Summary
The process of affinity maturation, whereby T and B cells bearing antigen receptors with optimal affinity to the relevant antigen undergo preferential expansion, is a key feature of adaptive immunity. Natural killer (NK) cells are innate lymphocytes capable of “adaptive” responses following cytomegalovirus (CMV) infection. However, whether NK cells are similarly selected on the basis of their avidity for cognate ligand is unknown. Here, we showed that NK cells with the highest avidity for the mouse CMV glycoprotein m157 were preferentially selected to expand and comprise the memory NK cell pool, whereas low avidity NK cells possessed greater capacity for interferon (IFN)-γ production. Moreover, we provide evidence for avidity selection occurring in human NK cells during human CMV infection. These results delineate how heterogeneity in NK cell avidity diversifies NK cell effector function during antiviral immunity, and how avidity selection may serve to produce the most potent memory NK cells.
Keywords: NK cells, viral infection, cytomegalovirus, heterogeneity, selection
Graphical Abstract
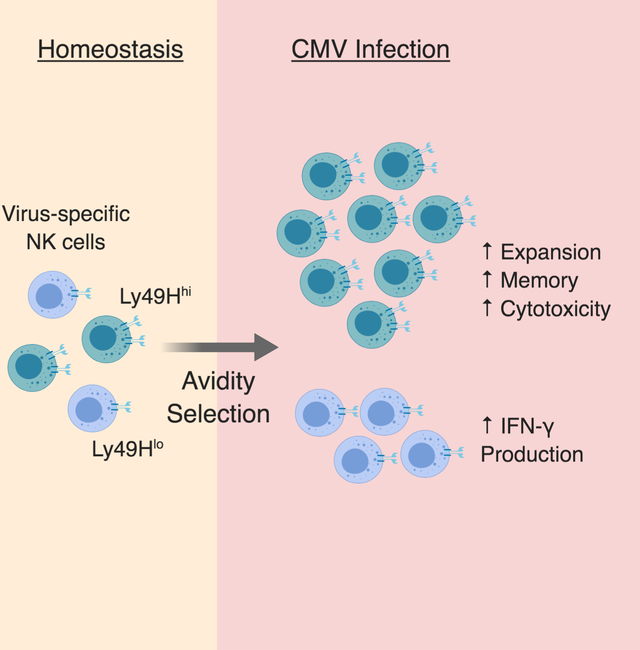
eTOC Blurb
NK cells are innate lymphocytes capable of “adaptive” responses following infection, but whether they undergo avidity selection is unknown. Adams et al. report that diversity in antigen receptor Ly49H expression drives NK cell functional heterogeneity during MCMV infection, with high avidity NK cells being selected to dominate the adaptive response.
Introduction
A fundamental feature of adaptive immunity is its specificity for a nearly infinite array of potential antigens (Goldrath and Bevan, 1999). During an infectious challenge, the repertoire of presented antigens and competition for these antigens shape the diversity of the effector lymphocyte population in a manner dependent on lymphocyte responsiveness to a given epitope (Johnson et al., 2016; Kedl et al., 2003; Kedl et al., 2000; Oberle et al., 2016). During a primary immune response, competition for antigen and resources results in outgrowth of CD8+ T cells bearing TCRs that recognize their cognate antigen with optimal affinity (Day et al., 2007; Trautmann et al., 2005). B cells iteratively mutate their BCR in the germinal center via somatic hypermutation until their BCR affinity is optimal for sensing antigen presented by follicular dendritic cells (FDCs), which provide high affinity B cells with necessary survival and proliferation signals, a process known as affinity maturation (Kim et al., 1981; Küppers et al., 1993; MacLennan, 1994; Muramatsu et al., 1999). Furthermore, T cell affinity maturation occurs during a secondary or recall response, a selective process in which the diversity of the epitopespecific CD8+ T cell TCR repertoire becomes restricted (Busch and Pamer, 1999). Thus, the adaptive immune system evolves in real-time by selecting lymphocytes best able to combat the specific pathogens encountered.
Natural killer (NK) cells are innate lymphocytes that play a critical, dominant role in herpesvirus control in both mice and humans (Biron et al., 1989; Bukowski et al., 1985; Etzioni et al., 2005). Although traditionally categorized under innate immunity, NK cells can exhibit “adaptive” antiviral responses to mouse cytomegalovirus (MCMV), including robust clonal proliferation and establishment of a long-lived population of memory cells with enhanced functionality and protective capacity following MCMV reinfection (Sun et al., 2009). In C57BL/6 mice, a subset of naïve NK cells bearing the activating receptor Ly49H undergo this adaptive response following recognition of MCMV-encoded m157, an MHC class I-like glycoprotein expressed on the infected cell surface (Arase et al., 2002; Daniels et al., 2001; Dokun et al., 2001; Sun et al., 2009). In both mice and humans, the combination of stocastically-expressed germline-encoded activating and inhibitory receptors generates NK cell diversity and heterogeneity at the population level (Horowitz et al., 2013; Lanier, 2005). Several groups have reported that heterogeneity within the naïve Ly49H+ NK cell pool affects the antiviral NK cell response, in particular the preferential expansion of unlicensed NK cells, NK cells with a history of recombination-activating gene (RAG) expression, and NK cells that lack expression of killer cell lectin-like receptor G1 (KLRG1) or the inhibitory receptor NKR-P1B (Kamimura and Lanier, 2015; Karo et al., 2014; Orr et al., 2010; Rahim et al., 2016). However, whether Ly49H+ NK cells undergo selection during generation of the effector and memory pool on the basis of their avidity for m157 remains unresolved. Here, we interrogated the effect that heterogeneity in antigen receptor expression has on Ly49H+ NK cell functional responses, and provide evidence that both mouse and human NK cells undergo avidity selection during CMV infection.
Results
MCMV m157 drives higher Ly49H expression within the antiviral NK cell pool
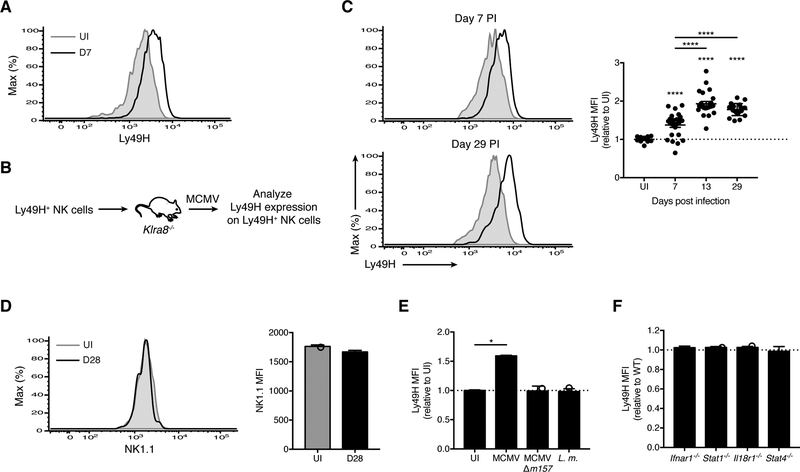
To investigate whether viral infection modulates the avidity of the virus-specific NK cell pool, we infected wild-type (WT) mice with MCMV and observed increased surface expression of Ly49H on Ly49H+ NK cells (Figures 1A and S1A). To track antigen-experienced NK cells longitudinally, we transferred Ly49H+ NK cells into Ly49H-deficient recipients (Klra8−/− mice) (Figure 1B). As the endogenous NK cells in Klra8−/− mice cannot engage MCMV m157 (Fodil-Cornu et al., 2008), only the transferred Ly49H+ NK cells undergo MCMV-driven clonal proliferation (Sun et al., 2009). Compared with the naïve Ly49H+ NK pool, effector and memory Ly49H+ NK cells displayed higher Ly49H expression (Figure 1C). In contrast, expression of NK1.1, which recognizes MCMV m12 (Aguilar et al., 2017), remained unchanged on memory NK cells (Figure 1D), indicating that the sustained change in the distribution of Ly49H expression on MCMV-driven NK cells may be unique among NK cell activating receptors. Neither infection with MCMV lacking m157 (MCMV-Δm157) nor with Listeria monocytogenes affected Ly49H expression on Ly49H+ NK cells (Figure 1E), indicating that m157 is required for this process. During MCMV infection, NK cells are exposed not only to m157 on infected cells, but also to an environment rich in proinflammatory cytokines (Biron and Tarrio, 2015). However, NK cells unresponsive to interleukin (IL)-12 (Stat4−/−), IL-18 (Il18r1−/−), and type I interferon (IFN) (Ifnar1−/− and Stat1−/−) expressed Ly49H comparably to WT effector NK cells following MCMV infection (Figures 1F and S1B), suggesting that these cytokines are individually dispensable for the MCMV-driven avidity changes. Thus, MCMV infection elicits an m157-dependent change in the distribution of Ly49H expression on effector and memory Ly49H+ NK cells.
