425 bp) (Pippin Prep; Sage Sciences, Beverly, MA) to reduce non-specific amplification products from host DNA. Final quality control checks and quantification were performed using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Sequencing was performed on the Illumina MiSeq platform (Illumina, San Diego, CA) using a 175-bp paired-end protocol according to the manufacturer’s specifications with addition of 5% PhiX.
16S rRNA sequence processing
Following standard quality control and disaggregation, reads were processed using a data curation pipeline in QIIME (Caporaso et al. 2010). First, paired-end reads were joined and size selected to reduce non-specific amplification. These reads were then grouped into operational taxonomic units (OTUs) based on sequence similarity. OTUs were subsequently assigned to an established taxonomy using the Greengenes database release 13_5 (McDonald et al. 2012). Approximately 30,000 reads per sample were obtained. OTUs with fewer than 8 reads and 20% prevalence were excluded from downstream analysis. Data was rarefied to the minimum library size using total sum scaling (TSS). MaAsLin (Morgan et al. 2012) was used for multivariate analysis to identify associations between microbiome abundance and factors of interest including alcohol exposure and various confounding factors (e.g. age and weight). LEfSe (Segata et al. 2011) was used for univariate analyses targeting alcohol exposure only. A maximum false discovery rate (corrected p-value significance threshold) determined using the Benjamini-Hochberg correction was set at 10%. Data visualization and initial statistical analysis was conducted using MicrobiomeAnalyst (Dhariwal et al. 2017).
Alpha richness and diversity from fecal samples collected from control rhesus macaques and animals with chronic access to alcohol before and after abstinence was calculated using the Chao1 (Chao and Mark 1993) and Shannon (Shannon 1948) measures. Differences between the groups were calculated using the non-parametric Mann-Whitney test for the unpaired control and alcohol (baseline or abstinent) groups and the Wilcoxon test for the paired alcohol groups before and after a period of abstinence.
Metabolomic analysis
Fecal samples collected as above were lyophilized and weight equivalents were used for untargeted metabolomic analyses by Metabolon, Inc (Morrisville, NC) using a UPLC-MS/MS and GC-MS platform. A full accounting of the commercialized process for isolating, identifying, and characterizing metabolites by Metabolon can be found elsewhere (Evans et al. 2009; Shin et al. 2014; Zierer et al. 2018). Briefly, samples are extracted using aqueous and organic solvents with added recovery controls. Small molecules are isolated first by methanol precipitation under vigorous shaking followed by centrifugation. Extract was then separated into an acidic positive-ion condition optimized for hydrophilic compounds, an acidic positive-ion condition optimized for hydrophobic compounds, a basic negative-ion condition with a C18 column, and a negative ionization following elution from an HILIC column. Following Metabolon procedure, fixed injection controls were added.
UPLC-MS/MS was performed using ACQUITY UPLC (Waters, Milford, MA) and a Finnigan LTQ FT mass spectrometer (Thermo Scientific, Waltham, MA). GC-MS was performed using a Finnigan TRACE DSQ GC/MS (Thermo Scientific, Waltham, MA). Compounds were identified by comparison to libraries of purified standards based on retention time/index (RI), mass to charge ratio (m/z), and chromatographic data (Dehaven et al. 2010; Evans et al. 2009). Pairwise comparisons between alcohol drinking animals before and after abstinence or alcohol drinking and control animals were calculated with paired t-tests or Welch’s two-sample t-tests respectively with Benjamini-Hochberg correction.
Results
The gut microbiome is stable in rhesus macaques across demographics after four years of age
Initial analyses explored whether there were significant differences in the gut microbiome resulting from demographic factors (e.g. age, body weight, history). Using LEfSe (Segata et al. 2011), no significant differences were found between the younger and older animals at any taxonomic level either when compared alone or in combination with alcohol drinking status (Table S1). MaAsLin (Morgan et al. 2012) was also used to investigate age*alcohol interactions, but again there were no significant effects detected. Age at time of study for alcohol drinkers was also inexorably linked to duration of alcohol exposure. Because no age difference was observed for alcohol drinkers before or after a period of abstinence, there was likewise no difference associated with duration of alcohol exposure. It remains formally possible that age and drinking duration have opposite effects that resulted in canceling each other out, but there was no evidence to suggest this to be the case.
Various other demographic factors were also considered including weight, birth location of animals, and housing room. No significant effect from any of these was expected and none was identified. Additional fecal microbiome data collected from a contemporary cohort of rhesus macaques (Yasuda et al. 2015) was also incorporated at this time and compared to the control animals in the present study. Although these animals had more variable backgrounds and were demographically more diverse, they had been housed at the same location for a minimum of two years and fed the same diet, but were older (ages 12–22) females. The raw reads generated from the previous study were incorporated into the same bioinformatics pipeline and processed concomitantly with the present samples. Again, no significant differences in microbiome composition were observed between these animals and the control animals of the present study. For the remaining analyses in the paper, all microbiome studies were conducted conflating the age and other demographic distinctions into the three primary groups: control animals, without any exposure to alcohol; alcohol drinking animals, animals with alcohol self-administration histories that were actively consuming alcohol at the time of fecal collection; and alcohol abstinent animals, the same animals with alcohol self-administration histories with samples collected following a five day abstinence period where alcohol was not available.
