Abstract
Lactococcus lactis is used as cell-factory and strain selections are regularly performed to improve production processes. When selection regimes only allow desired phenotypes to survive, for instance by using antibiotics to select for cells that do not grow in a specific condition, the presence of more resistant subpopulations with a wildtype genotype severely slows down the procedure. While the food grade organism L. lactis is not often exposed to antibiotics we characterized its response to ampicillin in more detail, to better understand emerging population heterogeneity and how this might affect strain selection procedures. Using growth-dependent viability assays we identified persister subpopulations in stationary and exponential phase. Growth-independent viability assays revealed a 100 times larger subpopulation that did not grow on plates or in liquid medium, but had an intact membrane and could maintain a pH gradient. Over one third of these cells restored their intracellular pH when we induced a temporary collapse, indicating that this subpopulation was metabolically active and in a viable but non-culturable state. Exposure of L. lactis MG1363 to ampicillin therefore results in a heterogeneous population response with different dormancy states. These dormant cells should be considered in survival-based strain selection procedures.
Subject terms: Physiology, Bacteria
Introduction
Lactococcus lactis is an important organism in the dairy industry and it is increasingly explored as a cell factory for food ingredients and as a delivery vehicle for bioactive molecules1,2. To improve the yield or titer of the compounds of interest or to decrease the amount of by-products, strain selections are regularly performed3,4. In these selections heterogeneity in the single cell response is relevant, especially in dominant selection systems in which mutants are selected based on the principle that only cells with a desired phenotype can survive5. If resistant subpopulations with a wild-type genotype also survive the imposed condition, they will severely slow down the selection process.
An example is that L. lactis produces high concentrations of lactate during food fermentations resulting in a low final pH, which is not always desired. We initially sought to select mutants that do not acidify the growth medium below a certain pH and intended to achieve this by simply exposing a partly acidified culture to ampicillin, thereby killing all cells that can grow in these conditions. However, we found high fractions of surviving cells which completely acidified the medium after re-growth, and which were not ampicillin resistant. L. lactis is a food grade organism and therefore not often exposed to ampicillin. Nevertheless, we characterized its response to ampicillin in more detail, to better understand the heterogeneity in its population response and how this might affect strain selection procedures.
When a bacterial population is exposed to a bactericidal antibiotic like ampicillin, most cells will die6. To survive the antibiotic treatment cells can acquire resistance through e.g. export of the antibiotic using multidrug-resistance pumps or through the expression of antibiotic degrading enzymes7. However, dormant, non-growing cells can also survive treatments with antibiotics that target growth-related processes8,9. These dormant cells can revive spontaneously or when conditions change, and therefore they play a role in chronic infections10–14. Two well-known dormancy states are the persister state and the viable but non-culturable (VBNC) state15.
Persisters were first described in 1944 by Joseph Bigger16. He showed that penicillin could not completely kill growing staphylococcal cultures and that surviving cells were as sensitive to penicillin as the parent culture. The addition of an antibiotic to a population with persisters results in bi-phasic killing curves, because growing cells die quickly while persisters die only when they spontaneously switch back to the growing state17–19. Persister subpopulations have been identified in several species, including Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, Candida albicans, Enterococcus faecalis and Mycobacterium tuberculosis12,20–22.
VBNC cells are metabolically active, but non-growing cells. Their presence is reported in over 100 species15,23. The VBNC state is induced by environmental stresses, but viable non-culturable cells are also reported to be stochastically present in cultures24,25. They can coexist with persisters and a dormancy continuum was suggested, in which cells first enter the persister state and thereafter the VBNC state24. A limited amount of studies analyzed the presence of VBNC cells in L. lactis populations. VBNC cells were found under carbohydrate starvation, pH stress and during retentostat cultivation26–29. To our knowledge their presence is not reported under any other (stress) conditions.
Unlike persisters, VBNC cells do not grow on agar plates or in liquid medium unless they are resuscitated24. To detect VBNC cells, growth-independent viability assays are used, which measure for instance membrane integrity or enzyme activity30,31. Another growth-independent viability criterion is the ability to maintain a pH gradient, which requires an intact cell membrane and metabolic activity32. The intracellular pH can be measured using intracellular compounds with a pH dependent fluorescence signal, such as fluorescein-based stains or fluorescent proteins like GFP32–35.
Here we characterized the response of L. lactis MG1363 to ampicillin. CFU counting revealed bi-phasic killing curves, indicating that L. lactis MG1363 forms persisters. LIVE/DEAD staining and GFP-based intracellular pH measurements showed that in addition to the persister fraction 100 times more cells were in a VBNC state after exposure to ampicillin. Given the observed heterogeneity in the single cells response, we discuss how the efficiency of dominant selection systems can be improved.
Materials and Methods
Construction of L. lactis MG1363_GFP
The gene coding for the green fluorescent protein (Dasher-GFP) was obtained from ATUM (previously DNA 2.0; Newark, CA, USA) in a pJ221-based E. coli vector. The gfp gene was inserted in Lactococcus lactis MG1363 integration vector pSEUDO::Pusp45-sfgfp(Bs)36 as an XhoI/BcuI restriction fragment replacing the resident sfgfp(BS) gene. The resulting vector pSEUDO::Pusp45-gfp was maintained in E. coli DH5α. L. lactis MG1363_GFP was obtained by a single-crossover integration of pSEUDO::Pusp45-gfp into the pseudo 10 locus on the chromosome of L. lactis MG1363. Integration was performed as described previously36.
