Abstract
Mitochondria play crucial roles in regulating metabolism and longevity. A body of recent evidences reveals that the gut microbiome can also exert significant effects on these activities in the host. Here, by summarizing the currently known mechanisms underlying these regulations, and by comparing mitochondrial fission-fusion dynamics with bacterial interactions such as quorum sensing, we hypothesize that the microbiome impacts the host by communicating with their intracellular relatives, mitochondria. We highlight recent discoveries supporting this model, and these new findings reveal that metabolite molecules derived from bacteria can fine-tune mitochondrial dynamics in intestinal cells and hence influence host metabolic fitness and longevity. This perspective mode of chemical communication between bacteria and mitochondria may help us understand complex and dynamic environment-microbiome-host interactions regarding their vital impacts on health and diseases.
Keywords: Mitochondria, microbiome, metabolites, metabolism, longevity
Graphical Abstract
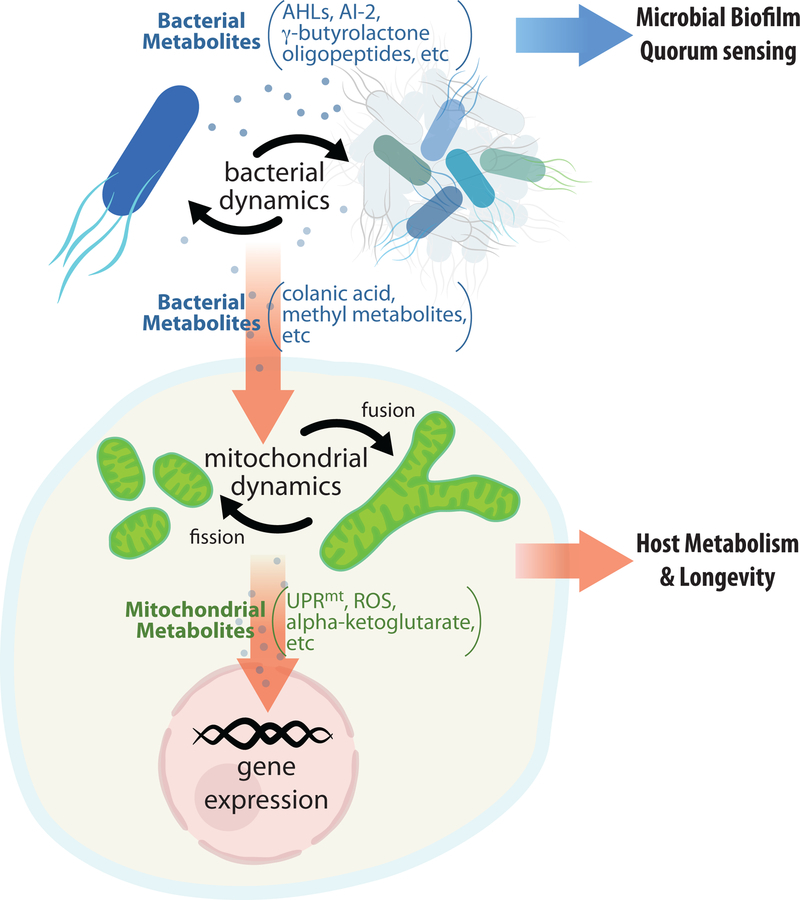
Gut microbiome orchestrates biological processes in the host by communicating with mitochondria, intracellular organelles that originated from bacteria. Reciprocating to environmental challenges, bacteria produce metabolites that can modulate mitochondrial dynamics to adjust intracellular activities. Together, the interplay between microbiome and mitochondria provides new insights into the regulation of host metabolic health and longevity.
Introduction:
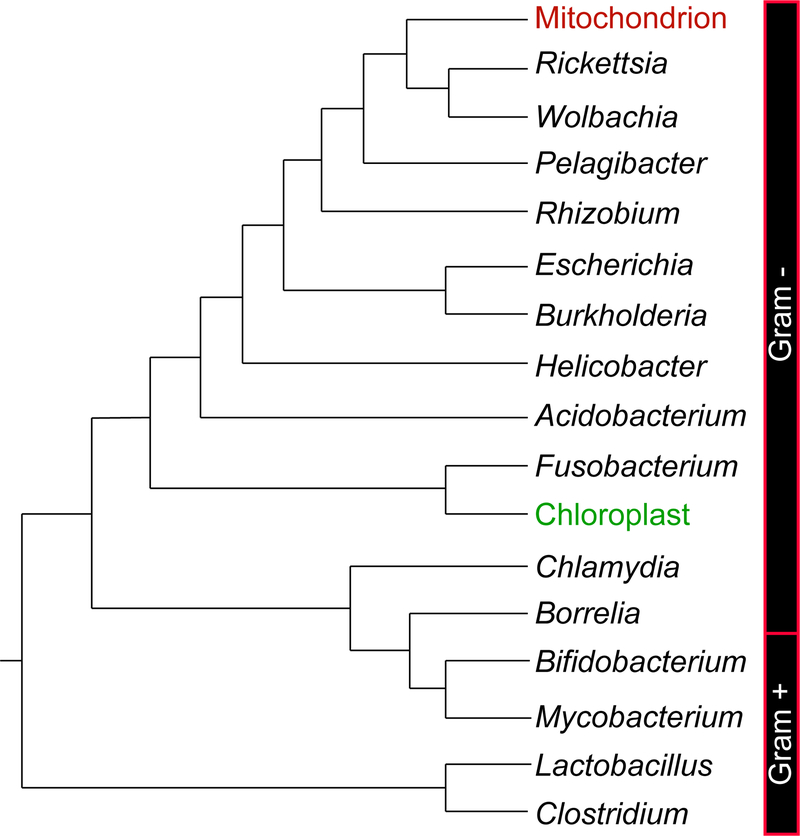
As described by the predominant endosymbiotic theory, the origin of intracellular organelles, mitochondria, can be traced back to bacterial cells that accidentally form a symbiotic relationship with some methanogenic archaea billions of years ago [1–3]. While relying on carbon organics provided by the host cells, the proto-mitochondria also conducted respiration to pay back with considerably more energy, leading to an enormous evolutionary advantage and eventually gave rise to nowadays thriving kingdom of eukaryotes. Numerous comparative molecular studies have demonstrated that mitochondria evolved from Rickettsiales bacteria [4–9] (Figure 1). In addition to this obligate endosymbiosis, eukaryotes possess their “external symbionts” - hundreds of bacterial species that colonize body surfaces and cavities [10]. The gut microbiome inhabiting the digestive tract is a good reprehensive of these symbionts, which shows stable composition over time in healthy adults [11] and exerts a substantial impact on the host physiology and pathology [12].
Figure 1. The cladogram of bacteria showing the origin of mitochondria.
This phylogenetic relationship among representative genera of clinical or ecological significance is inferred from previous comparative molecular studies [8–9], with mitochondria highlighted in red. Green color shows chloroplasts, result of another endosymbiosis event from some cyanobacteria. Side bars label the division between Gram negative and positive taxa.
