Abstract
Primary intraosseous rhabdomyosarcomas are extremely rare. Recently two studies reported 4 cases of primary intraosseous rhabdomyosarcoma with EWSR1/FUS-TFCP2 gene fusions, associated with somewhat conflicting histologic features, ranging from spindle to epithelioid. In this study we sought to further investigate the pathologic and molecular abnormalities of a larger group of intraosseous rhabdomyosarcomas by a combined approach using targeted RNA sequencing analysis and fluorescence in-situ hybridization (FISH). We identified 7 cases, 3 males and 4 females, all in young adults, age range 20–39 years (median 27 years). Three cases involved the pelvis, 2 involved the femur and 1 each involved the maxilla and the skull. Molecular studies identified recurrent gene fusions in all 7 cases tested, including: a novel MEIS1-NCOA2 fusion in 2 cases, EWSR1-TFCP2 in 3 cases and FUS-TFCP2 gene fusions in 1 case. One case showed a FUS gene rearrangement, without a TFCP2 gene abnormality by FISH. The MEIS1-NCOA2 positive cases were characterized by a more primitive and fascicular spindle cell appearance, while the EWSR1/FUS rearranged tumors had a hybrid spindle and epithelioid phenotype, with more abundant eosinophilic cytoplasm and mild nuclear pleomorphism. Immunohistochemically, all tumors were positive for desmin and myogenin (focal). In addition, 4 tumors with TFCP2 associated gene fusions also co-expressed ALK and Cytokeratin. In conclusion, our results suggest a high incidence of gene fusions in primary rhabdomyosarcomas of bone, with two molecular subsets emerging, defined by either MEIS1-NCOA2 or EWSR1/FUS-TFCP2 fusions, showing distinct morphology and immunophenotype. Additional studies with larger numbers of cases and longer follow-up data are required to definitively evaluate the biologic behavior of these tumors and to establish their relationship to other spindle cell rhabdomyosarcoma genetic groups.
Keywords: rhabdomyosarcoma, intraosseous, MEIS1, NCOA2, EWSR1, FUS, TFCP2
INTRODUCTION
Primary intraosseous rhabdomyosarcomas are rare, with only few case reports being described, more often involving the head and neck bones, and less commonly the long bones or pelvis.(1–8) Recently, Watson et al.(9) reported a group of 3 rhabdomyosarcomas in young adult females (age range 16–38 yrs) involving the pelvis, chest wall and sphenoid bones that showed a distinctive epithelioid to spindle cell morphology and had EWSR1-TFCP2 or FUS-TFCP2 gene fusions. Dashti et al.(10) subsequently reported an additional case of a skeletal rhabdomyosarcoma of the mandible of a 72 year-old woman showing a spindle cell morphology and a FUS-TFCP2 gene fusion.
In this study, we further investigate a larger group of intraosseous rhabdomyosarcomas by applying targeted RNA sequencing and FISH methodology and correlate with their clinicopathologic features.
MATERIALS AND METHODS
Patient Selection and Histologic Diagnosis
Archival and personal consultation material (CRA) from patients with a diagnosis of rhabdomyosarcoma involving bone were retrieved from the pathology files at Memorial Sloan Kettering Cancer Center. Morphology was reviewed, and diagnosis was confirmed by immunohistochemical stains for desmin and myogenin. Cases with material available for ancillary cytogenetic / molecular studies were selected for the study. The study was approved by the Institutional Review Board at Memorial Sloan Kettering Cancer Center.
FISH
FISH on interphase nuclei from paraffin-embedded 4-micron sections was performed applying custom probes using bacterial artificial chromosomes (BAC), covering and flanking genes MEIS1, NCOA2, EWSR1, FUS, and TFCP2. BAC clones were chosen according to UCSC genome browser (http://genome.ucsc.edu), see Supplementary Table 1 and as previously described.(11) The BAC clones were obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution. The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Newton, MA). A positive score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with incomplete set of signals were omitted from the score.
ARCHER Assay
Archer FusionPlex™®-targeted RNA sequencing analysis was performed in one case (case 1). The sarcoma fusion assay is a targeted RNA-based panel that utilizes the Archer Anchored Multiplex PCR (AMP™) technology and next generation sequencing to detect gene fusions in sarcoma samples. The Archer FusionPlex™® Sarcoma panel targets 26 genes known to be recurrently involved in rearrangements associated with these tumors. RNA is extracted from formalin-fixed paraffin-embedded (FFPE) tumor material followed by cDNA synthesis. cDNA undergoes end repair, dA tailing, and ligation with Archer Molecular Barcode Adapters. SPRI-cleaned ligated fragments are subject to two rounds of PCR amplifications using two sets of gene specific primers (GSP1 used in PCR1 and a nested GSP2 pool used in PCR2) and a primer complementary to the adapter. At the end of two PCR steps, the final targeted amplicon library is quantified. Templating was performed on an Ion Chef™ and sequencing was performed on an Ion Torrent™ S5 XL instrument (Thermo Fisher). The Archer™ analysis software V5.1 was used for data analysis to identify the fusion.
