Abstract
T follicular helper (TFH) cells are crucial for effective humoral immunity by providing the required signals to cognate B cells and promoting germinal center (GC) formation. Many intrinsic and extrinsic factors have been reported to be involved in the multistage, multifactorial differentiation process of TFH cells. By comparing gene expression between TFH cells and TH1 cells based on published GEO data, we found selective and high expression of sclerostin domain-containing protein 1 (SOSTDC1) in TFH cells but not in TH1 cells; however, it is unclear whether SOSTDC1 is important for the differentiation and/or function of TFH cells. Using a mouse model of acute lymphocytic choriomeningitis virus (LCMV) infection, we confirmed the selective expression of SOSTDC1 in TFH cells compared to that in TH1 cells, but the ablation of SOSTDC1 did not affect TFH cell differentiation or effector function. Thus, our results indicate that the SOSTDC1 protein is merely a specific marker of TFH cells but does not play a functional role in the differentiation of TFH cells during acute viral infection.
Keywords: Acute infection, TFH cell, SOSTDC1
Introduction
CD4+ T cells initiate a multi-step transcriptional program when viral pathogens are recognized, thereby directing the differentiation of CD4+ T cells into TH1 and follicular helper T (TFH) cells that coordinate to clear the viral infection [1,2]. TH1 cells depend on the transcription factor T-bet for lineage-specific differentiation [3,4] and secrete inflammatory cytokines, primarily IFN-γ, to mediate effector immune function. However, the differentiation of TFH cells is mediated primarily by the lineage-determining transcription factor Bcl6 [5-7]. TFH cells highly express chemokine receptor CXCR5 to enable their homing to B cell follicles within secondary lymphoid tissues [8-12], where TFH cells interact with germinal center (GC) B cells and provide the essential signals to GC B cells for the ensuing development of high-affinity and long-lived plasma cells and memory B cells [13-15].
Apart from the master transcription factors, a network of cytokines also participates in controlling the differentiation of CD4+ T cells during acute viral infection. For example, IL-12, mainly produced by antigen-presenting cells, induces TH1 cell differentiation in a paracrine manner [16]. IFN-γ produced by TH1 cells through an autocrine mechanism can also promote TH1 cell differentiation via activating STAT1 [17-19]. Additionally, IL-4, which can be produced by TH2 cells, can promote TH2 cell differentiation through activating STAT6 in an autocrine signalling manner [20-22]. In addition, IL-2, mainly produced by TH1 cells, is important for the differentiation of TH2 cells in vitro through activating STAT5 in a paracrine manner [23,24]. For TH17 cells, TGF-β, with IL-6-mediated STAT3 activation, promotes TH17 cell differentiation [25,26]. Additionally, the IL-2-mTORC1 signalling axis orchestrates the reciprocal balance between the TH1 and TFH cell fates by promoting TH1 while inhibiting TFH cell differentiation. Similarly, TFH cells also regulate themselves and other cells through autocrine and paracrine manners. For example, IL-21 and IL-4 which were produced by TFH cells can contribute to GC formation, affinity maturation and immunoglobulin class switching in a paracrine manner. IL-21 can also promote TFH cell survival in an autocrine manner [27]. Whether any secretory proteins produced by TFH cells-in addition to the cytokines involved in TFH cell differentiation-could influence TFH cell differentiation and effector function remains largely unknown.
Sclerostin domain-containing protein 1 (SOSTDC1) is a secreted protein also known as Ectodin, USAG-1 and WISE. It is an important regulator of cell signalling and participates in various developmental processes, including hair follicle and trigeminal ganglion formation, limb morphogenesis and tooth development [28-31]. SOSTDC1 is also coupled with some disease processes, such as adult renal cancer, paediatric Wilms’ tumours, and breast and gastric cancer [32-35]. Its gene has been identified as a candidate tumour suppressor gene in these types of tumours [34]. SOSTDC1 participates in these physiological and pathological processes mainly due to the regulation of the bone morphogenetic protein (BMP) and Wnt signalling pathways [36,37]. BMP is an important ligand that can activate various pathways involved in cell differentiation and proliferation. Binding of BMP to BMP receptors can phosphorylate receptor-regulated Smad proteins (Smad-1, -5, and -8), which associate with Smad-4 and form the Smad complex [38,39]. Then, this complex enters the nucleus and, in turn, induces the transcription of an array of cell regulatory factors related to proliferation and differentiation, such as p53, p21 and Bcl2 [40,41]. In addition, SOSTDC1 can inhibit the interaction of BMP with its receptors by directly binding to BMP, thus limiting BMP activity [42]. The influence of SOSTDC1 on the Wnt signalling pathway has different manifestations [43]. SOSTDC1 can decrease Wnt signalling by blocking the binding of Wnt8 to LRP6 receptors [44]. Other reports have suggested that secretory SOSTDC1 exerts either inhibitory or activating effects, while the form localized in the endoplasmic reticulum (ER) is exclusively inhibitory [45].
Despite the profound effects of SOSTDC1 on organ development and tumour formation, whether SOSTDC1 regulates T cell immune responses is not clear. One study showed that SOSTDC1 expression is higher in TFH cells than in naïve CD4+ T cells on day 7 post sheep red blood cell (SRBC) immunization [46]. A previous study in our laboratory also suggested that the mRNA level of SOSTDC1 in TFH cells was significantly higher than that in TH1 cells on day 8 after lymphocytic choriomeningitis virus (LCMV) Armstrong infection. However, the expression level of SOSTDC1 in TCF-1-null TFH cells was dramatically decreased [2]. Despite the high transcription levels of SOSTDC1 in TFH cells, whether SOSTDC1 functions to regulate the differentiation and effector functions of TFH cells remains unknown.
Here, we first observed abundant SOSTDC1 expression at both the mRNA and protein levels in TFH cells relative to that in TH1 cells in a model of acute LCMV infection. Next, we used a conditional knockout system to investigate the putative regulation of SOSTDC1 on the TFH cell response to acute viral infection. Our results indicated that deletion of Sostdc1 specifically in CD4+ T cells did not affect the differentiation of TFH cells. In addition, SOSTDC1-deficient TFH cells also showed normal auxiliary function to B cells. In addition, we noted the normal immune responses of TH1, Foxp3+ Treg and follicular regulatory T (Tfr) cells in this model. Taken together, our findings demonstrate that SOSTDC1 serves as an indicator but not a regulator of TFH cell differentiation during acute viral infection.
