Abstract
Copper ion-catalyzed oxidation of yeast SOD1 (ySOD1) was examined to determine early oxidative modifications, including oxidation of a crucial disulfide bond, and the structural and functional repercussions of these events. The study used distinct oxidative conditions: Cu2+/H2O2, Cu2+/H2O2/AscH− and Cu2+/H2O2/glucose. Capillary electrophoresis experiments and quantification of protein carbonyls indicate that ySOD1 is highly susceptible to oxidative modification and that changes can be detected within 0.1 min of the initiation of the reaction. Oxidation-induced structural perturbations, characterized by circular dichroism, revealed the formation of partially-unfolded ySOD1 species in a dose-dependent manner. Consistent with these structural changes, pyrogallol assay indicates a partial loss of enzymatic activity. ESI-MS analyses showed seven distinct oxidized ySOD1 species under mild oxidation within 0.1 min. LC/MS analysis after proteolytic digestion demonstrated that the copper-coordinating active site histidine residues, His47 and His49, were converted into 2-oxo-histidine. Furthermore, the Cu and Zn bridging residue, His64 is converted into aspartate/asparagine. Importantly, the disulfide-bond Cys58-Cys147 which is critical for the structural and functional integrity of ySOD1 was detected as being oxidized at Cys147. We propose, based on LC/MS analyses, that disulfide-bond oxidation occurs without disulfide bond cleavage. Modifications were also detected at Met85 and five surface-exposed Lys residues. Based on these data we propose that the Cys58-Cys147 bond may act as a sacrificial target for oxidants and protect ySOD1 from oxidative inactivation arising from exposure to Cu2+/H2O2 and auto-inactivation during extended enzymatic turnover.
Keywords: Amyotrophic lateral sclerosis, Disulfide oxidation, Metal-ion catalyzed oxidation, Oxidative stress, Protein carbonyls, Superoxide dismutase
Graphical abstract
Highlights
-
•
Oxidation of yeast superoxide dismutase (ySOD1) by Cu2+/H2O2 is examined.
-
•
Rapid modification of His, Met, Cys and Lys residues detected by LC-MS methods.
-
•
Oxidation of active site His residues and partial protein unfolding are early events.
-
•
The Cys58-Cys147 disulfide bond is oxidized and may act as a sacrificial target.
-
•
Excess exogenous Cu2+ decreases protein damage and can reverse loss of activity.
1. Introduction
Recent discussions on the underlying neurotoxic mechanisms of various neurological disorders including ALS [1] have been dominated by the ‘oxidative stress’ hypothesis. This theory is persuasive in the case of metalloenzymes as the hypothesis requires redox-active metal ions to initiate the oxidative reactions that generate reactive oxidants (so-called ‘reactive oxygen species’, ROS) which promote damage to the proteins [2,3]. Mounting evidence suggests that the combined effects of oxidative damage and subsequent aggregation of Cu/Zn-Superoxide dismutase-1 (SOD1) contribute to the neurodegeneration in the case of ALS [[4], [5], [6]].
SOD1 is an indispensable homo-dimeric metalloenzyme that catalyzes the dismutation of the superoxide anion, O2•−, to O2 and H2O2 and thereby protects cells against O2•− -dependent damage [[7], [8], [9]]. However, the presence of O2•− and H2O2 within the enzyme active site also makes SOD1 susceptible to damage, metal ion release, and self-inactivation [10]. Cu(I) reacts rapidly with H2O2 producing oxygen radicals [11] that can damage and inactivate SOD1 [10,12]. Such radical attack may release free copper ions and result in increased generation of radicals and damage to other proteins.
ALS is a complex, multifactorial and devastating late-onset motor neuronal disease [1,13] where the majority (~90%) of cases are sporadic (SALS), with only ~10% due to familial ALS (FALS) [14]. The complexity of ALS is further increased by a phenotypic overlap between SALS and FALS, making these difficult to distinguish clinically [4,15]. Over 185 mutations covering 153 amino acids of SOD1 have been described to date and associated with FALS cases [16]. While FALS-associated genetic risk factors provide significant clues about the disease mechanism [17], ca. 66% of FALS cases are related to mutations in ~15 genes, the most common being SOD1 (20% of FALS cases) [14,16], followed by FUS and UBQLN2 [16]. The mechanism by which these mutations exert neural damage in FALS cases, and the trigger for increased oxidative stress in SALS cases, remain unclear. However, by analogy with other diseases like Alzheimer's [18,19] and Parkinson's disease [20], it has been suggested that ALS could be initiated by misfolded conformations of SOD1, with these leading to aggregation and inclusion bodies [10,21,22]. Mutant SOD1 proteins have been detected with reduced stability [23,24], reduced metal ion content [25], or a combination of these [26]. Mutant SOD1 proteins have also been suggested to generate reactive oxygen species (ROS) or reactive nitrogen species (RNS) as a result of the exposure of the copper ion due to misfolding, with this then participating in oxidative reactions. Furthermore, a more open conformation of SOD1 mutants, compared to wild-type, may enable interactions with substrates other than superoxide at the active site and resulting in aberrant oxidative reactions [4]. Both familial and sporadic cases of ALS have been reported with increased levels of oxidatively-damaged SOD1 protein [27]. Multiple studies have provided evidence for increased oxidative stress in the spinal cords of ALS patients, compared to controls, as detected by elevated levels of protein carbonyls [27,28], 3-nitrotyrosine [29] and 8-hydroxy-2′-deoxyguanosine (8-OHdG) [6]. Due to a large number of known SOD1 mutations [16] and the link of SOD1 to metal ion dyshomeostasis [6,30] a clear association between oxidative stress and disease onset of ALS has been established [4,28,31].
SOD1 harbors a stable intra-subunit disulfide bond which is an uncommon structural motif in proteins present in the cytoplasm of cells which contain multiple reducing species [32]. This disulfide bond has been suggested to maintain stability and quaternary structure [21,33,34]. Although the requirement of the disulfide bond for the enzymatic activity of SOD1 is actively debated [10,35], it has been suggested that this bond is required for full catalytic activity as it anchors a positively-charged Arg side chain that guides O2•− into the active site [3,36]. A recent study of a C57S human SOD1 mutant, which lacks this disulfide bond, showed that this protein has only 10% of the activity of wild-type SOD1 [35]. Furthermore, the disulfide bond in ALS-associated mutants is suggested to be more susceptible to chemical reduction compared to the wild-type protein [37], leading to a higher degree of dimer destabilization and aggregation. In addition, immature ALS SOD1 mutants with a reduced disulfide-bond have been suggested to play a significant role in the disease [38]. Intact disulfide-containing apo SOD1 has been shown to be less prone to form amyloid fibrils compared to disulfide-reduced apo SOD1, and low concentrations of disulfide-reduced apo SOD1 have also been reported to trigger fibrilization of apo SOD1 with an intact disulfide bridge [39,40].
Several molecular studies have focused on elucidating the metal ion coordination, activity, mutation and structural determinants of metal-ion binding to SOD1, and the role of metal ion–catalyzed oxidation (MCO) on SOD1 aggregation/oligomerization [2,39,[41], [42], [43], [44], [45], [46], [47], [48], [49]]. The significance of the disulfide bond and the impact of oxidative modification of this species on SOD1 structure and function has been studied [35,45,50,51]. However, the early events in SOD1 oxidation and the explicit impact of oxidative events on the disulfide bond remain to be clarified.
Previous studies have shown that H2O2 alone can induce misfolding of SOD1 [2,52]. In this study, we examine the effect of metal ion-catalyzed oxidation using three distinct oxidation systems: Cu2+/H2O2, Cu2+/H2O2/AscH− and Cu2+/H2O2/glucose. The oxidation induced by these systems has been examined at various concentrations, and at various time points ranging from 0.1 to 180 min. Furthermore, the integrity of the disulfide bond is investigated at 0.1 and 60 min under mild oxidative conditions. Saccharomyces cerevisiae Cu/Zn SOD1 (ySOD1) was chosen for study as this has only two Cys residues which form the disulfide bond. Glucose was included in some reaction systems because specific lysine (Lys) residues on the SOD1 protein have been reported to undergo glycation (Maillard) reactions, and SOD1 inactivation has been reported to occur both in vivo as well as in vitro as a result of glycation [53,54].
2. Materials and methods
2.1. Reagents
Copper-zinc superoxide dismutase (Cu/Zn-SOD1; EC1.15.1.1) from the yeast Saccharomyces cerevisiae (ySOD1) was kindly donated by Carl-Biotech A/S (Copenhagen, Denmark) [3]. The N-terminal methionine of the ySOD1 protein was cleaved off during purification. Stabilizer-free hydrogen peroxide (H2O2, 30%) and copper (II) chloride dihydrate (CuCl2 × 2H2O) was supplied by Merck Chemicals GmbH (Darmstadt, Germany). Ethylenediaminetetraacetic acid disodium salt (2Na-EDTA) and 3-(N-morpholino) propane-sulfonic acid (MOPS) were obtained from BDH Ltd. (Poole, England). l-Ascorbic acid and glucose were supplied by Sigma-Aldrich (St. Louis, MO, USA). All chemicals and reagents were of the highest purity grade (>99.5%).
2.2. Solution and protein preparations
Unless noted otherwise, all reactions were buffered with 20 mM MOPS-NaOH (MOPS, pH 7.4) made from the free acid and NaOH. Stock solutions of oxidants (CuCl2 and H2O2), reductant (ascorbate (AscH−) and glucose and the chelator (2Na-EDTA) were prepared using MilliQ water. The stability of the H2O2 stock solution was monitored using its UV absorbance at 265 nm and a molar extinction coefficient of 10 M−1 cm−1 [55]. Purified and lyophilized wild-type ySOD1, previously stored at −80 °C, was freshly prepared in MOPS. ySOD1 concentrations were determined from UV absorbance (Agilent UV–visible ChemStation) at 258 nm using ε258 = 0.38 mg mL−1 cm−1 [56]. Solutions were filtered using 0.2 μm syringe filters before use.
2.3. Metal-ion catalyzed oxidation of ySOD1
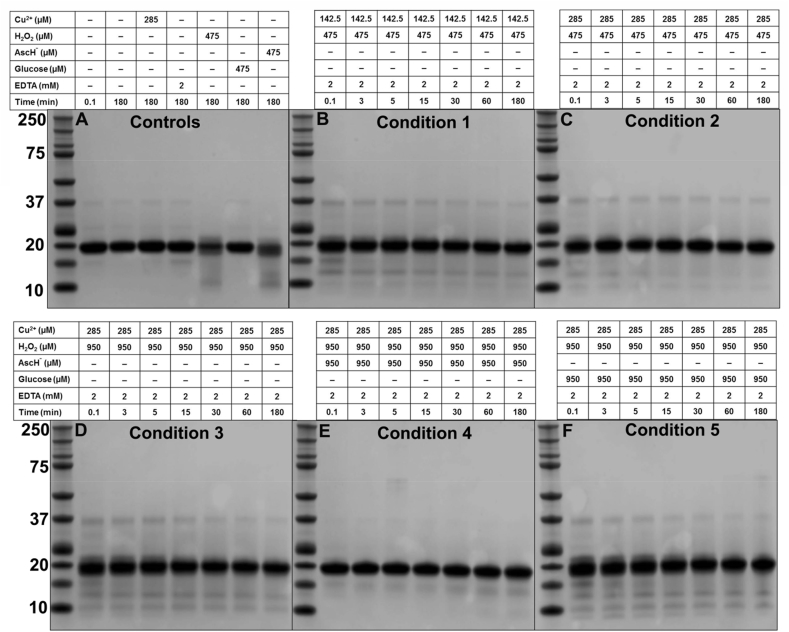
Wild-type ySOD1 (final concentration 59 μM) was treated with five oxidation systems (Conditions 1–5) consisting of Cu2+ (142.5 or 285 μM), H2O2 (475 or 950 μM), AscH− (950 μM) and glucose (950 μM) at the molar ratios described in Table 1. After addition and rapid mixing, samples were transferred to a 37 °C heating block with aliquots withdrawn after 3, 5, 15, 30, 60, and 180 min, with these added to a solution of 2 mM EDTA to quench the reaction [20]. For the first time point (0.1 min) the reactions were mixed at 22 °C without heating. Control reactions including non-oxidized ySOD1, or ySOD1 with individual separate components of the oxidation systems (Cu2+, H2O2, AscH−, glucose, EDTA) were prepared and removed for analysis at 0.1 min and after 180 min at 37 °C (Table 1). All samples used for analyses were aliquoted and stored at −80 °C as single-use samples (i.e. no freeze-thaw cycles) after flash freezing in liquid nitrogen.
Table 1.
Molar ratio and experimental conditions used in Cu/Zn-SOD1 oxidation.
| Samples | Final concentrations in reaction mixture (μM) |
EDTA (mM)a | Molar ratios |
Time (min) | ||||
|---|---|---|---|---|---|---|---|---|
| ySOD1 | CuCl2 | H2O2 | AscH- | Glucose | (ySOD1:Cu2+:H2O2:AscH−: Glucose) | |||
| Control 1 | 59 | 0 | 0 | 0 | 0 | 0 | 1: 0: 0: 0: 0 | 0.1 and 180 |
| Control 2 | 59 | 285 | 0 | 0 | 0 | 0 | 1: 4.8: 0: 0: 0 | 0.1 and 180 |
| Control 3 | 59 | 0 | 950 | 0 | 0 | 0 | 1: 0: 16: 0: 0 | 0.1 and 180 |
| Control 4 | 59 | 0 | 0 | 950 | 0 | 0 | 1: 0: 0: 16: 0 | 0.1 and 180 |
| Control 5 | 59 | 0 | 0 | 0 | 950 | 0 | 1: 0: 0: 0: 16 | 0.1 and 180 |
| Control 6 | 59 | 0 | 0 | 0 | 0 | 2 | 1: 0: 0: 0: 0 + 2 mM EDTA | 0.1 and 180 |
| Condition 1 | 59 | 142.5 | 475 | 0 | 0 | 2 | 1: 2.4: 8: 0: 0 | 0.1, 3, 5, 15, 30, 60, and 180 |
| Condition 2 | 59 | 285 | 475 | 0 | 0 | 2 | 1: 4.8: 8: 0: 0 | 0.1, 3, 5, 15, 30, 60, and 180 |
| Condition 3 | 59 | 285 | 950 | 0 | 0 | 2 | 1: 4.8: 16: 0: 0 | 0.1, 3, 5, 15, 30, 60, and 180 |
| Condition 4 | 59 | 285 | 950 | 950 | 0 | 2 | 1: 4.8: 16: 16: 0 | 0.1, 3, 5, 15, 30, 60, and 180 |
| Condition 5 | 59 | 285 | 950 | 0 | 950 | 2 | 1: 4.8: 16: 0: 16 | 0.1, 3, 5, 15, 30, 60, and 180 |
Added to terminate the Cu2+-catalyzed reaction after incubation for the time period given in the last column.