In particular, mitochondria and the gut microbiome both play crucial roles in regulating host metabolism and longevity. Mitochondria and bacteria also share commonalities in terms of inter-communications. Our recent findings further reveal chemical communications between bacteria and host mitochondria, and the specific involvement of these communications in the control of metabolic and aging processes. Inspired by these findings, we tentatively propose the hypothesis that mitochondria still possess the remnant abilities of communicating with extracellular bacteria, and the impacts of symbiotic bacteria on the host act largely through modulating mitochondrial activities. We also describe the merits of using the nematode Caenorhabditis elegans as a model system in deciphering this microbiome-mitochondria communication.
Mitochondria with bacterial origin signal to the nucleus
The endosymbiotic relationship between mitochondria and eukaryotic cells have been continuously strengthened by evolutionary pressure, resulting in an extensive gene transfer between the two genomes [13,14]. Now in eukaryotic cells, a vast majority of mitochondrial proteins are encoded in the nucleus and transported into mitochondria for maturation following cytoplasmic translation. Mitochondria coordinate many vital metabolic functions such as fatty acid oxidation and oxidative phosphorylation, and serve as the powerhouse to carry out ATP production. Interestingly, these metabolic functions are also associated with the signaling role of mitochondria in the control of nuclear activities. Retrograde signals from mitochondria act through various transcriptional and epigenetic factors to actively modulate gene expression in the nucleus, such as alpha-ketoglutarate metabolites, reactive oxygen species (ROS), and mitochondrial unfolded protein responses (UPRmt) [15] (Figure 2). In particular, UPRmt senses the perturbation of the protein-folding environment in mitochondria, and directs the translocation of transcription factors into the nucleus to activate expressions of specific chaperones and proteases [16–18]. This UPRmt signaling process helps restore organelle functionality of mitochondria under different stress conditions [19,20], and consequently plays crucial roles in the regulation of organismal longevity [21,22].
Figure 2. Molecular communications between bacteria and mitochondria are vital for symbiotic bacteria to regulate metabolism and longevity in the eukaryotic host.
Bacteria communicate with each other through biofilm formation and quorum sensing that can be mediated by specific metabolites, such as AHLs (acyl-homoserine lactones), AI-2 (furanosyl borate diester), oligopeptides, and Υ-butyrolactones. On the other hand, mitochondria undergo organellar fission and fusion and communicate through these dynamic processes. Interestingly, new discoveries reveal that mitochondrial fission-fusion dynamics in the eukaryotic host cell can be regulated by chemical signals from symbiotic bacteria in the form of colanic acid and methyl metabolites. These metabolite-mediated cross-kingdom communications are crucial for host metabolism and longevity.
Mitochondrial dynamics regulates metabolism and longevity
Mitochondria are highly dynamic organelles. Although generally depicted as discrete organelles, multiple mitochondria frequently interconnect with each other, forming a large intracellular reticulum and mixing their membrane, matrix, and nucleoid contents [23–25]. At the same time, mitochondria divide constantly to facilitate organelle degradation and recycling [26,27]. A number of GTPases mediate mitochondrial dynamics: Mitofusin (Mfn1 and Mfn2) dimeric complexes and OPA1 mediate the connection between adjacent mitochondria to facilitate fusion [28,29]; while DRP1 forms ring-like structures to constrict mitochondria where the fission of organelles occurs [30,31]. The matter and information exchange resulting from mitochondrial fusion helps alleviate negative influences from impaired individual organelles [32,33]. On the other hand, selective degradation of damaged mitochondria through mitophagy requires mitochondrial fission [34–37]. Overall, these fusion and fission events keep mitochondria in a dynamic balance and ensure their quality and quantity controls, which are crucial for maintaining a healthy functional mitochondrial network.
Mitochondrial fission-fusion dynamics is tightly linked to mitochondrial bioenergetic functions and metabolic health of cells and organisms [38]. Generally, mitochondrial fusion can greatly increase efficiency of ATP synthesis [39]. In cultured cells, nutrient withdrawal has been found to promote mitochondrial elongation through inhibiting mitochondrial fission by PKA-mediated DRP1 phosphorylation [39,40] or through facilitating mitochondrial fusion by MFN1 deacetylation [41]. This starvation-induced mitochondrial elongation increases ATP synthesis capacity and efficiency, which sustains the energy demand required under nutrient limited environments [39]. In contrast, excess nutrients can lead to mitochondrial fragmentation. For example, glucose overload promotes mitochondrial fragmentation in a DRP1 dependent manner [42], and animals feeding on a high fat diet display reduced levels of MFN2 and enhanced mitochondrial fragmentation [43–46]. Thus, mitochondrial dynamics can be actively influenced by environmental signals and coupled with cellular metabolic status [44,47].
Mitochondrial dynamics is also closely associated with the aging process [48–50]. With senescence, mitochondrial dynamics tends to shift toward fission more than fusion in most of the tissues [50–52], which is likely resulted from cumulated damages in mitochondria. The fragmentation of mitochondrial network facilitates mitophagy as a protective mechanism [53,54]. However, with advancing age, mitochondrial biogenesis becomes less effective [55], and the ability to maintain the plasticity of fission-fusion dynamics declines. As a result, superfused and swollen mitochondrial morphology is often detected [56–60], which is proposed to compensate for both quality and quantity decreases [48].
Interestingly, manipulation of mitochondrial dynamics is sufficient to modulate both glucose and lipid metabolic homeostasis systemically, and also influences organism longevity. For example, mitochondrial fragmentation driven by MFN2 deletion in muscle and hepatic cells disturbs glucose homeostasis and leads to obesity in aged animals [61]. In contrast, impairment of mitochondrial fission by DRP1 deletion in liver protects animals from high-fat-diet-induced obesity and metabolic disorders [62]. On the other hand, alterations in mitochondrial electron transport chain (ETC) activities modulate lifespan and healthspan in a variety of organisms, through interplaying with diverse longevity regulatory mechanisms such as insulin and mTOR signaling, dietary restriction, and autophagy [63–67]. In Caenorhabditis elegans, dietary restriction-AMPK-mediated longevity is associated with alterations in mitochondrial fission-fusion dynamics and consequent changes in peroxisome activities [52,68]. It is also shown that mitochondrial fusion is essential for the lifespan extension conferred by reduced insulin/IGF-1 signaling [69]. More recently, studies in C. elegans and Drosophila melanogaster further discover that a mild induction of mitochondrial fission specifically in intestinal cells is sufficient to promote organism longevity systemically [70,71]. Therefore, mitochondria communicate through their fission-fusion dynamics, which safeguards the homeostasis of these essential cellular organelles and plays a pivotal role in the control of metabolic health and longevity [72].
