Abstract
Long non-coding RNAs (lncRNAs) function as critical regulator in human cancers. However, the biological regulatory mechanisms of lncRNAs in Ewing’s sarcoma are still elusive. This study tries to investigate the clinical significance and pathological role of lncRNA SOX2 overlapping transcript (SOX2OT) in Ewing’s sarcoma progression. SOX2OT was identified to be up-regulated in Ewing’s sarcoma tissue and cells. In vitro, SOX2OT knockdown suppressed Ewing’s sarcoma cells proliferation and invasion, and triggered apoptosis. In vivo xenograft assays, SOX2OT knockdown significantly inhibited Ewing’s sarcoma growth. With the help of bioinformatics analysis and luciferase assay, SOX2OT was validated to harbor miR-363, acting as miRNA sponge or competing endogenous RNA (ceRNA). Furthermore, FOXP4 was validated to be the target protein of miR-363. Western blot and RT-PCR confirmed that SOX2OT was positively correlated with FOXP4 protein via sponging miR-363, forming a negative cascade regulation. In conclusion, our study realizes that SOX2OT acted as oncogene in the tumorigenesis of Ewing’s sarcoma, suggesting the SOX2OT/miR-363/FOXP4 pathway in Ewing’s sarcoma.
Keywords: Ewing’s sarcoma, SOX2OT, miR-363, FOXP4
Introduction
Ewing’s sarcoma is one of the most common malignant tumors in children. In fact, Ewing’s sarcoma is the high widespread solid tumors [1,2]. The incidence of Ewing’s sarcoma causes serious influence on children’s health [3]. Every year, Ewing’s sarcoma triggers thousands of fatality rate [4]. Therefore, it is urgently necessary to investigate the pathogenesis and tumorigenesis of Ewing’s sarcoma.
Long noncoding RNAs (lncRNAs) are a type of transcription products with 200 nt length in human genome [5,6]. The roles of lncRNAs in human diseases have been renewedly recognized in decades, especially in human cancers [7]. LncRNAs are implicated in series of molecular processes, including differentiation, proliferation, metastasis, and transcriptional regulation. For instance, lncRNA LINC00460 is significantly up-regulated in meningeoma tissues and cells and it promotes MMP-9 expression through targeting miR-539, thereby accelerating the pathogenesis [8]. LncRNA UCA1 antagonizes miR-26a and recovers its target PETN to alleviate VSMCs proliferation against atherosclerosis [9].
In several human cancers, lncRNA SOX2OT has been reported to function as oncogenic molecular, such as esophageal squamous cell carcinoma (ESCC) [10], colorectal cancer [11], gastric cancer [12]. In the present study, we investigate the expression prolife and biological roles of lncRNA SOX2OT in Ewing’s sarcoma carcinogenesis. We identify that SOX2OT is overexpressed in Ewing’s sarcoma samples and cells, and the knockdown of SOX2OT impair the proliferation and invasion in cellular levels.
Materials and methods
Tissue samples collection
Ewing’s sarcoma tissues and adjacent normal tissues were obtained from patients at People’s Hospital of Nanjing Medical University. The Institute Research Medical Ethics Committee approved this study.
Cell culture
Ewing’s sarcoma cell lines (SK-NEP-1, SK-ES-1) were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China), and then cultured in DMEM medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin (Baomanbio) and 100 mg/ml streptomycin (Baomanbio). Cell were cultured in a humidified atmosphere (37°C, 5% CO2).
siRNA transfection
The siRNAs or mimics targeting lncRNA SOX2OT were offered by Guangzhou Ribo bioCo. Cells were transiently transfected with siRNA-SOX2OT, inhibitor (50 nM) or mimics (50 nM) (Table S1) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated from Ewing’s sarcoma tissue and cell lines with TRIzol reagent (Takara). The qualified extraction was identified according to the ratio of OD260/280 using spectrophotometer. RNA (2 μg) was reverse-transcribed into complementary DNA (cDNA). The real-time quantitative PCR was conducted with the SYBR Premix Ex Taq™ II kit (Takara, Shiga, Japan). The target genes’ expression was calculated using the 2-ΔΔCt method. Reaction condition was set at 37°C for 30 minutes and 5 seconds at 85°C and then maintained at 4°C. Primers sequences were as in Table S1.