Figure 1. MCMV-driven changes in Ly49H expression within the antiviral NK cell pool are m157-dependent.
(A) Histograms of Ly49H expression on splenic Ly49H+ NK cells (gating strategy in Figure S1A) from uninfected (UI) and MCMV-infected WT mice at day 7 PI (D7). Data are representative of at least five independent experiments with 5–10 mice per experiment.
(B-C) Experimental schematic (B). Ly49H+ NK cells were transferred into Klra8−/− mice, and their phenotype analyzed following MCMV infection of the recipients. (C) Histograms of Ly49H expression on Ly49H+ NK cells from blood of UI WT mice (shaded gray) versus at day 7 PI (top left) and day 29 PI (bottom left) (black lines). Quantification of Ly49H median fluorescence intensity (MFI) on Ly49H+ NK cells at indicated days PI relative to UI WT mice bled on the same day (right). Data are representative of three independent experiments with 10–25 mice per experiment.
(D) As in (C), histograms (left) and MFI (right) of NK1.1 on Ly49H+ NK cells from blood of UI WT mice and at day 28 PI. Data are representative of at least three independent experiments with 3–9 mice per experiment.
(E) WT mice were infected with MCMV, MCMV-Δm157, Listeria monocytogenes (L.m.) or UI. Data is represented as Ly49H MFI on Ly49H+ NK cells from blood at day 7 PI for indicated infections relative to UI. Data are representative of two independent experiments with 2–5 mice per group.
(F) As in (B), except splenocytes from WT and indicated knock-out (KO) mice were co-transferred. Data is represented as Ly49H MFI on KO Ly49H+ NK cells relative to WT from blood at day 7 PI. Data are representative of two to four independent experiments with 2–5 mice per group. Groups with a ratio < 1 were compared against 1 using a one sample t test.
Groups were compared using an unpaired, two-tailed Student’s t test (D) or against 1 using a one sample t test (E). In (C), each timepoint was compared against 1 using a one sample t test, and against each other using a paired two-tailed t test. Data are presented as the mean ± SEM. *p < 0.05; ****p < 0.0001. See also Figure S1.
Avidity selection shapes the effector and memory NK cell pool during MCMV infection
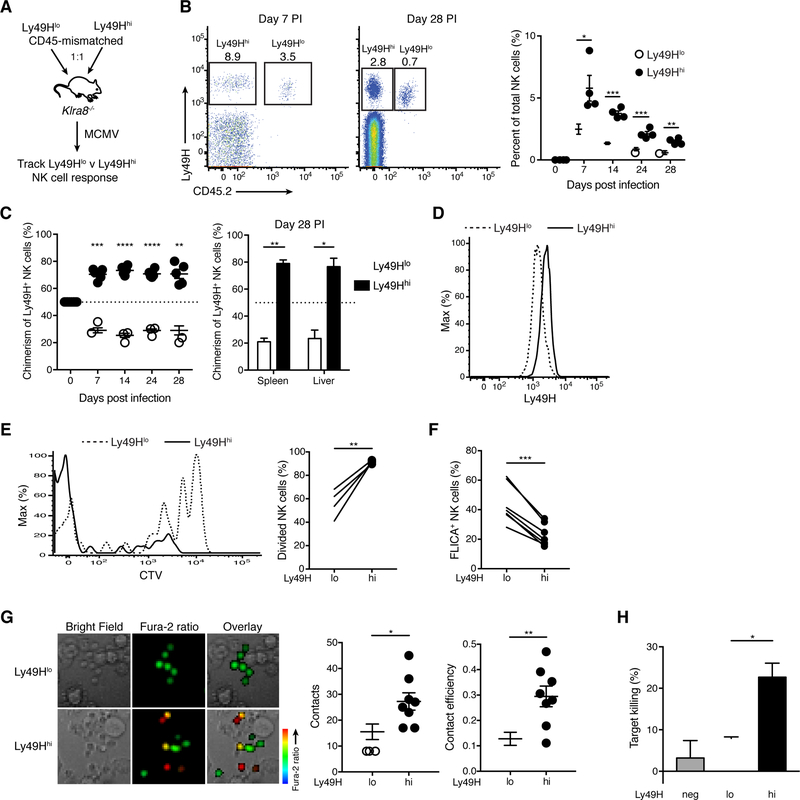
We reasoned that one explanation for a population increase in Ly49H could be preferential expansion of Ly49H+ NK cells with greater baseline Ly49H expression (i.e. avidity selection). To test this hypothesis, we purified equal numbers of Ly49H+ NK cells with different Ly49H receptor abundance (Ly49Hlo and Ly49Hhi, defined as Ly49H+ NK cells in the bottom or top ~20% of Ly49H median fluorescence intensity (MFI), respectively) from congenically distinct WT mice, co-transferred them into MCMV-infected Ly49H-deficient recipients, and longitudinally analyzed their activation and expansion (Figures 2A and S1C). Although both Ly49Hlo and Ly49Hhi NK cells clonally expanded and formed memory (Figures 2B and 2C), the magnitude of their responses differed, regardless of their congenic markers (Figure S2A). Ly49Hhi NK cells expanded more robustly and outnumbered Ly49Hlo NK cells at 7 days post infection (PI) (the peak of the antiviral response) and preferentially contributed to the memory NK cell pool in peripheral blood and various tissues at 28 days PI (Figures 2B and 2C). The ratio of Ly49Hhi to Ly49Hlo NK cells did not diverge further during contraction (Figure 2C), suggesting the greater abundance of Ly49Hhi memory NK cells is due to their expansion advantage. Even in a non-competitive setting, when Ly49Hlo and Ly49Hhi NK cells were transferred into separate animals, Ly49Hhi NK cells produced a larger effector and memory population (Figure S2B), indicating that NK cell avidity for viral ligand regulates intrinsic aspects of the antiviral NK cell response. Despite their differential expansion, Ly49Hlo and Ly49Hhi NK cells matured similarly (Figure S2C). They also retained their relative differences in Ly49H expression following infection (Figure 2D), suggesting that Ly49H expression is clonally maintained. Naïve NK cells co-expressing both Ly49H and Ly49D exhibited reduced abundance of each receptor compared to naïve NK cells expressing either Ly49H or Ly49D alone (Figure S2D). Thus, Ly49D expression may contribute to establishing heterogeneity in Ly49H expression within the naïve NK cell pool.
Figure 2. Ly49H+ NK cells undergo avidity selection during MCMV infection.
(A-D) Experimental schematic (A). Equal numbers of splenic Ly49Hlo and Ly49Hhi NK cells, purified from congenically distinct WT mice, were co-transferred into Klra8−/− mice, and their responses tracked following MCMV infection of the recipients. (B) Flow plots gated on NK cells from blood at day 7 PI (left) and spleen at day 28 PI (middle). Quantification of percent Ly49Hlo and Ly49Hhi NK cells within total NK cells in blood at indicated days PI (right). (C) The percentage of Ly49Hlo and Ly49Hhi NK cells within transferred Ly49H+ NK cells in blood at indicated days PI (left) and in indicated organs at day 28 PI (right). (D) Histograms of Ly49H expression on Ly49Hlo and Ly49Hhi NK cells from blood at day 7 PI. Data are representative of two independent experiments with 3–5 mice per experiment.
(E) WT splenocytes were labeled with CTV and transferred into congenically distinct WT mice prior to MCMV infection. Histograms of CTV in splenic Ly49Hlo and Ly49Hhi NK cells at day 7 PI (left). Quantification of indicated NK cell populations that have divided at least once (right). Data are representative of three independent experiments with 4 mice per experiment.
(F) Percent of splenic Ly49Hlo and Ly49Hhi NK cells staining positive for FLICA at day 4 PI. Data are pooled from two independent experiments with 3–4 mice per experiment.
(G) Purified splenic Ly49Hlo and Ly49Hhi NK cells were loaded with Fura-2AM and co-cultured with Ba/F3–m157 cells. Live-cell imaging was then performed. Representative images from the time-lapse analysis (left). Scale bar = 10 μM. Quantification of the number of contacts between NK cells and target cells (middle) and contact efficiency (right), defined as the proportion of target cell contacts that resulted in an NK cell Ca2+ flux (change in NK cell color from green to yellow or red). Data are representative of two experiments with 4 replicates per group per experiment.
(H) Purified splenic Ly49H−, Ly49Hlo, and Ly49Hhi NK cells were incubated with Ba/F3–m157 target cells (CTVhi) and Ba/F3 control cells (CTVlo) at 10:1:1 ratio (effector: target: control) for 6 hours. Quantification of target cell killing by indicated NK cell populations compared to control wells lacking NK cells. Data are representative of three independent experiments with 3 replicates per group per experiment. Ly49Hlo and Ly49Hhi NK cells were compared using an unpaired, two-tailed Student’s t test.
