Abstract
Single-copy loss-of-function mutations in the progranulin gene (PGRN) underlie the neurodegenerative disease frontotemporal lobar degeneration, while homozygous loss-of-function of PGRN results in the lysosomal storage disorder, neuronal ceroid lipofuscinosis. Despite evidence that normal PGRN levels are critical for neuronal health, the function of this protein is not yet understood. Here, we show that PGRN stimulates the in vitro maturation of the lysosomal aspartyl protease cathepsin D (CTSD). CTSD is delivered to the endolysosomal system as an inactive precursor (proCTSD) and requires sequential cleavage steps via intermediate forms to achieve the mature state (matCTSD). In co-immunoprecipitation experiments PGRN interacts predominantly with immature pro- and intermediate forms of CTSD. PGRN enhances in vitro conversion of proCTSD to matCTSD in a concentration-dependent manner. Differential scanning fluorimetry shows a destabilizing effect induced by PGRN on proCTSD folding (ΔTm = −1.7°C at a 3:1 molar ratio). We propose a mechanism whereby PGRN binds to proCTSD, destabilizing the propeptide from the enzyme catalytic core and favoring conversion to mature forms of the enzyme. Further understanding of the role of PGRN in CTSD maturation will assist in the development of targeted therapies for neurodegenerative disease.
Keywords: progranulin, cathepsin D, maturation, propeptide, neurodegeneration, lysosome, protease
INTRODUCTION
Progranulin (PGRN) is a highly conserved glycoprotein implicated in diverse biological processes1; 2; 3. Loss-of-function mutations in one copy of the progranulin gene (PGRN) cause the adult-onset neurodegenerative disease, frontotemporal lobar degeneration (FTLD)4; 5, whereas homozygous null mutations result in the childhood-onset lysosomal storage disorder, neuronal ceroid lipofuscinosis (NCL)6. In addition, PGRN is over-expressed in cancerous cells, where it can promote tumor growth and metastasis7; 8; 9. This suggests an important role for PGRN in regulating cellular growth, lysosome function and neuronal health. Indeed, loss of PGRN results in dysfunctional lysosomes10; 11; 12, and PGRN repletion has a beneficial effect on disease pathology in animal models of both Alzheimer’s13 and Parkinson’s disease14.
Cathepsin D (CTSD) is a lysosomal aspartyl protease with nearly ubiquitous tissue expression15; 16. In striking parallel to PGRN, CTSD loss-of-function is implicated in lysosomal storage disorders 17; 18 and neurodegeneration 19; 20. In addition, it is over-expressed and secreted from multiple cancerous cell types21; 22, where it stimulates invasion and metastasis 23; 24; 25. CTSD is synthesized as an inactive pro-protein (proCTSD, ~52kDa) and undergoes several proteolytic processing steps to a mature active protease (matCTSD, ~30kDa) in the lysosomal compartment26. These include removal of a forty-four amino acid propeptide26 and the cleavage of the single-chain protein into a ~30kDa heavy chain and a ~15kDa light chain, held together in the mature enzyme through non-covalent interactions26; 27; 28.
The propeptide of CTSD is required for proper folding of the proenzyme, lysosomal targeting and ensures the enzyme remains inactive until activation in the appropriate environment29. It obstructs access to the active site cleft, thereby preventing protease activity30; 31, although there is evidence of proCTSD activity on small peptides32. The propeptide can be removed by cysteine proteases in vivo 28; 33; 34 and in an autocatalytic manner triggered by acidification in vitro 32; 35; 36. An exon 2 polymorphism within the propeptide region of CTSD can disrupt normal maturation of proCTSD and has been associated with neurodegenerative disease20; 37; 38. This Ala38Val substitution results in higher proCTSD secretion, reduced maturation of the enzyme38 and increased levels of beta-amyloid39 and tau protein37 in Alzheimer’s disease.
Recently, several studies have reported an interaction between PGRN and CTSD40; 41; 42, and have proposed that PGRN may stimulate CTSD protease activity43; 44. Indeed, CTSD activity is reduced in both mouse and patient-derived cell models of PGRN deficiency41; 43; 44; 45. Despite these advances, the molecular basis of the CTSD/PGRN interaction remains elusive. In this study, we demonstrate that PGRN induces a destabilizing effect on proCTSD which promotes its maturation to the enzymatically active mature form. We propose a kinetic model whereby PGRN binds the CTSD propeptide to destabilize its interaction with the enzyme catalytic core. Our results suggest a potential mechanism for PGRN deficiency contributing to lysosomal dysfunction and neuronal cell loss.
RESULTS AND DISCUSSION
PGRN binds predominantly to pro-forms of CTSD in a co-immunoprecipitation assay
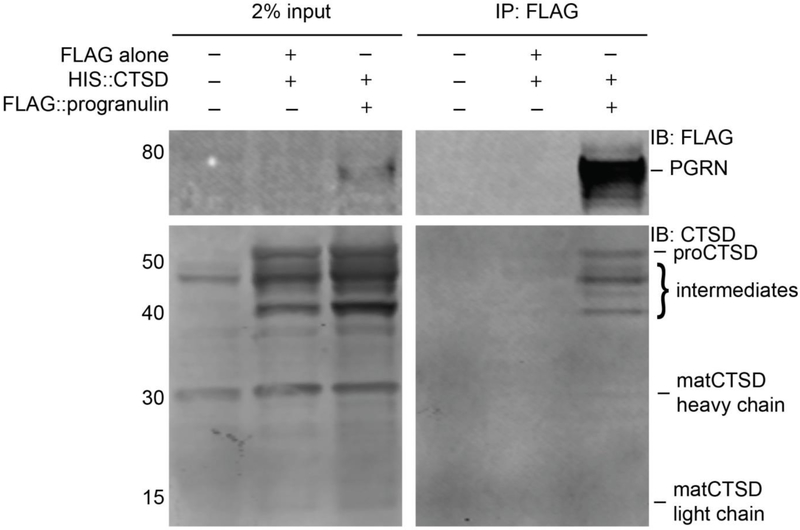
Several recent studies have shown that PGRN can bind to CTSD, where it may function to regulate CTSD stability and protease activity41; 43; 44. We confirmed this interaction through the expression and co-immunoprecipitation of tagged PGRN and proCTSD in HEK293FT cells (Figure 1). Here, we noted that primarily pro- (~52kDa) and intermediate (~40-50kDa) forms of CTSD bind to PGRN, rather than the final mature forms of CTSD, consisting of the heavy (~30kDa) and light (~15kDa) chains. This was a first indication that PGRN may bind to CTSD primarily via the propeptide and could thereby effectively function in regulating the maturation of proCTSD to matCTSD. Such a preferential interaction with proCTSD has also been shown for prosaposin46, a lysosomal protein of modulatory structure like PGRN 47 which is known to interact with PGRN48; 49.