Strains and media
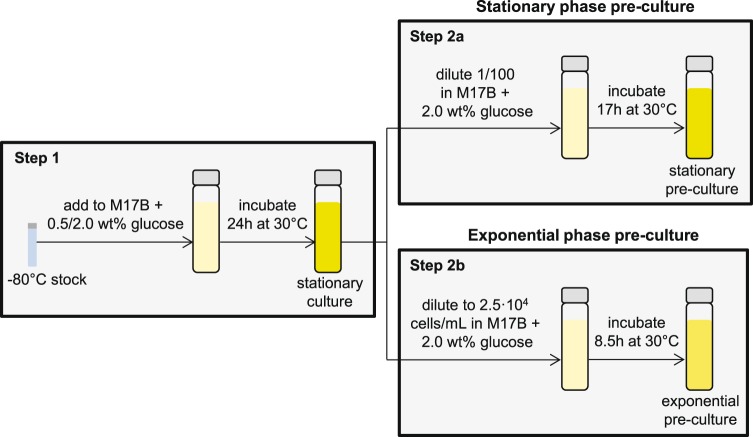
L. lactis MG136337 and L. lactis MG1363_GFP were cultured in M17 broth supplemented with 2.0 wt% glucose, unless otherwise indicated. M17 supplemented with 1.0 wt% glucose and 1.1 wt% agarose was used for plate counting. Cultures and plates were incubated at 30 °C. As suggested by Harms et al. we standardized the pre-culture procedure (Fig. 1) and used exponential phase pre-cultures in balanced growth (13 generations in excess glucose)38. 10 or 100 µg/mL ampicillin (Applichem, Darmstadt, Germany) was directly added to exponential pre-cultures; stationary pre-cultures were diluted 1/100 in fresh medium with ampicillin. Heat-killed cells were prepared by incubating stationary pre-cultures in phosphate buffered saline (PBS) of pH 7.5 at 80 °C for 20 minutes. E. coli DH5α was used for cloning and grown in LB medium at 37 °C with shaking or on LB medium solidified with 1.5 wt% agar supplemented with 100 µg/mL erythromycin.
Figure 1.
Overview of the pre-culture procedure. In step 1 2.0 wt% glucose was used for the stationary phase pre-culture and 0.5 wt% glucose for the exponential phase pre-culture.
Growth assays
To measure if cells could grow samples were centrifuged for 2 minutes at a relative centrifugal force (rcf) of 16,000 g. Pellets were resuspended and diluted in PBS (pH 7.5) and plated in duplicate. Plate counts were determined after at least 48 hours of incubation. Alternative growth assays were done in liquid medium using the most probable number (MPN) procedure. 16 replicates of a 10-fold dilution series were scored for growth after 7 days of incubation. MPN counts were determined as described earlier39.
Flow cytometry measurements
An Accuri C6 flow cytometer (BD Biosciences, San Jose, CA, US) was used for flow cytometry measurements.
To determine the number of intact cells we used the LIVE/DEAD BacLight bacterial viability kit L7007 (Molecular Probes, Eugene, OR, US) according to the manufacturer’s instructions for flow cytometry measurements. To determine the thresholds to separate live- and dead-stained cells a calibration curve was prepared with mixtures of living and heat- or ethanol-killed cells.
For intracellular pH measurements we centrifuged samples for 2 minutes at an rcf of 16,000 g and we resuspended the pellets in PBS with 0.5 wt% glucose at a desired pH (4.0–8.0). Glucose was added to keep the cells in an energized state, allowing metabolically active cells to maintain a pH gradient40. Below pH 5.5 PBS has a low buffering capacity, we therefore monitored the pH of the samples to ensure that it was constant. Cells were uncoupled by adding 1 µM valinomycin (a K+ transporter) and 2 µM nigericin (a H+ K+ antiporter) to the sample, followed by an incubation for 5 minutes at room temperature. Valinomycin and nigericin together cycle K+ and transport H+, ensuring that the intracellular pH equals the buffer pH40,41. To determine which cells could maintain a pH gradient we set a fluorescence threshold based on the fluorescence of uncoupled cells measured at pH 7.0.
Persister-model fit
We fitted the persister model of Balaban et al. to our experimental data17. The persister percentage at t = 0 h was estimated by extrapolating the second phase of the killing curve, in which persisters switch to the growing state, to t = 0 h.
Results
Membrane pH gradients can be determined using GFP
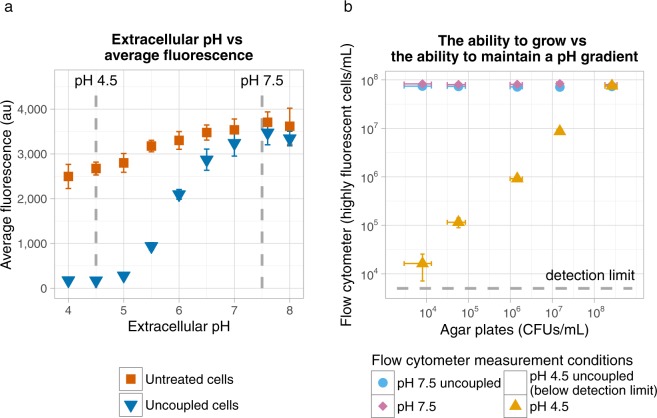
Metabolically active L. lactis cells are reported to maintain a pH gradient over their membrane: at low extracellular pH they can keep their intracellular pH above the extracellular pH40,42. The intracellular pH can be measured with GFP, as its fluorescence intensity decreases with a decreasing pH33,34. To test if cells that maintain a pH gradient over their membrane show a higher GFP signal than cells without a pH gradient, we compared the fluorescence of stationary L. lactis MG1363_GFP cells with cells in which the pH gradient was dissipated by valinomycin and nigericin (uncoupled cells). At pH 7.5 we observed that both uncoupled and untreated stationary cells were highly fluorescent, while at pH 4.5 uncoupled cells gave a 15 times lower fluorescence signal than untreated cells (Fig. 2a).
Figure 2.
(a) GFP signal of L. lactis MG1363 at different extracellular pH values. Stationary L. lactis MG1363_GFP cells were measured before and after uncoupling their pH gradient. Some error bars (SEM, n = 3) are too small to be visible. (b) Correlation between the ability to grow and the ability to maintain a pH gradient. For mixtures of living (stationary) and heat-killed cells we measured the ability to grow on agar plates and the ability to maintain a pH gradient by looking at their GFP fluorescence at pH 4.5. As controls we measured the fluorescence at pH 7.5 and the fluorescence after uncoupling the pH gradient. Some error bars (SD, n = 3) are too small to be visible.