Groups were compared using a paired, two-tailed t test (B, E, F), an unpaired, two-tailed Student’s t test (G) or against 50 using a one sample t test (C). Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figures S1 and S2.
We also evaluated whether avidity maturation (i.e. upregulation of Ly49H within an individual NK cell) and avidity selection occur concomitantly. Ly49Hlo NK cells increased their expression of Ly49H, most notably during the contraction phase, with Ly49H expression increasing 2- to 3-fold compared to that of naïve Ly49Hlo NK cells (Figure S2E). In contrast, MCMV-driven Ly49Hhi NK cells did not considerably increase their Ly49H expression relative to naïve Ly49Hhi NK cells (Figure S2E), suggesting there may be an “upper limit” for Ly49H expression on NK cells.
To investigate the mechanism behind differential expansion of Ly49Hlo and Ly49Hhi NK cells, we transferred bulk splenocytes labeled with Cell Trace Violet (CTV) into WT mice. Following MCMV infection, Ly49Hhi NK cells divided more efficiently (Figure 2E). Furthermore, during MCMV-driven expansion, Ly49Hhi NK cells demonstrated lower caspase activity as assessed by fluorescent labeled inhibitors of caspases (FLICA) staining (Figure 2F). Collectively, these data suggest that NK cell avidity regulates the proliferative capacity and survival of a given Ly49H+ NK cell during MCMV infection, which underlies the greater expansion and subsequent memory pool establishment of Ly49Hhi NK cells.
NK cell avidity regulates m157-mediated activation and cytotoxicity
These data raised the possibility that Ly49H density affects NK cell engagement with infected cells (i.e. avidity) or the magnitude of Ly49H-dependent signaling. To test this, we performed live cell imaging of Ly49Hlo and Ly49Hhi NK cells co-cultured with Ba/F3 cells expressing m157 (Ba/F3–m157). Ly49H receptor ligation results in phosphorylation of its adaptor DAP12, which recruits and activates Syk and zeta chain of T cell receptor associated protein kinase 70 (ZAP70), initiating their signaling cascades and resulting in increased intracellular calcium (Lanier, 2008). To visualize productive target cell encounters that resulted in activation-induced Ca2+ flux, NK cells were labeled with the cell permeable Fura-2 calcium indicator. Ly49Hhi NK cells made more contacts with Ba/F3–m157 cells than did Ly49Hlo NK cells, and a greater percentage of Ly49Hhi NK cell contacts resulted in NK cell activation and Ca2+ flux (Figure 2G). These results suggest that Ly49Hhi NK cells may be preferentially recruited into the antiviral response.
Given that NK cell cytotoxicity requires activating receptor engagement, we speculated that NK cell cytotoxic function is dependent on avidity for cognate ligand. To assess this, we separately co-cultured Ly49H−, Ly49Hlo, and Ly49Hhi NK cells with equal numbers of target Ba/F3–m157 and control Ba/F3 cells labeled differentially with CTV. Although Ly49Hlo cells were capable of killing Ba/F3–m157 target cells, Ly49Hhi cells possessed the most potent cytotoxic function (Figures 2H, S2F and S2G). The enhanced functionality of Ly49Hhi NK cells was strictly Ly49H-dependent, as a greater percentage of Ly49Hhi NK cells produced IFN-γ and degranulated only in response to Ly49H ligation ex vivo, but not NK1.1 ligation or phorbol 12- myristate 13-acetate (PMA) and ionomycin (Figure S2H), excluding a developmentally-determined general heightened reactivity of these cells. Collectively, these data indicate that higher Ly49H expression precipitates greater Ly49H-dependent activation and effector function (i.e. functional avidity) in the presence of target cells expressing cognate ligand.
Ly49H density specifies differential effector functions of Ly49H+ NK cells during early MCMV infection
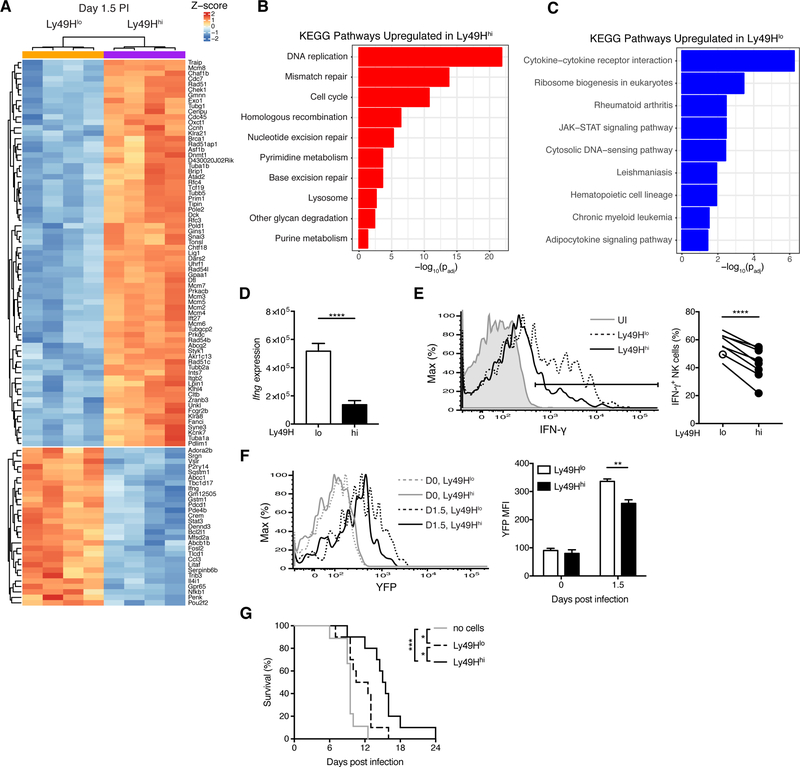
The detection of viral ligand-bearing infected cells by Ly49H+ NK cells is an essential signal for NK cell-mediated host protection, yet MCMV also induces a highly inflammatory environment rich in IL-12, IL-18, and type I interferon (Biron and Tarrio, 2015). These cytokines can lead to nonspecific activation of NK cells, promote NK cell effector function (e.g. IFN-γ production), and program their adaptive responses (Biron and Tarrio, 2015; Dokun et al., 2001; Madera et al., 2016; Madera and Sun, 2015; Sun et al., 2012). To comprehensively understand how avidity regulates the NK cell response to MCMV, we performed RNA-seq on splenic Ly49Hlo and Ly49Hhi NK cells during early MCMV infection (day 1.5 PI). Of the 1941 differentially expressed genes (padj < 0.05), 1232 transcripts were upregulated and 709 downregulated in Ly49Hhi NK cells (Table S1), the top 100 of which are shown in Figure 3A. KEGG pathway analysis of differentially expressed genes revealed an enrichment of cell cycle control, DNA replication, and genome integrity maintenance genes in Ly49Hhi NK cells (Figures 3B and S3A), consistent with their proliferative advantage (Figure 2E). In contrast, genes upregulated in Ly49Hlo NK cells were primarily involved with cytokine and JAK-STAT signaling (Figures 3C and S3B). The extent and nature of the transcriptional differences underscore the diversity within the Ly49H+ NK cell compartment and the divergence of their cellular activities within the earliest days of infection.
Figure 3. Ly49Hlo NK cells produce more IFN-γ during early MCMV infection.
(A-D) Splenic Ly49Hlo and Ly49Hhi NK cells were sorted for RNA-seq at day 1.5 PI (4 replicates each). (A) Heat map and hierarchical clustering of top 100 differentially expressed genes by p-value. (B-C) Gene ontology analysis of differential KEGG pathways for genes significantly (padj < 0.05) upregulated in Ly49Hhi NK cells (B) and upregulated in Ly49Hlo NK cells (C). Their respective p values are shown. (D) Quantification of RNA-seq reads mapping to the Ifng locus. P value was calculated in DESeq2 and adjusted for testing multiple hypotheses.
(E) Histograms of intracellular IFN-γ expression in splenic Ly49H+ NK cells from UI and MCMV-infected WT mice at day 1.5 PI (left). Quantification of percent IFN-γ+ NK cells within indicated NK cell populations (right). Data are representative of at least five independent experiments with 3–15 mice per experiment.
(F) As in (E), except UI or MCMV-infected Ifng-IRES-YFP mice at day 1.5 PI. Histograms (left) and quantification of YFP MFI (right) before and after MCMV infection. Data are representative of two independent experiments with 2–3 mice per time point per experiment.
(G) Kaplan-Meier survival curves of Rag2−/− Il2rg−/− mice that received either no cells, 50,000 purified Ly49Hlo NK cells, or 50,000 purified Ly49Hhi NK cells 2 days prior to MCMV infection. Data are pooled from two independent experiments with 4–5 mice per group per experiment.
