Abstract
Methylketones are broadly distributed in nature and perform a variety of functions. Most microorganisms are thought to produce methylketone by abortive β-oxidation of fatty acid catalytic metabolism. However, two methylketone synthetase genes in wild tomatoes are reported to synthesize methylketone using intermediates of the fatty acids biosynthetic pathway. In our previous study on Trojan horse-like interactions between the bacterium Bacillus nematocida B16 and its host worm, the chemical 2-heptanone was found to be an important attractant for the hosts. So here we used this model to investigate the genes involved in synthesizing 2-heptanone in microorganisms. We identified a novel methylketone synthase gene yneP in B. nematocida B16 and found enhancement of de novo fatty acid synthesis during 2-heptanone production. Interestingly, a homolog of yneP’ existed in the non-pathogenic species Bacillus subtilis 168, a close relative of B. nematocida B16 that was unable to lure worms, but GC-MS assay showed no 2-heptanone production. However, overexpression of yneP’ from B. subtilis in both heterologous and homologous systems demonstrated that it was not a pseudogene. The transcriptional analysis between those two genes had few differences under the same conditions. It was further shown that the failure to detect 2-heptanone in B. subtilis 168 was at least partly due to its conversion into 6-methyl-2-heptanone by methylation. Our study revealed methylketone biosynthesis of Bacillus species, and provided a co-evolution paradigm of second metabolites during the interactions between pathogenic/non-pathogenic bacteria and host.
Keywords: methylketone biosynthesis, 2-heptanone, pathogenic bacterium, interaction between pathogen and host, co-evolution, Bacillus sp.
Introduction
Methylketones are broadly distributed in natural environments and can be produced by a variety of bacteria, fungi, plants, insects, and even mammals. In bacteria, methylketones, such as 2-heptanone, 2-butanone, 2-petanone, and 3-methyl-1-butanone, are detected in the volatile organic compounds (VOCs) of Lactobacillus casei (Gallegos et al., 2017). Staphylococcus carnosus produces 2-pentanone and a highly autolytic strain of Lactobacillus helveticus is reported to enhance the levels of methylketones during cheese ripening (Fadda et al., 2002; Hannon et al., 2006). In fungi, both 2-heptanone and 2-petanone are detected from a range of species, from unicellular yeast to filamentous fungi (Sunesson et al., 1996). In plants, short-chain methylketones (C5–C11) are aromatic substances, commonly enriched in essential oil from leaves and fruits; while medium-chain methylketones (C7–C15) provide resistance to insects (Schwab et al., 2008; Fischer et al., 2013). In animals, methylketones are used as the pheromones or allomone. For example, 2-heptanone functions as honey bee’s alarm pheromone to guide other bees to attack enemies (Torto et al., 2007). Some small arthropods were reportedly paralyzed by 2-heptanone after bitten by honeybees (Papachristoforou et al., 2012). A higher concentration of 2-heptanone in the urine of male Rattus norvegicus was found to be more attractive to the females (Zhang et al., 2008).
Despite their extensive distributions and diverse roles, relatively little is known about the biosynthesis of methylketones. In bacteria, it was reported that the production of odd-numbered methylketones via decarboxylation of even-numbered fatty acids (Yu et al., 2000; Dickschat et al., 2005; Maddula et al., 2009). Studies in fungi found fatty acids were oxidized into β-ketoacids followed by β-keto-acyl-decarboxylase to produce methylketones (Kinsella and Hwang, 1976; Qian et al., 2002). But the spores of Penicillum roqueforti catalyzed the production of 2-undecanone, 2-heptanone, and 2-pentanone from short-chain alkyl esters of lauric acid, octanoic, hexanoic in an aqueous-organic two-phase system, respectively (Park et al., 2000). Acetone is the common methylketone found in mammals and is produced by oxidation of butyric acid (Kalapos, 1999). Collectively, it is widely believed that odd-numbered methylketones are synthesized from their corresponding even-numbered fatty acids via abortive β-oxidation (Schulz and Dickschat, 2007). However, it was recently shown that in wild tomatoes (Solanum habrochaites) that could produce methylketones, the gene expressional levels involved in fatty acid synthesis were much higher than those of fatty acid beta oxidation pathway (Fridman et al., 2005; Yu and Pichersky, 2014). Furthermore, two methylketone synthetase genes shMKSI and shMKSII have been identified to catalyze the substrate β-ketoacyl-ACP (β-ketoacyl-acyl carrier protein), which was one of the intermediate products in fatty acid synthesis, into the products of methylketone (Fridman et al., 2005). But whether the methylketone synthetase genes exist in the other organisms, and if they do, whether they work similarly as in wild tomatoes remains to be elucidated.
In our previous study about the interactions between the bacterial pathogen Bacillus nematocida B16 strain and its host nematode, a Trojan horse-like strategy was demonstrated necessary for the bacterial pathogenesis, in which the VOCs, including 2-heptanone, were secreted as the attractants to lure worms (Niu et al., 2010). In this study, we used this bacterium B. nematocida B16 to investigate the methylketone biosynthesis. A novel methylketone synthase gene yneP in B. nematocida B16 was identified and found to be involved in the production of 2-heptanone. In addition, we compared B. nematocida B16 with a close relative B. subtilis 168 and found that the latter bacterium also had a similar methyl ketone synthase but that it lacked both 2-heptanone and attractive ability for nematodes. We further traced the potential reasons responsible for the differences of 2-heptanone production between them.
Materials and Methods
Bacterial Strains, Plasmids, Primers, and Grow Conditions
The strains and plasmids used in this study are listed in Table 1. The primers used in this study are listed in Table 2. Escherichia coli DH5α was used as the host strain for the construction of plasmids. E. coli BL21 was used to express the target proteins. C. elegans was grown on NGM medium (50 mM NaCl, 20 g/L of agar, 2.5 g/L of peptone, 1.0 mM cholesterol, 1.0 mM CaCl2, 1.0 mM MgSO4, and 25 mM potassium phosphate at pH 6.0) seeded with E. coli OP50. Then the well-fed adult C. elegans were prepared by washing three times with M9 buffer and once with water for the following experiments.
Table 1.