Materials and methods
Mice, virus and immunization
Sostdc1 fl/fl mice were obtained from Cyagen Biosciences (Santa Clara, USA) and the PCR genotyping primers are: F: 5’-TGGCGTTCTTTCCTGGGTAGTGA-3’, R: 5’-TAGACATCTGTGTGAGCAATTCCAT-3’. Cd4-Cre transgenic and C57BL/6J (CD45.2+) mice were obtained from The Jackson Laboratory. SMARTA (CD45.1+) T cell receptor transgenic mice and the LCMV Armstrong strain were provided by R. Ahmed (Emory University). Sostdc1 fl/fl mice were bred with Cd4-Cre transgenic mice to generate Sostdc1 fl/fl Cd4-Cre mice. All mice were on a C57BL/6 background and housed under specific-pathogen free (SPF) conditions. Mice were infected with 2 × 105 plaque-forming units (PFU) of LCMV Armstrong virus at 6-10 weeks of age, and both sexes were included without randomization or “blinding”. For all analyses, at least 3 animals of each group were matched for age and sex. All immunized mice were housed in accordance with institutional biosafety regulations of the Third Military Medical University. All mouse experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committees of the Third Military Medical University.
Flow cytometry and antibodies
Single-cell suspensions of spleen were prepared from mice infected with LCMV Armstrong virus. For surface staining for flow cytometry, cells were incubated with a saturating amount of monoclonal antibodies against CD4 (RM4-5, Biolegend; 1:200), CD8 (53-6.7, BD Biosciences; 1:200), CD44 (IM7, eBioscience; 1:100), ICOS (C398.4, Biolegend; 1:100), PD-1 (RMP1-30, eBioscience; 1:100), CTLA-4 (UC10-4B9, Biolegend; 1:50), Live/Dead (L10119, Life Technologies; 1:200), CD138 (281-2, BD Biosciences; 1:50), B220 (RA3-6B2, eBioscience; 1:200), PNA (FL-1071, Vector Labs; 1:100), FAS (JO2, BD Biosciences; 1:50), CD19 (6D5, Biolegend; 1:100), CD45.1 (A20, Biolegend; 1:100), and SLAM (TC15-12F12.2, Biolegend; 1:100). Staining was performed for 30 min at 4°C in phosphate-buffered saline (PBS) containing 2% fetal bovine serum (FBS, weight/volume). CXCR5 staining was performed as previously described [47]; cells were stained with purified rat anti-mouse CXCR5 (2G8, BD Bioscience; 1:100) antibody at 4°C for 1 h, followed by biotin-streptavidin-conjugated goat anti-rat IgG (112-065-143, Jackson Immunoresearch; 1:300) on ice for 30 min. Finally, cells were washed and stained with streptavidin (25-4317-82, eBioscience; 1:200) and other surface antibodies on ice for 30 min in PBS supplemented with 2% FBS, 1% bovine serum albumin (BSA) and 2% normal mouse serum. Staining of intranuclear Bcl6 (K112-91, BD Bioscience; 1:50), TCF-1 (C46C7, Cell Signalling Technology; 1:400) and Foxp3 (FJK-16s, eBiosciences; 1:100) was performed with Foxp3/Transcription Factor Staining Buffer (005523; eBioscience). Stained cells were evaluated by FACS Canto II flow cytometry (BD Bioscience), and the flow cytometry data were analysed with FlowJo software (Tree Star).
Adoptive cell transfer and cell sorting
A total of 2 × 104 naïve SMARTA (CD45.1+) cells were adoptively transferred into naïve C57BL/6J (CD45.2+) recipient mice, which were infected intraperitoneally with 2 × 105 PFU of LCMV Armstrong virus the next day. To isolate CD4+ T cells, lymphocytes isolated from naïve and SMARTA-transferred mice on day 8 post LCMV Armstrong infection were subjected to lineage depletion using biotin-conjugated antibodies (anti-B220 (RA3-6B2), anti-CD11c (N418), anti-CD11b (M1/70), anti-TER119 (TER119), anti-NK1.1 (PK136), and anti-CD8 (53-6.7) for sorting CD4+ T cells, Biolegend) coupled with Beaver Beads Mag500 Streptavidin Matrix (22302; Beaver). The naïve CD4+ T cells (CD4+CD44-CD62L+), TFH cells (CD4+CD44+ SLAM-CXCR5+), and TH1 cells (CD4+CD44+CXCR5-SLAM+) were sorted by a FACS Aria II cell sorter (BD Biosciences).
Retroviral transduction
The Sostdc1 coding sequences were inserted into MIGR1 (MSCV-IRES-GFP) vectors as previously reported [2]. Retroviral vectors were transfected into 293T cells along with the pCLeco plasmid to generate recombinant retrovirus in culture supernatants. SMARTA cells were activated by intravenous injection of 200 µg of GP61-77 peptide. Eighteen hours later, pre-activated SMARTA cells were purified by biotin-conjugated antibodies coupled with Beaver Beads as described above. Then, the SMARTA cells were “spin-infected” with freshly harvested retrovirus supernatants for 90 minutes at 37°C by centrifugation (2100 rpm) in the presence of 20 ng/ml IL-2 (130-098-221, Miltenyi Biotec) and 8 ug/ml polybrene (H9268, Sigma-Aldrich). Retrovirus-transduced SMARTA cells were transferred into recipient mice, which were infected with LCMV Armstrong the next day.