Cell Counting Kit-8 (CCK-8) and colony formation assay
The cell proliferation potential was detected using CCK-8 assay. Cells (5 × 103 per well) were seeded into 96-well plate. Then, CCK-8 solution (10 μl) was treated the cells after transfection of 48 h. Absorbance value was tested by microplate spectrophotometer at 450 nm from triplicate groups. Colony formation assay was conducted as previous described [12].
Flow cytometry analysis
Flow cytometry apoptosis assay was performed and detected as previously reported. Briefly, Ewing’s sarcoma cells (SK-NEP-1, SK-ES-1) were transfected with siRNAs, and then seeded in 6-well plates at 1 × 103 cells per well. After incubation of 24 hour, cells were harvested and washed by PBS for two times. SK-NEP-1 and SK-ES-1 were re-suspended in Annexin V-FITC (5 μl) and propidium iodide (PI, 5 μl) by Annexin V-FITC Apoptosis Detection Kit (Invitrogen, Carlsbad, Calif, USA). FACS Calibur (BD, USA) was used to measure the apoptotic cells.
Transwell invasion assay
Invasion assay was carried using Transwell chambers assay (Corning, NY, USA) which was pre-coated by Matrigel (BD, Franklin Lakes, NJ, USA) on 24 well plates. Briefly, the transfected cells (1 × 105 per well) were seeded on the upper chambers, supplementing DMEM without serum. Besides, lower chamber was added with DMEM medium with 10% serum. After 24 h, cells invaded through the filter, which was fixed in 4% paraformaldehyde, following the staining with crystal violet. The invasive cells were counted and quantitated in 5 different areas under a microscope.
Western blot analysis
Lysates was extracted from cells using RIPA buffer (CST, Danvers, MA, USA) containing protease inhibitor. Proteins were isolated by sodium dodecyl sulfate (SDS)-polyacrylamide gels (PAGE) and transferred no PVDF membranes (Millipore, Darmstadt, Germany). Membranes were blocked at room temperature for more than 30 min, and then incubated with primary antibodies (anti-FOXP4, 1:1000 dilution, Abcam), following the incubation of horseradish peroxidase-conjugated secondary antibody (Beyotime). Finally, membranes were treated with ECL and graphed by X-ray film processor.
Cell cytoplasm/nucleus fraction isolation
RNA was isolated from the nuclear and cytoplasm fractions using Cytoplasmic Nuclear RNA Purification Kit (Norgen, Belmont, CA, USA), according to the manufacturer’s instructions.
Luciferase reporter gene assay
The luciferase reporters cloned with SOX2OT 3’-UTR sequence, including wild type targeting miR-363 binding sites or mutant type, were generated. Then, 293T cells were cultured and co-transfected with above recombinant luciferase reporter and miR-363 mimics or controls. Renilla luciferase activity acted as internal control. After 48 hours, luciferase activity was tested with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
In vivo xenograft assay
This study was approved by the ethical committee of People’s Hospital of Nanjing Medical University, and all these male nude mice (BALB/c, 4-week-old) were got from its laboratory animal center. SK-NEP-1 cells were subcutaneously injected into mice at the concentration of 100 µl containing 5 × 106 cells. After 4 weeks, tumour volume was calculated using the equation: 0.5 × length × width2.
Statistical analysis
Variable data were presented as the mean ± standard deviations (SD) or standard errors. Statistical test was calculated using SPSS (SPSS Inc., Chicago, IL, USA) by student’s t-test or one-way ANOVA. P < 0.05 was considered as the statistically differences.