To compare the ability to grow and the ability to maintain a pH gradient, we measured mixtures of living and heat-killed cells. At pH 7.5 living and heat-killed cells showed a high fluorescence signal, indicating that the heat-treatment did not denature the GFP and cells did not lyse (Fig. 2b). At pH 4.5 we found no significant difference between the number of highly fluorescent cells per mL and the CFUs per mL at different ratios of living and heat-killed cells (paired t-test, p = 0.56). We subsequently evaluated whether highly fluorescent cells indeed maintained a pH gradient by adding valinomycin and nigericin to the cells. At pH 7.5 uncoupling did not affect the number of highly fluorescent cells per mL, but at pH 4.5 it dropped below the detection limit. These results confirm that highly fluorescent cells at pH 4.5 maintained a pH gradient. Together these results show that for stationary and heat-killed L. lactis MG1363 the presence of a pH gradient correlated well with the cell’s ability to grow.
Ampicillin-treated L. lactis MG1363 cultures show bi-phasic killing curves
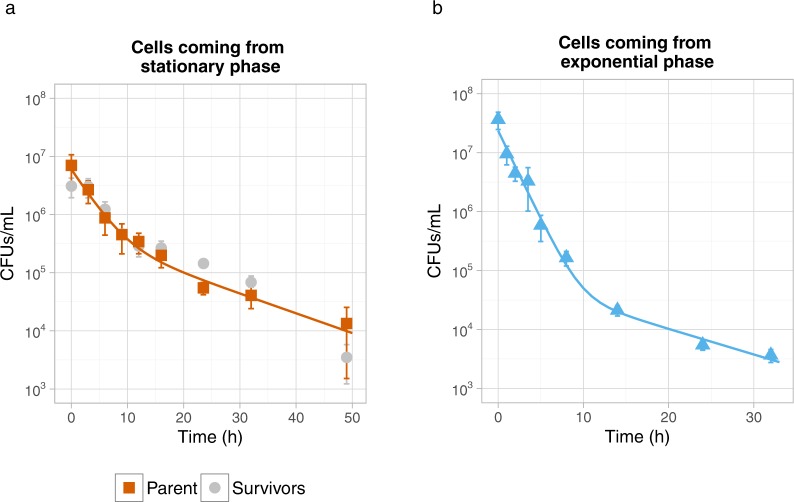
Exposing L. lactis MG1363 to ampicillin showed that a considerable fraction of the cells survived. We subsequently determined the CFU counts in time and found bi-phasic exponential killing curves (Fig. 3). After 48 hours of ampicillin exposure we re-grew the survivors and exposed them again to ampicillin, to determine if these cells were mutants that acquired ampicillin resistance. Re-grown survivors also showed bi-phasic killing (Fig. 3a), indicating that the bi-phasic behavior was not caused by genetic changes, but rather by phenotypic heterogeneity. We fitted the persister model of Balaban et al. to our data17. Based on the model the estimated persister fractions were 7.6 ± 3.6% in a stationary phase culture and 0.32 ± 0.17% in an exponential phase culture.
Figure 3.
Killing curves of ampicillin treated L. lactis MG1363. Stationary (a) and exponential (b) L. lactis MG1363 cells were diluted in medium with ampicillin and their ability to grow was measured on agar plates. Surviving cells from culture (a) (Parent) were propagated in medium without ampicillin and a second killing curve was recorded (Survivors). Some error bars (SD, n = 3) are too small to be visible. The solid line indicates the fit of a persister-model17.
Presence of toxin/antitoxin systems in L. lactis MG1363
Previous studies proposed toxin/antitoxin (TA) systems as a mechanism for persister formation43. The TA database TADB2.0 predicts five type II TA pairs in L. Lactis MG136344, from which two are 100% identical at the protein level. The predicted toxins contain Xre domains, the predicted antitoxins contain RelE, COG2856 and Bro domains. A phylogenetic analysis with experimentally validated TA proteins retrieved from the TADB2.0 database was performed44. This analysis showed that the L. lactis MG1363 proteins fall within clades of experimentally validated TA systems. However, sequence identities for many of the proteins are low (typically <25%) and it therefore remains unclear if these predicted TA systems are active in L. lactis MG1363.
Ampicillin-treated L. lactis MG1363 cultures contain viable but non-culturable cells
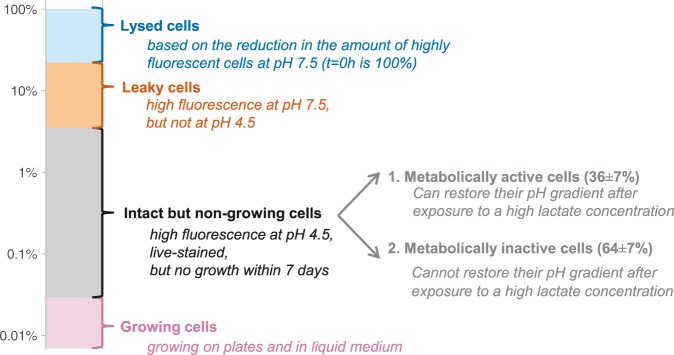
To determine the viability of ampicillin-treated L. lactis MG1363 with a growth-independent assay we measured the GFP fluorescence of cells and analyzed if they could maintain a pH gradient. We only analyzed the response to ampicillin for cells coming from stationary phase, because the persister fraction in exponential cultures is low and we approached the detection limit of our GFP-based assay under these conditions. After 48 hours of exposure to ampicillin the concentration of highly fluorescent cells in the cell suspension at pH 7.5 was reduced to 22 ± 4% compared to the start of the experiment, suggesting that 78% of the cells lysed (Fig. 4). Within this 22 ± 4% two subpopulations were present. 18 ± 3% of the cells were highly fluorescent at pH 7.5, but not at pH 4.5. These cells could not maintain a pH gradient, similar to what was observed for the heat-killed cells in Fig. 2b. The remaining 3.5 ± 0.8% of the cells showed high fluorescence also at pH 4.5. These cells could maintain a pH gradient, indicating that they had an intact membrane and suggesting that they were metabolically active. However, only 0.029 ± 0.004% of the cells could grow on agar plates, suggesting that cells entered the VBNC state. We tested four alternative hypotheses that could explain why we found 100-fold more cells with a high fluorescence signal than cells that could grow: (1) the GFP-based assay over-estimates the number of cells with an intact membrane, (2) the used ampicillin concentration is too low to completely kill all dividing cells, (3) highly fluorescent cells can grow, but not on agar plates, 4) highly fluorescent cells are dead, but they have a high intracellular pH because their membrane is still intact.
