Abstract
Treatment of prostate cancer (PC) by androgen suppression promotes the emergence of aggressive variants that are androgen receptor- (AR-) independent. Here we identify the transcription factor ONECUT2 (OC2) as a master regulator of AR networks in metastatic castration-resistant prostate cancer (mCRPC). OC2 acts as a survival factor in mCRPC models, suppresses the AR transcriptional program by direct regulation of AR target genes and the AR licensing factor FOXA1, and activates genes associated with neural differentiation and progression to lethal disease. OC2 appears active in a substantial subset of human prostate adenocarcinoma and neuroendocrine tumors. Inhibition of OC2 by a newly identified small molecule suppresses metastasis in mice. These findings suggest that OC2 displaces AR-dependent growth and survival mechanisms in many cases where AR remains expressed, but where its activity is bypassed. OC2 is also a potential drug target in the metastatic phase of aggressive PC.
INTRODUCTION
Aggressive PC variants are poorly understood and associated with rapid treatment resistance, metastasis, and death1. Although the precise clinical, pathologic, and molecular features of these variants continue to be refined, the AR, considered to be the primary oncoprotein in PC and mCRPC, is often heterogeneously expressed, even under conditions of AR gene amplification2. In prostate tumors expressing AR, resistance to hormonal therapies may occur through clonal selection, adaptation to decreased androgen, or intracrine mechanisms3. Although many mCRPC appear to rely on AR activity, even when the androgen axis is pharmacologically suppressed, recent studies suggest that alternate transcriptional pathways emerge in lethal disease4. For example, in contrast to primary PC, in one mCRPC patient population, AR gene expression signatures are inversely correlated with signatures of cell proliferation5. AR also exerts a tumor- and metastasis suppressor function in basal-like PC6. These observations show that disease progression is compatible with reduced AR activity.
Here we describe the results of experimental testing of a bioinformatics model that identified the atypical homeobox protein ONECUT2 (HNF6β/OC-2/OC2 hereafter, OC2) as a highly active transcription factor (TF) in mCRPC. OC2 and the paralog ONECUT1 play a role in in liver, pancreatic, and neuronal development7–9. A role for OC2 in cancer is not well defined, and studies of OC2 activity in PC are limited. One report identified OC2 mRNA in urine of PC patients10 and a PC risk-associated genetic variant, which modifies expression of the lncRNA PCAT1, was recently shown to be associated with OC2 activity11. In this study, we demonstrate that OC2 functions as a master regulator and survival factor that controls transcriptional networks that emerge in aggressive PC variants. We further demonstrate that OC2 can be targeted with a small molecule that inhibits mCRPC metastasis.
RESULTS
Computational modeling predicts OC2 as a key transcriptional regulator in mCRPC
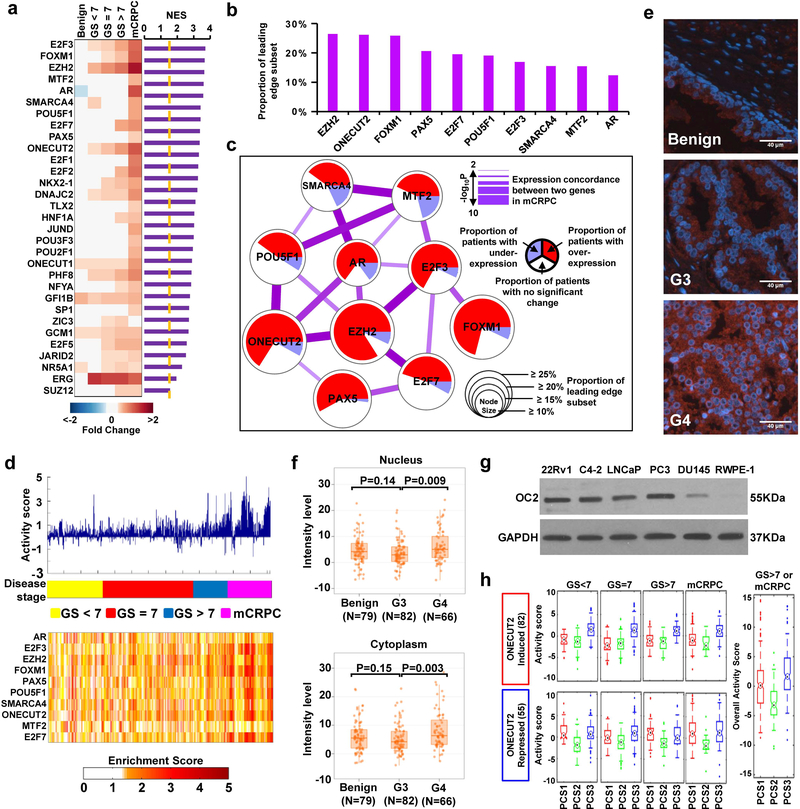
We recently described a resource developed from the assembly of 38 transcriptome datasets from 2,115 PC cases, including 260 samples of mCRPC12. This dataset was used as a discovery (DISC) cohort to identify key TFs in mCRPC using master regulator analysis (see Methods). Out of 402 TFs included in the model, we identified 31 TFs as significantly active in mCRPC compared to high-grade primary tumors (Fig. 1a), while 7 TFs were calculated to be significantly down-regulated (Supplementary Fig. 1a). The 10 most active TFs identified in this procedure are ranked in Fig. 1b. EZH2 was ranked first, with the largest fraction of target genes showing significant correlation with EZH2 gene expression. Next, we constructed a network model that considers the correlation between the expression of the 10 TFs and expression of their target genes, as well as pairwise interactions between them. The model (Fig. 1c) has a number of interesting features: 1) it contains TFs known to be active in PC (EZH2, AR, FOXM1, and E2F3); 2) the predicted activity of OC2 is comparable to EZH2, a known driver of mCRPC13 (Fig. 1b,c); 3) OC2 is predicted to be networked with other key TFs, such as POU5F1 (Oct-4), PAX5, AR and EZH2; and 4) predicted OC2 activity is greater than AR activity. Overall activity of this network is relatively high in mCRPC compared to other disease categories (Fig. 1d, N=1,321). Principal component analysis indicated the network model has high discriminatory accuracy in distinguishing mCRPC from localized disease (Supplementary Fig. 1b). Quantitative analysis of OC2 immunostaining intensity, using a PC tissue microarray (TMA) containing benign prostate, low- (Gleason pattern 3, G3) and high-grade (G4) cancers, showed that nuclear and cytoplasmic OC2 protein levels were increased in aggressive disease (Fig. 1e,f). Consistent with this, in the DISC cohort (N=2,115), OC2 mRNA expression increased gradually from benign prostate tissue to mCRPC, and this pattern was distinct from the AR, where increased expression was only observed in mCRPC (Supplementary Fig.1c).
Figure 1. OC2 is predicted to be active in mCRPC.
(a) Up-regulated TFs in mCRPC in the DISC cohort. The heatmap displays TF expression level in 5 disease categories. GS = Gleason sum score. Purple bars represent normalized enrichment score (NES), a statistical measure of TF activity. (Benign N=794, GS<7 N=328, GS=7 N=530, GS>7 N=203, mCRPC N=260).
(b) Ranking of the 10 most active TFs (from Fig. 1a) in mCRPC from the DISC cohort.
(c) A network model describing activity and association of highly active TFs in mCRPC. TFs are identified as nodes. Node size = activity. Node color = proportion of patients where the TF is up- or down-regulated, or unchanged. Edges = degree of interaction. Edge thickness = concordance in RNA expression.
(d) The bar plot (top) displays summary of TF activation across disease categories. The heatmap (bottom) shows activation of individual TFs across disease categories.
(e and f) Representative images and quantitation of OC2 expression (red) in benign (N=79), Gleason Grade 3 (G3, N=82) and Gleason Grade 4 (G4, N=66) PC tissues. Tissue sections are stained using a fluorescent antibody detection system and nuclei visualized with DAPI (blue). Scale bar 40μm. The boxes show the 25th-75th percentile range and the center line is the median. Whiskers show 1.5 times the inter-quartile range (IQR) from the 25th or 75th percentile values. P values obtained from Wilcoxon two-tailed rank-sum test.
(g) Immunoblot showing endogenous OC2 levels in prostate cell lines. Representative blots from two independent experiments. Full-length blots are presented in Supplementary Fig. 8.
(h) Inferred activity of OC2 in PCS1–3 subtypes12 plotted by disease category. Upper and lower panels show inducible and repressive activity of OC2, respectively. Right panel shows overall OC2 activity in GS>7 and mCRPC tumors. Red= PCS1, green=PCS2, blue=PCS3. The boxes show the 25th-75th percentile range and the center mark is the median. Whiskers show 1.5 times IQR from the 25th or 75th percentile values. Data points beyond the whiskers are displayed using dots. (GS<7 & PCS1, N=28; GS<7 & PCS2, N=158; GS<7 & PCS3, N=142; GS=7 & PCS1, N=98; GS=7 & PCS2, N=201; GS=7 & PCS3, N=231; GS>7 & PCS1, N=79; GS>7 & PCS2, N=52; GS>7 & PCS3, N=72; mCRPC & PCS1, N=172; mCRPC & PCS2, N=17; mCRPC & PCS3, N=71; GS>7 or mCRPC & PCS1, N=251; GS>7 or mCRPC & PCS2, N=69; GS>7 or mCRPC & PCS3, N=143).
Evidence of OC2 activity in mCRPC
OC2 protein is expressed in metastatic, castration-resistant 22Rv1, C4–2, and PC3 PC cell lines, in weakly metastatic, androgen-dependent LNCaP PC cells, and at detectable but substantially lower levels in moderately metastatic, castration-resistant DU145 PC cells (Fig. 1g). Non-tumorigenic RWPE-1 PC cells are negative. To study the role of OC2 in aggressive PC, we performed gene expression profiling using 22Rv1 and LNCaP cell lines. OC2 was enforced in LNCaP and 22Rv1 or silenced in 22Rv1 (Supplementary Fig. 1d). OC2-induced and –repressed genes were identified with false discovery rate (FDR) <0.05 and fold-change (FC) ≥2 (Supplementary Fig. 1e). OC2-induced genes were enriched in pathways related to cell motion, neuronal differentiation, and mesenchymal cell differentiation (Supplementary Fig. 1f), whereas OC2-repressed genes were enriched in pathways corresponding to AR activity, β3- and β1-integrin cell surface interactions, and syndecan-4-mediated signaling (Supplementary Fig. 1g). These data suggest that OC2 interacts with the AR transcriptional program. To explore this further, OC2-regulated genes were compiled into an OC2 activity gene signature (Supplementary Table 1), which we used to infer OC2 activity in transcriptome datasets. The signature was used to calculate a Z-score14 of OC2-induced and –repressed activity in each tumor in the DISC cohort. Hierarchical clustering of cases in the DISC cohort showed that OC2 activation is positively correlated with signatures reflecting activation of rat sarcoma viral oncogene homolog (RAS), de-repression of genes regulated by polycomb repression complexes (PRC), epithelial-mesenchymal transition (MES), proneural (PN), and aggressive PC (AV), and a negative correlation with the AR signature (AR) (Supplementary Fig. 1h). OC2 activity was also assessed using the PCS1–3 classification scheme12 (Fig. 1h). Among these 3 subtypes, the PCS1 luminal subtype is consistently more aggressive, and PCS1 luminal and PCS3 basal subtypes are overrepresented in mCRPC in comparison to PCS212. OC2 activity was highest in PCS1 and PCS3 tumors across the disease course, including Gleason sum score >7 and mCRPC tumors. These data suggest that OC2 functions in luminal as well as basal PC.
The above results imply that OC2 can operate under conditions of suppressed AR activity. Consistent with this, in mCRPC tumors from the DISC cohort (N=260), AR activity was significantly decreased in tumors with high OC2 expression (Supplementary Fig. 1i). Similarly, OC2 expression was induced in a model where long-term androgen deprivation resulted in castration resistance15 and was highly elevated in the KRas-active/Pten-null transgenic model16, where AR expression is suppressed (Supplementary Fig. 1j). In contrast, in the LNCaP-abl mCRPC model, characterized by hyperactive AR, OC2 expression was suppressed (Supplementary Fig. 1j). Using a different approach, OC2 expression in the entire DISC cohort was positively correlated with expression of AR-repressed gene activity17 (Supplementary Fig. 1k). Taken together, these results suggest that OC2 is active when AR activity is repressed.
