Abstract
The mitochondrial unfolded protein response (UPRmt) is rapidly gaining attention. While the CHOP (ATF4/5) axis of the UPRmt was the first to be described, other axes have subsequently been reported. Validation of this complex pathway in C. elegans has been extensively studied. However, validation of the UPRmt in mouse models of disease known to implicate mitochondrial reprogramming or dysfunction, such as cancer and neurodegeneration, respectively, is only beginning to emerge. This review summarizes recent findings and highlights the major role of the superoxide dismutase SOD1 in the communication between the mitochondria and the nucleus in these settings. While SOD1 has mostly been studied in the context of familial amyotrophic lateral sclerosis (fALS), recent studies suggest that SOD1 may be a potentially important mediator of the UPRmt and converge to emphasize an increasingly vital role of SOD1 as a therapeutic target in cancer.
Keywords: ROS, mitochondria, UPRmt, SOD1, SOD2, SIRT3, estrogen receptor, cancer, ALS, neurodegeneration
Introduction
A common denominator between cancer and neurodegeneration is the elevation in oxidative stress through the formation of reactive oxygen species (ROS). As the activity of the electron transport chain, embedded in the mitochondrial inner membrane, is responsible for the production of the majority of ROS, mitochondria are especially prone to the effects of ROS. On one hand, increased ROS levels act as signaling molecules to promote cancer formation 1,2. Notably, this increase in ROS facilitates the switch from oxidative phosphorylation to glycolysis through the stabilization of HIF1α 3,4. On the other hand, oxidative stress leads to damage to lipids, DNA, and proteins and contributes to protein misfolding and complex misassembly5. Therefore, adaptive mechanisms including increased expression of antioxidant machinery, mitochondrial proteases and chaperones, mitochondrial biogenesis and mitophagy must be activated in order to restore mitochondrial fitness and avoid cellular death.
In recent years, the mitochondrial unfolded protein response (UPRmt) has gained much attention as a complex response that orchestrates several signaling cascades in parallel to coordinate the up-regulation of these adaptive mechanisms. Since multiple sources of mitochondrial stress have now been reported to activate the UPRmt and several axes have been described, suggestion to rename this response, the integrated mitochondrial stress response has obtained popularity. However, for the purpose of this review, we will refer to this pathway as the UPRmt.
The mitochondrial unfolded protein response (UPRmt)
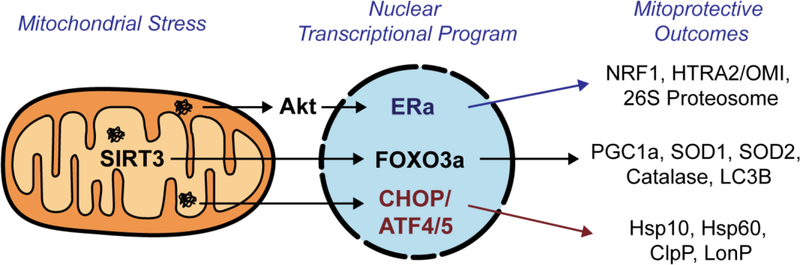
The UPRmt was originally discovered by the Hoogenraad group using overexpression of the matrix misfolded protein OTC delta6,7. Using this system, they described a retrograde signaling axis from the mitochondria to the nucleus mediated through the transcription factor CHOP, resulting in the induction of mitochondrial proteases and chaperones such as ClpP, HSP10, and HSP60 7–12 (Figure 1). Following this original finding of the CHOP axis of the UPRmt in mammalian cells, much of the subsequent studies of the UPRmt were performed in C. elegans using HSP60 as a reporter of UPRmt activation. These important studies identified ATFS-1 and the DVE-1/UBL5 complex as transcriptional activators of the UPRmt 11–16. More recently, ATF5 was identified as the mammalian homolog of ATFS-117. Since ATF5 works downstream of CHOP16, the finding of ATF5 in the setting of the UPRmt nicely complements the seminal finding of the Hoogenraad group. This axis of the UPRmt has been extensively reviewed18–26.
Figure 1. The mitochondrial unfolded protein response (UPRmt).
Collectively, the three parallel axes of the UPRmt coordinate a global mito-protective program against mitochondrial stress by activating antioxidant machinery, increasing protein quality control, inducing mitochondrial biogenesis, and promoting mitophagy.
In mammalian cells, our group reported a SIRT3-axis of the UPRmt (UPRmt-SIRT3) using the same model system as the Hoogenraad group. We also reported that this axis is independent of the CHOP-axis, as inhibition of CHOP did not affect the SIRT3-axis and conversely inhibition of SIRT3 did not affect activation of markers of the CHOP-axis27. We found that the SIRT3-axis orchestrates the induction of antioxidant genes, notably the dismutase SOD2, mitochondrial biogenesis, and mitophagy18,19,27–29 (Figure 1). A strikingly similar sirtuin axis of the UPRmt, leading to activation of SOD2, has also been discovered in C. elegans, suggesting the evolutionary conservation of this signaling pathway30.
We also reported an additional axis of the UPRmt, which is regulated by the estrogen receptor alpha (ERα)31. Rather than promoting the activation of mitochondrial matrix chaperones and proteases, we found that this axis results in the activation of Akt by ROS, leading the phosphorylation of the ERα and the transcription of the IMS protease HTRA2/OMI, the mitochondria biogenesis transcription factor NRF1, as well as the up-regulation of the activity of the proteasome31 (Figure 1). Further, in contrast to the discovery of the other axes of the UPRmt, the ERα-axis was discovered using overexpression of misfolded proteins in the intermembrane space (IMS) of the mitochondria.