Groups were compared using a paired, two-tailed t test (E, F) or the Log-rank (Mantel-Cox) test with correction for testing multiple hypotheses (G). Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figures S2 and S3 and Table S1.
To validate and extend our RNA-seq results, we focused on several differentially expressed genes notable for their role in NK cells and regulation by proinflammatory cytokines. Transcript (Il2ra) and protein (CD25) of the high-affinity α subunit of the IL-2 receptor were both more robustly upregulated in Ly49Hlo NK cells during MCMV infection (Figures S3C and S3D). Furthermore, consistent with higher quantities of Ifng (encoding IFN-γ) transcripts in Ly49Hlo NK cells (Figure 3D), a greater percentage of Ly49Hlo NK cells produced IFN-γ during MCMV infection (Figure 3E), and Ly49Hlo NK cells produced more IFN-γ than Ly49Hhi NK cells on a per-cell basis (Figure S3E). In Ifng-IRES-YFP reporter mice (“GREAT” mice), Ly49Hlo NK cells more robustly upregulated Ifng transcript during MCMV infection (Figure 3F). However, prior to infection, there were no differences in baseline Ifng transcript (Figures 3F and S3F) or their ability to produce IFN-γ following IL-12 and IL-18 stimulation ex vivo (Figure S2H), suggesting that infection-specific differences in the regulation of the Ifng locus accounted for the observed phenotype. Although bulk Ly49H− and Ly49H+ NK cells are thought to express similar IFN-γ at this early time point (Dokun et al., 2001), the “population average” across Ly49H+ NK cells concealed considerable heterogeneity in the Ly49H+ NK cell compartment (Figure S3G). Thus, the extent of Ly49H signaling in vivo during MCMV infection regulates NK cell effector properties.
Ly49Hlo and Ly49Hhi NK cells had similar maturation profiles (by CD11b and CD27 subsets) despite modest differences in KLRG1 (Figure S3H), and similar proportions of both NK cell populations expressed NKG2A (Figure S3I), one of the inhibitory receptors licensed in C57BL/6 mice. A greater proportion of Ly49Hhi NK cells expressed Ly49C/I (Figure S3I), yet consistent with an earlier report (Orr et al., 2010), Ly49C/I expression did not effect IFN-γ production in vivo during MCMV infection (Figure S3J). Thus, the functional and transcriptional differences between Ly49Hlo and Ly49Hhi NK cells during MCMV infection are likely due to infection-specific, rather than developmental differences. Furthermore, Ly49Hlo and Ly49Hhi NK cells displayed similar abundance of phosphorylated STAT4 (pSTAT4), comparable IL-18 receptor alpha (IL-18Ra), and transcripts encoding IL-12 receptor chains (Figures S3K and S3L). Thus, Ly49H-mediated regulation of proinflammatory cytokine responsiveness is likely occurring further downstream of cytokine receptor expression and STAT4 phosphorylation.
To understand whether the functional differences between these two NK cell subsets have a physiological role in host defense, we compared protection of susceptible Rag2−/− Il2rg−/− mice by Ly49Hlo or Ly49Hhi NK cells. Although Ly49Hlo NK cells provided some protection against MCMV (compared to mice that did not receive NK cells), Ly49Hhi NK cells significantly extended the survival of susceptible hosts beyond that of Ly49Hlo NK cells (Figure 3G). Collectively, these results reveal that the avidity-dependent functional differences between Ly49Hlo and Ly49Hhi NK cells regulate the protective antiviral responses mounted by NK cells.
Human NK cells show evidence for avidity selection during HCMV infection
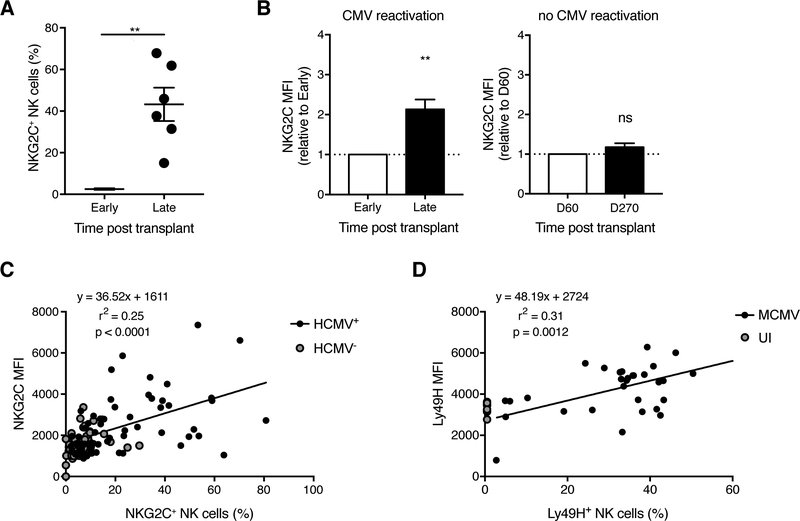
Analogous to the Ly49H+ NK cell response against MCMV in mice, CD94/NKG2C+ NK cells are expanded in the peripheral blood of human cytomegalovirus (HCMV)-seropositive healthy individuals (Gumá et al., 2004; Lopez-Vergès et al., 2011), as well as in recipients of solid organ and hematopoietic stem cell transplants (HSCT) who reactivate HCMV (Della Chiesa et al., 2012; Foley et al., 2012; Horowitz et al., 2015; Lopez-Vergès et al., 2011; Muccio et al., 2016). HCMV-encoded UL40 peptides loaded onto HLA-E were recently shown to activate human NKG2C+ NK cells in a peptide-specific manner (Hammer et al., 2018), comparable to the interaction between MCMV m157 and Ly49H in mice. To determine whether HCMV infection similarly modulates the avidity of human NKG2C+ NK cells, we analyzed NK cells from peripheral blood of T cell-depleted HSCT recipients. In the transplant setting, the timing of HCMV infection (or reactivation in patients previously reported to be HCMV-seropositive) can be estimated, which enabled longitudinal analysis of NK cells in the same patient. Consistent with previous reports (Foley et al., 2012; Horowitz et al., 2015; Muccio et al., 2016), NKG2C+ NK cells were expanded in HSCT patients following HCMV detection (Figure 4A). Phenotypic analysis revealed higher NKG2C expression on the expanded NKG2C+ NK cell population from HCMV-reactivating patients, but not on the unexpanded population of NKG2C+ NK cells from patients who remained HCMV-seronegative post-transplant (Figure 4B). This evidence supports the notion that NK cell avidity selection during CMV infection may be conserved between mouse and human.
Figure 4. HCMV reactivation in HSCT recipients drives higher NKG2C expression on human NKG2C+ NK cells.
(A-B) Blood was drawn from T cell-depleted HSCT recipients in the year following transplant (every ~30–90 days). HCMV infection status was determined by viral qPCR. “Early” and “Late” are paired samples from the same HCMV-reactivating transplant recipients. “Early” refers to 15–60 days post transplant depending on the recipient, and “Late” includes the window between 200 days to 1 year post transplant. The data are presented as such to synchronize the timing of adaptive NKG2C+ NK cell appearance, which was recipient-dependent. It was expansion of the NKG2C+ NK cell population that dictated the observed kinetic profile of NKG2C MFI. (A) Percentage of NKG2C+ NK cells among CD3−CD56+ cells (gating strategy in Figure S4). (B) Quantification of NKG2C MFI on NKG2C+ NK cells from transplant recipients who reactivated (left) or did not reactivate (right) HCMV. Groups were compared using a paired, two-tailed t test (A) or against 1 using a one sample t test (B). Data are presented as the mean ± SEM. ns, not significant; **p < 0.01.
(C) Blood was drawn from healthy HCMV-seropositive and HCMV-seronegative donors. Correlation between NKG2C MFI and percentage of NKG2C+ NK cells. Linear regression was performed on HCMV+ samples. P value represents the likelihood of a non-zero slope.
(D) Experimental design as in Figure 1B. Correlation between Ly49H MFI and the Ly49H+ NK cell expansion (i.e. percentage of Ly49H+ NK cells among total NK cells) at day 7 PI. UI WT mice were assigned 0% expansion. Linear regression was performed on MCMV-infected samples. P value represents the likelihood of a non-zero slope.
See also Figure S4.
In a larger healthy human cohort, HCMV-seronegative donors had minimal variation in NKG2C expression and NKG2C+ NK cell percentage, whereas HCMV-seropositive donors displayed a positive linear relationship between NKG2C expression and percentage of NKG2C+ NK cells (Figure 4C). Similarly, during the mouse Ly49H+ NK cell response to MCMV, Ly49H expression strongly correlated with expansion of the Ly49H+ NK cell population at the peak of the antiviral response (Figure 4D). These data support the idea that the degree of virus-driven NK cell expansion and the extent of NK cell avidity selection are closely linked processes in both mice and humans.