Figure 4.
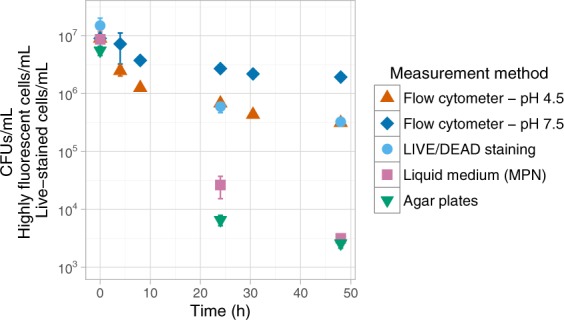
Growth and maintenance of a pH gradient in ampicillin treated L. lactis MG1363. We diluted stationary L. lactis MG1363_GFP cells in medium with ampicillin and measured their ability to grow in liquid medium and on agar plates (shown as CFUs/mL) and their ability to maintain a pH gradient in time (shown as highly fluorescent cells/mL). LIVE/DEAD staining was used to determine the amount of L. lactis MG1363 cells that have an intact membrane after ampicillin exposure (shown as live-stained cells/mL). Some error bars (SD, n = 3) are too small to be visible.
To independently confirm the results of the GFP-based assay we also performed LIVE/DEAD staining on cells that were exposed to ampicillin. As L. lactis MG1363_GFP is already highly fluorescent we used its parental strain L. lactis MG1363 for this assay. After 24 and 48 hours of ampicillin exposure the number of live-stained cells was similar to the number of cells that could maintain a pH gradient (Fig. 4), confirming that the fraction of cells with an intact membrane is much higher than the number of cells that can grow.
The second option considered was that the used ampicillin concentration was too low to completely kill all dividing cells. We hypothesized that increasing the ampicillin concentration would lower the number of cells that maintained a pH gradient, bringing it to the level of the number of cells that could grow. To test this hypothesis we diluted stationary L. lactis MG1363_GFP cells in medium supplemented with 100 µg/mL instead of 10 µg/mL ampicillin, and measured their ability to grow and to maintain a pH gradient. After 48 hours of exposure to 100 µg/mL ampicillin the difference between the number of cells that maintained a pH gradient and the number of cells that could grow was larger than it was for 10 µg/mL ampicillin (10 µg/mL ampicillin: 3.5 ± 0.8% maintained a pH gradient and 0.029 ± 0.004% could grow; 100 µg/mL ampicillin: 15 ± 1% maintained a pH gradient and 0.035 ± 0.016% could grow). These results showed that increasing the ampicillin concentration did not reduce the difference between the number of cells that maintained a pH gradient and the number of cells that could grow, it even increased the difference. This indicates that the observed 100-fold difference in the ability to grow and the ability to maintain a pH gradient is not caused by using a too low ampicillin concentration.
The third option considered was that cells that maintained a pH gradient had difficulties growing on agar plates. However, after 48 hours of exposure to ampicillin we found no significant difference in growth on agar plates and in liquid medium (Fig. 4), confirming that cells that maintained a pH gradient were unable to grow on both agar plates and in liquid medium.
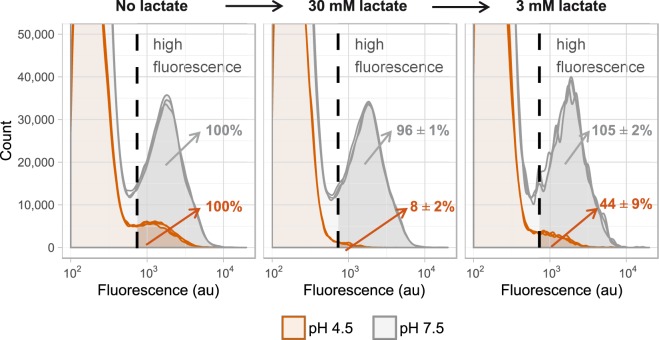
The fourth option considered was that the membrane of ampicillin-treated cells was intact, while the cells were actually dead. To test if the cell wall of the cells was damaged we placed ampicillin-treated cells in demineralized water for 2.5 hours, but the number of cells with an intact membrane remained constant (data not shown). If cells were only weakly permeable to protons this would allow metabolically inactive cells to have a high fluorescence signal at pH 4.5, because the cytosol is not yet acidified before the cells are measured. To test this hypothesis we analyzed in three steps if cells that were exposed to ampicillin for 48 hours could restore their pH gradient after temporary exposure to a high lactate concentration (Fig. 5). In the first step the GFP signal was measured in absence of lactate and the observed number of highly fluorescent cells was set to 100%. At pH 4.5 these are the cells that can maintain a pH gradient. In the second step 30 mM lactate was added to the sample. At pH 4.5 this led to weak acid uncoupling: the intracellular pH became similar to the extracellular pH and 92 ± 2% of the cells lost their high fluorescence signal. When the lactate concentration was diluted from 30 mM to 3 mM in the third step, the number of highly fluorescent cells increased from 8 ± 2% to 44 ± 9%, indicating that 36 ± 7% of the cells that maintained a pH gradient at pH 4.5 could restore their intracellular pH. When cells were not treated with ampicillin all cells restored their pH gradient after the lactate exposure (data not shown). Overall these results indicate that around 1.2% (36% of 3.5%) of the ampicillin-treated cells were metabolically active while only 0.029 ± 0.004% of the cells could grow, confirming that ampicillin-treated L. lactis MG1363 cultures contain VBNC cells.
