Abstract
BACKGROUND:
Previous research on autism risk (ASD), developmental regulatory (DevReg), and central nervous system (CNS) genes suggests they tend to be large in size, enriched in nested repeats, and mutation intolerant. The relevance of these genomic features is intriguing yet poorly understood.
OBJECTIVE:
In this study, we investigated the feature landscape of these gene groups to discover structural themes useful in interpreting their function, developmental patterns, and evolutionary history.
METHODS:
ASD, DevReg, CNS, housekeeping, and whole genome control (WGC) groups were compiled using various resources. Multiple gene features of interest were extracted from NCBI/UCSC Bioinformatics. Residual Variation Intolerance Scores (RVIS), Exome Aggregation Consortium pLI (ExAC pLI) scores, and copy number variation data from Decipher were used to estimate variation intolerance. Gene age and protein-protein interactions (PPI) were estimated using Ensembl and EBI Intact databases respectively.
RESULTS:
Compared to WGC, ASD, DevReg, and CNS genes are longer, produce larger proteins, maintain greater numbers/density of conserved noncoding elements (CNE) and transposable elements (TE), produce more transcript variants, and are comparatively variation intolerant. After controlling for gene size, mutation tolerance, and clinical association, ASD genes still retain many of these same features. In addition, we also found that ASD genes that are extremely mutation intolerant have larger PPI networks.
CONCLUSIONS:
These data support many of the recent findings within the field of autism genetics but also expand our understanding of the evolution of these broad gene groups, their potential regulatory complexity, and the extent to which they interact with the cellular network.
Keywords: retroelements, DNA transposons, genes, developmental, central nervous system
LAY SUMMARY
Autism risk genes are more ancient compared to other genes in the genome. As such, they exhibit physical features related to their age, including long gene and protein size and regulatory sequences that help to control gene expression. They share many of these same features with other genes that are expressed in the brain and/or are associated with prenatal development.
INTRODUCTION
Most genes within the human genome carry the impression of hundreds of millions of years of evolutionary history. Features such as the basic structure, size, and component parts of a gene all provide a record reflective of function and the selective forces driving that gene over evolutionary timescales. This record can help us clarify species relatedness, better understand gene regulation, and even predict probable mutation patterns [Nikaido et al., 2001; Sironi et al., 2005a; De Smith et al., 2008]. Though early Drosophila researchers originally defined the gene as the smallest unit of inheritance, it is nevertheless a gestalt of numerous component parts, which are themselves subject to selective forces [Portin & Wilkins, 2017].
Previous research suggests that genes associated with autism spectrum disorder (ASD) maintain structural features that may be useful in understanding their functions, mutation propensity, and evolutionary histories. For instance, King et al. [2013] reported that the inhibition of topoisomerase 1 specifically reduced the expression of extremely long genes (200,000+ bp) within human neurons. In particular, they found this group was enriched in autism risk genes, suggesting ASD genes are longer than expected. That same research group went on to study the effects of topoisomerase inhibition on synaptic genes, many of which overlap autism risk, finding that synaptic protein expression and synaptic transmission were significantly impaired [Mabb et al., 2014]. Many of the long neuronal genes implicated in ASD are downstream targets of risk genes such as the Rett’s-linked, MECP2, and Fragile X syndrome-associated, FMR1, both of which have recently been identified by our group as major hubs linking sizable gene clusters associated with syndromic autism [Gabel et al., 2015; Ouwenga et al., 2015; Casanova et al., 2018].
Surprisingly, in contrast to reports such as King et al. [2013], Krishnan et al. [2016] found that likely gene disrupting (LGD) mutations do not disproportionately target long, brain-expressed genes, suggesting conflicting evidence that we hope to address with the current work.
Long genes, particularly those involved in the central nervous system and developmental regulation, are enriched in conserved noncoding elements (CNE) [Sironi et al., 2005a]. What is more, the proteins of genes with high CNE density are more highly conserved (i.e., mutation intolerant) across species and are functionally complex, the latter a feature characteristic of long ancient genes that form major hubs in the eukaryotic genome [Ekman et al., 2006]. The size overlap between these genes and those of ASD risk genes suggest the latter may maintain many of the same features, a hypothesis that the following work supports.
Here we argue that understanding a gene’s structure and deep history provides context within which human development and related pathologies can be viewed. For instance, the realization that overall gene size shares links with both gene function and regulatory complexity allows us to predict that long genes, such as those associated with ASD, may contain a plethora of CNE that are potential targets for deleterious mutations [Sironi et al., 2005a; King et al., 2013]. In fact, scientists have recently found that some autistic probands exhibit enrichment of de novo mutations within putative regulatory sites in and near recognized major effect genes [Turner et al., 2016; Takata et al., 2016; Williams et al., 2018]. Short et al. [2018] have also estimated that within neurodevelopmental patient groups without diagnostic coding variants, 1–3% harbor de novo mutations in regulatory elements that are involved in fetal brain development. Only a small percentage (0.15%) of these mutations behave in a dominant fashion, suggesting comparatively lower penetrance and etiological complexity.
The integration of evolutionary theory into autism genomics can complement modern clinical research, enriching our understanding of these genes and the selective forces that drive change (or stasis) within them. As we will show in the following material, ASD, central nervous system, and developmental regulatory genes all maintain a similar feature landscape, which provides important clues as to their evolutionary histories, their functions (both as individual gene products and as cogs within the larger cellular network), and their propensities for mutation. It is our hope that this area of research may eventually yield context from which one can make clinically relevant predictions about the genome and human development.