Figure 1. PGRN binds predominantly to pro- and intermediate forms of CTSD.
Pulldown of proCTSD with PGRN expressed in HEK293FT cells. Immunoprecipitation was performed against the FLAG epitope on FLAG-tagged PGRN. Immunoblotting was performed with anti-FLAG and anti-CTSD antibodies. Immunoblots are representative of three independent experiments.
PGRN reduces the melting temperature of proCTSD through a destabilizing effect
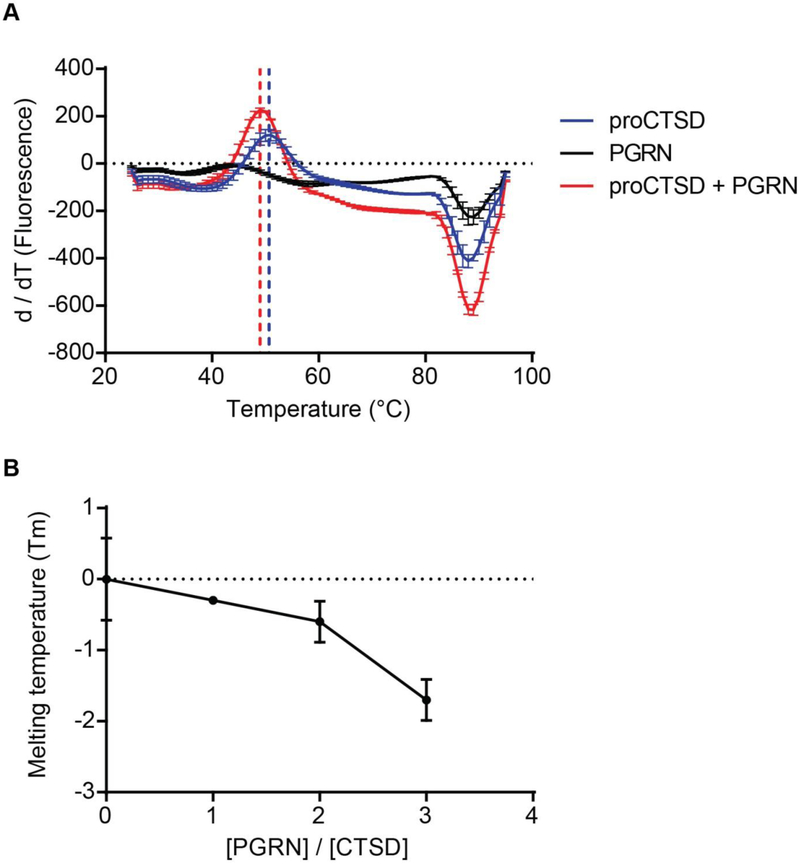
As our immunoprecipitation results suggest a specific interaction between PGRN and proCTSD, we sought to evaluate how this interaction impacts the conformational stability of proCTSD in vitro. Differential scanning fluorimetry (DSF) is a technique that can measure the melting temperature (Tm), or stability, of recombinant proteins alone or in complexes. We first confirmed the purity and correct molecular weight of commercially available recombinant PGRN and proCTSD proteins by silver stain and SDS-PAGE (Figure S1). We next performed DSF on these proteins at neutral pH to assess their stability in the absence of auto-activation of proCTSD to matCTSD. When subjected to DSF, PGRN alone did not show an unfolding transition on increasing temperature (Figure 2A and Figure S2). This suggests that recombinant PGRN is thermally stable, as expected from its highly disulfide-bonded structure50; 51. ProCTSD alone showed an unfolding transition at a Tm of 50.7°C (Figure 2A and Figure S2). The addition of PGRN to proCTSD at a 3:1 molar ratio caused a significant destabilizing effect on the Tm of proCTSD (ΔTm = −1.7°C) (Figure 2A). Lower molar ratios of PGRN to proCTSD (2:1 and 1:1) resulted in a concentration-dependent temperature shift of −0.6°C and −0.3°C, respectively (Figure 2B and Figure S2).
Figure 2. PGRN reduces the melting temperature of proCTSD through a destabilizing effect.
(A) Differential scanning fluorimetry (DSF) was used to obtain fluorescent intensity curves versus temperature, and the curve derivatives are plotted for recombinant proteins: 1.5μM HIS-tagged proCTSD alone (blue), 4.5μM HIS-tagged PGRN alone (black), and 1.5μM proCTSD with 4.5μM PGRN (red). DSF was performed at neutral pH 7.4. Assays were run in triplicate and values plotted are mean ± SEM. (B) Melting temperature, Tm, for PGRN:proCTSD complex at increasing molar ratios of PGRN.