Figure 5.
Metabolic activity of ampicillin treated L. lactis MG1363. Stationary L. lactis MG1363_GFP cells were diluted in medium with ampicillin (n = 3). After 48 hours (left panel) cells were temporarily exposed to high lactate levels (middle panel). At pH 4.5 this reduced the GFP fluorescence due to weak acid uncoupling. When the lactate concentration was lowered the amount of highly fluorescent cells increased from 8 ± 2% to 44 ± 9% (right panel). This shows that 36 ± 7% of the cells could restore the high fluorescence signal, indicating that these cells were metabolically active.
In total we used four different viability assays to analyze the response of L. lactis MG1363 to ampicillin. Figure 6 summarizes results obtained after 48 hours of ampicillin exposure.
Figure 6.
Schematic overview of the response of L. lactis MG1363 to ampicillin. Stationary L. lactis MG1363 or MG1363_GFP cells were diluted in medium with ampicillin. After 48 hours of ampicillin exposure we analyzed their viability by looking at their membrane integrity, their ability to grow in liquid medium and on agar plates, and their ability to maintain and build-up a pH gradient. This figure summarizes the combined results of these assays.
Discussion
Dormant bacterial cells can survive treatments with antibiotics that target growth-related processes and they can therefore cause chronic infections and food spoilage. In a different context they can influence mutant selections. In this study we analyzed the response of L. lactis MG1363 to ampicillin using four different viability assays, which led to the identification of several subpopulations (Fig. 6).
To our knowledge this is the first study that shows that L. lactis MG1363 can form a persister subpopulation. Harms et al. showed that antibiotic treatments can induce prophages, resulting in cell-lysis due to phages instead of the antibiotic38. The strains used in this study are “phage-cured”: they contain 2 prophages and 4 prophage remnants, but induction has not been reported37. We therefore do not expect that phage induction plays a role in our experiments.
We observed that after an ampicillin treatment only 0.029% of the cells could grow in liquid medium or on agar plates, while 1.2% showed metabolic activity based on GFP fluorescence measurements (Fig. 6). These findings indicate the presence of VBNC cells in ampicillin-treated L. lactis MG1363 populations. Previous studies found VBNC L. lactis cells during carbohydrate starvation, pH stress and in retentostat cultivations26–29. Our results show that exposure of L. lactis to ampicillin can also lead to VBNC cells. From 24 to 48 hours of ampicillin exposure we observed a decrease in the number of cells that maintained a pH gradient (Fig. 4), indicating that the number of VBNC cells decreased. Because VBNC cells do not spontaneously switch back to the growing state this decrease cannot be caused by killing of the cells by ampicillin15, but by a so far unidentified mechanism.
When L. lactis MG1363 is exposed to ampicillin it shows a heterogeneous population response with several dormancy states. Coexistence of persisters and VBNC cells is also described in E. coli and Vibrio vulnificus24,25. Toxin/antitoxin (TA) systems were for a long time widely accepted to play a role in persister formation and Ayrapetyan et al. proposed a dormancy continuum: with increasing toxin to antitoxin ratios cells first become persisters and then viable but non-culturable43. However, recent studies found no direct correlation between TA systems and bacterial persistence, and the role of these modules is therefore unclear at this moment38,45–47. In L. lactis AbiQ is reported to be a type III TA system, but this gene is located on a native L. lactis plasmid and is therefore not present in the plasmid-free L. lactis MG1363 used here48.The TA database TADB2.0 predicted five type II TA pairs in L. lactis MG136344, but it remains to be determined if they are functional.
Next to TA systems the ppGpp-mediated stringent response is reported to play a role in persister formation in Gram-negative E. coli and P. aeruginosa18,38,49,50. In E. coli high ppGpp levels are also reported to induce the VBNC state51. In a dormancy continuum ppGpp could therefore be a trigger for both the persister and the VBNC state in Gram-negatives. For Gram-positives this connection is less clear. In the Gram-positive S. aureus Conlon et al. found no effect of the stringent response on persister formation46, and to our knowledge there is no correlation reported between ppGpp and the VBNC state in Gram-positives. In the Gram-positive L. lactis high ppGpp concentrations are reported to increase acid resistance52, but the effect on antibiotic resistance, persister formation and the VBNC state is unknown.
Recent studies report that in Gram-positives and Gram-negatives persisters are formed when the intracellular ATP concentration of cells drops46,47. However, in VBNC cells ATP levels are reported to be similar or higher than in culturable cells53–56. This difference in ATP concentration in persister and VBNC cells contradicts the dormancy continuum and points towards different mechanisms behind the persister state and the VBNC state. In this study we observed that after 48 hours of ampicillin exposure 1.2% of the cells could quickly restore their intracellular pH when we induced a temporary pH decrease due to weak acid uncoupling (Fig. 5). Lactic acid bacteria have three main systems to remove protons from their cytosol and increase their intracellular pH: ATPase proton pumping, arginine deiminase activity that results in production of the base NH3, and glutamate decarboxylase activity that results in proton consumption in the cytoplasm57,58. In our experiment L. lactis MG1363 restored its intracellular pH in absence of arginine and glutamate and in presence of glucose, leaving ATPase proton pumping as the only mechanism to increase their intracellular pH. Our results therefore indicate that cells that restored their intracellular pH were not ATP depleted. The cells that had a high fluorescence before the lactate treatment, but could not restore their intracellular pH might be cells with a low ATP content or dead cells with an intact membrane. Because our assay cannot distinguish between these two states and because the persisters are 100 fold less abundant than the VBNC cells, we cannot draw conclusions about the ATP content of persisters. Our data does however suggest that VBNC L. lactis MG1363 cells are not ATP depleted, similar to what was shown for different organisms in previous studies53–56.