Discussion
Our study provides a mechanistic understanding of how avidity for viral ligand regulates the functional contribution of an NK cell during MCMV infection. Although NK cells of varying avidities were recruited during early MCMV infection, similar to CD8+ T cells with a range of TCR affinities (Zehn et al., 2009), high avidity NK cells preferentially expanded. Concurrent with our studies using NK cell populations sorted on the basis of Ly49H surface density, Grassmann et al. used retrogenic color barcoding to track single NK cell-derived responses (Grassmann et al., 2019). In accordance with our findings, they also observed that Ly49H expression in individual NK cell clones correlated with the degree of their clonal expansion. Thus, the major conclusions from both of these complementary studies are in agreement. Furthermore, it appeared that effector and memory Ly49Hlo NK cells expressed more Ly49H than they do as naïve NK cells, suggesting that avidity maturation (i.e. upregulation of Ly49H within an individual NK cell) and avidity selection may be occurring concomitantly. Together these processes may contribute to the generation of a memory NK cell pool with heightened specificity for MCMV re-encounter (Sun et al., 2009).
Previous in vivo imaging studies have described that TCR transgenic CD8+ T cell priming by cognate peptide-loaded DCs occurs in three phases: 1) transient serial encounters with DCs, 2) stable contacts with DCs, and 3) CD8+ T cell motility and proliferation (Mempel et al., 2004). Stable contacts between CD8+ T cells and DCs are required for the full differentiation program. These stronger and longer interactions program CD8+ T cell clonal expansion, and are dependent on the absolute number of TCR-pMHC interactions (Henrickson et al., 2008). Furthermore, high affinity TCR ligands and prolonged antigen exposure dictate the duration and magnitude, respectively, of the CD8+ T cell expansion (Prlic et al., 2006; Zehn et al., 2009). Our data reveal that many requirements for naïve CD8+ T cell and NK cell priming and clonal expansion are conserved. Although MCMV infection activated both Ly49Hlo and Ly49Hhi NK cells, these cells were transcriptionally distinct during the earliest days of infection. We speculate that the number of activating receptor-cognate ligand interactions regulates the strength or duration of contact between an NK cell and an infected cell, which plays a fundamental role in programming the response of that cell.
Despite the greater potential of Ly49Hhi NK cells for expansion and cytotoxicity, Ly49Hlo NK cells were the primary IFN-γ-producing NK cells during the early effector response, suggesting a division of labor between these two cell populations. One possibility is that an NK cell must allocate cellular and metabolic resources for the competing demands of IFN-γ secretion and expansion, or that strong Ly49H receptor ligation may antagonize NK cell responsiveness to proinflammatory cytokines. Indeed, MCMV-driven memory NK cells, which have greater Ly49H surface density, have diminished bystander responses to heterologous infection (Min-Oo and Lanier, 2014). Thus, strong Ly49H activation precipitates both immediate and heritable changes in IFN-γ production.
Ly49Hlo and Ly49Hhi NK cells maintained their relative differences in Ly49H density following MCMV-driven proliferation, suggesting clonal maintenance of Ly49H expression. Given that Klra8 (encoding Ly49H) was the most differentially expressed gene ranked by p- value between Ly49Hlo and Ly49Hhi NK cells at day 1.5 PI (Table S1), we hypothesize that Ly49H expression is transcriptionally maintained during infection. Since the effector function profile of an NK cell during MCMV infection hinges on its Ly49H expression, further investigation into the full complement of factors (of which, Ly49D may be one such variable) that regulate and maintain Ly49H expression during development, homeostasis, and infection is warranted.
Analogous to avidity selection of mouse NK cells during MCMV infection, we demonstrated that the NKG2C+ NK cell pool that arose in HSCT recipients following HCMV infection or reactivation expressed more NKG2C. NKG2C zygosity has been reported to influence surface receptor density and NKG2C+ NK cell numbers in HCMV+ subjects (Muntasell et al., 2013), and could contribute to the range in these parameters in our transplant patient cohort that reactivated HCMV. Future work is required to determine the molecular mechanisms behind avidity selection of human NK cells, and how avidity affects human NK cell repertoire diversity based on prior pathogen experience (Strauss-Albee et al., 2015). Nevertheless, our findings collectively indicate that avidity selection may be a general mechanism in NK cell biology conserved between mouse and human, likely resulting from the extensive co-evolution of NK cells with CMV, and can inform vaccination strategies to elicit memory NK cells with the greatest avidity for viral ligands.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joseph Sun (sunj@mskcc.org).
Experimental Model and Subject Details
Mice
All mice used in this study were housed and bred under specific pathogen-free conditions at Memorial Sloan Kettering Cancer Center (MSKCC), and handled in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC). The following mouse strains were used in this study: C57BL/6 (CD45.2), B6.SJL (CD45.1), Klra8−/− (Ly49H-deficient) (Fodil-Cornu et al., 2008), Stat4−/−, Il18r1−/−, Ifnar1−/−, Stat1−/−, Ifng-IRES-YFP (GREAT), and Rag2−/− Il2rg−/− mice. Experiments were conducted using age- and gender-matched mice in accordance with approved institutional protocols. Mice used in this study were of both genders and were 6–8 weeks of age at the time of experimentation.
Primary Human Cells
Peripheral blood samples were collected from allogeneic bone marrow transplantation patients and healthy human donors following approval from the MSKCC Institutional Review Board, and donors provided informed, written consent. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll centrifugation. Additional PBMCs were isolated from buffy coats obtained from healthy volunteer donors via the New York Blood Center (NYBC, http://nybloodcenter.org/). The MSKCC IRB waived the need for additional research consent for anonymous NYBC samples. Donors were of both genders and ranged in age between 12 and 76 years old. PBMCs were cryopreserved in fetal bovine serum with 10% DMSO. HCMV serostatus was provided by NYBC.
Virus
MCMV (Smith strain) was serially passaged through BALB/c hosts three times, and then salivary gland viral stocks were prepared with a dounce homogenizer for dissociating the salivary glands of infected mice 3 weeks after infection.
Bacteria
Frozen stocks of Listeria monocytogenes were grown in brain-heart infusion (BHI) broth. Bacteria culture samples were grown to mid-log phase, measured by optical density (A600), and diluted in PBS for injection.
Method Details
In Vivo Virus Infection
Adoptive co-transfer studies (both competitive and non-competitive) were performed by transferring 105 Ly49Hlo NK cells and/or 105 Ly49Hhi NK cells, purified from spleens of congenically distinct WT mice (CD45.1 or CD45.2), into Klra8−/− mice 1 day prior to MCMV infection. Recipient mice in adoptive (co-)transfer studies were infected with MCMV by intraperitoneal (i.p.) injection of 7.5 × 102 plaque-forming units (PFU) in 0.5 mL.
Survival studies were performed by transferring either no cells, 5×104 purified splenic Ly49Hlo NK cells, or 5×104 purified splenic Ly49Hhi NK cells into Rag2−/− Il2rg−/− mice 2 days prior to MCMV infection. Recipient mice in survival studies were infected with 7.5 × 103 PFU MCMV in 0.5 mL by i.p. injection.
In several experiments, WT mice were directly infected with MCMV (7.5 × 103 PFU) or MCMV-Δm157 (105 PFU) (Bubic et al., 2004) in 0.5 mL by i.p. injection.
In Vivo Bacterial Infection
Mice were infected with priming doses equivalent to 2000–5000 colony forming units (CFU) by tail vein injection.
Lymphocyte Isolation
Spleens were dissociated using glass slides and filtered through a 100-μm strainer. To isolate lymphocytes from liver, the tissue was physically dissociated using a glass tissue homogenizer and purified using a discontinuous gradient of 40% over 60% Percoll. Red blood cells in blood, spleen, and liver were lysed using ACK lysis buffer.
Flow Cytometry and Cell Sorting
Cell surface staining of single-cell suspensions from various organs was performed using fluorophore-conjugated antibodies (BD Biosciences, eBioscience, BioLegend, Tonbo Biosciences, Beckman Coulter, Miltenyi). Intracellular staining was performed by fixing and permeabilizing with the eBioscience Foxp3 Transcription Factor Staining Set (Thermo Fisher) for staining intranuclear proteins and cytokines, or with formaldehyde and methanol for staining phosphorylated STAT proteins.