We noted that the unfolding curve for the proCTSD:PGRN complex presented with a larger change in fluorescence than for proCTSD alone (ΔFprocTSD:PGRN > 1500a.u.; ΔFprocTSD < 1000a.u.) (Figure S2A), suggesting an increase in exposure of proCTSD hydrophobic residues and cooperativity in unfolding in the presence of PGRN. Interestingly, a similar mechanism of action has been proposed for sulfated polysaccharides on both aspartyl31 and cysteine proteases52; 53; 54, whereby destabilization of the propeptide favors its cleavage. These negatively charged compounds are hypothesized to interact with Arg3 and Arg11 residues of the CTSD propeptide, reducing their electrostatic interaction with residues Asp181 and Asp12 of the enzyme catalytic core31.
PGRN increases the conversion rate of proCTSD to matCTSD
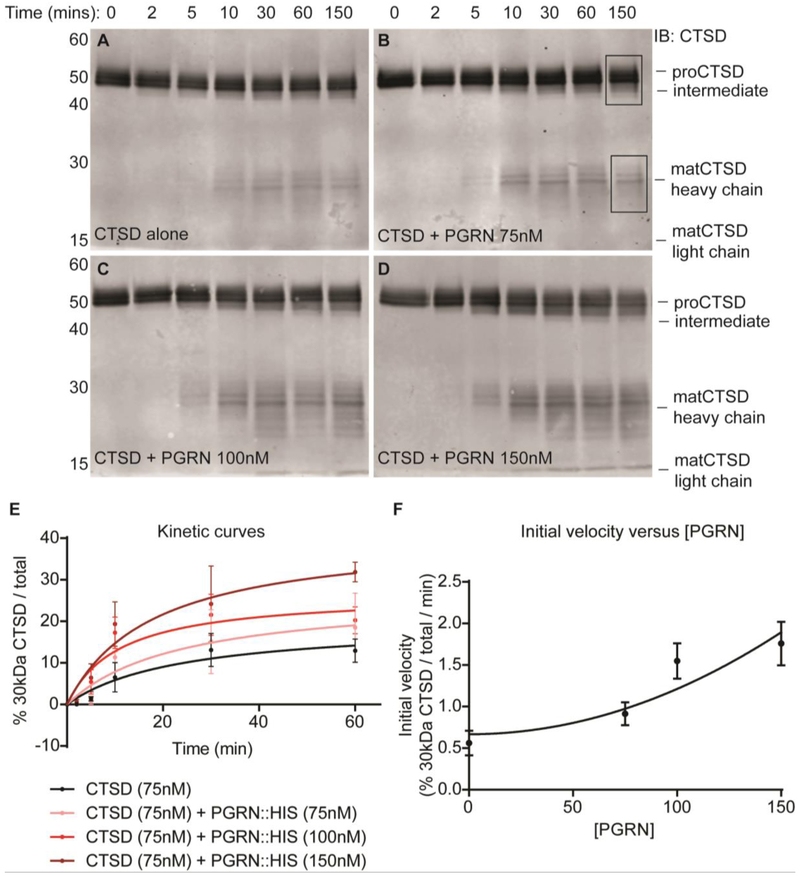
Given that PGRN binds to and destabilizes proCTSD, we next evaluated a potential role for PGRN in proCTSD maturation at an acidic pH of 3.4. In the presence of PGRN we noted an increase in the formation of matCTSD (Figure 3A-D). We observed the same result with a differentially-tagged recombinant PGRN purchased from an alternate source (Figure S3). To test for a concentration-dependent effect of PGRN on proCTSD conversion to matCTSD, we estimated the kinetics of maturation from the immunoblot signals. First, we assessed whether there was a concentration-dependent conversion of proCTSD to matCTSD in the absence of PGRN. Indeed, we observed a concentration-dependence in the maturation of proCTSD alone (Figure S4). Calculation of the initial velocities (V0) of these reactions with increasing proCTSD concentration demonstrated that V0 increases non-linearly with proCTSD concentration, consistent with a quadratic relationship (Appendix 1), as predicted from an intermolecular activation mechanism (Figure S4). We next calculated V0 for proCTSD maturation in the presence of increasing concentrations of PGRN. We found that V0 increased with increasing PGRN concentration (Figure 3E-F), confirming a concentration-dependent increase in proCTSD maturation in the presence of PGRN. Both PGRN and proCTSD undergo direct trafficking to the lysosome via the mannose-6-phosphate receptor pathway49; 55; 56. It is possible that at least part of this trafficking may occur in complex together. On reaching the lysosome, this may allow PGRN to regulate the maturation and/or activity of the protease. Such a role for PGRN in proCTSD maturation may help to explain the impaired CTSD activity observed in models of PGRN deficiency41; 43; 44. Indeed, CTSD activity (Vmax) was increased by approximately 50% in the presence of PGRN at a 3:1 molar ratio (Figure S5).
Figure 3. PGRN increases the conversion rate of proCTSD to matCTSD.
(A-D) In vitro maturation time-course assays for recombinant HIS-tagged proCTSD at pH 3.4 in the (A) absence (n = 4) and presence of (B) 75nM (n = 3), (C) 100nM (n = 3) and (D) 150nM (n = 3) HIS-tagged PGRN. Membranes were immunoblotted with anti-CTSD antibody. Boxed regions in panel B indicate the signal measured for quantification of % 30kDa CTSD / total. (E) Michaelis-Menten kinetic curves for the conversion of proCTSD to matCTSD. (F) Correlation between initial velocity of reaction (V0) and PGRN concentration.