Bacteria live in a community
The community of bacteria is highly dynamic and interactive. Although conventionally considered unicellular and isolated, bacteria do communicate and cooperate with each other, resembling those cells in multicellular organisms. One typical example in point is myxobacteria. To survive harsh environments, multiple myxobacterial cells can aggregate to form ‘swarms’ by contact-mediated signaling, for the sake of better moving, feeding and reproducing [73]. In fact, the interactions between bacterial cells are not limited to a single taxonomical group, as evidenced by formation of biofilms ubiquitously found on our planet, either on abiotic surfaces or in the animal gut. A great variety of bacteria secret a matrix of extracellular polymeric substances that helps adhesion and links cells together to form a colonial group [74], a process typically triggered by unfavorable environmental factors such as antibiotics [75]. Once embedded in this complex biofilm structure, bacterial cells undergo a lot of behavioral changes, differentially regulate many genes, and frequently exchange their genetic materials [76,77]. As a result, biofilms provide not only protection, but also opportunities of communication among otherwise isolated bacterial cells.
Moreover, virtually all bacterial species constitutively produce diffusible chemical signals to alter gene expression of others, referred to as quorum sensing. These signal molecules include certain oligopeptides, N-acyl homoserine lactones (AHLs), and autoinducer-2 (AI-2, furanosyl borate diester) that stimulate synthesis and release of themselves among different cells [78,79]. Quorum sensing implies a response to population density, allowing multiple bacterial cells to adjust their growth and activities accordingly. Moreover, this communication is required for multiple bacteria to synchronize their gene expression so that macroscopic effects can be achieved by these tiny organisms. For instance, quorum sensing plays a central role in the production of bioluminescence and in the biofilm formation [80,81]. Although quorum sensing takes place mostly among members of the same species, it is intriguing to note the existence of interspecies communications via quorum sensing. For example, Escherichia coli encodes proteins of the LuxR family for detection of AHLs, a quorum-sensing signal released only by other microbes [82]. Moreover, AI-2 is a universal signal mediating interspecies quorum sensing because it is secreted and perceived by a great variety of bacteria [79]. Thus, a bacterial community is highly dynamic and communicative, not only comprising diverse species but also adjusting their activities, sending and receiving signals constantly.
Microbiome influences host metabolism and longevity
Gut microbiome, consisting mainly of bacteria, inhabits the digestive track of the host. An ever-growing body of evidences suggests that the composition and metabolism of the gut microbiome influence metabolic health and aging. First of all, gut bacteria generate metabolic products that directly act on the host. They are responsible for the synthesis of various vitamins to maintain metabolic health of the host [83]. Gut bacteria can also break down many carbohydrates that are otherwise non-digestible, and ferment them into short-chain fatty acids as nutrients to the host, which regulate fatty acid, cholesterol and glucose metabolism [84,85]. Without them, germ-free mice are significantly leaner than normal mice [86–88]. In addition, primary bile acids are processed by gut bacteria into secondary bile acids that feed back to the liver and influence lipogenesis, gluconeogenesis and insulin sensitivity in the host [89–92]. Moreover, changes in the host’s diet, lifestyle, and medication with antibiotics and other drugs dramatically influence transcriptomic, proteomic and biochemical profiles of gut bacteria [93–96]. These bacterial changes in turn modulate the susceptibility of the host to environmental insults, dietary intervention and diseases.
Human gut microbiome is dominated by two major bacterial phyla, Firmicutes and Bacteroidetes [97]. It is intriguing that both diet- and genetic-induced obesity are associated with a reduction in Bacteroidetes and a proportional increase of Firmicutes [98–101]. These obese animals also display less diverse microbiome [102,103]. Conversely, the phylogenetic composition of gut bacteria can determine the onset and progression of obesity, through modulating the efficiency of energy uptake [100,104] and inflammatory responses in the host [105–111]. On the other hand, dietary inputs not only reshape the phylogenetic structure of gut microbiome, but also reprogram gene expression and metabolite production in these bacteria [96,112]. The regulatory loop among the environment, gut microbiome and the host is dynamic and complex, which can be mediated by different signaling mechanisms [113,114].
Similarly, changes in gut microbiome are also associated with the aging process in the host [115]. The reduction of bacterial number and diversity especially that of Bifidobacterium spp. and Bacteroides spp. in the elderly has been reported [116–118]. Furthermore, the microbiome composition is significantly correlated with increased frailty and age-related chronic conditions among old individuals, and diet-driven microbiome alterations have been shown to improve health in elderly people [119]. Studies in model systems show that the growth, proliferation and diversity of gut bacteria are good predictors of longevity [120,121]. Moreover, transplantation of gut microbiome from young to middle-aged killifish prolongs lifespan and healthspan [122]. Not only different bacterial species, but also individual bacterial genes are correlated with the longevity regulation in the host. Specific bacterial mutants have been shown to play a causative role in prolonging host lifespan and healthspan [123–126]. Interestingly, some of these beneficial effects are directly linked to specific bacterial metabolites [123–126]. Therefore, an active chemical communication between gut bacteria and the host is essential for organism fitness during aging.
Evidences emerge for a communication between microbiome and mitochondria
Because of their critical roles in determining metabolic health and longevity, both mitochondria and gut bacteria become hot targets in biological and medical investigations. However, studying them with the cellular resolution at the organism level is highly challenging, especially in mammalian models. First, fixation of mammalian tissues for microscopic observations undoubtedly interferes with the regulation of mitochondrial dynamics, and may give limited or misleading results. Secondly, mammalian microbiome is complex, composed of 300 to 1000 bacterial species with a total number even exceeding that of host cells [127–129]. Plus tremendous individual compositional variations [130,131], isolating any bacterial components for causative or mechanistic analyses would be extremely hard.
The nematode C. elegans has been extensively used as a model organism. These soil-dwelling worms exhibit many merits for laboratory manipulations, including their short life time, low price in rearing, known genome as well as availability of numerous mutants and transgenic lines. Most importantly, some innate features of C. elegans make them ideal for studying mitochondrial and bacterial activities that influence host physiology. First of all, their bodies are colorless and transparent. After fluorescently labelling of mitochondrial-targeting sequences, mitochondrial dynamics can be directly viewed in vivo with high resolution at the organism level [26]. With a rapid technical progress on lattice light-sheet microscopy [132], an even more detailed real-time super-resolution monitoring of mitochondrial structure and dynamics becomes feasible using C. elegans [133].
Furthermore, the gut of C. elegans is naturally colonized by a complex community of commensal bacteria whose composition is influenced but distant from the environment [134,135]. This relationship between the host and microbiome resembles that in humans, implicating a valid model. More remarkably, a strain of C. elegans has been tamed in laboratories [115]. This strain is reared monoxenically, feeding on and accommodating in the gut a single bacterial clone, therefore carries accurately defined microbiome. At the same time of enjoying these merits, one should also take necessary cautions to interpret conclusions obtained using C. elegans, partly because the typical bacteria used in laboratories, E. coli OP50, only colonize aging individuals and this colonization is associated with bacterial pathogenesis [120,121,136]. However, with the simple manipulation of gut bacteria and some proved conservation [137], C. elegans still represents an ideal model for studies of microbiome-host interaction. Evidently, utilizing the system of C. elegans and their symbiotic bacteria, a lot of insights on microbe-host interactions have been gained [137,138], especially the microbial contributions to metabolism and aging [139–142].