RESULTS
Clinicopathologic Features
Seven cases were identified in 3 males and 4 females, with an age range of from 20 to 39 years (median – 27 years; mean – 28.5 years). The tumors were located in the iliac bone (3), femur (2), maxilla and skull. One patient (case 5) presented with a large mass involving the maxilla and masticator space with extensive extension into the maxillary and sphenoidal sinuses, orbit and clivus. Staging work-up in this patient revealed synchronous distant metastasis to the femur, which was confirmed histologically by core biopsy. Morphologically, all cases showed a predominant spindle cell morphology arranged in intersecting fascicles. None of the cases showed evidence of rhabdomyoblastic differentiation. A detailed morphologic description is provided below based on the two molecular subgroups. Immunohistochemical stains showed diffuse reactivity for desmin and focal to patchy positivity for myogenin.
Novel MEIS1-NCOA2 gene fusion correlates with a primitive spindle cell phenotype
ARCHER Fusionplex study performed in one case identified a novel MEIS1-NCOA2 gene fusion (case 1, Figure 1). The fusion transcript consisted of MEIS1 exon 6 fused to NCOA2 exon 12. FISH studies confirmed these findings, showing break-apart signals in both MEIS1 and NCOA2 genes. Additional FISH screening showed one additional case positive for MEIS1 and NCOA2 gene rearrangements (case 2).
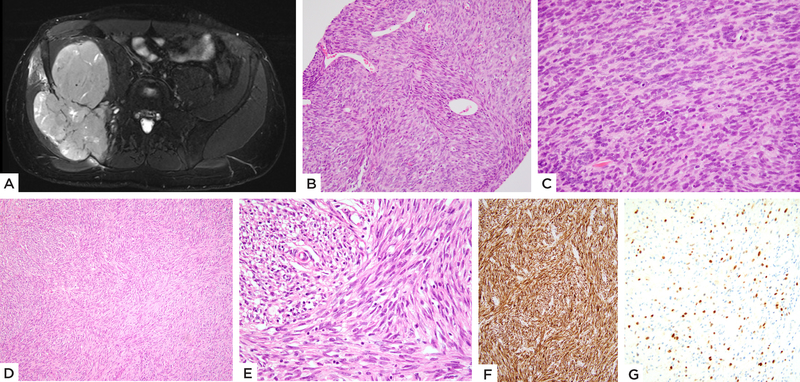
Figure 1: MEIS1-NCOA2 positive iliac bone rhabdomyosarcoma.
(case 1, 22/M). A) MRI showing a large tumor centered in the iliac bone with extraosseous extension. B) Pre-therapy biopsy shows a highly cellular spindle cell neoplasm with scattered branching vessels and insignificant stromal component; C) higher power with monomorphic spindle cells arranged in a streaming pattern. D, E) Resected tumor s/p chemotherapy reveals spindle cells in intersecting fascicles and scant eosinophilic cytoplasm. Immunohistochemical stains show F) diffuse positivity for desmin and G) focal Myogenin staining.
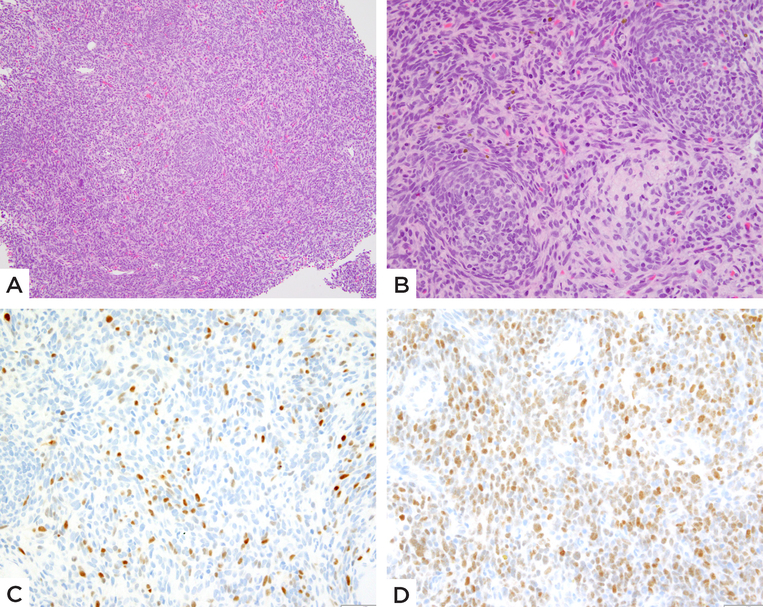
Cases 1 and 2 showed similar clinical presentations of large tumors centered in the iliac bone with associated destruction of the bone and extensive soft tissue extension. (Figure 1A). No definite osteoid or chondroid matrix was identified on imaging. Case 1, on the initial biopsy, showed dense cellular fascicles of primitive appearing spindle cells with scant cytoplasm and ovoid hyperchromatic nuclei. The tumor was associated with a branching hemangiopericytoma-like vascularity in some areas. (Figure 1B–C) The post-therapy en-bloc resection revealed therapy-related changes in the form of hyalinization, with the residual areas of viable tumor showing spindle cells in intersecting fascicles. (Figure 1D–E) Case 2 showed primitive spindle cells with scant cytoplasm arranged in short fascicles. Focally the tumor cells had a more ovoid to rounded appearance, displaying an unusual whorling or micronodular pattern. (Figure 2) Mitotic activity was markedly increased, with more than 15 mitotic figures (MF) per 10 high power fields (HPF) in both cases (18 MF/10 HPFs in case 1 and 50 MF/10 HPFs in case 2). Necrosis was noted in Case 1. Immunohistochemically, Cases 1 and 2 showed diffuse positivity for desmin and focal positivity for myogenin. No cytokeratin or ALK positivity was identified in either of the cases. Case 1 was treated with 4 cycles of neo-adjuvant chemotherapy followed by surgical resection and additional 4 cycles of chemotherapy and has no evidence of disease 8 months since diagnosis.