A kinetic model for the role of PGRN in proCTSD maturation
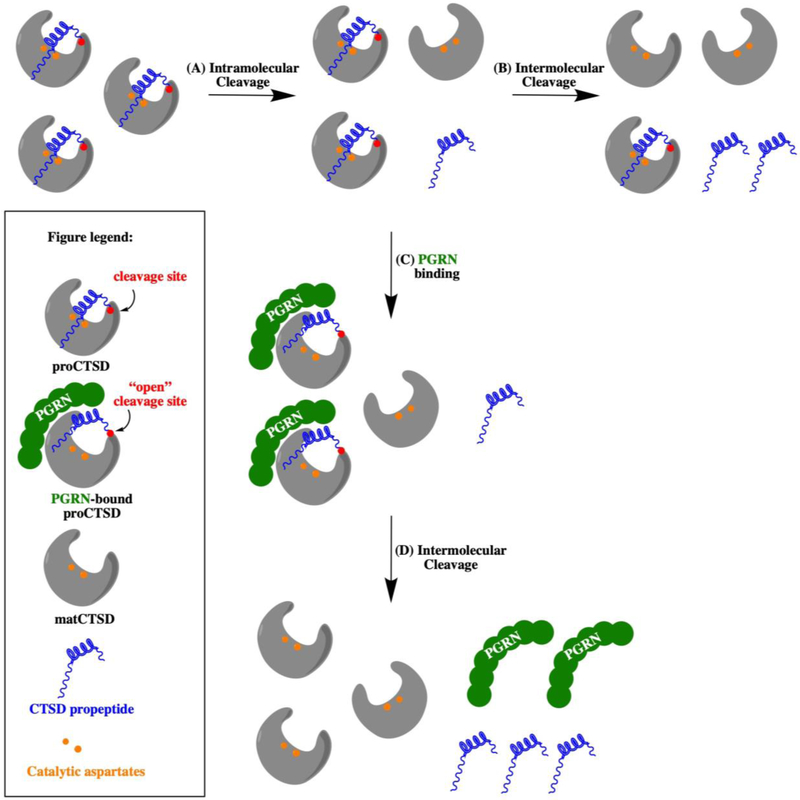
ProCTSD undergoes in vitro maturation to its heavy (~30kDa) and light (~15kDa) chains at acidic pH, a process that involves the cleavage of the 44 residue propeptide32. Initially, the first molecules of proCTSD are processed to matCTSD through a slow intramolecular cleavage mechanism32. The newly formed matCTSD then accelerates the maturation process further by cleaving the propeptide from proCTSD through an intermolecular activation mechanism32. We developed a simple kinetic model for the role of PGRN in converting proCTSD to matCTSD (Appendix 1) and hypothesize that destabilizing the interaction of the propeptide with the enzyme catalytic core facilitates its cleavage by matCTSD molecules (Figure 4).
Figure 4. A model for the role of PGRN in the conversion of proCTSD to matCTSD.
(A) ProCTSD undergoes an autocatalytic activation mechanism for the formation of initial matCTSD molecules at a low rate. (B) Once formed, matCTSD can convert proCTSD to matCTSD through an intermolecular cleavage of the propeptide. (C) PGRN binds around the propeptide region of proCTSD to destabilize its interaction with the enzyme catalytic core, (D) facilitating propeptide cleavage by matCTSD. Catalytic aspartyl residues are represented as orange dots, the propeptide in blue and the propeptide cleavage site in red.
Our kinetic model suggests that the concentration of matCTSD in cells would depend on the concentration of PGRN (Appendix 1). As such, loss-of-function mutations in PGRN would be expected to impair the maturation of proCTSD to matCTSD and thus reduce protease activity. Such a reduction in CTSD enzymatic activity has been observed in several recent studies41; 43; 44; 45. Despite these observed impairments in CTSD enzymatic activity, studies also report an increase in pro-CTSD and/or matCTSD expression levels on loss of PGRN43; 44; 57; 58; 59. It is possible that these changes in CTSD expression level represent a compensatory transcriptional response to promote lysosome function60; 61. Such cellular compensatory responses highlight the importance of utilizing in vitro assays to isolate the direct molecular effect of PGRN on CTSD. Furthermore, CTSD expression is nearly ubiquitous but there are certain cell types where progranulin expression is low (e.g. neurons, muscle, adipose, soft tissue). As CTSD can undergo a slow auto-activation mechanism by itself at acidic pH, it is likely that the presence of PGRN for CTSD maturation is not an absolute requirement in all cell types under all conditions. PGRN expression may perhaps be a regulatory mechanism to “tune up” CTSD activity. Furthermore, PGRN is a secreted glycoprotein, and therefore cell autonomous expression is not required in order for PGRN to affect protease function. We also cannot exclude other mechanisms regulating CTSD maturation, such as other proteases. Despite this, the data presented here suggest that the impairment of CTSD activity observed in cell models of PGRN deficiency may result from a role for PGRN in proCTSD maturation.
CONCLUSION
In this work, we investigated the nature of the interaction between PGRN and proCTSD and discovered a function for PGRN in promoting the in vitro maturation of this aspartyl protease. DSF studies highlight a potential mechanism for this, whereby PGRN may bind the propeptide region of proCTSD, destabilizing its interaction with the CTSD catalytic core. This may increase accessibility for intermolecular cleavage of the inhibitory propeptide, promoting formation of the active matCTSD. Further investigation will be required to determine which domains of the PGRN molecule mediate the interaction with, and stimulate the maturation of, proCTSD. A potential role for PGRN in lysosomal aspartyl protease maturation will aid in understanding of the mechanism underlying neurodegenerative diseases that are linked to PGRN loss-of-function.
METHODS
HEK293FT transfections and co-immunoprecipitation assays
The cDNAs for human progranulin (a generous gift from Professor Robert Farese) and proCTSD (GenScript #OHu26913) were sub-cloned into the pSecTag2B vector (ThermoFisher, #V90020). Progranulin was expressed with an N-terminal FLAG tag and proCTSD was expressed with an N-terminal His tag. HEK293FT cells (3x106) were seeded into 10cm plates and grown to 60% confluency. 5μg total DNA was transfected (Opti-MEM, ThermoFisher, #31985070, X-tremeGENE HP DNA Transfection Reagent, Sigma Aldrich, #6366236001). Cells were collected forty-eight hours later and re-suspended in lysis buffer with protease inhibitor (50mM Tris pH 7.4 (Teknova #T5074), 150mM NaCl (Fisher #S271-3), 1% Nonidet P-40 (Thermo Scientific #85124), cOmplete protease inhibitor (Roche, #04693124001). Cells were lysed for one hour on ice, centrifuged at 20,000g for 20 minutes and the supernatant was collected. The protein concentration of samples was measured by BCA. Immunoprecipitation was carried out against the FLAG tag on progranulin (Sigma Aldrich, #M8823).