Using C. elegans, our recent studies bring light into the interaction between bacteria and mitochondrial dynamics in the gut of the host. These communications are mediated by chemical signals from intestinal bacteria (Figure 2). In one case, a cluster of bacterial metabolites including betaine, methionine and homocysteine initiate a signaling cascade that triggers the NR5A nuclear receptor and activates hedgehog signaling to regulate mitochondrial fission-fusion balance in intestinal cells. This bacteria-mitochondria communication ultimately regulates fat storage homeostasis in the host [112]. In another, a slime polysaccharide named colanic acid, a major biofilm component of E. coli, is secreted from intestinal bacteria. After entering host cytoplasm via endocytosis, colanic acid increases the fragmentation of intestinal mitochondria in a DRP-1 dependent fashion, as well as enhances UPRmt mediated by the transcription factor ATFS-1 in response to mitochondrial stress. These signaling effects of bacterial colanic acid on mitochondrial dynamics and UPRmt consequently lead to lifespan extension and protection against age-associated pathologies, like germline tumor progression and toxic amyloid-beta accumulation, in the host [123]. Together, these results consistently show that mitochondria undergo chemical communication with bacteria, a process modulating metabolic and senescent states of eukaryotic cells.
This view can also be strengthened by our unbiased search for bacterial factors involved in the regulation of host longevity [123]. This genome-wide analysis implicates YceO and LsrC, two bacterial proteins important for biofilm formation and AI-2 transport, and several proteins controlling the level of colanic acid [123]. AI-2 is a key quorum-sensing molecule that has been recorded to interact with eukaryotic cells [143,144], although no mechanisms have been specified. As undergoing exclusive intracellular lives, mitochondria likely preserve their capabilities of, and possibly are responsible for perceiving quorum-sensing signals. This view gains recent supports from the fact that a signaling molecule secreted by Pseudomonas aeruginosa accumulates within mitochondria and regulates cellular functions. In this bacteria-mitochondria interaction, P. aeruginosa secrets N-(3-oxo-dodecanoyl)-L-homoserine lactone (3OC12), which is hydrolyzed by lactonase paraoxonase 2 in mitochondria to attenuate its toxicity. The hydrolyzed form, 3OC12 acid then stays in mitochondria and mediates calcium release and stress signaling through intracellular acidification [145]. Collectively, based on these new discoveries, it is reasonable to hypothesize that microbiome may affect the host by directly interacting with mitochondria through bacterial metabolites and specific signaling mechanisms.
Future perspectives
With our findings and other sporadic evidences, a model that merges functionalities of mitochondria and microbiome is not merely plausible, but also probable. Within the context of our model, we propose that mitochondria are the mediators for this cross-domain chemical dialogue. This is not to deny the existence of indirect communications from bacteria to mitochondria, such as by regulating nuclear gene expression. But following the Law of Parsimony, a conserved and wide-spread direct interaction is most likely. A systematic search for signaling molecules sent by gut microbiome, transporting mechanisms across the plasma membrane, and receptors on mitochondrial outer membrane would be essential to confirm this communication.
This model may help us understand many aspects of physiological and pathological regulations by host-microbiome interactions. For example, gut microbiome has been indicated to play vital roles in a number of neurological disorders [146]. Clearly, these bacterial effects on neurons have to occur via cell non-autonomous mechanisms. Other than a way of indiscriminately releasing certain molecules into body fluid, we propose that the dialogue between bacteria and mitochondria likely is the underlying mode of action. On one hand, patients with mutations in several mitochondrial dynamics regulators display neurological symptoms [147–150]; likewise, neurodegeneration and many other diseases have been linked to dysregulation of mitochondrial dynamics [151]. On the other hand, mitochondria within different cells have been shown to communicate with each other, resulting in a cell non-autonomous effect [152]. Hence, the proposed crosstalk exhibits high explanatory power for the function of microbiome in modulating systemic responses of the host, as exampled by metabolism and aging.
There have been quite a few biological phenomena following a ‘non-Darwinian’ pattern lacking mechanistic explanations. Interesting to note, mitochondria are inherited maternally, and the founding colonies of microbiome in newborns are also from a maternal source [153]. It would be intriguing to hypothesize that the microbiome-mitochondria axis also plays a role in mediating those maternal effects. By many means, a chemical dialogue across the cell membrane, orchestrated by mitochondria and symbiotic bacteria, is promising to broaden our views on biological sciences. We anticipate an era in that the mitochondria-microbiome communication is fully characterized, which would shed great light on improving metabolic health and healthy aging.
Acknowledgements
We thank two anonymous reviewers for their comments and suggestions on writing. The authors of this review have been supported by HHMI (M.C.W.), March of Dimes Foundation (M.C.W.), Welch Foundation (M.C.W.), and NIH (R01AG045183, DP1DK113644, R01AT009050; M.C.W.)