Western blotting
A total of 5 × 105 cells were washed twice in pre-cooled PBS and lysed in RIPA buffer (Thermo Scientific) containing phenylmethanesulfonyl fluoride and protease inhibitor cocktail (Cell Signaling Technology). Protein lysates were run on 12% SDS-PAGE gels (Beyotime) and transferred to polyvinylidene difluoride membranes (Millipore) after electrophoresis. Membranes were blocked in PBS supplemented with 0.1% Tween 20 and 5% BSA for 2 h. Membranes were then incubated with anti-SOSTDC1 (PA5-72000, Thermo Scientific; 1:1000) or anti-β-actin (8H10D10, Cell Signaling Technology; 1:1000) primary antibodies at 4°C overnight, followed by a 2 h incubation with HRP-conjugated anti-rabbit IgG antibody (124398, Jackson ImmunoResearch). Finally, ECL (Beyotime) was used to visualize proteins.
Quantitative RT-PCR
To compare the Sostdc1 gene expression levels in TFH cells from WT and Sostdc1fl/fl Cd4-Cre mice on day 8 post LCMV Armstrong infection, CD4+CD25-GITR-CD44+CXCR5+ TFH cells were sorted and lysed in TRIzol LS reagent (10296; Life Technologies). Total RNA was extracted and reverse transcribed with a RevertAid H Minus First-Strand cDNA Synthesis Kit (K1632; Thermo Scientific). The expression levels were measured using a QuantiNova SYBR Green PCR Kit (208054; Qiagen) on a CFX96 Touch Real-Time System (Bio-Rad). The sequences of the Sostdc1 primer pairs are as follows: Sostdc1 Forward, 5’-GAGGCAGGCATTTCAGTAGC-3’ and Sostdc1 Reverse, 5’-GTATTTGGTGGACCGCAGTT-3’. The β-actin expression level was calculated for normalization.
Immunohistochemistry
Fresh spleens from WT and Sostdc1fl/fl Cd4-Cre mice on day 8 post LCMV Armstrong infection were harvested and soaked in OCT, then snap-freeze it on liquid nitrogen. Sections 5-10 μm in thickness were cut with Leica Cryostat, mounted on Superfrost Plus glass slides. Before staining, thaw the slides at room temperature and fix in cold acetone for 10 min. Dehydrate slides in PBS (containing with 1% BSA) for 10 min, then PBS rinse 4 times. Block slides with blocking buffer (PBS + 5% BSA) for 40 min in wet chamber. Sections were then stained with PE-conjugated CD4 (RM4-5; BD Biosciences; 1:50), Alexa Fluor 647-conjugated IgD (11-26c. 2a; eBioscience; 1:50) and FITC-conjugated GL7 (GL7; BD Biosciences; 1:50) antibodies. Rinse slides in PBS for 4 times, and briefly rinse sections in ddH2O and wait until almost dry. Finally, coverslips were mounted on slides with the Prolong Antifade Kit (P-7481; Life Technologies) and examined under Zeiss LSM 800 confocal fluorescence microscope. The images were analysed with ImageJ software.
Statistical analysis
Statistical analysis was conducted with Prism 6.0 software (GraphPad). An unpaired two-tailed non-parametric t-test with a 95% confidence interval was used to assess between-group differences. For the retroviral transduction assay, a paired two-tailed t-test with a 95% confidence interval was used to calculate the P values. Significance was defined as P < 0.05.
Results
SOSTDC1 expression is abundant in TFH cells but not in TH1 cells or naïve CD4+ T cells
Virus-specific naïve CD4+ T cells mainly differentiate into TFH and TH1 cells under conditions of acute viral infection [2,48,49]. From the published data sets (GEO accession code GSE65693), we found that Sostdc1 is one of the top 20 genes upregulated in TFH cells compared with TH1 cells and naïve cells (Figure 1A). In addition, similar results were reported in an article by Youn Soo Choi et al [50] (data not shown). To further confirm the expression of SOSTDC1 in these CD4+ T cell subsets, we adoptively transferred congenically marked (CD45.1+) naïve SMARTA transgenic CD4+ T cells (with a T cell antigen receptor specific for the epitope on LCMV glycoprotein (amino acids 66-77)) into wild-type C57BL/6J (CD45.2+) recipients, which were infected with LCMV Armstrong the next day. On day 8 post infection, antigen-specific TFH cells and TH1 cells were sorted and analysed with naïve SMARTA cells. Similarly, TFH cells expressed higher SOSTDC1 expression at both the mRNA and protein levels than naïve CD4+ T cells and TH1 cells, as shown by the reverse transcription quantitative PCR (RT-PCR) assay and immunoblot analysis, respectively (Figure 1B, 1C). Taken together, these results demonstrate that SOSTDC1 was selectively expressed in TFH cells compared with TH1 cells following LCMV Armstrong infection.
Figure 1.