Results
SOX2OT expression level was elevated in Ewing’s sarcoma specimens
Firstly, we measured the clinical expression profile of lncRNA SOX2OT in 25 paired Ewing’s sarcoma specimens and their matched normal tissue. RT-PCR revealed that SOX2OT expression was dramatically elevated in Ewing’s sarcoma specimens compared with the paired normal tissue (Figure 1A). Meanwhile, the matched analysis of these Ewing’s sarcoma specimens showed that SOX2OT almost up-regulated in every pair of Ewing’s sarcoma specimens and matched adjacent normal tissue (Figure 1B). Overall, our data illustrated that SOX2OT expression level was elevated in Ewing’s sarcoma specimens, indicating the potential oncogenic role of SOX2OT in Ewing’s sarcoma tumorigenesis.
Figure 1.
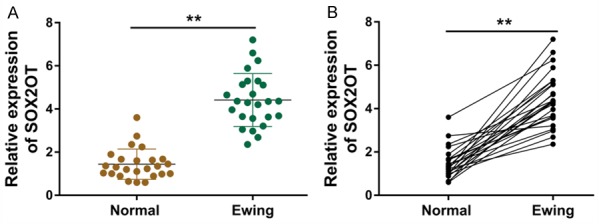
SOX2OT expression level was elevated in Ewing’s sarcoma specimens. A. RT-PCR revealed the expression of SOX2OT in Ewing’s sarcoma specimens and the paired normal tissue. B. Matched analysis of these Ewing’s sarcoma specimens showed the SOX2OT expression in every pair of Ewing’s sarcoma specimens and matched adjacent normal tissue. **P < 0.01 presented the statistic difference respectively compared with normal tissues.
LncRNA SOX2OT silencing suppressed the proliferation of Ewing’s sarcoma cells in vitro
The elevated expression of lncRNA SOX2OT in the Ewing’s sarcoma samples inspires the thought that SOX2OT might function as an oncogenic RNA in the tumorigenesis. RT-PCR revealed that SOX2OT expression was significantly enhanced in Ewing’s sarcoma cell lines (SK-NEP-1, SK-ES-1) (Figure 2A). Three different siRNAs were transfected into Ewing’s sarcoma cells to silence its expression, showing that only one siRNA could effectively suppress the expression quantity (Figure 2B). Then, the proliferation assays, including CCK-8 assay ad colony formation assay, were performed using Ewing’s sarcoma cells. The cell proliferation was significantly reduced after the si-SOX2OT transfection using CCK-8 assay, comparing with the control transfection (Figure 2C). Subsequently, the cellular colony formation number of Ewing’s sarcoma cells was markedly decreased in the si-SOX2OT transfection group, comparing with that of controls (Figure 2D, 2E). In conclusion, these changes triggered by si-SOX2OT transfection indicated the tumor inhibiting of SOX2OT silencing on Ewing’s sarcoma cells in vitro.
Figure 2.

LncRNA SOX2OT silencing suppressed the proliferation of Ewing’s sarcoma cells in vitro. A. SOX2OT expression was measured by RT-PCR in Ewing’s sarcoma cell lines (SK-NEP-1, SK-ES-1). Mesenchymal stem cells (MSC) act as control. B. Three different siRNAs were transfected into Ewing’s sarcoma cells to silence SOX2OT expression. C. The cell proliferation was tested using CCK-8 assay after the si-SOX2OT transfection or not. D, E. Cellular colony formation assay revealed the number of Ewing’s sarcoma cells in cells transfected with the si-SOX2OT or controls. All results were expressed as the means ± SD of three independent experiments (*P < 0.05, **P < 0.01).
LncRNA SOX2OT silencing impaired the Ewing’s sarcoma cells’ invasion, accelerated the apoptosis and inhibited the tumor growth in vivo
To validate the oncogenic role of SOX2OT on Ewing’s sarcoma cells more convincingly, we performed further assays to confirm it. Flow cytometry analysis revealed that SOX2OT silencing elevated the apoptotic cells in Ewing’s sarcoma cell lines (SK-NEP-1, SK-ES-1) (Figure 3A, 3B). Transwell invasion assay illustrated that SOX2OT silencing impaired the invaded cells in Ewing’s sarcoma cell lines (SK-NEP-1, SK-ES-1) (Figure 3C, 3D). In vivo xenograft assay showed that stable SOX2OT silencing inhibited the volume and weight of in vivo tumor in mice comparing with the control transfection (Figure 3E, 3F). In conclusion, data revealed that lncRNA SOX2OT silencing impaired the Ewing’s sarcoma cells’ invasion, accelerated the apoptosis and inhibited the tumor growth in vivo.
