Abstract
We combined behavioral survey data from the HIV Prevention Trials Network 068 study with phylogenetic information to determine if cluster membership was associated with characteristics of young women and their partners. Clusters were more likely to involve young women from specific villages and schools, indicating some localized transmission.
Keywords: HIV transmission, Phylogenetic analysis, Adolescent Girls and Young Women, South Africa
Summary
We identified HIV phylogenetic clustering among young women in South Africa and determined that viral cluster membership was associated with village, school and wealth, but not with other characteristics.
Introduction
Successful prevention of HIV transmission requires in-depth knowledge of local HIV epidemics, including transmission patterns among high-risk groups. Even within generalized epidemics in Southern Africa, some sub-groups are disproportionately affected by a high risk of HIV acquisition. Such is the case for young women in rural South Africa, where HIV prevalence is 3–4 times higher than similarly aged young men.1 In addition, migration is prevalent in Southern Africa, including in our study site in the rural northeast region near the Mozambique border.2 Characteristics such as migration for work and contact with older partners may affect HIV acquisition among young women, but the contribution of these factors to local HIV transmission networks is uncertain.3,4
Phylogenetic analysis provides information about HIV genetic networks and can identify putative transmission chains involving more than one young woman through shared sexual partners.5 Such analyses can help assess if HIV transmission occurs in social circles, through age-disparate relationships, in certain geographic areas, or through extensive migration of male partners. We previously characterized viral clustering among young women in South Africa, finding high level of HIV diversity, suggesting that migration is an important contributor to HIV transmission in rural South Africa6; however, behavioral factors were not assessed. In this report, we evaluated socio-behavioral associations with cluster membership including newly detected HIV infections from a follow-up visit. Socio-behavioral information included measures of migration, partner characteristics, and geographic residence.
Methods
Study population
We analyzed samples and data from the HIV Prevention Trials Network (HPTN) 068 study, a randomized trial where young women and their households were given a cash transfer to reduce HIV acquisition. The study enrolled 2,533 young women aged 13 to 20 years who were unmarried, not pregnant, and attending high school grades 8 to 11 in the Bushbuckridge sub-district of Mpumalanga province, South Africa. Young women enrolled in the study were seen annually from 2011 to 2015 until study completion or they graduated from high school. Annual study visits included an Audio Computer-Assisted Self-Interview (ACASI) with young women and HIV testing for the young women who were HIV-uninfected at the previous visit. More information on the study design and main trial results are reported elsewhere.7,8 A post-intervention follow-up visit was also conducted one to two years after the young women exited the main trial (2015–2017).
During the study period, 288 participants acquired HIV infection. Genotyping was successful for 231 (80.2%) of the total 288 HIV cases. Our previous report included analysis of HIV sequences from 201 women (68 infected at enrollment, 92 infected in the main study, 41 infected in the follow-up study).6 This report includes sequences from 30 additional women who seroconverted in the follow-up study. The annual ACASI survey collected self-reported information on demographics, risk behaviors, and partner characteristics. As proxies for migration, we examined self-reported characteristics including young women’s report of moving in the last 12 months or having slept away from home at least once a week, and having a partner that lives outside the village or province. For baseline and post-intervention visits, we used survey information from the same visit; for incident infections, we used survey information collected at the visit prior to HIV diagnosis. Among girls who were members of a cluster, all girls had complete school information but 8/51 were out of school at the time of infection.
Phylogenetic analysis
HIV pol sequences from study participants were aligned to HXB2 and were manually edited with stripping of gapped positions.11 Sequences with a high fraction of ambiguous nucleotides (>5%) were excluded. We assessed regional diversity by evaluating clades that involved sequences sampled outside of South Africa. We conducted a BLAST search to identify the 10 sequences in the Los Alamos National Laboratory (LANL) HIV database (http://www.hiv.lanl.gov) that were most closely related to each study sequence; only sequences in LANL with known locations were used in the phylogenetic analysis. A maximum-likelihood (ML) tree was constructed in PhyML3.0 with the general time-reversible model of nucleotide substitution to evaluate clades involving study sequences.12 Statistical support of clades was assessed with local support values (Shimodaira-Hasegawa-like test [SH-test]). We defined closely related clusters as clades involving ≥2 study sequences with pairwise divergence of ≤0.020 nucleotide substitutions to another study sequence,13,14 which is <0.3% quantile of all pairwise comparisons between sequences (Figure 2 and 3, Supplementary Content). Clusters were visualized in R using the iGraph Package.15 Pairwise genetic distances were evaluated with the Ape Package using the Tamura-Nei 93-substitution model.