OC2 negatively regulates the AR and its transcriptional program
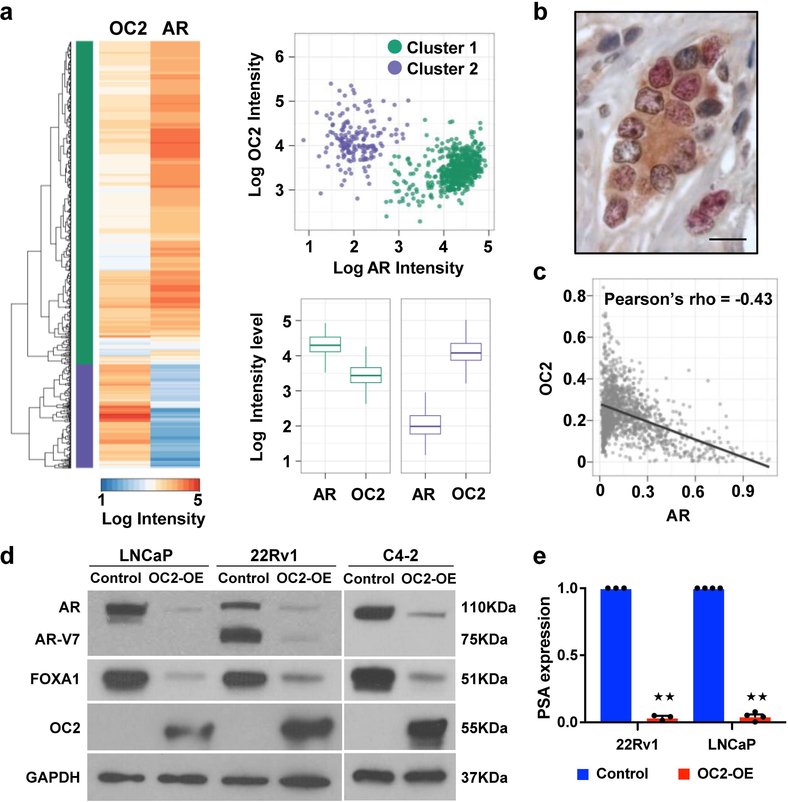
Because OC2 expression and inferred activity correlate inversely with AR activity, we measured AR and OC2 levels in individual nuclei in 6 cases of high-grade PC by multiplex immunofluorescence (IF) microscopy using anti-OC2 and -AR antibodies. Hierarchical clustering indicates that nuclei can be separated into two groups: high OC2/low AR, and low OC2/high AR (Fig. 2a). We next performed the same analysis on 35 cases of high-grade PC. OC2 expression was detected in the cytoplasm and nucleus of AR-positive and AR-negative cancer cells (Fig. 2b). Nuclear OC2 and AR expression was inversely correlated in cells expressing both OC2 and AR (Fig. 2c).
Figure 2. Inverse relationship between OC2 and AR nuclear localization in radical prostatectomy specimens.
(a) Multiplex IF staining with anti-OC2 and anti-AR antibodies in 6 cases of high-grade PC. Heatmap and dendrogram of nuclear staining intensities of AR and OC2, using the Euclidean distance and complete linkage method in an unsupervised cluster analysis, identifies two cell populations: AR high/OC2 low and AR low/OC2 high. Scatter plot of individual nuclei separated by intensity levels of AR and OC2. Cluster 1 (C1, green) represents high AR/low OC2 cells (N=1,109) and cluster 2 (C2, purple) represents low AR/high OC2 cells (N=199). Boxplots of intensity levels of nuclear AR and OC2 in C1 (left) or C2 (right). The boxes show the 25th-75th percentile range and the center line is the median. Whiskers show 1.5 times the IQR from the 25th or 75th percentile values. Left boxplot: P=2.1×10−191, right boxplot: P=2.3×10−66, Wilcoxon two-tailed rank-sum test.
(b) Representative IHC image of OC2 and AR in high-grade PC. OC2=red, AR=brown. Scale bar 20μm.
(c) Scatter plot of AR and OC2 nuclear intensity from 35 TMA cores of high-grade PC. Each dot represents one nucleus (N=1,373). Pearson’s method, P=4.8×10−64.
(d) AR and FOXA1 protein in PC cell lines after enforced expression of OC2. Representative blots from two independent experiments. Full-length blots are presented in Supplementary Fig. 8.
(e) PSA mRNA expression after enforced expression of OC2. The mean + S.E.M. from three to four independent experiments is shown. Unpaired two-tailed Student´s t-test, for 22Rv1 ★★P=8.73×10−8; for LNCaP ★★P=1.11×10−9.
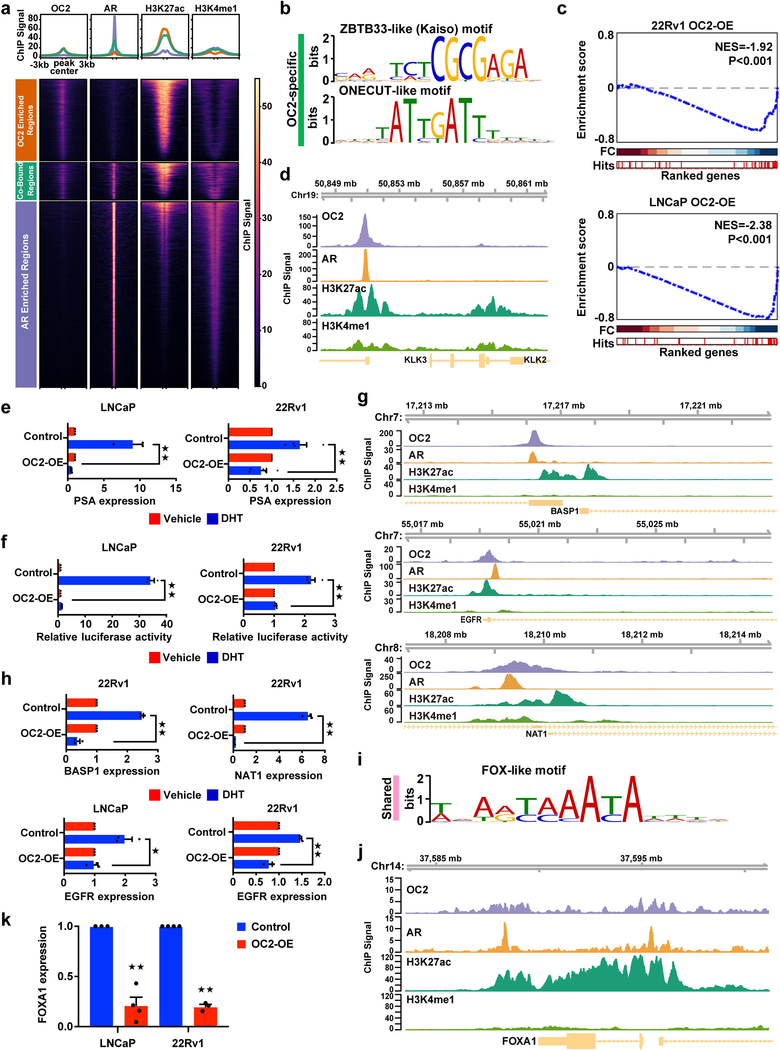
The above evidence suggests that OC2 suppresses AR activity. Consistent with this, when OC2 expression was enforced in LNCaP, 22Rv1 and C4–2 cells, AR mRNA and protein were down-regulated, including AR-V7, a common truncated AR splice variant18 (Fig. 2d and Supplementary Fig. 2a). The AR-regulated genes PSA/KLK3, KLK2 and EHF were repressed by OC2 (Fig. 2e and Supplementary Fig. 2b). In order to determine whether OC2 directly regulates the AR transcriptional program, or whether the effect of OC2 is a consequence of AR down-regulation, we performed OC2 ChIP-seq using the 22Rv1 cell line (Fig. 3a), which expresses relatively high levels of OC2. In an analysis of two experimental replicates, 8,131 OC2 peaks were observed, 67% of which were in promoters, 10% in intergenic regions distal to transcriptional start sites (TSS), and the remainder in untranslated regions, exons, introns, and immediately downstream of gene bodies (Supplementary Fig. 3a). The consensus motif with highest enrichment of solo OC2 binding sites was a ZBTB33-like (Kaiso) motif (E-value=1.6×10−540), and the second-ranked motif was a ONECUT-like motif (E-value=9.8 ×10−207) (Fig. 3b). Using the OC2-regulated transcriptome, as defined in LNCaP and 22Rv1 from enforced OC2 expression and OC2 depletion, endogenous OC2 bound to the promoters of 25% of the perturbed genes (1,791 out of 7,095). Endogenous OC2 bound to promoter regions of 474 genes that were positively regulated by enforced OC2 in 22Rv1 cells, and to the promoters of 594 genes that were negatively regulated. These results suggest that OC2 directly activates as well as suppresses genes where it binds to promoter regions.
Figure 3. OC2 suppresses the AR transcriptional program.
(a) The normalized ChIP-seq signal across merged OC2 replicates (N=2 biologically independent experiments), and individual samples of AR, H3K27ac and H3K4me1 in the peak set representing AR-enriched regions, OC2-enriched regions, and co-bound regions is shown. The number of peaks represented in each heatmap are: OC2-enriched regions (4,927 peaks), co-bound regions (2,151 peaks), and AR-enriched regions (10,726 peaks). The upper panel represents the average ChIP signal across each peak set.
(b) Top binding motifs in OC2-specific peaks.
(c) GSEA plot of an androgen response hallmark signature showing negative enrichment of AR target genes in 22Rv1 (upper panel) and LNCaP (lower panel) cells with enforced OC2 expression. Three independent microarray experiments were performed per condition.
(d) OC2 and AR binding to the PSA/KLK3 core enhancer (AREIII).
(e and f) PSA mRNA levels (e) and PSA enhancer luciferase reporter activity (f) in LNCaP and 22Rv1 cells with enforced OC2 expression, treated for 24h with 10nM DHT or vehicle.
(g) OC2 and AR binding at the promoters of BASP1, EGFR and NAT1.
(h) BASP1, EGFR and NAT1 mRNA levels in 22Rv1 and/or LNCaP cells with enforced OC2 expression, treated for 24h with 10nM DHT or vehicle.
(i) FOX-like motif recognized uniquely in OC2-AR shared regions.
(j) OC2 binding to FOXA1.
(k) FOXA1 mRNA levels after enforced expression of OC2.
For (d), (g) and (j) the browser views are representative of the results obtained in two biologically independent ChIP-seq experiments.
For (e), (f), (h) and (k) the mean + S.E.M. from three to four independent experiments is shown. Unpaired two-tailed Student´s t-test. In (e) for LNCaP ★★P=0.003 and for 22Rv1 ★★P=0.002; in (f) for LNCaP ★★P= 1.32×10−5 and for 22Rv1 ★★P= 7.93×10−5; in (h) for BASP ★★P= 2.23×10−5, for NAT1 ★★P=1.40×10−5, for EGFR in LNCaP ★P=0.02 and for EGFR in 22Rv1 ★★P=0.0006; in (k), for LNCaP ★★P=6.65×10−5 and for 22Rv1 ★★P= 4.37×10−6.
To determine whether OC2 and AR share common regulatory targets, we compared these OC2 ChIP-seq data with published AR ChIP-seq data obtained from 22Rv1 cells19. The AR consensus binding motif was the most common motif observed for AR, followed by the FOXA1 consensus motif (Supplementary Fig. 3b). We identified 2,151 peaks (26% of OC2 peaks) that were significantly bound by both OC2 and AR (Fig. 3a), however a majority of the OC2 peaks did not overlap with AR-binding regions, suggesting that OC2 can function independently of the AR. OC2/AR shared sites and OC2 solo sites were predominantly in transcriptionally active regions, based on co-localization with H3K27ac. This pattern was distinct from AR-enriched/OC2-depleted regions, which were largely distal to TSS but co-localized with well-positioned H3K4me1 enhancer marks (Fig. 3a, Supplementary Fig. 3c). OC2 potentially regulates over 500 TF genes based on OC2 binding to gene promoters (Supplementary Fig. 3d), consistent with the master regulator network model (Fig. 1c).