Figure 2: MEIS1-NCOA2 positive iliac bone rhabdomyosarcoma.
(case 2, 39/M). A) Low power showing a densely cellular primitive neoplasm. B) High power reveals round to ovoid and short spindle cells arranged in vague micronodules or whorling patterns. Immunohistochemical stains show C) scattered multifocal Myogenin positivity and D) relatively diffuse MyoD1 staining.
EWSR1/FUS-TFCP2 gene fusions were associated with a more variable spindle and epithelioid morphology with eosinophilic cytoplasm
The remaining 5 cases were then tested for EWSR1, FUS and TFCP2 gene abnormalities by FISH. Three cases showed an EWSR1-TFCP2 gene fusion (cases 3, 4 and 5) and one case was positive for FUS-TFCP2 fusion (case 6). One case showed a FUS gene rearrangement without abnormalities detected in TFCP2 or NCOA2 genes (case 7).
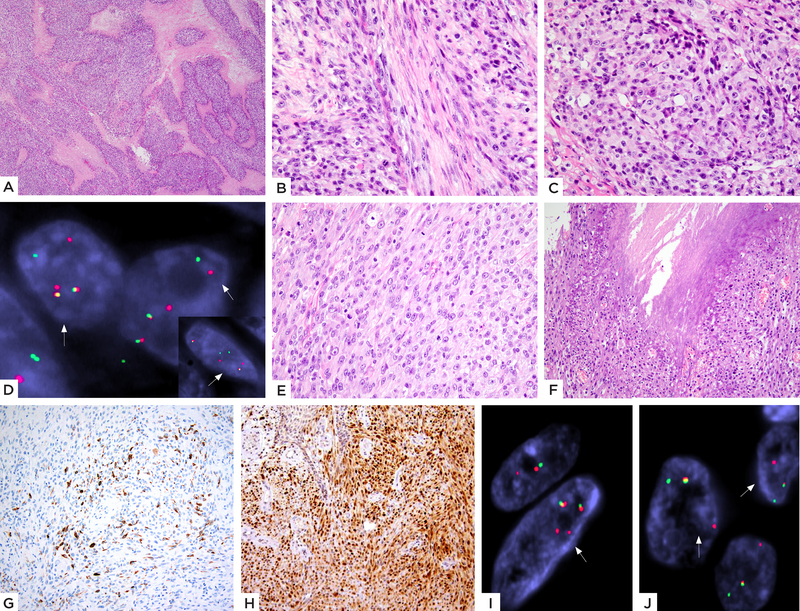
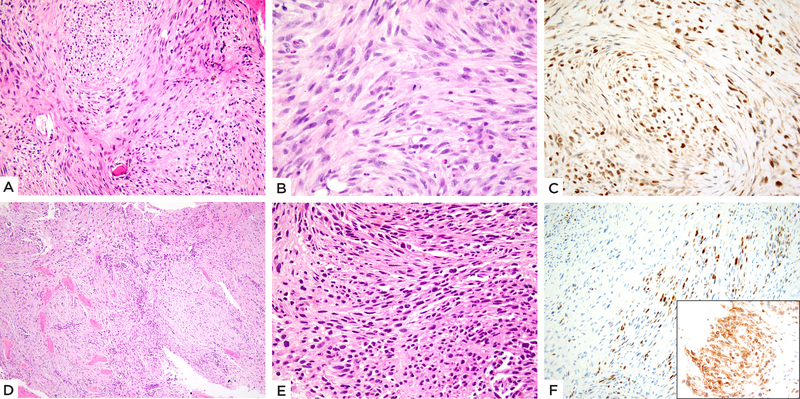
The morphology of the TFCP2-associated group was distinct from the NCOA2-fusion positive subset, showing tumor cells with more abundant light eosinophilic cytoplasm arranged in short fascicles or sheets. Similar to the first group, the TFCP2 positive lesions showed only mild nuclear pleomorphism and lacked overt rhabdomyoblastic differentiation. Overall, the tumors showed scant stromal component and a paucity of vascular spaces. The predominant growth pattern included plump spindle cells with moderate amount of eosinophilic cytoplasm, arranged in short fascicles or vague storiform growth. However, distinctly epithelioid cells were noted in 3 cases (cases 3, 4 and 7), showing centrally located nuclei with vesicular chromatin and prominent nucleoli. (Figure 4) The epithelioid cells merged imperceptibly with the spindle cell areas. (Figure 4) The remaining 2 cases (cases 5 and 6) showed a relatively pure spindle cell morphology arranged in fascicles infiltrating the bony trabeculae. The lesional cells showed slender fusiform to ovoid nuclei and moderate amount of fibrillary, pale eosinophilic cytoplasm. (Figure 5) Mitotic activity was highly elevated in all except one case (case 6), with >15 MFs/10 HPFs and necrosis was seen in all 3 cases with mixed spindle and epithelioid features.