Strains and plasmids used in this study.
| Strain or plasmid | Genotype/description | Source or references |
|---|---|---|
| Strains | ||
| B. nematocida B16 | Wild Type | Laboratory stock |
| B. subtilis 168 | Wild Type | Laboratory stock |
| E. coli DH5α | Clone strain | TaKaRa |
| E. coli BL21 | E. coli strain for heterologously expressing protein | Laboratory stock |
| E. coli EyneP | BL21 heterologously expressing yneP of B. nematocida | This study |
| E. coli EytpA | BL21 heterologously expressing ytpA of B. nematocida | This study |
| E. coli EyneP’ | BL21 heterologously expressing yneP of B. subtilis | This study |
| B. subtilis EyneP’ | B.subtilis 168 homologously expression yneP of B. subtilis | This study |
| BSP1 | B.subtilis 168 amyE::PyneP-627bp-lacZ(Cmr) | This study |
| BSP2 | B.subtilis 168 amyE::PyneP-518bp-lacZ(Cmr) | This study |
| BSP3 | B.subtilis 168 amyE::PyneP-383bp-lacZ(Cmr) | This study |
| BSP4 | B.subtilis 168 amyE::PyneP-356bp-lacZ(Cmr) | This study |
| BSP5 | B.subtilis 168 amyE::PyneP-217bp-lacZ(Cmr) | This study |
| BSP6 | B.subtilis 168 amyE::PyneP-198bp-lacZ(Cmr) | This study |
| BNP1 | B.nematocida B16 amyE::PyneP-628bp-lacZ(Cmr) | This study |
| BNP2 | B.nematocida B16 amyE::PyneP-491bp-lacZ(Cmr) | This study |
| BNP3 | B.nematocida B16 amyE::PyneP-444bp-lacZ(Cmr) | This study |
| BNP4 | B.nematocida B16 amyE::PyneP-229bp-lacZ(Cmr) | This study |
| BNP5 | B.nematocida B16 amyE::PyneP-205bp-lacZ(Cmr) | This study |
| BNP6 | B.nematocida B16 amyE::PyneP-200bp-lacZ(Cmr) | This study |
| Plasmids | ||
| pMD-19T | Cloning vector in E. coli | TaKaRa |
| pMD-18T | Cloning vector in E. coli | TaKaRa |
| pET-28a | Expressional vector in E. coli BL21 | Laboratory stock |
| pET28a-yneP | Recombinant expressional plasmid for expressing yneP of B. nematocida | This study |
| pET28a-ytpA | Recombinant expressional plasmid for expressing ytpA of B. nematocida | This study |
| pET28a-yneP’ | Recombinant expressional plasmid for expressing yneP’ of B. subtilis | This study |
| pIs284 | Insertion vector to amyE, chloramphenicol resistance, lacZ | From Mitsuo Ogura |
| pDG148 | Expressional vector in B. subtilis | From Francois Denizot |
| pDG148-yneP’ | Expressional plasmid for homologously expressing yneP in B. subtilis | This study |
| pCP115 | Gene knockout plasmid in Bacillus sp. | Bacillus Genetic Stock Center |
| pCP115-yneP | yneP gene knockout plasmid in B. nematocida | This study |
Table 2.
The primers used in this study.
| Primer | Nucleotide sequence (5′→3′) | Functions |
|---|---|---|
| FytpA | GGATCCATGTGGACTTGGAAAGCAG | pET28a-ytpA |
| RytpA | CTCGAGTCAAATATACTGATCTGTAAAG | pET28a-ytpA |
| FyneP | GGATCCTTGCATGTGTCAAAAAAAG | pET28a-yneP |
| RyneP | CTCGAGTTATTTTTTTGCCTTTTCGT | pET28a-yneP |
| FyneP’ | GGATCCTTGCATGTGTCAAAAAAAG | pET28a-yneP’ |
| RyneP’ | CTCGAGCTATTTTTTGGCCTTTTCAT | pET28a-yneP’ |
| FbsyneP’ | GTCGACTTGCATGTATCAAAAAAA | pDG148-yneP’ |
| RbsyneP’ | GCATGCCTAGTGGTGATGGTGATGATGTTTTTTGGCCTTTTCATAT | pDG148-yneP’ |
| BSP1 | GAATTCCATTATCACCTCAATCAT | Reporter gene upstream 1 |
| BSP2 | GAATTCATAAAAACATTGATATTTAC | Reporter gene upstream 2 |
| BSP3 | GAATTCAACTGCAGCAAAAGAC | Reporter gene upstream 3 |
| BSP4 | GAATTCGAGAAAAACGTTTACTAC | Reporter gene upstream 4 |
| BSP5 | GAATTCGGCTTCGTTATCAAAAA | Reporter gene upstream5 |
| BSP6 | GAATTCAACACGTGGAAAATTTG | Reporter gene upstream 6 |
| BSPRV | GGATTCTAACGAGATCAACAGGAA | Reporter gene downstream |
| BNP1 | GAATTCTCACATGATTGGCTTTCC | Reporter gene upstream 1 |
| BNP2 | GAATTCTAAAAACATTGATATTTACTTATG | Reporter gene upstream 2 |
| BNP3 | GAATTCTGTTTTGCGTCAGGTAGA | Reporter gene upstream 3 |
| BNP4 | GAATTCTTGTGAGAAAATTGTGATGG | Reporter gene upstream 4 |
| BNP5 | GAATTCTTCTATTAAGGTTCTTTTGG | Reporter gene upstream 5 |
| BNPRV | GGATTCTTTATCAGGGTCTCCGCCGA | Reporter gene downstream |
The underlined sequences mean the restriction enzyme cutting sites.
Plasmids Construction, Subcloning, and Transformation
To confirm the functions of candidate genes in methylketone synthesis, the full opening reading frames (ORF) of target genes were amplified via PCR using the primers of Exp FyneP and RyneP for the yneP gene, FytpA and RytpA for the ytpA gene in B. nematocida B16, and FyneP’ and RyneP’ for the yneP’ gene in B. subtilis 168. Those DNA fragments obtained from gel recovery were ligated to the pMD-19T vector respectively, and then the recombinant plasmid was transformed into E. coli DH5α competent cells. After the positive transformants were validated by PCR reactions and DNA sequencing, they were digested with BamHI and XohI at the primer-incorporated restriction sites, and then inserted into a BamHI/XohI digested pET28a vector to obtain the recombinant plasmids pET28a-yneP, pET28a-ytpA, and pET28a-yneP’. Those three recombinant plasmids were finally transformed into the competent cells of E. coli BL21 again to express the target proteins.
In order to over-express yneP’ in its native host B. subtilis 168, the yneP’ ORF from B. subtilis 168 was amplified using primers FbsyneP’ and RbsyneP’ that had the primer-incorporated restriction sites of SalI and SphI. The vector pDG148 was used to construct the recombinant plasmid pDG148-yneP’ and this recombinant plasmid was finally transformed into competent cells of B. subtilis 168.