Out of the 2,151 regions with shared OC2 and AR binding peaks, 809 were situated within gene promoters. To address the functional relationship between OC2 and AR at these shared sites, we analyzed the promoters of 101 genes from an androgen response hallmark gene signature20. OC2 bound to 48 of these genes (48/101, P=2.0×10−11). This represents a relative enrichment of 3.2-fold over the expected ratio of 14.8% when comparing the total of 809 shared peaks out of 5,448 total OC2 promoter peaks (P=7.4×10−17). Of these 48 genes, 41 contained shared OC2-AR sites in the promoters (85%), based on a comparison with AR binding sites in the ReMap database21, which incorporates multiple datasets from a wide range of cell types, including 5 datasets from 3 PC cell lines (VCaP, LNCaP and LNCaP-abl). We next performed gene set enrichment analysis (GSEA) with the 48-gene set of OC2-AR targets in the androgen response hallmark signature using the transcriptome datasets obtained from OC2 overexpression or silencing. Enforced OC2 expression significantly repressed the majority of the 48 OC2-AR target genes in both 22Rv1 and LNCaP (Fig. 3c), while OC2 silencing exhibited modest de-repression of these genes (Supplementary Fig. 3e). The inverse relationship between OC2 and AR gene expression was confirmed in another AR-regulated gene set from CRPC tissues4. OC2 ChIP-seq peaks were noted in promoter regions of 323 out of 1,367 genes in this gene set, with an enrichment score similar to the previous gene set (Supplementary Fig. 3f). ChIP-seq showed OC2 binding to the AR promoter and to an AR intron (not shown); this was confirmed by ChIP-qPCR (Supplementary Fig. 3g), consistent with OC2 acting as a direct repressor of AR gene expression. Taken together, these findings indicate that OC2 and AR act in functional opposition at promoter sites throughout the genome.
Prostate specific antigen (PSA/KLK3) is a classic regulatory target of AR22. We observed a strong OC2 ChIP-seq peak in the well-characterized AREIII23 KLK3 enhancer region (Fig. 3d). This peak overlapped with AR binding. In the presence of dihydrotestosterone (DHT), ectopic OC2 suppressed endogenous PSA gene expression and activity of a PSA-enhancer/luciferase construct (Fig. 3e,f). Similar to PSA, the androgen-responsive genes EGFR, NAT1 and BASP1 also showed OC2 and AR binding in the promoters (Fig. 3g). In the presence of DHT, EGFR, NAT1 and BASP1 mRNA were lower in the ectopic OC2 condition compared to controls (Fig. 3h). OC2 and AR co-binding to some of these regions was confirmed by ChIP-qPCR (Supplementary Fig. 3h), and formation of a complex between endogenous OC2 and AR was demonstrated by co-immunoprecipitation (Supplementary Fig. 3i). These results are consistent with a direct antagonistic effect of OC2 on a subset of the AR-regulated transcriptome.
OC2 activates an mCRPC transcriptional program
The single enriched motif in the OC2-AR shared regions was a FOX-like motif (E-value=2.7×10−143, Fig. 3i). Enrichment of this motif only occurred in regions with OC2 and AR binding and not at OC2 solo binding sites (Fig. 3b). FOXA1 is a pioneer factor involved in positioning AR on chromatin24. These results suggest that a subset of genes is jointly regulated by AR, FOXA1 and OC2. Because we observed suppression of AR by OC2, we examined whether OC2 might also inhibit FOXA1. Endogenous OC2 bound the FOXA1 promoter and gene body (Fig. 3j). Enforced OC2 repressed FOXA1 mRNA and protein expression in LNCaP, 22Rv1 and C4–2 cells (Fig. 3k, Fig. 2d).
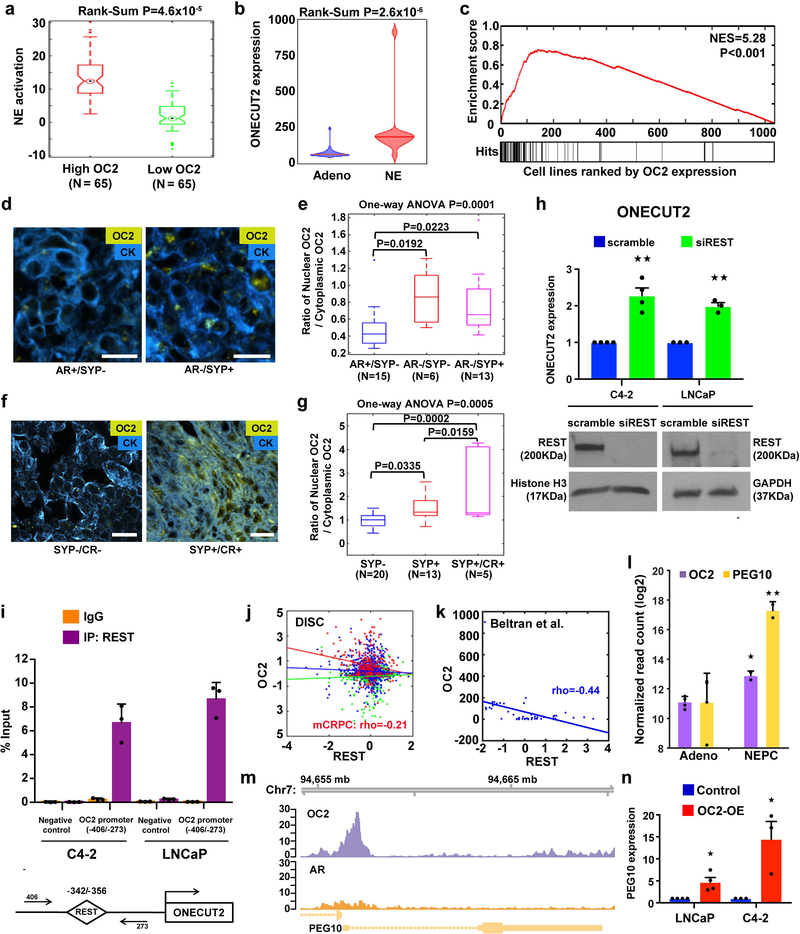
A recent report identified FOXA1 as an inhibitor of neuroendocrine (NE) differentiation that is lost with progression to mCRPC25. The observation that OC2 represses AR and FOXA1 suggests that OC2 may play a role in the emergence of NE properties in CRPC. Consistent with this, NE differentiation, assessed using a published signature26, in mCRPC tumors in the DISC cohort was elevated in tumors with high OC2 (Fig. 4a). OC2 depletion in 22Rv1 cells resulted in attenuation of NE differentiation, evaluated using the same signature (Supplementary Fig. 4a). OC2 expression is significantly increased in human NE-CRPC in comparison to Adeno-CRPC26 (Fig. 4b). We noted relatively high OC2 expression in cell lines of neural or NE derivation in the 1,063 cancer cell lines of the Cancer Cell Line Encyclopedia (CCLE)27 (Supplementary Fig. 4b). To quantify this observation, we applied running sum statistics to the CCLE dataset. We obtained a positive correlation between OC2 levels and cell lines with a neural phenotype, as designated by CCLE27–30 (Fig. 4c). Synaptophysin (SYP) is a marker commonly used in clinical practice to identify NE differentiation in CRPC31. OC2 protein was detected in mCRPC specimens (N=34) (Fig. 4d). In these cases, nuclear OC2 was seen in AR-positive cells (Supplementary Fig. 4c), however the OC2 nuclear to cytoplasmic ratio was higher in AR-negative/SYP-positive tumors and AR-negative/SYP-negative tumors in comparison to AR-positive/SYP-negative tumors32 (Fig. 4e). In addition, in patient-derived xenografts from the LuCaP xenograft compendium33 (N=38), the OC2 nuclear to cytoplasmic ratio was higher in SYP-positive compared to SYP-negative tumors, and further increased in castration-resistant (CR)/SYP-positive xenografts (Fig. 4f,g and Supplementary Fig. 4d). Consistent with this, enforced OC2 upregulated genes associated with NE differentiation (NSE, SYP and CHGA) and with neuronal differentiation (NFASC, TUBB2B, LHX2, RET, CAV2, TGFB1, and MTSS1) (Supplementary Fig. 4e,f), and promoted neural morphological features (Supplementary Fig. 4g).
Figure 4. OC2 activates a lethal transcriptional program.
(a) NE activation score in mCRPC tumors with high (N=65) and low (N=65) OC2 expression (top and bottom quartiles) from the DISC cohort. The boxes show the 25th-75th percentile range and the center line is the median. Whiskers show 1.5 times the IQR from the 25th or 75th percentile values. Data points beyond the whiskers are displayed using dots. Wilcoxon two-tailed rank-sum test, P=4.6×10−5.
(b) Violin plot of OC2 gene expression in a CRPC cohort from Beltran et al.26. Wilcoxon two-tailed rank-sum test was performed to compare NE tumors (N=15) and adenocarcinoma (Adeno, N=34). Distribution of OC2 expression shown by kernel density. The horizontal line within the violin represents the median. Wilcoxon two-tailed rank-sum test, P=2.6×10−6.
(c) The enrichment plot depicts sample-wise enrichment of the set of cancer cell lines (N=1,063) in the CCLE with NE or neural properties, sorted by OC2 expression.
(d and e) Representative images and quantitation of OC2 expression in mCRPC specimens annotated for AR and synaptophysin (SYP) expression status. IF detection of OC2 (gold) and cytokeratin (blue). AR-positive/SYP-negative (AR+/SYP-) N=15, AR/SYP-N=6 and AR-/SYP+ N=13. Scale bar 20μm.
(f and g) Representative images and quantitation of OC2 expression in LuCaP human PC xenografts annotated for SYP expression and castration resistance (CR): SYP-N=20, SYP+ N=13 and SYP+/CR+ N=5. Scale bar 20μm.
For (e) and (g) the ratio of nuclear OC2 to cytoplasmic OC2 is shown on the Y-axis and cytoplasm was defined by a cytokeratin mask. The boxes show the 25th-75th percentile range and the center line is the median. Whiskers show 1.5 times the IQR from the 25th or 75th percentile values.
(h) OC2 mRNA levels in C4–2 and LNCaP cell lines after depletion of REST. The mean + S.E.M. from three to four independent experiments is shown. Unpaired two-tailed Student´s t-test, for C4–2★★P=0.002; for LNCaP ★★P=0.001.
(i) REST binding to the OC2 promoter. Data show mean + S.D. from triplicates. Results are representative of two independent experiments.
(j) Correlation of OC2 and REST expression levels in the DISC cohort. No significant correlation was observed in benign (green, N=673) and primary (blue, N=1,061) tumors, while mCRPC tumors (red, N=260) show significant inverse correlation between OC2 and REST. Colored lines represent regression lines of the respective data points. Pearson’s method, P=0.001.
(k) Inverse relationship of OC2 and REST expression, from Beltran et al.26. Regression line in blue. Pearson’s method, P=2.9×10−36.
(l) OC2 and PEG10 mRNA expression with NE transdifferentiation in the LTL331 model35. (Adeno N=4, NEPC N=2). Unpaired two-tailed Student´s t-test followed by Storey’s FDR correction method, ★FDR=0.031, ★★ FDR=0.005.
(m) Genome browser view of endogenous OC2 binding to the PEG10 promoter in 22Rv1 cells. The result shown is representative of the two biologically independent ChIP-seq experiments performed.
(n) PEG10 mRNA expression after enforced expression of OC2. The mean + S.E.M. from three to four independent experiments is shown. Unpaired two-tailed Student´s t-test, LNCaP★P=0.012, C4–2 ★P=0.029.
Loss of REST, a master repressor of neuronal differentiation, has been implicated in NE-CRPC34. The ONECUT2 promoter contains a conserved REST binding site (Supplementary Fig. 4h), suggesting that REST is a direct ONECUT2 repressor. Depletion of REST resulted in up-regulation of OC2 mRNA levels in C4–2, LNCaP and DU145 cells (Fig. 4h; Supplementary Fig. 4i). ChIP-qPCR demonstrated direct binding of REST at the ONECUT2 promoter (Fig. 4i). In the DISC cohort, we found an inverse correlation between OC2 and REST gene expression in mCRPC (N=260, r=−0.21), but not in benign (N=673, r=−0.08) or locally confined prostate tumors (N=1,061, r=−0.01) (Fig. 4j). An inverse correlation (r=−0.44) was also seen in a separate NEPC cohort26 (Fig. 4k). Consistent with this, REST mRNA expression dramatically declined and OC2 mRNA increased with transition from adenocarcinoma to NEPC in the LTL331 NE human xenograft transdifferentiation model35 (Supplementary Fig. 4j). OC2 correlated inversely with REST mRNA expression in a cohort of human PC organoids36 (Supplementary Fig. 4k). Notably, relative OC2 expression was highest and REST expression lowest in MSK-PCa4, an organoid line derived from a treatment-induced NE tumor. Similar to results described above from other datasets, the inferred OC2 activation signature correlated positively with an AR repressed gene signature17, and OC2 activation correlated inversely with the hallmark AR gene signature in these organoids. These results indicate that REST is a disease-relevant repressor of OC2 expression.