Flow cytometry and cell sorting were performed on the LSR II and Aria II cytometers (BD Biosciences), respectively. Data were analyzed with FlowJo software (Tree Star). Flow cytometry of mouse lymphocytes was performed using the following fluorophore-conjugated antibodies: CD3ε (17A2), TCRβ (H57–597), CD19 (6D5), F4/80 (BM8.1), NK1.1 (PK136), Ly49H (3D10), CD45.1 (A20), CD45.2 (104), CD11b (M1/70), CD27 (LG.7F9), KLRG1 (2F1), Ly49D (4E5), Ly49C/I (5E6), NKG2A/C/E (20D5), CD25 (PC61), IFN-γ (XMG1.2), CD107a (1D4B), STAT4 pY693 (38/p-Stat4), IL-18Ra (P3TUNYA), and Fixable Viability Dye. Staining of human PBMCs was performed using: CD3 (UCHT1), CD56 (N901), NKG2C (REA205), and LIVE/DEAD as viability marker.
Apoptosis was evaluated by caspase activity staining using the carboxyfluorescein FLICA poly caspase assay kit (Bio-Rad). NK cell proliferation was analyzed by labeling cells with 5 μM CellTrace Violet (CTV, Thermo Fisher) prior to transfer, and CTV labeling was performed according to manufacturer protocol.
NK Cell Enrichment and Purification
NK cells (TCRβ−CD3ε−CD19−F4/80−CD45+NK1.1+) were enriched from spleens of pooled C57BL/6 mice by negative selection over BioMag goat anti-rat IgG beads (Qiagen) coated with rat anti-mouse CD8α, CD4, CD19, and Ter-119 antibodies (Bio X Cell, clones 2.43, GK1.5, 1D3, and TER-119 respectively) before being sorted to high purity on an Aria II cytometer (BD Biosciences).
Ex Vivo Stimulation of Lymphocytes
104 purified splenic Ly49Hlo or Ly49Hhi NK cells were stimulated for 4 hours in RPMI containing 10% fetal bovine serum with 20 ng/mL recombinant mouse IL-12 (R&D Systems) plus 10 ng/mL IL-18 (MBL), 10 ng/mL PMA (Sigma) plus 1 g/mL Ionomycin (Sigma), or 25 g/mL of plate-bound anti-mouse NK1.1 (PK136, BioLegend) or anti-mouse Ly49H (3D10, BioLegend). Cells were cultured in media alone as a negative control.
Ca2+ Imaging of NK Cells
Purified splenic NK cells (TCRβ−CD3ε−CD19−F4/80−CD45+NK1.1+) with different Ly49H expression were loaded with 5 μg/ml Fura-2AM for 30 min and transferred into colorless RPMI (no phenol red) containing 5% FCS. 8 × 104 NK cells were mixed 1:4 with Ba/F3–m157 cells and imaged in 8-well chamber slides (Thermo Fisher Scientific) using an inverted fluorescence video microscope (IX-81; Olympus) fitted with a 20×, 0.75 NA objective lens (Olympus). A Xe lamp (DG-4; Sutter Instrument) was used for fluorophore excitation, and data were collected on an electron-multiplying charge-coupled device camera (ImagEM; Hamamatsu Photonics). Time-lapse recordings were made using SlideBook software (Intelligent Imaging Innovations). One brightfield image and one Fura-2 image were taken every 20 s for 20 min.
Ex Vivo Killing Assay
Ba/F3 control cells and Ba/F3–m157 target cells (Arase et al., 2002) were labeled differentially with CTV (Ba/F3, CTVlo; Ba/F3–m157, CTVhi). 5 × 103 of each cell line were mixed with 5 × 104 purified splenic NK cells with different Ly49H expression, or without NK cells (control condition). Effector and target cells were co-cultured for 6 hours at 37°C in RPMI-1640 containing 10% FBS. After 6 hours, cells were stained with propidium iodide prior to flow cytometry. The percentages of target cell killing were determined using the following formula, adapted from (Viant et al., 2017): 100 −([(% Ba/F3–m157 cells/% Ba/F3 cells)]/[(% Ba/F3–m157 cells/% Ba/F3 cells) control] × 100). For this formula, only CTV+ cells within live cells were considered. The percentage of propidium iodide-staining target cells was determined from total CTVhi cells.
RNA Sequencing
Ly49Hlo and Ly49Hhi NK cells were sorted from the spleens of WT mice 1.5 days post MCMV infection. Spleens from three infected mice were pooled to achieve ~5×104 cells of each population (one paired replicate). RNA from cells suspended in Trizol was extracted with chloroform. Isopropanol and linear acrylamide were added, and the RNA was precipitated with 75% ethanol. Samples were resuspended in RNase-free water. After RiboGreen quantification and quality control by Agilent BioAnalyzer, 2ng total RNA with RNA integrity numbers ranging from 8.1 to 10 underwent amplification using the SMART-Seq v4 Ultra Low Input RNA Kit (Clonetech catalog # 63488), with 12 cycles of PCR. Subsequently, 10ng of amplified cDNA were used to prepare libraries with the KAPA Hyper Prep Kit (Kapa Biosystems KK8504) using 8 cycles of PCR. Samples were barcoded and run on a HiSeq 4000 in a 50bp/50bp paired end run, using the HiSeq 3000/4000 SBS Kit (Illumina). An average of 38 million paired reads were generated per sample and the percent of mRNA bases per sample ranged from 79% to 81%.
Quantification and Statistical Analysis
RNA Sequencing Analysis
Paired-end reads were trimmed for adaptors and removal of low-quality reads using Trimmomatic (v.0.36) (Bolger et al., 2014). Trimmed reads were mapped to the Mus musculus genome (mm10 assembly) and counted at transcript-level using quasi-mapping approach by Salmon (v0.10.2) (Patro et al., 2017). These transcript-level estimates were then summarized at gene-level using tximport (v1.9.12) (Soneson et al., 2015). Differential expression (DE) analysis was executed with DESeq2 (v1.12.17) (Love et al., 2014) using UCSC knownGene model as a reference annotation. Genes were considered to be differentially expressed between two groups (Ly49Hhi versus Ly49Hlo) if their adjusted p-values were less than 0.05. Gene Ontology (GO) analysis was performed using goseq (v1.33.0) (Young et al., 2010) on KEGG pathway database (KEGG.db v3.2.3) with a cutoff of log2 fold change > 0 or < 0 for DE upregulated or downregulated genes, respectively.
Statistical Analysis
For graphs, data are shown as mean ± SEM, and unless otherwise indicated, statistical differences were evaluated using an unpaired, two-tailed Student’s t test, assuming equal sample variance. For experiments in which Ly49Hlo and Ly49Hhi NK cells were compared within the same mouse, a paired, two-tailed t test was used. Statistical differences in survival were determined by the Log-rank (Mantel-Cox) test. p < 0.05 was considered significant. Graphs were produced and statistical analyses were performed using GraphPad Prism.
Data Availability
RNA-seq data comparing Ly49Hlo and Ly49Hhi NK cells during early MCMV infection (day 1.5 PI) are available in the Gene Expression Omnibus (GEO) under accession code GSE129490.