The IMS of the mitochondria
As the mitochondria are composed of the inner membrane and outer membrane, the presence of such double membranes results in two sub-compartments; the space in between these membranes, therefore referred to as the intermembrane space (IMS) and the matrix. Most studies on the mitochondria focus on the matrix or components of the outer and inner membranes. Notably, the inner membrane is central to the study of the electron transport chain, and the outer membrane is important for regulation of mitochondrial fusion and fission32. The matrix has also been intensively studied due to its role in the Krebs cycle, amino acid metabolism, and mitochondrial DNA transcription and translation32. The IMS, on the other hand, is largely considered as a passive storage compartment for pro-apoptotic proteins. In contrast, the IMS comprises over one hundred proteins, accounting for approximately ten percent of the mitochondrial proteome33. These proteins are important in various functions such as ROS detoxification, ROS-mediated signaling, oxidative protein folding, protein import, electron transport chain assembly, transport of metabolites, and metabolism processes32,34. Collectively, these observations indicate that the IMS plays an active role in many mitochondrial processes and suggest that defects in the IMS have the potential to impact the entire organelle.
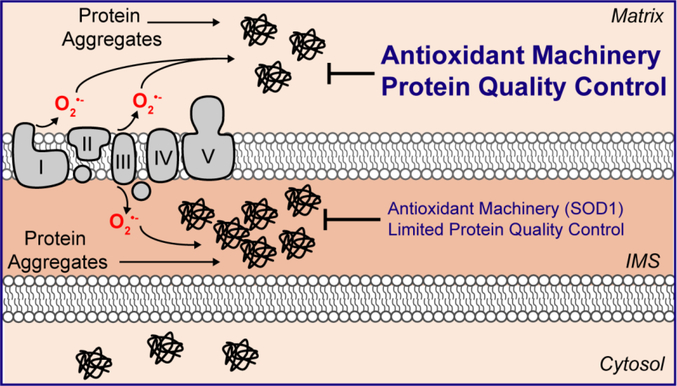
Given that the inner membrane of the mitochondria is the site of oxidative phosphorylation and that ROS is produced on both sides the electron transport chain35, ROS accumulation also occurs in the IMS. In addition, oxidative protein folding in the IMS also contributes to ROS levels34. Collectively, the combined effects of the electron transport chain and oxidative folding make the IMS the most oxidative environment of any cellular compartment. Further, the IMS does not contain heat shock proteins and has a limited number of proteases involved in protein quality controls32,33. Since ROS can lead to oxidation of cysteine residues, misfolding, and aggregation36, the IMS appears uniquely susceptible to the accumulation of misfolded proteins (Figure 2). The detrimental effect of the accumulation of misfolded proteins in the IMS is best illustrated by the role of misfolded copper/zinc superoxide dismutase (SOD1) in the IMS in familial ALS.
Figure 2. The IMS of the mitochondria is highly susceptible to proteotoxic stress.
The IMS of the mitochondria has limited protein quality control mechanism and poor antioxidant capacity, which makes this sub-compartment prone to the accumulation of misfolded proteins.
The role of SOD1
SOD1 is an abundant antioxidant enzyme responsible for the conversion of superoxide to hydrogen peroxide36. SOD1 is ubiquitously expressed in many cellular compartments, including the IMS of the mitochondria37–41. In contrast, the manganese superoxide SOD2 localizes exclusively to the matrix of the mitochondria42. The function of superoxide dismutases was first discovered by McCord and Fridovich43, who established the landmark study in support of the oxidative toxicity theory. Subsequent studies revealed that SOD1 is a dimeric enzyme, containing a copper ion and zinc ion in its catalytic core and that its main function is to convert superoxide (O2•−) into hydrogen peroxide (H2O2)44. The O2•− radical is directed to the catalytic site through a positively charged channel, where the dismutation reaction is catalyzed by the copper ion45. The copper chaperone CCS facilitates copper insertion and disulfide bridge formation in SOD146. Copper/zinc insertion and proper formation of the SOD1 homodimer are important for its enzymatic activity47. SOD1 is predominantly a cytosolic protein, but is also localized in the nucleus, and the IMS of the mitochondria37–41.
Many studies suggest that SOD1 plays a key role in protecting cells against oxidative damage36,48–50. For instance, overexpression of SOD1 in yeast confers resistance to oxidative stress51. Conversely, ablation of SOD1 in yeast leads to oxidative damage in the mitochondria52–54. In mammals, SOD1 knockout mice demonstrate widespread oxidative damage49,55–57 and studies in normal human fibroblasts showed that knockdown of SOD1 induces senescence58. Emerging studies have also demonstrated that SOD1 is at the center of nutrient and oxygen sensing59,60. The Culotta group showed that SOD1 is important for transmitting signals from glucose and oxygen to repress respiration59. Another study showed that nutrient deprivation increases SOD1 activity to reduce oxidative damage and promote cell survival60. They report that mTORC1 regulates SOD1 activity through phosphorylation in response to nutrients60. In addition, they show that SOD1 activation promotes cell survival and tumor growth in the ischemic tumor microenvironment and protects against cell death by the chemotherapeutic agent cisplatin60. Taken together, SOD1 and its localization may play important roles in maintaining ROS levels below a critical threshold compatible with cell viability.
SOD1-G93A in familial ALS (fALS)
Amyotrophic lateral sclerosis (ALS), also known as Lou Gehrig’s disease, is a fatal, neurodegenerative disease characterized by damage of motor neurons of the spinal cord, which leads to paralysis. While sporadic cases of no known etiology account for the majority of ALS cases, mutations in SOD1 are responsible for approximately 20% of familial ALS (fALS)61,62. More than 180 different causative mutations in the SOD1 gene have been identified in fALS patients63. It is widely accepted that SOD1-linked fALS is partly caused by a toxic gain-of-function of the SOD1 mutant rather than loss of function62. This interpretation is supported by the fact SOD1 mutants without enzymatic activity also cause ALS64.