The above results suggest that OC2 contributes to NE differentiation in mCRPC. Recently, the placental gene PEG10 was identified as a driver of NEPC in the LTL331 transdifferentiation model35. PEG10 is repressed by AR in PC cells, but during conditions of androgen suppression PEG10 is markedly up-regulated. In the LTL331 model, OC2 expression increased in parallel to PEG10 during the transition from adenocarcinoma to NEPC (Fig. 4l). Consistent with this finding, PEG10 and OC2 expression in NEPC26 were highly correlated (Spearman’s rho=0.58, P<0.001; not shown). OC2 binds to the PEG10 promoter (Fig. 4m), and binding to this region was further confirmed by surface plasmon resonance (SPR) using a dsDNA probe corresponding to the OC2 motif in the PEG10 promoter (KD~1nM, Supplementary Fig. 4l). Enforced OC2 increased PEG10 mRNA levels (Fig. 4n), indicating that OC2 is a direct upstream activator of PEG10 transcription. Collectively, these data position OC2 between REST and PEG10 during transdifferentiation of adenocarcinoma to NEPC.
Targeting OC2 inhibits CRPC metastasis
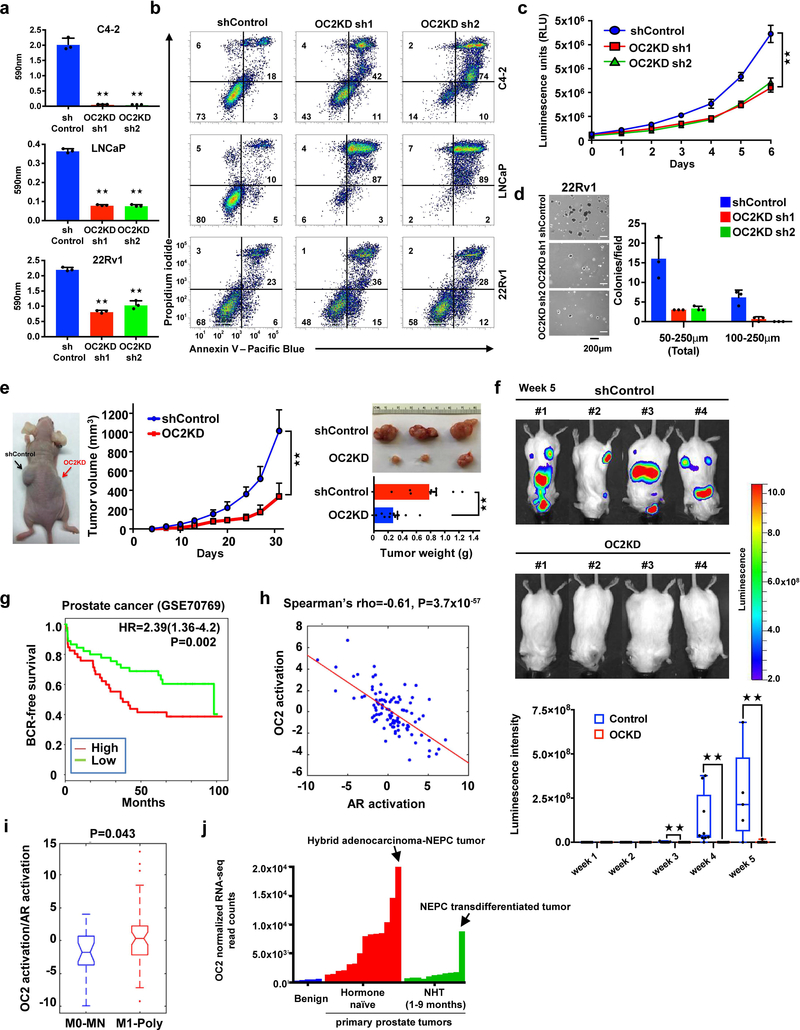
To evaluate the function of OC2 in metastasis of CRPC, we depleted OC2 expression in multiple PC cell lines. OC2 silencing by lentiviral infection with two independent shRNAs caused extensive apoptosis in LNCaP and C4–2 cells (Fig. 5a,b and Supplementary Fig. 5a). This result was also seen with a third independent shRNA (data not shown). OC2 depletion using siRNA oligos caused similar effects in AR-negative PC3 cells (Supplementary Fig. 5b). While OC2 loss caused cytotoxicity in 30% of 22Rv1 cells, which express high OC2 levels among the cell lines examined, the surviving OC2deficient cells could be propagated and studied experimentally. Depletion of OC2 resulted in growth inhibition and impaired the ability of 22Rv1 cells to form colonies in soft agar (Fig. 5c,d). Depletion of OC2 using siRNA oligos suppressed growth in 22Rv1, LNCaP, C4–2, and PC3 cells (Supplementary Fig. 5c). To analyze the consequences of OC2 silencing in vivo, we subcutaneously engrafted 22Rv1 cells expressing control and OC2 shRNAs into immunodeficient nude mice and tracked tumor growth. Consistent with the in vitro results, OC2 depletion resulted in reduced tumor burden and tumor weight (Fig. 5e). In order to evaluate the effect of OC2 depletion on metastasis, we injected luciferase-tagged 22Rv1/shOC2 or 22Rv1/shControl cells intracardially into SCID mice and monitored tumor growth by bioluminescence imaging. We observed reduction of metastatic growth at weeks 3–5 (Fig. 5f). Taken together, these findings demonstrate that OC2 depletion inhibits 22Rv1 growth and metastasis.
Figure 5. OC2 promotes mCRPC tumor growth and metastasis.
(a) Cell survival 4 days after start of OC2 depletion. The mean + S.D. from three independent experiments is shown. Unpaired two-tailed Student´s t-test, C4–2: sh1★★P= 7.58×10−5, sh2★★P= 7.48×10−5; LNCaP: sh1★★P=1.57×10−6, sh2★★P=2.03×10−6, 22Rv1: sh1★★P=5.42×10−6, sh2★★P= 0.0002.
(b) OC2 depletion results in cell death 4 days after infection, measured by flow cytometry and double staining with Annexin V-FITC and propidium iodide (PI). These experiments were repeated independently three times with similar results.
(c) Growth in cell culture of 22Rv1 cells after OC2 depletion. The mean +/− S.D. from three independent experiments is shown. Unpaired two-tailed Student´s t-test, sh1★★ P=0.0006, sh2★★P=0.0009.
(d) Growth in soft agar of 22Rv1 cells after OC2 depletion. The mean + S.D. from triplicates is shown. The result shown is representative of two independent experiments.
(e) Subcutaneous 22Rv1 tumor growth after OC2 depletion (left). Appearance of tumors after 31 d (top right). Tumor volume (middle) and weight (bottom right) are shown as mean + S.D. (N=10 per mice group). Unpaired two-tailed Student´s t-test, tumor volume ★★P=0.005, tumor weight ★★P=0.0002.
(f) Effect of OC2 silencing on 22Rv1 metastasis following intracardiac injection. Bioluminescence images of mice bearing metastatic tumors 5 weeks after injection (top) and graphical representation of normalized bioluminescence signals of tumor development (bottom). N=9 mice per group. The boxes show the 25th-75th percentile range and the center line is the median. Whiskers include the smallest and largest values. Wilcoxon two-tailed rank-sum test, in week 3 ★★P= 0.023, in week 4 ★★P= 0.009, in week 5 ★★P=0.0017.
(g) Association of BCR-free survival with OC2 expression. High (N=46) and low (N=45). Cox proportional hazard regression, two-sided test, P=0.002.
(h) Inverse correlation between OC2 activation and AR activation based upon mRNA expression in tumor foci of diagnostic prostate needle biopsies (N=99). Spearman’s rank correlation test, P=3.7×10−57.
(i) Ratio of OC2 activation to AR activation in diagnostic prostate needle biopsies (M0-MN (N=23) and M1-Poly (N=52)). M1-Poly = patients with concurrent polymetastases (>5 bone or extrapelvic lymph node metastases and/or visceral metastases) at the time of diagnostic biopsy. M0-NM = high-grade prostate tumor biopsies from patients that remained locally confined (no metastatic progression) with average follow-up of 56 months. The boxes show the 25th-75th percentile range and the center line is the median. Whiskers show 1.5 times the IQR from the 25th or 75th percentile values. Data points beyond the whiskers are displayed using dots. Wilcoxon two-tailed rank-sum test, P=0.043.
(j) Expression of OC2 mRNA in benign and high risk primary prostate tumors from Wyatt et al.37. NHT= neo-adjuvant hormone therapy.
Consistent with the possibility of a role for OC2 in human PC metastasis, Kaplan-Meier analysis of two human PC cohorts showed that high expression of OC2 mRNA is associated with biochemical recurrence (Fig. 5g and Supplementary Fig. 5d). In these cohorts, the tissues were obtained before treatment of localized PC, demonstrating that elevated OC2 expression can occur prior to androgen deprivation therapy (ADT) and confers risk of disease progression. To further investigate the relationship between OC2 activation and AR activation in patients at highest risk of developing CRPC, we compared transcriptome (RNA-seq) data from diagnostic prostate needle biopsies obtained from patients with high-grade non-metastatic (M0-NM; metastasis-free with long-term follow-up) and metastatic (M1-Poly; patients presenting with concurrent poly-metastases) disease. An inverse correlation between OC2 and AR activation was identified (Fig. 5h), and tumors demonstrating high OC2 activity and low AR activity were significantly more abundant in the M1-Poly group (Fig. 5i). OC2 mRNA was elevated in M1-Poly specimens in comparison to M0-NM specimens (Supplementary Fig. 5e). In contrast, AR expression did not differ between these groups (Supplementary Fig. 5f). In a separate cohort37, OC2 mRNA expression was also higher in high risk primary prostate tumors from treatment-naïve patients compared to benign prostate tissue and to tumors from patients treated with neo-adjuvant ADT (Fig. 5j). Among these patients, OC2 expression was highest overall in a hybrid Adeno-NE tumor and, in the neoadjuvant ADT group, in a NEPC transdifferentiated tumor.