ProCTSD thermal denaturation by DSF
DSF measurements λex = 490nm, λex 610nm) were performed in 96-well plates (USA Scientific #1402-9590) on a Biorad CFX96 instrument. Thermal denaturation assays (70μl) for 1.5μM proCTSD (R&D Systems Inc., #1014-AS) were measured in the presence and absence of PGRN (R&D Systems, #2420-PG-050) in 50mM Tris-HCl (Teknova #T5074), 150mM NaCl (Fisher #S271-3) pH 7.4. SyproOrange (Sigma #S5692) was added to all wells. Thermal denaturation curves were recorded over a temperature range from 25–95°C with 1°C min−1 increments. All measurements were carried out in triplicate.
Time-dependent immunoblotting for proCTSD maturation
Assays were run in 20μl of 100mM sodium citrate buffer (pH 3.4). 75nM of recombinant cathepsin D (R&D Systems, #1014-AS) was incubated in the absence or presence of 75, 100 or 150nM recombinant progranulin (R&D Systems, #2420-PG-050). Reactions were incubated at 37°C and then immediately terminated through the addition of 4 x LDS (Thermo #NP0007) and 10% sample reducing agent (Thermo #NP0009). Samples were boiled at 70°C for 10 minutes, analyzed by SDS PAGE on 10% gradient Bis Tris gels (Thermo #NP0302BOX) and transferred to PVDF membrane (Bio-Rad #1620177). Membranes were immunoblotted with an anti-cathepsin D primary antibody (R&D Systems Inc., #AF1014, 1:250 dilution) and a donkey anti-goat secondary antibody (LI-COR IRDye 800CW, #925-32214, 1:10,000 dilution). Membranes were imaged on the Licor Odyssey system and quantified in Licor Image Studio Lite version 5.2 software. Background-corrected signal for 30kDa CTSD bands were divided by total signal (30kDa + 50kDa bands) to obtain a fraction of conversion. This was plotted against time and points were fitted to Michaelis-Menten equation. Initial velocities were calculated using the first four time points of incubation (0, 2, 5 and 10 minutes). Linear regression analysis was performed in GraphPad Prism Version 6. Initial velocities were plotted against proCTSD or PGRN concentration. Silver staining of commercial recombinant protein was carried out using the SilverQuest Silver Staining Kit (Thermo Fisher #LC6070). The anti-paragranulin antibody was made in-house (GRN P.1, epitope TRCPDGQFCPVACCLDPGGASYSCCRPLLD, 1:500 dilution) and used with a goat anti-mouse secondary antibody (LI-COR IRDye 800CW, #925-32210, 1:10,000 dilution).
In vitro cathepsin D activity assays
Fluorescent protease activity assays were performed in triplicate in black, flat-bottom 384 well plates (Greiner #781091). Assays were run for 2 hours in 15μl of sodium acetate buffer (pH 3.5). 20nM of cathepsin D (R&D Systems Inc., #1014-AS) was used for all assays with 5μM of a fluorescent substrate that was identified previously (Lys(7-methoxycoumarin-4-acetic acid) Arg-Gly-Leu-Tyr-Phe-Ile-Thr-His-Lys(dinitrophenol))62. Assays were run in the absence or presence of 65nM progranulin (R&D Systems Inc., #2420-PG-050). Fluorescent substrate cleavage was monitored every 30 seconds with a Biotek Synergy HT plate reader using excitation and emission wavelengths of 328nm and 393nm, respectively. Linear regression analysis was performed in GraphPad Prism Version 6 on 20 consecutive time points to calculate the maximal CTSD enzymatic activity in the presence of unlimited substrate (maximal velocity, Vmax). Data were normalized to the CTSD alone condition.
Supplementary Material
Highlights.
Interaction of progranulin and pro-cathepsin D was investigated in vitro.
Progranulin binds to pro- and intermediate forms of cathepsin D in co-immunoprecipitation experiments.
Progranulin stimulates the maturation of pro-cathepsin D to its mature form in vitro at acidic pH.
We propose a kinetic model for the stimulation of pro-cathepsin D maturation by progranulin, involving destabilization of the cathepsin D propeptide.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health R21NS082709 and R01NS095257, the Alzheimer’s Disease Research Center, the Consortium for Frontotemporal Dementia Research, the UCSF School of Medicine Irene Perstein Award, the John Douglas French Alzheimer’s Foundation and the Brain Research Foundation (A.W.K.). We also thank The James and Barbara Knuppe Family Foundation for research support. We thank Joseph Lobel for technical advice with DSF assays, and members of the Kao lab for helpful discussions.
Appendix 1
We developed a simple kinetic model for the role of PGRN in converting proCTSD to matCTSD. We propose a multi-step mechanism involving the destabilization of the propeptide, where openCTSD = proCTSD with destabilized propeptide:
Therefore, matCTSD can be generated through:
| 1) |
| 2) |
Here, we assume the destabilized propeptide may not interfere with CTSD activity and we consider that 1) and 2) may have the kinetic rate constants 2k1 and k1, respectively. We also assume that proOPEN is catalytically active, with similar activity to matCTSD, i.e. removing the auto-inhibition of the propeptide can occur reversibly by conformational changes, as well as irreversibly by cleavage.
For 1):
For 2):
Summing 1) and 2) to calculate total matCTSD produced:
| 3) |
In addition, proCTSD will convert to openCTSD with a kinetic rate constant of k2:
| 4) |
Where:
| 5) |
Considering equilibrium:
Substituting for equation 5) into equation 3):
| 6) |
At the initiation of the reaction, [matCTSD] → 0, therefore:
| 7) |
Therefore, the rate of production of matCTSD is predicted to depend quadratically on the concentration of proCTSD. The observed kinetics (Figure S4) appears to be even “steeper” than quadratic, but it is clearly not linear.