Statistical analysis of characteristics
We examined descriptive characteristics of HIV-infected young women in a cluster versus not and, among those in a cluster, characteristics of cluster membership. We used a chi square test to test for differences between groups. For continuous variables, we calculated the intraclass correlation statistic as a measure of cluster membership. For categorical measures, we estimated the probability of having the same value for a randomly-chosen pair of girls in the same cluster, over the probability of having the same value for a randomly-chosen pair of girls overall. In the case of no association with cluster membership, we expected this ratio to be 1; an association with cluster membership would lead to a number greater than 1. Confidence intervals were calculated using the bootstrap standard deviation from 200 full samples (with replacement) from the observed data.16,17
Formula: Cluster membership statistic = Pcluster/Poverall where
where N = number of girls, nclust =number of clusters, mh = size of cluster h, Yi = outcome (school/village) for girl i and I(Yi = Yj) is equal to 1 if Yi = Yj and 0 otherwise.
Findings
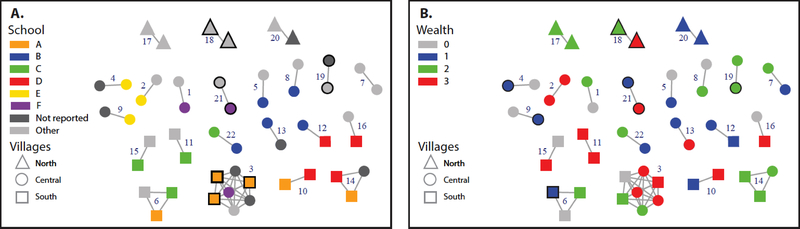
Of 231 (80.2%) participants with HIV sequence data, 228 were included in the phylogenetic analysis. Three sequences had ambiguity fractions >5% and were excluded. Among study sequences, nearly all (227/228) were subtype C; one was subtype A. There were 841 unique references identified by the BLAST search. In the ML tree, a high degree of viral diversity was noted, including within clades containing only South African sequences (Figure 1, Supplementary Content). While the majority of sequences in all clades were from South Africa, several clades included a high proportion of sequences from Botswana. In total, 22% (51/228) of sequences were identified in 22 clusters based on ≤2% pairwise divergence from at least one other study sequence (Figure 1). Nineteen clusters were in pairs (n=2 sequences); two clusters included three sequences and one cluster included seven sequences.
Figure 1:
HIV transmission clusters involving sequences from 51 young women sampled in the Bushbuckridge sub-district of Mpumalanga province, South Africa. Clusters are shown grouped by region of village location (some clusters involve more than one region). Clusters are labeled by cluster id number. Node shape corresponds to individuals connected by village location; edges (lines) represent linkages ≤0.02 nucleotide substitutions per site based on Tamura-New 93 model. Bold outlines indicate that clusters include ≥2 individuals (sequences) from the same village. Nodes are color coded by A) schools and B) wealth quantile (3=lowest to 0=highest) that included three or more persons in the network (A-F). Schools that are not reported indicate girls out of school and other refers to other school that are not A-F.
Among all, being in a cluster was associated with the village of residence (p=0.0469) and grade (p=0.0225), where young women in higher grades were more likely to be part of a cluster Among girls that were part of a cluster, cluster membership was associated with village (p=0.0131) but not with other characteristics.
Although over 27 schools were represented in the survey, 37% (n=19) of clustered persons were at four schools and 22% of all HIV infections were at these schools; of 25 villages represented, 39% (n=20) of clustered persons were in three villages, while 25% of all HIV infections were in these villages. We found multiple localized chains; School B was part of 5 clusters and Schools A, C, and D were all part of four clusters (Figure 1). Eight young women that were part of a cluster had infections out of school; the cluster membership statistic was similar after excluding these girls. Among girls in a cluster, the highest percentage were from the lowest quartile (N=16, 31%) which represented 24% of all HIV cases. In our calculated measure of cluster membership, we observed excess co-clustering by school (Cluster membership statistic (CMS): 1.73; 95% confidence intervals [CI]: 1.14, 2.62), wealth quartile (CMS: 1.51; 95% CI: 1.09,2.07), and village (CMS: 2.13; 95% CI: 1.31, 3.47) (Table 1). We did not find any clustering by other factors examined. However, the sample sizes for the clusters were small, which may have limited our ability to detect associations with these factors.