Supplementary Material
Table S1. Differentially Expressed Genes between Ly49Hlo and Ly49Hhi NK Cells at Day 1.5 PI, Related to Figure 3
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Mouse CD3ε (clone 17A2) | Tonbo Biosciences | Cat#25–0032; RRID:AB_2621619 |
| Anti-Mouse TCRβ (clone H57–597) | BioLegend | Cat#109220; RRID:AB_893624 |
| Anti-Mouse CD19 (clone 6D5) | BioLegend | Cat#115530; RRID:AB_830707 |
| Anti-Mouse F4/80 (clone BM8.1) | BioLegend | Cat#123117; RRID:AB_893489 |
| Anti-Mouse NK1.1 (clone PK136) for flow cytometry | Tonbo Biosciences | Cat#65–5941; RRID:AB_2621910 |
| Anti-Mouse Ly49H (clone 3D10) for flow cytometry | eBioscience | Cat#11-5886-81; RRID:AB_1257160 |
| Anti-Mouse CD45.1 (clone A20) | BioLegend | Cat#110729; RRID:AB_1134170 |
| Anti-Mouse CD45.2 (clone 104) | BioLegend | Cat#109821; RRID:AB_493730 |
| Anti-Mouse/Human CD11b (clone M1/70) | BioLegend | Cat#101223; RRID:AB_755985 |
| Anti-CD27 (clone LG.7F9) | eBioscience | Cat#14-0271-81; RRID:AB_467182 |
| Anti-Mouse KLRG1 (clone 2F1) | Tonbo Biosciences | Cat#50–5893; RRID:AB_2621800 |
| Anti-Mouse Ly49D (clone 4E5) | BioLegend | Cat#138308; RRID:AB_10639939 |
| Anti-Mouse Ly49C and Ly49I (clone 5E6) | BD Biosciences | Cat#553277; RRID:AB_394751 |
| Anti-Mouse NKG2A/C/E (clone 20D5) | BD Biosciences | Cat#740549; RRID:AB_2740251 |
| Anti-Mouse CD25 (clone PC61) | BioLegend | Cat#102016; RRID:AB_312865 |
| Anti-Mouse IFN gamma (clone XMG1.2) | Tonbo Biosciences | Cat#20–7311; RRID:AB_2621616 |
| Anti-Mouse CD107a (clone 1D4B) | BioLegend | Cat#121611; RRID:AB_1732051 |
| Anti-Stat4 pY693 (clone 38/p-Stat4) | BD Biosciences | Cat#558137; RRID:AB_397052 |
| Anti-Mouse CD218a (IL-18Ra) (clone P3TUNYA) | eBioscience | Cat#48-5183-80; RRID:AB_2574068 |
| Anti-Human CD3 (clone UCHT1) | BD Biosciences | Cat#563852; RRID:AB_2744391 |
| Anti-Human CD56 (clone N901) | Beckman Coulter | Cat#A82943; RRID:AB_2750853 |
| Anti-Human CD159c (NKG2C) (clone REA205) | Miltenyi | Cat#130-117-398; RRID:AB_2727933 |
| InVivoMab Anti-Mouse CD8α (clone 2.43) for NK cell enrichment | Bio X Cell | Cat#BE0061; RRID:AB_1125541 |
| InVivoMab Anti-Mouse CD4 (clone GK1.5) for NK cell enrichment | Bio X Cell | Cat#BE0003–1; RRID:AB_1107636 |
| InVivoMab Anti-Mouse CD19 (clone 1D3) for NK cell enrichment | Bio X Cell | Cat#BE0150; RRID:AB_10949187 |
| InVivoMab Anti-Mouse Ter-119 (clone TER-119) for NK cell enrichment | Bio X Cell | Cat#BE0183; RRID:AB_10949625 |
| LEAF™ Purified Anti-Mouse NK-1.1 (clone PK136) for plate-bound stimulation | BioLegend | Cat#108712; RRID:AB_ 313399 |
| LEAF™ Purified Anti-Mouse Ly49H (clone 3D10) for plate-bound stimulation | BioLegend | Cat#144704; RRID:AB_ 2561551 |
| Bacterial and Virus Strains | ||
| Mouse Cytomegalovirus (MCMV) | J. Sun (PI) | Smith Strain |
| MCMV-Δm157 | U. Koszinowski (PI) (Bubić et al., 2004) | N/A |
| Listeria monocytogenes | J. Sun (PI) | N/A |
| Biological Samples | ||
| Healthy human donor buffy coats | New York Blood Center | http://nybloodcenter.org/ |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant Mouse IL-12 Protein | R&D Sytems | Cat#419-ML |
| Recombinant Mouse IL-18 | MBL | Cat#B002–5 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Cat#P8139 |
| Ionomycin calcium salt from Streptomyces conglobatus (Ionomycin) | Sigma-Aldrich | Cat#I0634 |
| Critical Commercial Assays | ||
| TRIzol™ Reagent | Thermo Fisher Scientific | Cat#15596026 |
| eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set | Thermo Fisher Scientific | Cat#00-5523-00 |
| BioMag Goat Anti-Rat IgG (NK cell enrichment) | Qiagen | Cat#310107 |
| CellTrace™ Violet Cell Proliferation Kit | Thermo Fisher Scientific | Cat#C34557 |
| FAM FLICA™ Poly Caspase Kit | Bio-Rad | Cat#ICT092 |
| Fura-2, AM, cell permeant | Thermo Fisher Scientific | Cat#F1201 |
| eBioscience™ Fixable Viability Dye eFluor™ 506 | eBioscience | Cat#65-0866-14 |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit, for 405 nm excitation | Thermo Fisher Scientific | Cat#L34966 |
| Deposited Data | ||
| Raw Data Files for RNA Sequencing (Ly49Hlo versus Ly49Hhi) | NCBI Gene Expression Omnibus | GSE129490 |
| Experimental Models: Cell Lines | ||
| Mouse: Ba/F3 cells | L. Lanier (PI) | N/A |
| Mouse: Ba/F3–m157 cells | L. Lanier (PI) (Arase et al., 2002) |
Derived from parental Ba/F3 cells |
| Experimental Models: Organisms/Strains | ||
| Mouse: WT or CD45.2: C57BL/6J | The Jackson Laboratory | Stock#000644; RRID:IMSR_JAX:000664 |
| Mouse: WT or CD45.1: B6.SJL-Ptprca Pepcb/BoyJ | The Jackson Laboratory | Stock#002014; RRID:IMSR_JAX:002014 |
| Mouse: Klra8−/− or Ly49H-deficient | S. Vidal (PI) (Fodil-Cornu et al., 2008) | N/A |
| Mouse: Stat4−/−, Ifnar1−/−, Stat1−/−, Il18r1−/− | J. Sun (PI) | N/A |
| Mouse: Ifng-IRES-YFP or GREAT: B6.129S4-Ifngtm3.1Lky/J | The Jackson Laboratory | Stock#017581; RRID:IMSR_ JAX:017581 |
| Mouse: Rag2−/− IL2rg−/−: C;129S4-Rag2tm1.1Flv Il2rgtm1.1Flv/J | The Jackson Laboratory | Stock#014593; RRID:IMSR_JAX:014593 |
| Software and Algorithms | ||
| Trimmomatic (v.0.36) | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| Salmon (v0.10.2) | Patro et al., 2017 | https://combine-lab.github.io/salmon |
| tximport (v1.9.12) | Soneson et al., 2015 | https://bioconductor.org/packages/release/bioc/html/tximport.html |
| DESeq2 (v.1.12.17) | Love et al., 2014 | http://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| goseq (v1.33.0) | Young et al., 2010 | https://bioconductor.org/packages/release/bioc/html/goseq.html |
| KEGG.db (v3.2.3) | https://bioconductor.org/packages/release/data/annotation/html/KEGG.db.html | https://bioconductor.org/packages/release/data/annotation/html/KEGG.db.html |
| R (v.3.5.1) | https://www.r-project.org/ | https://www.r-project.org/ |
| SlideBook software (v6.0.15) | Intelligent Imaging Innovations | https://www.intelligent-imaging.com/slidebook |
Highlights:
Ly49H receptor density drives diversity in NK cell function during MCMV infection
NK cells undergo avidity selection during CMV infection
Ly49Hhi NK cells possess greater potential for cytotoxic and adaptive responses
Ly49Hlo NK cells possess greater potential for IFN-γ production
Acknowledgments
We thank members of the Sun lab, Georg Gasteiger, and Christin Friedrich for experimental assistance, and members of the MSKCC NK club for discussions. We thank Lewis Lanier for helpful discussions and comments on the manuscript. We thank Simon Grassmann and Veit Buchholz for sharing their manuscript with us before publication. Graphical abstract was created with created with BioRender. N.M.A., D.L., and B.T.J. were supported by an MSTP grant from the NIGMS of the NIH (T32GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program). N.M.A. was also supported by an F30 Predoctoral Fellowship from the NIH NIAID (F30 AI136239). M.H. was supported by NIH grant AI087644. K.C.H. was supported by NIH grant AI123658 and CA023766. J.C.S. was supported by the Ludwig Center for Cancer Immunotherapy, the American Cancer Society, the Burroughs Wellcome Fund, and the NIH (AI100874, AI130043, and P30CA008748).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Aguilar OA, Berry R, Rahim MM, Reichel JJ, Popović B, Tanaka M, Fu Z, Balaji GR, Lau TN, Tu MM, et al. (2017). A viral immunoevasin controls innate immunity by targeting the prototypical natural killer cell receptor family. Cell 169, 58–71. [DOI] [PubMed] [Google Scholar]
- Arase H, Mocarski ES, Campbell AE, Hill AB, and Lanier LL (2002). Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296, 1323–1326. [DOI] [PubMed] [Google Scholar]
- Biron CA, Byron KS, and Sullivan JL (1989). Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med 320, 1731–1735. [DOI] [PubMed] [Google Scholar]
- Biron CA, and Tarrio ML (2015). Immunoregulatory cytokine networks: 60 years of learning from murine cytomegalovirus. Med. Microbiol. Immunol 204, 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubic I, Wagner M, Krmpotic A, Saulig T, Kim S, Yokoyama WM, Jonjic S, and Koszinowski UH (2004). Gain of virulence caused by loss of a gene in murine cytomegalovirus. J Virol 78, 7536–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowski JF, Warner JF, Dennert G, and Welsh RM (1985). Adoptive transfer studies demonstrating the antiviral effect of natural killer cells in vivo. J. Exp. Med 161, 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch DH, and Pamer EG (1999). T cell affinity maturation by selective expansion during infection. J. Exp. Med 189, 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels KA, Devora G, Lai WC, O’Donnell CL, Bennett M, and Welsh RM (2001). Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J. Exp. Med 194, 29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day EK, Carmichael AJ, ten Berge IJ, Waller EC, Sissons JG, and Wills MR (2007). Rapid CD8+ T cell repertoire focusing and selection of high-affinity clones into memory following primary infection with a persistent human virus: human cytomegalovirus. Journal of immunology 179, 3203–3213. [DOI] [PubMed] [Google Scholar]