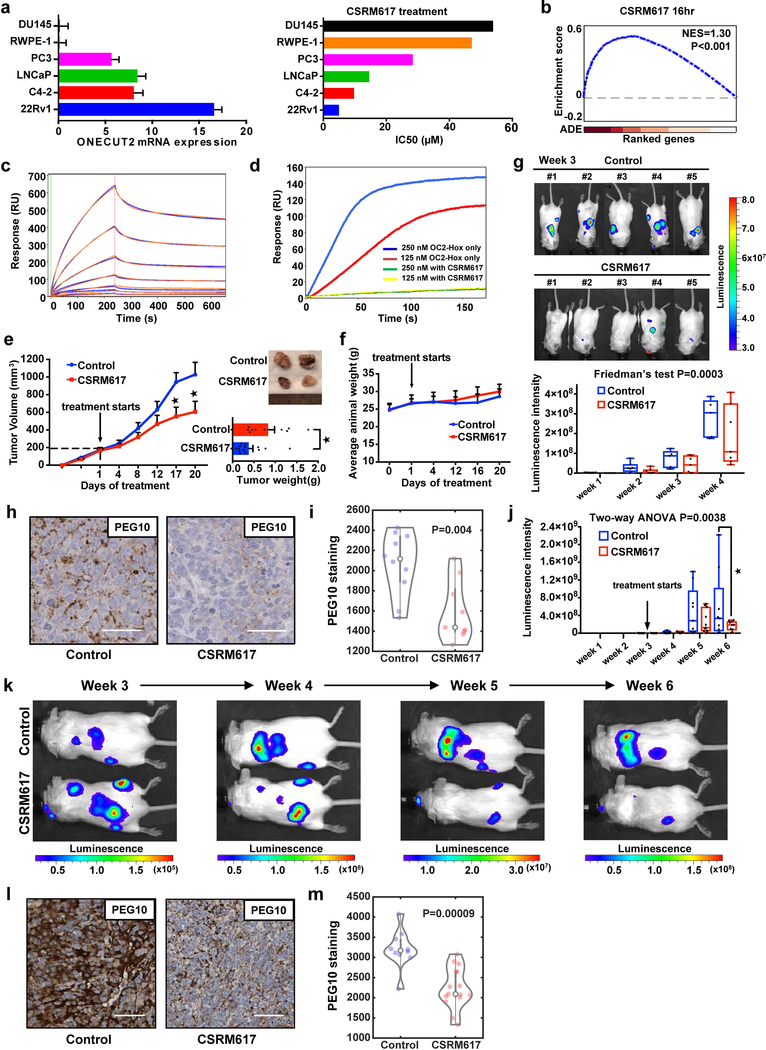
These results suggest that OC2 might be a clinically relevant therapeutic target in the context of metastatic PC. To identify small molecule inhibitors of OC2 we used structure-based drug design. We constructed a 3-dimensional model of the OC2-HOX domain and screened a chemical library consisting of 500,000 small molecules. One compound, CSRM617 (Supplementary Fig. 6a), predicted to bind to OC2-HOX (Supplementary Fig. 6b), was biologically active in experimental tests and was selected for further study. CSRM617 inhibited cell growth and induced apoptosis in vitro in several PC cell lines that expressed moderate to high levels of OC2 (Supplementary Fig. 6c–f), phenocopying the effects observed when OC2 was depleted with shRNA or siRNA (Fig. 5a,b and Supplementary Fig. 5a–c). Notably, OC2 RNA expression levels in the various cell lines correlated with the IC50, indicating that cells with low OC2 expression were less responsive to treatment with the compound (Fig. 6a). To evaluate the specificity of CSRM617 for genes regulated by OC2, we generated gene expression microarray data from 22Rv1 cells treated with the compound for 4, 6 and 16 h. For this analysis, the OC2 target gene set was defined by OC2 binding to the gene promoters in the ChIP-seq data set. GSEA showed significant perturbation of OC2 target genes at all three time points (Fig. 6b and Supplementary Fig. 6g). As described above, we identified PEG10 as a gene directly regulated by OC2 as a result of binding of the protein to the PEG10 promoter (Fig. 4m and Supplementary Fig. 4l). In response to CSRM617, PEG10 mRNA was down-regulated in a time-dependent manner (Supplementary Fig. 6h).
Figure 6. Inhibition of OC2 with a small molecule.
(a) Relative OC2 mRNA levels in 6 prostate cell lines determined by RT-qPCR (left). ACTB and GAPDH expression levels were used for normalization. Data show mean + S.D. from triplicates. Results are representative of two independent experiments. The IC50 values for compound CSRM617 shown are the mean from two independent experiments (right).
(b) Enrichment plot of OC2 target genes perturbed by 10μM CSRM617 at 16 h in 22Rv1 cells. ADE = Absolute differential expression (N=3 independent microarray experiments).
(c) Dose-dependent binding of CSRM617 (1.56μM to 100μM) to OC2-HOX is shown in blue. 2:2 Langmuir model simulation used to derive kinetic parameters of the bimolecular interactions shown in orange.
(d) SPR competition assay showing compound CSRM617 (100μM) inhibiting OC2-HOX binding to DNA at two different concentrations protein 250nM (blue) and 125nM (red). For (c) and (d) these experiments were repeated independently three times with similar results.
(e) 22Rv1 cells were implanted subcutaneously. When tumors reached 200mm3, mice were randomized and received vehicle or 50 mg/Kg CSRM617 daily (Control N=11 tumors, CSRM617 N=14). Appearance of tumors after 20 d (top right). Tumor volume (left) and tumor weight (bottom right) are shown as mean + S.E.M. (Unpaired two-tailed Student´s t-test, tumor volume 1st★P=0.02, 2nd★P=0.037, tumor weight ★P=0.011).
(f) CSRM617 treatment did not affect animal weight (mice from (Fig. 6e)). Mean ± S.D. is shown (N=8 mice per group).
(g) 22Rv1-luciferase labeled cells were injected intracardially; two days after injection mice were randomized (N=7 per group) to receive vehicle or 50 mg/Kg CSRM617 daily. Bioluminescence images (BLI) of mice bearing metastatic tumors 3 w after the injection (top) and graphical representation of normalized BLI signals, weeks 1–4 (bottom). The boxes show the 25th-75th percentile range and the center line is the median. Whiskers include the smallest and largest values. Friedman’s two-tailed test, P=0.0003.
(h and i) Representative PEG10 IHC staining and quantification in adrenal metastases from the experiment in Fig. 6g. Scale bar 50μm. Control (N=10 microscope fields) and CSRM617 (N=9). Kernel density estimation shows the distribution of PEG10 staining. The open circle indicates the median. The thick and thin vertical grey lines represent the IQR and 95% confidence interval, respectively. Wilcoxon two-tailed rank-sum test, P=0.004.
(j) 22Rv1-luciferase labeled cells were injected intracardially. Once metastases were established, mice were randomized (N=11 per group) to receive vehicle or 50 mg/Kg CSRM617 daily. Graphical representation of normalized BLI signals of mice bearing metastatic tumors 6 weeks after the injection. The boxes show the 25th-75th percentile range and the center line is the median. Whiskers include the smallest and largest values. Two-way ANOVA (P=0.0038), followed by post-hoc Dunnett’s test at week 6 (★P=0.0158).
(k) Representative result from (Fig. 6j) showing one control and one treated subject with CSRM617 in weeks 3 to 6.
(l and m) Representative PEG10 IHC staining and quantification in adrenal metastases from the experiment in Fig. 6j. Scale bar 50μm. Control (N=10 microscope fields) and CSRM617 (N=15). Kernel density estimation shows the distribution of PEG10 staining. The open circle indicates the median. The thick and thin vertical grey lines represent the IQR and 95% confidence interval, respectively. Wilcoxon two-tailed rank-sum test, P=9×10−5.
Next we used SPR to test whether CSRM617 binds directly to OC2. The OC2-HOX domain (aa 330–485) was expressed in a bacterial system and it purified as a dimer in the absence of DNA (not shown). As mentioned above, the dimeric protein bound with high affinity to a dsDNA probe corresponding to the experimentally determined OC2 binding motif in the PEG10 promoter (Supplementary Fig. 4l). CSRM617 bound to OC2 in a dose-dependent manner (KD=7.43μM) (Fig. 6c), demonstrating that the compound binds the OC2-HOX domain directly. Furthermore, in a competitive SPR assay, CSRM617 significantly inhibited OC2-HOX binding to immobilized DNA (Fig. 6d).
To test the effect of CSRM617 in vivo, 22Rv1 cells were implanted subcutaneously in nude mice. Treatment with CSRM617 in small tumors (50mm3) showed significant reduction of tumor volume and weight (data not shown). To assess the efficacy of the compound in a setting that better mimics the clinical scenario, when tumors reached a size of 200mm3, mice were randomized and treated daily with CSRM617 or vehicle control (2.5% DMSO in PBS) intraperitoneally at a dose of 50 mg/Kg/d. The treatment group showed significant reduction of tumor volume and weight (Fig. 6e). CSRM617 did not affect body weight, suggesting the compound was well tolerated (Fig. 6f). To determine whether CSRM617 is effective against PC metastases, luciferase-tagged 22Rv1 cells were injected intracardially in SCID mice and two days later the animals were randomized and treated daily (50 mg/Kg) with the compound or vehicle. CSRM617 elicited a significant reduction in the onset and growth of diffuse metastases (Fig. 6g). To test for evidence of bioactivity of CSRM617 using an independent metric, PEG10 protein levels were quantitatively measured in tumors in both the experimental and control groups. PEG10 protein was significantly down-regulated in tumors from mice treated with CSRM617 (Fig. 6h,i). To assess the efficacy of the compound after metastases are already established, 22Rv1-luciferase tagged cells were injected intracardially in SCID mice. Mice that developed metastases (Supplementary Fig. 6i) were randomized (N=11 per group) and treated daily (50 mg/Kg) with the compound or vehicle. CSRM617 elicited a significant reduction of diffuse metastases (Fig. 6j). A representative result showing one control and one treated subject is shown in Fig. 6k. PEG10 protein levels were quantitatively measured in tumors in both the experimental and control groups. PEG10 protein was similarly down-regulated in tumors from mice treated with CSRM617 (Fig. 6l,m).
DISCUSSION
This study identified ONECUT2/HNF6β (OC2) as a central regulatory node in a subset of aggressive PC where AR activity is relatively suppressed. OC2 emerged from an unbiased computational analysis that evaluated over 400 TFs in a large cohort of mCRPC. To our knowledge, this is the first master regulator TF network assembled specifically within the lethal phase of PC progression. We incorporated comprehensive TF-target relationships, available from public ChIP-seq and ChIP-on-chip databases, and pair-wise expression correlations between expression levels of TFs and their target genes. This procedure identified TFs known to play important roles in CRPC (AR, EZH2), less studied TFs in CRPC (FOXM1, E2F3), and several TFs that are largely uncharacterized in CRPC (MTF2, E2F7, and PAX5). The outcome of the functional validation studies of OC2 suggests that the network we describe here is valuable in nominating other novel drivers of mCRPC. Interestingly, OC2 expression is significantly associated with poor clinical outcome in cancer types other than prostate, including breast, gastric, colon, clear cell renal, brain, and lung cancer (Supplementary Fig. 7a), suggesting it may play a role more widely in aggressive cancers. Consistent with this concept, KEGG Pathway Enrichment of OC2 binding to chromatin using ChIP-seq data identifies several other tumor types as potentially relevant (Supplementary Fig. 7b). Notably, the most significant association with the OC2-AR shared sites was PC.
In PC, our findings suggest that OC2 operates as a genome-wide suppressor of AR activity, a survival factor and a driver of a neural differentiation gene expression program within the context of adenocarcinoma. The AR is frequently referred to as the most significant PC oncoprotein, even in mCRPC, and is still the primary focus of novel drug design. AR activation under castrate conditions has been shown to arise from gene amplification38, androgen synthesis or retention within the tumor3, and collaboration with signal transduction pathways and other TFs39. Aggressive variants emerge following hormone therapy and these tumors express variable levels of AR and, in some instances, NE features40. Histologically defined small cell (SC) NEPC mostly arises prior to treatment, and represents a less frequent cause of mCRPC because of its low incidence (<5% of mCRPC)38. Transitions from adenocarcinoma to NEPC occur in cell and xenograft models35,41. Our findings point to an important role for OC2 in the transition from AR-dependent adenocarcinoma toward AR-indifferent CRPC: 1) OC2 is a direct activator of PEG10, which was recently identified as a master regulator of the transition from adenocarcinoma to NEPC35; 2) The ONECUT2 promoter is repressed by REST, a TF lost during progression to NEPC34; 3) OC2 inhibits expression of the AR pioneer factor, FOXA124, which is down-regulated during the transition from adenocarcinoma to NEPC25; 4) Using a recently developed system for classifying PC12, OC2 activity was highest in the PCS3 subtype, which exhibits a relatively heightened NE differentiation program as evaluated by signature analysis. PCS3 represents about 1/3 of mCRPCs evaluated, suggesting that activation of NE differentiation can occur without full progression to NEPC. 5) Finally, the majority of OC2 binding regions on chromatin do not overlap with AR binding sites, suggesting that OC2 can function independently of the AR. Consistent with this, OC2 depletion in AR-negative PC3 cells suppressed cell growth. Taken together, our findings suggest the existence of an mCRPC subtype, characterized by continued AR expression, reduced AR activity, and emergence of NE features, wherein OC2 operates as a survival factor.
Inhibition of the AR axis by OC2 suggests that tumor cells with increased OC2 expression could be selected following treatment with ADT. We present data demonstrating elevated OC2 mRNA expression in pre-treatment diagnostic needle biopsy tissue of patients with de novo polymetastatic PC, suggesting the presence of OC2 activity in PC cells prior to therapy or metastatic dissemination. Because reports by others have associated OC2 with cell migration42, and our results link OC2 to tumor cell survival, emergence of OC2 activity in a primary tumor may be an important driver of PC metastasis prior to treatment. This hypothesis should be tested in future studies.