METHODS
Data Compilation
Gene, transcript, and protein sequence data were extracted from NCBI (ftp://ftp.ncbi.nlm.nih.gov) and from UCSC Genome Bioinformatics [Karolchik et al., 2004]. A gene was included in the study if and only if it:
was attested in both databases,
was present in human reference genome (hg19/GRCh37),
had at least one transcript with a validated reference sequence,
and that transcript coded for a protein product.
Protein-coding genes were determined from the knownGenePep annotation on UCSC Genome Browser and therefore includes also predicted proteins.
The initial gene set for the whole genome control (WGC) was composed of all genes fulfilling the above criteria (N = 19,015 genes). The ASD gene set was compiled using a combination of the SFARI database (categories 1–2 and syndromic ratings) and the data described in Casanova et al. [2016], the latter comprising a collection of largely syndromic major effect genes associated with autism (N = 157 genes) [Abrahams et al., 2013]. Comparison intellectual disability (ID) genes (unassociated with autism) were borrowed from Casanova et al. [2016, 2018] (N = 152). All genes even weakly associated with autism, including single case reports, have been removed providing a “pure” list of ID genes (Supplementary Material 1, tab “ID_genes”). Syndromic and nonsyndromic gene subgroups were compiled according to SFARI annotation (category 1–2, syndromic). In addition, genes that were reported by Casanova et al. [2016] that were not already rated “syndromic” by SFARI (or were not included in the 1–2 categories) were placed in the syndromic subgroup since all of these genes are strongly associated with autism-linked genetic syndromes (syndromic N = 119, nonsyndromic N = 38) (Supplementary Materials 1, tab “SyndromicVsNonsyndromic”).
Meanwhile, a developmental regulatory (DevReg) gene set was compiled using the Gene Ontology (GO) term, Regulation of Developmental Process (GO: 0050793; N = 2,175 genes) [Gene Ontology Consortium, 2015]. Likewise, the central nervous system (CNS) gene set was derived from GO terms beginning with “central nervous system” (set C) or were categorized under terms that were subsumed under the same (N = 790 genes). And, finally, the housekeeping (HK) gene list was borrowed from Eisenberg and Levanon [2013] (N = 3,804 genes). In total, there were five overlapping preliminary gene groups: WGC, ASD, DevReg, CNS, and HK. Analyses were duplicated using both overlapping and non-overlapping gene sets, the latter to ensure reproducibility of the findings (Table 1).
Table 1. Gene Groups & Frequencies.
A list of non-overlapping gene groups and their frequencies.
| Gene Types (non-overlapping) | Frequency | Percent |
|---|---|---|
| ASD | 71 | 0.37 |
| ASD + CNS | 14 | 0.07 |
| ASD + CNS + HK | 2 | 0.01 |
| ASD + DevReg | 21 | 0.11 |
| ASD + DevReg + CNS | 14 | 0.07 |
| ASD + DevReg + CNS + HK | 5 | 0.03 |
| ASD + DevReg + HK | 5 | 0.03 |
| ASD + HK | 25 | 0.13 |
| CNS | 369 | 1.94 |
| CNS + HK | 74 | 0.39 |
| DevReg | 1,487 | 7.82 |
| DevReg + CNS | 313 | 1.65 |
| DevReg + CNS + HK | 38 | 0.20 |
| DevReg + HK | 292 | 1.54 |
| HK | 3,360 | 17.67 |
| WGC | 12,925 | 67.97 |
All genes were identified internally by NCBI Gene ID. Quantities derived from these databases included natural log gene length, number of transcripts (considering only transcripts with a validated reference sequence), and natural log of protein length coded by the canonical (longest) transcript. Identification of intronic TE was performed using the UCSC RepeatMasker [Karolchik et al., 2004]. Intergenic, exonic, and promoter TEs and non-TE repeat classes, such as microsatellites, and unclassified repeats were excluded due to small numbers and different selective pressures on insertion and retention. For TE-specific analyses, genes with a TE count of zero were also removed. Meanwhile, intronic CNE were abstracted from the Multiz alignment of hg19 and 99 other vertebrate genomes [Blanchette et al., 2004]. Following Sironi et al. [2005a], we considered only eutherian [alignment of hg19 and mm10 (Mus musculus)] downstream intronic sequences. The first intron was discarded as per Sironi et al. [2005a], as the presence of increased regulatory content is well recognized within these gene regions. Allelic tolerance was estimated using the Residual Variation Intolerance Score (RVIS) developed by Petrovski et al. [2013] and Exome Aggregation Consortium (ExAC) pLI scores were acquired from the ExAC Database for additional exploratory analyses concerning loss-of-function (LOF) tolerance [Lek et al., 2016].
In order to identify protein-protein interactions (PPI), all genes were queried against the EBI Intact database (https://www.ebi.ac.uk/intact; PMID:24234451) on 3/12/2019. Only unique human (txid9606) interactors classified as “physical association” or “direct interaction” with an intact-miscore greater than or equal to 0.5 were counted [Orchard et al., 2013]. In terms of general gene age, Ensembl was used to estimate the oldest known eukaryotic homolog via the gene tree feature [Herrero et al., 2016]. All genes were then assigned to one of three nominal groups relative to their oldest known eukaryotic homologs: 1) “single-celled eukaryotes,” 2) “bilateria,” and 3) “chordates and younger.”