Figure 4: EWSR1-TFCP2 fusion positive rhabdomyosarcoma with a mixed spindle and epithelioid histology.
(A-D) Case 3 (27/F, skull): A) Low power showing a cellular neoplasm with geographic areas of necrosis. High power showing B) spindle cells with pale eosinophilic cytoplasm arranged in intersecting fascicles at 90-degrees and C) areas with distinct epithelioid features. D) FISH studies show break-apart signals for both TFCP2 and EWSR1 (inset) genes (red, centromeric, green, telomeric). (E-J) Case 4 (33/F, maxilla): E) Cellular neoplasm composed of plump ovoid to short spindle cells with moderately abundant pale eosinophilic cytoplasm, arranged in solid sheets or streaming pattern and F) areas of necrosis. Immunohistochemical stains show G) patchy positivity for desmin and H) relatively diffuse nuclear staining for MyoD1. FISH analysis showed the presence of break-apart split signals for both I) EWSR1 and J) TFCP2 genes (red, centromeric; green, telomeric)
Figure 5: TFCP2 fusion positive subset showing mainly spindle cell morphology.
(A-C) EWSR1-TFCP2 fusion positive femur rhabdomyosarcoma (case 5, 20/M). A) Cellular neoplasm involving the bone and completely replacing marrow spaces with spindle cells arranged in intersecting fascicles or vague storiform, with B) pale eosinophilic cytoplasm and ovoid to fusiform nuclei with high mitotic activity. C) Immunohistochemical stain shows diffuse MYOD1 positivity. (D-F) FUS-TFCP2 positive iliac bone tumor (case 6, 37/F). D) Low power showing a cellular neoplasm diffusely involving bone. E) High power reveals short spindle cells arranged in short fascicles. F) Immunohistochemical stain shows focal Myogenin positivity (Inset: diffuse ALK immunoreactivity).
Immunohistochemically, all cases showed expression of desmin and focal positivity for myogenin. MyoD1 expression was noted in all 5 cases, showing a diffuse pattern of staining in 4. All of the tumors except for Case 7 showed expression for pan-cytokeratin and ALK (Figure 5).
Follow-up data was available in 2 cases. In case 4, the patient had surgical resection of the maxillary tumor and had no evidence of disease at 108 months. Case 7 developed multiple pulmonary metastasis after 14 months and is currently alive with disease at 30 months following diagnosis. The other 3 cases were relatively recent cases.
DISCUSSION
In a recent study, Watson et al. (9) identified novel gene fusions in primary skeletal rhabdomyosarcoma involving TFCP2 gene fused with either EWSR1 or FUS. In their study investigating a series of 184 undifferentiated/small round cell sarcomas, 3 such cases of intraosseous rhabdomyosarcoma were identified in young adult females (age range 16–38 yrs) involving the pelvis, chest wall and sphenoid bone. Histologically, the tumors were described as having an epithelioid morphology, arranged in sheets or short fascicles, in a partially sclerotic stroma. Immunohistochemically, apart from the positivity for muscle markers desmin, myogenin and MyoD1, they also showed positivity for cytokeratin and cytoplasmic positivity for ALK. Gene expression profiles also showed an overexpression of ALK and TERT. Subsequently, Dashti et al. reported a case of a mandibular lesion in a 72 year-old man with an identical FUS-TFCP2 gene fusion and noted that the morphology was that of a spindle cell rhabdomyosarcoma with long cellular ‘fibrosarcoma-like’ fascicles of tumor cells. This case also showed co-expression of ALK along with the myogenic markers.(10)
In the current study, we investigate a larger series of 7 intra-osseous rhabdomyosarcomas, to further characterize the molecular and pathologic features of this clinical subset and to establish any relationship to other known subtypes of rhabdomyosarcomas. Remarkably, all cases tested were positive for a gene fusion abnormality, distinct from any other types of rhabdomyosarcoma. The most prevalent genetic alteration were fusions involving either EWSR1 or FUS with TFCP2 gene, detected in 5 of the 7 (71%) cases. Additionally, our results identify a novel molecular subtype, characterized by the presence of recurrent MEIS1-NCOA2 fusion in 2 cases. Regardless of the gene fusion type, the clinical presentation was variable, affecting long bones of extremity, pelvic flat bones or head and neck. The radiological findings (when available in selected cases) suggested a locally destructive growth pattern in the head and neck and the pelvic tumors with tumors centered in the bone and showing extensive extra-osseous extension.