In summary, we demonstrated that OC2 is a master regulator of lethal PC that may be an important therapeutic target in as many as 1/3 of mCRPC tumors. We demonstrate that OC2 can be inhibited with a small molecule that suppresses human mCRPC metastasis in a xenograft model. Our study supports the conclusion that many lethal PCs, characterized histologically as adenocarcinoma, are comprised of cells with partial activation of an NE differentiation program in which OC2 plays a major role.
METHODS
Identification of key TFs.
In order to identify key transcriptional regulators that drive PC progression, we performed a master regulator analysis based on a GSEA procedure. A compendium of TF and TF-target gene interaction information was collected from databases of genome-wide ChIP data in human (ChIPBase (http://rna.sysu.edu.cn/chipbase/), Amadeus (http://acgt.cs.tau.ac.il/amadeus/), and hmChIP (http://jilab.biostat.jhsph.edu/database/cgi-bin/hmChIP.pl )) and curated databases containing genes that share a TF binding site (ChEA (http://amp.pharm.mssm.edu/ChEA2/), TRED (https://cb.utdallas.edu/cgibin/TRED/tred.cgi?process=home ), and MSigDB (http://software.broadinstitute.org/gsea/msigdb )). In order to reflect context dependent TF-target interactions in mCRPC, we selected interactions with significant correlation between TF and target genes based on Spearman’s rank correlation coefficient in 260 mCRPC transcriptome profiles. TF-target enrichment score (ES) was calculated using a Kolmogorov-Smirnov (K-S) running sum statistic for each TF. We randomly permuted the sample labels and repeated ES calculation for 10,000 times to compute the ES null distribution. Statistical significance of the ES was computed by comparing the observed ES to the ES null distribution. Normalized enrichment score (NES) was computed by dividing the observed ES by the average of values >0 from the ES null. 31 TFs were selected based on their high expression and significant enrichment of their target genes in the mCRPC cohort.
Construction of the network model.
To construct a network model of TFs operating in mCRPC, we employed the 10 TFs with the largest number of leading edge subsets in the target genes. Interactions between the top 10 TFs were initially assigned by the protein-protein interaction (PPI) information from STRING database (http://stringdb.org/). We then estimated the strength of the interactions among the 10 TFs by quantifying the expression concordance using the Chi-square test. The TF network model was reconstructed and visualized using Cytoscape software (http://cytoscape.org/), along with the TF expression in mCRPC, significance of their target gene regulation, and interaction strength between TFs.
Clinical outcome analysis.
We studied the relationships between the OC2 expression and BCR-free survival, metastasis-free survival, overall survival, and relapse-free survival in cancer types shown in Fig. 5g, Supplementary Fig. 5d and Supplementary Fig. 7a. SurvExpress (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp ) and PROGgeneV2 (http://watson.compbio.iupui.edu/chirayu/proggene/database/?url=proggene) tools were used to perform Kaplan-Meier analysis and Cox proportional hazard regression. OC2 expression and survival outcomes were correlated to assess significant associations. Samples were stratified by OC2 expression level at median value of all the samples in each cohort. ‘Breast cancer 10 cohorts’ indicates the breast meta-base build from SurvExpress database, which merges 10 breast cancer cohorts generated by the same Affymetrix microarray platform. For every comparison, a corresponding P-value < 0.05 was considered significant.
Correlating OC2 expression and neural/NE tumor derivation in human cancer cell lines.
Normalized OC2 mRNA expression intensities across 1,063 cancer cell lines was downloaded from the Cancer Cell Line Encyclopedia (CCLE) data portal (http://www.broadinstitute.org/ccle/home). The 1,063 cancer cell line datasets were ranked according to the normalized OC2 intensities. K-S test was applied to test whether high expression of OC2 is correlated with neural/NE tumor derivation. The 85 neural/NE cancer cell lines were identified using the annotation from CCLE data portal, IGRhCellID database, and the literature27–30. A permutation test strategy was used to determine the statistical significance of a particular K-S statistic. The gene expression data were randomly shuffled 10,000 times to compute the empirical P-value.
Prostate cell lines culture.
Prostate cancer cell lines were obtained from the American Type Culture Collection. LNCaP, C4–2 and 22Rv1 were grown in RPMI-1640 media (Gibco) supplemented with 10% FBS and penicillin/streptomycin. PC3 and DU145 were maintained in DMEM media (Gibco) supplemented with 10% FBS and penicillin/streptomycin. RWPE-1 were grown in Keratinocyte Serum Free Medium (Gibco) supplemented with 0.05 mg/ml BPE (bovine pituitary extract) and 5 ng/mL EGF (human recombinant epidermal growth factor). All cell lines tested negative for mycoplasma contamination.
Antibodies.
Anti-OC2 HPA057058, rabbit, polyclonal, Lot#R78318, from Sigma was used for the ChIP-Seq and ChIP-qPCR experiments (4μg per 30μg of chromatin); anti-OC2 21916–1-AP, rabbit, polyclonal, Lot#00016559, from Proteintech was used for WB (1:1000); anti-OC2 ab28466, rabbit, polyclonal, from Abcam (1:400) and anti-OC2 LSC499462, Lot#118218, from LSBio (1:200), were used for the IHC and IF studies; antiAR N-20, rabbit, polyclonal, Lot#I1014, from Santa Cruz was used for ChIP-qPCR (4μg per 30μg of chromatin), co-immunoprecipitation (5μg per 500μg of protein) and WB (1:500), anti-AR #5153, clone D6F11, rabbit, monoclonal, from Cell Signaling was used for IHC (1:500); anti-PEG10 HPA051038, rabbit, polyclonal, from Sigma was used for IHC (1:500); anti-REST 17–10456, rabbit, polyclonal, Lot#2665779, from EMD Millipore was used for WB (1:1000) and ChIP-qPCR (4μg per 30μg of chromatin). The following antibodies were used for WB: anti-GAPDH 14C10, rabbit, monoclonal, Lot#4, from Cell Signaling (1:5000); anti-B-Actin 12262S, mouse, monoclonal, Lot#1, from Cell Signaling (1:5000); anti-FOXA1 ab23738, rabbit, polyclonal, Lot#GR176970–1 from Abcam (1:1000); anti-Histone H3 3H1, rabbit, monoclonal, Lot#1, from Cell Signaling (1:5000); anti-Caspase-9 #9502, rabbit, polyclonal, Lot#8, from Cell Signaling (1:1000); anti-Caspase-3 31A1067, mouse, monoclonal, Lot#AB022811A-08, from Novus Biologicals (1:1000); anti-PARP #9542, rabbit, polyclonal, Lot#13, from Cell Signaling (1:1000).
Lentiviral constructs.
Lentivirus were created by co-transfection of the Delta 8.9 packaging plasmid and the pCMV-VSVG plasmid (Addgene 8454) into Lenti-XTM-293T cells (Clontech) using Lipofectamine 2000 (Invitrogen). Medium was changed every 24 h, and the 48 and 72 h supernatants were filtered through a 0.45μm filter syringe for the transduction of the recipient cells. The OC2 overexpression plasmid was obtained by cloning the full-length OC2 cDNA (NM_004852) into the pLenti-C-Myc-DDK-IRES-Puro (Origene). For the knockdown of OC2, three validated shRNA clones (TRCN0000013443, TRCN0000013445, TRCN000235576) in the vector pLKO1 were purchased from Sigma. A non-mammalian shRNA Control Plasmid (TRC2-pLKO-puro non-target shRNA #1, Sigma) was used as a control. Cells were subsequently transduced in the presence of 5μg/mL of polybrene and selected on 5μg/ml puromycin to create the stable line.
siRNA transfection.
OC2 (#SR306292) and REST (#SR304036) siRNA pools and Universal scrambled negative control siRNA duplexes (Origene) were used according to manufacturers’ instructions. A second set of REST siRNA pools (L-006466–00) and control pools (D-001810–10) were purchased from Dharmacon. Briefly, cells were transfected with siRNA (100nM) for 48 to 72 h with Lipofectamine RNAiMAX Reagent (Thermo Fisher Scientific).
Apoptosis quantification.
Cells were harvested and stained with Pacific Blue–Annexin V and propidium iodide (PI) (BioLegend) according to the manufacturers’ instructions. Apoptotic cells were detected by flow cytometry using an LSRFortessa (BD Biosciences; excitations at 355, 405, 488, 561, 640 nm) and data were analyzed with FlowJo v. 7.6.4. Cells were gated by FSC-A x SSC-A to exclude debris and then by FSC-H x FSC-W following SSC-H x SSC-W to exclude cell doublets. Examples of the gating strategy are shown in Supplementary Fig. 7c.
Gene expression microarrays.
Total RNA was extracted using the RNeasy Kit (Qiagen) and hybridized to the Human U133plus2.0 Array Hybridization (Affymetrix) at the UCLA Clinical Microarray Core. The probe intensities were normalized using the quantile normalization method43. Gene expression datasets were deposited into the GEO database (GSE97549 and GSE97548). For the comparisons 1) OC2 OE/Control in 22Rv1, 2) OC2 OE/control in LNCaP, 3) OC2 KD/control in 22Rv1, 4) CSRM617 treated in 4h/Vehicle treated, 5) CSRM617 treated in 6h/Vehicle treated, and 6) CSRM617 treated in 16h/Vehicle treated, we used the integrative statistical method44. Briefly, for each gene, an adjusted P-value was computed by performing a two-tailed T-test and log2 median ratio test using the empirical distributions that were estimated by random permutations of the samples. The two sets of P-values from the individual tests were combined into overall P-values using Stouffer’s method and FDR was computed from the overall P-values using Storey’s method45. Finally, the DEGs were selected with the ones having FDR<0.05 and FC>2. We then defined two sets of DEGs that are up- or down-regulated by OC2 perturbations, which are corresponding to up- and downregulated targets of OC2 in PC cells. The cellular processes represented by the up- and down-regulated targets by OC2 were identified as the cellular processes and pathways having P<0.05 from DAVID v6.8 software (https://david.ncifcrf.gov/).
Quantitative PCR.
Total RNA was extracted (RNeasy Kit, Qiagen) and subjected to One-Step Real Time RT-PCR in a 7900HT Sequence Detection System with MultiScribe Reverse Transcriptase, RNase Inhibitor, TaqMan Gene Expression Master Mix (Applied Biosystems) and the Taqman probes for ONECUT2 (Hs00191477_m1), AR (Hs00171172_m1), PSA (Hs02576345_m1), KLK2 (Hs01091576_m1), EHF (Hs00171917_m1), FOXA1 (Hs04187555_m1), NSE (Hs00157360_m1), CHGA (Hs00900370_m1), SYP (Hs00300531_m1), NFASC (Hs00391791_m1), TUBB2B (Hs00603550_g1), LHX2 (Hs00180351_m1), CAV2 (Hs00184597_m1), TGFB1 (Hs00998133_m1), MTSS1 (Hs00207341_m1), RET (Hs01120030_m1), PEG10 (Hs00248288_s1), EGFR (Hs01076090_m1), BASP1 (Hs00932356_s1) and NAT1 (Hs00265080_s1). Data were analyzed using the 2−ΔΔCT method. ACTB and GAPDH TaqMan probes (Hs01060665_g1 and Hs02758991_g1, respectively) were used for normalization.
ChIP-qPCR and ChIP-seq.
ChIP-qPCR and ChIP-seq experiments were performed with the ChIP-IT High Sensitivity kit from Active Motif. Briefly, 80% confluent 22Rv1, LNCaP or C4–2 cells were cross-linked with 1% formaldehyde at room temperature for 10 min. Chromatin was sonicated to 100–500 bp in a Bioruptor UCD-200 (Diagenode) and incubated overnight with 4 μg of antibody (anti-OC2 HPA057058, Sigma; anti-REST 17–10456, EMD Millipore; anti-AR N-20, Santa Cruz or IgG control). After RNase A and Proteinase K treatment, ChIP DNA was purified with the column system provided with the kit. For targeted ChIP, eluted DNA was used for quantitative PCR with the specific primers listed in Supplementary Table 2. A negative primer set that amplifies a 78 base pair fragment from a gene desert on human chromosome 12 (Human Negative Control Primer Set 1, Active Motif) was used as a control. For the ChIP-seq experiments, libraries were prepared with the Kapa Hyper DNA kit (Kapa Biosystems; PCR conditions: 98°C for 45 sec; 98°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec, for 7 cycles; 72°C for 1 min) at the UCLA Clinical Microarray Core, followed by direct sequencing using the Illumina HiSeq3000 system.