In this study we looked at the response of L. lactis MG1363 to ampicillin. β-lactam antibiotics like ampicillin disturb the balance between peptidoglycan synthesis and hydrolysis in growing cells, resulting in cell lysis59,60. In VBNC cells the peptidoglycan structure is changed and induction of peptidoglycan hydrolysis plays a role in the resuscitation of these cells30,61. Because ampicillin targets processes related to the VBNC state, there could be a direct relation between ampicillin and the presence of VBNC cells. We also exposed L. lactis MG1363 to heat-stress in this study, and we found no significant difference between the number of cells that maintained a pH gradient over their membrane and the number of CFUs (Fig. 2), indicating that L. lactis MG1363 does not form VBNC cells in response to heat-stress. It would be interesting to expose L. lactis MG1363 to other stress conditions in future studies and analyze the presence of persisters and VBNC cells, to see if our observations are specific for ampicillin or part of a more general stress response.
An antimicrobial that is produced by L. lactis itself is the peptide nisin, which kills Gram-positive bacteria by inhibiting cell-wall biosynthesis and by forming pores in the membrane62,63. Nisin has the GRAS status and it is commonly used as a food preservative because several pathogenic bacteria like Listeria monocytogenes are nisin sensitive64. Nisin is reported to kill S. aureus persisters65, but L. monocytogenes forms nisin-tolerant persisters66. To our knowledge there are no studies looking at the effect of nisin on the VBNC state in bacteria. L. lactis MG1363 could be a good model organism to study the effect of nisin on dormancy, because this strain is nisin sensitive and it can form both persisters and VBNC cells.
This study shows that L. lactis MG1363 forms persister and VBNC cells in stationary and exponential phase. The presence of these dormant subpopulations can reduce the efficiency of survival-based strain selections for microbial cell-factories. In L. lactis MG1363 the observed persister fraction is significantly lower at a high growth rate (exponential growth on glucose) compared to the stationary phase, similar to what is observed in other studies on persistence17,67. This result matches previous observations that at a low growth rate cells are more stress-resistant and prepared for changes in the environment68,69. We therefore expect that low efficiency of survival-based selection strategies through culture heterogeneity can be prevented by using fast growing cells.
VBNC cells themselves could also be interesting as cell-factories. These cells are non-growing, but they are reported to be metabolically active, to express genes and to exchange compounds with their environment31. These cells might therefore give high product yields, because growth and product formation are uncoupled and there is no carbon loss to biomass, similar to retentostats70,71.
VBNC cells can survive antimicrobial treatments because they are dormant, but they can start growing when conditions change30,61,72. Because VBNC cells do not spontaneously grow, viability measurements using agar plates might underestimate the potential viability of a culture, which is relevant in food safety and during antibiotic treatments. In those cases viability should not only be assessed by looking at growth, but also by looking at metabolic activity.
Acknowledgements
We thank Frank Bruggeman, Patrick van Delden, Daan de Groot and Patrick Janssen for fruitful discussions and technical assistance and we thank Bas Teusink for critically reading the manuscript. The plasmid pJ221 harboring Dasher-GFP was kindly provided by ATUM (Newark, CA, USA). R.J.v.T., H.B. and E.Z. were partly financed by the Netherlands Organisation for Scientific Research (NWO), as part of the research programme TTW with project number 13858. A.S. was supported by the FP7 EU grant BachBerry.
Author Contributions
R.J.v.T. and H.B. conceived the study, designed the experiments and wrote the paper. R.J.v.T. and E.Z. carried out the experiments. O.P.K. and A.S. constructed the strain L. lactis MG1363_GFP and helped to improve the manuscript.
Data Availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Competing Interests
H.B. is employed by NIZO Food Research, a contract research organization. NIZO Food Research had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. R.J.v.T., E.Z., O.P.K. and A.S. declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Song AAL. In, L. L. A., Lim, S. H. E. & Rahim, R. A. A review on Lactococcus lactis: from food to factory. Microb. Cell Fact. 2017;16:1–15. doi: 10.1186/s12934-016-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morello E, et al. Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J. Mol. Microbiol. Biotechnol. 2007;14:48–58. doi: 10.1159/000106082. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, et al. Finding the needle in the haystack—the use of microfluidic droplet technology to identify vitamin-secreting lactic acid bacteria. MBio. 2017;8:1–12. doi: 10.3391/mbi.2017.8.1.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachmann H, Pronk JT, Kleerebezem M, Teusink B. Evolutionary engineering to enhance starter culture performance in food fermentations. Curr. Opin. Biotechnol. 2015;32:1–7. doi: 10.1016/j.copbio.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Derkx PMF, et al. The art of strain improvement of industrial lactic acid bacteria without the use of recombinant DNA technology. Microb. Cell Fact. 2014;13:S5. doi: 10.1186/1475-2859-13-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 7.Khan A, Miller WR, Arias CA. Mechanisms of antimicrobial resistance among hospital-associated pathogens. Expert Rev. Anti. Infect. Ther. 2018;16:269–287. doi: 10.1080/14787210.2018.1456919. [DOI] [PubMed] [Google Scholar]