Finally, we performed an analysis to determine to what extent our experimental gene groups fell near or within common and rare copy number variations (CNV). Both overlapping and non-overlapping versions of the experimental gene groups were assessed. CNV and their general population frequencies were pulled from Decipher (https://decipher.sanger.ac.uk/about#downloads/data) [Firth et al., 2009]. The data included the CNV, its chromosomal location, and the number of deletion/duplication observations and population frequency. Deletions/duplications whose frequency was greater than zero but less than or equal to 1% were considered “rare” (deletion N = 20,808; duplication N = 13,206). Deletions/duplications whose population frequency was greater than 1% were labeled “common” (deletion N = 21,691; duplication N = 6,355). Because the original gene list used within this study had a build of GRCh37 and Decipher used GRCh38, genes that were not included in both builds were dropped from the analysis (see Supplementary Materials 1, Tab “CNV”). Size-matched controls were generated for each experimental gene list according to size. Each experimental gene was randomly paired with a control gene whose gene size fell within ± 10% of the experimental gene and likewise did not overlap any of the other groups. Each subsequent control gene was removed from the remaining list of potential genes. The potential control list was reset after the completion of each list, such that duplicates may exist across size-matched control lists. (This same method was used to build the ExAC pLI-matched controls (± 10% of the pLI score) used for other analyses.)
Statistical Analyses
Several sets of analyses were performed. The first analysis using WGC involves mutually exclusive gene sets (Table 1) and was analyzed using one-way ANOVAs. In the second, each of our overlapping gene sets of interest (ASD, DevReg, CNS, HK) was compared to WGC using two-sample t-tests (gene and protein lengths) and two-sample Wilcoxon rank sum tests (transcript number, total/relative TE, total/relative CNE, and RVIS). This set of data analyses were conducted in R Statistical Software and significance was set at α = 0.05.
The second set of analyses utilized both size- and ExAC pLI-matched control genes for each of the experimental gene lists. Once again this was performed on both overlapping and mutually exclusive gene sets. In addition, two-sample t-tests (gene and protein lengths) and two-sample Wilcoxon rank sum tests (transcript number, total/relative TE, total/relative CNE, RVIS, PPI, ExAC pLI, and homolog age) were again used and analyzed via JASP (α = 0.05).
CNV data were analyzed using a Poisson distribution (Decipher cohort N = 28,428).
RESULTS
Group Differences in the Gene Landscape
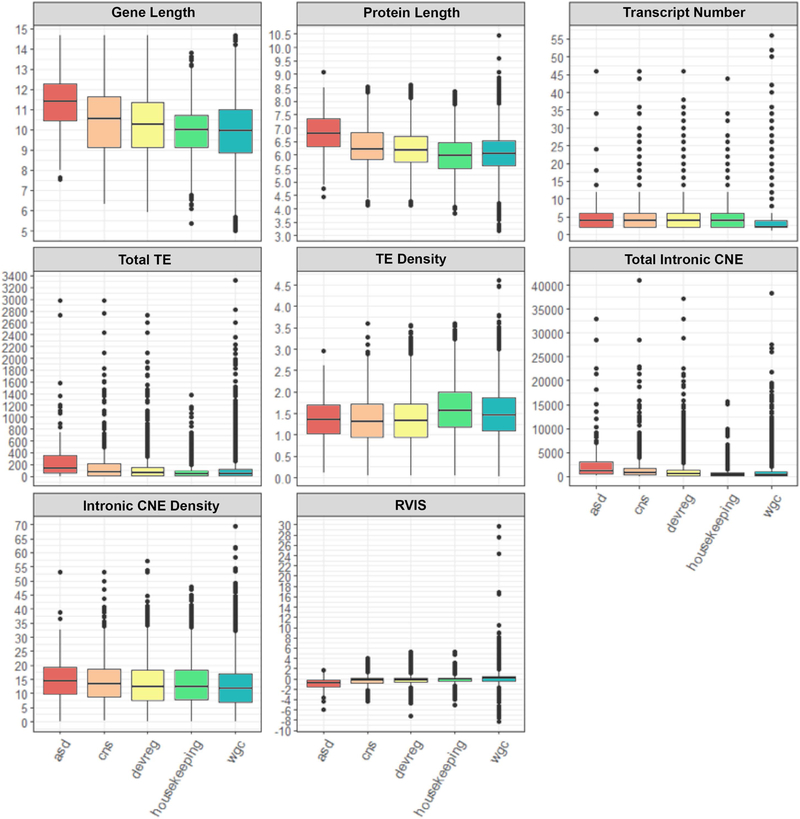
The distribution of gene and amino acid lengths differed significantly across groups, the former having been reported previously by King et al. [2013] with respect to autism risk genes (F scores between 37.5374 and 67.0384, p < 0.001) (Fig. 1, Table 2). Post hoc analysis utilizing two-sample t-tests indicated that ASD, DevReg, and CNS genes and proteins were each significantly longer than those of WGC (ASD/DevReg/CNS all p < 0.0001), while HK genes and proteins were significantly shorter (gene p = 0.0151, protein p < 0.0001). In addition, all experimental gene groups, including HK genes, appear to produce more transcript variants than WGC, suggesting potential regulatory complexity within these functional and clinical groups (ASD p = 0.0015, DevReg p = 0.0015, CNS p < 0.0001, HK p < 0.0001). It should be noted, as with most of the data that will be presented here, there was considerable overlap across groups. Therefore, although the experimental gene groups are larger/smaller on average and produce varying numbers of transcripts, not all associated genes differ dramatically from the controls. The standard deviations presented within Table 2 attest to that fact.
Figure 1.
Boxplots illustrating the distribution for variables of interest across the different gene groups. (Gene and protein lengths represent the logarithmic value rather than absolute lengths.) Abbreviations: autism spectrum disorder (ASD), central nervous system (CNS), developmental regulatory (devreg), whole genome control (WGC), transposable elements (TE), conserved noncoding elements (CNE), residual variance intolerance score (RVIS).