Morphologically, the two tumors displaying MEIS1-NCOA2 fusions showed a rather primitive growth of monomorphic spindle cells with scant cytoplasm arranged in long intersecting fascicles. The histologic appearance was somewhat reminiscent to congenital/infantile spindle cell rhabdomyosarcoma or to other undifferentiated spindle cell sarcomas, such as monophasic synovial sarcomas, BCOR-family of tumors, etc. MEIS1-NCOA2 gene fusion was recently reported by our group in two undifferentiated spindle cell sarcomas of the kidney (12), which occurred in a 72 year-old woman and a 21 year-old man. One of the cases in our study (Case 2) showed tumor cells arranged in a whorled pattern, similar to the renal case# 1 from the study by Argani et al.(12). By RNA sequencing, the fusion transcripts in the kidney tumors were slightly different, including MEIS1 exon 7 fused to NCOA2 exon 13/14, while the current case showed MEIS1 exon 6 fused to NCOA2 exon 12. None of the renal tumors showed evidence of skeletal muscle differentiation. NCOA2 related gene fusions have been described in a number of both benign and malignant mesenchymal neoplasms, including a subset of congenital / infantile spindle cell rhabdomyosarcoma. (13, 14) In the setting of infantile spindle cell rhabdomyosarcoma, NCOA2 gene partners include critical transcription factors or co-activators involved in muscle development and regulation, such as SRF, VGLL2 and TEAD1. Rare cases of alveolar rhabdomyosarcoma have also been associated with PAX3-NCOA2 gene fusion. (15) NCOA2 gene fusions have also been reported in soft tissue angiofibroma with AHRR-NCOA2 and GTF21-NCOA2 fusions (16, 17) and mesenchymal chondrosarcoma with HEY1-NCOA2 fusion.(18) In the recent study by Argani et al., although the MEIS1-NCOA2 positive kidney tumor clustered together with the HEY1-NCOA2 mesenchymal chondrosarcoma and away from the NCOA2-positive soft tissue angiofibroma by unsupervised hierarchical clustering, no increase in expression level of HEY1 or AHRR was noted.(12)
MEIS1 (myeloid ectopic viral integration site-1), on 2p14 locus, encodes for a 390 amino acid protein, which is a member of the three amino acid loop extension (TALE) homeodomain transcription factor family. MEIS1 regulates target gene activation by association with Hox transcription factors. MEIS1 plays a role in cardiac regeneration, stem cell function and tumorigenesis.(19, 20) MEIS1 also functions as a tumor suppressor in some neoplasms, such as clear cell renal cell carcinoma, prostatic adenocarcinoma and esophageal squamous cell carcinoma. (21) MEIS1 gene amplification has been reported in a subset of malignant peripheral nerve sheath tumors (22) and neuroblastomas. (23) MEIS1 is overexpressed in ovarian cancers (24) and acts as co-factor with HOXA9 in driving acute leukemias, particularly aggressive MLL-associated leukemias (25) and chemotherapy-resistant leukemias. (26–28)
In the second molecular group of intraosseous rhabdomyosarcomas harboring recurrent EWSR1/ FUS-TFCP2 fusions, tumors had a similar histologic appearance as described by Watson et al. and Dashti et al. (9, 10), showing plump spindle cells with moderate amount of fibrillary eosinophilic cytoplasm, arranged in short fascicles, with scattered foci of more epithelioid growth. Immunohistochemically, all of the cases in our study stained for desmin, myogenin and MyoD1. Co-expression of ALK was noted in cases with EWSR1 / FUS-TFCP2 gene fusion, but not in the MEIS1-NCOA2 molecular group. This finding is in keeping with the cases reported by Watson et al. (9) and Dashti et al.(10) where ALK expression was consistently reported in all cases. ALK expression has been previously reported in a subset of alveolar and embryonal rhabdomyosarcoma.(29–31) In our study, aberrant cytokeratin positivity was noted in all 4 cases with TFCP2 gene fusions, but not in the NCOA2-positive group.