The exact mechanism by which SOD1-linked fALS mediates its effect remains incompletely understood but mitochondrial dysfunction has been shown to play a role65–67. In addition, mutant SOD1 has been shown to cause abnormalities in multiple organelles, such as the endoplasmic reticulum68 and peroxisomes65 in addition to the mitochondria65–67,69–73. Mutant SOD1 also affects neuronal cytoskeleton integrity, which potentially impairs normal trafficking along axons and dendrites74. Further, mutant SOD1 suppresses protein quality control75, increases production of free radicals76, activates pro-apoptotic signaling77,78, deregulates multiple signaling pathways79, and affects levels of critical transporters and receptors, such as astrocytic glutamate transporter and Glur280. SOD1-linked fALS has also been demonstrated to be a cell non-autonomous disease, in which the mutant protein has been shown to be toxic in multiple cell types, such as glia and astrocytes, and mutant SOD1 is also secreted, which may stimulate neuroinflammatory responses, and the resulting effects are likely additive81,82.
The most commonly studied mutation in SOD1 is the G93A amino acid substitution83. The SOD1-G93A transgenic mouse model ubiquitously overexpresses the mutant SOD1 under the control of endogenous SOD1 promoter. SOD1-G93A mice recapitulate phenotypes associated with fALS in humans. They experience rapid motor neuron degeneration, resulting in onset of paralysis and depending on genetic background, die within four to five months of age83. Work from several groups in this mouse model has provided key insights into the role of mitochondrial dysfunction in SOD1-linked fALS. In the mitochondria, mutant SOD1 accumulates on the outer membrane where it interacts with important mitochondrial proteins such as Bcl2 and VDAC69,70. In vitro studies have also demonstrated that mitochondrial localization of mutant SOD1 impairs mitochondrial morphology and leads to cell death66,67.
Mutant SOD1 has also been shown to accumulate in the IMS, where it has the potential to alter import, assembly, and maturation of mitochondrial proteins47,84. In agreement, mutant SOD1 specifically targeted to the IMS was shown to cause mitochondrial fragmentation and impaired mitochondrial dynamics and these lead to impairment in neuritic processes47,84. In addition, a new mouse model that expresses IMS-targeted mutant SOD1-G93A has been shown to develop similar ALS phenotypes, such as motor neuron defects and mitochondrial abnormalities, however symptoms develop much later85. These findings support a pathogenic role for accumulation of mutant SOD1 in the IMS.
SOD1-G93A in familial ALS and the UPRmt
We have recently reported that IMS fraction of mutant SOD1 plays an important role in activation of the UPRmt 86. We reported that the UPRmt-CHOP axis is transiently activated in the spinal cord G93A-SOD1 mice in the late symptomatic phase. Most striking, we observed a significant sex difference in the activation of the UPRmt-CHOP, where it was only significantly activated in female G93A-SOD1 mice but not in males86. In addition, we found that the female specific activation of the UPRmt extended to the early activation of the UPRmt-ERα axis. This was demonstrated by elevation in Akt phosphorylation, upregulation of NRF1, OMI, and proteasome activity in the spinal cord of female G93A-SOD1 mice86. The up-regulation of the proteasome correlated with a decrease in total ubiquitinated proteins in the spinal cords of female mice. Further, to ascertain whether the IMS fraction of mutant SOD1 was responsible for the activation of the UPRmt, analysis of the UPRmt was performed in the IMS-targeted G93A-SOD1 model. We found that in these mice, NRF1 was activated during the presymptomatic phase of the disease in both males and females, but only females showed an upregulation of OMI in the symptomatic phase86. These observations suggest that IMS-localized mutant SOD1 is sufficient to activate the UPRmt-ERα in vivo. We further confirmed that ERα is required for protective effects of the UPRmt by crossing the ERα knockout mice to the G93A-SOD1 mice. We found that the absence of ERα prevents the activation of the UPRmt-ERα 86. However, under these conditions, upregulation of the UPRmt-CHOP axis was observed, suggesting a compensatory mechanism of the activation of one axis in absence of another. The findings of our study provide the first demonstration of the activation of the UPRmt-ER axis in an in vivo model of fALS.
Activation of the UPRmt has also recently been identified in a novel mouse model of neurodegeneration. Work from the Manfredi lab has demonstrated that mutant CHCHD10 mice develop accumulation of misfolded proteins in affected tissue, and that this accumulation selectively aggregated in the mitochondria87. Similar to other models of neurodegeneration, this proteotoxic stress led to disruption of mitochondrial morphology and function87. Using RNA sequencing, they found that diseased tissue of CHCHD10 mutant mice show increased expression of CHOP and ATF587. In addition, they also found that expression of various subunits of the ETC were downregulated, which further supports the notion that upregulation of the UPRmt leads to a decrease in oxidative phosphorylation to limit oxidative stress87. These findings support the hypothesis that the UPRmt plays an important role in maintaining mitochondrial proteostasis and integrity. Interestingly, female mice in this model showed delayed onset of disease, similar to the sex difference observed in the G93A fALS model. Whether the ERα axis of the UPRmt plays a role in this sex difference in this new model has yet to be determined.
While the interest in SOD1 has largely been in the context of fALS, an emerging body of work is shedding light on the role of SOD1 in cancer biology.