Abbreviations:
- ROS
reactive oxygen species
- UPRmt
mitochondrial unfolded protein response
- GTPase
guanosine triphosphate hydrolase
- MFN
mitofusin
- OPA
optic atrophy protein
- DRP1
dynamin related protein 1
- PKA
protein kinase A
- ETC
electron transport chain
- mTOR
mammalian target of rapamycin
- AMPK
AMP activated protein kinase
- IGF-1
insulin-like growth factor 1
- AHL
N-acyl homoserine lactone
- AI-2
autoinducer 2
- NR5A
nuclear receptor 5A
- 3OC12
N-(3-oxo-dodecanoyl)-L-homoserine lactone
References
- 1.Embley TM & Martin W (2006) Eukaryotic evolution, changes and challenges. Nature 440, 623–630. [DOI] [PubMed] [Google Scholar]
- 2.Sagan L (1967) On the origin of mitosing cells. J. Theor. Biol 14, 255–274. [DOI] [PubMed] [Google Scholar]
- 3.Martin W & Müller M (1998) The hydrogen hypothesis for the first eukaryote. Nature 392, 37–41. [DOI] [PubMed] [Google Scholar]
- 4.Mileykovskaya E & Dowhan W (2009) Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim. Biophys. Acta 1788, 2084–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeth K & Thein M (2010) Porins in prokaryotes and eukaryotes: common themes and variations. Biochem. J 431, 13–22. [DOI] [PubMed] [Google Scholar]
- 6.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UC, Podowski RM, Näslund AK, Eriksson AS, Winkler HH & Kurland CG (1998) The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–140. [DOI] [PubMed] [Google Scholar]
- 7.Manuell AL, Quispe J & Mayfield SP (2007) Structure of the chloroplast ribosome: novel domains for translation regulation. PLoS Biol 5, e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B & Bork P (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283–1287. [DOI] [PubMed] [Google Scholar]
- 9.Ferla MP, Thrash JC, Giovannoni SJ & Patrick WM (2013) New rRNA gene-based phylogenies of the Alphaproteobacteria provide perspective on major groups, mitochondrial ancestry and phylogenetic instability. PloS One 8, e83383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sears CL (2005) A dynamic partnership: celebrating our gut flora. Anaerobe 11, 247–251. [DOI] [PubMed] [Google Scholar]
- 11.De La Cochetière MF, Durand T, Lepage P, Bourreille A, Galmiche JP & Doré J (2005) Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J. Clin. Microbiol 43, 5588–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Lee I-S, Braun C & Enck P (2016) Effect of Probiotics on Central Nervous System Functions in Animals and Humans: A Systematic Review. J. Neurogastroenterol. Motil 22, 589–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Archibald JM (2015) Endosymbiosis and Eukaryotic Cell Evolution. Curr. Biol. CB 25, R911–921. [DOI] [PubMed] [Google Scholar]
- 14.Martin WF, Garg S & Zimorski V (2015) Endosymbiotic theories for eukaryote origin. Philos. Trans. R. Soc. Lond. B. Biol. Sci 370, 20140330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quirós PM, Mottis A & Auwerx J (2016) Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol 17, 213–226. [DOI] [PubMed] [Google Scholar]
- 16.Tatsuta T & Langer T (2009) AAA proteases in mitochondria: diverse functions of membrane-bound proteolytic machines. Res. Microbiol 160, 711–717. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT & Hoogenraad NJ (2002) A mitochondrial specific stress response in mammalian cells. EMBO J 21, 4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM & Haynes CM (2012) Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ & Haynes CM (2014) Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 516, 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Runkel ED, Baumeister R & Schulze E (2014) Mitochondrial stress: balancing friend and foe. Exp. Gerontol 56, 194–201. [DOI] [PubMed] [Google Scholar]
- 21.Lee S-J, Hwang AB & Kenyon C (2010) Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. CB 20, 2131–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riera CE & Dillin A (2015) Tipping the metabolic scales towards increased longevity in mammals. Nat. Cell Biol 17, 196–203. [DOI] [PubMed] [Google Scholar]
- 23.Hales KG & Fuller MT (1997) Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90, 121–129. [DOI] [PubMed] [Google Scholar]
- 24.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM & Spelbrink JN (2003) Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell 14, 1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legros F, Malka F, Frachon P, Lombès A & Rojo M (2004) Organization and dynamics of human mitochondrial DNA. J. Cell Sci 117, 2653–2662. [DOI] [PubMed] [Google Scholar]
- 26.Labrousse AM, Zappaterra MD, Rube DA & van der Bliek AM (1999) C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4, 815–826. [DOI] [PubMed] [Google Scholar]
- 27.Mozdy AD, McCaffery JM & Shaw JM (2000) Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol 151, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE & Chan DC (2003) Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol 160, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishihara N, Eura Y & Mihara K (2004) Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell Sci 117, 6535–6546. [DOI] [PubMed] [Google Scholar]
- 30.Pitts KR, Yoon Y, Krueger EW & McNiven MA (1999) The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol. Biol. Cell 10, 4403–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smirnova E, Griparic L, Shurland DL & van der Bliek AM (2001) Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakada K, Inoue K, Ono T, Isobe K, Ogura A, Goto YI, Nonaka I & Hayashi JI (2001) Inter-mitochondrial complementation: Mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat. Med 7, 934–940. [DOI] [PubMed] [Google Scholar]
- 33.Chen H, Chomyn A & Chan DC (2005) Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem 280, 26185–26192. [DOI] [PubMed] [Google Scholar]
- 34.Mao K, Wang K, Liu X & Klionsky DJ (2013) The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev. Cell 26, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rambold AS, Kostelecky B, Elia N & Lippincott-Schwartz J (2011) Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. U. S. A 108, 10190–10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Twig G, Elorza A, Molina AJA, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE & Shirihai OS (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Twig G & Shirihai OS (2011) The interplay between mitochondrial dynamics and mitophagy. Antioxid. Redox Signal 14, 1939–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picard M & Turnbull DM (2013) Linking the metabolic state and mitochondrial DNA in chronic disease, health, and aging. Diabetes 62, 672–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomes LC & Scorrano L (2011) Mitochondrial elongation during autophagy: a stereotypical response to survive in difficult times. Autophagy 7, 1251–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rambold AS, Kostelecky B & Lippincott-Schwartz J (2011) Together we are stronger: fusion protects mitochondria from autophagosomal degradation. Autophagy 7, 1568–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J-Y, Kapur M, Li M, Choi M-C, Choi S, Kim H-J, Kim I, Lee E, Taylor JP & Yao T-P (2014) MFN1 deacetylation activates adaptive mitochondrial fusion and protects metabolically challenged mitochondria. J. Cell Sci 127, 4954–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu T, Robotham JL & Yoon Y (2006) Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc. Natl. Acad. Sci. U. S. A 103, 2653–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jheng H-F, Tsai P-J, Guo S-M, Kuo L-H, Chang C-S, Su I-J, Chang C-R & Tsai Y-S (2012) Mitochondrial fission contributes to mitochondrial dysfunction and insulin resistance in skeletal muscle. Mol. Cell. Biol 32, 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liesa M & Shirihai OS (2013) Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab 17, 491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu R, Jin P, Yu L, Wang Y, Han L, Shi T& Li X (2014) Impaired mitochondrial dynamics and bioenergetics in diabetic skeletal muscle. PloS One 9, e92810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Putti R, Sica R, Migliaccio V & Lionetti L (2015) Diet impact on mitochondrial bioenergetics and dynamics. Front. Physiol 6, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wai T & Langer T (2016) Mitochondrial Dynamics and Metabolic Regulation. Trends Endocrinol. Metab. TEM 27, 105–117. [DOI] [PubMed] [Google Scholar]