TFCP2 (also known as LSF or LBP1) was first described as an activator of the Late SV40 promoter and later found to bind globins and HIV-1 promoters (32, 33) It is ubiquitously expressed and plays determinant roles in lineage-specific gene expression and cell cycle regulation. (34) TFCP2 is overexpressed in hepatocellular carcinomas, (35) and is inhibited by small molecules leading to cell cycle arrest; (36) whereas in melanoma, TFCP2 is underexpressed, and its re-expression induces growth arrest. (37) Although, in the study by Watson et al.,(9) these tumors did not cluster together with other rhabdomyosarcoma types, interestingly, a number of up-regulated genes in the FUS/EWSR1–TFCP2-positive tumors were involved in muscle biology, such as cholinergic receptor nicotinic subunit alpha 1, delta and gamma, and sarcoglycan alpha. (9) From a therapeutic aspect, recently developed small molecules which inhibit the TFCP2 DNA-binding activity (36, 38) retained in the fusion protein, may be effective in inhibiting the EWSR1–TFCP2 function.
Primary osseous rhabdomyosarcomas are exceptionally rare, with relatively few cases reported to date. (1–8) Involvement of the head and neck bones (mandible, maxilla and temporal) appears to be more frequent than long bones. One case in our study presented with a large mass involving the maxilla with extensive extension into the maxillary and sphenoidal sinuses and clivus. However, the incidence of intra-osseous, ‘gnathic rhabdomyosarcoma’ might be difficult to establish, as medical oncologists have traditionally classified these lesions as para-meningeal primary tumors arising in the paranasal sinuses, with secondary invasion into the maxilla. Although most cases reported were diagnosed as embryonal rhabdomyosarcoma, rare cases of spindle cell rhabdomyosarcoma with fibrosarcoma-like pattern (6) have also been documented. The recent report of mesenchymal chondrosarcoma with rhabdomyoblastic differentiation (39) also presents a diagnostic pitfall as these cases may be misdiagnosed as spindle cell rhabdomyosarcoma, especially on a biopsy. In the study by Watson et al.(9) one of the 3 cases reported was in the pelvic region. In our study, 3 of the 7 cases were located in the iliac region, suggesting that pelvic bones might also be a preferred site for this disease.
In the study by Watson et al.(9) all 3 patients followed a highly aggressive course, succumbing to disease in less than 5 months. The patient reported by Dashti et al.(10) was a recent case with no significant follow-up. In our study, follow-up was available only on two patients, one with FUS gene rearrangement who developed pulmonary metastasis at 14 months and was alive with disease at 30 months and the other with EWSR1-TFCP2 gene fusion who had no evidence of disease at 108 months. The remaining 5 cases were relatively recent with limited follow-up. One of these recent cases with EWSR1-TFCP2 gene fusion presented with synchronous bone metastasis from a maxillary primary.
In conclusion, we report the largest series of intraosseous spindle cell rhabdomyosarcoma to date and expand their morphologic and molecular spectrum. Despite their lack of overt rhabdomyoblastic on histology, the tumors showed unequivocal evidence of skeletal muscle markers expression by immunohistochemistry. First, primary skeletal rhabdomyosarcomas have a predilection for young adults and for either pelvic or gnathic bones. Second, all cases harbored a gene fusion by molecular assessment, showing either a MEIS1-NCOA2 fusion or a TFCP2 fusion with either EWSR1 or FUS genes. The 2 molecular subsets appear to correlate with distinct phenotypes, the MEIS1-NCOA2 fusion being associated with a primitive fascicular spindle cell growth, while the more common EWSR1/FUS-TFCP2 fusion with a more variable spindle to epithelioid histology, and pale eosinophilic cytoplasm. Additional studies with larger numbers of cases and longer follow-up data are required to definitively evaluate the biologic behavior of these tumors and to determine whether they represent a variant of spindle cell RMS or a stand-alone subtype of rhabdomyosarcomas. Larger series will also establish if indeed these genetic signatures represent a hallmark for the intra-osseous location or similar cases can occur at various anatomic sites.
Supplementary Material
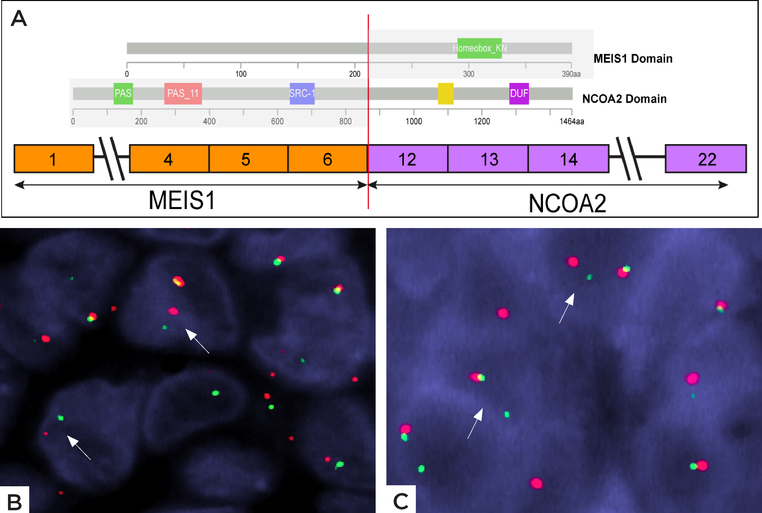
Figure 3: Schematic diagram of MEIS1-NCOA2 fusion.