Figure 3.
LncRNA SOX2OT silencing impaired the Ewing’s sarcoma cells’ invasion, accelerated the apoptosis and inhibited the tumor growth in vivo. A, B. Flow cytometry analysis revealed the apoptotic cells in Ewing’s sarcoma cell lines (SK-NEP-1, SK-ES-1). C, D. Transwell invasion assay illustrated the invaded cells in Ewing’s sarcoma cell lines (SK-NEP-1, SK-ES-1). E, F. In vivo xenograft assay showed the volume and weight of in vivo tumor in mice comparing with the control transfection. All results were expressed as the means ± SD of three independent experiments (*P < 0.05, **P < 0.01).
miR-363 acted as the target RNA of SOX2OT and was negatively correlated with it
Subcellular fractionation analysis found that the subcellular location of SOX2OT in SK-NEP-1 cells was on the cytoplasm, proposing the possibility of post-transcriptional regulation (Figure 4A). Online bioinformatics tools (https://lncipedia.org/db/transcript/) presented that miR-363 acted as the target RNA of SOX2OT, and SOX2OT harbored the complementary binding sites with miR-363 at 3’-untranslated regions (UTR) (Figure 4B). Luciferase reporter assays revealed that the luciferase activity was decreased when miR-363 was co-transfected with SOX2OT wild type (Figure 4C). Moreover, the transfection of si-SOX2OT could markedly up-regulate miR-363 expression in SK-NEP-1 and SK-ES-1 cells comparing with the controls transfection (Figure 4D). Therefore, results revealed that miR-363 acted as the target RNA of SOX2OT and was negatively correlated with it.
Figure 4.

miR-363 acted as the target RNA of SOX2OT and was negatively correlated with it. A. Subcellular fractionation analysis showed the subcellular location (cytoplasm or nuclear) of SOX2OT in SK-NEP-1 cells. B. Online bioinformatics tools (https://lncipedia.org/db/transcript/) presented the complementary binding sites with miR-363 and SOX2OT at 3’-untranslated regions (UTR). C. Luciferase reporter assays revealed the luciferase activity in SK-NEP-1 cells when co-transfected with miR-363 and SOX2OT wild type or mutant. D. The level of miR-363 was measured by RT-PCR. All results were expressed as the means ± SD of three independent experiments (*P < 0.05, **P < 0.01).
SOX2OT positively modulated FOXP4 expression via miR-363
Although the data had indicated that SOX2OT sponged miRNA-363, as a miRNA sponge, in Ewing’s sarcoma cells, the potential functional protein of them was still unclear. Thus, we further investigated the downstream of SOX2OT and miRNA-363. Online bioinformatics tools (TargetScan, http://www.targetscan.org/vert_71/) presented that miR-363 targeted the 3’-UTR of FOXP4 mRNA (Figure 5A). Luciferase reporter assays revealed that the luciferase activity was decreased when miR-363 was co-transfected with FOXP4 wild type (Figure 5B). RT-PCR revealed that FOXP4 mRNA was significantly downregulated in SK-NEP-1 and SK-ES-1 cells when transfected with si-SOX2OT (Figure 5C). Western blot illustrated that FOXP4 protein expression was significantly decreased in SK-NEP-1 cells when transfected with si-SOX2OT (Figure 5D, 5E). Furthermore, in SK-NEP-1 cells, the transfection of miR-363 inhibitor enhanced the FOXP4 protein expression, while the co-transfection of miR-363 inhibitor and si-SOX2OT recused the FOXP4 protein expression (Figure 5F, 5G). In conclusion, we draw a conclusion that SOX2OT positively modulated FOXP4 expression via miR-363.