The gut microbiome is substantially altered in rhesus macaques following chronic alcohol self-administration
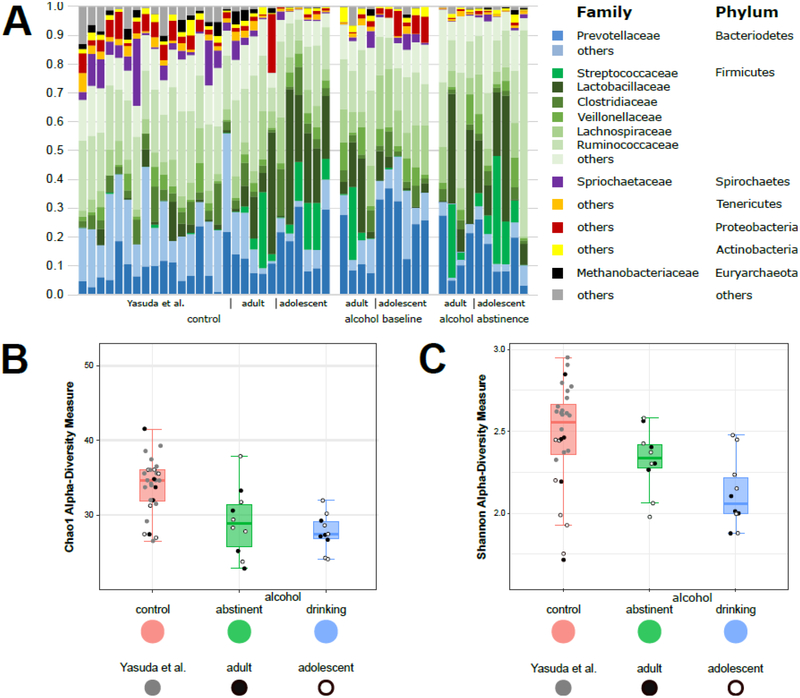
The gut microbiome of individual animals did not vary systematically and meaningfully within specific groups (Fig 1A). Overall, animals showed similar patterns with the greatest abundance of Bacteroidetes and Firmicutes. Tenericutes, Actinobacteria, Proteobacteria, Spirochaetes, and other phyla were irregularly distributed among animals and at much lower levels. Taxa richness and evenness, measured by alpha-diversity, was significantly different among the three groups (Fig 1B and 1C). The greatest diversity was observed in control animals, while alcohol drinking animals showed the most restricted microbial communities. Following abstinence diversity was somewhat recovered in animals with a history of alcohol drinking, but still not to the levels seen by control animals.
Figure 1.
Influence of alcohol on the gut microbiome in rhesus macaques. (A) Family-level relative abundance of intestinal microbiota in the stool of rhesus macaques without exposure to alcohol (left), with long-term free access to alcohol (right), and following a period of abstinence from alcohol (middle). (B) Alpha diversity from fecal samples collected from control rhesus macaques and animals with chronic access to alcohol before and after abstinence using the Chao1 measure (control vs. alcohol abstinent: two-tailed Mann-Whitney p = 0.0076; control vs. alcohol drinking: two-tailed Mann-Whitney p = 0.0003; alcohol drinking vs. alcohol abstinent: two-tailed Wilcoxon 0.2983). (C) Alpha diversity using the Shannon measure (control vs. alcohol abstinent: two-tailed Mann-Whitney p = 0.0414; control vs. alcohol drinking: two-tailed Mann-Whitney p = 0.0047; alcohol drinking vs. alcohol abstinent: two-tailed Wilcoxon 0.0969).
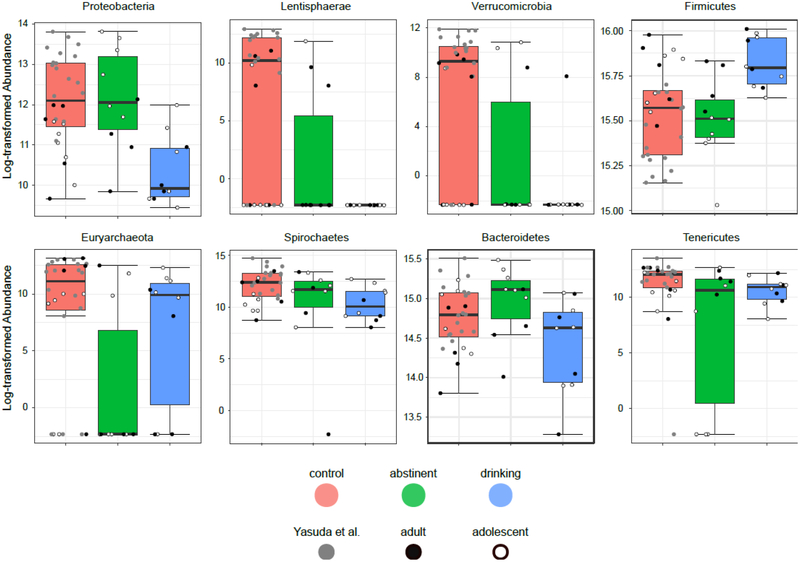
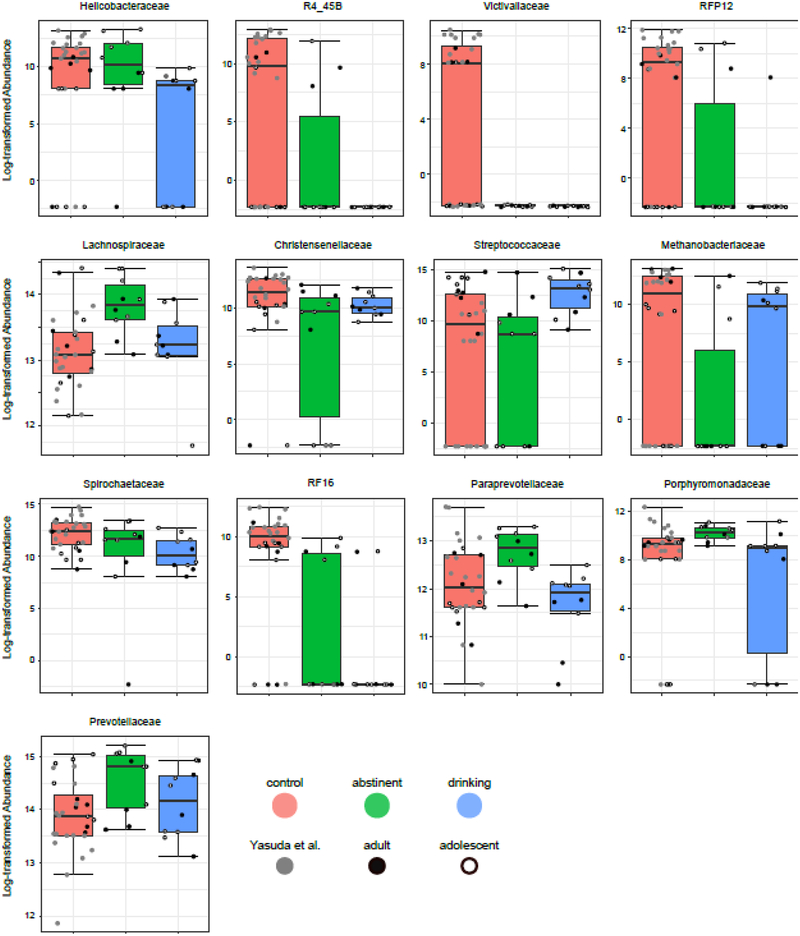
Beyond the general composition of the microbiome, specific taxa also were observed to be differentially expressed between the groups. Univariate analyses implemented using LEfSe (Segata et al. 2011) identified taxa significantly differentially expressed among the three group (Table 2). Following correction for multiple comparisons, 8 phyla (Fig 2) and 13 families (Fig 3) were identified as showing significant group effects. Broadly speaking, three patterns were observed: alcohol-ever, similar relative abundance in alcohol drinking and abstinent animals but different from controls; alcohol-present, similar relative abundance in controls and alcohol abstinent animals but different from active alcohol drinking animals; and recovering, intermediate relative abundances in alcohol abstinent animals between significantly different controls and alcohol drinking animals