The mitochondria and oxidative stress in cancer
Cancer cells are characterized by increased levels of ROS. While elevated levels of ROS can promote cancer cell proliferation by activating signaling pathways such as PI3K/AKT, excessive ROS can lead to cell death1. This increase in ROS, in addition to the switch to glycolysis, known as the Warburg effect, have led to the common misconception that in cancer cells mitochondria are non-essential. However, the importance of mitochondrial reprogramming to provide the metabolites required for the production of amino acids and lipids as the building blocks of rapidly dividing cells, is now appreciated. In the matrix, ROS levels are limited by antioxidant machinery regulated by the deacetylase SIRT319. Notably, SIRT3 deacetylates and activates the manganese superoxide SOD288. SOD2, similar to SOD1, catalyzes the conversion of superoxide to hydrogen peroxide. Additional mitochondrial matrix enzymes such as glutathione peroxidases, peroxiredoxins, and catalase reduce hydrogen peroxide to water. Numerous studies have focused on the role of the mitochondrial matrix antioxidant machinery in many diseases, including cancer5,52,89–91.
The potential importance of SOD1 on mitochondrial function has been highlighted by recent work from the Haigis lab that showed that SIRT3 levels are decreased in 87% of breast cancers92. Considering that SIRT3 is important for the activation of SOD2, the observation that SIRT3 levels are low in breast cancer cells is in agreement with the reduced SOD2 activity in cancer. As a result, levels of ROS in the mitochondria of cancer cells are elevated. In addition, mutations in subunits of the electron transport chain in cancers also contribute to the accumulation of ROS in these cells 19,28,29. One logical extension of these findings is that increasing levels of ROS in the IMS, which has limited antioxidant capacity, would likely lead to excessive oxidative stress in the mitochondria93. Several reports have now supported a critical role of SOD1 in cancer.
SOD1 in cancer
Several groups have identified SOD1 as a potential therapeutic target in cancer. SOD1 was the top hit in an extensive high throughput small molecule screen in lung cancer cells, by the Varmus group. They identified LCS-1 (lung cancer screen-1) among 189,000 small molecules as a selective inhibitor of lung cancer cell growth94,95. Using affinity proteomics and gene expression analysis, they identified SOD1 as the target of LCS-195. Their findings revealed that inhibition of SOD1 by siRNA inhibited growth of LCS-1 sensitive cell lines, and conversely, SOD1 overexpression increased lung cancer cell proliferation and decreased apoptosis95. More importantly, the Varmus group also found that LCS-1 is a potent growth inhibitor in most cell lines in the NCI-60 human tumor cell line panel, including breast cancer cell lines95. The findings support the previous finding that SOD1 is a potent anti-cancer target in leukemia96. Huang et al. reported that inhibition of SOD1 leads to accumulation of superoxide and oxidative damage and subsequent selective death of myeloid leukemia cell lines96. A recent study similarly demonstrated that inhibition of SOD1 decreases cell proliferation and enhanced the effects of the common leukemia chemotherapy agent cytarabine97. These data suggest an essential role for SOD1 in adaptation to oxidative stress in cancer cells.
In agreement with these findings, the Chandel group also showed that SOD1 is a potent therapeutic target in cancer. They confirmed that SOD1 inhibition by with shRNA or the SOD1 inhibitor ATN-224 induces cell death in various non-small-cell lung cancer cells lines, and had minimal effects on normal lung epithelial cells98. They found that inhibition of SOD1 by ATN-224 increases superoxide levels, which decreases the enzymatic activity of glutathione peroxidase, resulting in elevated hydrogen peroxide levels. They further showed that increased hydrogen peroxide levels leads to p38 MAPK activation and subsequently results in a decrease in the anti-apoptotic factor MCL198. It was also recently reported that SOD1 inhibition in nasopharyngeal carcinoma inhibits cell growth and promotes apoptosis99. These observations suggest that one mechanism by which SOD1 inhibition reduces cancer cell growth is through apoptosis.
Our group has shown that treatment of breast cancer cells with LCS-1 leads to increased mitochondrial superoxide and altered mitochondrial morphology as indicated by increased fragmentation and matrix swelling100. These observations suggested that the mitochondrial IMS fraction of SOD1 may be of particular importance in cancer cells. Collectively, these results suggest that SOD1 may be a potent target in cancer, however, since all experimental evidence arose from work performed in vitro, a definitive demonstration of the role of SOD1 in cancer awaited evidence in vivo.
SOD1 is essential for oncogene-driven proliferation but not normal proliferation
The observation that SIRT3 and consequently SOD2 activity are decreased in 87% of breast cancers92 led us to hypothesize that the resulting elevation in mitochondrial oxidative stress may be counterbalanced by SOD1 upregulation. Consistent with this idea, we found that SOD1 is overexpressed in a panel of breast cancer cell lines while SIRT3 is decreased compared to the non-malignant cell line MCF10A, which show high SIRT3 and low SOD1 expression levels100. We confirmed this inverse relationship of SIRT3 and SOD1 in panel of human breast tumor specimens by immunohistochemistry100. Further, we found that SOD1 is overexpressed in tumors of three independent mouse models of breast cancer (MMTV-Wnt, MMTV-erbB2, and MMTV-Myc), compared to normal mammary ducts that showed no detectable SOD1 levels100. Therefore, these results suggest that the upregulation SOD1 occurs frequently in breast cancer regardless of the subtype or the oncogene driving tumor progression.