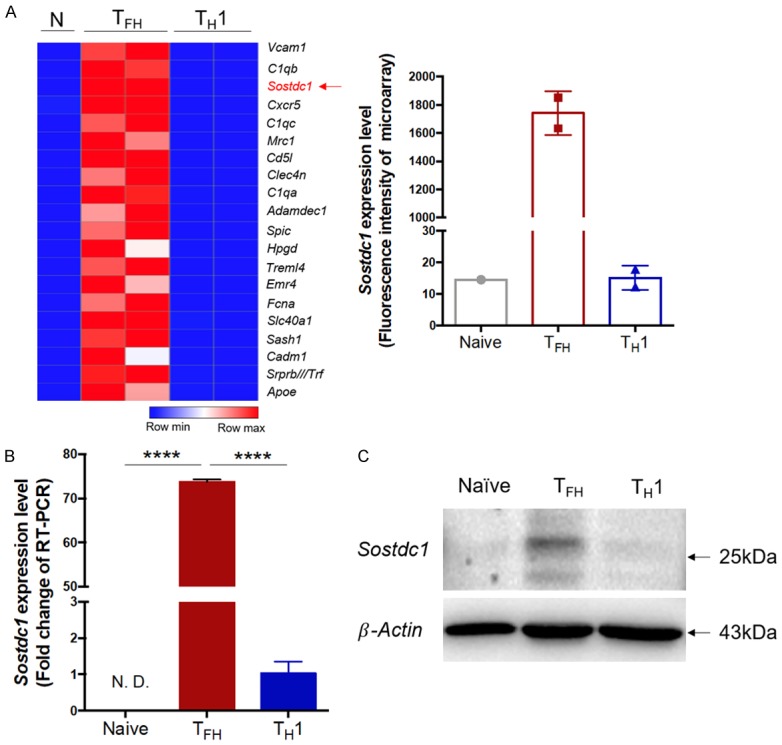
SOSTDC1 expression is abundant in TFH cells but not in TH1 cells or naïve CD4+ T cells. Naïve SMARTA (CD45.1+) cells were transferred into C57BL/6J (CD45.2+) mice followed by LCMV Armstrong infection the next day. On day 8 after infection, CD45.1+ TFH (CD4+CD44+SLAM-CXCR5+) and TH1 (CD4+CD44+SLAM+CXCR5-) cells were sorted and analysed with naïve SMARTA cells (CD4+CD62L+CD44-). A. Heat map of the top 20 genes with higher expression in TFH cells than in TH1 cells and naïve cells (left). The fluorescence intensity of the Sostdc1 probe in TFH, TH1 and naïve cells (right) from previous microarray data from our laboratory (GEO accession code GSE65693). B. Quantitative RT-PCR analysis of Sostdc1 gene expression in TFH cells and TH1 cells in the spleen of wild-type (CD45.2+) mice adoptively transferred with SMARTA cells (CD45.1+) on day 8 post infection with LCMV Armstrong and in naïve CD4+ T cells (CD62L+CD44-). C. Immunoblot analysis of SOSTDC1 and β-actin (loading control) expression in naïve, TFH and TH1 cells. N. D., not detected. ****P < 0.0001 (unpaired two-tailed t-test; the error bars in A and B indicate the standard errors).
SOSTDC1 is not required for TFH cell differentiation during acute viral infection
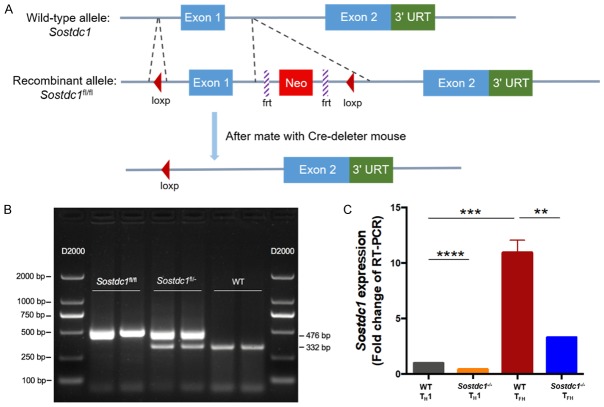
The high expression level of SOSTDC1 in TFH cells prompted us to investigate whether SOSTDC1 plays a functional role in the differentiation of TFH cells. We generated Sostdc1 fl/fl Cd4-Cre mice (called “Sostdc1 -/- mice” here) by crossing Sostdc1 fl/fl mice (in which the loxP sites were knocked in in the flanking regions of the first exon of the Sostdc1 alleles, Figure 2A) with Cd4-Cre mice (which had transgenic expression of Cre recombinase from the T cell-specific Cd4 promoter). The genotyping data indicated the expected genotypes (Figure 2B). To assess the knockout efficiency in the Sostdc1 -/- mice, on day 8 after LCMV Armstrong infection, we sorted the TFH and TH1 cells from both the WT and Sostdc1 -/- groups and found that Sostdc1 RNA expression was apparently abrogated in the TFH cells from Sostdc1 -/- mice (Figure 2C).
Figure 2.
The generation and validation of Sostdc1 fl/fl Cd4-Cre mice. (A) Schematic map showing the generation of Sostdc1 fl/fl mice. The PCR genotyping results for the Sostdc1 fl/fl, Sostdc1 fl/- and Sostdc1 -/- (WT) mice combined with marker (D2000) are shown in (B). (C) Expression of SOSTDC1 in TFH and TH1 cells in spleens from Sostdc1 fl/fl Cd4-Cre (Sostdc1 -/-) and wild-type (WT) mice 8 days after infection with LCMV Armstrong. The bars in C represent the standard errors. The p value in (C) was calculated by an unpaired t-test; **P < 0.01, ***P < 0.001, and ****P < 0.0001. The data are representative of the results of three independent experiments with at least three or four mice per group.