Expression of Methylketone Synthase
Selected positive transformants were cultured by incubation overnight at 37°C with shaking in 5 ml of LB supplemented with 50 μg/ml kanamycin. After a 3 ml overnight culture was transferred into 300 ml of fresh LB medium and grown until an OD600 reached 0.3, 0.1 mM IPTG was then added to induce the protein expression for 4 h. The supernatant was discarded by centrifugation at 8000 rpm for 10 min. The precipitate was suspended with 20 mM Tris–HCl (pH 8.0) followed by sonication on the condition of 5s-on, 7s-off for 10 min. After that, the protein samples from both the supernatant and the precipitant were collected again by centrifugation at 8000 rpm for 10 min at 4°C and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Western Blot
After the same amount of protein samples were separated by SDS-PAGE, the unstained gels were electro-transferred onto immune-blot polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Hercules, CA, United States). Following blocked with 5% skim milk in PBST buffer for 1 h, an anti-his antibody (TransGen Biotech) was added at a 1:2000 dilution and incubated overnight. After washed three times with PBST buffer, the PVDF membrane was treated with goat anti-mouse IgG alkaline phosphatase-conjugated secondary antibody at a dilution of 1:1000 (Bio-Rad) for 2 h and washed again. The position of the bound antibodies on the membrane was detected by submerging it in a solution containing nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP), following a standard protocol.
β-Galactosidase Assay
Plasmid pIs284 that contained a lacZ reporter gene was used for assaying the transcriptional levels. A series of nested fragments from yneP promoter region were fused to a promoter-less lacZ gene in pIS284. The truncated promoter regions were digested with HindIII and EcoRI, and then inserted into the similarly digested pIs284 plasmid. Finally, the successfully constructed plasmids, including BSP1, BSP2, BSP3, BSP4, BSP5, BSP6, BNP1, BNP2, BNP3, BNP4, BNP5, and BNP6, were transformed into B. subtilis 168 or B. nematocida B16, respectively. After the bacterial strains were cultured in LB medium for 30 h, β-galactosidase activities were determined with GENMED-galactosidase assay kit.
SPME-GC-MS Analysis
A headspace solid-phase microextraction (SPME) method in combination with GC-MS was employed to detect methylketones, including 2-heptanone, in different bacterial cultures. The methods of sample preparation and detection were similar to our previous reports (Niu et al., 2010). Briefly, bacterial cultures were placed in 15 ml vials with a magnetic stirrer and extracted with the extraction head (CAD-PDMS 75 μm, Supelco). The fiber was exposed to the headspace above the sample at 65°C for 60 min followed by inserting into the GC injector of Clarus 500 GC-MS System (PE Co., Waltham, MA, United States) and desorbing at 250°C for 2 min. DM-5 column (DM-5 30 m × 0.32 mm × 1 μm) (Chrompack, Mid-delburg, Netherlands) was used in our GC-MS analysis. The initial oven temperature was 50°C, held for 2 min, ramped 4°C/min to 180°C and then 6°C/min to 280°C, and held for 10 min. The volatile components were detected by mass spectrometry with electron impact ionization at 70 eV, with a continuous scan for mass to charge ratio (m/z) from 35 to 550. Compounds were identified by matching the mass spectra with standards in the NBS 2005 library and Nist 2005 library.
The bacterial strains for SPME-GC-MS assay include B. nematocida B16, B. subtilis 168, E. coli BL21 with heterologously expressed YneP and YtpA, and B. subtilis 168 overexpressing YneP’.
Real-Time PCR Assays
The total RNA was isolated using an RNA extracting kit (Tiangen, China) following the treatment of DNaseI to avoid DNA contamination. RNAclean Kit (BioTeck, China) was then employed to further purify the total RNA. RNA concentration was determined by measuring absorbance at 260 nm using a UV spectrophotometer. After random-primed cDNAs were generated, qPCR analysis was performed with SYBR Green JumpStart Taq Ready Mix for qPCR kit (Sigma-Aldrich Co.) following manufacturer’s instructions. The partial sequence of 16S rRNA was used as an internal control. The PCR amplification used 40 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 40 s on ABI PRISM 7000 Real-Time PCR. The real-time PCR experiments were repeated three times for each reaction using independent RNA sample.
Bioassay for Nematocidal Activity
The test bacterial strains were grown overnight at 37°C with 200 rpm shaking in 3 ml YPD (Yeast Extract/Peptone/Dextrose) medium. Then each 100 μl bacterial culture was diluted with M9 medium to 600 μl and transferred into 16-well plates. Seeded with L4 stage hermaphrodite nematodes (30–60 per well), the infection assay was performed at 25°C for different time points. Mortalities of nematodes were defined as the ratio of dead nematodes over the total number of tested nematodes. All infection experiments were performed in triplicates and were repeated at least three times.
Chemotaxis Assays
Chemotaxis assay was performed based on the modified method reported previously (Stein and Murphy, 2014). In the chemotaxis assays, we used 10 cm-diameter tissue culture Petri dishes containing 10 ml of 1.6% agar each. Two marks were made on the back of the plate at opposite sides of the plate about 0.5 cm from the edge of the agar. The tested bacteria and the control (E. coli P50) were at the attractant source and the counter-attractant source, respectively. At the same time, 1 μl sodium azide with the concentration of 1 M was placed to anesthetize animals within about a 0.5 cm radius of the attractant or counter-attractant. After the tested worms of C. elegans were placed near the plate center, the numbers of animals at the attractant and the counter-attractant areas were determined within 1 h. The chemotaxis index was calculated as (animal numbers at attractant - animal numbers at counter-attractant)/total animal number.
Statistical Analysis
All the data were expressed as the mean ± standard deviation (SD). Statistical comparisons were performed by a one-way analysis of variance (ANOVA) followed by Dunnet-t test.
Results
A Novel Methylketone Synthetase YneP in B. nematocida B16
To investigate that the potential methylketone synthetase existed in our bacterial strain, we aligned shMKSI (AY701574) and shMKSII (EU883793) from wild tomatoes with the annotated genome of B. nematocida B16 at amino acid level, and two genes of ytpA and yneP were identified as having the highest sequence identity to shMKSI and shMKSII. The sequences of ytpA and yneP were then submitted in GenBank with accession number of MH687930 and MH687929, respectively. Compared to the 265-amino acid shMKSI and the 149-amino acid shMKSII, the full ORF of ytpA was 777 bp and encoded 259 amino acids, which was annotated as a putative lysophospholipase. The other gene yneP was 414 bp long and encoded a 138-amino acid hypothetical protein with unknown function. Between them, YtpA shared 31% identity and 54% positive amino acid sequence with shMKSI (Figure 1A), and YneP shared 26% identity and 52% positive with shMKSII (Figure 1B).
FIGURE 1.
Sequence alignment of the YtpA gene to shMKSI from Lycopersicon hirsutum f. glabratum (A) and YneP to shMKSII (B).