Table 2.
Phyla and families significantly (FDR < 0.10) different in control, alcohol-drinking, and alcohol-abstinent rhesus macaques.
| Alcohol | |||||||
|---|---|---|---|---|---|---|---|
| Phylum | Family | P-value | FDR | Control | Abstinent | Drinking | LDAscc |
| Proteobacteria | 0.0021 | 0.0082 | 299194 | 344045 | 48834 | 5.17 | |
| Helicobacteraceae | 0.0047 | 0.0292 | 96206 | 143037 | 4726 | 4.84 | |
| Lentisphaerae | 0.0008 | 0.0049 | 95193 | 7876 | 630 | 4.67 | |
| R4_45B | 0.0009 | 0.0105 | 87092 | 7876 | 630 | 4.64 | |
| Verrucomicrobia | 0.0006 | 0.0049 | 31056 | 9767 | 0 | 4.19 | |
| Victivallaceae | 0.0043 | 0.0292 | 8102 | 0 | 0 | 3.61 | |
| RFP12 | 0.0006 | 0.0097 | 31056 | 9767 | 0 | 4.19 | |
| Firmicutes | 0.0029 | 0.0087 | 5737015 | 5575929 | 7508822 | 5.99 | |
| Lachnospiraceae | 0.0052 | 0.0292 | 596588 | 1108066 | 603970 | 5.41 | |
| Christensenellaceae | 0.0216 | 0.0648 | 178009 | 43478 | 32136 | 4.86 | |
| Streptococcaceae | 0.0229 | 0.0648 | 351067 | 299307 | 1008192 | 5.55 | |
| Eury archaeota | 0.0361 | 0.0467 | 155505 | 45684 | 48204 | 4.74 | |
| Methanobacteriaceae | 0.0454 | 0.0909 | 145828 | 42848 | 39067 | 4.73 | |
| Spirochaetes | 0.0272 | 0.0408 | 448285 | 203214 | 82861 | 5.26 | |
| Spirochaetaceae | 0.0272 | 0.0712 | 448285 | 203214 | 82861 | 5.26 | |
| Bacteroidetes | 0.0389 | 0.0467 | 2808421 | 3454316 | 2092628 | 5.83 | |
| RF16 | 0.0002 | 0.0060 | 52435 | 3151 | 1260 | 4.41 | |
| Paraprevotellaceae | 0.0111 | 0.0419 | 246647 | 386894 | 138626 | 5.09 | |
| Porphyromonadaceae | 0.0073 | 0.0354 | 22842 | 35602 | 12917 | 4.05 | |
| Prevotellaceae | 0.0417 | 0.0909 | 1332478 | 2405797 | 1632325 | 5.73 | |
| Tenericutes | 0.0169 | 0.0337 | 177446 | 73094 | 64272 | 4.75 | |
Figure 2.
Influence of alcohol on gut taxa in rhesus macaques at the Phylum level. For each phylum, a box-and-whisker plot is shown of the log-transformed abundance observed in control (left), alcohol-abstinent (center), and alcohol-drinking (right) rhesus macaques. Animals from the Yasuda et al. study are shown in gray, animals from the older cohort in solid black, and animals from the younger cohort in white with black outline. Significance was determined using LEfSe (Segata et al. 2011) and is shown in Table 2.