2.4. Monitoring ySOD1 oxidation by capillary electrophoresis
Capillary electrophoresis (CE) experiments were performed on Hewlett-Packard 3DCE apparatus (Agilent) equipped with a UV–Vis diode array detector (200–600 nm) attached to a temperature-controlled water bath. Separations were performed on a 75 cm × 0.005 cm fused silica capillary. The pH, the ionic strength of the buffer and the voltage were adapted to avoid any modification in the ySOD1 charge distribution as well as the Joule effect [57]. The CE running conditions were: temperature of capillary cassette 25 °C, vial holder temperature 15 °C, voltage 25 kV with a typical current of 55 μA. The separation buffer consisted of 50 mM phosphoric acid (Na) (pH 2.1). Capillaries were rinsed at 93000 Pa with 1 M NaOH for 10 min followed by 10 min with the Milli-Q water before initial use, and prior to each run, the capillary was rinsed with 0.1 M NaOH followed by the separation buffer for 3 min each. All samples were injected at 5000 Pa for 10 s and plugged into the capillary by immediate subsequent injection of separation buffer under the same conditions. On-column detection was at 220 nm. CE control and data acquisition were performed using ChemStation software (Agilent Technologies).
2.5. Quantification of carbonyl formation on ySOD1
Protein carbonyls were quantified using the OxiSelect™ protein carbonyl fluorometric assay kit (Cell Biolabs, San Diego, CA, USA), as described in the manufacturer's instructions, except with the washing and dilution steps as described previously [20]. Samples (100 μL) along with a series of standards in duplicate were placed into an optical bottom 96-well black fluorescence microtiter plate (Thermo Scientific Nunc, Thermo Fisher). End-point fluorescence spectra were recorded using λex 485 nm, λem 530 nm on a Molecular Devices SpectraMax M2 instrument (Sunnyvale, CA, USA). With the exception of incubation steps at defined temperatures, all other steps were performed either on ice or using a cooled centrifuge. Prior to carbonyl quantification, the concentration of soluble protein was estimated by the BCA assay [58] with the data reported as mol of carbonyls per mol protein.
2.6. ySOD1 activity
ySOD1 activity was assessed by its ability to inhibit pyrogallol autoxidation [59,60]. Briefly, ySOD1 samples (0.3 μg recombinant protein) in pH 8.2, 50 mM Tris-HCl buffer containing 1.1 mM DPTA was added to pyrogallol (0.2 mM from a stock solution in 10 mM HCl) in a total volume of 3 mL, with the absorbance decrease of pyrogallol was monitored at 420 nm. The SOD activity corresponding to percentage inhibition of pyrogallol autoxidation was calculated by the following formula:
Where;
Vp = Auto-oxidation rate of pyrogallol (control)
Vs = Auto-oxidation rate of samples (native and oxidized samples)
Vt = Total reaction volume (mL)
Vs = Volume of enzyme used for the assay (mL)
n = Dilution fold of the SOD sample
0.5 = Factor for 50% inhibition
One unit of SOD activity was defined as the amount required for inhibiting pyrogallol autoxidation by 50% per min, with specific activities calculated for each sample.
2.7. Monitoring structural changes by circular dichroism and SDS-PAGE gel analysis
Far-UV (178 − 260 nm) CD spectra were collected at 25 °C on a Jasco J-815 spectropolarimeter equipped with a Peltier-element-controlled thermostat. All CD measurements were performed with a spectral bandwidth of 2 nm, a digital integration time of 4 s, and a scanning speed of 20 nm min−1 using a 50 μm path length cell. The final spectrum was obtained by subtracting the spectrum of the sample buffer (20 mM MOPS-NaOH buffer, pH 7.4) from the mean sample spectrum of three individual scans using Jasco Spectra Analysis software, with a Savitzky-Golay algorithm of convolution width 11 applied.
SDS-PAGE was employed to examine oxidation-induced oligomerization. Control samples were collected at 0.1 min and 180 min and compared to ySOD1 incubated with Cu2+, H2O2, AscH−, and EDTA alone for 180 min, or aliquots from complete reaction systems collected at 0.1, 3, 5, 15, 30, 60, and 180 min. Sample containing 37.5 μM ySOD1 were heated to 95 °C for 10 min. Molecular mass markers (Invitrogen pre-stained marker 161–036) and samples were electrophoresed using 4–12% NuPAGE Bis-Tris mini acrylamide gels (Invitrogen, Carlsbad, CA, USA), using the suppliers’ protocol (200 V, 40 min) and stained with Coomassie Brilliant Blue.
2.8. Electrospray ionization mass spectrometry (ESI-MS) analysis of intact ySOD1
ESI-MS analyses were performed in the positive ion mode on a SolariX XR Bruker mass spectrometer coupled to a Dionex Ultimate 3000 high-performance liquid chromatography system without a column. Samples (5 μL) were diluted with 100 μL MeOH (containing 0.1% formic acid) and 100 μL deionized H2O (containing 0.1% formic acid). Samples were injected using a HyStar_LC system with a flow rate of 3 μl min−1. The MS was operated at a drying gas temperature of 200 °C and a capillary voltage of 4000 V. The in-source collision energy was 60 V and the lower and upper excitation event limits were 150 and 4000 m/z respectively. Data acquisition and data analysis were carried out using Compass ftms control and Compass data analysis software, respectively.
2.9. Identification of modified sites by proteolytic digestion and LC-MS/MS
Samples of ySOD1 (0.93 mg mL−1) was subjected to proteolysis with: LysC + Trypsin; LysC + GluC + Trypsin; or GluC alone. Samples of ySOD1 (10 μL) were added to 40 μL 8 M urea in 100 mM Tris/HCl pH 8.0 followed by addition of 1 μL LysC (0.2 μg μL−1; Promega) and 3 h incubation at 21 °C. Subsequently, 350 μL of 21.4 mM Tris/HCl pH 8.0 was added, together with either 2 μL trypsin (0.1 μg μL−1; Promega) or 2 μL trypsin +2 μL GluC (0.1 μg μL−1; Promega) followed by incubation at 21 °C overnight. Samples of 10 μL ySOD1 (0.93 mg mL−1) subjected to proteolysis with GluC only, were added to 40 μL 8 M urea in 100 mM Tris/HCl pH 8.0, 350 μL 21.4 mM Tris/HCl pH 8.0 and 2 μL GluC (0.1 μg μL−1) followed by incubation at 21 °C overnight. For reduction and alkylation, 100 μL of acidified peptide digest samples were neutralized with 200 μL 1 M Tris pH 7.0 followed by addition of 20 μL 200 mM DTT and incubation for 1 h at 21 °C. Then, 20 μL of 400 mM iodoacetamide was added followed by incubation for 1 h at 21 °C in the dark. All samples were then subjected to solid-phase extraction using StageTip C18 reverse-phase discs packed into pipette tips (Thermo Fischer Scientific) as described previously [61].
Samples were analyzed on a Bruker Impact II ESI-QTOF (Bruker Daltonics) mass spectrometer with an on-line Dionex Ultimate 3000 chromatography system (Thermo Fisher Scientific) equipped with an Acclaim Pepmap C18 Column (15 cm, 300 μm ID). Peptides were eluted using a solvent gradient over 55 min, using acetonitrile with 0.1% formic acid at a flow rate of 5 μL min−1. The MS scan range was 150–1500 m/z with a sampling rate of 1 Hz followed by intensity-based data-dependent MS/MS (TOP3). Database searches with the “dependent peptide” [62,63] feature were performed with MaxQuant v1.6.1.0 [64,65] using the following parameters: enzyme: GluC and/or Trypsin, with three missed cleavages; 1% peptide-level false discovery rate; mass tolerance: 0.07 and 0.005 Da (first and main searches, respectively); MS/MS mass tolerance: 40 ppm (first and main searches); and “dependent peptide” bin size: 0.0065 Da. For alkylated samples, carbamidomethyl (cysteine) was included as a fixed modification. Subsequent MaxQuant database searches without the “dependent peptide” feature were performed with the same parameters, but including variable modifications corresponding to the mass-shifts detected in the “dependent peptide” search.
2.10. Statistical analysis
Data are presented as the mean ± standard deviations of three replicates from three independent experiments unless otherwise stated. Statistical analysis was carried out using the STDEV.P function in Excel.
3. Results
3.1. Low concentrations of H2O2 induce oxidative damages to native ySOD1
Incubation of 950 μM H2O2 alone with ySOD1 for 0.1 min did not result in significant alterations to the capillary electrophoresis electropherograms (Fig. S1A), or the CD spectra of the protein (Fig. S1B). However, samples analyzed after 180 min incubation showed a complete loss of the native protein peak in the CE electropherograms (Fig. S1A), and unfolding of the ySOD1 secondary structure as detected by CD (Fig. S1B). Inclusion of EDTA (1 mM) in these incubations, to prevent oxidation initiated by trace metal ions, had only marginal effects on the CE and CD data, suggesting that these structural changes are due to a direct interaction of H2O2 with the ySOD1 (Fig. S1) in agreement with previous data [2,66,67].
3.2. Oxidation alters the overall structure of ySOD1
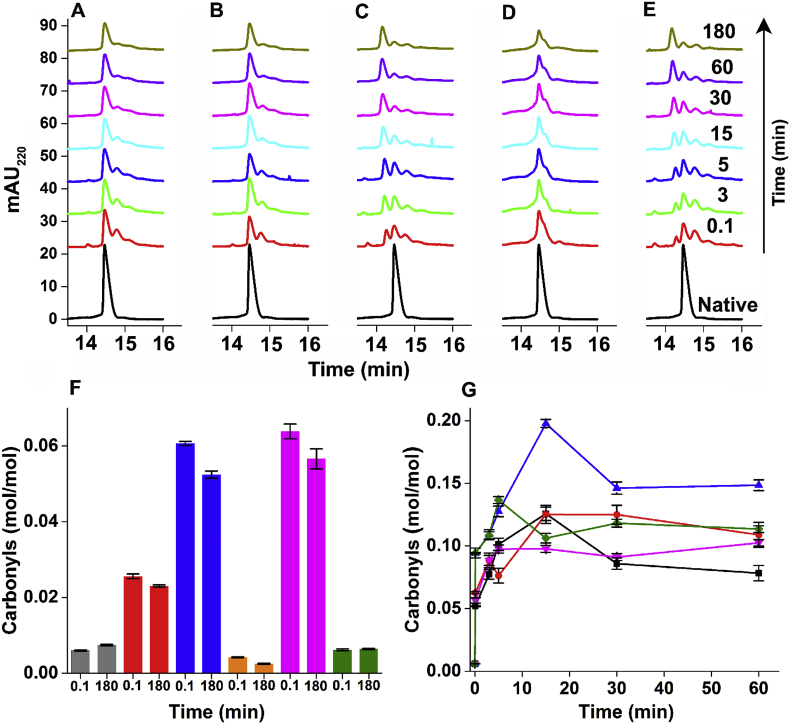
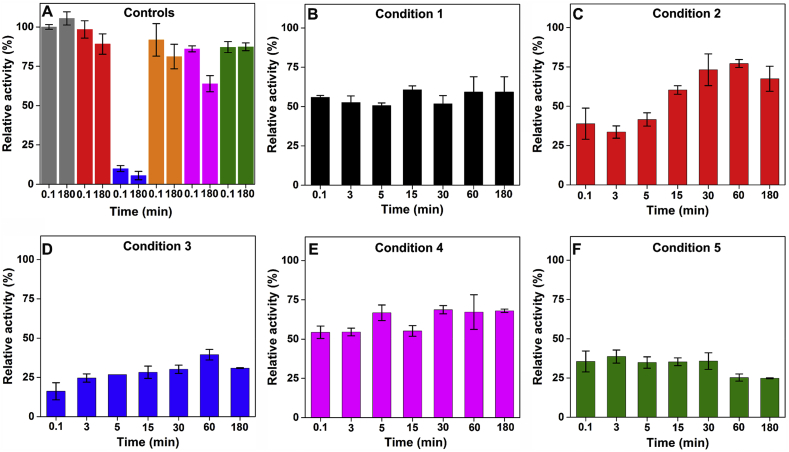
ySOD1 was subjected to oxidation using five different conditions as indicated in the Materials and methods and Table 1 for seven time points (0.1, 3, 5, 15, 30, 60 and 180 min; Fig. 1), then subjected to CE analysis. The samples from the earliest time point (0.1 min; Fig. 1A) after exposure to condition 1 (142.5 μM Cu2+ and 475 μM H2O2) showed ~50% loss of intensity of the parent protein peak, and the formation of two new peaks with migration times of ~14.8 and 15.2 min, indicating rapid modification of ySOD1. With the same oxidation conditions and longer incubation periods, the overall pattern of the peaks remained the same, but with a decrease in intensity (Fig. 1A) and a complete loss of resolution of the peaks at 14.8 and 15.2 min by 60 min. With a higher concentration of Cu2+ (285 μM; condition 2, Fig. 1B), the overall peak behavior and migration times were similar to condition 1. With higher concentrations of H2O2 (950 μM; condition 3, Fig. 1C) more significant changes were observed, with nearly 75% loss of the parent protein peak detected at the earliest time point (0.1 min; Fig. 1C) and detection of a third product peak at 14.3 min along with the peaks at 14.8 and 15.2 min observed in conditions 1 and 2. At longer incubation times (60 min and 180 min; Fig. 1C), the intensity of the peak at 14.2 min increased concomitantly with almost complete loss of the peaks from native ySOD1 and the species detected at 14.8 and 15.2 min. The broadening of the ySOD1 peaks with increasing reaction time is ascribed to the presence of a heterogeneous mixture of modified ySOD1 species.
Fig. 1.