- 8.Lewis K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 9.Shah D, et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fauvart M, de Groote VN, Michiels J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J. Med. Microbiol. 2011;60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 11.Lewis K. Persister cells. Annu. Rev. Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 12.LaFleur MD, Qi Q, Lewis K. Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob. Agents Chemother. 2010;54:39–44. doi: 10.1128/AAC.00860-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulcahy LR, Burns JL, Lory S, Lewis K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 2010;192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Defraine V, Fauvart M, Michiels J. Fighting bacterial persistence: Current and emerging anti-persister strategies and therapeutics. Drug Resist. Updat. 2018;38:12–26. doi: 10.1016/j.drup.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Ayrapetyan M, Williams T, Oliver JD. Relationship between the viable but nonculturable state and antibiotic persister cells. J. Bacteriol. 2018;200:e00249–18. doi: 10.1128/JB.00249-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bigger JW. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet. 1944;244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 17.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science (80-.). 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 18.Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 19.Carvalho, G., Balestrino, D., Forestier, C. & Mathias, J.-D. How do environment-dependent switching rates between susceptible and persister cells affect the dynamics of biofilms faced with antibiotics? npj Biofilms Microbiomes6 (2018). [DOI] [PMC free article] [PubMed]
- 20.Möker N, Dean CR, Tao J. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J. Bacteriol. 2010;192:1946–1955. doi: 10.1128/JB.01231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keren I, Minami S, Rubin E, Lewis K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. MBio. 2011;2:e00100–11. doi: 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaca AO, et al. Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond beyond the stringent response. MBio. 2013;4:e00646–13. doi: 10.1128/mBio.00646-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinto D, Santos MA, Chambel L. Thirty years of viable but nonculturable state research: unsolved molecular mechanisms. Crit. Rev. Microbiol. 2015;41:61–76. doi: 10.3109/1040841X.2013.794127. [DOI] [PubMed] [Google Scholar]
- 24.Ayrapetyan M, Williams TC, Baxter R, Oliver JD. Viable but nonculturable and persister cells coexist stochastically and are induced by human serum. Infect. Immun. 2015;83:4194–4203. doi: 10.1128/IAI.00404-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orman MA, Brynildsen MP. Establishment of a method to rapidly assay bacterial persister metabolism. Antimicrob. Agents Chemother. 2013;57:4398–4409. doi: 10.1128/AAC.00372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganesan B, Stuart MR, Weimer BC. Carbohydrate starvation causes a metabolically active but nonculturable state in Lactococcus lactis. Appl. Environ. Microbiol. 2007;73:2498–2512. doi: 10.1128/AEM.01832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuart MR, Chou LANSZU, Weimer BC. Influence of carbohydrate starvation and arginine on culturability and amino acid utilization of Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 1999;65:665–673. doi: 10.1128/aem.65.2.665-673.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arbi AE, Ghorbal S, Delacroix-Buchet A, Bouix M. Assessment of the dynamics of the physiological states of Lactococcus lactis ssp. cremoris SK11 during growth by flow cytometry. J. Appl. Microbiol. 2011;111:1205–1211. doi: 10.1111/j.1365-2672.2011.05114.x. [DOI] [PubMed] [Google Scholar]
- 29.Mastrigt O, van, Abee T, Lillevang SK, Smid EJ. Quantitative physiology and aroma formation of a dairy Lactococcus lactis at near-zero growth rates. Food Microbiol. 2018;73:216–226. doi: 10.1016/j.fm.2018.01.027. [DOI] [PubMed] [Google Scholar]
- 30.Ramamurthy T, Ghosh A, Pazhani GP, Shinoda S. Current perspectives on viable but non-culturable (VBNC) pathogenic bacteria. Front. Public Heal. 2014;2:103. doi: 10.3389/fpubh.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Mendis N, Trigui H, Oliver JD, Faucher SP. The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 2014;5:1–1. doi: 10.3389/fmicb.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breeuwer P, Abee T. Assessment of viability of microorganisms employing fluorescence techniques. Int. J. Food Microbiol. 2000;55:193–200. doi: 10.1016/S0168-1605(00)00163-X. [DOI] [PubMed] [Google Scholar]
- 33.Han J, Burgess K. Fluorescent indicators for intracellular pH. Chem. Rev. 2009;110:2709–2728. doi: 10.1021/cr900249z. [DOI] [PubMed] [Google Scholar]
- 34.Kneen M, Farinas J, Li Y, Verkman AS. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys. J. 1998;74:1591–1599. doi: 10.1016/S0006-3495(98)77870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shinoda H, et al. Acid-tolerant monomeric GFP from Olindias formosa. Cell Chem. Biol. 2018;25:330–338.e7. doi: 10.1016/j.chembiol.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 36.Overkamp W, et al. Benchmarking various green fluorescent protein variants in Bacillus subtilis, Streptococcus pneumoniae, and Lactococcus lactis for live cell imaging. Appl. Environ. Microbiol. 2013;79:6481–6490. doi: 10.1128/AEM.02033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wegmann U, et al. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 2007;189:3256–3270. doi: 10.1128/JB.01768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harms A, Fino C, Sørensen MA, Semsey S, Gerdes K. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. MBio. 2017;8:1–18. doi: 10.1128/mBio.01964-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jarvis B, Wilrich C, Wilrich P-T. Reconsideration of the derivation of most probable numbers, their standard deviations, confidence bounds and rarity values. J. Appl. Microbiol. 2010;109:1660–1667. doi: 10.1111/j.1365-2672.2010.04792.x. [DOI] [PubMed] [Google Scholar]
- 40.Molenaar D, Abee T, Konings WN. Continuous measurement of the cytoplasmic pH in Lactococcus lactis with a fluorescent pH indicator. Biochim. Biophys. Acta. 1991;1115:75–83. doi: 10.1016/0304-4165(91)90014-8. [DOI] [PubMed] [Google Scholar]
- 41.Kessler RJ, et al. Uncouplers and the molecular mechanism of uncoupling in mitochondria. Proc. Natl. Acad. Sci. USA. 1977;74:2241–2245. doi: 10.1073/pnas.74.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nannen NL, Hutkins RW. Intracellular pH effects in lactic acid bacteria. J. Dairy Sci. 1991;74:741–746. doi: 10.3168/jds.S0022-0302(91)78219-2. [DOI] [Google Scholar]