In an attempt to examine whether ytpA and yneP function as methylketone synthase, we tried to construct ytpA or yneP gene knockout strain in B. nematocida B16. Though the pCP115-yneP and pCP115-ytpA recombinant plasmids for knocking-out ytpA and yneP genes were successfully constructed, the positive transformants of B. nematocida B16 were never obtained (data not shown). Thus, heterologous expressions of those two genes were further carried out in E. coli BL21, and the changes of methylketone production were determined using SPME-GC-MS analysis. First, the SDS-PAGE analysis confirmed that the recombinant proteins of YtpA and YneP were expressed and had the molecular weight of 29.5 and 16.1 kDa as expected (Figure 2A). Then, we compared the VOCs produced by the E. coli BL21 with a blank vector, E. coli BL21 with ytpA and E. coli BL21 with yneP. The results from SPME-GC-MS revealed that 2-pentanone, 2-hexanone, 2-heptanone, 2-octanone, 2-nonanone, 2-decanone, 2-undecanone, 2-tridecanone were detected in the VOCs of E. coli BL21 when YneP was overexpressed (Figure 2B). The peak area of methylketones with odd-numbered carbon were more prominent, such as 2-pentanone, 2-heptanone, and 2-nonanone with peak areas reaching 6.66, 6.21, and 4.8%, respectively (Table 3). Furthermore, the peak area of 2-heptanone far exceeded the 0.16% in the positive control of B. nematocida B16 (Figure 2C). In contrast, methylketones were undetectable under the same condition in E. coli BL21 with YtpA overexpression, which was similar to the result from the negative control of wild type E. coli BL21 (Figure 2B). Altogether, our current results suggest that yneP, but not ytpA, functions as the methylketone synthase.
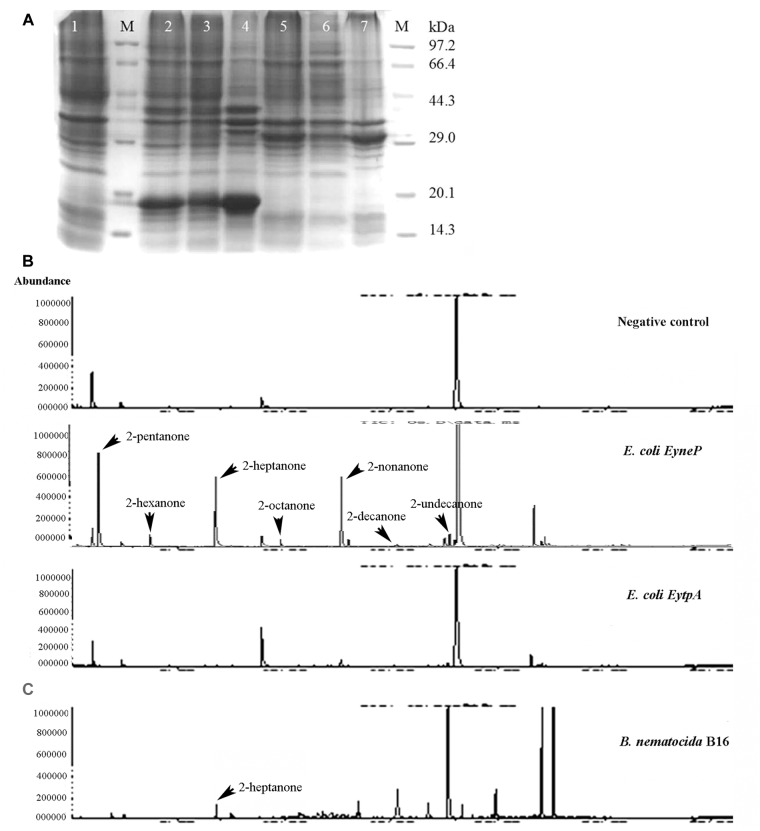
FIGURE 2.
YneP heterologously expressed in E. coli synthesizes a variety of methylketone with medium chain length and functions as a methylketone synthase. (A) SDS-PAGE confirmed expression of the recombinant proteins of YtpA and YneP in E. coli BL21. Lane M represented the protein Marker; Lane 1–7 showed the negative control of E. coli BL21 with blank vector pET28a, E. coli BL21 overexpressing yneP, the supernatant of E. coli BL21 containing yneP, the precipitate of E. coli BL21 containing yneP, E. coli BL21 overexpressing ytpA, the supernatant of E. coli BL21 containing ytpA, the precipitate of E. coli BL21 containing ytpA, respectively. Arrowhead and arrow represented the proteins of YneP and YtpA expressed in E. coli BL21, respectively. (B) GC-MS analyses of the VOCs produced by the negative control E. coli BL21 with blank vector, E. coli BL21 with YtpA overexpression and E. coli BL21 with YneP overexpression. (C) GC-MS analyses of the VOCs produced by B. nematocida B16 strain.
Table 3.
SPME-GC-MS analyzed the production of methylketones in E. coli BL21 heterologously expressing yneP.
| RT | Peak area (%) | Compounds | CAS | Quality |
|---|---|---|---|---|
| 1.423 | 6.66 | 2-Pentanone (C5) | 000107-87-9 | 91 |
| 3.805 | 1.08 | 2-Hexanone (C6) | 000591-78-6 | 91 |
| 6.828 | 6.21 | 2-Heptanone (C7) | 000110-43-0 | 91 |
| 9.840 | 0.49 | 2-Octanone (C8) | 000111-13-7 | 93 |
| 12.646 | 4.80 | 2-Nonanon (C9) | 000821-55-6 | 97 |
| 15.234 | 0.14 | 2-Decanone (C10) | 000693-54-9 | 92 |
| 17.651 | 0.73 | 2-Undecanone (C11) | 000112-12-9 | 94 |
| 22.051 | 0.62 | 2-Tridecanone (C13) | 000593-08-8 | 93 |
RT, retention time; CAS, chemical abstract service number; Quality, credibility score.
Enhancement of de novo Fatty Acid Synthesis During 2-Heptanone Production
Based on the fact that the methylketone synthetases shMKSI and shMKSII in wild tomatoes catalyze the substrate β-ketoacyl-ACP (β-ketoacyl-acyl carrier protein) which is one of the intermediate products in fatty acid synthesis (Fridman et al., 2005), it is reasonable to speculate that the methylketones in B. nematocida B16 are also synthesized via the de novo fatty acid synthesis pathway rather than the abortive degradation pathway. To test this hypothesis, we fused the promoter region of yneP to the reporter plasmid pIs284 to drive the expression of β-galactosidase. We determined the transcriptional levels of yneP by assaying β-galactosidase activities when induced by the different carbon sources including glucose, ethanol, and glycerol. Our results showed that, compared to the control in LB medium, the activities of β-galactosidase ranged from high to low levels in the following order: glycerol, glucose, and ethanol (Figure 3A). Considering that glycerol is closer than glucose to pyruvic acid in the glycolysis pathway, our result implied that the methylketones in B. nematocida B16 were likely synthesized through the de novo fatty acid synthesis pathway.