Time-dependent capillary electrophoresis electropherograms and carbonyl quantifications of ySOD1 under various oxidative conditions. Overlayed CE electropherogram profiles of 59 μM of ySOD1 subjected to oxidation using (A) Condition 1, (B) Condition 2, (C) Condition 3, (D) Condition 4, and (E) Condition 5. Absorbance detection was carried at 220 nm. (F) Protein carbonyls determined for all control reactions including unoxidized (native) ySOD1 (grey), mixture of ySOD1 with Cu2+ (red), H2O2 (blue), EDTA (orange), AscH− (pink) and glucose (green) incubated at two time–points, 0.1 min and 180 min. (G) Time–course of protein carbonyl formation at six–time points (0.1, 3, 5, 15, 30, and 60 min). Condition 1 (black line), condition 2 (red line), condition 3 (blue line), condition 4 (pink line), and, Condition 5 (green line). Protein carbonylation levels are given in mol/mol and the data points and error bars represent the mean and standard deviation of triplicate technical replicates from two independent experiments. Except for 0.1 min, all other MCO reaction mixtures were incubated at 37 °C for the time shown. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The presence of AscH− in the reaction mixture (condition 4; i.e. condition 3 with 950 μM AscH−) resulted in a more rapid decrease in intensity of the peak from native ySOD1 (Fig. 1D). The 0.1 min time point sample showed a 50% loss of intensity, a loss of peak symmetry and the presence of a shoulder; this peak envelope then showed a slow decrease in intensity up to 180 min (Fig. 1D). The presence of glucose in the reaction mixture (i.e. condition 3 with 950 μM glucose) resulted in rapid changes to the parent ySOD1 peak (Fig. 1E), with the overall migration pattern being similar to condition 3. A decrease in the parent protein peak intensity of ca. 75% was detected after 0.1 min incubation, together with several discrete peaks with migration times of 13.7, 14.3, 14.8 and 15.2 min. At longer incubation times the native ySOD1 peak along with the peaks at 13.7, 14.8 and 15.2 min decreased in intensity, while the peak at 14.3 min increased in intensity (Fig. 1E).
The electrophoretic mobility of the control samples is illustrated in the Supplementary Data and Fig. S1. Untreated native ySOD1 protein migrated as a single peak (Fig. S2B) and no changes were observed after 180 min at 37 °C (Fig. S2C). Incubation of the ySOD1 with Cu2+, glucose or EDTA alone for 180 min at 37 °C had no effect on peak intensity, resolution or migration (Figs. S2D, S2F, and S2H). However, almost complete loss of native of the native ySOD1 peak was observed in the presence of H2O2, and nearly 50% loss in the presence of AscH− (Figs. S2E and S2G), indicating that modification of ySOD1 can occur in the absence of added redox-active metal ions.
3.3. Metal-ion catalyzed oxidation induces formation of protein carbonyls in ySOD1
The carbonyl levels detected for the control samples (Fig. 1F, Table S1) ranged from 0.006 to 0.06 mol carbonyl per mol ySOD1. Incubation of ySOD1 with H2O2 or AscH− alone gave carbonyl levels of ca. 0.06 mol/mol (Fig. 1F, Table S1). With condition 1 (142.5 μM Cu2+ and 475 μM H2O2), a higher level of carbonylation was detected, with a maximum yield of carbonyls of 0.126 mol/mol detected at 15 min (Fig. 1G). This value decreased at longer incubation times (e.g. 0.086 mol/mol at 30 min; Fig. 1G, Fig. S3, Table S2). With a higher Cu2+ concentration (285 μM; condition 2), the 15 min sample showed a similar yield of carbonyls (0.125 mol/mol; Fig. 1G) with this level maintained for the remaining incubation period (Fig. 1G, Table S2). With higher concentrations of both Cu2+ and H2O2 (285 μM and 950 μM, respectively; condition 3), the highest yield of carbonyls was observed (0.198 mol/mol at 15 min; Fig. 1G, Table S2). However, despite the concentration-dependent increase in carbonylation, steady-state or reduced levels of carbonylation were observed after 15 min. The addition of AscH− (950 μM) reduced the concentration of protein carbonyls detected, with a maximum concentration of 0.112 mol/mol after 180 min incubation, whereas after 15 min incubation only 0.097 mol/mol of carbonyls (Figs. 1G and S3, and Table S2) were formed. The presence of glucose in the oxidation system (950 μM; condition 5) resulted in 0.094–0.103 mol/mol of carbonyls with the highest levels of carbonyls detected after 15 min of incubation (0.137 mol/mol; Figs. 1G and S3, and Table S2).
3.4. Ascorbate protects ySOD1 from formation of dimers and truncated species
Analysis of control samples (Fig. 2A) indicated the presence of monomeric ySOD1 as a single band at ~17.5 kDa, together with very low levels of a species assigned to a ySOD1 dimer at ~35 kDa. Incubation of ySOD1 with H2O2 alone (Fig. 2A) or AscH− (Fig. 2A) resulted in the detection of a broad smear on the gels between 10 and 20 kDa consistent with the presence of a large number of heterogenous modified forms of the protein. These data are consistent with the multiple peaks detected by CE analysis, and the increased carbonyl levels detected for these samples. For ySOD1 exposed to reaction system 1 (Fig. 2B), a dimeric band similar to that seen with native ySOD1 was detected but at higher intensity, but increasing incubation time did not alter the levels of this species (Fig. 2B), however, two weak protein bands at lower apparent mass (~12 and ~10 kDa), possibly due to the formation of truncated/fragmented species, were detected at longer reaction times (Fig. 2B). At higher levels of Cu2+ (285 μM) a more pronounced dimeric band was detected, together with a reduced intensity of the lower mass species (Fig. 2C). However, with high levels of both Cu2+ (285 μM) and H2O2 (950 μM) increased intensities of the fragment bands at ~12 and ~10 kDa were detected, together with a time-dependent loss of the dimer protein band (Fig. 2D). ySOD1 samples incubated under condition 4 in the presence of AscH− (Fig. 2E), showed an almost complete loss of the dimer and truncated SDS bands detected. In contrast, the presence of glucose gave rise to higher levels of the dimer species, as well as multiple discrete lower molecular mass bands at ~15, ~12, ~10, and ~8 kDa. The intensity of these bands decreased with increasing incubation time (Fig. 2F).
Fig. 2.
SDS–PAGE analyses of control and oxidized ySOD1 under different oxidative conditions. Samples were prepared as indicated in the Materials and methods. Control and oxidized ySOD1 samples under various conditions are organized on each SDS gel (A to F). Whereas gel (A) organized with control samples; lane M, molecular mass markers; lane – 1, native ySOD1; lane – 2, ySOD1 incubated at 37 °C for 180 min; lane – 3, ySOD1 incubated with Cu2+ at 37 °C for 180 min; lane – 4, ySOD1 incubated with EDTA at 37 °C for 180 min; lane – 5, ySOD1 incubated with H2O2 at 37 °C for 180 min; lane – 6, ySOD1 incubated with glucose at 37 °C for 180 min; lane – 7, ySOD1 incubated with AscH− at 37 °C for 180 min. (B) Condition 1, (C) condition 2, (D) condition 3, (E) condition 4, and (F) condition 5. The left-hand side lane on each gel contains molecular mass markers.
3.5. Oxidation induces changes in the secondary structure of ySOD1
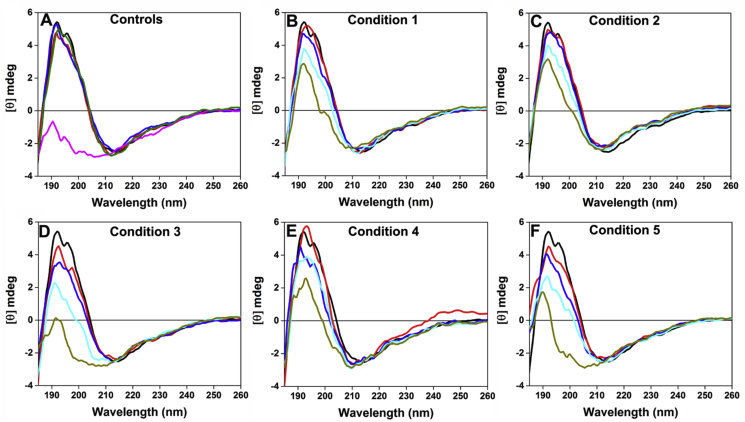
Far-UV CD spectra for control and selected oxidized samples are shown in Fig. 3 whereas an overlay of CD spectra showing all controls and oxidized samples are presented in Fig. S4. With the exception of ySOD1 mixed with H2O2, all control samples showed a positive band at approximately 195 nm and a broad minimum at ~210 nm, typical of a β-sheet-rich protein structure. In contrast, incubation with 950 μM H2O2 alone (Fig. 3A) resulted in a complete loss of the positive band at 195 nm and additional changes between 222 and 225 nm. For reaction condition 1, the positive band at 195 nm showed a decrease in intensity with increasing reaction time (Fig. 3B) and especially over the first 30 min (Fig. S4B). When the Cu2+ concentration was raised in condition 2, perturbations in the CD spectra at 222 nm increased, whereas changes in the positive band at 195 nm were similar to those detected with condition 1 (Figs. 3C and S4C). The most marked changes were observed with the highest Cu2+ and H2O2 concentrations (condition 3) with a marked, time-dependent loss of the positive band at 195 nm, and a shift in the CD-signal towards 200 nm, observed, indicative of an increased unfolding of the ySOD1 secondary structure as a function of time (Figs. 3D and S4D). In the presence of AscH− (condition 4) a time-dependent decrease in the positive band at 195 nm was detected, but to a much smaller extent than seen with condition 3, and the shift of the CD-signal towards 200 nm was absent (Figs. 3E and S4E). An increase in a positive band at wavelengths above 230 nm was also detected at 180 min. In the presence of glucose (condition 5), the spectral changes were similar to those observed in condition 3, though to a less marked extent at 180 min, with significant decreases at 195 nm and a shift towards 200 nm detected (Figs. 3F and S4F).
Fig. 3.
Overlay of far-UV CD spectra for selected control and oxidized ySOD1 under different oxidative conditions. (A) CD spectra for the control reactions including 59 μM of untreated ySOD1 in 20 mM MOPS buffer pH 7.4 (black line), ySOD1 incubated at 37 °C for 180 min (red line). Mixture of 59 μM ySOD1 incubated at 37 °C for 180 min with 285 μM Cu2+ (blue line), 950 μM H2O2 (magenta line), and 2 mM EDTA (olive line), separately. (B) Condition 1, (C) condition 2, (D) condition 3, (E) condition 4, and (F) condition 5. The color lines in panel B to F shows untreated ySOD1in black line whereas 0.1 min (red), 5 min (blue), 15 min (cyan), and 180 min (dark yellow). Except for untreated ySOD1 and 0.1 min reactions, all other MCO reaction mixtures were incubated at 37 °C. Complete overlay of all spectra including AscH− and glucose controls and oxidation data obtained after 0.1, 3, 5, 15, 30, 60 and 180 min, are shown in Supplementary Fig. S3. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Metal-ion catalyzed oxidation impacts on the enzymatic activity of ySOD1
Native ySOD1 showed 11.6 kU/mg of specific enzyme activity when assayed using pyrogallol (Fig. 4A, Table S3). The activity detected for ySOD1 incubated with one component of the reaction system (Cu2+, H2O2, AscH−, glucose, EDTA individually) ranged between 65 and 98% of the native activity (Fig. 4A), with the exception of the H2O2 system which resulted in a 90% loss of activity at 0.1 min, and a further small decline by 180 min (Fig. 4A).
Fig. 4.
The time-dependent activity of ySOD1 of control and oxidized ySOD1 under different oxidative conditions. The native ySOD1 showed 11.6 kU/mg of specific enzyme activity when subjected to the pyrogallol auto-oxidation assay. This value was considered to be 100% ySOD1 activity for the comparison with other control and oxidized samples. (A) ySOD1 activity determined for all control reactions including unoxidized (native) ySOD1 (grey), mixture of ySOD1 with Cu2+ (red), H2O2 (blue), EDTA (orange), AscH− (pink) and glucose (green) incubated at two time–points, 0.1 min and 180 min. (B) Condition 1, (C) condition 2, (D) condition 3, (E) condition 4, and (F) condition 5. Error bars represent the mean and standard deviation of data from three independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
With reaction condition 1 (142.5 μM Cu2+ and 475 μM H2O2), ySOD1 activity was reduced to 55% at the earliest time point (0.1 min) and this activity remains constant for the entire 180 min period (Fig. 4B, Table S4). With a higher Cu2+ concentration (285 μM) lower levels of activity (33–39% of control) were detected at 0.1 and 3 min, but the activity recovered at longer time points to give a plateau value of ~75% of the native ySOD1 activity (Fig. 4C, Table S4). Condition 3 (285 μM Cu2+ and 950 μM H2O2) resulted in the lowest levels of activity at the 0.1 min time point (~16%) with the activity again recovering to a modest extent at longer times (Fig. 4D, Table S5). In the presence of AscH− (950 μM), similar data was obtained to that seen with condition 2, with an activity of ~50% at all time points (Fig. 4E, Table S4). The presence of glucose (950 μM) resulted in a low level of activity after 0.1 min (~35%) and a further slow decline with increasing incubation time (Fig. 4F, Table S4).