If PGRN binds the propeptide, in the presence of PGRN;
| 8) |
where:
| 9) |
Combining equations 9) and 5):
| 10) |
Combining equations 10) and 7):
| 11) |
Therefore, the production of matCTSD is also predicted to depend quadratically on the concentration of PGRN which is consistent with Figure 3F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bateman A & Bennett HP (2009). The granulin gene family: from cancer to dementia. Bioessays 31, 1245–54. [DOI] [PubMed] [Google Scholar]
- 2.Cenik B, Sephton CF, Kutluk Cenik B, Herz J & Yu G (2012). Progranulin: a proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J Biol Chem 287, 32298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kao AW, McKay A, Singh PP, Brunet A & Huang EJ (2017). Progranulin, lysosomal regulation and neurodegenerative disease. Nat Rev Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, Cannon A, Dwosh E, Neary D, Melquist S, Richardson A, Dickson D, Berger Z, Eriksen J, Robinson T, Zehr C, Dickey CA, Crook R, McGowan E, Mann D, Boeve B, Feldman H & Hutton M (2006). Mutations in progranulin cause taunegative frontotemporal dementia linked to chromosome 17. Nature 442, 916–9. [DOI] [PubMed] [Google Scholar]
- 5.Cruts M, Kumar-Singh S & Van Broeckhoven C (2006). Progranulin mutations in ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Curr Alzheimer Res 3, 485–91. [DOI] [PubMed] [Google Scholar]
- 6.Smith KR, Damiano J, Franceschetti S, Carpenter S, Canafoglia L, Morbin M, Rossi G, Pareyson D, Mole SE, Staropoli JF, Sims KB, Lewis J, Lin WL, Dickson DW, Dahl HH, Bahlo M & Berkovic SF (2012). Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet 90, 1102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Z & Bateman A (1999). Progranulin gene expression regulates epithelial cell growth and promotes tumor growth in vivo. Cancer Res 59, 3222–9. [PubMed] [Google Scholar]
- 8.Tangkeangsirisin W & Serrero G (2004). PC cell-derived growth factor (PCDGF/GP88, progranulin) stimulates migration, invasiveness and VEGF expression in breast cancer cells. Carcinogenesis 25, 1587–92. [DOI] [PubMed] [Google Scholar]
- 9.Monami G, Emiliozzi V, Bitto A, Lovat F, Xu SQ, Goldoni S, Fassan M, Serrero G, Gomella LG, Baffa R, Iozzo RV & Morrione A (2009). Proepithelin regulates prostate cancer cell biology by promoting cell growth, migration, and anchorage-independent growth. Am J Pathol 174, 1037–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed Z, Sheng H, Xu Y, Lin W, Innes A, Gass J, Yu X, Hou H, Chiba S, Yamanouchi K, Leissring M, Petrucelli L, Nishihara M, Hutton M, McGowan E, Dickson D & Lewis J (2010). Accelerated Lipofuscinosis and Ubiquitination in Granulin Knockout Mice Suggests a Role for Progranulin in Successful Aging Am J Pathol 177, 311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petkau TL, Neal SJ, Milnerwood A, Mew A, Hill AM, Orban P, Gregg J, Lu G, Feldman HH, Mackenzie IR, Raymond LA & Leavitt BR (2012). Synaptic dysfunction in progranulin-deficient mice. Neurobiol Dis 45, 711–22. [DOI] [PubMed] [Google Scholar]
- 12.Wils H, Kleinberger G, Pereson S, Janssens J, Capell A, Van Dam D, Cuijt I, Joris G, De Deyn PP, Haass C, Van Broeckhoven C & Kumar-Singh S (2012). Cellular ageing, increased mortality and FTLD-TDP-associated neuropathology in progranulin knockout mice. J Pathol 228, 67–76. [DOI] [PubMed] [Google Scholar]
- 13.Minami SS, Min SW, Krabbe G, Wang C, Zhou Y, Asgarov R, Li Y, Martens LH, Elia LP, Ward ME, Mucke L, Farese RV Jr. & Gan L (2014). Progranulin protects against amyloid beta deposition and toxicity in Alzheimer's disease mouse models. Nat Med 20, 1157–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Kampen JM, Baranowski D & Kay DG (2014). Progranulin gene delivery protects dopaminergic neurons in a mouse model of Parkinson's disease. PLoS One 9, e97032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benes P, Vetvicka V & Fusek M (2008). Cathepsin D--many functions of one aspartic protease. Crit Rev Oncol Hematol 68, 12–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaidi N, Maurer A, Nieke S & Kalbacher H (2008). Cathepsin D: a cellular roadmap. Biochem Biophys Res Commun 376, 5–9. [DOI] [PubMed] [Google Scholar]
- 17.Steinfeld R, Reinhardt K, Schreiber K, Hillebrand M, Kraetzner R, Bruck W, Saftig P & Gartner J (2006). Cathepsin D deficiency is associated with a human neurodegenerative disorder. Am J Hum Genet 78, 988–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyynela J, Sohar I, Sleat DE, Gin RM, Donnelly RJ, Baumann M, Haltia M & Lobel P (2000). A mutation in the ovine cathepsin D gene causes a congenital lysosomal storage disease with profound neurodegeneration. EMBO J 19, 2786–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidson Y, Gibbons L, Pritchard A, Hardicre J, Wren J, Tian J, Shi J, Stopford C, Julien C, Thompson J, Payton A, Thaker U, Hayes AJ, Iwatsubo T, Pickering-Brown SM, Pendleton N, Horan MA, Burns A, Purandare N, Lendon CL, Neary D, Snowden JS & Mann DM (2006). Genetic associations between cathepsin D exon 2 C-->T polymorphism and Alzheimer's disease, and pathological correlations with genotype. J Neurol Neurosurg Psychiatry 77, 515–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papassotiropoulos A, Bagli M, Kurz A, Kornhuber J, Forstl H, Maier W, Pauls J, Lautenschlager N & Heun R (2000). A genetic variation of cathepsin D is a major risk factor for Alzheimer's disease. Ann Neurol 47, 399–403. [PubMed] [Google Scholar]