- Della Chiesa M, Falco M, Podestà M, Locatelli F, Moretta L, Frassoni F, and Moretta A (2012). Phenotypic and functional heterogeneity of human NK cells developing after umbilical cord blood transplantation: a role for human cytomegalovirus? Blood 119, 399–410. [DOI] [PubMed] [Google Scholar]
- Dokun AO, Kim S, Smith HR, Kang HS, Chu DT, and Yokoyama WM (2001). Specific and nonspecific NK cell activation during virus infection. Nat. Immunol 2, 951–956. [DOI] [PubMed] [Google Scholar]
- Etzioni A, Eidenschenk C, Katz R, Beck R, Casanova JL, and Pollack S (2005). Fatal varicella associated with selective natural killer cell deficiency. J. Pediatr 146, 423–425. [DOI] [PubMed] [Google Scholar]
- Fodil-Cornu N, Lee SH, Belanger S, Makrigiannis AP, Biron CA, Buller RM, and Vidal SM (2008). Ly49h-deficient C57BL/6 mice: a new mouse cytomegalovirus-susceptible model remains resistant to unrelated pathogens controlled by the NK gene complex. J. Immunol 181, 6394–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, Lopez-Vergès S, Lanier LL, Weisdorf D, and Miller JS (2012). Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood 119, 2665–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldrath AW, and Bevan MJ (1999). Selecting and maintaining a diverse T-cell repertoire. Nature 402, 255–262. [DOI] [PubMed] [Google Scholar]
- Grassmann S, Pachmayr LO, Leube J, Mihatsch L, Andrae I, Flommersfeld S, Oduro J, Cicin-Sain L, Schiemann M, Flossdorf M, and Buchholz VR (2019). Single-cell fate mapping reveals clonal dynamics of adaptive NK-cell responses. Immunity [DOI] [PubMed] [Google Scholar]
- Gumá M, Angulo A, Vilches C, Gómez-Lozano N, Malats N, and López-Botet M (2004). Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104, 3664–3671. [DOI] [PubMed] [Google Scholar]
- Hammer Q, Ruckert T, Borst EM, Dunst J, Haubner A, Durek P, Heinrich F, Gasparoni G, Babic M, Tomic A, et al. (2018). Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nature immunology 19, 453–463. [DOI] [PubMed] [Google Scholar]
- Henrickson SE, Mempel TR, Mazo IB, Liu B, Artyomov MN, Zheng H, Peixoto A, Flynn MP, Senman B, Junt T, et al. (2008). T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat. Immunol 9, 282–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz A, Guethlein LA, Nemat-Gorgani N, Norman PJ, Cooley S, Miller JS, and Parham P (2015). Regulation of adaptive NK cells and CD8 T cells by HLA-C correlates with allogeneic hematopoietic cell transplantation and cytomegalovirus reactivation. J. Immunol 195, 4524–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE, et al. (2013). Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 5, 208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LR, Weizman OE, Rapp M, Way SS, and Sun JC (2016). Epitope-specific vaccination limits clonal expansion of heterologous naive T cells during viral challenge. Cell Rep 17, 636–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, and Lanier LL (2015). Homeostatic control of memory cell progenitors in the natural killer cell lineage. Cell Rep 10, 280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karo JM, Schatz DG, and Sun JC (2014). The RAG recombinase dictates functional heterogeneity and cellular fitness in natural killer cells. Cell 159, 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedl RM, Kappler JW, and Marrack P (2003). Epitope dominance, competition and T cell affinity maturation. Curr. Opin. Immunol 15, 120–127. [DOI] [PubMed] [Google Scholar]
- Kedl RM, Rees WA, Hildeman DA, Schaefer B, Mitchell T, Kappler J, and Marrack P (2000). T cells compete for access to antigen-bearing antigen-presenting cells. J. Exp. Med 192, 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Davis M, Sinn E, Patten P, and Hood L (1981). Antibody diversity: somatic hypermutation of rearranged VH genes. Cell 27, 573–581. [DOI] [PubMed] [Google Scholar]
- Küppers R, Zhao M, Hansmann ML, and Rajewsky K (1993). Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO J 12, 4955–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LL (2005). NK cell recognition. Annu. Rev. Immunol 23, 225–274. [DOI] [PubMed] [Google Scholar]
- Lanier LL (2008). Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol 9, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Vergès S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, Houchins JP, Miller S, Kang SM, Norris PJ, et al. (2011). Expansion of a unique CD57⁺ NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 108, 14725–14732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan IC (1994). Germinal centers. Annu. Rev. Immunol 12, 117–139. [DOI] [PubMed] [Google Scholar]
- Madera S, Rapp M, Firth MA, Beilke JN, Lanier LL, and Sun JC (2016). Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J. Exp. Med 213, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madera S, and Sun JC (2015). Cutting edge: stage-specific requirement of IL-18 for antiviral NK cell expansion. J. Immunol 194, 1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mempel TR, Henrickson SE, and Andrian UHV (2004). T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature 427, 154–159. [DOI] [PubMed] [Google Scholar]
- Min-Oo G, and Lanier LL (2014). Cytomegalovirus generates long-lived antigen-specific NK cells with diminished bystander activation to heterologous infection. J. Exp. Med 211, 2669–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muccio L, Bertaina A, Falco M, Pende D, Meazza R, Lopez-Botet M, Moretta L, Locatelli F, Moretta A, and Chiesa MD (2016). Analysis of memory-like natural killer cells in human cytomegalovirus-infected children undergoing αβ+T and B cell-depleted hematopoietic stem cell transplantation for hematological malignancies. Haematologica 101, 371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntasell A, Lopez-Montanes M, Vera A, Heredia G, Romo N, Penafiel J, Moraru M, Vila J, Vilches C, and Lopez-Botet M (2013). NKG2C zygosity influences CD94/NKG2C receptor function and the NK-cell compartment redistribution in response to human cytomegalovirus. European journal of immunology 43, 3268–3278. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, and Honjo T (1999). Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem 274, 18470–18476. [DOI] [PubMed] [Google Scholar]
- Oberle SG, Hanna-El-Daher L, Chennupati V, Enouz S, Scherer S, Prlic M, and Zehn D (2016). A minimum epitope overlap between infections strongly narrows the emerging T cell repertoire. Cell Rep 17, 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MT, Murphy WJ, and Lanier LL (2010). ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat. Immunol 11, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nature methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prlic M, Hernandez-Hoyos G, and Bevan MJ (2006). Duration of the initial TCR stimulus controls the magnitude but not functionality of the CD8+ T cell response. J. Exp. Med 203, 2135–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim MM, Wight A, Mahmoud AB, Aguilar OA, Lee SH, Vidal SM, Carlyle JR, and Makrigiannis AP (2016). Expansion and protection by a virus-specific NK cell subset lacking expression of the inhibitory NKR-P1B receptor during murine cytomegalovirus infection. J. Immunol 197, 2325–2337. [DOI] [PubMed] [Google Scholar]
- Soneson C, Love MI, and Robinson MD (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss-Albee DM, Fukuyama J, Liang EC, Yao Y, Jarrell JA, Drake AL, Kinuthia J, Montgomery RR, John-Stewart G, Holmes S, and Blish CA (2015). Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Sci Transl Med 7, 297ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Beilke JN, and Lanier LL (2009). Adaptive immune features of natural killer cells. Nature 457, 557–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Madera S, Bezman NA, Beilke JN, Kaplan MH, and Lanier LL (2012). Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J. Exp. Med 209, 947–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann L, Rimbert M, Echasserieau K, Saulquin X, Neveu B, Dechanet J, Cerundolo V, and Bonneville M (2005). Selection of T cell clones expressing high-affinity public TCRs within Human cytomegalovirus-specific CD8 T cell responses. Journal of immunology 175, 6123–6132. [DOI] [PubMed] [Google Scholar]
- Viant C, Guia S, Hennessy RJ, Rautela J, Pham K, Bernat C, Goh W, Jiao Y, Delconte R, Roger M, et al. (2017). Cell cycle progression dictates the requirement for BCL2 in natural killer cell survival. J. Exp. Med 214, 491–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, and Oshlack A (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome biology 11, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehn D, Lee SY, and Bevan MJ (2009). Complete but curtailed T-cell response to very low-affinity antigen. Nature 458, 211–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Differentially Expressed Genes between Ly49Hlo and Ly49Hhi NK Cells at Day 1.5 PI, Related to Figure 3
Data Availability Statement
RNA-seq data comparing Ly49Hlo and Ly49Hhi NK cells during early MCMV infection (day 1.5 PI) are available in the Gene Expression Omnibus (GEO) under accession code GSE129490.