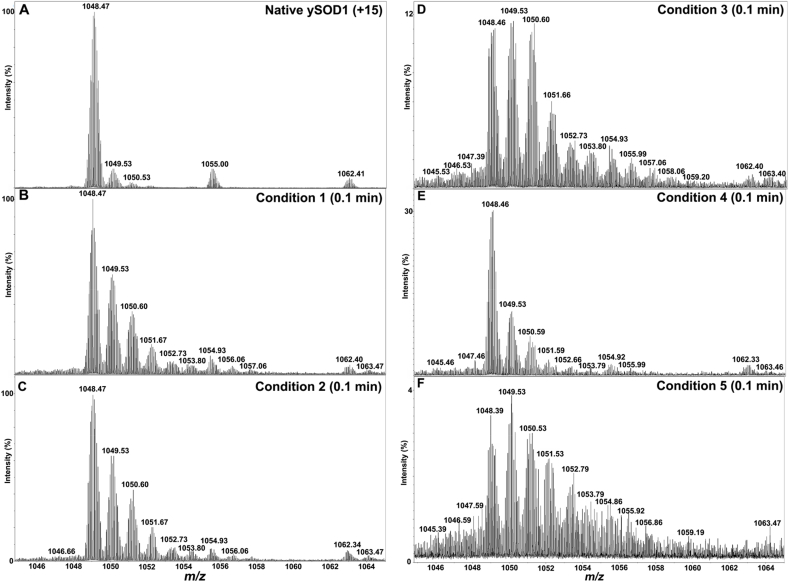
3.7. ESI–MS analysis of intact ySOD1 indicates formation of multiple oxygenated species
ESI–MS analysis of native ySOD1 showed a charge envelope from +12 to +18 (Fig. S5), with the +15-charge base peak state (monoisotopic m/z 1048.47; Fig. 5A), yielding a deconvoluted mass of 15712.91 Da consistent with the theoretical mass of the disulfide-linked species lacking the N-terminal methionine (15712.81 Da). In addition, four minor species were detected with experimental m/z values of 1049.53, 1050.53, 1055.00, and 1062.41 (Fig. 5A, Table S5) with these assigned to the addition of 1O (+16.05 Da), 2O–H (+31.07 Da), H2SO4 (+98.03 Da) and MOPS (+209.17 Da), respectively.
Fig. 5.
Positive ion ESI–MS spectra of native and oxidized ySOD1 showing +15 charged species under various oxidative conditions. (A) Native ySOD1, (B) Oxidized ySOD1 for 0.1 min under condition 1, (C) oxidized ySOD1 for 0.1 under condition 2, (D) oxidized ySOD1 for 0.1 min under condition 3 (E) oxidized ySOD1 for 0.1 min under condition 4 and (F) oxidized ySOD1 for 0.1 min under condition 5. The m/z values indicated are monoisotopic masses.
With ySOD1 exposed to reaction condition 1 (285 μM Cu2+ and 475 μM H2O2), shifts in the mass spectrum consistent with the presence of multiple oxidized ySOD1 species were detected after 0.1 min (Fig. 5B). In addition to the prominent peak from unmodified ySOD1 (see above), seven distinct signals were observed with experimental monoisotopic m/z values of 1049.53 (15728.86 Da, +16 Da, i.e. one oxygen, 1O), 1050.60 (15744.87 Da, +32 Da, 2O), 1051.67 (15760.59 Da, +48 Da, 3O), 1052.73 (15776 Da, +64 Da, 4O), 1053.80 (15792.84 Da, +80 Da, 5O). Detailed analysis of the mass spectra representing monoisotopic m/z value of 1054.93 (15809.82 Da, +97 Da) suggested overlapping features from a mixture of 96 Da (6O and 1H) +98 Da (H2SO4) species. An additional species was observed with m/z 1056.06 (15826.82 Da, +114 Da, 7O and 2H), with the addition of 2 hydrogen atoms in this species consistent with cleavage of the disulfide bond with the incorporation of 7 O atoms. Additional adducts of protonated ySOD1 with MOPS (m/z 1062.41) and protonated and oxidized ySOD1 with MOPS (m/z 1063.47) were also detected (Fig. 5B, Table S6). Further incubation under the same reaction conditions (condition 1) yielded identical mass shift patterns, and loss of 1–2 hydrogen atoms were also observed (Fig. S6A- S6D; Table S6). With the higher Cu2+ concentrations of condition 2 (Table 1), the monoisotopic m/z distributions were identical to those detected from condition 1 at reaction times of 0.1 min (Fig. 5C), 3 min and, 5 min (Figs. S7A and S7B and, Table S7). The spectra detected from the 15- and 60-min samples, despite showing similar ions assigned to the incorporation of 1–7 oxygen atoms, were of much lower relative intensity (Figs. S7C and S7D and, Table S7). Low-intensity mass peaks with a mass loss of −15 Da, – 30 Da and – 47 Da were also detected (Fig. S7, Table S7)
The results from condition 3 (high Cu2+ and high H2O2) are shown in Fig. 5D, and Fig. S8 and corresponding masses are summarized in Table S8. The mass spectra analyzed at each time point show major peaks related to both unmodified ySOD1 and oxygen atom adducts with a significant loss of ion intensity. With the addition of AscH− a similar m/z distribution of oxygen atom adducts to ySOD1 was detected with moderate loss of the ion intensities as a function of time (Figs. 5E and S9, and Table S9). The presence of glucose in the reaction mixture (condition 5) resulted in a marked loss of ion intensities from both unmodified and modified ySOD1 species (Figs. 5F and S10 and Table S10). The corresponding masses summarized in Table S10 are consistent with the presence of native ySOD1 with one to seven oxygen atoms added. However, in the presence of glucose, the MS signal also indicates the formation of a hydroxyl group with a gain of 1 H as well as cleavage of the disulfide bond.
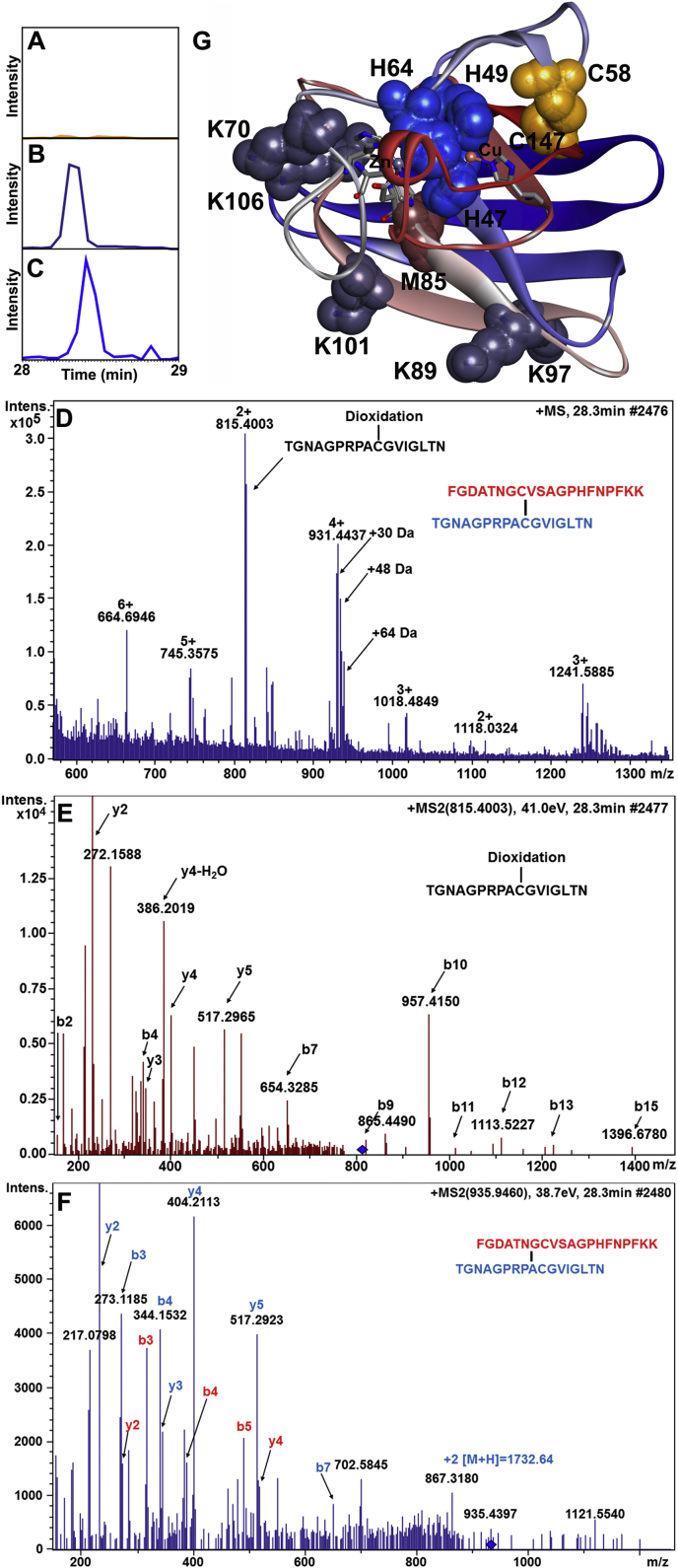
3.8. Peptide mass mapping reveals sites of modification in ySOD1
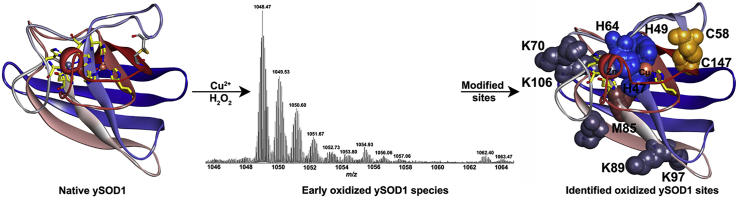
Protein digests were analyzed by LC-MS/MS to identify specifically modified peptides in ySOD1 exposed to the mild oxidation system (condition 1) for 0.1 and 60 min as well as in untreated ySOD1. The mass shifts detected were mainly associated with modifications at His, Met, Lys and the Cys residues from the disulfide bond (Table 2). Specific residues detected as modified include the Cu ion coordinating His residues (H47, H49, and H64), a number of exposed Lys (K70, K97, K101 and K106), Met (M85) and the Cys residues (C147 and C58) (Fig. 6) from the intramolecular disulfide bond; most of these modifications, including oxidation of C147 to sulfinic and sulfonic acids were only detected in the modified samples and not the controls (Fig. 6A–C). The formation of the C147-derived species may occur as a result of secondary reactions following oxidation of the initial disulfide to thiosulfinate or thiosulfonate forms. Assessment of the LC-MS data from proteolytic digests performed under non-reducing conditions revealed several oxidized forms of the disulfide-containing dipeptide, but it was not possible to unambiguously assign these mass shifts. However, we observed that the single peptide containing an oxidation of C147 to a sulfinic acid (138TGNAGPRPAC*GVIGLTN154) co-elutes with signals displaying spectral features of a disulfide-linked peptide (Fig. 6D–F). This may indicate that the disulfide bond is subjected to “in-source” reduction in the mass spectrometer, a phenomenon reported previously [68].
Table 2.
Oxidatively-modified peptides in ySOD1 identified by LC-MS/MS-based detection. The peptides were generated by proteolytic digestion of reduced and alkylated ySOD1 with either trypsin or GluC or a mixture the two enzymes as described in the methods section. LC-MS/MS peptide mapping was carried out on control (untreated) and Cu2+/H2O2 treated ySOD1 after 0.1 and 60 min incubations. Amino acid residues are numbered according to UniProtKB database id (P00445) for Saccharomyces cerevisiae (strain ATCC 204508/S288c) (Baker's yeast) SOD1.*GluC #Trypsin*# Trypsin/GluC.
| Sites | Sequence position/Peptide$ | Ctrl | 0.1 min | 60 min | Δm | Comp. | Products/Modifications |
|---|---|---|---|---|---|---|---|
| H47 | 35IAGNSPNAERGFHIHE50* | – | ✓ | – | 15.99 | O(1) | 2-oxo histidine |
| H49 | 35IAGNSPNAERGFHIHE50* | – | ✓ | – | 15.99 | O(1) | 2-oxo histidine |
| H49 | 35IAGNSPNAERGFHIHE50* | – | ✓ | – | 47.98 | O(3) | Trioxidation |
| H49 | 44RGFHIHE50* | – | ✓ | – | 16 | O(1) | 2-oxo histidine |
| H49 | 44RGFHIHE50* | – | ✓ | – | 47.99 | O(3) | Trioxidation |
| H64 | 51FGDATNGCVSAGPHFNPFKKTHGAPTDE78* | – | ✓ | – | −23.02 | H(-1) C(-2) N(-1) O(1) | His-> Asn |
| H64 | 51FGDATNGCVSAGPHFNPFKKTHGAPTDE78* | – | ✓ | – | −22.03 | H(-2) C(-2) N(-2) O(2) | His-> Asp |
| H64 | 45GFHIHEFGDATNGCVSAGPHFNPFKK70# | – | ✓ | ✓ | −22.02 | H(-2) C(-2) N(-2) O(2) | His-> Asp |
| H64 | 45GFHIHEFGDATNGCVSAGPHFNPFKK70# | – | ✓ | ✓ | −23.02 | H(-1) C(-2) N(-1) O(1) | His-> Asn |
| H64 | 51FGDATNGCVSAGPHFNPFKK70*# | – | – | ✓ | −23.02 | H(-1) C(-2) N(-1) O(1) | His-> Asn |
| K70 | 70KTHGAPTDEVR80*# | – | ✓ | – | 27.01 | H C N | |
| K70 | 70KTHGAPTDEVR80# | – | – | ✓ | 27.01 | H C N | |
| M85 | 79VRHVGDMGNVKTDE92* | ✓ | ✓ | ✓ | 15.99 | O(1) | Methionine sulfoxide |
| M85 | 81HVGDMGNVK89# | ✓ | ✓ | ✓ | 15.99 | O(1) | Methionine sulfoxide |
| M85 | 81HVGDMGNVK89*# | – | – | ✓ | 15.99 | O(1) | Methionine sulfoxide |
| M85 | 81HVGDMGNVK89*# | – | – | ✓ | 31.99 | O(2) | Methionine sulfone |
| M85 | 81HVGDMGNVKTDENGVAK97*# | ✓ | ✓ | ✓ | 15.99 | O(1) | Methionine sulfoxide |
| M85 | 79VRHVGDMGNVK89*# | – | ✓ | ✓ | 15.99 | O(1) | Methionine sulfoxide |
| M85 | 79VRHVGDMGNVK89*# | – | ✓ | ✓ | 31.99 | O(2) | Methionine sulfone |
| M85 | 79VRHVGDMGNVKTDE92*# | – | ✓ | ✓ | 16.00 | O(1) | Methionine sulfoxide |
| K97 | 90TDENGVAKGSFKDSLIK106# | – | – | ✓ | 14.97 | H(-3) N(-1) O(2) | |
| K97 | 90TDENGVAKGSFK101# | – | – | ✓ | −19.04 | H(-5) N(-1) | |
| K97 | 90TDENGVAKGSFKDSLIK106# | – | – | ✓ | −19.04 | H(-5) N(-1) | |
| K97 | 90TDENGVAKGSFK101*# | – | – | ✓ | −19.04 | H(-5) N(-1) | |
| K97 | 90TDENGVAKGSFK101*# | – | – | ✓ | 14.96 | H(-3) N(-1) O(2) | |
| K101 | 98GSFKDSLIK106 | – | ✓ | – | 58.01 | H(2) C(2) O(2) | Carboxymethyllysine |
| K101 | 98GSFKDSLIK106 | – | ✓ | – | 58.01 | H(2) C(2) O(2) | Carboxymethyllysine |
| K106 | 102DSLIKLIGPTSVVGR116# | – | – | ✓ | −31.04 | H(-5) C(-1) N(-1) | Lys->Pro |
| K106 | 102DSLIKLIGPTSVVGR116# | – | – | ✓ | −13.03 | H(-3) C(-1) N(-1) O(1) | |
| K106 | 102DSLIKLIGPTSVVGR116*# | – | – | ✓ | −31.04 | H(-5) C(-1) N(-1) | Lys->Pro |
| K106 | 102DSLIKLIGPTSVVGR116*# | – | – | ✓ | −13.03 | H(-3) C(-1) N(-1) O(1) | |
| C147 | 138TGNAGPRPACGVIGLTN154# | – | ✓ | ✓ | −25.03 | $Dioxidation | |
| C147 | 138TGNAGPRPACGVIGLTN154# | – | ✓ | ✓ | −9.04 | $Trioxidation | |
| C147 | 138TGNAGPRPACGVIGLTN154*# | – | ✓ | ✓ | −25.03 | $Dioxidation | |
| C147 | 138TGNAGPRPACGVIGLTN154*# | – | – | ✓ | −9.04 | $Trioxidation |
$ Peptides identified in samples subjected to proteolysis with GluC*, trypsin# or a combination of GluC and trypsin *#.