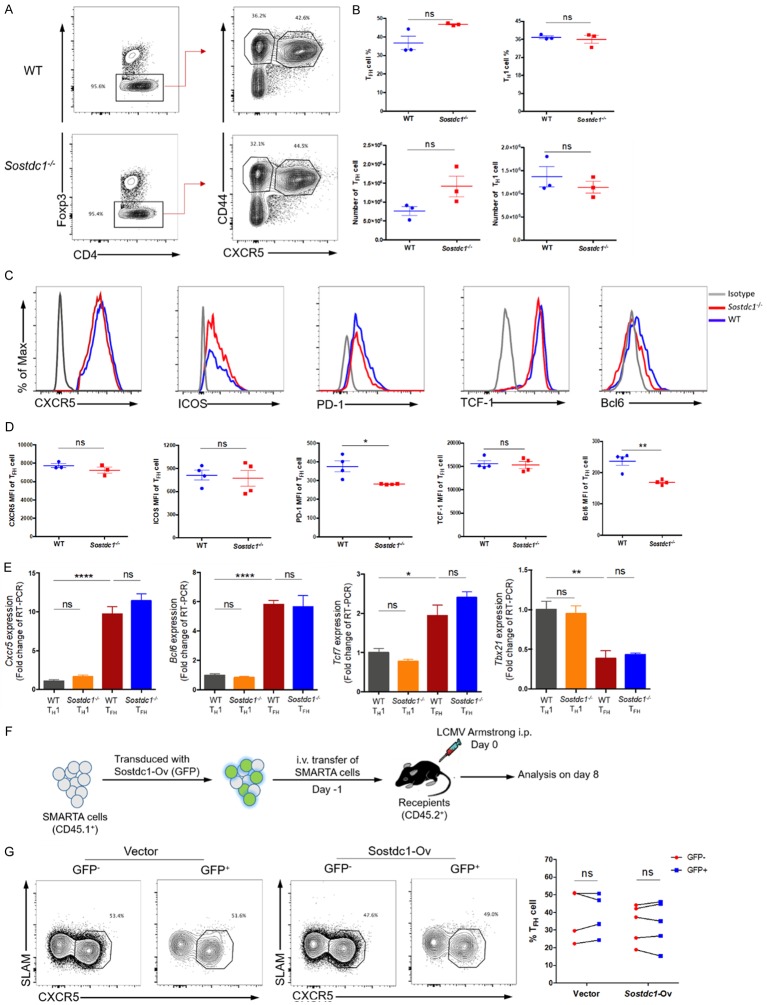
On day 8 after infection with LCMV Armstrong, we observed a comparable frequency and number of TFH cells (CD4+Foxp3-CD44+CXCR5+) and TH1 cells (CD4+Foxp3-CD44+CXCR5-) in Sostdc1 -/- and WT mice (Figure 3A, 3B). In addition, we assessed the expression of TFH cell-associated molecules and found that except for the expression of PD-1 and Bcl6, which was decreased in Sostdc1 -/- mice, the expression of other molecules, such as CXCR5, ICOS and TCF-1 was not significantly different between the Sostdc1 -/- and WT groups (Figure 3C, 3D). We further confirmed this phenotype by RT-PCR and found that the RNA expression levels of Cxcr5, Bcl6, Tcf7 and Tbx21 in TFH and TH1 cells were comparable between the WT and Sostdc1 -/- groups (Figure 3E). Together, these data demonstrate that the absence of SOSTDC1 did not affect the differentiation of TFH cells. Next, we wondered whether SOSTDC1 overexpression would influence the commitment of TFH cells in response to acute viral infection. To this end, we used a retroviral system to greatly enhance SOSTDC1 expression in activated SMARTA cells, which were then transferred into recipient mice and infected with the LCMV Armstrong strain the next day (Figure 3F). The assessment of SLAM-CXCR5+ TFH cells on day 8 post infection showed no difference between the GFP- and GFP+ SMARTA cell populations in the SOSTDC1 overexpression group (Figure 3G). Taken together, these data reveal that SOSTDC1 was not required for the fate of TFH cell differentiation during acute viral infection.
Figure 3.
SOSTDC1 is not required for TFH cell differentiation during acute viral infection. (A) Flow cytometry of CD4+ T cells in the spleens of either WT mice (WT) or Sostdc1 fl/fl Cd4-Cre mice (Sostdc1 -/-) on day 8 after LCMV Armstrong infection. The percentages of CD4+Foxp3- T cells (left), CD44+CXCR5+ T (TFH) cells and CD44+CXCR5- T (TH1) cells (right) in the WT (upper) and Sostdc1 -/- (lower) groups are indicated, and the statistical data are shown in (B). (C) Summary of CXCR5, ICOS, PD-1, TCF-1 and Bcl6 expression in TFH cells from the WT and Sostdc1 -/- groups calculated by subtracting the MFI of the isotype controls. (D) Statistical data for the expression levels of CXCR5, ICOS, PD-1, TCF-1 and Bcl6 in the TFH cells described in (A). (E) Quantitative RT-PCR analysis of Cxcr5, Bcl6, Tcf7 and Tbx21 gene expression in the TH1 and TFH cells described in A normalized to the corresponding expression levels in WT TH1 cells. (F) A brief summary of the experimental strategy is shown. SMARTA cells transduced with (GFP+) or not transduced (GFP-) with retrovirus expressing empty vector or a Sostdc1 overexpression plasmid were then transferred into recipients, which were subsequently infected with LCMV Armstrong the next day. (G) Flow cytometric analysis (left and middle) of the proportion of SLAM-CXCR5+ TFH cells in both GFP+ SMARTA cells and GFP- SMARTA cell populations on day 8 post infection as described in (F); the statistical data are shown to the right. The numbers adjacent to the outlined areas indicate the proportion of each cell type (A, G). The bars represent the standard errors (B, D, E). The p value was calculated by an unpaired t-test in (B, D, E) and by a paired t-test in (G); *P < 0.05, **P < 0.01 and ****P < 0.0001. ns, no significance. The bars in (B, D) and (E) represent the standard errors. The data are representative of the results of three independent experiments with at least three or four mice per group.
SOSTDC1-deficient TFH cells show normal effector functions
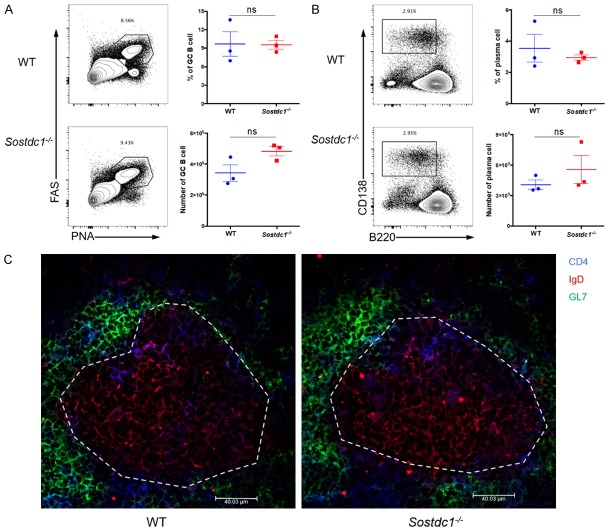
Although SOSTDC1 was not essential for TFH differentiation, it might be involved in helping B cell responses in a paracrine manner after secretion from TFH cells. Thus, we also investigated the GC reaction in Sostdc1 -/- and WT mice on day 8 after LCMV Armstrong infection. We observed a comparable frequency and number of GC B cells (characterized by high expression of FAS (CD95) and peanut agglutinin (PNA)) and plasma cells (CD138hiB220lo) in the Sostdc1 -/- and WT groups (Figure 4A, 4B). Furthermore, we performed an immunohistochemistry (IHC) experiment. The morphology of GCs in the Sostdc1 -/- and WT groups was not different (Figure 4C). Taken together, these results indicate that SOSTDC1 deficiency in TFH cells did not impair the effector function of these cells on the GC formation, which allows normal humoural immunity ability under conditions of acute viral infection.