Following these observations, we aimed at validating the role of SOD1 in breast cancer in vivo. We crossed the MMTV-Wnt and inducible MMTV-rtTa/TetO-NeuNT mice, which develop mammary tumors at high frequency, with SOD1 knockout mice. In both models, we found that absence of SOD1 abolishes mammary tumor formation101. Moreover, we show that genetic ablation of SOD1 is specific to oncogene-driven proliferation as hyperproliferation of the mammary gland ductal tree normally observed during pregnancy is not affected in absence of SOD1101. Further, the architecture and physiological functions of highly proliferative normal tissues such as the skin and the gut were also not affected. These results suggest that SOD1 is necessary for oncogene-driven proliferation but not normal proliferation.
We reasoned that increased mitochondrial ROS may be the factor that distinguishes oncogene–driven proliferation from normal proliferation. Indeed, we found that mitochondrial ROS is elevated upon oncogene activation but not during proliferation of the ductal epithelial cells during pregnancy. We then investigated the mode of cell death in absence of SOD1. Consistent with findings by other groups, we found that genetic ablation or pharmacological inhibition of SOD1 in cancer cells induces apoptosis. In addition, we found that SOD1 is critical to evade oncogene-induced senescence during transformation101. Finally, we validated our findings in human breast cancer patients. We found that high expression of SOD1 is associated with worse clinical outcomes, regardless of breast cancer subtype. These findings indicate that SOD1 may be essential for cancer cells to survive the elevation in oxidative stress observed during transformation and has the potential to be a selective target for cancer therapy.
Overexpression of SOD1 in the nucleus and the IMS may participate in the induction of the mitochondrial unfolded protein response
An incidental finding in our study of SOD1 in human primary breast cancers was the observation of a significant increase in SOD1 in both the mitochondria and the nucleus. Interestingly, SOD1 has been shown to bind the estrogen receptor alpha (ERα) when bound to DNA102. The interaction of SOD1 with ERα was also reported to increase the transcriptional activity of ERα102. It is therefore tempting to hypothesize that SOD1 may have multiple functions: first, its canonical function as a dismutase, in limiting oxidative damage; second, a signaling function through its accumulation in the IMS and the activation of the UPRmt-ERα axis; and third, a role in the enhancement of the transcriptional activity of the ERα in the nucleus. Collectively, these functions will place SOD1 as a key regulator of the communication between the nucleus and the mitochondria.
Concluding Remarks
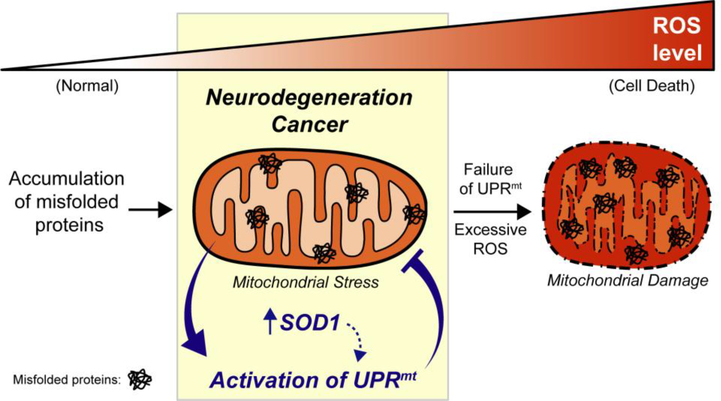
The interest in the study of the UPRmt in cancer and neurodegenerative diseases is mounting. Our work in both fALS and cancer suggests that the IMS of the mitochondria may be an important sub-compartment to signal mitochondrial stress to the nucleus. The unique oxidative environment of the IMS combined with the absence of chaperones and the minimal number of proteases involved in protein quality control in this sub-compartment, synergize to prime the IMS for proteotoxic stress. We suggest that as a result, the IMS is ideal as a signaling sub-compartment. Our work indicates that SOD1 may play a key role in this mitochondria-nucleus communication signaling (Figure 3). Further, the involvement of the ERα in the UPRmt and the finding of sex differences in its activation highlight the fact that much remains to be understood in this pathway and that its activation may be tissue and sex specific. Future investigations will be required to shed light on these possibilities.
Figure 3. Crosstalk between SOD1 and the mitochondrial UPR in neurodegeneration and cancer.
Upregulation of SOD1 and activation of the UPRmt protects cells from mitochondrial damage and cell death in models of neurodegeneration and cancer. It is possible that SOD1 plays a critical role in mito-nuclear communication as both a signaling function through its accumulation in the IMS and through enhancement of transcriptional activity of ERα, leading to activation of the UPRmt.
Highlights.