$The analysis is based on the reduced and alkylated samples and therefore we used carbamidomethylation (+57 Da) as a fixed modification of cysteine. This has to be taken into consideration for the cysteine residues. For example, a cysteine mass shift of −25 Da, means 57–25 = +32 Da relative to unmodified cysteine.
Fig. 6.
Oxidative modification sites of the ySOD1 protein. Extracted ion chromatograms of the doubly charged precursor ion with m/z 815.4 matching the peptide 138TGNAGPRPACGVIGLTN154 with dioxidation at C147 in SOD digested with trypsin and Glu-C. (A) Untreated native ySOD1, MCO samples exposed to condition 1 for 0.1 min (B) and, 60 min (C). (D) Mass spectrum of co-eluting species matching the peptide 138TGNAGPRPACGVIGLTN154 with dioxidation at C147 (m/z 815.4) and the disulphide bonded peptide 45FGDATNGCVSAGPHFNPFKK70-S-S-138TGNAGPRPACGVIGLTN154 with mass shifts of +30 Da (m/z 930.43), +48 Da (m/z 934.94) and +64 Da (m/z 938.94) in SOD sample exposed to MCO for 0.1 min and digested with trypsin and GluC. (E) MS/MS spectrum of the doubly charged precursor ion with m/z 815.4. (F) MS/MS spectrum of the doubly charged precursor ion with m/z 934.94. (G) Oxidized residues are shown as spheres where the active site histidine residues (H47, H49, and H64) are in blue color, whereas disulfide-linked cysteine (C58 and C147) residues are in yellow color, oxidized methionine (M85) in magenta color and lysine (K70, K89, K97, K101, and K106) residues are in grey color. Amino acid residues are numbered according to UniProtKB database id (P00445) for Saccharomyces cerevisiae (strain ATCC 204508/S288c) (Baker's yeast) SOD1. The ySOD1 structure is from Saccharomyces cerevisiae (strain ATCC 204508/S288c) (PDB ID code 1B4L [3]) as rendered in Discovery Studio visualizer 4.0 (51). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Two major mechanisms have been suggested to rationalize the toxicity of SOD1-associated FALS mutations. These are: i) structural destabilization of the SOD1 architecture including decreased stability of SOD1 monomer and increased dimer destabilization and, ii) altered catalytic properties. Increasing experimental evidence suggests that the molecular interplay of oxidative stress with a decreased integrity of the disulfide bond could be of critical importance in initiating unfolding and aggregation of SOD1 [23,28,69]. The current study therefore examined the hypothesis that early oxidative modification of ySOD1, including oxidation of the crucial disulfide bond, may perturb the structural and functional properties of this enzyme.
4.1. H2O2 derived oxidative damages to native ySOD1 is a slow process
Compared to previous studies where mM concentrations of H2O2 and long incubation periods were employed to induced structural and functional perturbations [2,52,70], we have shown that exposure of ySOD1 to 950 μM H2O2 alone for 180 min at 37 °C could induce significant oxidative modifications, and that the metal ion chelator EDTA had only a marginal effect on these H2O2-derived changes (Fig. S1). Although, the H2O2 levels used in this study (950 μM) do not mimic physiological steady-state levels (10–100 nM) [71], the use of excess H2O2 allows the study of accelerated reactions over a reasonable experimental time scale, that may mimic the long term development of the human disease, where continued exposure to lower levels of H2O2 will occur over many years. Previous studies that have proposed that H2O2 can rapidly reduce enzyme-bound Cu2+ to Cu+ [66,67], with this followed by a Fenton-like reaction in the presence of additional H2O2 [67], resulting in oxidation of the active site His residues and metal-ion release [2,72]. In line with these previous observations of oxidative modifications [66,67,72] including structural rearrangements [2,52,70], our study indicates that H2O2 alone can increase the yield of protein carbonyls, decrease enzyme activity and induce structural rearrangements (vide infra). However, it is important to note that these effects of H2O2 on ySOD1 observed at long incubation times may be of limited biological relevance as H2O2 is very rapidly turned over by multiple protective enzymes in vivo (e.g. by members of glutathione peroxidases, peroxiredoxins and catalase) [73]. However, it is worth noting that H2O2 can transverse membranes freely [74], and this factor, combined with the high abundance of SOD1 protein in motor neurons and the long half-life of this protein (~1.5 years) [75], may allow the long-term accumulation of oxidative SOD1 damage. Furthermore, there is emerging evidence that both metal-ion free (apo) and metal-deficient SOD1 mutants may contribute to SOD1's toxicity [76,77], and it has been suggested that exogenous supply of copper to these species may reduce SOD1 pathogenicity [78]. Thus, studies on the early oxidation events arising from the use of a complete system (Cu2+/H2O2), as reported in the current study, may help shed light on the molecular interplay of Cu2+ and H2O2 in SOD1-associated ALS progression and prevention.
4.2. Early oxidative events result in alterations in the net charge and conformation of ySOD1
The electropherograms from conditions 1 and 2 (Fig. 1A and B) indicate a rapid and significant loss of native ySDO1 at the earliest time examined (0.1 min) and the presence of two additional peaks with increased retention time, indicating the presence of protein species with different charge and/or conformation. These species disappeared with increasing incubation time. Oxidative conditions with high levels of Cu2+ and H2O2 (condition 3, Fig. 1C) and with the addition of glucose (condition 5; condition 3 + glucose, Fig. 1F) resulted in different electropherograms with a new peak detected at shorter retention times indicating a different heterogeneous mixture of oxidized ySOD1 species under these conditions. Interestingly, the peaks with decreased retention time showed a continuous increase in the peak intensity over the entire incubation period and likely represent new specific modifications (Fig. 1C and E). The presence of AscH− in condition 4 had a marked effect (Fig. 1D), with a decrease in the native ySOD1 peak and formation of a peak different to those observed in the absence of AscH−. AscH− is known to have both pro- and anti-oxidant effects on Cu2+-catalyzed oxidative reactions [20], and has been suggested to limit non-specific oxidation of bovine SOD1 [79] and α–Synuclein [20]. Overall, these data indicate that a mixture of oxidized ySOD1 species are produced in rapid succession during the reaction. The changes in retention time in response to oxidation may be attributed to altered net charge and/or conformation of ySOD1, and it is interesting to note that the ALS-associated SOD1 mutations G37R, H46R, G85R, D90A, G93R, E100G, E110K, and D101 N are reported to decrease the overall net charge whereas V7E tend to increase the net charge [39,69]. Furthermore, heterodimerization studies on some of these mutations have observed changes in the electrophoretic mobility [80]. By analogy to the properties of ALS associated mutations, we suggest that early oxidative modifications of wild-type ySOD1 may be a unifying mechanism that links sporadic and familial cases.
4.3. Carbonylation represents a broad marker of oxidative stress in ySOD1
The quantification data for carbonyl formation show the maximum yield is generated within 15 min of initiation of the reactions under all the conditions examined (Table S2), indicating that the generation of these modifications on ySOD1 is rapid. The highest carbonyl yield (~0.2 mol carbonyl/mol ySOD1) was obtained with the highest concentrations of Cu2+ and H2O2 (condition 3; Tables 1 and S2 and Fig. 1G). With the same concentration of Cu2+ but half the H2O2, the carbonyl levels were reduced to ~0.13 mol carbonyl/mol ySOD1 suggesting that the H2O2 concentration is a major driver of the extent of oxidative modification of ySOD1. This conclusion is supported by the significant increase in carbonyls detected when H2O2 alone was incubated with ySOD1, compared to ySOD1 with Cu2+ alone (Table S2, Fig. 1F). These observations are in agreement with a previous study where the exogenous supply of Cu2+, Fe2+, or metal chelator DTPA did not alter H2O2-driven carbonylation of bovine SOD1 (bSOD1) [42]. Under the conditions studied here, the carbonyl levels detected were higher in the presence of Cu2+ alone, compared to its absence, but the carbonyl yield was not sensitive to the addition of higher levels of Cu2+ over the range studied (Fig. S3).
A previous study of bSOD1 has reported maximum carbonylation in reaction mixtures containing 30 μM Cu2+, 1 mM H2O2, and 5 mM AscH− [42]. However, we found that 285 μM Cu2+, 950 μM H2O2, and 950 μM AscH− resulted in lower levels of carbonyls throughout the entire incubation period (Table S2, Fig. 1G) when compared to condition 3. The difference in carbonyl levels may be due to differences in the stoichiometry of the oxidants and reductants used [81]. Nevertheless, the low level of carbonyls detected in the presence of AscH− are consistent with its role as an oxidant scavenger as described earlier [20,82,83]. In the presence of glucose (condition 5), slightly higher levels of carbonyls were detected throughout the entire incubation period (Table S2, Fig. 1G) compared to condition 4. This may be due to glycation reactions, which can give rise to adducted sugar-derived carbonyls on the protein, or a release of Cu ions from the ySOD1 and subsequent enhanced Fenton-like reactions [84]. In all conditions, the carbonyl levels detected either attained a steady-state level after 10–15 min incubation or decreased at longer incubation times. Similar behavior has been reported for α-synuclein [20], bovine serum albumin (BSA) [85] and β-lactoglobulin [86]. This may be due to further oxidation of these species into carboxylic acids via reaction with H2O2 or auto-oxidation [87], or as a result of the reaction of the carbonyls with nucleophiles (e.g. Lys) to give cross-links and Schiff base species [86]. Thus, the relatively high numbers of Lys residues (10) present in ySOD1 may participate in secondary reactions that limit a further increase in carbonyl production over time. Although protein carbonyl levels are a widely accepted biomarker of oxidative stress, the current data confirm previous suggestions [88] that these species should only be considered as a broad (generic) marker of damage. Furthermore, it is well established that other amino acid side-chains, including sulfur- (Cys, Met), and aromatic residues (Trp, Tyr, His) can be more rapidly and readily oxidized [20,73], thus, an increase in absolute carbonyl concentration may represent a late event in disease-derived protein dysfunction.
4.4. Oxidation-induced modifications result in partially unfolded ySOD1 species
Small changes were seen for the ySOD1 samples examined by SDS–PAGE (Fig. 2). The observation of low-intensity dimer bands under condition 1, and a slight increase in the intensity of these bands under harsher conditions (condition 2, 3 and 5) suggests that non-reducible covalent crosslinks are formed between SOD1 monomers. The same conditions also resulted in the detection of several low-intensity bands of lower mass than the parent ySOD1. The detection of these species are consistent with studies that have mapped the fragmentation pattern of bSOD1in the presence of Cu2+ and H2O2 [42] as well as glucose [53] in the case of hSOD1. These data indicate that μM concentrations of H2O2 or glucose, in the presence of Cu2+, are sufficient to induce fragmentation of ySOD1, in addition to the mM concentrations employed previously [2,42,53]. The presence of AscH− (condition 4) inhibited both the dimer and fragment bands of ySOD1, an observation in contrast to the Cu2+-dependent AscH−/H2O2-mediated fragmentation observed with bSOD1 [42].
Further insight into the structural changes induced by oxidation was obtained through far-UV CD spectroscopy. Our data support previous observations that H2O2 alone can induce partial unfolding of SOD1 (Figs. 3A and S1B) and that this occurs with considerably lower concentrations than those employed previously [2,89]. However, the presence of Cu2+ or AscH− with the same amount of H2O2, resulted in less significant structural changes (vide infra). The gradual decline in positive intensity between 190 and 210 nm observed under all conditions (Fig. 3B–F) is indicative of major alterations to the secondary structure of the protein, and this was more pronounced with increased incubation time as well as with increased concentrations of oxidants and reductants, i.e., harsher oxidative conditions. These structural rearrangements induced by oxidation appear to be distinct from the fully unfolded state and are of interest as mounting evidence indicates that partially folded SOD1 subunits are a likely common factor in ALS pathology [23,90]. The structural destabilization hypothesis, which rationalizes the widespread distribution of ALS mutation sites, provides a unifying mechanism of toxicity [23]. Whilst protein carbonylation may contribute to this structural destabilization, the data indicate that a maximum of 15% of the ySOD1 molecules are carbonylated under the conditions employed. Although a recent computational simulation study combined with thermodynamic integration suggests that oxidative damage to even a single residue at the dimeric interface might destabilize the SOD1 homodimer and monomer, and result in partial unfolding [24], it is unlikely that carbonyl formation is the sole cause of the observed changes in structure.