- 48.Seo AY, Joseph A-M, Dutta D, Hwang JCY, Aris JP & Leeuwenburgh C (2010) New insights into the role of mitochondria in aging: mitochondrial dynamics and more. J. Cell Sci 123, 2533–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ikeda Y, Sciarretta S, Nagarajan N, Rubattu S, Volpe M, Frati G & Sadoshima J (2014) New insights into the role of mitochondrial dynamics and autophagy during oxidative stress and aging in the heart. Oxid. Med. Cell. Longev 2014, 210934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Regmi SG, Rolland SG & Conradt B (2014) Age-dependent changes in mitochondrial morphology and volume are not predictors of lifespan. Aging 6, 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rana A, Oliveira MP, Khamoui AV, Aparicio R, Rera M, Rossiter HB & Walker DW (2017) Promoting Drp1-mediated mitochondrial fission in midlife prolongs healthy lifespan of Drosophila melanogaster. Nat. Commun 8, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weir HJ, Yao P, Huynh FK, Escoubas CC, Goncalves RL, Burkewitz K, Laboy R, Hirschey MD & Mair WB (2017) Dietary Restriction and AMPK Increase Lifespan via Mitochondrial Network and Peroxisome Remodeling. Cell Metab 26, 884–896.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen H & Chan DC (2009) Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum. Mol. Genet 18, R169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skulachev VP (2001) The programmed death phenomena, aging, and the Samurai law of biology. Exp. Gerontol 36, 995–1024. [DOI] [PubMed] [Google Scholar]
- 55.Reznick RM, Zong H, Li J, Morino K, Moore IK, Yu HJ, Liu Z-X, Dong J, Mustard KJ, Hawley SA, Befroy D, Pypaert M, Hardie DG, Young LH & Shulman GI (2007) Aging-associated reductions in AMP-activated protein kinase activity and mitochondrial biogenesis. Cell Metab 5, 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murakoshi M, Osamura Y & Watanabe K (1985) Mitochondrial alterations in aged rat adrenal cortical cells. Tokai J. Exp. Clin. Med 10, 531–536. [PubMed] [Google Scholar]
- 57.Tandler B & Hoppel CL (1986) Studies on giant mitochondria. Ann. N. Y. Acad. Sci 488, 65–81. [DOI] [PubMed] [Google Scholar]
- 58.Terman A & Brunk UT (2005) The aging myocardium: roles of mitochondrial damage and lysosomal degradation. Heart Lung Circ 14, 107–114. [DOI] [PubMed] [Google Scholar]
- 59.Lee S, Jeong S-Y, Lim W-C, Kim S, Park Y-Y, Sun X, Youle RJ & Cho H (2007) Mitochondrial fission and fusion mediators, hFis1 and OPA1, modulate cellular senescence. J. Biol. Chem 282, 22977–22983. [DOI] [PubMed] [Google Scholar]
- 60.Yoon Y-S, Yoon D-S, Lim IK, Yoon S-H, Chung H-Y, Rojo M, Malka F, Jou M-J, Martinou J-C & Yoon G (2006) Formation of elongated giant mitochondria in DFO-induced cellular senescence: involvement of enhanced fusion process through modulation of Fis1. J. Cell. Physiol 209, 468–480. [DOI] [PubMed] [Google Scholar]
- 61.Sebastián D, Hernández-Alvarez MI, Segalés J, Sorianello E, Muñoz JP, Sala D, Waget A, Liesa M, Paz JC, Gopalacharyulu P, Orešič M, Pich S, Burcelin R, Palacín M & Zorzano A (2012) Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homeostasis. Proc. Natl. Acad. Sci. U. S. A 109, 5523–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Ishihara T, Ibayashi Y, Tatsushima K, Setoyama D, Hanada Y, Takeichi Y, Sakamoto S, Yokota S, Mihara K, Kang D, Ishihara N, Takayanagi R & Nomura M (2015) Disruption of mitochondrial fission in the liver protects mice from diet-induced obesity and metabolic deterioration. Diabetologia 58, 2371–2380. [DOI] [PubMed] [Google Scholar]
- 63.Sohal RS, Ku HH, Agarwal S, Forster MJ & Lal H (1994) Oxidative damage, mitochondrial oxidant generation and antioxidant defenses during aging and in response to food restriction in the mouse. Mech. Ageing Dev 74, 121–133. [DOI] [PubMed] [Google Scholar]
- 64.Pan Y & Shadel GS (2009) Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density. Aging 1, 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang CC, Chen D, Lee SS & Walter L (2011) The dynamin-related protein DRP-1 and the insulin signaling pathway cooperate to modulate Caenorhabditis elegans longevity. Aging Cell 10, 724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weber TA & Reichert AS (2010) Impaired quality control of mitochondria: aging from a new perspective. Exp. Gerontol 45, 503–511. [DOI] [PubMed] [Google Scholar]
- 67.Lee SS, Lee RYN, Fraser AG, Kamath RS, Ahringer J & Ruvkun G (2003) A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet 33, 40–48. [DOI] [PubMed] [Google Scholar]
- 68.Burkewitz K, Zhang Y & Mair WB (2014) AMPK at the nexus of energetics and aging. Cell Metab 20, 10–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaudhari SN & Kipreos ET (2017) Increased mitochondrial fusion allows the survival of older animals in diverse C. elegans longevity pathways. Nat. Commun 8, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McQuibban GA, Lee JR, Zheng L, Juusola M & Freeman M (2006) Normal mitochondrial dynamics requires rhomboid-7 and affects Drosophila lifespan and neuronal function. Curr. Biol. CB 16, 982–989. [DOI] [PubMed] [Google Scholar]
- 71.Rana A, Rera M & Walker DW (2013) Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc. Natl. Acad. Sci. U. S. A 110, 8638–8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sebastián D, Palacín M & Zorzano A (2017) Mitochondrial Dynamics: Coupling Mitochondrial Fitness with Healthy Aging. Trends Mol. Med 23, 201–215. [DOI] [PubMed] [Google Scholar]
- 73.Sozinova O, Jiang Y, Kaiser D & Alber M (2005) A three-dimensional model of myxobacterial aggregation by contact-mediated interactions. Proc. Natl. Acad. Sci. U. S. A 102, 11308–11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall-Stoodley L, Costerton JW & Stoodley P (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol 2, 95–108. [DOI] [PubMed] [Google Scholar]
- 75.Karatan E & Watnick P (2009) Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. MMBR 73, 310–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.An D & Parsek MR (2007) The promise and peril of transcriptional profiling in biofilm communities. Curr. Opin. Microbiol 10, 292–296. [DOI] [PubMed] [Google Scholar]
- 77.Molin S & Tolker-Nielsen T (2003) Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol 14, 255–261. [DOI] [PubMed] [Google Scholar]
- 78.Day WA & Maurelli AT (2001) Shigella flexneri LuxS quorum-sensing system modulates virB expression but is not essential for virulence. Infect. Immun 69, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miller MB & Bassler BL (2001) Quorum sensing in bacteria. Annu. Rev. Microbiol 55, 165–199. [DOI] [PubMed] [Google Scholar]