The mitochondrial unfolded protein response (UPRmt) is rapidly gaining attention. However, validation in mouse models of neurodegenerative diseases and cancer is only beginning to emerge. This review focuses on the recent studies describing the UPRmt in mouse models of familial ALS and breast cancer, with a special focus on the role of the dismutase SOD1 in these settings.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schieber M & Chandel NS ROS function in redox signaling and oxidative stress. Current Biology 24, 453–462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D’Autréaux B & Toledano MB ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol 8, 813–824 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Finley LWS & Haigis MC Metabolic regulation by SIRT3: implications for tumorigenesis. Trends Mol Med 18, 516–523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandel NS et al. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J. Biol. Chem 275, 25130–8 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Cross CE et al. Oxygen radicals and human disease. Ann. Intern. Med 107, 526–45 (1987). [DOI] [PubMed] [Google Scholar]
- 6.Martinus RD et al. Selective Induction of Mitochondrial Chaperones in Response to Loss of the Mitochondrial Genome. Eur. J. Biochem 240, 98–103 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Zhao Q et al. A mitochondrial specific stress response in mammalian cells. EMBO J. 21, 4411–9 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aldridge JE, Horibe T & Hoogenraad NJ Discovery of Genes Activated by the Mitochondrial Unfolded Protein Response (mtUPR) and Cognate Promoter Elements. PLoS One 2, e874 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horibe T & Hoogenraad NJ The Chop Gene Contains an Element for the Positive Regulation of the Mitochondrial Unfolded Protein Response. PLoS One 2, e835 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan MT & Hoogenraad NJ Mitochondrial-Nuclear Communications. Annu. Rev. Biochem 76, 701–722 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Haynes CM, Petrova K, Benedetti C, Yang Y & Ron D ClpP Mediates Activation of a Mitochondrial Unfolded Protein Response in C. elegans. Dev. Cell 13, 467–480 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Haynes CM, Yang Y, Blais SP, Neubert TA & Ron D The Matrix Peptide Exporter HAF-1 Signals a Mitochondrial UPR by Activating the Transcription Factor ZC376.7 in C. elegans. Mol. Cell 37, 529–540 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benedetti C, Haynes CM, Yang Y, Harding HP & Ron D Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics 174, 229–39 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nargund AM et al. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nargund AM, Fiorese CJ, Pellegrino MW, Deng P & Haynes CM Mitochondrial and Nuclear Accumulation of the Transcription Factor ATFS-1 Promotes OXPHOS Recovery during the UPRmt. Mol. Cell 58, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teske BF et al. CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol. Biol. Cell 24, 2477–90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiorese CJ et al. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr. Biol 26, 2037–2043 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenny TC & Germain D From discovery of the CHOP axis and targeting ClpP to the identification of additional axes of the UPRmt driven by the estrogen receptor and SIRT3. J. Bioenerg. Biomembr 49, 297–305 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Kenny TC, Manfredi G & Germain D The Mitochondrial Unfolded Protein Response as a Non-Oncogene Addiction to Support Adaptation to Stress during Transformation in Cancer and Beyond. Front. Oncol 7, 159 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiorese CJ & Haynes CM Integrating the UPR mt into the mitochondrial maintenance network. Crit. Rev. Biochem. Mol. Biol 52, 304–313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Münch C The different axes of the mammalian mitochondrial unfolded protein response. BMC Biol. 16, 81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Germain D Toward the identification and the targeting of key players of the mitochondrial unfolded protein response (UPRmt) in cancer. J. Bioenerg. Biomembr 49, 291 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qureshi MA, Haynes CM & Pellegrino MW The mitochondrial unfolded protein response: Signaling from the powerhouse. (2017). doi: 10.1074/jbc.R117.791061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng P & Haynes CM Mitochondrial dysfunction in cancer: Potential roles of ATF5 and the mitochondrial UPR. Semin. Cancer Biol 47, 43–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melber A & Haynes CM UPRmt regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res. 28, 281–295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naresh NU & Haynes CM Signaling and Regulation of the Mitochondrial Unfolded Protein Response. (2019). doi: 10.1101/cshperspect.a033944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papa L & Germain D SirT3 Regulates the Mitochondrial Unfolded Protein Response. Mol. Cell. Biol 34, 699–710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenny TC et al. Selected mitochondrial DNA landscapes activate the SIRT3 axis of the UPRmt to promote metastasis. Oncogene 36, 4393–4404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenny TC & Germain D mtDNA, Metastasis, and the Mitochondrial Unfolded Protein Response (UPRmt). Front. Cell Dev. Biol. 5, 37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mouchiroud L et al. The NAD+/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell 154, 430–441 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papa L & Germain D Estrogen receptor mediate a distinct mitochondrial unfolded protein response. JCS 1396–1402 (2011). doi: 10.1242/jcs.078220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Germain D in Advances in cancer research 130, 211–256 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Herrmann JM & Riemer J The Intermembrane Space of Mitochondria. Antioxid. Redox Signal 13, 1341–1358 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Chatzi A, Manganas P & Tokatlidis K Oxidative folding in the mitochondrial intermembrane space: A regulated process important for cell physiology and disease. Biochim. Biophys. Acta 1863, 1298–306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy MP How mitochondria produce reactive oxygen species. Biochem. J 417, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Branicky R, Noë A & Hekimi S Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol 217, jcb.201708007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang LY, Slot JW, Geuze HJ & Crapo JD Molecular immunocytochemistry of the CuZn superoxide dismutase in rat hepatocytes. J. Cell Biol 107, 2169–79 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liou W et al. Distribution of CuZn superoxide dismutase in rat liver. Free Radic. Biol. Med 14, 201–7 (1993). [DOI] [PubMed] [Google Scholar]