- 21.Bossard N, Descotes F, Bremond AG, Bobin Y, De Saint Hilaire P, Golfier F, Awada A, Mathevet PM, Berrerd L, Barbier Y & Esteve J (2003). Keeping data continuous when analyzing the prognostic impact of a tumor marker: an example with cathepsin D in breast cancer. Breast Cancer Res Treat 82, 47–59. [DOI] [PubMed] [Google Scholar]
- 22.Vetvicka V, Vetvickova J, Hilgert I, Voburka Z & Fusek M (1997). Analysis of the interaction of procathepsin D activation peptide with breast cancer cells. Int J Cancer 73, 403–9. [DOI] [PubMed] [Google Scholar]
- 23.Vignon F, Capony F, Chambon M, Freiss G, Garcia M & Rochefort H (1986). Autocrine growth stimulation of the MCF 7 breast cancer cells by the estrogenregulated 52 K protein. Endocrinology 118, 1537–45. [DOI] [PubMed] [Google Scholar]
- 24.Vetvicka V, Vektvickova J & Fusek M (1994). Effect of human procathepsin D on proliferation of human cell lines. Cancer Lett 79, 131–5. [DOI] [PubMed] [Google Scholar]
- 25.Vashishta A, Ohri SS, Proctor M, Fusek M & Vetvicka V (2006). Role of activation peptide of procathepsin D in proliferation and invasion of lung cancer cells. Anticancer Res 26, 4163–70. [PubMed] [Google Scholar]
- 26.Erickson AH, Conner GE & Blobel G (1981). Biosynthesis of a lysosomal enzyme. Partial structure of two transient and functionally distinct NH2-terminal sequences in cathepsin D. J Biol Chem 256, 11224–31. [PubMed] [Google Scholar]
- 27.Conner GE & Richo G (1992). Isolation and characterization of a stable activation intermediate of the lysosomal aspartyl protease cathepsin D. Biochemistry 31, 1142–7. [DOI] [PubMed] [Google Scholar]
- 28.Gieselmann V, Hasilik A & von Figura K (1985). Processing of human cathepsin D in lysosomes in vitro. J Biol Chem 260, 3215–20. [PubMed] [Google Scholar]
- 29.Stoka V, Turk V & Turk B (2016). Lysosomal cathepsins and their regulation in aging and neurodegeneration. Ageing Res Rev 32, 22–37. [DOI] [PubMed] [Google Scholar]
- 30.Dunn BM (2002). Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem Rev 102, 4431–58. [DOI] [PubMed] [Google Scholar]
- 31.Masa M, Maresova L, Vondrasek J, Horn M, Jezek J & Mares M (2006). Cathepsin D propeptide: mechanism and regulation of its interaction with the catalytic core. Biochemistry 45, 15474–82. [DOI] [PubMed] [Google Scholar]
- 32.Wittlin S, Rosel J, Hofmann F & Stover DR (1999). Mechanisms and kinetics of procathepsin D activation. Eur J Biochem 265, 384–93. [DOI] [PubMed] [Google Scholar]
- 33.Samarel AM, Ferguson AG, Decker RS & Lesch M (1989). Effects of cysteine protease inhibitors on rabbit cathepsin D maturation. Am J Physiol 257, C1069–79. [DOI] [PubMed] [Google Scholar]
- 34.Laurent-Matha V, Derocq D, Prebois C, Katunuma N & Liaudet-Coopman E (2006). Processing of human cathepsin D is independent of its catalytic function and auto-activation: involvement of cathepsins L and B. J Biochem 139, 363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasilik A, von Figura K, Conzelmann E, Nehrkorn H & Sandhoff K (1982). Lysosomal enzyme precursors in human fibroblasts. Activation of cathepsin D precursor in vitro and activity of beta-hexosaminidase A precursor towards ganglioside GM2. Eur J Biochem 125, 317–21. [DOI] [PubMed] [Google Scholar]
- 36.James MN & Sielecki AR (1986). Molecular structure of an aspartic proteinase zymogen, porcine pepsinogen, at 1.8 A resolution. Nature 319, 33–8. [DOI] [PubMed] [Google Scholar]
- 37.Riemenschneider M, Blennow K, Wagenpfeil S, Andreasen N, Prince JA, Laws SM, Forstl H & Kurz A (2006). The cathepsin D rs17571 polymorphism: effects on CSF tau concentrations in Alzheimer disease. Hum Mutat 27, 532–7. [DOI] [PubMed] [Google Scholar]
- 38.Touitou I, Capony F, Brouillet JP & Rochefort H (1994). Missense polymorphism (C/T224) in the human cathepsin D pro-fragment determined by polymerase chain reaction--single strand conformational polymorphism analysis and possible consequences in cancer cells. Eur J Cancer 30A, 390–4. [DOI] [PubMed] [Google Scholar]
- 39.Papassotiropoulos A, Lewis HD, Bagli M, Jessen F, Ptok U, Schulte A, Shearman MS & Heun R (2002). Cerebrospinal fluid levels of beta-amyloid(42) in patients with Alzheimer's disease are related to the exon 2 polymorphism of the cathepsin D gene. Neuroreport 13, 1291–4. [DOI] [PubMed] [Google Scholar]