Figure 3.
Influence of alcohol on gut taxa in rhesus macaques at the Family level. For each family, a box-and-whisker plot is shown of the log-transformed abundance observed in control (left), alcohol-abstinent (center), and alcohol-drinking (right) rhesus macaques. Animals from the Yasuda et al. study are shown in gray, animals from the older cohort in solid black, and animals from the younger cohort in white with black outline. Significance was determined using LEfSe (Segata et al. 2011) and is shown in Table 2.
“Alcohol-ever” taxa were characterized by consistent differences between control and alcohol-drinking animals regardless of abstinence periods. The patterns suggest that taxa abundance is reduced following exposure to alcohol and does not rebound. This could be due to the fact that taxa are effectively eradicated or simply that growth is slow compared to the time scales that were considered. There were perhaps surprisingly few of these taxa (i.e. phylum: Euryarchaeota, Tenericutes, Lentisphaerae, Verrucomicrobia; family: R4_45B, Victivallaceae, RFP12, Christensenellaceae, Methanobacteriaceae, RF16). Many more taxa were entirely recovered with abstinence, “alcohol-current” taxa dependent only on alcohol drinking status at time of sample collection (i.e. phylum: Proteobacteria, Firmicutes, Bacteroidetes; family: Helicobacteraceae, Streptococcaceae), or were at intermediate levels during the abstinence period, “recovering” taxa (i.e. phylum: Spirochaetes; family: Spirochaetaceae). Most of these taxa were less abundant in animals drinking alcohol compared to the controls or even the same animals following a period of abstinence. The primary exception to this was the phylum Firmicutes, particularly the family Streptococcaceae, which was both of high absolute and relative abundance in alcohol drinking animals.
The findings from individual taxa reflect the previous understandings generated from the diversity analyses. In alcohol drinking animals, abundance in most taxa is reduced and Firmicutes dominates the microbiome, while in control animals there are both more taxa represented and they are more abundant. The ratio of Firmicutes to Bacteroidetes, one common measure in gut microbiome studies, is elevated in alcohol drinking animals compared to both control animals and following a period of abstinence. It is perhaps relevant that despite the absence of statistically significant major effects there did appear to be more change occurring in the younger cohort of animals than in the older cohort (Fig S1). It is tempting to relate this back to length of alcohol exposure, although the study remains underpowered and lacks the necessary design to be confident in this interpretation.
Within both of the major phyla, Firmicutes and Bacteroidetes, there is also evidence of a remodeling of communities at lower levels. In Firmicutes the dominant taxa, Streptococcaceae, is significantly elevated in alcohol drinking animals. This decreases following a period of abstinence and Lachnospiraceae becomes significantly elevated. This greater abundance of Lachnospiraceae is not observed in control animals, however, although there a significantly higher level of Christensenellaceae is observed than in animals with any history of alcohol drinking. Within Bacteroidetes, the patterns of taxa recovery following an abstinence period are also observed at the family level. RF16 remains significantly lower in animals with an alcohol history even after a period of abstinence, while Bacteroidaceae returns to control levels. Interestingly, Paraprevotellaceae, Porphyromonadaceae, and Prevotellaceae all show elevated levels following a short period of abstinence that are not seen when comparing control animals with the alcohol drinking animals under their normal, alcohol available, circumstances.
Across the various taxa and analyses two commonalities stand out. First, the microbiome of animals drinking alcohol is reduced in complexity and dominated by Firmicutes. Almost every other taxa is more abundant in control animals, resulting in a significantly greater diversity overall. Second, the microbiome of alcohol drinking animals following a period of abstinence tends to represent a middle ground between when they are actively drinking alcohol and control animals without any prior alcohol exposure.
The fecal metabolome of rhesus macaques is significantly altered by alcohol drinking and abstinence
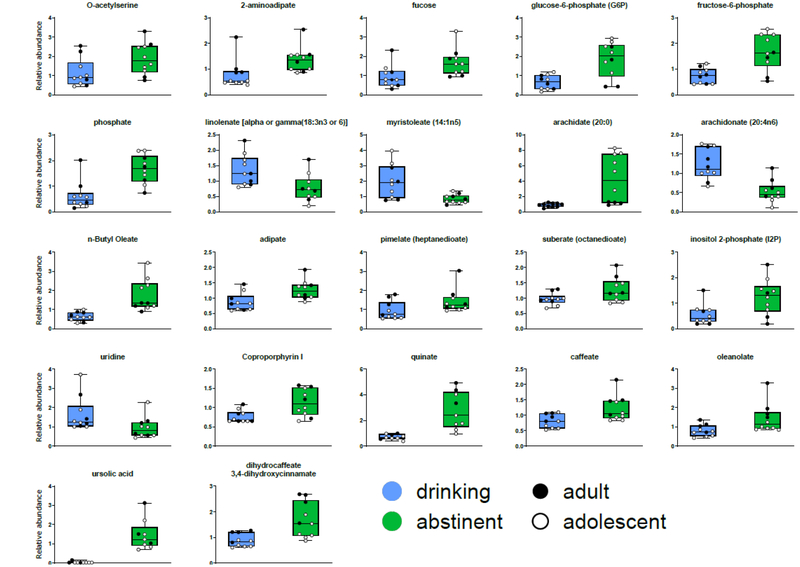
The relative levels of 410 biochemical compounds were identified in this study (Table S2). For each compound, control animals and animals exposed to alcohol were compared using a Welch’s two-sample t-test. After correction for multiple testing, there were no statistically significant different metabolites between the groups. However, when the same metabolites were compared using paired t-tests between animals while drinking compared to a period of abstinence, 22 metabolites were significantly changed (Table 3, Figure 4).