A second potential contributing factor is oxidative modifications to the amino acids of the metal ion binding loop regions. The Greek key β-barrel [91] fold of SOD1 has been suggested to be important for the high structural stability of the enzyme, and its rapid catalysis [23]. In fact, an ALS-associated β-barrel residue FALS mutant (H43R, hSOD1 numbering) has been shown to rapidly form fibrillar aggregates due to perturbation of the β-barrel fold and increased monomerization [92]. In the current study, we have identified conversion of the two β-barrel active site His residues (H47 and H49; H46 and H48 in hSOD1) into 2-oxo-His. A third His (H64; H63 in hSOD1) outside the β-barrel, which forms a bridge between Cu and Zn ions, was also observed to be converted into Asp or Asn (Fig. 6 G-H, Table 2). These His modifications are consistent with previous studies [2,22,52,70,89]. These active-site His modifications have been shown to result in the release of the catalytically-active metal ions, and subsequent unfolding and aggregation of SOD1 [2,22]. To the best of our knowledge, the Zn ion coordinating His residue, H64, has not been reported previously to be oxidized by MCO. The modification of these three His residues (H47, H49, and H64), which are critical to the structure and catalytic activity [23] could make both the Cu and Zn metal ions labile, and promote SOD1 aggregation through metal ion loss [22].
A third contributory factor to the protein destabilization maybe the observed modifications to the electrostatic potential of the SOD1 surface which has been suggested to maintain hydrogen bonding and salt-bridge interactions [23]. The modification of five exposed Lys residues (K70, K89, K97, K101, and K106; Fig. 6G, Table 2) may disrupt this surface electrostatic potential and promote aberrant interactions that nucleate aggregation. It is well established that various stressors and mutations can destabilize native protein structures [93]. Although early studies suggested that fully unfolded protein molecules could form aggregates [94,95], recent evidence indicates that partially unfolded states may be the key drivers of aggregate [96,97] and fibril [98,99] formation. Thus, a modest increase in the population of partially-unfolded species may enhance the aggregation rate. Moreover, the toxic gain-of-function in ALS has been proposed to occur via a partially unfolded intermediate of SOD1 protein lacking metal-ion cofactors [100]. Thus, the detection in this study of partially unfolded ySOD1 conformations under various oxidative conditions may be of biological significance.
4.5. Cu ion acts as a sacrificial substrate and partially folded ySOD1 species retain enzymatic function
Consistent with the significant conformational changes that result from oxidant exposure, the enzymatic activity of ySOD1 was found to be reduced by around 50% under the mildest oxidative conditions examined (condition 1, Fig. 4B). This rapid loss of enzyme activity may be attributed to the active site modifications discussed above. However, in condition 2, which included exogenous Cu2+, an immediate loss of 60% activity was observed followed by an increase in the activity after 5–60 min (Fig. 4C). Such a reversal in ySOD1 enzymatic activity (Table S4) is likely to be due to a reconstitution of Cu ions as these are present in excess (Table 1). The addition of 142.5 μM Cu2+ in condition 1 had less pronounced effects on the structure and activity (~50% of native ySOD1 after 30 min; Fig. 3, Fig. 4B) than that induced by 950 μM H2O2 alone (Fig. 3, Fig. 4A). This trend of Cu2+ protecting against H2O2 was more distinct with higher concentrations of Cu2+ (285 μM) in condition 2, with both a higher level of ySOD1 activity (>70% of native ySOD1 after 30 min, Fig. 4C), and less pronounced effects on secondary structure, detected (Fig. 3C). Molecules including xanthine, azide, imidazole, urate, and formate have been reported to protect SOD1 against H2O2 induced inactivation, by either acting as a sacrificial target or by reducing histidinyl radical intermediates [67,72]. Thus, it is conceivable that free Cu2+ in the reaction mixture reacts directly with excess H2O2, and thereby prevents reaction of H2O2 with enzyme-bound Cu2+. This suggestion is supported by the data from condition 3 where the presence of 285 μM Cu2+ resulted in the retention of 30% of the native ySOD1 activity compared to only ~5% with 950 μM H2O2 alone after 180 min (Fig. 4A and D). Copper-ion loading of metal-ion deficient SOD1 has been proposed as a therapeutic strategy against SOD1-associated ALS [101], and our observations lend support to this hypothesis. Recent studies have employed diacetyl-bis (4-methylthiosemicarbazonato)copper (II) complexes (CuIIATSM) to deliver copper to the spinal cord tissue of Cu-deficient SOD1G37R mice. It was postulated that the exogenous copper supplied by CuIIATSM to partially folded intermediates of SOD1, might protect ALS transgenic models expressing human Copper-Chaperone-for-SOD [101]. Further studies have shown that CuIIATSM treatment can reduce the pool of Cu-deficient SOD1 and increase the level of fully metallated (holo) SOD1 [102]. Improving the metal-ion content of SOD1 may therefore represent a valid therapeutic intervention against ALS induced by SOD1 mutations [[101], [102], [103], [104]].
Although site-specific modification was investigated only for the mildest oxidation conditions, it is conceivable that the loss of ySOD1 activity in conditions 3 and 5 (Fig. 4D and F) also stems from the destabilization of the protein structure as detected in the CE and CD analyses. Furthermore, the presence of AscH− (950 μM, condition 4) protected against loss of SOD1 activity, indicating a protective role of AscH− against oxidative protein dysfunction [20]. Although the loss-of-function paradigm is not considered to be causative for ALS, a number of FALS mutant SOD1 species have ~50% reduced activity in fibroblast, lymphoblastoid and red blood cells [44,105]. Moreover, a loss-of-function would result in impaired clearance of O2•− by less active ySOD1 and may result in i) increased oxidative stress and, ii) a disease-modifying toxic gain-of-function.
4.6. Disulfide bond oxidation: a protective mechanism against oxidant damage?
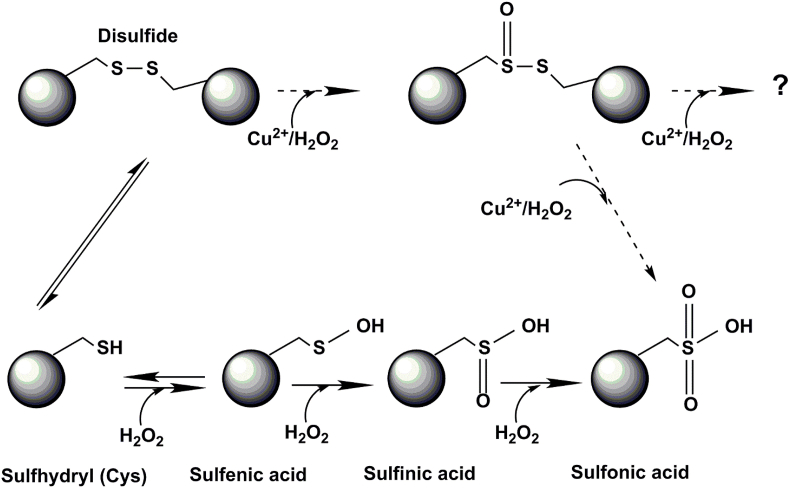
The C58–C147 disulfide bond is an important structure stabilizing factor in ySOD1 [35]. The disulfide bond is positioned ~10 Å from the copper binding site in ySOD1 (see PDB structure 1B4L), with C147 more solvent exposed than C58. The LC-MS peptide mapping data (Table 2) obtained under condition 1 indicates that C147, but not C58, is subject to oxidation within 0.1 min, though this does not result in a massive loss of enzymatic activity. ESI–MS spectra of intact ySOD1 show that a series of distinct oxidation products have already been generated at the 0.1 min time point (Fig. 5B), with the first 7 oxidized peaks observed (for the +15-charge state) having monoisotopic masses consistent with the addition of 1–7 oxygen atoms. Based on peptide mapping by LC-MS/MS these oxidation products are attributed to oxidation of M85, C147 and possibly H47 and H49 (Table 2). The extent of oxidation detected by ESI-MS after 0.1 min under condition 3 is ~90% of the parent protein, however, ~25% enzymatic activity was retained, indicating that not all modifications result in a loss of activity. Earlier studies have reported that cleavage of the disulfide bridge reduces the activity to 5–10% of the native SOD [35]. The retained activity of ySOD1 after MCO under condition 3 could indicate that the disulfide bridge is not immediately cleaved by MCO. Consistent with this suggestion, analysis of the LC-MS data from proteolytic digests performed under non-reducing conditions revealed several oxidized forms of the disulfide-containing dipeptide, but it was not possible to unambiguously assign these mass shifts. However, the observation that a peptide with oxidation of C147 to a sulfinic acid (138TGNAGPRPAC*GVIGLTN154) co-elutes with signals displaying spectral features of a disulfide-linked peptide (Fig. 6D–F), may indicate that cleavage of the disulfide bond occurs by in-source reduction in the mass spectrometer, a phenomenon that has been reported previously, and not during the initial MCO. These data, therefore, indicate that disulfide bond oxidation may occur without cleavage of the -S-S- linkage in line with studies carried out on low-molecular-mass disulfide-containing model compounds and small peptides, where thiosulfinate and thiosulfonate (i.e. RSS(O)R′ and RSS(O)2R’) formation has been detected [[106], [107], [108]]. We, therefore, propose that this disulfide bond besides being a stabilizing factor in ySOD1 also potentially protects ySOD1 against destructive oxidation by acting as a sink for oxidative damage without a concomitant loss of enzymatic activity. The disulfide bond, like M85, may, therefore, be an evolutionary-conserved protective mechanism in ySOD1 that makes the enzyme more resistant to oxidative damage. The proposed steps for the oxidation of the disulfide bond are shown in Fig. 7. The idea of having protecting Met and Cys residues in SOD is not new [109], but the observation that a disulfide bond might work in the same way is novel.
Fig. 7.
Proposed mechanism for the oxidation of Cysteine in the disulfide bond of ySOD1. LC-MS data suggests that the first modifications by MCO on ySOD1 might oxidize C147 in the C58–C147 disulfide bond without cleaving this bond. Further oxidation of the intact disulfide bond is not ruled out.
5. Conclusions
This study shows that although SOD1 plays a major role in protection against oxidative damage in cells, it can also be readily modified by moderate levels of oxidants. H2O2, the product of SOD1 catalysis, can induce severe structural perturbations at pathophysiological concentrations. We have for the first time identified H64 (H63 in hSOD1) as one of the major targets of copper ion catalyzed oxidation. This residue which acts as a ligand to both the Cu and Zn ions is located outside of the active site β-barrel and is relatively exposed. Modification of this residue could result in loss of both metal ions and promote SOD1 dysfunction and unfolding. By analogy with ALS-causing SOD1 mutations, which primarily affect the structural integrity of SOD1 protein [44] and initiate unfolding cascades, our data indicate that early oxidation-induced modifications may result in the formation of partially unfolded SOD1 species, and contribute to a vicious cycle of loss-of-function and SOD1 aggregation through seeding. We propose that the ability of ySOD1 to retain its function whilst continually exposed to reactive oxidants may be attributed to its evolutionary-conserved disulfide bond which acts as a sink for oxidation equivalents and ensures the structural and functional integrity of ySOD1. These data may have bearing on the molecular events occurring in SOD1-associated ALS cases.