Table 1:
Measure of categorical cluster membership for self-reported characteristics of HIV-infected young women and their partners captured in the ACASI survey.
| Variable | Pcluster (95% CI) | Poverall (95% CI) | Cluster Membership statistic*: Pcluster/Poverall | (95% CI) |
|---|---|---|---|---|
| Partner age difference >=5 years (yes/no) | 0.80 (0.57, 1.14) | 0.70 (0.56, 0.85) | 1.14 | (0.80,1.63) |
| Any alcohol use (yes/no) | 0.89 (0.60, 1.32) | 0.82 (0.69, 0.95) | 1.09 | (0.74,1.58) |
| Partner HIV+ (yes/no) | 0.76 (0.53, 1.09) | 0.79 (0.65, 0.93) | 0.96 | (0.68,1.37) |
| Grade | 0.22 (0.14, 0.34) | 0.18 (0.11, 0.24) | 1.24 | (0.82,1.86) |
| Partner does not live in the same village as young woman (yes/no) | 0.41 (0.29, 0.60) | 0.49 (0.046, 0.53) | 0.83 | (0.58,1.20) |
| Partner out of school (yes/no) | 0.43 (0.41, 0.62) | 0.49 (0.46, 0.52) | 0.88 | (0.62,1.26) |
| Prevalent, incident in main study, incident post-intervention | 0.47 (0.32, 0.71) | 0.39 (0.30, 0.48) | 1.22 | (0.82,1.80) |
| School where young woman was enrolled | 0.07 (0.04, 0.11) | 0.04 (0.01, 0.06) | 1.73 | (1.14,2.62) |
| Young woman sleeps away from home at least once in a given week (yes/no) | 0.83 (0.58, 1.16) | 0.89 (0.77, 1.00) | 0.93 | (0.67,1.28) |
| Village of residence** | 0.09 (0.05, 0.16) | 0.04 (0.01, 0.07) | 2.13 | (1.31, 3.47) |
| Household wealth quartiles | 0.37 (0.26, 0.52) | 0.25 (0.21, 0.28) | 1.51 | (1.09,2.07) |
Pcluster is the probability of having the same characteristic for a randomly chosen pair of girls in the same cluster, Poverall is the probability of having the same characteristic for a randomly chosen pair of girls in the study. The cluster membership statistic is Pcluster/Poverall.
For the cluster membership statistic, we used actual village of residence; these were grouped into areas for figure 1.
Discussion
We identified that cluster membership was related to school, village, and wealth quartile. These associations involved multiple transmission chains and suggest localized sexual networks with shared partnerships. Similar geographic location, including village and school, may be a driver of HIV transmission for young women in this age group. Cluster membership was not related to age-disparate relationships, alcohol use or any measures of migration. Of note, the number of young women who were in the same cluster was very small. Lack of clustering may be an artifact of small sample sizes and low sampling fraction rather than a lack of association with these characteristics.
Samples were not available from male partners of the young women in the study. Therefore, we are unable to link the characteristics of those men directly to the young women who were infected. Instead, our study relied on inferring that phylogenetically-linked young women may have shared a common partner and, given the genetic distance threshold, indirect connections may be greater than one degree. Phylogenetic analyses confirming sexual transmission in HIV serodisordant couples indicates that this assumption is likely valid.18 Additionally, behavioral characteristics were self-reported and may be misreported. ACASI was used to minimize social desirability bias in the reporting of sensitive behaviors.19 Lastly, many girls at the post-intervention visit had graduated from school (65% N=65/100) limiting cluster membership by school.
Clustering in this study was typically between pairs, although we did find three larger clusters and associations between cluster membership and geographical characteristics. Our results indicate that most transmissions were not related to each other and multiple viral introductions through migration or external sexual partnerships are common.6 Additionally, we show that the larger clusters included multiple girls from the same village or school, and that some rapid, localized transmission occurred in the area. Future studies could expand this analysis by collecting phylogenetic and behavioral information directly from the male partners of young women, and by using larger sample sizes.
Supplementary Material
Acknowledgments
Conflicts of Interests and Source of Funding: Funding support for the HPTN was provided by the National Institute of Allergy and Infectious Diseases (NIAID), the National Institute of Mental Health (NIMH), and the National Institute on Drug Abuse (NIDA) of the National Institutes of Health (NIH; award numbers UM1AI068619 [HPTN Leadership and Operations Center], UM1AI068617 [HPTN Statistical and Data Management Center], and UM1AI068613 [HPTN Laboratory Center]. The study was also funded under R01MH087118 and R24 HD050924 to the Carolina Population Center. Additional funding was provided by the Division of Intramural Research, NIAID, and NIH. The Agincourt Health and Socio-Demographic Surveillance System is supported by the School of Public Health University of the Witwatersrand and Medical Research Council, South Africa, and the UK Wellcome Trust (grants 058893/Z/99/A; 069683/Z/02/Z; 085477/Z/08/Z; and 085477/B/08/Z). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We have no conflicts of interest to declare.