- 39.Crapo JD, Ouryf T, Rabouille C, Slot JW & Chang L-Y Copper,zinc superoxide dismutase is primarily a cytosolic protein in human cells (antioxidant enzyme/peroxlsm/catalase/hnmunocytocemistry). Cell Biology 89, (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okado-Matsumoto A & Fridovich I Subcellular distribution of superoxide dismutases (SOD) in rat liver. Cu,Zn-SOD in mitochondria. J. Biol. Chem 276, 38388–38393 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Sturtz LA, Diekert K, Jensen LT, Lill R & Culotta VC A Fraction of Yeast Cu,Zn-Superoxide Dismutase and Its Metallochaperone, CCS, Localize to the Intermembrane Space of Mitochondria. J. Biol. Chem 276, 38084–38089 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Slot JW, Geuze HJ, Freeman BA & Crapo JD Intracellular localization of the copper-zinc and manganese superoxide dismutases in rat liver parenchymal cells. Lab. Invest 55, 363–71 (1986). [PubMed] [Google Scholar]
- 43.McCord J, F. I. Superoxide Dismutase, an enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem 244, 6049–6055 (1969). [PubMed] [Google Scholar]
- 44.Fridovich I Superoxide Radical and Superoxide Dismutases. Annu. Rev. Biochem 64, 97–112 (1995). [DOI] [PubMed] [Google Scholar]
- 45.Culotta VC et al. The copper chaperone for superoxide dismutase. J. Biol. Chem 272, 23469–72 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Furukawa Y, Torres AS & O’Halloran TV Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS. EMBO J. 23, 2872–2881 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawamata H & Manfredi G Import, maturation, and function of SOD1 and its copper chaperone CCS in the mitochondrial intermembrane space. Antioxid. Redox Signal 13, 1375–84 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dukan S & Nyström T Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J. Biol. Chem 274, 26027–32 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Muller FL et al. Absence of CuZn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy. Free Radic. Biol. Med 40, 1993–2004 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Elchuri S et al. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene 24, 367–380 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Wu C-Y, Steffen J & Eide DJ Cytosolic Superoxide Dismutase (SOD1) Is Critical for Tolerating the Oxidative Stress of Zinc Deficiency in Yeast. PLoS One 4, e7061 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung W-H Unraveling new functions of superoxide dismutase using yeast model system: Beyond its conventional role in superoxide radical scavenging. J. Microbiol 55, 409–416 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Gralla EB & Valentine JS Null mutants of Saccharomyces cerevisiae Cu,Zn superoxide dismutase: characterization and spontaneous mutation rates. J. Bacteriol 173, 5918–20 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wawryn J, Krzepiłko A, Myszka A & Biliński T Deficiency in superoxide dismutases shortens life span of yeast cells. Acta Biochim. Pol 46, 249–53 (1999). [PubMed] [Google Scholar]
- 55.Sentman M-L et al. Phenotypes of Mice Lacking Extracellular Superoxide Dismutase and Copper-and Zinc-containing Superoxide Dismutase *. (2005). doi: 10.1074/jbc.M510764200 [DOI] [PubMed] [Google Scholar]
- 56.Jang YC et al. Increased superoxide in vivo accelerates age-associated muscle atrophy through mitochondrial dysfunction and neuromuscular junction degeneration. FASEB J. 24, 1376–90 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y et al. A new role for oxidative stress in aging: The accelerated aging phenotype in Sod1−/− mice is correlated to increased cellular senescence. Redox Biol. 11, 30–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blander G, Machado De Oliveira R, Conboy CM, Haigis M & Guarente L Superoxide Dismutase 1 Knock-down Induces Senescence in Human Fibroblasts*. (2003). doi: 10.1074/jbc.M307146200 [DOI] [PubMed] [Google Scholar]
- 59.Reddi AR & Culotta VC SOD1 Integrates Signals from Oxygen and Glucose to Repress Respiration. Cell 152, 224–235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsang CK et al. SOD1 Phosphorylation by mTORC1 Couples Nutrient Sensing and Redox Regulation Article SOD1 Phosphorylation by mTORC1 Couples Nutrient Sensing and Redox Regulation. Mol. Cell 70, 502–515.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosen DR et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 (1993). [DOI] [PubMed] [Google Scholar]
- 62.Andersen PM Amyotrophic lateral sclerosis associated with mutations in the CuZn superoxide dismutase gene. Curr. Neurol. Neurosci. Rep 6, 37–46 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Keskin I et al. Effects of Cellular Pathway Disturbances on Misfolded Superoxide Dismutase-1 in Fibroblasts Derived from ALS Patients. PLoS One 11, e0150133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agar J & Durham H Relevance of oxidative injury in the pathogenesis of motor neuron diseases. Amyotroph. Lateral Scler. Other Mot. Neuron Disord 4, 232–242 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Higgins CMJ, Jung C & Xu Z ALS-associated mutant SODIG93A causes mitochondrial vacuolation by expansion of the intermembrane space by involvement of SODI aggregation and peroxisomes. BMC Neurosci. 4, 16 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hervias I, Beal MF & Manfredi G Mitochondrial dysfunction and amyotrophic lateral sclerosis. Muscle Nerve 33, 598–608 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Magrané J, Cortez C, Gan W-B & Manfredi G Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum. Mol. Genet 23, 1413–1424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kikuchi H et al. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc. Natl. Acad. Sci 103, 6025–6030 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pedrini S et al. ALS-linked mutant SOD1 damages mitochondria by promoting conformational changes in Bcl-2. Hum. Mol. Genet 19, 2974–2986 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Israelson A et al. Misfolded Mutant SOD1 Directly Inhibits VDAC1 Conductance in a Mouse Model of Inherited ALS. Neuron 67, 575–587 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferri A et al. Familial ALS-superoxide dismutases associate with mitochondria and shift their redox potentials. Proc. Natl. Acad. Sci 103, 13860–13865 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cozzolino M et al. Oligomerization of Mutant SOD1 in Mitochondria of Motoneuronal Cells Drives Mitochondrial Damage and Cell Toxicity. Antioxid. Redox Signal 11, 1547–1558 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Vijayvergiya C, Beal MF, Buck J & Manfredi G Mutant Superoxide Dismutase 1 Forms Aggregates in the Brain Mitochondrial Matrix of Amyotrophic Lateral Sclerosis Mice. J. Neurosci 25, 2463–2470 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Couillard-Després S et al. Protective effect of neurofilament heavy gene overexpression in motor neuron disease induced by mutant superoxide dismutase. Proc. Natl. Acad. Sci. U. S. A 95, 9626–30 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kabashi E & Durham HD Failure of protein quality control in amyotrophic lateral sclerosis. Biochim. Biophys. Acta - Mol. Basis Dis 1762, 1038–1050 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Beckman JS, Estévez AG, Crow JP & Barbeito L Superoxide dismutase and the death of motoneurons in ALS. Trends Neurosci. 24, S15–S20 (2001). [DOI] [PubMed] [Google Scholar]
- 77.Raoul C et al. Motoneuron death triggered by a specific pathway downstream of Fas. potentiation by ALS-linked SOD1 mutations. Neuron 35, 1067–83 (2002). [DOI] [PubMed] [Google Scholar]
- 78.Yoshihara T et al. Differential expression of inflammation- and apoptosis-related genes in spinal cords of a mutant SOD1 transgenic mouse model of familial amyotrophic lateral sclerosis. J. Neurochem 80, 158–67 (2002). [DOI] [PubMed] [Google Scholar]
- 79.Breckenridge DG, Germain M, Mathai JP, Nguyen M & Shore GC Regulation of apoptosis by endoplasmic reticulum pathways. Oncogene 22, 8608–8618 (2003). [DOI] [PubMed] [Google Scholar]
- 80.Shaw PJ & Eggett CJ Molecular factors underlying selective vulnerability of motor neurons to neurodegeneration in amyotrophic lateral sclerosis. J. Neurol 247 Suppl, I17–27 (2000). [DOI] [PubMed] [Google Scholar]
- 81.Ilieva H, Polymenidou M & Cleveland DW Non–cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J. Cell Biol 187, 761–772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boillée S, Vande Velde C & Cleveland DW ALS: A Disease of Motor Neurons and Their Nonneuronal Neighbors. Neuron 52, 39–59 (2006). [DOI] [PubMed] [Google Scholar]
- 83.Gurney ME et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264, 1772–5 (1994). [DOI] [PubMed] [Google Scholar]
- 84.Kawamata H & Manfredi G Different regulation of wild-type and mutant Cu,Zn superoxide dismutase localization in mammalian mitochondria. Hum. Mol. Genet 17, 3303–3317 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Igoudjil A et al. In Vivo Pathogenic Role of Mutant SOD1 Localized in the Mitochondrial Intermembrane Space. J. Neurosci 31, 15826–15837 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Riar AK et al. Sex specific activation of the ERα axis of the mitochondrial UPR (UPRmt) in the G93A-SOD1 mouse model of familial ALS. Hum. Mol. Genet 26, 1318–1327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anderson CJ et al. ALS/FTD mutant CHCHD10 mice reveal a tissue-specific toxic gain-of-function and mitochondrial stress response. Acta Neuropathol. 1–19 (2019). doi: 10.1007/s00401-019-01989-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schlicker C et al. Substrates and Regulation Mechanisms for the Human Mitochondrial Sirtuins Sirt3 and Sirt5. J. Mol. Biol 382, 790–801 (2008). [DOI] [PubMed] [Google Scholar]
- 89.Schumacker PT Reactive oxygen species in cancer cells: Live by the sword, die by the sword. Cancer Cell (2006). doi: 10.1016/j.ccr.2006.08.015 [DOI] [PubMed] [Google Scholar]
- 90.Fukai T & Ushio-Fukai M Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid. Redox Signal 15, 1583–606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Che M, Wang R, Li X, Wang HY & Zheng XFS Expanding roles of superoxide dismutases in cell regulation and cancer. Drug Discovery Today (2016). doi: 10.1016/j.drudis.2015.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Finley LWS et al. SIRT3 Opposes Reprogramming of Cancer Cell Metabolism through HIF1α Destabilization. Cancer Cell 19, 416–428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Papa L, Manfredi G & Germain D SOD1, an unexpected novel target for cancer therapy. Genes Cancer 5, 15–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Somwar R, Shum D, Djaballah H & Varmus H Identification and Preliminary Characterization of Novel Small Molecules That Inhibit Growth of Human Lung Adenocarcinoma Cells. J. Biomol. Screen 14, 1176–1184 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Somwar R et al. Superoxide dismutase 1 (SOD1) is a target for a small molecule identified in a screen for inhibitors of the growth of lung adenocarcinoma cell lines. Proc. Natl. Acad. Sci 108, 16375–16380 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang P, Feng L, Oldham EA, Keating MJ & Plunkett W Superoxide dismutase as a target for the selective killing of cancer cells. Nature 407, 390–395 (2000). [DOI] [PubMed] [Google Scholar]
- 97.Chen Y-L & Kan W-M Down-regulation of superoxide dismutase 1 by PMA is involved in cell fate determination and mediated via protein kinase D2 in myeloid leukemia cells. Biochim. Biophys. Acta - Mol. Cell Res 1853, 2662–2675 (2015). [DOI] [PubMed] [Google Scholar]
- 98.Glasauer A, Sena LA, Diebold LP, Mazar AP & Chandel NS Targeting SOD1 reduces experimental non-small-cell lung cancer. J. Clin. Invest 124, 117–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li S et al. Disrupting SOD1 activity inhibits cell growth and enhances lipid accumulation in nasopharyngeal carcinoma. Cell Commun. Signal 16, 28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Papa L, Hahn M, Marsh EL, Evans BS & Germain D SOD2 to SOD1 switch in breast cancer. J. Biol. Chem 289, 5412–5416 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gomez ML, Shah N, Kenny TC, Jenkins EC & Germain D SOD1 is essential for oncogene-driven mammary tumor formation but dispensable for normal development and proliferation. Oncogene In Press, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rao AK, Ziegler YS, McLeod IX, Yates JR & Nardulli AM Effects of Cu/Zn superoxide dismutase on estrogen responsiveness and oxidative stress in human breast cancer cells. Mol. Endocrinol 22, 1113–24 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]