Table 2. Gene Feature Means & Standard Deviations.
Means and standard deviations of gene features of interest across non-overlapping gene groups. (CNE = intronic conserved noncoding elements; TE = intronic transposable elements; RVIS = residual variation intolerance score; bp = base pairs; aa = amino acids.)
| Gene Group | Gene Length | Protein Length | Transcript Number | Total CNE/gene | CNE Density | Total TE/gene | TE Density | RVIS |
|---|---|---|---|---|---|---|---|---|
| Autism | 210,984 bp (± 251,959 bp) | 1,485 aa (±1,395 aa) | 5.1 (± 5.6) | 2,865 (± 3,791) | 13.45 (± 7.73) | 285 (± 327) | 1.4 (± 0.5) | −1.0572 (± 1.2955) |
| Developmental Regulatory (DevReg) | 72,405 bp (± 148,533 bp) | 640 aa (± 567 aa) | 4.6 (± 4.1) | 881 (± 2,162) | 10.34 (± 8.99) | 101 (± 195) | 1.2 (± 0.7) | −0.2051 (± 0.9314) |
| Central Nervous System (CNS) | 97,375 bp (± 191,505 bp) | 765 aa (± 813 aa) | 4.4 (± 4.1) | 1,263 (± 3,135) | 10.67 (± 9.60) | 130 (± 246) | 1.1 (± 0.7) | −0.3133 (± 0.9577) |
| Housekeeping (HK) | 38,505 bp (± 59,367 bp) | 479 aa (± 363 aa) | 4.3 (± 3.2) | 452 (± 817) | 11.46 (± 8.60) | 64 (± 96) | 1.5 (± 0.7) | −0.1678 (± 0.6164) |
| Whole Genome Control (WGC) | 58,465 bp (± 115,992 bp) | 575 aa (± 633 aa) | 3.8 (± 3.2) | 646 (± 1,512) | 9.10 (± 8.59) | 88 (± 162) | 1.3 (± 0.8) | 0.1180 (± 1.0840) |
There is evidence that long genes are believed to be regulatorily and functionally complex [Sironi et al., 2005a; Sironi et al., 2005b; Neduva & Russell, 2005]. Potentially underlying some of that complexity are noncoding elements, some of which are conserved or even “ultraconserved” across species, clades, and stem groups [Elgar & Vavouri, 2008; Schwaiger et al., 2014; Polychronopoulos et al., 2017]. Because ASD, DevReg, and CNS genes are on average longer while HK genes tend to be rather short, we studied the number and density of intronic CNE across gene groups. We found that, as expected by gene size, all gene groups significantly differed from one another in both relative and total CNE content: ASD, DevReg, and CNS genes appear to contain greater relative and total CNE content compared to WGC (all p < 0.0001), while total count within HK genes is comparatively low (p < 0.0001). However, relative density of CNE in the introns of HK genes was higher than WGC, suggesting that these genes may be more complex than their size would otherwise indicate—a finding that matches, for instance, the increase in transcript variations derived from these genes.
CNE are often derived from the insertion, retention, and exaptation of transposable elements (TE) [Xie et al., 2006]. We therefore looked at total and relative densities of these elements within the introns of our respective gene groups. Similar to CNE, total TE count was significantly enriched in ASD (p < 0.001), DevReg (p = 0.0067), and CNS gene sets (p < 0.0001), while it was low in HK genes (p < 0.0001). In contrast to CNE, however, relative density of TE was lower in DevReg and CNS genes, higher in HK genes (p < 0.0001), and did not significantly differ in autism genes compared to the other groups (p = 0.4506). Therefore, TE density seems to vary strongly by gene size across gene groups, suggesting that mechanisms peculiar to long genes select against higher density despite greater TE content, potentially adding to the larger gene lengths we see in these functional and clinical groups.
Perhaps counter to intuition, long genes are relatively mutation intolerant [Sironi et al., 2005a; Han et al., 2018]. This propensity is partly mechanistic but also a reflection of gene function [Niu & Yang, 2011]. Given the variation in gene length across the gene groups, we investigated genes’ tolerance to allelic variation using RVIS and found that the RVIS distribution among the groups significantly differed (Kruskal Wallis, x2 = 499.4761, p < 0.0001). As one would expect given their average gene size, ASD, DevReg, and CNS genes were all relatively intolerant to variation compared to WGC. In addition, in spite of their tendency for a small gene size, HK genes were also intolerant—an effect perhaps the result of their basal cellular function and, as will be discussed later, their older ages [Eisenberg & Levanon, 2003].