Figure 5.

SOX2OT positively modulated FOXP4 expression via miR-363. A. Online bioinformatics tools (TargetScan, http://www.targetscan.org/vert_71/) presented the binding within miR-363 and 3’-UTR of FOXP4 mRNA. B. Luciferase reporter assays revealed the luciferase activity when co-transfected with miR-363 and FOXP4 wild type or mutant. C. RT-PCR showed the FOXP4 mRNA in SK-NEP-1 and SK-ES-1 cells when transfected with si-SOX2OT. D, E. Western blot illustrated the FOXP4 protein expression in SK-NEP-1 cells when transfected with si-SOX2OT. F, G. Western blot illustrated the FOXP4 protein expression in SK-NEP-1 cells when transfected with miR-363 inhibitor or si-SOX2OT. All results were expressed as the means ± SD of three independent experiments (*P < 0.05, **P < 0.01).
Discussion
More and more literature have demonstrated the important role of long noncoding RNA (lncRNA) in series of human diseases, such as nervous system disease, endocrine disorder, and multiple cancers [13-17]. Ewing’s sarcoma is one of the most common renal malignant tumors in children. Up to now, there is limited reported literature and published research about the lncRNA and Ewing’s sarcoma. Thus, it is necessary to devote energy into this field.
In this study, our team put effort into the clinical characteristic and molecular phenotypic of lncRNA SOX2OT in the Ewing’s sarcoma. Clinically, lncRNA SOX2OT abundance was statistically high-regulated in Ewing’s sarcoma specimens, besides, its quantity was also enriched in cells. The overexpression level of SOX2OT in Ewing’s sarcoma might function as a cancer activator. In other type of human cancers, lncRNA SOX2OT has been indicated to accelerate the carcinogenesis. For instance, lncRNA SOX2OT expression was significantly associated with worse overall survival and related to clinical stage and distant metastasis, might be a promising prognostic factor in various cancers [18]. LncRNA Sox2ot upregulation is significantly associated with lymph node invasion, TNM stage and recurrence, moreover Sox2ot is correlated with malignant biological behaviors, including proliferation, migration and invasion [19].
The hypothesis of competing endogenous RNA (ceRNA) is the mainstream theory for the lncRNAs. This novel regulatory mechanism masterly illustrated the crosstalk of lncRNA and functional protein (or mRNA) through competing binding with miRNA. In other words, lncRNAs negatively sponge the miRNA enrichment and miRNAs negatively regulated with mRNA, thereby, lncRNAs positively promote mRNA expression. In this study, we found that lncRNA SOX2OT functions as the sponge of miR-363 in Ewing’s sarcoma cells and tumorigenesis. Then, we discover the downstream protein of miR-363, FOXP4, which is a type transcription factor and the member of FOX family [20,21].
Our results confirmed the regulatory pathway of SOX2OT/miR-363/FOXP4 in Ewing’s sarcoma tumorigenesis, which is the first time to identify the regulatory mechanism of SOX2OT in Ewing’s sarcoma. The ceRNA is constituted with lncRNA-miRNA-mRNA. For example, lncRNA LINC00339 is significantly up-regulated in NSCLC tissue and cells, facilitating the tumorigenesis of NSCLC progression by sponging miR-145 via targeting miR-145 [22]. In colorectal cancer, miR-145 is confirmed to be the target of SOX21-AS1, moreover, MYO6 acts as one of the downstream protein of miR-145, forming the SOX21-AS1/miR-145/MYO6 [23].
Forkhead-box (FOX) gene family consists of more than 43 members [24,25]. The transcription factors of FOX family also epigenetically regulate the protein gene expression in cells, thereby modulating the cellular progression [26,27]. In this study, we found that SOX2OT promotes FOXP4 protein expression via sponging and inhibiting miR-363. The molecular function of FOXP4 has been identified to be oncogenic gene in human cancer [28,29]. In breast cancer, FOXP4 promotes the cell proliferation, migration and invasion via accelerating its downstream target Snail [30]. Thus, FOXP4 exerts the tumor promoting effects in the Ewing’s sarcoma tumorigenesis, which is promoting by lncRNA SOX2OT.