A) ARCHER results revealed a fusion transcript composed of MEIS1 exon 6 fused to NCOA2 exon 12. The proteins domains of MEIS1 and NCOA2 are illustrated above. B) FISH studies showing break-apart signals of B) NCOA2 gene and C) MEIS1 gene (red, centromeric; green telomeric).
ACKNOWLEDGEMENTS
The authors would like to thank Jordana Shapiro for preparation of composite figures. They also thank the following pathologists and oncologists who kindly contributed case material and / or clinical follow-up information when available: Dr. Alfredo Enrique Martelo, Columbia.
Supported by: P50 CA 140146-01 (CRA), Cycle for Survival (CRA), Slifka Foundation (CRA), St Baldrick Foundation (CRA).
REFERENCES
- 1.Andrade CR, Trento GDS, Jeremias F, et al. Rabdomyosarcoma of the Mandible: An Uncommon Clinical Presentation. J Craniofac Surg. 2018;29:e221–e224. [DOI] [PubMed] [Google Scholar]
- 2.Cemiloglu R, Tekalan SA, Patiroglu T, et al. Rhabdomyosarcoma of the temporal bone: clinical report. Arch Otorhinolaryngol. 1987;244:195–197. [DOI] [PubMed] [Google Scholar]
- 3.Chasin WD. Rhabdomyosarcoma of the temporal bone. Ann Otol Rhinol Laryngol Suppl. 1984;112:71–73. [DOI] [PubMed] [Google Scholar]
- 4.de Melo ACR, Lyra TC, Ribeiro ILA, et al. Embryonal rhabdomyosarcoma in the maxillary sinus with orbital involvement in a pediatric patient: Case report. World J Clin Cases. 2017;5:440–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iatrou I, Theologie-Lygidakis N, Schoinohoriti O, et al. Rhabdomyosarcoma of the maxillofacial region in children and adolescents: Report of 9 cases and literature review. J Craniomaxillofac Surg. 2017;45:831–838. [DOI] [PubMed] [Google Scholar]
- 6.Oda Y, Tsuneyoshi M, Hashimoto H, et al. Primary rhabdomyosarcoma of the iliac bone in an adult: a case mimicking fibrosarcoma. Virchows Arch A Pathol Anat Histopathol. 1993;423:65–69. [DOI] [PubMed] [Google Scholar]
- 7.Williams TP, Vincent SD. Embryonal rhabdomyosarcoma of the mandible. Journal of oral and maxillofacial surgery : official journal of the American Association of Oral and Maxillofacial Surgeons. 1987;45:441–443. [DOI] [PubMed] [Google Scholar]
- 8.Lucas DR, Ryan JR, Zalupski MM, et al. Primary embryonal rhabdomyosarcoma of long bone. Case report and review of the literature. Am J Surg Pathol. 1996;20:239–244. [DOI] [PubMed] [Google Scholar]
- 9.Watson S, Perrin V, Guillemot D, et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol. 2018;245:29–40. [DOI] [PubMed] [Google Scholar]
- 10.Dashti NK, Wehrs RN, Thomas BC, et al. Spindle cell rhabdomyosarcoma of bone with FUS-TFCP2 fusion: confirmation of a very recently described rhabdomyosarcoma subtype. Histopathology. 2018;73:514–520. [DOI] [PubMed] [Google Scholar]
- 11.Antonescu CR, Zhang L, Chang NE, et al. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Argani P, Reuter VE, Kapur P, et al. Novel MEIS1-NCOA2 Gene Fusions Define a Distinct Primitive Spindle Cell Sarcoma of the Kidney. Am J Surg Pathol. 2018;42:1562–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alaggio R, Zhang L, Sung YS, et al. A Molecular Study of Pediatric Spindle and Sclerosing Rhabdomyosarcoma: Identification of Novel and Recurrent VGLL2-related Fusions in Infantile Cases. Am J Surg Pathol. 2016;40:224–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosquera JM, Sboner A, Zhang L, et al. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer. 2013;52:538–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sumegi J, Streblow R, Frayer RW, et al. Recurrent t(2;2) and t(2;8) translocations in rhabdomyosarcoma without the canonical PAX-FOXO1 fuse PAX3 to members of the nuclear receptor transcriptional coactivator family. Genes Chromosomes Cancer. 2010;49:224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Y, Moller E, Nord KH, et al. Fusion of the AHRR and NCOA2 genes through a recurrent translocation t(5;8)(p15;q13) in soft tissue angiofibroma results in upregulation of aryl hydrocarbon receptor target genes. Genes Chromosomes Cancer. 2012;51:510–520. [DOI] [PubMed] [Google Scholar]
- 17.Panagopoulos I, Gorunova L, Viset T, et al. Gene fusions AHRR-NCOA2, NCOA2-ETV4, ETV4-AHRR, P4HA2-TBCK, and TBCK-P4HA2 resulting from the translocations t(5;8;17)(p15;q13;q21) and t(4;5)(q24;q31) in a soft tissue angiofibroma. Oncol Rep. 2016;36:2455–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Motoi T, Khanin R, et al. Identification of a novel, recurrent HEY1-NCOA2 fusion in mesenchymal chondrosarcoma based on a genome-wide screen of exon-level expression data. Genes Chromosomes Cancer. 2012;51:127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aksoz M, Turan RD, Albayrak E, et al. Emerging Roles of Meis1 in Cardiac Regeneration, Stem Cells and Cancer. Curr Drug Targets. 2018;19:181–190. [DOI] [PubMed] [Google Scholar]