Figure 4.
SOSTDC1-deficient TFH cells show normal effector functions. (A) Flow cytometric analysis of cells in the spleens of either WT mice (WT) or Sostdc1 fl/fl Cd4-Cre mice (Sostdc1 -/-) on day 8 after LCMV Armstrong infection. The percentages of PNAhiFAShi GC B cells (gate on CD19+B220+) are indicated for the WT (upper) and Sostdc1 -/- (lower) groups (left), and the statistical data are shown (right). (B) Flow cytometric analysis (left) and statistical data (right) of CD138hiB220lo plasma cells (gated on CD19+) are indicated for the WT (upper) and Sostdc1 -/- (lower) groups. (C) Immunohistochemical staining of the spleens to visualize germinal centers of mice in the WT (left) and Sostdc1 -/- (right) groups on day 8 after LCMV Armstrong infection, the blue represent for the CD4+ T cells, the red represent for the IgD+ B cells and the green represent for the GL7+ B cells, whiter dotted lines indicate the margin of GCs. The numbers adjacent to the outlined areas indicate the proportion of each cell type (A, B). The bars represent the standard errors (A, B). The p value was calculated by an unpaired t-test (A, B). ns, no significance. The data are representative of the results of three independent experiments with at least three mice per group.
Roles of SOSTDC1 in Foxp3+ Treg cells and Tfr cells
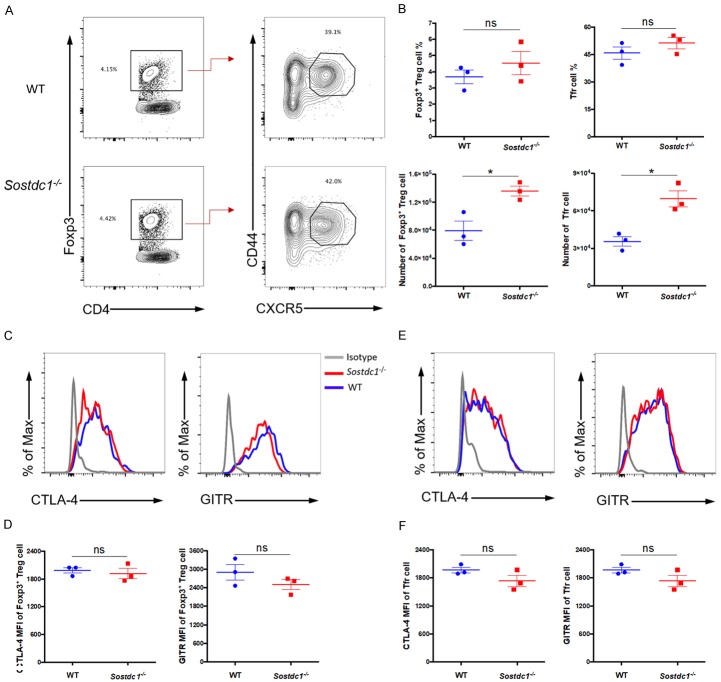
Follicular regulatory T (Tfr) cells are a unique subset of Foxp3+CD4+ regulatory T (Foxp3+ Treg) cells that suppress an excessive GC reaction by acting on both TFH cells and GC B cells. Previously published microarray data showed that Tfr cells express SOSTDC1 at higher levels than naïve CD4+ T cells [46]. Therefore, we speculated that SOSTDC1 may play a role in the differentiation of Tfr cells. To test this hypothesis, we compared the Foxp3+ Treg and Tfr populations between Sostdc1 -/- and WT mice on day 8 after LCMV Armstrong infection. We noted a comparable frequency of both Foxp3+ Treg cells (CD4+Foxp3+) and follicular regulatory T (Tfr) cells (CD4+Foxp3+CD44+CXCR5+) in the Sostdc1 -/- and WT groups, although the number of cells in these two populations was slightly increased in the Sostdc1 -/- group (Figure 5A, 5B). We also assessed the expression of the receptors CTLA-4 and GITR, which are functional markers of Foxp3+ Treg and Tfr cells. Notably, the mean fluorescence intensities (MFIs) corresponding to CTLA-4 and GITR expression did not differ between the Sostdc1 -/- and WT groups (Figure 5C-F). Taken together, these data suggest that similar to the findings in TFH cells, SOSTDC1 was dispensable for the fate of Tfr and Foxp3+ Treg cell differentiation during acute viral infection.
Figure 5.
Roles of SOSTDC1 in Foxp3+ Treg cells and Tfr cells. (A) Flow cytometric analysis of CD4+ T cells in the spleens of either WT mice (WT) or Sostdc1 fl/fl Cd4-Cre mice (Sostdc1 -/-) on day 8 after LCMV Armstrong infection. (B) The percentages and numbers of CD4+Foxp3+ T (Foxp3+ Treg) cells and CD4+Foxp3+CD44+CXCR5+ T (Tfr) cells in the WT and Sostdc1 -/- groups are indicated. Summary of CTLA-4 and GITR expression in Foxp3+ Treg cells (C) and Tfr cells (E) in the WT and Sostdc1 -/- groups as calculated by subtracting the MFI of the isotype controls. The statistical data for (C) and (E) are shown in (D) and (F), respectively. The numbers adjacent to the outlined areas indicate the proportion of each cell type (A). The bars represent the standard errors (B, D, F). The p value was calculated by an unpaired t-test (B, D, F); *P < 0.05. ns, no significance. The data are representative of the results of three independent experiments with at least three mice per group.