Table 3.
Fecal biochemicals significantly (FDR < 0.05) different between alcohol drinking animals before and after 5 day abstinence
| Mean Values | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Alcohol | |||||||||
| Biochemical | Platform | PubChem | CAS | Abstinent | Drinki ng | P-value | FDR | ||
| ursolic acid | GC/MS | 64945 | 77-52-1 | 3.385 | 0.038 | 0.0000 | 0.0006 | ||
| Isobar: dihydrocaffeate, 3,4-dihydroxycinnamate | GC/MS | 1.679 | 0.885 | 0.0000 | 0.0034 | ||||
| uridine | LC/MS Neg | 6029 | 58-96-8 | 0.952 | 1.641 | 0.0001 | 0.0078 | ||
| inositol 2-phosphate (I2P) | GC/MS | 1.249 | 0.554 | 0.0001 | 0.0101 | ||||
| phosphate | GC/MS | 1061 | 7664-38-2 | 1.657 | 0.623 | 0.0002 | 0.0135 | ||
| caffeate | GC/MS | 689,043 | 331-39-5 | 1.219 | 0.812 | 0.0002 | 0.0135 | ||
| n-Butyl Oleate | GC/MS | 5354342 | 142-77-8 | 1.757 | 0.639 | 0.0004 | 0.0180 | ||
| quinate | GC/MS | 77-95-2 | 5.382 | 0.733 | 0.0004 | 0.0180 | |||
| adipate | GC/MS | 196 | 124-04-9 | 1.269 | 0.872 | 0.0004 | 0.0180 | ||
| suberate (octanedioate) | LC/MS Pos | 10457 | 505-48-6 | 1.277 | 0.972 | 0.0008 | 0.0324 | ||
| oleanolate | GC/MS | 10494 | 508-02-1 | 1.416 | 0.797 | 0.0013 | 0.0405 | ||
| fucose | GC/MS | 3034656 | 2438-80-4 | 1.675 | 0.944 | 0.0013 | 0.0405 | ||
| fructose-6-phosphate | GC/MS | 103213-47-4 | 1.658 | 0.736 | 0.0014 | 0.0405 | |||
| 2-aminoadipate | GC/MS | 469 | 542-32-5 | 1.370 | 0.789 | 0.0015 | 0.0405 | ||
| O-acetylserine | GC/MS | 66638-22-0 | 1.879 | 1.134 | 0.0015 | 0.0405 | |||
| myristoleate (14:1n5) | LC/MS Neg | 5281119 | 544-64-9 | 0.834 | 1.960 | 0.0017 | 0.0430 | ||
| pimelate (heptanedioate) | GC/MS | 385 | 111-16-0 | 1.436 | 0.950 | 0.0021 | 0.0490 | ||
| linolenate [alpha or gamma; (18:3n3 or 6)] | LC/MS Neg | 0.773 | 1.352 | 0.0022 | 0.0490 | ||||
| Coproporphyrin I | LC/MS Neg | 69477-27-6 | 1.130 | 0.768 | 0.0023 | 0.0490 | |||
| arachidate (20:0) | GC/MS | 10467 | 506-30-9 | 4.238 | 0.871 | 0.0025 | 0.0497 | ||
| glucose-6-phosphate (G6P) | GC/MS | 103192-55-8 | 1.841 | 0.651 | 0.0027 | 0.0497 | |||
| arachidonate (20:4n6) | LC/MS Neg | 444899 | 506-32-1 | 0.526 | 1.222 | 0.0027 | 0.0497 | ||
Figure 4.
Influence of alcohol on fecal metabolites in rhesus macaques. A box-and-whisker plot of scaled abundance is show in alcohol-drinking (left), and alcohol-abstinent (right) rhesus macaques. Animals from the older cohort in solid black and animals from the younger cohort in white with black outline. Significant main-effect of alcohol exposure was calculated a paired t-test with Benjamini-Hochberg correction for multiple test comparisons and is shown in Table 3.
More than one third of the metabolites significantly changed following abstinence were fatty acids, a significant overrepresentation compared to all detectable metabolites (8 out of 22 compared to 60 out of 410; p = 0.0079, Fishers Exact test). Glucose-6-phosphate and fructose-6-phosphate, part of energy utilization upstream of the Krebs cycle, show patterns of decreased levels in animals while drinking alcohol compared to following a period of abstinence. Unlike the pattern observed among the fatty acids, there is also a significantly higher relative concentration detected in control animals compared to alcohol drinking. Again, the glycolysis pathway is overrepresented among the overrepresented among significantly different metabolites (2 out of 22 compared to 7 out of 410; p = 0.049, Fishers Exact test), though this does not survive correction for multiple tests.
These results are impacted greatly by intra-group variation. A two way repeated measures ANOVA showed that only a paucity of the metabolites varied significantly by age or age * alcohol interaction (Table S3). More generally, however, the alcohol drinking animals following abstinence show much higher levels of variation than either of the other two groups or when compared between groups. While these patterns hold for both the younger cohort of animals and the older cohort of animals, they are more pronounced in the younger animals (Fig 4). This suggests that the effects of the abstinence period, whether the lack of alcohol presence in particular or the change in relative dietary caloric intake more generally, has a stronger effect on younger animals. Apart from the alcohol intake, there is also an observed increase in free amino acids in older animals compared to the younger animals. This likely reflects an age related difference in protein degradation that is independent of alcohol exposure.