In conclusion, we conclude that the enhanced expression level of lncRNA SOX2OT is closely correlated with the malignant biological behaviors of Ewing’s sarcoma. SOX2OT positively accelerates FOXP4 expression via sponging miR-363, providing a valuable treatment strategy for the Ewing’s sarcoma.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Morlote D, Harada S, Lindeman B, Stevens TM. Adamantinoma-like ewing sarcoma of the thyroid: a case report and review of the literature. Head Neck Pathol. 2019 doi: 10.1007/s12105-019-01021-5. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almomen A, Alkhatib A, Alazzeh G, Weber MA, Papakonstantinou O, Nikodinovska VV, Vanhoenacker FM. Ewing’s sarcoma and primary osseous lymphoma: spectrum of imaging appearances. Case Rep Pathol. 2019;23:36–57. doi: 10.1055/s-0038-1676125. [DOI] [PubMed] [Google Scholar]
- 3.Aldandan A. Pediatrics ewing’s sarcoma of the sinonasal tract: a case report and literature review. Head Neck Pathol. 2019;2019:8201674. doi: 10.1155/2019/8201674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.PDQ Pediatric Treatment Editorial Board. PDQ Cancer Information Summaries. Bethesda (MD): National Cancer Institute (US); 2002. Ewing Sarcoma Treatment (PDQ(R)): Patient Version. [PubMed] [Google Scholar]
- 5.Lv L, Jia JQ, Chen J. The lncRNA CCAT1 upregulates proliferation and invasion in melanoma cells via suppressing miR-33a. Oncol Res. 2018;26:201–208. doi: 10.3727/096504017X14920318811749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Y, Jin JG, Mi WY, Zhang SR, Meng Q, Zhang ST. Long noncoding RNA UCA1 targets miR-122 to promote proliferation, migration, and invasion of glioma cells. Oncol Res. 2018;26:103–110. doi: 10.3727/096504017X14934860122864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, Li JY, Tian FZ, Zhao G, Hu H, Ma YF, Yang YL. LncRNA NEAT1 promotes growth and metastasis of cholangiocarcinoma cells. Oncol Res. 2017;26:879–888. doi: 10.3727/096504017X15024935181289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xing H, Wang S, Li Q, Ma Y, Sun P. Long noncoding RNA LINC00460 targets miR-539/MMP-9 to promote meningioma progression and metastasis. Biomed Pharmacother. 2018;105:677–682. doi: 10.1016/j.biopha.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Tian S, Yuan Y, Li Z, Gao M, Lu Y, Gao H. LncRNA UCA1 sponges miR-26a to regulate the migration and proliferation of vascular smooth muscle cells. Gene. 2018;19:30682–30686. doi: 10.1016/j.gene.2018.06.031. [DOI] [PubMed] [Google Scholar]
- 10.Shafiee M, Aleyasin SA, Vasei M, Semnani SS, Mowla SJ. Down-regulatory effects of miR-211 on long non-coding RNA SOX2OT and SOX2 genes in esophageal squamous cell carcinoma. Cell J. 2016;17:593–600. doi: 10.22074/cellj.2016.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S, Xu B, Yan D. Enhanced expression of long non-coding RNA Sox2ot promoted cell proliferation and motility in colorectal cancer. Minerva Med. 2016;107:279–286. [PubMed] [Google Scholar]
- 12.Qu F, Cao P. Long noncoding RNA SOX2OT contributes to gastric cancer progression by sponging miR-194-5p from AKT2. Exp Cell Res. 2018;19:30288–30295. doi: 10.1016/j.yexcr.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Jing H, Qu X, Liu L, Xia H. A novel long noncoding RNA (lncRNA), LL22NC03-N64E9.1, promotes the proliferation of lung cancer cells and is a potential prognostic molecular biomarker for lung cancer. Med Sci Monit. 2018;24:4317–4323. doi: 10.12659/MSM.908359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X, Zhang P, Zhu H, Li S, Chen X, Shi L. Long noncoding RNA FEZF1-AS1 indicates a poor prognosis of gastric cancer and promotes tumorigenesis via activation of Wnt signaling pathway. Biomed Pharmacother. 