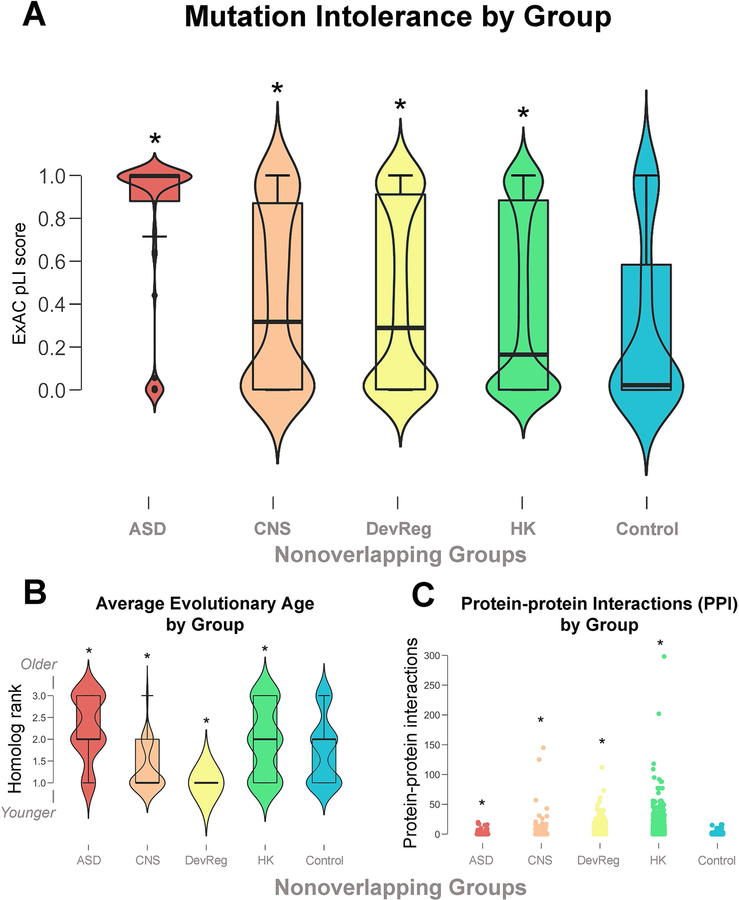
Interestingly, Petrovski et al. [2015] have shown that variation intolerant genes tend to be dosage sensitive, exhibiting LOF mutations primarily only in association with disease states. We found that all gene groups, including HK genes, are relatively mutation intolerant (experimental pLI mean = 0.4151–0.7552, control pLI mean = 0.2902, W = 5023.5–214,062, p < 0.0001) (Fig. 2a). This was especially the case for the ASD genes (mean = 0.7552), suggesting unique epigenetic patterning may make these genes particularly vulnerable to LOF mutations. This matches well with our previous findings of autosomal dominant enrichment in genetic syndromes with high rates of autism comorbidity, suggesting a striking pattern of haploinsufficiency in major effect genes [Casanova et al., 2016]. Petrovski et al. [2015] also reported that certain non-coding regions (promoter, 3’ UTR, 5’ UTR) of these same genes are also resistant to allelic variation (ncRVIS). Although we didn’t investigate this aspect of autism risk genes, CNE enrichment in combination with low RVIS/high ExAC pLI scores suggests that select non-coding regions in risk genes may be similarly intolerant and worthy of further study.
Figure 2.
(A) ExAC pLI scores across non-overlapping groups. Higher pLI scores indicate greater sensitivity to loss-of-function mutations. All gene groups, including controls, exhibit an hourglass formation suggesting genes tend to be either very mutation tolerant or intolerant with fewer genes falling intermediate. (B) Average evolutionary age across non-overlapping groups. Both autism spectrum disorder (ASD) and housekeeping (HK) genes appear to be significantly older than central nervous system (CNS) genes, developmental regulatory (DevReg) genes, and controls. (C) Number of protein-protein interactions (PPI) according to non-overlapping groups. All experimental gene groups significantly differed from size- and ExAC pLI-matched controls as indicated with an asterisk.
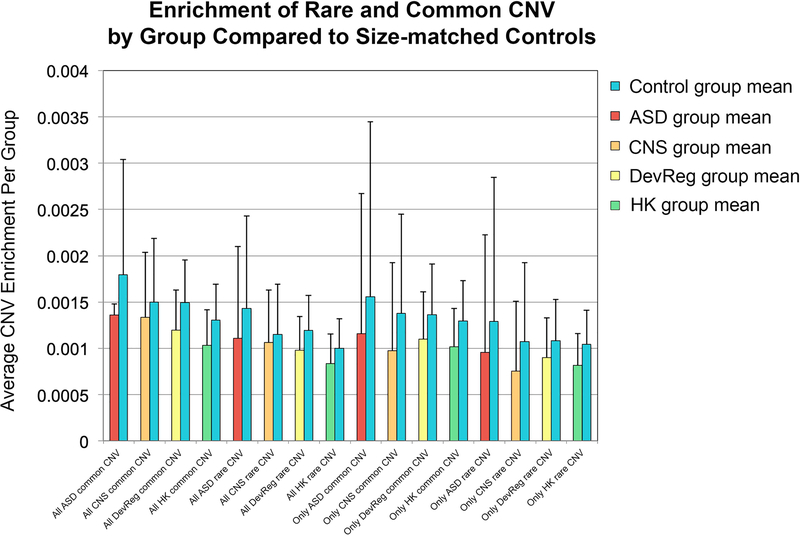
We extended this line of research by investigating overlap of common and rare copy number variants (CNV) with our genes of interest, given that previous studies have reported enrichment for de novo CNV in autism [Sebat et al., 2007]. We found that the average likelihood of rare and common CNVs overlapping size-matched controls is significantly greater than the likelihood of overlap with genes within any of the experimental groups (all p < 0.05) (Fig. 3). In addition, this enrichment was consistent across both “overlapping” and “non-overlapping” experimental gene groups. These results further support overall variation intolerance in the ASD, DevReg, CNS, and HK gene groups. (See Supplementary Materials 1, Tabs “CNV,” “ExAC_pLI,” and “RVIS” for full statistical results).
Figure 3.