Pipeline for OC2 and AR peak classification.
ChIP-seq data were processed in accordance with the AQUAS transcription factor and histone ChIP-seq processing pipeline [https://www.encodeproject.org/chip-seq/]. The “optimal” peak set was used for downstream analysis. Briefly, relaxed (with very low significance threshold) peaks were called with SPP46, and then input to IDR, in order to control the irreproducible discovery rate to <0.05. After mapping to hg38 with BWA47 and removing duplicates, there were >65 million high-quality reads for all ChIP-seq replicates. We used SPP48 to calculate the cross-correlation QC metrics and MACS2 + IDR to obtain the rescue ratio and self-consistency ratio for each cell line. We then used the DiffBind package in R/Bioconductor to assess differential binding of OC2 and AR in genomic regions. Within DiffBind, we calculated the read counts from each of our samples in the union of the IDR called peaks, and proceeded to assess differential binding of the OC2 and AR within this set. Differential binding was assessed with DESeq2 at a FDR < 0.05. Shared peaks were identified as those not found in differentially enriched regions, but with overlapping peaks. These regions were visualized with deepTools249 and placed in genomic context with the ChIPSeeker package50, and analyzed for gene ontology with the DOSE package51. Motifs were identified using MEME-Chip52 within each peak set. Anti-AR, Anti-H3K4me1, and Anti-H3K27ac datasets from 22Rv1 cells were obtained from GEO (GSE85558). The ChIP-seq data were deposited into the GEO database (GSE97551).
Co-immunoprecipitation.
These experiments were performed as previously described53. Briefly, LNCaP were seeded and after 48 h in charcoal stripped media, cells were treated with 10nM DHT or vehicle (100% ethanol) for additional 24 h. Nuclear proteins extracts were pre-cleared and incubated with an AR antibody (N-20, rabbit, polyclonal, Lot#I1014, Santa Cruz) at 4°C overnight. AR-binding proteins were next precipitated with agarose beads and subjected to western blot.
Western blot analysis.
Cell lysates were separated on 4–20% SDS-PAGE (Bio-Rad Laboratories) and transferred to nitrocellulose or PVDF membranes. The membranes were blocked with 5% non-fat dry milk and subsequently incubated with the pertinent primary antibody overnight. Membranes were subsequently washed with PBST (0.1% Tween-20) and incubated with an HRP-conjugated secondary antibody (GE Healthcare Bio-Sciences). After washing with PBST, the protein bands were detected by chemiluminescence (Thermo Fisher Scientific). Entire blots are shown in Supplementary Fig. 8,9.
Proliferation assays.
To assay viability, cells were plated at a density of 1,000 cells per well, in triplicate, in a 96 well plate. Twenty-four hours after, cells were harvested at time 0 h (T0) using a luminometric assay (CellTiter-Glo Luminescent Cell Viability Assay, Promega) and subsequently harvested daily for six consecutive days. Cell viability was also assayed by crystal violet staining. Briefly, cells (150,000 cells per well in 6-well plates) were seeded and transduced as indicated above. Cells were then fixed in 4% formaldehyde, washed and subsequently stained with 0.1% crystal violet. Quantitation was performed by extracting the crystal violet dye with 10% acetic acid and measuring the absorbance at 590 nm on a SpectraMax M2 Microplate reader (Molecular Devices). The anchorage-independent growth of tumor cells was assayed by their ability to form colonies in agar plates. Soft agar assays were performed in triplicate in six-well plates. Ten thousand cells per well were seeded in RPMI containing 0.4% low-melting agarose on bottom agar containing 0.7% low-melting agarose in RPMI medium. After 15 days, colonies were stained with iodonitrotetrazolium chloride and counted. Finally, for the colony assay, 1,000 cells were plated in 6 well plates and after 24 h, cells were treated with vehicle (1% DMSO) or compound CSRM617 at 10, 20 and 40μM. Two weeks later, cells were fixed with 4% paraformaldehyde for 10 min at room temperature, and then stained with crystal violet for 15 min. Wells were scanned and the total area occupied by the colonies was quantified by ImageJ.
Dose response curves and IC50.
Cells were plated at a density of 1,000 cells per well, in triplicate, in a 96 well plate. The next day, T0 reading was collected and cells were exposed to compound CSRM617 or vehicle (1% DMSO) at 2-fold concentration ranging from 100μM to 0.2μM. Viability was measured 48 hours post treatment using CellTiter-Glo Luminescent Cell Viability Assay (Promega). Dose response curves and IC50 values were generated using GraphPad Prism version 6 Software.
Luciferase assays.
For the PSA promoter assay, the pGL3 vector containing the androgen responsive 548-bp PSA promoter region (−541 to +7) preceded by 1450 bp of the PSA enhancer region (−5322 to −3873) (PSA-luc; provided by Dr. Gerhard Coetzee at University of Southern California) was used. LNCaP or 22Rv1 cells were co-transfected with this luciferase construct, the OC2 overexpression vector or empty vector and the pRL-SV40 vector (Promega) with Lipofectamine 2000. Six hours after transfection cells were washed, maintained overnight in phenol-free RPMI supplemented with 10% charcoal-stripped FBS (Gibco) and treated for 24 hours with 10nM DHT (Sigma). Luciferase activity was measured using the Dual-Luciferase Assay System (Promega).
Animal studies.
All experimental protocols and procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Cedars-Sinai Medical Center. All relevant ethical regulations, standards, and norms were rigorously adhered to. 4–5 week-old male CrTac:NCr-Foxn1nu nude mice and C.B-Igh-1b/GbmsTac-Prkdcscid-Lystbg N7 SCID/Beige mice were purchased from Taconic and maintained under specific pathogen-free conditions. For the subcutaneous experiments, 750,000 cells were resuspended in a total volume of 50μL of PBS containing Matrigel (1:1 v/v; BD Bioscience) and injected into the flanks of nude mice. Tumor volume was measured and calculated using the formula V = 1/2 × length × width2. For the OC2 inhibitor treatment study with the CSRM617 compound, mice bearing 22Rv1 tumors with a mean volume of 200mm3 were randomly divided into vehicle control (2.5% DMSO in PBS) or compound CSRM617 (50 mg/Kg) groups and subjected to intraperitoneal injection daily. For the metastasis experiments, luciferase-tagged 22Rv1 cells (1×106 cells) were injected intracardially into 6–8 week-old SCID/Beige mice. Bioluminescence imaging was performed weekly to monitor tumor metastasis using a Xenogen IVIS Spectrum Imaging System (PerkinElmer). At the end of the animal studies, tumor tissues were harvested from euthanized mice and fixed with 4% formaldehyde for histological analysis.
Immunohistochemistry (IHC) and immunofluorescence.
Supplementary Fig. 7d shows a representative image of OC2 staining with the anti-OC2 Abcam antibody in PC tissue. TMAs were: 1) a prostate cancer TMA from the Cedars-Sinai Biobank (Fig. 1e,f and Fig. 2a–c); 2) TMA-83 comprised of mCRPC cases from the rapid autopsy program at the University of Washington32 (Fig. 4d,e; and Supplementary Fig. 4c); and 3) LuCaP xenograft TMAs-78.1 and −89A from the University of Washington33 (Fig. 4f,g; and Supplementary Fig. 4d). Each core in the TMA 83, 78.1 and 89A was annotated for presence or absence of expression of synaptophysin (SYP) and AR using immunohistochemistry and microscopic evaluation. Prior to analysis of primary prostate cancers in the TMA from Cedars-Sinai, cancer and normal glands were manually outlined by a pathologist (B.S.K.). DAPI or cytokeratin was used to distinguish nuclear and cytoplasmic compartments in cancer cells. Intensity levels of OC2 were determined in the nucleus and cytoplasm of cancer cells by averaging staining intensities across all cancer cells in a TMA core. The background intensity in the stroma was subtracted in cases of primary prostate cancer. Wilcoxon rank-sum test was used to calculate the significance of differences in staining intensities between groups (normal, G3, G4).
For workup and validation of OC2 antibodies tissue sections of pancreas, gallbladder and large cell NE carcinoma were used. Each staining experiment was controlled by including a positive and negative control slide. Results were evaluated by comparison to positive and negative controls. Staining results in positive and negative controls were consistent across multiple staining experiments. The specificity of the OC2 antibodies ((Abcam ab28466, LS-Bio LS-C499462 and Sigma HPA057058 used by the Human Protein Atlas consortium) was confirmed by staining of consecutive tissue sections from the same block with all three antibodies. Staining patterns in prostate cancer, gallbladder and large cell NE cancer were similar between the three antibodies (data not shown). Immunofluorescent detection of OC2 was carried out with the OPAL 4-color kit (Perkin-Elmer) or with the Ventana Discovery Cy5 reagent kit (Ventana Medical Systems). TMA slides were first blocked with 3% H2O2 for 10 min, then treated with animal-free protein blocker (Vector Laboratories) for 15 min, and then incubated over-night at 4°C with the anti-OC2 primary antibody diluted in Antibody Dilution Buffer (Ventana Medical Systems). The next day, the TMA slide was incubated with EnVision+ System – HRP labeled polymer goat anti-rabbit secondary antibody (Dako) for 30 min at room temperature followed by incubation with OPAL-Cy3 fluorophore for 10 min, or alternatively with the Cy5 labeling kit. For slides labeled with Cy5, the cytokeratin 8/18 antibody cocktail was used to identify the cytoplasm of cancer cells. Slides were stained with DAPI and covered by Prolong Gold antifade solution prior to coverslipping for imaging. The TMA slide was imaged and digitized using the TissueFAX fluorescent slide scanner (TissueGnostics).
For double IHC with AR and OC2 antibodies, slides were first stained with OC2 as described above, except that Tyramide amplification was used together with ImmPACT DAB Peroxidase (HRP) and Red Alkaline Phosphatase (Red AP) chromogens (Vector Laboratories). Slides were first stained for OC2. Next, the OC2 antibodies were denatured and stripped off the slide using the Biocare Decloaker with pH6 Citrate buffer. Slides were treated with antigen retrieval solution pH 8, blocked and incubated with anti-AR antibody (#5153, clone D6F11 Cell Signaling) at 1:500 dilution. After signal amplification and development of the red chromogen, slides were stained with hematoxylin and surfaced by a cover slip. Digital images of fields of view were captured by the Vectra-II slide scanner (PerkinElmer). Cancer areas in digital images of individual TMA cores were annotated by a pathologist (B.S.K). Nuclear outlines were determined based on hematoxylin staining. AR and OC2 intensities in nuclear areas were measured. Intensity values of AR and OC2 were log2 transformed. An unsupervised hierarchical clustering of cells was performed based on AR and OC2 intensity values using Euclidean distance and complete linkage methods.
To measure expression of PEG10, tissues on glass slides were stained with the PEG10 antibody (Sigma HPA051038). Slides underwent heat retrieval with CC1 buffer (Ventana) for 40 min. and 3% H2O2 for 10 min. After blocking nonspecific binding with animal-free protein blocker for 15 min and with avidin/biotin block for 5 min, slides were incubated with the PEG10 antibody at 1:1000 dilution for 32 min at 37°C. The antibody signal was amplified as described above with tyramide and antibody binding was visualized using the VECTOR-ImmPACT DAB Peroxidase (HRP) substrate kit. Slides were scanned on the Aperio Slide Scanner (Leica Biosystems) and the average staining intensity was determined using the software of the instrument (Aperio ImageScope).
Computer model of OC2 and drug discovery.