FIGURE 3.
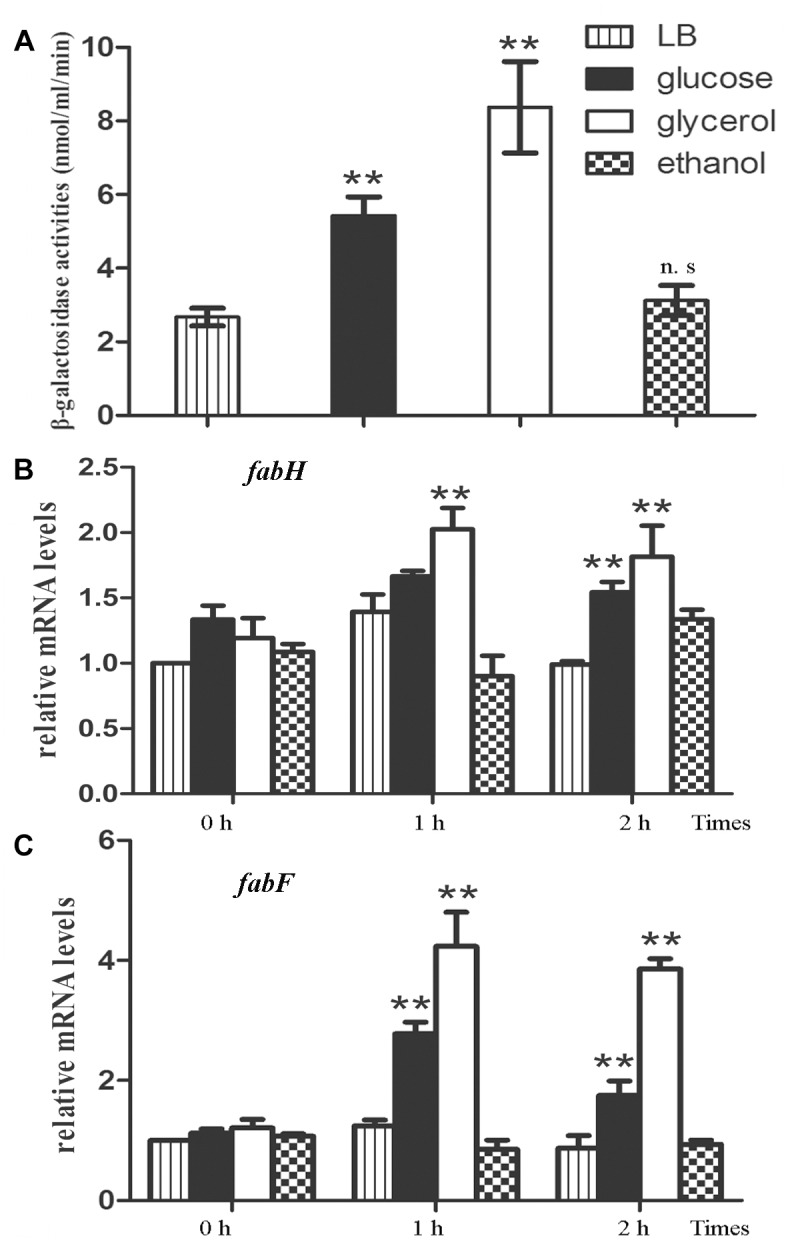
The methylketones in B. nematocida B16 are synthesized during de novo fatty acid synthesis. (A) The transcriptional levels of yneP gene were determined by assaying β-galactosidase activities when induced by the different carbon sources. (B,C) qPCR was used to determine the relative mRNA levels of fabH and fabF, the key enzymes in de novo fatty acid synthesis. ∗∗P < 0.05.
To further validate our hypothesis, the transcriptional levels of β-ketoacyl-ACP synthase II (fabF) and β-ketoacyl-ACP synthase III (fabH), two key enzymes in the de novo fatty acid synthesis, were determined again under the above carbon sources using qPCR. Between them, FabH catalyzes the quintessential ketoacyl synthase reaction with malonyl ACP and acetyl CoA, and FabF catalyzes the related reaction. As expected, the relative mRNA levels of fabH and fabF in glycerol-contained medium were the highest among the three media (followed by glucose and ethanol) and increased by about two or fourfolds within 2 h, respectively (Figure 3B,C).
Absence of 2-Heptanone in VOCs From the Non-pathogenic B. subtilis 168
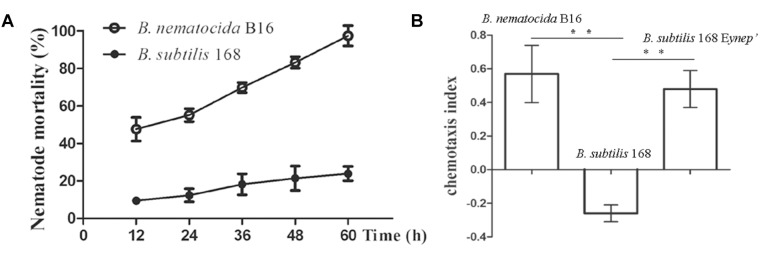
The saprobiont bacterium B. subtilis 168 was one of the closest relatives of B. nematocida in the Bacillus genus based on the 16s rDNA phylogenetic tree, but it obviously had little nematocidal activity compared to the strain of B. nematocida (Figure 4A) (Deng et al., 2013). Therefore, we tested whether B. subtilis 168 could attract nematodes or produce the attractant 2-heptanone. In chemotaxis assays, the worms immediately oriented their movements and then they congregated toward B. nematocida B16. The chemotaxis index of B. nematocida B16 was about 0.57 ± 0.17, which meant that our strain showed a stronger attractive capability for nematodes compared to the counter-attractant E. coli OP50 (Figure 4B). Contrarily, the bacterium B. subtilis 168 showed little ability to lure nematodes with the chemotaxis index of −0.26 ± 0.05 (Figure 4B). The VOCs produced by B. nematocida B16 and B. subtilis 168 were further assayed. Our results demonstrated the presence of 2-heptanone in B. nematocida B16 (Figure 5E), but it was not detected in B. subtilis 168 (Figure 5F).
FIGURE 4.
The non-pathogenic bacterium B. subtilis 168 shows little ability to kill or attract nematodes. (A) The nematotoxic activities of B. nematocida B16 and B. subtilis 168. (B) The attractive capabilities for C. elegans in the different bacterial strains of B. nematocida B16, B. subtilis 168, and B. subtilis 168 homolously expressing yneP’. ∗∗P < 0.05.