Discussion
This study further validates the rhesus macaque as an animal model of the human gut microbiome. The predominance of the phyla Firmicutes and Bacteroidetes and the relative sparsity of Actinobacteria and Verrucomicrobia found in the current study in fecal samples is consistent with the lumenal content of previously studies of the macaque gut microbiome (Yasuda et al. 2015). Similarly, the more specific patterns of taxa presence and abundance at the family level were consistent with those observed in both previous studies of macaques (Handley et al. 2012; McKenna et al. 2008; Yasuda et al. 2015) and humans on more traditional (i.e. non-Western) diets (Yatsunenko et al. 2012). This parallel also extends to the effects of alcohol consumption with the reductions in microbiome diversity and increase in relative abundance of Firmicutes in rhesus macaques similar to previous studies of humans with alcohol use disorders (Leclercq et al. 2014; Mutlu et al. 2012).
The use of rhesus macaques allows for controlled interventions that can be difficult in humans because of confounding factors. Here we tested a relatively short five day abstinence period and observed a largely intermediate phenotype, measures of alpha-diversity were intermediate between those seen for controls and during active drinking and the Firmicutes to Bacteroidetes ratio normalized. Relatively few taxa remained at similar, significantly lower, levels than controls in alcohol drinking animals before and after the abstinence period. It remains unproven if these differences would more completely normalize given a longer period of abstinence, but we would hypothesize that they would. This suggests that the drinking procedure employed here reflects a more moderate consumption pattern.
Differences in the fecal metabolome were largely associated with shifts from drinking to abstinence, although it seems likely that this study was statistically underpowered to detect differences from control animals. The dominant changes in metabolites that were observed were associated with fatty acids and metabolites from the glycolysis pathway. The effects of microbiome on behavior are believed to be mediated, at least in part, from changes in small molecules resulting from microbial metabolism (Cryan and Dinan 2012; Sampson and Mazmanian 2015), though the specific changes observed have not previously been demonstrated to have any effects. It is possible, rather, that they are reflective of a shift in dietary homeostasis. While the overall diets of both alcohol consuming and control animals were roughly isocaloric and animals in both groups maintained similar weights without consistent between group deviations including during periods of drinking and abstinence, the relative contribution of calories from differing sources was variable.
These studies offer both a validation of human studies of the effects of alcohol consumption on the microbiome and a well-controlled model for further investigations. In human studies, a separation was observed during withdrawal with some individuals showing greater microbiome dysbiosis that coupled with increased psychological symptoms associated with relapse (Leclercq et al. 2014). The reason for these disparate groups was not immediately obvious; the differences were not correlated with amount of alcohol consumed or history of alcohol consumption. A rhesus macaque model offers the opportunity to investigate factors that may be driving these differences. Secondary insults to the microbiome may have additional additive effects that make a more severe form of alcohol dependence more likely. In particular, a Western diet may exaggerate or predispose the gut microbiome towards dysbiosis (Ley et al. 2006). This model may also present a more translationally relevant opportunity to test interventional approaches to alcohol use disorders such as probiotics or fecal transplantation. More generally, it allows for the dissociation the direct effects of alcohol from co-occurring behavioral and physiological confounds.
Conclusion
Here we demonstrate that chronic consumption of alcohol has specific effects on the microbiome of rhesus macaques characterized by a decrease in microbial diversity with enrichment of relative abundance of Firmicutes. These can be attributed directly to the alcohol consumption and are not the result of concomitant changes to diet or other environmental factors, but reflect a microbiota and gastrointestinal activity common to primates and translationally relevant to humans. Surprisingly, these effects were partially if not wholly ameliorated following a relatively short 5-day period of abstinence, suggesting that the specific effects observed here are the direct effects of alcohol. While there changes may be specific to the conditions of this model of chronic but moderate-level alcohol consumption, it offers an important perspective on the variables at play in the investigation of the role the gut microbiome plays in alcohol use and its potential as a therapeutic target.
Supplementary Figure 1. Comparison of relative abundances of Firmicutes (left) and Bacteriodetes (right) in individual animals while self-administering alcohol and after a 5-day period of abstinence. Older and younger cohorts are identified by color.
Supplementary Figure 2. Comparison of scaled abundance of metabolites with a significant main effect of alcohol exposure in individual animals while self-administering alcohol and after a 5-day period of abstinence. Older and younger cohorts are identified by color.
Supplementary Material
Acknowledgments
The authors would like to thank the New England Primate Research Center Division of Veterinary Resources and all the animal care technicians for their assistance with sample collection. We would also like to thank Kristin Waraska and the Biopolymers Facility in the Harvard Medical School Department of Genetics and Ashley Johnson and Michael Garrett at the University of Mississippi Medical Center Genomics Core for support with next generation sequencing.
Funding
These studies were funded by grant from the National Institutes of Health: OD011103, AA019688 (EJV), AA016179 (DMP), and AA016828 (DMP). The work performed through the University of Mississippi Medical Center Molecular and Genomics Facility is supported, in part, by funds from the National Institute of General Medical Sciences: GM103476, GM104357, and GM121334. The content of the manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
All animals studies were performed in accordance with the guidelines of the National Academies Guide for Care and Use of Laboratory Animals, 8th Edition (National Research Council 2011). All protocols were approved by the Harvard Medical School Institutional Animal Care and Use Committee prior to commencement of animal studies.