- 20.Blasi F, Bruckmann C, Penkov D, et al. A tale of TALE, PREP1, PBX1, and MEIS1: Interconnections and competition in cancer. Bioessays. 2017;39. [DOI] [PubMed] [Google Scholar]
- 21.Zhu J, Cui L, Xu A, et al. MEIS1 inhibits clear cell renal cell carcinoma cells proliferation and in vitro invasion or migration. BMC cancer. 2017;17:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel AV, Chaney KE, Choi K, et al. An ShRNA Screen Identifies MEIS1 as a Driver of Malignant Peripheral Nerve Sheath Tumors. EBioMedicine. 2016;9:110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones TA, Flomen RH, Senger G, et al. The homeobox gene MEIS1 is amplified in IMR-32 and highly expressed in other neuroblastoma cell lines. European journal of cancer. 2000;36:2368–2374. [DOI] [PubMed] [Google Scholar]
- 24.Crijns AP, de Graeff P, Geerts D, et al. MEIS and PBX homeobox proteins in ovarian cancer. European journal of cancer. 2007;43:2495–2505. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Qin YZ, Yang S, et al. Meis1 is critical to the maintenance of human acute myeloid leukemia cells independent of MLL rearrangements. Ann Hematol. 2017;96:567–574. [DOI] [PubMed] [Google Scholar]
- 26.Collins CT, Hess JL. Deregulation of the HOXA9/MEIS1 axis in acute leukemia. Curr Opin Hematol. 2016;23:354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosales-Avina JA, Torres-Flores J, Aguilar-Lemarroy A, et al. MEIS1, PREP1, and PBX4 are differentially expressed in acute lymphoblastic leukemia: association of MEIS1 expression with higher proliferation and chemotherapy resistance. J Exp Clin Cancer Res. 2011;30:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong P, Iwasaki M, Somervaille TC, et al. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes & development. 2007;21:2762–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corao DA, Biegel JA, Coffin CM, et al. ALK expression in rhabdomyosarcomas: correlation with histologic subtype and fusion status. Pediatr Dev Pathol. 2009;12:275–283. [DOI] [PubMed] [Google Scholar]
- 30.Gasparini P, Casanova M, Villa R, et al. Anaplastic lymphoma kinase aberrations correlate with metastatic features in pediatric rhabdomyosarcoma. Oncotarget. 2016;7:58903–58914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pillay K, Govender D, Chetty R. ALK protein expression in rhabdomyosarcomas. Histopathology. 2002;41:461–467. [DOI] [PubMed] [Google Scholar]
- 32.Swendeman SL, Spielholz C, Jenkins NA, et al. Characterization of the genomic structure, chromosomal location, promoter, and development expression of the alpha-globin transcription factor CP2. J Biol Chem. 1994;269:11663–11671. [PubMed] [Google Scholar]
- 33.Yoon JB, Li G, Roeder RG. Characterization of a family of related cellular transcription factors which can modulate human immunodeficiency virus type 1 transcription in vitro. Mol Cell Biol. 1994;14:1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veljkovic J, Hansen U. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene. 2004;343:23–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoo BK, Emdad L, Gredler R, et al. Transcription factor Late SV40 Factor (LSF) functions as an oncogene in hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2010;107:8357–8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajasekaran D, Siddiq A, Willoughby JL, et al. Small molecule inhibitors of Late SV40 Factor (LSF) abrogate hepatocellular carcinoma (HCC): Evaluation using an endogenous HCC model. Oncotarget. 2015;6:26266–26277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goto Y, Yajima I, Kumasaka M, et al. Transcription factor LSF (TFCP2) inhibits melanoma growth. Oncotarget. 2016;7:2379–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant TJ, Bishop JA, Christadore LM, et al. Antiproliferative small-molecule inhibitors of transcription factor LSF reveal oncogene addiction to LSF in hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2012;109:4503–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Folpe AL, Graham RP, Martinez A, et al. Mesenchymal chondrosarcomas showing immunohistochemical evidence of rhabdomyoblastic differentiation: a potential diagnostic pitfall. Hum Pathol. 2018;77:28–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.