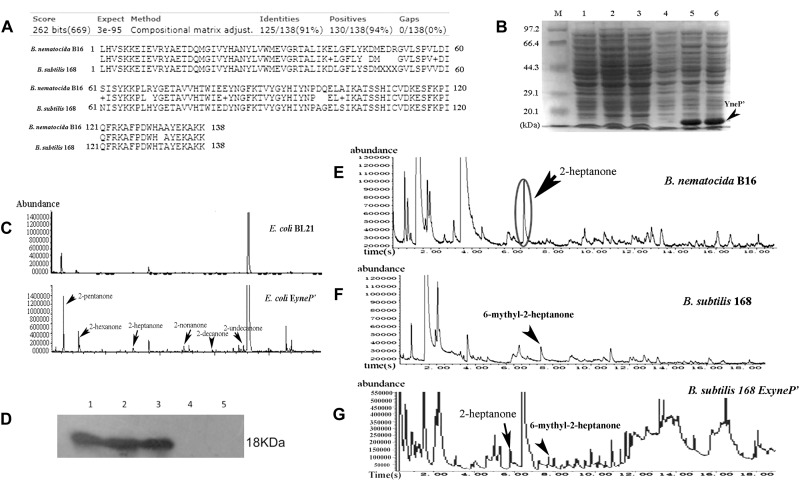
FIGURE 5.
The yneP’ gene in the non-pathogenic B. subtilis 168 has the methylketone synthetase activity but absence of 2-heptanone in the VOCs from B. subtilis 168. (A) The YneP’ in B. subtilis 168 showed highly homologous to YneP in B. nematocida B16. (B) SDS-PAGE confirmed expression of the recombinant protein of YneP’ in E. coli BL21. Lane M, the protein Marker; lane 1 and 2, the negative control of E. coli BL21 with blank vector; lane 3 and 4, the strain E. coli BL21 with ExyneP’ without IPTG induction; lane 5 and 6, the heterologous expression of YneP’ in E. coli BL21 after induced with 0.1 mM IPTG. The molecular size of target protein is about 18 kDa. (C) GC-MS analyses of the VOCs from E.coli ExyneP’ and the negative control. (D) Western blot to the homolously expressed YneP’ with 6 × his tag using anti-his antibody. Lane 1–3 represented the three positive transformants of B. subtilis 168 ExyneP’; Lane 4 and 5 represented the wild type B. subtilis 168 and B. subtilis 168 with the blank vector pDG148. (E–G) Comparisons of the VOCs among B. nematocida B16, B. subtilis 168, and B. subtilis ExyneP’. Among those three strains, the peak of 2-heptanone was detected in B. nematocida B16 but absence in B. subtilis 168. Instead, a peak of 6-methyl-2-heptanone could be detected in B. subtilis 168. But 2-heptanone in VOCs reappeared when overexpressing yneP’ in B. subtilis 168.
A Homology yneP’ With Methylketone Synthetase Exists in B. subtilis 168
Since neither attractive capability nor 2-heptanone was found in B. subtitis 168, we speculated that the homolog of yneP either does not exist or if it exists, it could be a pseudogene. To address this issue, we aligned yneP of B. nematocida B16 with the genome of B. subtilis 168 and found that YneP’ in B. subtilis 168 shared 91% identity and 94% positive amino acid sequence with that of B. nematocida B16 (Figure 5A). To determine whether B. subtilis 168 yneP’ had the same synthase activity, the ORF of yneP’ was amplified, cloned into pET28a, and expressed in E. coli BL21 with a molecular weight as we had expected (Figure 5B). Next, we verified its activity as a methylketone synthase by GC-MS. Compared with the negative control of E. coli BL21, a variety of methylketons were identified, which meant that the heterologously expressed YneP’ of B. subtilis 168 indeed functioned in methylketone biosynthesis (Figure 5C).
To further confirm its activity in vivo, a homologous expression of YneP’ fused with 6 × his tag (B. subtilis 168 EyneP’) was achieved in B. subtilis 168. Production of the target protein was validated by Western blot using anti-his antibody (Figure 5D). Measuring the VOCs produced by B. subtilis 168 EyneP’ using GC-MS, the peak of 2-heptanone became more obvious compared to the negative control (Figure 5G). Furthermore, in chemotaxis assay, B. subtilis 168 overexpressing yneP’ showed a stronger attractive capability for nematodes (Figure 4B), though the chemotaxis index was still lower than B. nematocida B16.
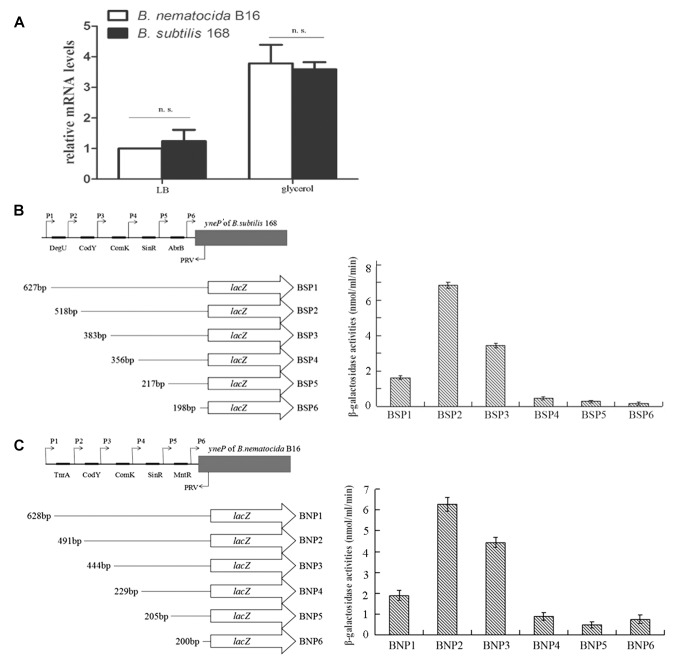
Metabolic Transformation but Not Transcriptional Level Responsible for the Difference of 2-Heptanone in the Two Bacteria
Based on our experimental evidence that the gene yneP’ of B. subtilis 168 had the activity of methylketone synthase, it was reasonable to speculate that the absence of 2-heptanone in this strain was likely due to the repressed expression of yneP’. To test this assumption, we firstly compared the mRNA levels of the methylketone synthase gene between B. nematocida B16 and B. subtilis 168 by qPCR. Our results showed the similar transcriptional levels in those two strains under the same conditions (Figure 6A).
FIGURE 6.
The transcriptional levels of yneP of B. nematocida B16 and yneP’ of B. subtilis 168. (A) qPCR detected the relative mRNA levels of yneP of B. nematocida B16 and yneP’ of B. subtilis 168 under the same conditions. n.s., P ≥ 0.05. (B) Assays to β-galactosidase activities in the series of yneP’::lacZ fusions. Schematic representation of the different yneP’::lacZ fusions used in this study. The filled arrows indicate the primers used for generating the various reporter fusions (P1–PRV), whereas the 5’ and 3’ end termini of the yneP’::lacZ fusions are denoted with their nucleotide positions relative to the initiator codon. The black oblongs indicate the binding motifs in the promoter of yneP’ of B. subtilis 168. (C) Assays to β-galactosidase activities in the series of yneP::lacZ fusions of B. nematocida B16.