Extent of overlap between the gene groups of interest (overlapping and non-overlapping) with rare/common copy number variants (CNV) compared to size-matched controls. All forms of CNV are, on average, comparatively enriched in all size-matched control groups compared to all experimental gene groups. (“All” = all genes within a given group regardless of overlap with other groups; “Only” = only genes within a given group that do not overlap other experimental gene groups; Autism spectrum disorder genes = ASD; central nervous system genes = CNS; developmental regulatory genes = DevReg; housekeeping genes = HK.)
Controlling for Gene Length, Mutation Propensity, & Clinical Association
Because several of our experimental gene groups vary significantly by length as well as mutation propensity, both of which may influence overall gene structure, it is important to control for these confounds (see Supplementary Materials 2, Tab “Genomewide_Correlations”). After doing so, we found that with the exception of TE total/density, ASD genes exhibited similarly significant patterns as before (gene/protein length, transcript number, CNE density, etc.) compared to size- and ExAC pLI-matched controls (p <0.0001–0.0056). In addition, with some minor exceptions, the same pattern of results was also maintained in the other gene groups, indicating that these patterns of structural enrichment cannot solely be explained by gene size and mutation propensity (see Supplementary Materials 1 for full results).
Kosmicki et al. [2017] reported on a subclass of de novo protein truncating variants (PTV) that are significantly enriched in individuals with ASD and which are also highly loss-of-function (LOF) intolerant. In order to further address whether any of the gene features studied here are related to LOF intolerance, we divided the ASD gene list according to those genes that fall above vs. below an ExAC pLI score of 0.9 as per methodology by Kosmicki et al. In doing so, we find that highly mutation intolerant ASD genes produce longer proteins than their more tolerant counterparts (t = −2.6964, p = 0.0156) and maintain a more extensive protein interaction network (Wilcoxon, W = 1,560.5, p = 0.0192). However, they do not differ in other features such as age, transcript number, and CNE and TE densities (all p = 1.000) (see Supplementary Materials 1, tab “ASD_LOF-intol” for full results). While these results are somewhat mixed, it does indicate that some of the features associated with the ASD gene group are not solely related to extreme mutation intolerance.
In order to address clinically-associated confounds, we also compared ASD risk genes to a group of syndromic ID-associated genes (not linked with autism), which we had compiled and reported in a previous study [Casanova et al., 2016, 2018]. We found that gene/protein lengths were significantly longer in the ASD genes compared to the ID genes (p < 0.0001). In addition, ASD genes differed from ID genes in all other respects with the exception of CNE density, TE density, and the number of PPI (dCNE W = 10,702, p = 1.000; dTE W = 11,836, p = 0.4592; PPI W = 9,042.5, p = 1.000). Interestingly, although PPI is more strongly related to pLI scores in the ASD group, PPI and pLI scores do not share the same intensive relationship amongst ID genes, suggesting high PPI may be characteristic of ID in general, regardless mutation tolerance or of the presence of autism.
Gene Age & Network Connectivity
We have previously reported that autism risk genes are, on average, an evolutionarily older class of genes, which partly relates to gene function [Casanova & Casanova, in press; Casanova, in press]. Because older genes experience more gene and protein interactions than younger ones, we hypothesized that ASD genes would follow this same trend [Capra et al., 2013]. In agreement with our previous findings, compared to size- and pLI-matched controls, ASD risk genes appear to be an evolutionarily older class of genes with the average age falling between the evolution of single-celled eukaryotes and the bilaterians (> 530 million years ago) (Wilcoxon, pLI W = 18,618, p < 0.001; size W = 18,933, p < 0.0001) (Fig. 2b). HK genes followed a similar age trend (Wilcoxon, W = 7,011.5, p < 0.0001), although were not quite as ancient as the ASD genes, most evolving during the early bilaterians. However, neither the CNS nor DevReg genes significantly differed from size- and pLI-matched controls in age, most having evolved at the time of the chordates or younger (Wilcoxon, CNS pLI W = 5,063, p = 0.8471, CNS size W = 5,093.5, p = 0.441; DevReg pLI W = 4,824.5, p = 0.4181, DevReg size W = 4,957, p = 0.7818).
As one might expect given their older ages, the protein products of ASD genes directly interact with a wider range of proteins than those of matched controls, suggesting ASD genes maintain a more extensive interaction network than their younger counterparts and may even function as hubs as described by Ekman et al. [2006] (Wilcoxon, pLI W = 12,894.5, p = 0.008; size W = 13,731, p < 0.0001) (Fig. 2c). Interestingly, the other experimental groups likewise exhibited significantly more PPI than size- and pLI-matched controls (all p < 0.0001), suggesting that while age may influence the number of PPI as reported by Capra et al. [2013], factors driving total PPI are highly complex (see Supplementary Materials 1 for full statistical results).
Applicability of Findings to Nonsyndromic Risk Genes
Approximately three-fourths of the risk genes used in this study are syndromically associated, although roughly one-fifth of those also overlap nonsyndromic categories. Given this overrepresentation of syndromic major effect genes, we cannot say whether the gene features identified as unique to this broad group of genes reliably describe the remainder of nonsyndromic minor effect genes [Parikshak et al., 2013]. However, to begin to address this question we compared our features of interest across syndromic and nonsyndromic subgroups. Syndromic and nonsyndromic subgroups did not significantly differ in gene size, protein length, transcript number, CNE density, TE density, variation tolerance (RVIS, ExAC pLI), PPI, or average gene age (all p = 0.1152–1.000). They did, however, differ mildly in total CNE and TE (p = 0.0108–0.0153), yet the relevance of these differences is uncertain. Although these data are preliminary and limited by small group size and do not include true minor effect genes, the results suggest that many of the features that typify the larger ASD gene group may apply to nonsyndromic genes as well. Further research is needed to address this possibility.