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- Baliunas DO, Taylor BJ, Irving H, Roerecke M, Patra J, Mohapatra S, Rehm J (2009) Alcohol as a risk factor for type 2 diabetes: A systematic review and meta-analysis Diabetes Care 32:2123–2132 doi: 10.2337/dc09-0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr T et al. (2018) Concurrent gut transcriptome and microbiota profiling following chronic ethanol consumption in nonhuman primates Gut microbes:1–19 doi: 10.1080/19490976.2018.1441663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertola A, Mathews S, Ki SH, Wang H, Gao B (2013) Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nature protocols 8:627–637 doi: 10.1038/nprot.2013.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschloo L, Vogelzangs N, Smit JH, van den Brink W, Veltman DJ, Beekman AT, Penninx BW (2011) Comorbidity and risk indicators for alcohol use disorders among persons with anxiety and/or depressive disorders: findings from the Netherlands Study of Depression and Anxiety (NESDA) Journal of affective disorders 131:233–242 doi: 10.1016/j.jad.2010.12.014 [DOI] [PubMed] [Google Scholar]
- Bull-Otterson L et al. (2013) Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment PLoS One 8:e53028 doi: 10.1371/journal.pone.0053028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG et al. (2010) QIIME allows analysis of high-throughput community sequencing data Nature methods 7:335–336 doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederbaum AI (2012) Alcohol metabolism Clin Liver Dis 16:667–685 doi: 10.1016/j.cld.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A, Mark CKY (1993) Stopping Rules and Estimation for Recapture Debugging with Unequal Failure Rates Biometrika 80:193–201 doi: 10.2307/2336768 [DOI] [Google Scholar]
- Chen Y et al. (2011) Characterization of fecal microbial communities in patients with liver cirrhosis Hepatology 54:562–572 doi: 10.1002/hep.24423 [DOI] [PubMed] [Google Scholar]
- Ciocan D, Rebours V, Voican CS, Wrzosek L, Puchois V, Cassard AM, Perlemuter G (2018) Characterization of intestinal microbiota in alcoholic patients with and without alcoholic hepatitis or chronic alcoholic pancreatitis Scientific reports 8:4822 doi: 10.1038/s41598-018-23146-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner KR, Pinquart M, Gamble SA (2009) Meta-analysis of depression and substance use among individuals with alcohol use disorders J Subst Abuse Treat 37:127–137 doi: 10.1016/j.jsat.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrao G, Bagnardi V, Zambon A, La Vecchia C (2004) A meta-analysis of alcohol consumption and the risk of 15 diseases Prev Med 38:613–619 doi: 10.1016/j.ypmed.2003.11.027 [DOI] [PubMed] [Google Scholar]
- Cresci GA (2015) The gut microbiome: a new frontier for alcohol investigation Alcoholism, clinical and experimental research 39:947–949 doi: 10.1111/acer.12732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, Dinan TG (2012) Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour Nature reviews Neuroscience 13:701–712 doi: 10.1038/nrn3346 [DOI] [PubMed] [Google Scholar]
- Dehaven CD, Evans AM, Dai H, Lawton KA (2010) Organization of GC/MS and LC/MS metabolomics data into chemical libraries Journal of cheminformatics 2:9 doi: 10.1186/1758-2946-2-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J (2017) MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data Nucleic acids research 45:W180–W188 doi: 10.1093/nar/gkx295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubinkina VB et al. (2017) Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease Microbiome 5:141 doi: 10.1186/s40168-017-0359-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois A, Natelson BH, van Eerdewegh P, Gardner JD (1977) Gastric emptying and secretion in the rhesus monkey Am J Physiol 232:E186–192 doi: 10.1152/ajpendo.1977.232.2.E186 [DOI] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E (2009) Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems Anal Chem 81:6656–6667 doi: 10.1021/ac901536h [DOI] [PubMed] [Google Scholar]
- Falk JL (1961) Production of polydipsia in normal rats by an intermittent food schedule Science 133:195–196 [DOI] [PubMed] [Google Scholar]
- Handley SA et al. (2012) Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome Cell 151:253–266 doi: 10.1016/j.cell.2012.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillemacher T, Bachmann O, Kahl KG, Frieling H (2018) Alcohol, microbiome, and their effect on psychiatric disorders Progress in neuro-psychopharmacology & biological psychiatry 85:105–115 doi: 10.1016/j.pnpbp.2018.04.015 [DOI] [PubMed] [Google Scholar]
- Jadhav KS et al. (2018) Gut microbiome correlates with altered striatal dopamine receptor expression in a model of compulsive alcohol seeking Neuropharmacology 141:249–259 doi: 10.1016/j.neuropharm.2018.08.026 [DOI] [PubMed] [Google Scholar]
- Kelly JR, Clarke G, Cryan JF, Dinan TG (2016) Brain-gut-microbiota axis: challenges for translation in psychiatry Annals of epidemiology 26:366–372 doi: 10.1016/j.annepidem.2016.02.008 [DOI] [PubMed] [Google Scholar]
- Kosnicki KL, Penprase JC, Cintora P, Torres PJ, Harris GL, Brasser SM, Kelley ST (2018) Effects of moderate, voluntary ethanol consumption on the rat and human gut microbiome Addict Biol doi: 10.1111/adb.12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Howitt MR, Garrett WS (2013) Exploring host-microbiota interactions in animal models and humans Genes Dev 27:701–718 doi: 10.1101/gad.212522.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S et al. (2014) Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity Proc Natl Acad Sci U S A 111:E4485–4493 doi: 10.1073/pnas.1415174111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Starkel P, Delzenne NM, de Timary P (2018) The gut microbiota: A new target in the management of alcohol dependence? Alcohol doi: 10.1016/j.alcohol.2018.03.005 [DOI] [PubMed] [Google Scholar]