The cis-acting elements within those two promoters were further analyzed by the reporter gene lacZ. Using the software DBTBS, the predicted transcription factor binding sites of DegU, CodY, ComK, SinR, and AbrB were found in the promoter region of B. subtilis 168 yneP’; while TnrA, CodY, ComK, SinR, and MntR were found within the promoter of B. nematocida B16 yneP (Figure 6B,C). After a series of reporter fusions containing the truncated promoter regions to lacZ were constructed (BSP1, BSP2, BSP3, BSP4, BSP5, BSP6 of B. subtilis 168 and BNP1, BNP2, BNP3, BNP4, BNP5, BNP6 of B. nematocida B16, respectively) and transformed into their host bacterial strains, β-galactosidase activity of each strain was measured when cultured in LB medium for 30 h. Our data showed that most of the cis-acting elements analyzed in B. subtilis 168 positively regulated the expression of yneP’ except for DegU (Figure 6B). To the yneP promoter of B. nematocida B16, TnrA and MntR negatively regulated yneP expression, but CodY, ComK, and SinR had positive effects (Figure 6C). Furthermore, the quantifications of β-galactosidase activities were similar between those two bacterial strains. Therefore, our results indicate that, under the same tested conditions, the transcription control of yneP’ in B. subtilis 168 was not more strongly suppressed than B. nematocida B16.
To further understand the reason for the lack of 2-heptanone in B. subtilis 168, we scanned each peak of GC-MS again and found a peak corresponding to 6-methyl-2-heptanone detected in both B. subtilis 168 and B. subtilis 168 EyneP’ (Figure 5F,G), but no appearance in the VOCs of B. nematocida B16. Our results suggest that 2-heptanone synthesized by the methylketone synthase YneP’ could be methylated and transformed into 6-methyl-2-heptanone upon its production in B. subtilis 168.
Discussion
The prevalence of methylketones with an odd number of carbons in nature suggests that they are most likely derived from decarboxylation of the corresponding β-ketoacids that are intermediates in either the biosynthesis or the degradation pathways of fatty acids. Of the two pathways, the spontaneous decarboxylation is more rationally expected since CoA esters and free β-ketoacids, the intermediates in fatty acid degradation are unstable. Wild tomatoes produce methylketones as repellants against its pests (Yu, 2013), in which two genes shMKSI and shMKSII are identified to be responsible for the production of methylketones during fatty acid biosynthesis (Yu et al., 2010). In this pathway, ShMKS2 hydrolyzes β-ketoacyl-A to release 3-ketoacids, and then ShMKS1 catalyzes the decarboxylation of 3-ketoacids to produce methylketones (Yu et al., 2010; Yu and Pichersky, 2014). So the shMKSI gene is epistatic to shMKSII in the pathway for methylketone biosynthesis. MKS homologs have also been found in other plants and non-plant organisms (Nikolau et al., 2008). However, there has been limited experimental evidence supporting these MKS homologs really have the synthetase activities of methylketones with different chain length. In the genome of B. nematocida B16, ytpA and yneP were identified as the homolog of shMKSI and shMKSII, respectively. The yneP gene was predicted as an unknown thioesterase, which belonged to a large group of hydrolytic enzyme superfamily with a “hot-dog”-like structure. Its activity as a methylketone synthase was verified when the methylketones with medium chain length were obtained in yneP-overexpressed E. coli but absence in the negative control. Furthermore, the data from transcriptional analyses also supported that the methylketones in B. nematocida B16 were synthesized during de novo fatty acid synthesis rather than via the degradation pathway. The other YtpA gene, though homologous with the tomato shMKSI, showed no obvious contribution to methylketone production. Therefore, in Bacillus sp., we hypothesize that yneP likely catalyzes β-ketoacyl-A, the intermediates of fat acid synthesis, into 3-ketoacids which are then further decarboxylated spontaneously. But the role of YtpA, the homolog of shMKSII in B. nematocida B16, remains to be confirmed.
In nature, 2-heptanone functions as an important allomone in the interaction between prey and predator. For example, when worker bees attack their enemies by sting, they often use the upper jaw to bite the enemy. Synchronously, the chemicals, such as 2-heptanone, are left on the enemy’s bodies to guide the other bees for attacking (Schearer and Boch, 1965). Furthermore, one yeast, Kodamaea ohmeri, carried by honeybees can produce more abundant 2-heptanone using the pollen collected in honeycomb, which attracts members of the intruder Aethina tumida and leads to disastrous damages for honeybees (Torto et al., 2007; Benda et al., 2015). Interestingly, our previous investigations have also illustrated that 2-heptanone produced by the bacterial pathogen B. nematocida B16 strain can successfully lure the worms and fulfill one of the prerequisites for a Trojan horse-like infection (Niu et al., 2010). But the evolutionarily closely related strain B. subtilis 168 showed little attraction for nematodes, even though the gene yneP’ had methylketone synthetase activity. The similar expressional levels of the homologs between those two bacteria form stark contrast to their vast differences in both 2-heptanone production and attractive capability. Our data further suggest that instead of producing 2-heptanone as a final product, 2-heptanone was likely converted into 6-methyl-2-heptanone in B. subtilis 168. This metabolic transformation may be an effective strategy to prevent the non-pathogenic B. subtilis 168 from being the food of nematodes. Our results provided a co-evolution paradigm of second metabolites during the interactions between pathogenic/non-pathogenic bacteria and host.
Author Contributions
XH and KZ designed the research. MZ, XX, YL, and PW carried out the experiments. KZ and SN analyzed the data. XH and MZ wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Natural Science Foundation Program of China (Grant No. 31460024), and the Department of Science and Technology of Yunnan Province (Grant No. 2014FA019).