- 43.Ayrapetyan M, Williams TC, Oliver JD. Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol. 2015;23:7–13. doi: 10.1016/j.tim.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Xie Y, et al. TADB 2.0: An updated database of bacterial type II toxin-antitoxin loci. Nucleic Acids Res. 2018;46:D749–D753. doi: 10.1093/nar/gkx1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goormaghtigh F, et al. Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. MBio. 2018;9:e00640–18. doi: 10.1128/mBio.00640-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conlon BP, et al. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 2016;1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 47.Shan Y, et al. ATP-dependent persister formation in Escherichia coli. MBio. 2017;8:e02267–16. doi: 10.1128/mBio.02267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samson JE, Spinelli S, Cambillau C, Moineau S. Structure and activity of AbiQ, a lactococcal endoribonuclease belonging to the type III toxin-antitoxin system. Mol. Microbiol. 2013;87:756–768. doi: 10.1111/mmi.12129. [DOI] [PubMed] [Google Scholar]
- 49.Amato SM, Orman MA, Brynildsen MP. Metabolic control of persister formation in Escherichia coli. Mol. Cell. 2013;50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen D, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science (80-.). 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boaretti M, del Mar Lleò M, Bonato B, Signoretto C, Canepari P. Involvement of rpoS in the survival of Escherichia coli in the viable but non-culturable state. Environ. Microbiol. 2003;5:986–996. doi: 10.1046/j.1462-2920.2003.00497.x. [DOI] [PubMed] [Google Scholar]
- 52.Rallu F, Gruss A, Ehrlich SD, Maguin E. Acid- and multistress-resistant mutants of Lactococcus lactis: Identification of intracellular stress signals. Mol. Microbiol. 2000;35:517–528. doi: 10.1046/j.1365-2958.2000.01711.x. [DOI] [PubMed] [Google Scholar]
- 53.Beumer PR, de Vries J, Rombouts FM. Campylobacter jejuni non-culturable coccoid cells. Int. J. Food Microbiol. 1992;15:153–163. doi: 10.1016/0168-1605(92)90144-R. [DOI] [PubMed] [Google Scholar]
- 54.Lindbäck, T., Rottenberg, M. E., Roche, S. M. & Rørvik, L. M. The ability to enter into an avirulent viable but non-culturable (VBNC) form is widespread among Listeria monocytogenes isolates from salmon, patients and environment. Vet. Res. 41 (2010). [DOI] [PMC free article] [PubMed]
- 55.Su X, et al. Identification, characterization and molecular analysis of the viable but nonculturable Rhodococcus biphenylivorans. Sci. Rep. 2015;5:18590. doi: 10.1038/srep18590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao F, et al. New insights into the formation of viable but nonculturable Escherichia coli O157:H7 induced by high-pressure CO2. MBio. 2016;7:1–11. doi: 10.1128/mBio.00961-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hutkins RW, Nannen NL. pH homeostasis in lactic acid bacteria. J. Dairy Sci. 1993;76:2354–2365. doi: 10.3168/jds.S0022-0302(93)77573-6. [DOI] [Google Scholar]
- 58.Cotter PD, Hill C. Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 2003;67:429–453. doi: 10.1128/MMBR.67.3.429-453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bayles KW. The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 2000;8:274–278. doi: 10.1016/S0966-842X(00)01762-5. [DOI] [PubMed] [Google Scholar]
- 60.Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J. Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinto D, Santos MA, Chambel L. Thirty years of viable but nonculturable state research: unsolved molecular mechanisms. Crit. Rev. Microbiol. 2013;41:61–76. doi: 10.3109/1040841X.2013.794127. [DOI] [PubMed] [Google Scholar]
- 62.Scherer KM, Spille JH, Sahl HG, Grein F, Kubitscheck U. The lantibiotic nisin induces lipid II aggregation, causing membrane instability and vesicle budding. Biophys. J. 2015;108:1114–1124. doi: 10.1016/j.bpj.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiedemann I, et al. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 2001;276:1772–1779. doi: 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- 64.Gharsallaoui A, Oulahal N, Joly C, Degraeve P. Nisin as a food preservative: part 1: Physicochemical properties, antimicrobial activity, and main uses. Crit. Rev. Food Sci. Nutr. 2016;56:1262–1274. doi: 10.1080/10408398.2013.763765. [DOI] [PubMed] [Google Scholar]
- 65.Kim W, et al. Identification of an antimicrobial agent effective against methicillin-resistant Staphylococcus aureus persisters using a fluorescence-based screening strategy. PLoS One. 2015;10:1–15. doi: 10.1371/journal.pone.0127640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu S, Yu PL, Flint S. Persister cell formation of Listeria monocytogenes in response to natural antimicrobial agent nisin. Food Control. 2017;77:243–250. doi: 10.1016/j.foodcont.2017.02.011. [DOI] [Google Scholar]
- 67.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 68.Hartke A, Bouche S, Gansel X, Boutibonnes P, Auffray Y. Starvation-induced stress resistance in Lactococcus lactis subsp. lactis IL1403. Appl. Environ. Microbiol. 1994;60:3474–3478. doi: 10.1128/aem.60.9.3474-3478.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ihssen J, Egli T. Global physiological analysis of carbon- and energy- limited growing Escherichia coli confirms a high degree of catabolic flexibility and preparedness for mixed substrate utilization. Environ. Microbiol. 2005;7:1568–1581. doi: 10.1111/j.1462-2920.2005.00846.x. [DOI] [PubMed] [Google Scholar]
- 70.Ercan O, et al. Physiological and transcriptional responses of different industrial microbes at near-zero specific growth rates. Appl. Environ. Microbiol. 2015;81:5662–5670. doi: 10.1128/AEM.00944-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Konopka A. Microbial Physiological Stat At Low Growth Rate in Natural and Engineered Ecosystems. Curr. Opin. Microbiol. 2000;3:244–247. doi: 10.1016/S1369-5274(00)00083-7. [DOI] [PubMed] [Google Scholar]
- 72.Wai SN, Mizunoe Y, Takade A, Yoshida SI. A comparison of solid and liquid media for resuscitation of starvation- and low-temperature-induced nonculturable cells of Aeromonas hydrophila. Arch. Microbiol. 2000;173:307–310. doi: 10.1007/s002030000142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.