Acknowledgments
We gratefully acknowledge financial support from the Danish Council for Independent Research Technology and Production Sciences (Grant no.: DFF| FTP 4005–00082 to IMM), the Novo Nordisk Foundation (Grant no.: NNF 13OC0004294 to MJD), and the Lundbeck Foundation postdoctoral fellowship (Grant no.: R231-2016-3276 to MKT).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101262.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Andersen J.K. Oxidative stress in neurodegeneration: cause or consequence? Nat. Med. 2004;10(Suppl):S18–S25. doi: 10.1038/nrn1434. [DOI] [PubMed] [Google Scholar]
- 2.Mulligan V.K., Kerman A., Laister R.C., Sharda P.R., Arslan P.E., Chakrabartty A. Early steps in oxidation-induced SOD1 misfolding: implications for non-amyloid protein aggregation in familial ALS. J. Mol. Biol. 2012;421(4–5):631–652. doi: 10.1016/j.jmb.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 3.Hart P.J., Balbirnie M.M., Ogihara N.L., Nersissian A.M., Weiss M.S., Valentine J.S., Eisenberg D. A structure-based mechanism for copper-zinc superoxide dismutase. Biochemistry. 1999;38(7):2167–2178. doi: 10.1021/bi982284u. [DOI] [PubMed] [Google Scholar]
- 4.Barber S.C., Shaw P.J. Oxidative stress in ALS: key role in motor neuron injury and therapeutic target. Free Radic. Biol. Med. 2010;48(5):629–641. doi: 10.1016/j.freeradbiomed.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 5.Bruijn L.I., Miller T.M., Cleveland D.W. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 6.Ihara Y., Nobukuni K., Takata H., Hayabara T. Oxidative stress and metal content in blood and cerebrospinal fluid of amyotrophic lateral sclerosis patients with and without a Cu, Zn-superoxide dismutase mutation. Neurol. Res. 2005;27(1):105–108. doi: 10.1179/016164105X18430. [DOI] [PubMed] [Google Scholar]
- 7.Fridovich I. Superoxide anion radical (O2-.), superoxide dismutases, and related matters. J. Biol. Chem. 1997;272(30):18515–18517. doi: 10.1074/jbc.272.30.18515. [DOI] [PubMed] [Google Scholar]
- 8.Fridovich I. Superoxide dismutases. An adaptation to a paramagnetic gas. J. Biol. Chem. 1989;264(14):7761–7764. [PubMed] [Google Scholar]
- 9.McCord J.M., Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J. Biol. Chem. 1969;244(22):6049–6055. [PubMed] [Google Scholar]
- 10.Valentine J.S., Hart P.J. Misfolded CuZnSOD and amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U. S. A. 2003;100(7):3617–3622. doi: 10.1073/pnas.0730423100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stadtman E.R., Berlett B.S. Reactive oxygen-mediated protein oxidation in aging and disease. Drug Metab. Rev. 1998;30(2):225–243. doi: 10.3109/03602539808996310. [DOI] [PubMed] [Google Scholar]
- 12.Valentine J.S., Doucette P.A., Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu. Rev. Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 13.Ferraiuolo L., Kirby J., Grierson A.J., Sendtner M., Shaw P.J. Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat. Rev. Neurol. 2011;7(11):616–630. doi: 10.1038/nrneurol.2011.152. [DOI] [PubMed] [Google Scholar]
- 14.Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J.P., Deng H.X. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 15.Andersen P.M., Nilsson P., Keranen M.L., Forsgren L., Hagglund J., Karlsborg M., Ronnevi L.O., Gredal O., Marklund S.L. Phenotypic heterogeneity in motor neuron disease patients with CuZn-superoxide dismutase mutations in Scandinavia. Brain. 1997;120(Pt 10):1723–1737. doi: 10.1093/brain/120.10.1723. [DOI] [PubMed] [Google Scholar]
- 16.Abel O., Powell J.F., Andersen P.M., Al-Chalabi A. ALSoD: a user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum. Mutat. 2012;33(9):1345–1351. doi: 10.1002/humu.22157. [DOI] [PubMed] [Google Scholar]
- 17.Andersen P.M., Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat. Rev. Neurol. 2011;7(11):603–615. doi: 10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- 18.Tiwari M.K., Kepp K.P. Kepp, β-Amyloid pathogenesis: chemical properties versus cellular levels. Alzheimers Dement. 2016;12(2):184–194. doi: 10.1016/j.jalz.2015.06.1895. [DOI] [PubMed] [Google Scholar]
- 19.Tiwari M.K., Kepp K.P. Pathogenic properties of Alzheimer's β-amyloid identified from structure-property patient-phenotype correlations. Dalton Trans. 2015;44(6):2747–2754. doi: 10.1039/c4dt03122a. [DOI] [PubMed] [Google Scholar]
- 20.Tiwari M.K., Leinisch F., Sahin C., Moller I.M., Otzen D.E., Davies M.J., Bjerrum M.J. Early events in copper-ion catalyzed oxidation of α-synuclein. Free Radic. Biol. Med. 2018;121:38–50. doi: 10.1016/j.freeradbiomed.2018.04.559. [DOI] [PubMed] [Google Scholar]
- 21.Hart P.J. Pathogenic superoxide dismutase structure, folding, aggregation and turnover. Curr. Opin. Chem. Biol. 2006;10(2):131–138. doi: 10.1016/j.cbpa.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 22.Martins D., English A.M. SOD1 oxidation and formation of soluble aggregates in yeast: relevance to sporadic ALS development. Redox Biol. 2014;2:632–639. doi: 10.1016/j.redox.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry J.J., Shin D.S., Getzoff E.D., Tainer J.A. The structural biochemistry of the superoxide dismutases. Biochim. Biophys. Acta. 2010;1804(2):245–262. doi: 10.1016/j.bbapap.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrov D., Daura X., Zagrovic B. Effect of oxidative damage on the stability and dimerization of superoxide dismutase 1. Biophys. J. 2016;110(7):1499–1509. doi: 10.1016/j.bpj.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abreu I.A., Cabelli D.E. Superoxide dismutases-a review of the metal-associated mechanistic variations. Biochim. Biophys. Acta. 2010;1804(2):263–274. doi: 10.1016/j.bbapap.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Casadio R., Vassura M., Tiwari S., Fariselli P., Luigi Martelli P. Correlating disease-related mutations to their effect on protein stability: a large-scale analysis of the human proteome. Hum. Mutat. 2011;32(10):1161–1170. doi: 10.1002/humu.21555. [DOI] [PubMed] [Google Scholar]
- 27.Ferrante R.J., Browne S.E., Shinobu L.A., Bowling A.C., Baik M.J., MacGarvey U., Kowall N.W., Brown R.H., Jr., Beal M.F. Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis. J. Neurochem. 1997;69(5):2064–2074. doi: 10.1046/j.1471-4159.1997.69052064.x. [DOI] [PubMed] [Google Scholar]
- 28.Shaw P.J., Ince P.G., Falkous G., Mantle D. Oxidative damage to protein in sporadic motor neuron disease spinal cord. Ann. Neurol. 1995;38(4):691–695. doi: 10.1002/ana.410380424. [DOI] [PubMed] [Google Scholar]
- 29.Beal M.F., Ferrante R.J., Browne S.E., Matthews R.T., Kowall N.W., Brown R.H., Jr. Increased 3-nitrotyrosine in both sporadic and familial amyotrophic lateral sclerosis. Ann. Neurol. 1997;42(4):644–654. doi: 10.1002/ana.410420416. [DOI] [PubMed] [Google Scholar]
- 30.Carrì M.T., Ferri A., Cozzolino M., Calabrese L., Rotilio G. Neurodegeneration in amyotrophic lateral sclerosis: the role of oxidative stress and altered homeostasis of metals. Brain Res. Bull. 2003;61(4):365–374. doi: 10.1016/s0361-9230(03)00179-5. [DOI] [PubMed] [Google Scholar]
- 31.Bozzo F., Mirra A., Carri M.T. Oxidative stress and mitochondrial damage in the pathogenesis of ALS: new perspectives. Neurosci. Lett. 2017;636:3–8. doi: 10.1016/j.neulet.2016.04.065. [DOI] [PubMed] [Google Scholar]
- 32.Kawamata H., Manfredi G. Import, maturation, and function of SOD1 and its copper chaperone CCS in the mitochondrial intermembrane space. Antioxidants Redox Signal. 2010;13(9):1375–1384. doi: 10.1089/ars.2010.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindberg M.J., Normark J., Holmgren A., Oliveberg M. Folding of human superoxide dismutase: disulfide reduction prevents dimerization and produces marginally stable monomers. Proc. Natl. Acad. Sci. U. S. A. 2004;101(45):15893–15898. doi: 10.1073/pnas.0403979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ray S.S., Nowak R.J., Strokovich K., Brown R.H., Walz T., Jr., Lansbury P.T., Jr. An intersubunit disulfide bond prevents in vitro aggregation of a superoxide dismutase-1 mutant linked to familial amytrophic lateral sclerosis. Biochemistry. 2004;43(17):4899–4905. doi: 10.1021/bi030246r. [DOI] [PubMed] [Google Scholar]
- 35.Sea K., Sohn S.H., Durazo A., Sheng Y., Shaw B.F., Cao X., Taylor A.B., Whitson L.J., Holloway S.P., Hart P.J., Cabelli D.E., Gralla E.B., Valentine J.S. Insights into the role of the unusual disulfide bond in copper-zinc superoxide dismutase. J. Biol. Chem. 2015;290(4):2405–2418. doi: 10.1074/jbc.M114.588798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borders C.L., Johansen J.T. Identification of ARG-143 as the essential arginyl residue in yeast Cu,Zn superoxide dismutase by use of a chromophoric arginine reagent. Biochem. Biophys. Res. Commun. 1980;96(3):1071–1078. doi: 10.1016/0006-291x(80)90061-3. [DOI] [PubMed] [Google Scholar]
- 37.Tiwari A., Hayward L.J. Familial amyotrophic lateral sclerosis mutants of copper/zinc superoxide dismutase are susceptible to disulfide reduction. J. Biol. Chem. 2003;278(8):5984–5992. doi: 10.1074/jbc.M210419200. [DOI] [PubMed] [Google Scholar]
- 38.Furukawa Y., O'Halloran T.V. Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo- and reduced form of SOD1, leading to unfolding and oxidative aggregation. J. Biol. Chem. 2005;280(17):17266–17274. doi: 10.1074/jbc.M500482200. [DOI] [PubMed] [Google Scholar]
- 39.Oztug Durer Z.A., Cohlberg J.A., Dinh P., Padua S., Ehrenclou K., Downes S., Tan J.K., Nakano Y., Bowman C.J., Hoskins J.L., Kwon C., Mason A.Z., Rodriguez J.A., Doucette P.A., Shaw B.F., Valentine J.S. Loss of metal ions, disulfide reduction and mutations related to familial ALS promote formation of amyloid-Like aggregates from superoxide dismutase. PLoS One. 2009;4(3) doi: 10.1371/journal.pone.0005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chattopadhyay M., Durazo A., Sohn S.H., Strong C.D., Gralla E.B., Whitelegge J.P., Valentine J.S. Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc. Natl. Acad. Sci. U. S. A. 2008;105(48):18663–18668. doi: 10.1073/pnas.0807058105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi J., Rees H.D., Weintraub S.T., Levey A.I., Chin L.S., Li L. Oxidative modifications and aggregation of Cu,Zn-superoxide dismutase associated with Alzheimer and Parkinson diseases. J. Biol. Chem. 2005;280(12):11648–11655. doi: 10.1074/jbc.M414327200. [DOI] [PubMed] [Google Scholar]
- 42.Uehara H., Luo S., Aryal B., Levine R.L., Rao V.A. Distinct oxidative cleavage and modification of bovine [Cu– Zn]-SOD by an ascorbic acid/Cu(II) system: identification of novel copper binding site on SOD molecule. Free Radic. Biol. Med. 2016;94:161–173. doi: 10.1016/j.freeradbiomed.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rakhit R., Chakrabartty A. Structure, folding, and misfolding of Cu,Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim. Biophys. Acta. 2006;1762(11–12):1025–1037. doi: 10.1016/j.bbadis.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Deng H.X., Hentati A., Tainer J.A., Iqbal Z., Cayabyab A., Hung W.Y., Getzoff E.D., Hu P., Herzfeldt B., Roos R.P. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science. 1993;261(5124):1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 45.Ding F., Dokholyan N.V. Dynamical roles of metal ions and the disulfide bond in Cu, Zn superoxide dismutase folding and aggregation. Proc. Natl. Acad. Sci. U. S. A. 2008;105(50):19696–19701. doi: 10.1073/pnas.0803266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J., Lee H., Lee J.H., Kwon D.Y., Genovesio A., Fenistein D., Ogier A., Brondani V., Grailhe R. Dimerization, oligomerization, and aggregation of human amyotrophic lateral sclerosis copper/zinc superoxide dismutase 1 protein mutant forms in live cells. J. Biol. Chem. 2014;289(21):15094–15103. doi: 10.1074/jbc.M113.542613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lelie H.L., Liba A., Bourassa M.W., Chattopadhyay M., Chan P.K., Gralla E.B., Miller L.M., Borchelt D.R., Valentine J.S., Whitelegge J.P. Copper and zinc metallation status of copper-zinc superoxide dismutase from amyotrophic lateral sclerosis transgenic mice. J. Biol. Chem. 2011;286(4):2795–2806. doi: 10.1074/jbc.M110.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lyons T.J., Liu H., Goto J.J., Nersissian A., Roe J.A., Graden J.A., Cafe C., Ellerby L.M., Bredesen D.E., Gralla E.B., Valentine J.S. Mutations in copper-zinc superoxide dismutase that cause amyotrophic lateral sclerosis alter the zinc binding site and the redox behavior of the protein. Proc. Natl. Acad. Sci. U. S. A. 1996;93(22):12240–12244. doi: 10.1073/pnas.93.22.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyons T.J., Nersissian A., Huang H., Yeom H., Nishida C.R., Graden J.A., Gralla E.B., Valentine J.S. The metal binding properties of the zinc site of yeast copper-zinc superoxide dismutase: implications for amyotrophic lateral sclerosis. J. Biol. Inorg. Chem. 2000;5(2):189–203. doi: 10.1007/s007750050363. [DOI] [PubMed] [Google Scholar]
- 50.Bouldin S.D., Darch M.A., Hart P.J., Outten C.E. Redox properties of the disulfide bond of human Cu,Zn superoxide dismutase and the effects of human glutaredoxin 1. Biochem. J. 2012;446(1):59–67. doi: 10.1042/BJ20120075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghosh S., Willard B., Comhair S.A.A., Dibello P., Xu W., Shiva S., Aulak K.S., Kinter M., Erzurum S.C. Disulfide bond as a switch for copper-zinc superoxide dismutase activity in asthma. Antioxidants Redox Signal. 2013;18(4):412–423. doi: 10.1089/ars.2012.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rakhit R., Cunningham P., Furtos-Matei A., Dahan S., Qi X.-F., Crow J.P., Cashman N.R., Kondejewski L.H., Chakrabartty A. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J. Biol. Chem. 2002;277(49):47551–47556. doi: 10.1074/jbc.M207356200. [DOI] [PubMed] [Google Scholar]
- 53.Arai K., Maguchi S., Fujii S., Ishibashi H., Oikawa K., Taniguchi N. Glycation and inactivation of human Cu-Zn-superoxide dismutase. Identification of the in vitro glycated sites. J. Biol. Chem. 1987;262(35):16969–16972. [PubMed] [Google Scholar]
- 54.Arai K., Iizuka S., Tada Y., Oikawa K., Taniguchi N. Increase in the glucosylated form of erythrocyte Cu-Zn-superoxide dismutase in diabetes and close association of the nonenzymatic glucosylation with the enzyme activity. Biochim. Biophys. Acta Gen. Subj. 1987;924(2):292–296. doi: 10.1016/0304-4165(87)90025-0. [DOI] [PubMed] [Google Scholar]