- Levine HG (1984) The alcohol problem in America: from temperance to alcoholism British journal of addiction 79:109–119 [DOI] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI (2006) Human gut microbes associated with obesity Nature 444:1022 doi:10.1038/4441022a10.1038/4441022ahttps://www.nature.com/articles/4441022a#supplementary-informationhttps://www.nature.com/articles/4441022a#supplementary-information [DOI] [PubMed] [Google Scholar]
- Lowe PP et al. (2017) Alcohol-related changes in the intestinal microbiome influence neutrophil infiltration, inflammation and steatosis in early alcoholic hepatitis in mice PLoS One 12:e0174544 doi: 10.1371/journal.pone.0174544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R (2012) Diversity, stability and resilience of the human gut microbiota Nature 489:220–230 doi: 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D et al. (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea ISME J 6:610–618 doi: 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna P et al. (2008) The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis PLoS Pathog 4:e20 doi: 10.1371/journal.ppat.0040020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC et al. (2012) Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment Genome Biol 13:R79 doi: 10.1186/gb-2012-13-9-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu E, Keshavarzian A, Engen P, Forsyth CB, Sikaroodi M, Gillevet P (2009) Intestinal dysbiosis: a possible mechanism of alcohol-induced endotoxemia and alcoholic steatohepatitis in rats Alcoholism, clinical and experimental research 33:1836–1846 doi: 10.1111/j.1530-0277.2009.01022.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu EA et al. (2012) Colonic microbiome is altered in alcoholism Am J Physiol Gastrointest Liver Physiol 302:G966–978 doi: 10.1152/ajpgi.00380.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (2011) Guide for the Care and Use of Laboratory Animals: Eighth Edition. The National Academies Press, Washington, DC: doi:doi: 10.17226/12910 [DOI] [Google Scholar]
- Peterson VL et al. (2017) Drunk bugs: Chronic vapour alcohol exposure induces marked changes in the gut microbiome in mice Behav Brain Res 323:172–176 doi: 10.1016/j.bbr.2017.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt DM, Rowlett JK (2012) Nonhuman primates in drug and alcohol addiction research In: Abee C, Mansfield K, Morris T, Tardif S (eds) Nonhuman Primates in Biomedical Research. Elsevier Press, Waltham, MA, pp 817–839 [Google Scholar]
- Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, Patra J (2009) Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders Lancet (London, England) 373:2223–2233 doi: 10.1016/s0140-6736(09)60746-7 [DOI] [PubMed] [Google Scholar]
- Sampson TR, Mazmanian SK (2015) Control of brain development, function, and behavior by the microbiome Cell host & microbe 17:565–576 doi: 10.1016/j.chom.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangi AN, Goel A, Singh A, Sasi A, Aggarwal R (2017) Faecal bacterial microbiota in patients with cirrhosis and the effect of lactulose administration BMC gastroenterology 17:125 doi: 10.1186/s12876-017-0683-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer EK et al. (2014) Little evidence of a role for the alpha1GABAA subunit-containing receptor in a rhesus monkey model of alcohol drinking Alcoholism, clinical and experimental research 38:1108–1117 doi: 10.1111/acer.12320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation Genome Biol 12:R60 doi: 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE (1948) A mathematical theory of communication The Bell System Technical Journal 27:379–423 doi: 10.1002/j.1538-7305.1948.tb01338.x [DOI] [Google Scholar]
- Shield KD, Parry C, Rehm J (2013) Chronic diseases and conditions related to alcohol use Alcohol Res 35:155–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SY et al. (2014) An atlas of genetic influences on human blood metabolites Nat Genet 46:543–550 doi: 10.1038/ng.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B, Irving HM, Baliunas D, Roerecke M, Patra J, Mohapatra S, Rehm J (2009) Alcohol and hypertension: gender differences in dose-response relationships determined through systematic review and meta-analysis Addiction (Abingdon, England) 104:1981–1990 doi: 10.1111/j.1360-0443.2009.02694.x [DOI] [PubMed] [Google Scholar]
- Vallender EJ, Miller GM (2013) Nonhuman primate models in the genomic era: a paradigm shift ILAR journal / National Research Council, Institute of Laboratory Animal Resources 54:154–165 doi: 10.1093/ilar/ilt044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian JA et al. (2001) Induction and maintenance of ethanol self-administration in cynomolgus monkeys (Macaca fascicularis): long-term characterization of sex and individual differences Alcoholism, clinical and experimental research 25:1087–1097 [PubMed] [Google Scholar]
- Vuong HE, Yano JM, Fung TC, Hsiao EY (2017) The Microbiome and Host Behavior Annual review of neuroscience 40:21–49 doi: 10.1146/annurev-neuro-072116-031347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G et al. (2018) Gut Microbiota and Relevant Metabolites Analysis in Alcohol Dependent Mice Front Microbiol 9:1874 doi: 10.3389/fmicb.2018.01874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan AW et al. (2011) Enteric dysbiosis associated with a mouse model of alcoholic liver disease Hepatology 53:96–105 doi: 10.1002/hep.24018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K et al. (2015) Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque Cell Host Microbe 17:385–391 doi: 10.1016/j.chom.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T et al. (2012) Human gut microbiome viewed across age and geography Nature 486:222–227 doi: 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierer J et al. (2018) The fecal metabolome as a functional readout of the gut microbiome Nat Genet 50:790–795 doi: 10.1038/s41588-018-0135-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.