DISCUSSION
The majority of ASD genes in this study are of major effect, exhibiting relatively strong penetrance for the autism phenotype [Abrahams et al., 2013; Casanova et al., 2016]. Seventy-one percent of the genes are extremely LOF intolerant (pLI > 0.9) as per Kosmicki et al. [2017], likely driving some of the significant findings reported in this study (e.g., longer proteins, more PPI). Other features, however, do not exhibit as strong a relationship with mutation intolerance, such as gene length, transcript number, CNE/TE density, and gene age.
Given both their phenotypic penetrance and variation intolerance, according to Parikshak et al. [2013] we may expect many of the ASD genes to be expressed during early corticogenesis and enriched in functions such as DNA binding and transcription regulation. This agrees with our earlier study [Casanova et al., 2016], which reported a functional enrichment of epigenetic regulators amongst many of the same genes. These genes are also strongly implicated in autism-associated genetic syndromes accompanied by widespread dysmorphic features, indicating that they play vital roles not just in corticogenesis but morphogenesis in general [Casanova et al., 2018].
The current work indicates that long, ancient genes tend to be variation intolerant, maintain a complex intragenic regulatory network, may be hubs within the cellular network, and are conserved regulators in animalian morphogenesis (including neurogenesis) [Parikshak et al., 2013; Casanova et al., 2018]. Major effect ASD genes epitomize this stereotype, most having evolved from single-celled eukaryotes or early bilaterians more than half a billion years ago, prior to the development of the proto-notochord in early chordates and long predating the central nervous system.
Although we have only begun to address whether the gene features studied here are particular to major effect ASD genes alone or include even minor effect genes, previous work by Krishnan et al. [2016] would suggest the former, finding that LGD variants in ASD do not disproportionately affect long, brain-expressed genes. Our work, however, indicates that major effect ASD genes do maintain some of the features reported here despite controlling for size and mutation rates. Interestingly, work by Krishnan et al. [2016] implies that minor effect genes may ultimately funnel into similar pathways and developmental stages within the brain. Further work is needed to determine if this is the case.
While ASD genes exhibit some featural overlap with CNS and DevReg genes, due most likely to their clinical association they are an extreme example along a broad continuum. Each of these gene groups is larger, produces longer amino acid sequences, exhibits an increased intronic density of CNE/TE, produces more transcripts, is more mutation intolerant, and maintains a larger PPI network than size-matched, pLI-matched, and whole genome controls. HK genes on the other hand are small compared to the average gene size and produce smaller proteins, yet are relatively mutation intolerant, produce more transcript variants, and exhibit increased CNE/TE density. In addition, similar to ASD genes, HK genes are ancient, having on average arisen during the early bilaterians. Unlike ASD genes, which typically exhibit tissue-specific expression patterns, HK genes are constitutively expressed across tissue types and are typically involved in more general metabolic processes [Eisenberg & Levanon, 2013].
The work presented here provides a brief glimpse into the evolutionary history of these special gene groups, particularly those associated with ASD. Their unique yet overlapping feature landscape affords a record from which we may view that history, as well as understand functional and mutational patterns relevant to research today. That knowledge, in turn, may provide the researcher with a significant advantage when designing clinically-relevant studies.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Autism Research Institute (ARI) and the NIH (R01 HD-65279). We would like to thank both agencies for their financial support. We would also like to thank the director of the ARI, Dr. Stephen Edelson.
Footnotes
CONFLICT OF INTEREST STATEMENT
All authors declare no biomedical financial interests or potential conflicts of interest.
Availability of materials
All raw data and supplementary materials are available at https://data.mendeley.com/datasets/sjctrd3r2k/1 (doi: 10.17632/sjctrd3r2k.1).
References
- Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, … Packer A (2013). SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Molecular Autism, 4, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AFA, Roskin KM, … Miller W (2004). Aligning multiple genomic sequences with the threaded blockset aligner. Genome Research 14: 708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Stolzer M, Durand D, & Pollard KS. (2013). How old is my gene? Trends in Genetics, 29, 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova EL Endless genes most beautiful: How molecular function follows form. Inference, in press.