- 80.Nealson KH, Platt T & Hastings JW (1970) Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol 104, 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Herzberg M, Kaye IK, Peti W & Wood TK (2006) YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol 188, 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Michael B, Smith JN, Swift S, Heffron F & Ahmer BM (2001) SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol 183, 5733–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Hara AM & Shanahan F (2006) The gut flora as a forgotten organ. EMBO Rep 7, 688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bergman EN (1990) Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev 70, 567–590. [DOI] [PubMed] [Google Scholar]
- 85.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J & Bakker BM (2013) The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res 54, 2325–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rabot S, Membrez M, Bruneau A, Gérard P, Harach T, Moser M, Raymond F, Mansourian R & Chou CJ (2010) Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 24, 4948–4959. [DOI] [PubMed] [Google Scholar]
- 87.Bäckhed F, Manchester JK, Semenkovich CF & Gordon JI (2007) Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. U. S. A 104, 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Woodard GA, Encarnacion B, Downey JR, Peraza J, Chong K, Hernandez-Boussard T & Morton JM (2009) Probiotics improve outcomes after Roux-en-Y gastric bypass surgery: a prospective randomized trial. J. Gastrointest. Surg. Off. J. Soc. Surg. Aliment. Tract 13, 1198–1204. [DOI] [PubMed] [Google Scholar]
- 89.de Aguiar Vallim TQ, Tarling EJ & Edwards PA (2013) Pleiotropic roles of bile acids in metabolism. Cell Metab 17, 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma K, Saha PK, Chan L & Moore DD (2006) Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Invest 116, 1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pols TWH, Noriega LG, Nomura M, Auwerx J & Schoonjans K (2011) The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J. Hepatol 54, 1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall H-U, Bamberg K, Angelin B, Hyötyläinen T, Orešič M & Bäckhed F (2013) Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17, 225–235. [DOI] [PubMed] [Google Scholar]
- 93.Benítez-Páez A, Belda-Ferre P, Simón-Soro A & Mira A (2014) Microbiota diversity and gene expression dynamics in human oral biofilms. BMC Genomics 15, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guo MS & Gross CA (2014) Stress-induced remodeling of the bacterial proteome. Curr. Biol. CB 24, R424–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liebeke M & Lalk M (2014) Staphylococcus aureus metabolic response to changing environmental conditions - a metabolomics perspective. Int. J. Med. Microbiol. IJMM 304, 222–229. [DOI] [PubMed] [Google Scholar]
- 96.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R & Gordon JI (2009) The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med 1, 6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M & Nageshwar Reddy D (2015) Role of the normal gut microbiota. World J. Gastroenterol 21, 8787–8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD & Gordon JI (2005) Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A 102, 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang C, Zhang M, Pang X, Zhao Y, Wang L & Zhao L (2012) Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J 6, 1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen Y-Y, Knight R, Ahima RS, Bushman F & Wu GD (2009) High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 137, 1716–1724.e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ, Mehrabian M, Ursell LK, He A, Castellani LW, Zinker B, Kirby M, Drake TA, Drevon CA, Knight R, Gargalovic P, Kirchgessner T, Eskin E & Lusis AJ (2013) Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab 17, 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R & Gordon JI (2009) A core gut microbiome in obese and lean twins. Nature 457, 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thaiss CA, Itav S, Rothschild D, Meijer M, Levy M, Moresi C, Dohnalová L, Braverman S, Rozin S, Malitsky S, Dori-Bachash M, Kuperman Y, Biton I, Gertler A, Harmelin A, Shapiro H, Halpern Z, Aharoni A, Segal E & Elinav E (2016) Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature 540, 544–551. [DOI] [PubMed] [Google Scholar]
- 104.Krajmalnik-Brown R, Ilhan Z-E, Kang D-W & DiBaise JK (2012) Effects of gut microbes on nutrient absorption and energy regulation. Nutr. Clin. Pract. Off. Publ. Am. Soc. Parenter. Enter. Nutr 27, 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Clarke SF, Murphy EF, Nilaweera K, Ross PR, Shanahan F, O’Toole PW & Cotter PD (2012) The gut microbiota and its relationship to diet and obesity: new insights. Gut Microbes 3, 186–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti J-F, Gibson GR, Casteilla L, Delzenne NM, Alessi MC & Burcelin R (2007) Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772. [DOI] [PubMed] [Google Scholar]
- 107.Hotamisligil GS (2006) Inflammation and metabolic disorders. Nature 444, 860–867. [DOI] [PubMed] [Google Scholar]
- 108.Tagliabue A & Elli M (2013) The role of gut microbiota in human obesity: recent findings and future perspectives. Nutr. Metab. Cardiovasc. Dis. NMCD 23, 160–168. [DOI] [PubMed] [Google Scholar]
- 109.Collado MC, Isolauri E, Laitinen K & Salminen S (2008) Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am. J. Clin. Nutr 88, 894–899. [DOI] [PubMed] [Google Scholar]
- 110.Santacruz A, Collado MC, García-Valdés L, Segura MT, Martín-Lagos JA, Anjos T, Martí-Romero M, Lopez RM, Florido J, Campoy C & Sanz Y (2010) Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br. J. Nutr 104, 83–92. [DOI] [PubMed] [Google Scholar]
- 111.Kalliomäki M, Collado MC, Salminen S & Isolauri E (2008) Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr 87, 534–538. [DOI] [PubMed] [Google Scholar]
- 112.Lin C-CJ & Wang MC (2017) Microbial metabolites regulate host lipid metabolism through NR5A-Hedgehog signalling. Nat. Cell Biol 19, 550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ussar S, Griffin NW, Bezy O, Fujisaka S, Vienberg S, Softic S, Deng L, Bry L, Gordon JI & Kahn CR (2015) Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell Metab 22, 516–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, Mao Y, Zhang X, Pang X, Wei C, Zhao G, Chen Y & Zhao L (2010) Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J 4, 232–241. [DOI] [PubMed] [Google Scholar]
- 115.Heintz C & Mair W (2014) You are what you host: microbiome modulation of the aging process. Cell 156, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mitsuoka T, Hayakawa K & Kimura N (1974) [The faecal flora of man. II. The composition of bifidobacterium flora of different age groups (author’s transl)]. Zentralblatt Bakteriol. Parasitenkd. Infekt. Hyg. Erste Abt. Orig. Reihe Med. Mikrobiol. Parasitol 226, 469–478. [PubMed] [Google Scholar]
- 117.Hopkins MJ & Macfarlane GT (2002) Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J. Med. Microbiol 51, 448–454. [DOI] [PubMed] [Google Scholar]
- 118.Woodmansey EJ, McMurdo MET, Macfarlane GT & Macfarlane S (2004) Comparison of compositions and metabolic activities of fecal microbiotas in young adults and in antibiotic-treated and non-antibiotic-treated elderly subjects. Appl. Environ. Microbiol 70, 6113–6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HMB, Coakley M, Lakshminarayanan B, O’Sullivan O, Fitzgerald GF, Deane J, O’Connor M, Harnedy N, O’Connor K, O’Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP & O’Toole PW (2012) Gut microbiota composition correlates with diet and health in the elderly. Nature 488, 178–184. [DOI] [PubMed] [Google Scholar]