Discussion
In response to acute viral infection, virus-specific CD4+ T cells are programmed to differentiate into either TFH cells or TH1 cells [2,48,49]. SOSTDC1 was highly expressed in TFH cells but not in TH1 cells during this bifurcated differentiation process. Therefore, it is tempting to speculate that the selective expression of SOSTDC1 may promote the TFH cell response to acute viral infection. Given that SOSTDC1 is a secretory protein, it might favour TFH differentiation in an autocrine manner. However, in this study, we found that the differentiation of TFH cells seemed to be independent of SOSTDC1 expression in a mouse model of LCMV Armstrong infection. Additionally, we found the normal GC response in Sostdc1 -/- mice, which indicated that TFH-derived SOSTDC1 does not act on B cells in a paracrine manner. Although TFH-derived SOSTDC1 does not appear to regulate TFH differentiation and effector function, we propose that it may be involved in regulating other TFH-associated physiological activities.
Similar to the case in TFH cells, high expression of SOSTDC1 has been reported in Tfr cells [46]. Therefore, we also examined the role of SOSTDC1 in Foxp3+ Treg and Tfr cells in this study. We did not observe a notable phenotype in Sostdc1 -/- mice compared to WT mice. Combined with the results for TFH and Foxp3+ Treg, Tfr, and TH1 cells, this finding indicates that it is reasonable to conclude that SOSTDC1 expressed by TFH and Tfr cells does not play a functional role in guiding the lineage commitment of these cells during LCMV Armstrong infection.
Moreover, SOSTDC1 is reported to be involved in tissue development and tumourigenesis-both physiologically and pathologically-primarily by modulating the activity of the BMP and Wnt signalling pathways. For example, in breast cancer and adult renal cancer, SOSTDC1 expression is downregulated. This downregulation attenuates the inhibition of the BMP and Wnt signalling pathways, which results in suboptimal control of tumour cells [33,51-53]. Considering the high expression level of SOSTDC1 in TFH cells, TFH cells may also be involved in anti-tumour activities by producing copious amounts of SOSTDC1. However, this hypothesis requires further investigation.
In conclusion, in this study, we clarify the effect of the SOSTDC1 protein on TFH cell differentiation and effector functions under conditions of acute viral infection for the first time. These findings provide novel insight into our understanding of the molecular mechanisms underlying TFH cell differentiation.
Acknowledgements
This study was supported by grants from the National Key Research Development Plan (No.2016YFA0502202 to L. Ye), the National Natural Science Foundation of China (No.31825011 to L. Ye; No.31700774 to L. Xu).
Disclosure of conflict of interest
None.
References
- 1.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu L, Cao Y, Xie Z, Huang Q, Bai Q, Yang X, He R, Hao Y, Wang H, Zhao T, Fan Z, Qin A, Ye J, Zhou X, Ye L, Wu Y. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nat Immunol. 2015;16:991–999. doi: 10.1038/ni.3229. [DOI] [PubMed] [Google Scholar]
- 3.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 4.Espinosa V, Rivera A. Cytokines and the regulation of fungus-specific CD4 T cell differentiation. Cytokine. 2012;58:100–106. doi: 10.1016/j.cyto.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJ, Parish CR, Mackay CR, Vinuesa CG. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Lu H, Chen T, Nallaparaju KC, Yan X, Tanaka S, Ichiyama K, Zhang X, Zhang L, Wen X, Tian Q, Bian XW, Jin W, Wei L, Dong C. Genome-wide analysis identifies Bcl6-controlled regulatory networks during T follicular helper cell differentiation. Cell Rep. 2016;14:1735–1747. doi: 10.1016/j.celrep.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J Exp Med. 2001;193:1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ansel KM, McHeyzer-Williams LJ, Ngo VN, McHeyzer-Williams MG, Cyster JG. In vivo-activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. J Exp Med. 1999;190:1123–1134. doi: 10.1084/jem.190.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, Forster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haynes NM, Allen CD, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 12.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 13.Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunol Rev. 2012;247:52–63. doi: 10.1111/j.1600-065X.2012.01124.x. [DOI] [PubMed] [Google Scholar]
- 14.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, Nurieva RI, Yan X, Chen P, van der Flier LG, Nakatsukasa H, Neelapu SS, Chen W, Clevers H, Tian Q, Qi H, Wei L, Dong C. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 2014;507:513–518. doi: 10.1038/nature12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O’Garra A, Murphy KM. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 17.Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O’Shea JJ. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci U S A. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 19.Martin-Fontecha A, Thomsen LL, Brett S, Gerard C, Lipp M, Lanzavecchia A, Sallusto F. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat Immunol. 2004;5:1260–1265. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 21.Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, Chu C, Quelle FW, Nosaka T, Vignali DA, Doherty PC, Grosveld G, Paul WE, Ihle JN. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 22.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19:739–748. doi: 10.1016/s1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- 24.Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, Hu-Li J, Zhu J, Paul WE. Interleukin 2 plays a central role in Th2 differentiation. Proc Natl Acad Sci U S A. 2004;101:3880–3885. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 26.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Yu D, Vinuesa CG. The elusive identity of T follicular helper cells. Trends Immunol. 2010;31:377–383. doi: 10.1016/j.it.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Ahn Y, Sims C, Murray MJ, Kuhlmann PK, Fuentes-Antras J, Weatherbee SD, Krumlauf R. Multiple modes of Lrp4 function in modulation of Wnt/beta-catenin signaling during tooth development. Development. 