References
- 1.Pettifor AE, Rees HV, Kleinschmidt I, Steffenson AE, MacPhail C, Hlongwa-Madikizela L, et al. Young people’s sexual health in South Africa: HIV prevalence and sexual behaviors from a nationally representative household survey. AIDS [Internet]. 2005;19(14):1525 Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16135907 [DOI] [PubMed] [Google Scholar]
- 2.Kahn K, Collinson MA, Xavier Gómez-olivé F, Mokoena O, Twine R, Mee P, et al. Profile: Agincourt health and socio-demographic surveillance system. Int J Epidemiol. 2012;41(4):988–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison A, Colvin CJ, Kuo C, Swartz A, Lurie M. Sustained High HIV Incidence in Young Women in Southern Africa: Social, Behavioral, and Structural Factors and Emerging Intervention Approaches. Curr HIV/AIDS Rep [Internet]. 2015. June;12(2):207–15. Available from: http://link.springer.com/10.1007/s11904-015-0261-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Oliveira T, Kharsany ABM, Gräf T, Cawood C, Khanyile D, Grobler A, et al. Transmission networks and risk of HIV infection in KwaZulu-Natal, South Africa: a community-wide phylogenetic study. Lancet HIV [Internet]. 2016;3018(16):1–10. Available from: http://linkinghub.elsevier.com/retrieve/pii/S2352301816301862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dennis AM, Herbeck JT, Brown AL, Kellam P, de Oliveira T, Pillay D, et al. Phylogenetic studies of transmission dynamics in generalized HIV epidemics: an essential tool where the burden is greatest? J Acquir Immune Defic Syndr [Internet]. 2014;67(2):181–95. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24977473%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4304655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sivay M, Hudelson S, Wang J, Agyei Y, Hamilton E, Selin A, et al. HIV diversity among young women in rural South Africa: HPTN 068 (in revision). PLoS One. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pettifor A, MacPhail C, Selin A, Gomez-Olive FX, Rosenberg M, Wagner RG, et al. HPTN 068: A Randomized Control Trial of a Conditional Cash Transfer to Reduce HIV Infection in Young Women in South Africa???Study Design and Baseline Results. AIDS Behav. 2016;20(9):1863–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pettifor A, MacPhail C, Hughes JP, Selin A, Wang J, Gómez-Olivé FX, et al. The effect of a conditional cash transfer on HIV incidence in young women in rural South Africa (HPTN 068): a phase 3, randomised controlled trial. Lancet Glob Heal [Internet]. 2016;(Hptn 068). Available from: http://linkinghub.elsevier.com/retrieve/pii/S2214109X16302534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collinson MA, Tollman SM, Kahn K. Migration, settlement change and health in post-apartheid South Africa: Triangulating health and demographic surveillance with national census data. Scand J Public Health [Internet]. 2007;35:77–84. Available from: http://sjp.sagepub.com/cgi/doi/10.1080/14034950701356401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahn K, Tollman SM, Collinson MA, Clark SJ, Twine R, Clark BD, et al. Research into health, population and social transitions in rural South Africa: Data and methods of the Agincourt Health and Demographic Surveillance System1. Scand J Public Health [Internet]. 2007;35(69 suppl):8–20. Available from: http://sjp.sagepub.com/content/35/69_suppl/8.abstract%5Cnhttp://sjp.sagepub.com/content/35/69_suppl/8.full.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guindon S, Gascuel O, Dufayard J-F, Lefort V, Anisimova M, Hordijk W. New Algorithms and Methods to Estimate Maximim-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst Biol [Internet]. 2010;59(3):307–21. Available from: http://www.atgc-montpellier.fr/download/papers/phyml_2010.pdf [DOI] [PubMed] [Google Scholar]
- 13.Hassan AS, Pybus OG, Sanders EJ, Albert J, Esbjörnsson J. Defining HIV-1 transmission clusters based on sequence data. Vol. 31, AIDS. 2017. p. 1211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wertheim JO, Kosakovsky Pond SL, Forgione LA, Mehta SR, Murrell B, Shah S, et al. Social and Genetic Networks of HIV-1 Transmission in New York City. PLoS Pathog. 2017;13(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Csárdi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Syst. 2006;1695:1–9. [Google Scholar]
- 16.Efron B, Tibshirani R. Bootstrap Methods for Standard Errors,Confidence Intervals, and Other Measures of Statistical Accuracy. Stat Sci. 1986;1(1):54–77. [Google Scholar]
- 17.Carpenter J, Bithell J. Bootstrap confidence intervals: When, which, what? A practical guide for medical statisticians. Stat Med. 2000;19(9):1141–64. [DOI] [PubMed] [Google Scholar]
- 18.Eshleman SH, Hudelson SE, Redd AD, Wang L, Debes R, Chen YQ, et al. Analysis of genetic linkage of HIV from couples enrolled in the HIV prevention trials network 052 trial. J Infect Dis. 2011; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison-Beedy D, Carey MP, Tu X. Accuracy of audio computer-assisted self-interviewing (ACASI) and self-administered questionnaires for the assessment of sexual behavior. AIDS Behav [Internet]. 2006;10(5):541–52. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2430922&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.