- 120.Garigan D, Hsu A-L, Fraser AG, Kamath RS, Ahringer J & Kenyon C (2002) Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161, 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Portal-Celhay C, Bradley ER & Blaser MJ (2012) Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC Microbiol 12, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Smith P, Willemsen D, Popkes M, Metge F, Gandiwa E, Reichard M & Valenzano DR (2017) Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. eLife 6e277014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Han B, Sivaramakrishnan P, Lin C-CJ, Neve IAA, He J, Tay LWR, Sowa JN, Sizovs A, Du G, Wang J, Herman C & Wang MC (2017) Microbial Genetic Composition Tunes Host Longevity. Cell 169, 1249–1262.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Larsen PL & Clarke CF (2002) Extension of life-span in Caenorhabditis elegans by a diet lacking coenzyme Q. Science 295, 120–123. [DOI] [PubMed] [Google Scholar]
- 125.Virk B, Correia G, Dixon DP, Feyst I, Jia J, Oberleitner N, Briggs Z, Hodge E, Edwards R, Ward J, Gems D & Weinkove D (2012) Excessive folate synthesis limits lifespan in the C. elegans: E. coli aging model. BMC Biol 10, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Virk B, Jia J, Maynard CA, Raimundo A, Lefebvre J, Richards SA, Chetina N, Liang Y, Helliwell N, Cipinska M & Weinkove D (2016) Folate Acts in E. coli to Accelerate C. elegans Aging Independently of Bacterial Biosynthesis. Cell Rep 14, 1611–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guarner F & Malagelada J-R (2003) Gut flora in health and disease. Lancet Lond. Engl 361, 512–519. [DOI] [PubMed] [Google Scholar]
- 128.Sender R, Fuchs S & Milo R (2016) Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 164, 337–340. [DOI] [PubMed] [Google Scholar]
- 129.Krych L, Hansen CHF, Hansen AK, van den Berg FWJ & Nielsen DS (2013) Quantitatively different, yet qualitatively alike: a meta-analysis of the mouse core gut microbiome with a view towards the human gut microbiome. PloS One 8, e62578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Conlon MA & Bird AR (2014) The impact of diet and lifestyle on gut microbiota and human health. Nutrients 7, 17–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK & Knight R (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang X, Fan J, Li L, Liu H, Wu R, Wu Y, Wei L, Mao H, Lal A, Xi P, Tang L, Zhang Y, Liu Y, Tan S & Chen L (2018) Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat. Biotechnol [DOI] [PubMed] [Google Scholar]
- 133.Liu T-L, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J, Kohrman AQ, Medwig TN, Dambournet D, Forster R, Cunniff B, Ruan Y, Yashiro H, Scholpp S, Meyerowitz EM, Hockemeyer D, Drubin DG, Martin BL, Matus DQ, Koyama M, Megason SG, Kirchhausen T & Betzig E (2018) Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Berg M, Zhou XY & Shapira M (2016) Host-Specific Functional Significance of Caenorhabditis Gut Commensals. Front. Microbiol 7, 1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dirksen P, Marsh SA, Braker I, Heitland N, Wagner S, Nakad R, Mader S, Petersen C, Kowallik V, Rosenstiel P, Félix M-A & Schulenburg H (2016) The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol 14, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Podshivalova K, Kerr RA & Kenyon C (2017) How a Mutation that Slows Aging Can Also Disproportionately Extend End-of-Life Decrepitude. Cell Rep 19, 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cabreiro F & Gems D (2013) Worms need microbes too: microbiota, health and aging in Caenorhabditis elegans. EMBO Mol. Med 5, 1300–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee W-J & Hase K (2014) Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol 10, 416–424. [DOI] [PubMed] [Google Scholar]
- 139.Nguyen TPT & Clarke CF (2012) Folate status of gut microbiome affects Caenorhabditis elegans lifespan. BMC Biol 10, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pang S & Curran SP (2014) Adaptive capacity to bacterial diet modulates aging in C. elegans. Cell Metab 19, 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Saiki R, Lunceford AL, Bixler T, Dang P, Lee W, Furukawa S, Larsen PL & Clarke CF (2008) Altered bacterial metabolism, not coenzyme Q content, is responsible for the lifespan extension in Caenorhabditis elegans fed an Escherichia coli diet lacking coenzyme Q. Aging Cell 7, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Brooks KK, Liang B & Watts JL (2009) The influence of bacterial diet on fat storage in C. elegans. PloS One 4, e7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wynendaele E, Verbeke F, D’Hondt M, Hendrix A, Van De Wiele C, Burvenich C, Peremans K, De Wever O, Bracke M & De Spiegeleer B (2015) Crosstalk between the microbiome and cancer cells by quorum sensing peptides. Peptides 64, 40–48. [DOI] [PubMed] [Google Scholar]
- 144.Götz-Rösch C, Sieper T, Fekete A, Schmitt-Kopplin P, Hartmann A & Schröder P (2015) Influence of bacterial N-acyl-homoserine lactones on growth parameters, pigments, antioxidative capacities and the xenobiotic phase II detoxification enzymes in barley and yam bean. Front. Plant Sci 6, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Horke S, Xiao J, Schütz E-M, Kramer GL, Wilgenbus P, Witte I, Selbach M & Teiber JF (2015) Novel Paraoxonase 2-Dependent Mechanism Mediating the Biological Effects of the Pseudomonas aeruginosa Quorum-Sensing Molecule N-(3-Oxo-Dodecanoyl)-L-Homoserine Lactone. Infect. Immun 83, 3369–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang Y & Kasper LH (2014) The role of microbiome in central nervous system disorders. Brain. Behav. Immun 38, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Waterham HR, Koster J, van Roermund CWT, Mooyer PAW, Wanders RJA & Leonard JV (2007) A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med 356, 1736–1741. [DOI] [PubMed] [Google Scholar]
- 148.Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J & Hamel CP (2000) Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet 26, 207–210. [DOI] [PubMed] [Google Scholar]
- 149.Abrams AJ, Hufnagel RB, Rebelo A, Zanna C, Patel N, Gonzalez MA, Campeanu IJ, Griffin LB, Groenewald S, Strickland AV, Tao F, Speziani F, Abreu L, Schüle R, Caporali L, La Morgia C, Maresca A, Liguori R, Lodi R, Ahmed ZM, Sund KL, Wang X, Krueger LA, Peng Y, Prada CE, Prows CA, Schorry EK, Antonellis A, Zimmerman HH, Abdul-Rahman OA, Yang Y, Downes SM, Prince J, Fontanesi F, Barrientos A, Németh AH, Carelli V, Huang T, Zuchner S & Dallman JE (2015) Mutations in SLC25A46, encoding a UGO1-like protein, cause an optic atrophy spectrum disorder. Nat. Genet 47, 926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Züchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schröder JM, Vance JM & Battologlu E (2004) Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet 36, 449–451. [DOI] [PubMed] [Google Scholar]
- 151.Wada J & Nakatsuka A (2016) Mitochondrial Dynamics and Mitochondrial Dysfunction in Diabetes. Acta Med. Okayama 70, 151–158. [DOI] [PubMed] [Google Scholar]
- 152.Durieux J, Wolff S & Dillin A (2011) The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Langhendries J-P (2006) [Early bacterial colonisation of the intestine: why it matters?]. Arch. Pediatr. Organe Off. Soc. Francaise Pediatr 13, 1526–1534. [DOI] [PubMed] [Google Scholar]