2017;144:2824–2836. doi: 10.1242/dev.150680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narhi K, Tummers M, Ahtiainen L, Itoh N, Thesleff I, Mikkola ML. Sostdc1 defines the size and number of skin appendage placodes. Dev Biol. 2012;364:149–161. doi: 10.1016/j.ydbio.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 30.Collette NM, Yee CS, Murugesh D, Sebastian A, Taher L, Gale NW, Economides AN, Harland RM, Loots GG. Sost and its paralog Sostdc1 coordinate digit number in a Gli3-dependent manner. Dev Biol. 2013;383:90–105. doi: 10.1016/j.ydbio.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shigetani Y, Howard S, Guidato S, Furushima K, Abe T, Itasaki N. Wise promotes coalescence of cells of neural crest and placode origins in the trigeminal region during head development. Dev Biol. 2008;319:346–358. doi: 10.1016/j.ydbio.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 32.Clausen KA, Blish KR, Birse CE, Triplette MA, Kute TE, Russell GB, D’Agostino RB Jr, Miller LD, Torti FM, Torti SV. SOSTDC1 differentially modulates Smad and beta-catenin activation and is down-regulated in breast cancer. Breast Cancer Res Treat. 2011;129:737–746. doi: 10.1007/s10549-010-1261-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blish KR, Wang W, Willingham MC, Du W, Birse CE, Krishnan SR, Brown JC, Hawkins GA, Garvin AJ, D’Agostino RB Jr, Torti FM, Torti SV. A human bone morphogenetic protein antagonist is down-regulated in renal cancer. Mol Biol Cell. 2008;19:457–464. doi: 10.1091/mbc.E07-05-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohshima J, Haruta M, Arai Y, Kasai F, Fujiwara Y, Ariga T, Okita H, Fukuzawa M, Hata J, Horie H, Kaneko Y. Two candidate tumor suppressor genes, MEOX2 and SOSTDC1, identified in a 7p21 homozygous deletion region in a Wilms tumor. Genes Chromosomes Cancer. 2009;48:1037–1050. doi: 10.1002/gcc.20705. [DOI] [PubMed] [Google Scholar]
- 35.Rajkumar T, Vijayalakshmi N, Gopal G, Sabitha K, Shirley S, Raja UM, Ramakrishnan SA. Identification and validation of genes involved in gastric tumorigenesis. Cancer Cell Int. 2010;10:45. doi: 10.1186/1475-2867-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laurikkala J, Kassai Y, Pakkasjarvi L, Thesleff I, Itoh N. Identification of a secreted BMP antagonist, ectodin, integrating BMP, FGF, and SHH signals from the tooth enamel knot. Dev Biol. 2003;264:91–105. doi: 10.1016/j.ydbio.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 37.Balemans W, Patel N, Ebeling M, Van Hul E, Wuyts W, Lacza C, Dioszegi M, Dikkers FG, Hildering P, Willems PJ, Verheij JB, Lindpaintner K, Vickery B, Foernzler D, Van Hul W. Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J Med Genet. 2002;39:91–97. doi: 10.1136/jmg.39.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koenig BB, Cook JS, Wolsing DH, Ting J, Tiesman JP, Correa PE, Olson CA, Pecquet AL, Ventura F, Grant RA, et al. Characterization and cloning of a receptor for BMP-2 and BMP-4 from NIH 3T3 cells. Mol Cell Biol. 1994;14:5961–5974. doi: 10.1128/mcb.14.9.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ten Dijke P, Yamashita H, Sampath TK, Reddi AH, Estevez M, Riddle DL, Ichijo H, Heldin CH, Miyazono K. Identification of type I receptors for osteogenic protein-1 and bone morphogenetic protein-4. J Biol Chem. 1994;269:16985–16988. [PubMed] [Google Scholar]
- 40.Miyazawa K, Shinozaki M, Hara T, Furuya T, Miyazono K. Two major Smad pathways in TGF-beta superfamily signalling. Genes Cells. 2002;7:1191–1204. doi: 10.1046/j.1365-2443.2002.00599.x. [DOI] [PubMed] [Google Scholar]
- 41.Derynck R, Zhang Y, Feng XH. Smads: transcriptional activators of TGF-beta responses. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 42.Yanagita M, Oka M, Watabe T, Iguchi H, Niida A, Takahashi S, Akiyama T, Miyazono K, Yanagisawa M, Sakurai T. USAG-1: a bone morphogenetic protein antagonist abundantly expressed in the kidney. Biochem Biophys Res Commun. 2004;316:490–500. doi: 10.1016/j.bbrc.2004.02.075. [DOI] [PubMed] [Google Scholar]
- 43.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 44.Itasaki N, Jones CM, Mercurio S, Rowe A, Domingos PM, Smith JC, Krumlauf R. Wise, a context-dependent activator and inhibitor of Wnt signalling. Development. 2003;130:4295–4305. doi: 10.1242/dev.00674. [DOI] [PubMed] [Google Scholar]
- 45.Guidato S, Itasaki N. Wise retained in the endoplasmic reticulum inhibits Wnt signaling by reducing cell surface LRP6. Dev Biol. 2007;310:250–263. doi: 10.1016/j.ydbio.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 46.Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, Fagarasan S, Liston A, Smith KG, Vinuesa CG. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, Xu L, Chen X, Hao Y, Wang P, Zhu C, Ou J, Liang H, Ni T, Zhang X, Zhou X, Deng K, Chen Y, Luo Y, Xu J, Qi H, Wu Y, Ye L. Follicular CXCR5-expressing CD8(+) T cells curtail chronic viral infection. Nature. 2016;537:412–428. doi: 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- 48.Marshall HD, Chandele A, Jung YW, Meng H, Poholek AC, Parish IA, Rutishauser R, Cui W, Kleinstein SH, Craft J, Kaech SM. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 2011;35:633–646. doi: 10.1016/j.immuni.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, Wu T, Iyer SS, Ahmed R. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 2013;38:805–817. doi: 10.1016/j.immuni.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, Love PE, Peng W, Xue HH, Crotty S. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol. 2015;16:980–990. doi: 10.1038/ni.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alarmo EL, Kuukasjarvi T, Karhu R, Kallioniemi A. A comprehensive expression survey of bone morphogenetic proteins in breast cancer highlights the importance of BMP4 and BMP7. Breast Cancer Res Treat. 2007;103:239–246. doi: 10.1007/s10549-006-9362-1. [DOI] [PubMed] [Google Scholar]
- 52.Clement JH, Raida M, Sanger J, Bicknell R, Liu J, Naumann A, Geyer A, Waldau A, Hortschansky P, Schmidt A, Hoffken K, Wolft S, Harris AL. Bone morphogenetic protein 2 (BMP-2) induces in vitro invasion and in vivo hormone independent growth of breast carcinoma cells. Int J Oncol. 2005;27:401–407. [PubMed] [Google Scholar]
- 53.Howe LR, Brown AM. Wnt signaling and breast cancer. Cancer Biol Ther. 2004;3:36–41. doi: 10.4161/cbt.3.1.561. [DOI] [PubMed] [Google Scholar]