References
- Benda N. D., Boucias D., Torto B., Teal P. (2015). Detection and characterization of Kodamaeaohmeri assciated with small hive beetle Aethina tumida infesting honeybee hives. J. Apic. Res. 47 194–201. 10.3827/ibra.1.47.3.07 [DOI] [Google Scholar]
- Deng X. D., Tian Y. X., Niu X., Shi H., Zhang H. B., et al. (2013). The ComP-ComA quorum system is essential for “Trojan horse” like pathogenesis in Bacillus nematocida. PLoS One 8:e76920. 10.1371/journal.pone.0076920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickschat J. S., Helmke E., Schulz S. (2005). Volatile organic compounds from arctic bacteria of the cytophaga-flavobacterium-bacteroides group: a retrobiosynthetic approach in chemotaxonomic investigations. Chem. Biodivers. 2 318–353. 10.1002/cbdv.200590014 [DOI] [PubMed] [Google Scholar]
- Fadda S. G., Lebert A., Talon R. (2002). Development of an enzymatic method to quantify methylKetones from bacterial origin. J. Agric. Food Chem. 50 2471–2474. 10.1021/jf011298v [DOI] [PubMed] [Google Scholar]
- Fischer D. C. H., Limberger R. P., Henriques A. T., Moreno P. R. H. (2013). Essential oils from fruits and leaves of Siparuna guianensis(Aubl) Tulasne from Southeastern Brazil. J. Essent. Oil Res. 17 101–102. 10.1080/10412905.2005.9698844 [DOI] [Google Scholar]
- Fridman E., Wang J., Iijima Y., Froehlich J. E., Gang D. R., Ohlrogge J., et al. (2005). Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell 17 1252–1267. 10.1105/tpc.104.029736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos J., Arce C., Jordano R., Arce L., Medina L. M. (2017). Target identification of volatile metabolites to allow the differentiation of lactic acid bacteria by gas chromatography-ion mobility spectrometry. Food Chem. 220 362–370. 10.1016/j.foodchem.2016.10.022 [DOI] [PubMed] [Google Scholar]
- Hannon J. A., Kilcawley K. N., Wilkinson M. G., Delahunty C. M., Beresford T. P. (2006). Flavour precursor development in cheddar cheese due to lactococcal starters and the presence and lysis of Lactobacillus helveticus. Int. Dairy J. 17 316–327. 10.1016/j.idairyj.2006.03.001 [DOI] [Google Scholar]
- Kalapos M. P. (1999). Possible physiological roles of acetone metabolism in humans. Med. Hypotheses 53 236–242. 10.1054/mehy.1998.0752 [DOI] [PubMed] [Google Scholar]
- Kinsella J. E., Hwang D. (1976). Biosynthesis of Flavors by Penicillium roqueforti. Biotechnol. Bioeng. 18 927–938. 10.1002/bit.260180706 [DOI] [Google Scholar]
- Maddula S., Blank L. M., Schmid A., Baumbach J. I. (2009). Detection of volatile metabolites of Escherichia coli by multi capillary column coupled ion mobility spectrometry. Anal. Bioanal. Chem. 394 791–800. 10.1007/s00216-009-2758-0 [DOI] [PubMed] [Google Scholar]
- Nikolau B. J., Perera M. A. D. N., Brachova L., Shanks B. (2008). Platform biochemicals for a biorenewable chemical industry. Plant J. 54 536–545. 10.1111/j.1365-313X.2008.03484.x [DOI] [PubMed] [Google Scholar]
- Niu Q. H., Huang X. W., Zhang L., Xu J. P., Yang D. M., We K. B., et al. (2010). A Trojan horse mechanism of bacterial pathogenesis against nematodes. Proc. Natl. Acad. Sci. U.S.A. 107 16631–16636. 10.1073/pnas.1007276107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papachristoforou A., Kagiava A., Papaefthimiou C., Termentzi A., Fokialakis N., Skaltsounis A. L., et al. (2012). The bite of the honeybee: 2-heptanone secreted from honeybee mandibles during a bite acts as a local anaesthetic in insects and mammals. PLoS One 7:e47432. 10.1371/journal.pone.0047432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park O. J., Holland H. L., Khan J. A., Vulfson E. N. (2000). Production of flavor ketones in aqueous-organic two-phase systems by using free and microencapsulated fungal spores as biocatalysts. Enzyme Microb. Technol. 26 235–242. 10.1016/s0141-0229(99)00140-4 [DOI] [PubMed] [Google Scholar]
- Qian M., Nelson C., Bloomer S. (2002). Evaluation of fat-derived aroma compounds in blue cheese by dynamic headspace GC/olfactometry-MS. J. Am. Oil Chem. Soc. 79 663–667. 10.1007/s11746-002-0540-4 [DOI] [Google Scholar]
- Schearer D. A., Boch R. (1965). 2-heptanone in the mandibular gland secrwtion of the honeybee. Nature 206:530. 10.1021/acs.jproteome.6b00526 [DOI] [PubMed] [Google Scholar]
- Schulz S., Dickschat J. S. (2007). Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep. 24 814–842. [DOI] [PubMed] [Google Scholar]
- Schwab W., Davidovich R. R., Lewinsohn E. (2008). Biosynthesis of plant-derived flavor compounds. Plant J. 54 712–732. 10.1111/j.1365-313X.2008.03446.x [DOI] [PubMed] [Google Scholar]
- Stein M. G., Murphy T. C. (2014). C. elegans positive olfactory associative memory is a molecularly conserved behavioral paradigm. Neurobiol. Learn. Mem. 115 86–94. 10.1016/j.nlm.2014.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunesson A. L., Sunesson C. N., Nilsson C., Andersson B., Blomquist G. (1996). Volatile metabolites produced by two fungal species cultivated on building materials. Ann. Occup. Hyg. 40 397–410. 10.1016/0003-4878(96)00002-6 [DOI] [PubMed] [Google Scholar]
- Torto B., Boucias D. G., Arbogast R. T., Tumlinson J. H., Teal P. E. (2007). Multitrophic interaction facilitates parasite–host relationship between an invasive beetle and the honey bee. Proc. Natl. Acad. Sci. U.S.A. 104 8374–8378. 10.1073/pnas.0702813104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. (2013). Two Methylketone Biosynthetic Enzymes from Wild Tomatoes. Ph.D. thesis. Ann Arbor, MI: University of Michigan. [Google Scholar]
- Yu G., Nguyen T. T. H., Guo Y. X., Schauvinhold I., Auldridge M. E., Bhuiyan N., et al. (2010). Enzymatic functions of wild tomato methylketone synthases 1 and 2. Plant Physiol. 154 67–77. 10.1104/pp.110.157073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Pichersky E. (2014). Heterologous expression of methylketone synthase1 and methylketone synthase2 leads to production of methylketones and myristic acid in transgenic plants. Plant Physiol. 164 612–622. 10.1104/pp.113.228502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Hamilton-Kemp T. R., Archbold D. D., Collins R. W., Newman M. C. (2000). Volatile compounds from Escherichia coli O157:H7 and their absorption by strawberry fruit. J. Agric. Food Chem. 48 413–417. [DOI] [PubMed] [Google Scholar]
- Zhang J. X., Sun L., Zhang J. H., Feng Z. Y. (2008). Sex- and gonad-affecting scent compounds and 3 male pheromones in the rat. Chem. Senses 33 611–621. 10.1093/chemse/bjn028 [DOI] [PMC free article] [PubMed] [Google Scholar]