- Casanova EL, & Casanova MF Evolution of autism risk genes. (Sokhadze T and Casanova MF, Eds.) In: Neuromodulation, Neurofeedback and Sensory Integration Approaches for Research and Treatment FNNR Publishers, in press. [Google Scholar]
- Casanova EL, Gerstner Z, Sharp JL, Casanova MF, & Feltus FA (2018). Widespread genotype-phenotype correlations in intellectual disability. Frontiers in Psychiatry, 9, e535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova EL, Sharp JL, Chakraborty H, Sumi NS, & Casanova MF (2016). Genes with high penetrance for syndromic and nonsyndromic autism typically function within the nucleus and regulate gene expression. Molecular Autism, 7, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smith AJ, Walters RG, Coin LJ, Steinfeld I, Yakhini Z, Sladek R, … Blakemore AI (2008). Small deletion variants have stable breakpoints commonly associated with alu elements. PLoS One, 3, e3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, & Levanon EY (2003) Human housekeeping genes are compact. Trends in Genetics, 19, 362–365. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, & Levanon EY (2013). Human housekeeping genes, revisited. Cell, 29, 569–574. [DOI] [PubMed] [Google Scholar]
- Ekman D, Light S, Björklund Å, & Elofsson A (2006). What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biology, 7, R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgar G, & Vavouri T (2008). Tuning in to the signals: Noncoding sequence conservation in vertebrate genomes. Trends in Genetics, 24, 344–352. [DOI] [PubMed] [Google Scholar]
- Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, … Carter NP (2009). DECIPHER: Database of chromosomal imbalance and phenotype in humans using ensemble resources. The American Journal of Human Genetics, 84, 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, … Greenberg ME (2015). Disruption of DNA-methylation-dependent long gene regression in Rett syndrome. Nature, 522, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium, The (2015). Gene Ontology Consortium: Going forward. Nucleic Acids Research, 43, D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero J, Muffato M, Beal K, Fitzgerald S, Gordon L, Pignatelli M, … Flicek P (2016). Ensembl comparative genomics resources. Database (Oxford), pii, bav096. [DOI] [PMC free article] [PubMed]
- Karolchik D, Hinrichs AS, Furye TS, Roskin KM, Sugnet CW, Haussler D, … Kent WJ (2004). The UCSC Table Browser data retrieval tool. Nucleic Acids Research, 32, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IF, Yandava CN, Mabb AM, Hsiao JS, Huang HS, Pearson BL, … Zylka MJ (2013). Topoisomerase facilitates transcription of long genes linked with autism. Nature, 501, 58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmicki JA, Samocha KE, Howrigan DP, Sanders SJ, Slowikowski K, Lek M, … Daly MJ (2017). Refining the role of de novo protein truncating variants in neurodevelopmental disorders using population reference samples. Nature Genetics, 49, 504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A, Zhang R, Yao V, Theesfeld CL, Wong AK, Tadych A, … Troyanskaya OG (2016). Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nature Neuroscience, 19, 1454–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, … Exome Aggregation Consortium. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabb AM, Kullman PH, Twomey MA, Miriyala J, Philpot BD, Zylka MJ (2014). Topoisomerase 1 inhibition reversibly impairs synaptic function. Proceedings of the National Academy of Sciences, 111, 17290–17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neduva V, & Russell RB (2005). Linear motifs: Evolutionary interaction switches. FEBS Letters, 579, 3342–3345. [DOI] [PubMed] [Google Scholar]
- Nikaido M, Matsuno F, Hamilton H, Brownell RL Jr, Cao Y, Din W, … Okada N (2001). Retrotransposon analysis of major cetacean lineages: The monophyly of toothed whales and the paraphyly of river dolphins. Proceedings of the National Academy of Sciences USA, 98, 7384–7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu D-K, & Yang Y-F (2011). Why eukaryotic cells use introns to ehance gene expression: Splicing reduces transcription-associated mutagenesis by inhibiting topoisomerase I cutting activity. Biology Direct, 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broaches-Carter F, … Duesbury M (2013). The MIntAct project—IntAct as a common curation platform for 11 molecular interaction. Nucleic Acids Research, 42, D358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwenga RL, Dougherty J (2015). Fmrp targets or not: Long, highly brain-expressed genes tend to be implicated in autism and brain disorders. Molecular Autism, 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, … Geschwind DH (2013). Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell, 155, 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovski S, Gussow AB, Wang Q, Halvorsen M, Han Y, Weir WH, … Goldstein DB (2015). The intolerance of regulatory sequence to genetic variation predicts gene dosage sensitivity. PLoS Genetics, 11, e1005492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovski S, Wang Q, Heinzen EL, Allen AS, & Goldstein DB (2013). Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genetics, 9, e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polychronopoulos D, King JWD, Nash AJ, Tan G, & Lenhard B (2017). Conserved non-coding elements: Developmental gene regulation meets genome organization. Nulceic Acids Research, 45, 12611–12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portin P, & Wilkins A (2017). The evolving definition of the term “gene.” Genetics, 205, 1353–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Schönauer A, Rendeiro AF, Pribitzer C, Schauer A, Gilles AF, … Technau U (2014). Evolutionary conservation of the eumetazoan gene regulatory landscape. Genome Research, 24, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, … Leotta A (2007). Strong association of de novo copy number variations with autism. Science, 316, 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short PJ, McRae JF, Gallone G, Sifrim A, Won H, Geschwind DH, … Hurles ME (2018). De novo mutations in regulatory elements in neurodevelopmental disorders. Nature, 555, 611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M, Menozzi G, Comi GP, Cagliani R, Bresolin N, & Pozzoli U (2005a). Analysis of intronic conserved elements indicates that functional complexity might represent a major source of negative selection on non-coding sequences. Human Molecular Genetics, 14, 2533–2546. [DOI] [PubMed] [Google Scholar]
- Sironi M, Menozzi G, Comi GP, Bresolin N, Cagliani R, & Pozzoli U (2005b). Fixation of conserved sequences shapes human intron size and influences transposon-insertion dynamics. Trends in Genetics, 21, 484–488. [DOI] [PubMed] [Google Scholar]
- Takata A, Ionita-Laza I, Gogos JA, Xu B, Karayiorgou M (2016). De novo synonymous mutations in regulatory elements contribute to the genetic etiology of autism and schizophrenia. Neuron, 89, 940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TN, Hormozdiari F, Duyzend MH, McClymont SA, Hook PW, Iossifov I, … Eichler EE (2016). Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. American Journal of Human Genetics, 98, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SM, An JY, Edson J, Watts M, Murigneux V, Whitehouse AJ, … Claudianos C (2018). An integrative analysis of non-coding regulatory DNA variations associated with autism spectrum disorder. Molecular Psychiatry, in press. [DOI] [PubMed]
- Xie X, Kamal M, & Lander ES (2006). A family of conserved noncoding elements derived from an ancient transposable element. Proceedings of the National Academy of Sciences, 103, 11659–11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.