- 55.Beers R.F., Jr., Sizer I.W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 1952;195(1):133–140. [PubMed] [Google Scholar]
- 56.Lyons T.J., Liu H.B., Goto J.J., Nersissian A., Roe J.A., Graden J.A., Cafe C., Ellerby L.M., Bredesen D.E., Gralla E.B., Valentine J.S. Mutations in copper-zinc superoxide dismutase that cause amyotrophic lateral sclerosis alter the zinc binding site and the redox behavior of the protein. Proc. Natl. Acad. Sci. U. S. A. 1996;93(22):12240–12244. doi: 10.1073/pnas.93.22.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xuan X., Li D. Joule heating effects on peak broadening in capillary zone electrophoresis. J. Micromech. Microeng. 2004;14(8):1171–1180. [Google Scholar]
- 58.Smith P.K., Krohn R.I., Hermanson G.T., Mallia A.K., Gartner F.H., Provenzano M.D., Fujimoto E.K., Goeke N.M., Olson B.J., Klenk D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 59.Marklund S., Marklund G. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur. J. Biochem. 1974;47:469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- 60.Borders C.L., Bjerrum M.J., Schirmer M.A., Oliver S.G. Characterization of recombinant Saccharomyces cerevisiae manganese-containing superoxide dismutase and its H30A and K170R mutants expressed in Escherichia coli. Biochemistry. 1998;37(32):11323–11331. doi: 10.1021/bi980167t. [DOI] [PubMed] [Google Scholar]
- 61.Rykær M., Svensson B., Davies M.J., Hägglund P. Unrestricted mass spectrometric data analysis for identification, localization, and quantification of oxidative protein modifications. J. Proteome Res. 2017;16(11):3978–3988. doi: 10.1021/acs.jproteome.7b00330. [DOI] [PubMed] [Google Scholar]
- 62.Savitski M.M., Nielsen M.L., Zubarev R.A. ModifiComb, a new proteomic tool for mapping substoichiometric post-translational modifications, finding novel types of modifications, and fingerprinting complex protein mixtures. Mol. Cell. Proteom. 2006;5(5):935–948. doi: 10.1074/mcp.T500034-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Nielsen M.L., Savitski M.M., Zubarev R.A. Extent of modifications in human proteome samples and their effect on dynamic range of analysis in shotgun proteomics. Mol. Cell. Proteom. 2006;5(12):2384–2391. doi: 10.1074/mcp.M600248-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 65.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 66.Hodgson E.K., Fridovich I. The interaction of bovine erythrocyte superoxide dismutase with hydrogen peroxide: chemiluminescence and peroxidation. Biochemistry. 1975;14(24):5299–5303. doi: 10.1021/bi00695a011. [DOI] [PubMed] [Google Scholar]
- 67.Hodgson E.K., Fridovich I. The interaction of bovine erythrocyte superoxide dismutase with hydrogen peroxide: inactivation of the enzyme. Biochemistry. 1975;14(24):5294–5299. doi: 10.1021/bi00695a010. [DOI] [PubMed] [Google Scholar]
- 68.Cramer C.N., Kelstrup C.D., Olsen J.V., Haselmann K.F., Nielsen P.K. Generic workflow for mapping of complex disulfide bonds using in-source reduction and extracted ion chromatograms from data-dependent mass spectrometry. Anal. Chem. 2018;90(13):8202–8210. doi: 10.1021/acs.analchem.8b01603. [DOI] [PubMed] [Google Scholar]
- 69.Shaw B.F., Valentine J.S. How do ALS-associated mutations in superoxide dismutase 1 promote aggregation of the protein? Trends Biochem. Sci. 2007;32(2):78–85. doi: 10.1016/j.tibs.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 70.Rakhit R., Crow J.P., Lepock J.R., Kondejewski L.H., Cashman N.R., Chakrabartty A. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J. Biol. Chem. 2004;279(15):15499–15504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 71.Sies H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: oxidative eustress. Redox Biol. 2017;11:613–619. doi: 10.1016/j.redox.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goldstone A.B., Liochev S.I., Fridovich I. Inactivation of copper, zinc superoxide dismutase by H2O2 : mechanism of protection. Free Radic. Biol. Med. 2006;41(12):1860–1863. doi: 10.1016/j.freeradbiomed.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 73.Davies M.J. Protein oxidation and peroxidation. Biochem. J. 2016;473(7):805–825. doi: 10.1042/BJ20151227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cardoso A.R., Chausse B., da Cunha F.M., Luevano-Martinez L.A., Marazzi T.B., Pessoa P.S., Queliconi B.B., Kowaltowski A.J. Mitochondrial compartmentalization of redox processes. Free Radic. Biol. Med. 2012;52(11–12):2201–2208. doi: 10.1016/j.freeradbiomed.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 75.Borchelt D.R., Wong P.C., Becher M.W., Pardo C.A., Lee M.K., Xu Z.S., Thinakaran G., Jenkins N.A., Copeland N.G., Sisodia S.S., Cleveland D.W., Price D.L., Hoffman P.N. Axonal transport of mutant superoxide dismutase 1 and focal axonal abnormalities in the proximal axons of transgenic mice. Neurobiol. Dis. 1998;5(1):27–35. doi: 10.1006/nbdi.1998.0178. [DOI] [PubMed] [Google Scholar]
- 76.Banci L., Bertini I., D'Amelio N., Libralesso E., Turano P., Valentine J.S. Metalation of the amyotrophic lateral sclerosis mutant glycine 37 to arginine superoxide dismutase (SOD1) apoprotein restores its structural and dynamical properties in solution to those of metalated wild-type SOD1. Biochemistry. 2007;46(35):9953–9962. doi: 10.1021/bi700620r. [DOI] [PubMed] [Google Scholar]
- 77.Hilton J.B., White A.R., Crouch P.J. Metal-deficient SOD1 in amyotrophic lateral sclerosis. J. Mol. Med. (Berl.) 2015;93(5):481–487. doi: 10.1007/s00109-015-1273-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tokuda E., Furukawa Y. Copper homeostasis as a therapeutic target in amyotrophic lateral sclerosis with SOD1 mutations. Int. J. Mol. Sci. 2016;17(5):636. doi: 10.3390/ijms17050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bridgewater J.D., Vachet R.W. Metal-catalyzed oxidation reactions and mass spectrometry: the roles of ascorbate and different oxidizing agents in determining Cu–protein-binding sites. Anal. Biochem. 2005;341(1):122–130. doi: 10.1016/j.ab.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 80.Shi Y., Acerson M.J., Abdolvahabi A., Mowery R.A., Shaw B.F. Gibbs energy of superoxide dismutase heterodimerization accounts for variable survival in amyotrophic lateral sclerosis. J. Am. Chem. Soc. 2016;138(16):5351–5362. doi: 10.1021/jacs.6b01742. [DOI] [PubMed] [Google Scholar]
- 81.Møller I.M., Rogowska-Wrzesinska A., Rao R.S. Protein carbonylation and metal-catalyzed protein oxidation in a cellular perspective. J Proteomics. 2011;74(11):2228–2242. doi: 10.1016/j.jprot.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 82.Nimse S.B., Pal D. Free radicals, natural antioxidants, and their reaction mechanisms. RSC Adv. 2015;5(35):27986–28006. [Google Scholar]
- 83.Hoey B.M., Butler J. The repair of oxidized amino acids by antioxidants. Biochim. Biophys. Acta. 1984;791(2):212–218. doi: 10.1016/0167-4838(84)90011-6. [DOI] [PubMed] [Google Scholar]
- 84.Ookawara T., Kawamura N., Kitagawa Y., Taniguchi N. Site-specific and random fragmentation of Cu,Zn-superoxide dismutase by glycation reaction. Implication of reactive oxygen species. J. Biol. Chem. 1992;267(26):18505–18510. [PubMed] [Google Scholar]
- 85.Mayo J.C., Tan D.X., Sainz R.M., Natarajan M., Lopez-Burillo S., Reiter R.J. Protection against oxidative protein damage induced by metal-catalyzed reaction or alkylperoxyl radicals: comparative effects of melatonin and other antioxidants. Biochim. Biophys. Acta. 2003;1620(1):139–150. doi: 10.1016/s0304-4165(02)00527-5. [DOI] [PubMed] [Google Scholar]
- 86.Yuan Q., Zhu X., Sayre L.M. Chemical nature of stochastic generation of protein-based carbonyls: metal-catalyzed oxidation versus modification by products of lipid oxidation. Chem. Res. Toxicol. 2007;20(1):129–139. doi: 10.1021/tx600270f. [DOI] [PubMed] [Google Scholar]
- 87.Monnier V.M., Nemet I., Sell D.R., Weiss M.F. Transition metals and other forms of oxidative protein damage in renal disease. In: Miyata T., Eckardt K.-U., Nangaku M., editors. Studies on Renal Disorders. Humana Press; Totowa, NJ: 2011. pp. 25–50. [Google Scholar]
- 88.Dalle-Donne I., Giustarini D., Colombo R., Rossi R., Milzani A. Protein carbonylation in human diseases. Trends Mol. Med. 2003;9(4):169–176. doi: 10.1016/s1471-4914(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 89.Uchida K., Kawakishi S. Identification of oxidized histidine generated at the active site of Cu,Zn-superoxide dismutase exposed to H2O2. Selective generation of 2-oxo-histidine at the histidine 118. J. Biol. Chem. 1994;269(4):2405–2410. [PubMed] [Google Scholar]
- 90.Kepp K.P. Genotype-Property patient-phenotype relations suggest that proteome exhaustion can cause amyotrophic lateral sclerosis. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0118649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Richardson J.S. β-Sheet topology and the relatedness of proteins. Nature. 1977;268(5620):495–500. doi: 10.1038/268495a0. [DOI] [PubMed] [Google Scholar]
- 92.DiDonato M., Craig L., Huff M.E., Thayer M.M., Cardoso R.M., Kassmann C.J., Lo T.P., Bruns C.K., Powers E.T., Kelly J.W., Getzoff E.D., Tainer J.A. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J. Mol. Biol. 2003;332(3):601–615. doi: 10.1016/s0022-2836(03)00889-1. [DOI] [PubMed] [Google Scholar]
- 93.Kazlauskas R. Engineering more stable proteins. Chem. Soc. Rev. 2018;47(24):9026–9045. doi: 10.1039/c8cs00014j. [DOI] [PubMed] [Google Scholar]
- 94.De Young L.R., Dill K.A., Fink A.L. Aggregation and denaturation of apomyoglobin in aqueous urea solutions. Biochemistry. 1993;32(15):3877–3886. doi: 10.1021/bi00066a006. [DOI] [PubMed] [Google Scholar]
- 95.Dong A., Prestrelski S.J., Allison S.D., Carpenter J.F. Infrared spectroscopic studies of lyophilization- and temperature-induced protein aggregation. J. Pharm. Sci. 1995;84(4):415–424. doi: 10.1002/jps.2600840407. [DOI] [PubMed] [Google Scholar]
- 96.Fink A.L. Protein aggregation: folding aggregates, inclusion bodies and amyloid. Fold. Des. 1998;3(1):R9–R23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 97.Fink A.L., Calciano L.J., Goto Y., Kurotsu T., Palleros D.R. Classification of acid denaturation of proteins: intermediates and unfolded states. Biochemistry. 1994;33(41):12504–12511. doi: 10.1021/bi00207a018. [DOI] [PubMed] [Google Scholar]
- 98.Booth D.R., Sunde M., Bellotti V., Robinson C.V., Hutchinson W.L., Fraser P.E., Hawkins P.N., Dobson C.M., Radford S.E., Blake C.C., Pepys M.B. Instability, unfolding and aggregation of human lysozyme variants underlying amyloid fibrillogenesis. Nature. 1997;385(6619):787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- 99.Jahn T.R., Parker M.J., Homans S.W., Radford S.E. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat. Struct. Mol. Biol. 2006;13(3):195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- 100.Kayatekin C., Zitzewitz J.A., Matthews C.R. Zinc binding modulates the entire folding free energy surface of human Cu,Zn superoxide dismutase. J. Mol. Biol. 2008;384(2):540–555. doi: 10.1016/j.jmb.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roberts B.R., Lim N.K.H., McAllum E.J., Donnelly P.S., Hare D.J., Doble P.A., Turner B.J., Price K.A., Chun Lim S., Paterson B.M., Hickey J.L., Rhoads T.W., Williams J.R., Kanninen K.M., Hung L.W., Liddell J.R., Grubman A., Monty J.-F., Llanos R.M., Kramer D.R., Mercer J.F.B., Bush A.I., Masters C.L., Duce J.A., Li Q.-X., Beckman J.S., Barnham K.J., White A.R., Crouch P.J. Oral treatment with CuII(atsm) increases mutant SOD1 in vivo but protects motor neurons and improves the phenotype of a transgenic mouse model of amyotrophic lateral sclerosis. J. Neurosci. 2014;34(23):8021. doi: 10.1523/JNEUROSCI.4196-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Williams J.R., Trias E., Beilby P.R., Lopez N.I., Labut E.M., Bradford C.S., Roberts B.R., McAllum E.J., Crouch P.J., Rhoads T.W., Pereira C., Son M., Elliott J.L., Franco M.C., Estévez A.G., Barbeito L., Beckman J.S. Copper delivery to the CNS by CuATSM effectively treats motor neuron disease in SODG93A mice co-expressing the Copper-Chaperone-for-SOD. Neurobiol. Dis. 2016;89:1–9. doi: 10.1016/j.nbd.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hilton J.B., Mercer S.W., Lim N.K., Faux N.G., Buncic G., Beckman J.S., Roberts B.R., Donnelly P.S., White A.R., Crouch P.J. Cu(II)(atsm) improves the neurological phenotype and survival of SOD1(G93A) mice and selectively increases enzymatically active SOD1 in the spinal cord. Sci. Rep. 2017;7:42292. doi: 10.1038/srep42292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McAllum E.J., Lim N.K., Hickey J.L., Paterson B.M., Donnelly P.S., Li Q.X., Liddell J.R., Barnham K.J., White A.R., Crouch P.J. Therapeutic effects of CuII(atsm) in the SOD1-G37R mouse model of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2013;14(7–8):586–590. doi: 10.3109/21678421.2013.824000. [DOI] [PubMed] [Google Scholar]
- 105.Saccon R.A., Bunton-Stasyshyn R.K.A., Fisher E.M.C., Fratta P. Is SOD1 loss of function involved in amyotrophic lateral sclerosis? Brain. 2013;136(Pt 8):2342–2358. doi: 10.1093/brain/awt097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Okamoto T., Akaike T., Sawa T., Miyamoto Y., van der Vliet A., Maeda H. Activation of matrix metalloproteinases by peroxynitrite-induced protein S-glutathiolation via disulfide S-oxide formation. J. Biol. Chem. 2001;276(31):29596–29602. doi: 10.1074/jbc.M102417200. [DOI] [PubMed] [Google Scholar]
- 107.Giles G.I., Tasker K.M., Collins C., Giles N.M., O'Rourke E., Jacob C. Reactive sulphur species: an in vitro investigation of the oxidation properties of disulphide S-oxides. Biochem. J. 2002;364(Pt 2):579–585. doi: 10.1042/BJ20011882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Packer L., Witt E.H., Tritschler H.J. Alpha-lipoic acid as a biological antioxidant. Free Radic. Biol. Med. 1995;19(2):227–250. doi: 10.1016/0891-5849(95)00017-r. [DOI] [PubMed] [Google Scholar]
- 109.Auclair J.R., Johnson J.L., Liu Q., Salisbury J.P., Rotunno M.S., Petsko G.A., Ringe D., Brown R.H., Bosco D.A., Agar J.N. Post-translational modification by cysteine protects Cu/Zn-superoxide dismutase from oxidative damage. Biochemistry. 2013;52(36):6137–6144. doi: 10.1021/bi4006122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.