Three-dimensional structure of human ONECUT2 (Uniprot: O95948) is not available and hence a structural model was built using homology modeling. OC2 consists of two DNA binding domains, a CUT domain and a homeodomain (HOX) near the C-terminus. Given the high sequence homology (90%) between rat and human OC1/Oc1 and OC2/Oc2, the homeodomain from the structure of rat Oc1 (PDB code: 2D5V)54 was identified as a suitable structural template for inhibitor design. An initial structural model for human OC2 homeobox domain was subsequently generated using SWISS-MODEL55,56. Prime module in Schrodinger (Schrodinger, Inc.) was additionally used to find an optimal conformation of the missing residues in the crystal structure. The final structural model was obtained after a cycle of minimization followed by a short 1.2ns molecular simulation. Putative binding site of small molecules was identified with site score 0.569 9 (Dscore 0.398). Subsequently, the model was subjected to an extended molecular simulation (2.4 ns) with and without DNA. Models were averaged over 0.5ns and the sitemap score was reevaluated for each ensemble. Two models with site score 0.9 was used for virtual screening using Gide using XP mode. For virtual screening chemicals from Hit2Lead (Chembridge Corporation) were prepared using ligprep and used to identify hits. Figures were produced using Maestro57 and PyMol (The PyMOL Molecular Graphics System, Version 1.8.2015). Using the three-dimensional model of OC2-HOX domain, we conducted a virtual screen of a chemical library consisting of 500,0000 small molecules (Chembridge). Stereochemistry of CSRM617 was not considered because it was not custom-synthesized and therefore the preparations we used are a racemic mixture.
IUPAC name for CSRM617:
1-{(E)-[(2,3,4-Trihydroxyphenyl)methylidene]hydrazino}−2-amino-3-hydroxy-1-propanone.
Surface plasmon resonance binding studies.
To measure binding features of CSRM617, the OC2-HOX domain was recombinantly produced. The purified OC2-HOX domain protein was first verified using a DNA probe corresponding to the experimentally determined OC2 binding motif in the PEG10 promoter ((F) 5’-TAGATCGATTTGCA-3’ and (R) 5’-TGCAAATCGATCTA-3’). Briefly, 5 ng of biotinylated dsDNA PEG10 probe was immobilized on a BioCap sensor chip (Pall ForteBio) in PBST buffer (pH 6.4) to a response unit of 300 RU. OC2-HOX was analyzed at 62.5 nM concentration. Next, binding of CSRM617 was measured. Briefly, 5μg/mL purified His-tag OC2 HOX binding domain was immobilized on a HisCap chip (Pall ForteBio) to 20,000 RU in PBST buffer (pH 6.4). CSRM617 was analyzed at serial doubling concentrations maximized at 100 μM. The sensorgram was fit using a 2:2 model (i.e. to a simple 2-site binding model) using QDAT (SensiQ, Inc.) and to calculate KD.
SPR competition assay.
To examine whether CSRM617 can block DNA binding to OC2-HOX, biotinylated dsDNA containing the PEG10 motif described above was immobilized on BioCap sensor chip in PBST buffer containing 5% DMSO (pH 6.4) to a response unit of 300 RU. OC2-HOX binding to the DNA was analyzed at two different protein concentrations of 250nM (blue) and 125nM (red), in the absence or presence of 100μM CSRM617.
OC2-HOX protein purification.
Recombinant OC2-HOX domain (L330-W485) of OC2 was produced using a bacterial expression system. Briefly, OC2-HOX was sub-cloned into a pET-His6-TEV_LIC vector (1B, Addgene, plasmid #29653) to include an N-terminal His-tag. The recombinant plasmid was transformed into Rosetta ™ (DE3) competent cells (Novagen) and OC2-HOX was expressed at 18°C overnight in Terrific Broth. Further purification was performed by Ni-NTA affinity chromatography using a HisTrap FF Crude column (GE Healthcare Life Sciences) and size exclusion chromatography using a Superdex 200 increase 10/300 GL column (GE Healthcare Life Sciences) to yield >95% pure protein.
Statistics and Reproducibility.
Statistically significant data in in vitro and in vivo assays were assessed by unpaired two-tailed Student’s t-test or Wilcoxson two-tailed rank sum test, unless otherwise noted. Tests for difference in the case of more than two groups were performed using one-way analysis of variance (ANOVA) where applicable using the Dunnett’s post-hoc test, unless otherwise indicated. Friedman’s test was performed to compute significance of time dependent changes by treatment. A permutation test strategy was used to determine the significance of overlap between OC2 target genes, where OC2 binds gene promoters, and total TF genes across the entire human genome. A total of 10,000 randomly selected genes were used to compute the empirical P value of the overlapping genes. We used the GraphPad Prism (Version 6), the MATLAB package including the Statistics toolbox (Mathworks, Natick, MA, USA), the R package (v.3.1 http://www.r-project.org/) for all statistical tests. Network visualization was implemented using Cytoscape (v.3.4 http://www.cytoscape.org/) with enhancedGraphics application.
Data Availability
Data generated or analyzed during this study are included in this published article (and its supplementary information files). ChIP-seq and microarray data generated in this study were deposited in GEO (GSE97551, GSE97548, GSE97549). The DISC cohort data are available at www.thepcta.org.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by Department of Defense W81XWH-16-1-0567 (M.R.F); 1R01CA143777 (M.R.F.); 1R01DK087806 (M.R.F.); Prostate Cancer Foundation Challenge Grant 17CHAL04 (I.P.G.); Spielberg Discovery Fund in Prostate Cancer Research (B.S.K. and M.R.F.); 2P01CA098912 (L.W.K.C.); Jean Perkins Foundation (I.P.G.); Movember/PCR GAP1 Unique TMAs Project (I.P.G.); DOD PC131996 (I.P.G.); DOD PC130244 (I.P.G.); NIH/NCI U54 CA143931 (I.P.G.); DOD W81XWH-14-1-0152 (S.Y.); Urology Care Foundation Research Scholar Award and Chesapeake Urology Associates Research Scholar Fund (M.R.); 2018 Donna and Jesse Garber Award for Cancer Research, Samuel Oschin Comprehensive Cancer Institute (SOCCI) CedarsSinai Medical Center (M.R.); Urology Care Foundation Research Scholars Program of AUA Western Section Research Scholar Fund (S.Y.). The authors are very appreciative of technical and conceptual contributions from N. Bhowmick, R. Matusik, V. Placencio, K. Kelly, M. Beshiri, W.R. Wiedemeyer, P.J. Aspuria, C. Spinelli and the Biobank & Translational Research Core at Cedars-Sinai Medical Center.
Footnotes
REFERENCES
- 1.Beltran H, et al. Aggressive variants of castration-resistant prostate cancer. Clin Cancer Res 20, 2846–2850 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gundem G, et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang KH, et al. Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proc Natl Acad Sci U S A 108, 13728–13733 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma NL, et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell 23, 35–47 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Kumar A, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 22, 369–378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niu Y, et al. Androgen receptor is a tumor suppressor and proliferator in prostate cancer. Proc Natl Acad Sci U S A 105, 12182–12187 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espana A & Clotman F Onecut factors control development of the Locus Coeruleus and of the mesencephalic trigeminal nucleus. Mol Cell Neurosci 50, 93–102 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Jacquemin P, Lannoy VJ, Rousseau GG & Lemaigre FP OC-2, a novel mammalian member of the ONECUT class of homeodomain transcription factors whose function in liver partially overlaps with that of hepatocyte nuclear factor-6. J Biol Chem 274, 2665–2671 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Vanhorenbeeck V, et al. Role of the Onecut transcription factors in pancreas morphogenesis and in pancreatic and enteric endocrine differentiation. Dev Biol 305, 685–694 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Leyten GH, et al. Identification of a Candidate Gene Panel for the Early Diagnosis of Prostate Cancer. Clin Cancer Res 21, 3061–3070 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Guo H, et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat Genet 48, 1142–1150 (2016). [DOI] [PubMed] [Google Scholar]
- 12.You S, et al. Integrated Classification of Prostate Cancer Reveals a Novel Luminal Subtype with Poor Outcome. Cancer Res 76, 4948–4958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu K, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338, 1465–1469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine DM, et al. Pathway and gene-set activation measurement from mRNA expression data: the tissue distribution of human pathways. Genome Biol 7, R93 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Antonio JM, Ma C, Monzon FA & Pflug BR Longitudinal analysis of androgen deprivation of prostate cancer cells identifies pathways to androgen independence. Prostate 68, 698–714 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Mulholland DJ, et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res 72, 1878–1889 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai C, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell 20, 457–471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu R, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res 69, 16–22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shukla S, et al. Aberrant Activation of a Gastrointestinal Transcriptional Circuit in Prostate Cancer Mediates Castration Resistance. Cancer Cell 32, 792–806 e797 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liberzon A, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffon A, et al. Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory landscape. Nucleic Acids Res 43, e27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cleutjens KB, et al. An androgen response element in a far upstream enhancer region is essential for high, androgen-regulated activity of the prostate-specific antigen promoter. Mol Endocrinol 11, 148–161 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Latham JP, Searle PF, Mautner V & James ND Prostate-specific antigen promoter/enhancer driven gene therapy for prostate cancer: construction and testing of a tissue-specific adenovirus vector. Cancer Res 60, 334–341 (2000). [PubMed] [Google Scholar]
- 24.Jin HJ, Zhao JC, Wu L, Kim J & Yu J Cooperativity and equilibrium with FOXA1 define the androgen receptor transcriptional program. Nat Commun 5, 3972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, et al. FOXA1 inhibits prostate cancer neuroendocrine differentiation. Oncogene 36, 4072–4080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beltran H, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 22, 298–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grozinsky-Glasberg S, Shimon I & Rubinfeld H The role of cell lines in the study of neuroendocrine tumors. Neuroendocrinology 96, 173–187 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Shiau CK, Gu DL, Chen CF, Lin CH & Jou YS IGRhCellID: integrated genomic resources of human cell lines for identification. Nucleic Acids Res 39, D520–524 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong C, Vosburgh E, Levine AJ, Cong L & Xu EY Human neuroendocrine tumor cell lines as a three-dimensional model for the study of human neuroendocrine tumor therapy. J Vis Exp, e4218 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parimi V, Goyal R, Poropatich K & Yang XJ Neuroendocrine differentiation of prostate cancer: a review. Am J Clin Exp Urol 2, 273–285 (2014). [PMC free article] [PubMed] [Google Scholar]
- 32.Roudier MP, et al. Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum Pathol 34, 646–653 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Nguyen HM, et al. LuCaP Prostate Cancer Patient-Derived Xenografts Reflect the Molecular Heterogeneity of Advanced Disease and Serve as Models for Evaluating Cancer Therapeutics. Prostate 77, 654–671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lapuk AV, et al. From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer. J Pathol 227, 286–297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akamatsu S, et al. The Placental Gene PEG10 Promotes Progression of Neuroendocrine Prostate Cancer. Cell Rep 12, 922–936 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Gao D, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyatt AW, et al. Heterogeneity in the inter-tumor transcriptome of high risk prostate cancer. Genome Biol 15, 426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson D, et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Epstein JI, et al. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am J Surg Pathol 38, 756–767 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou M, et al. Transdifferentiation as a Mechanism of Treatment Resistance in a Mouse Model of Castration-resistant Prostate Cancer. Cancer Discov 7, 736–749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Y, et al. MiR-429 inhibits cells growth and invasion and regulates EMT-related marker genes by targeting Onecut2 in colorectal carcinoma. Mol Cell Biochem 390, 19–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 43.Bolstad BM, Irizarry RA, Astrand M & Speed TP A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Hwang D, et al. A data integration methodology for systems biology. Proc Natl Acad Sci U S A 102, 17296–17301 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Storey JD & Tibshirani R Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100, 9440–9445 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kharchenko PV, Tolstorukov MY & Park PJ Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26, 1351–1359 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kharchenko PV, Tolstorukov MY & Park PJ Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26, 1351–1359 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramirez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu G, Wang LG & He QY ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Yu G, Wang LG, Yan GR & He QY DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 31, 608–609 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37, W202–W208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rotinen M, et al. Estradiol induces type 8 17beta-hydroxysteroid dehydrogenase expression: crosstalk between estrogen receptor alpha and C/EBPbeta. J Endocrinol 200, 85–92 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Iyaguchi D, Yao M, Watanabe N, Nishihira J & Tanaka I DNA recognition mechanism of the ONECUT homeodomain of transcription factor HNF-6. Structure 15, 75–83 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Arnold K, Bordoli L, Kopp J & Schwede T The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research 42, W252–W258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morin A, et al. Collaboration gets the most out of software. Elife 2 :e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated or analyzed during this study are included in this published article (and its supplementary information files). ChIP-seq and microarray data generated in this study were deposited in GEO (GSE97551, GSE97548, GSE97549). The DISC cohort data are available at www.thepcta.org.