- 40.Beel S, Moisse M, Damme M, De Muynck L, Robberecht W, Van den Bosch L, Saftig P & Van Damme P (2017). Progranulin functions as a cathepsin D chaperone to stimulate axonal outgrowth in vivo. Human Molecular Genetics 26, 2850–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X, Paushter DH, Feng T, Pardon CM, Mendoza CS & Hu F (2017). Regulation of cathepsin D activity by the FTLD protein progranulin. Acta Neuropathol 134, 151–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valdez C, Wong YC, Schwake M, Bu G, Wszolek ZK & Krainc D (2017). Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients. Hum Mol Genet 26, 4861–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beel S, Moisse M, Damme M, De Muynck L, Robberecht W, Van Den Bosch L, Saftig P & Van Damme P (2017). Progranulin functions as a cathepsin D chaperone to stimulate axonal outgrowth in vivo. Hum Mol Genet 26, 2850–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valdez C, Wong YC, Schwake M, Bu G, Wszolek ZK & Krainc D (2017). Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients. Hum Mol Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward ME, Chen R, Huang HY, Ludwig C, Telpoukhovskaia M, Taubes A, Boudin H, Minami SS, Reichert M, Albrecht P, Gelfand JM, Cruz-Herranz A, Cordano C, Alavi MV, Leslie S, Seeley WW, Miller BL, Bigio E, Mesulam MM, Bogyo MS, Mackenzie IR, Staropoli JF, Cotman SL, Huang EJ, Gan L & Green AJ (2017). Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gopalakrishnan MM, Grosch HW, Locatelli-Hoops S, Werth N, Smolenova E, Nettersheim M, Sandhoff K & Hasilik A (2004). Purified recombinant human prosaposin forms oligomers that bind procathepsin D and affect its autoactivation. Biochem J 383, 507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Brien JS & Kishimoto Y (1991). Saposin proteins: structure, function, and role in human lysosomal storage disorders. FASEB J 5, 301–8. [DOI] [PubMed] [Google Scholar]
- 48.Nicholson AM, Finch NA, Almeida M, Perkerson RB, van Blitterswijk M, Wojtas A, Cenik B, Rotondo S, Inskeep V, Almasy L, Dyer T, Peralta J, Jun G, Wood AR, Frayling TM, Fuchsberger C, Fowler S, Teslovich TM, Manning AK, Kumar S, Curran J, Lehman D, Abecasis G, Duggirala R, Pottier C, Zahir HA, Crook JE, Karydas A, Mitic L, Sun Y, Dickson DW, Bu G, Herz J, Yu G, Miller BL, Ferguson S, Petersen RC, Graff-Radford N, Blangero J & Rademakers R (2016). Prosaposin is a regulator of progranulin levels and oligomerization. Nat Commun 7, 11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X, Sun L, Bastos de Oliveira F, Qi X, Brown WJ, Smolka MB, Sun Y & Hu F (2015). Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J Cell Biol 210, 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tolkatchev D, Malik S, Vinogradova A, Wang P, Chen Z, Xu P, Bennett HP, Bateman A & Ni F (2008). Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities. Protein Sci 17, 711–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tolkatchev D, Ng A, Vranken W & Ni F (2000). Design and solution structure of a well-folded stack of two beta-hairpins based on the amino-terminal fragment of human granulin A. Biochemistry 39, 2878–86. [DOI] [PubMed] [Google Scholar]
- 52.Almeida PC, Nantes IL, Chagas JR, Rizzi CC, Faljoni-Alario A, Carmona E, Juliano L, Nader HB & Tersariol IL (2001). Cathepsin B activity regulation. Heparin-like glycosaminogylcans protect human cathepsin B from alkaline pH-induced inactivation. J Biol Chem 276, 944–51. [DOI] [PubMed] [Google Scholar]
- 53.Rozman J, Stojan J, Kuhelj R, Turk V & Turk B (1999). Autocatalytic processing of recombinant human procathepsin B is a bimolecular process. FEBS Lett 459, 358–62. [DOI] [PubMed] [Google Scholar]
- 54.Yasuda Y, Li Z, Greenbaum D, Bogyo M, Weber E & Bromme D (2004). Cathepsin V, a novel and potent elastolytic activity expressed in activated macrophages. J Biol Chem 279, 36761–70. [DOI] [PubMed] [Google Scholar]
- 55.Fortenberry SC, Schorey JS & Chirgwin JM (1995). Role of glycosylation in the expression of human procathepsin D. J Cell Sci 108 ( Pt 5), 2001–6. [DOI] [PubMed] [Google Scholar]
- 56.von Figura K & Hasilik A (1986). Lysosomal enzymes and their receptors. Annu Rev Biochem 55, 167–93. [DOI] [PubMed] [Google Scholar]
- 57.Gotzl JK, Colombo AV, Fellerer K, Reifschneider A, Werner G, Tahirovic S, Haass C & Capell A (2018). Early lysosomal maturation deficits in microglia triggers enhanced lysosomal activity in other brain cells of progranulin knockout mice. Mol Neurodegener 13, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gotzl JK, Mori K, Damme M, Fellerer K, Tahirovic S, Kleinberger G, Janssens J, van der Zee J, Lang CM, Kremmer E, Martin JJ, Engelborghs S, Kretzschmar HA, Arzberger T, Van Broeckhoven C, Haass C & Capell A (2014). Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol 127, 845–60. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka Y, Suzuki G, Matsuwaki T, Hosokawa M, Serrano G, Beach TG, Yamanouchi K, Hasegawa M & Nishihara M (2017). Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum Mol Genet. [DOI] [PubMed] [Google Scholar]
- 60.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E & Ballabio A (2009). A gene network regulating lysosomal biogenesis and function. Science 325, 473–7. [DOI] [PubMed] [Google Scholar]
- 61.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC & Ballabio A (2011). TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivry SL, Sharib JM, Dominguez DA, Roy N, Hatcher SE, Yip-Schneider MT, Schmidt CM, Brand RE, Park WG, Hebrok M, Kim GE, O'Donoghue AJ, Kirkwood KS & Craik CS (2017). Global Protease Activity Profiling Provides Differential Diagnosis of Pancreatic Cysts. Clin Cancer Res 23, 4865–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.