2017;96:1103–1108. doi: 10.1016/j.biopha.2017.11.113. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Dun Y, Zhou S, Huang XH. LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells proliferation and invasion by targeting miR-133a-3p and activating Wnt/beta-catenin signaling pathway. Biomed Pharmacother. 2017;96:1216–1221. doi: 10.1016/j.biopha.2017.11.096. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Zi Y, Wang W, Li Y. Long noncoding RNA MEG3 inhibits cell proliferation and metastasis in chronic myeloid leukemia via targeting miR-184. Oncol Res. 2018;26:297–305. doi: 10.3727/096504017X14980882803151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Yang J, Xia Y, Fan Q, Yang KP. Long noncoding RNA NEAT1 promotes proliferation and invasion via targeting miR-181a-5p in non-small cell lung cancer. Oncol Res. 2018;26:289–296. doi: 10.3727/096504017X15009404458675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song X, Yao H, Liu J, Wang Q. The prognostic value of long noncoding RNA SOX2OT expression in various cancers: a systematic review and meta-analysis. Clin Chim Acta. 2018;484:52–59. doi: 10.1016/j.cca.2018.05.038. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Li J, Ji D, Leng K, Xu Y, Huang L, Jiang X, Cui Y. Overexpressed long noncoding RNA SOX2OT predicts poor prognosis for cholangiocarcinoma and promotes cell proliferation and invasion. Gene. 2018;645:131–136. doi: 10.1016/j.gene.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 20.Estruch SB, Graham SA, Quevedo M, Vino A, Dekkers DHW, Deriziotis P, Sollis E, Demmers J, Poot RA, Fisher SE. Proteomic analysis of FOXP proteins reveals interactions between cortical transcription factors associated with neurodevelopmental disorders. Hum Mol Genet. 2018;27:1212–1227. doi: 10.1093/hmg/ddy035. [DOI] [PubMed] [Google Scholar]
- 21.Yin Z, Ding H, He E, Chen J, Li M. Up-regulation of microRNA-491-5p suppresses cell proliferation and promotes apoptosis by targeting FOXP4 in human osteosarcoma. Cell Prolif. 2017;50 doi: 10.1111/cpr.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan Y, Haiying G, Zhuo L, Ying L, Xin H. Long non-coding RNA LINC00339 facilitates the tumorigenesis of non-small cell lung cancer by sponging miR-145 through targeting FOXM1. Biomed Pharmacother. 2018;105:707–713. doi: 10.1016/j.biopha.2018.06.022. [DOI] [PubMed] [Google Scholar]
- 23.Wei AW, Li LF. Long non-coding RNA SOX21-AS1 sponges miR-145 to promote the tumorigenesis of colorectal cancer by targeting MYO6. Biomed Pharmacother. 2017;96:953–959. doi: 10.1016/j.biopha.2017.11.145. [DOI] [PubMed] [Google Scholar]
- 24.Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, Bond CS, Nakagawa S, Pierron G, Hirose T. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol Cell. 2018;70:1038–1053. e1037. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 25.Sun J, Lian M, Ma H, Wang R, Ma Z, Wang H, Zhai J, Meng L, Feng L, Bai Y, Cui X, Fang J. Competing endogenous RNA network analysis of CD274, IL10 and FOXP3 coexpression in laryngeal squamous cell carcinoma. Mol Med Rep. 2018;17:3859–3869. doi: 10.3892/mmr.2017.8307. [DOI] [PubMed] [Google Scholar]
- 26.Yin Z, Ding H, He E, Chen J, Li M. Up-regulation of microRNA-491-5p suppresses cell proliferation and promotes apoptosis by targeting FOXP4 in human osteosarcoma. Cell Prolif. 2017;50:65–75. doi: 10.1111/cpr.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu G, Niu F, Humburg BA, Liao K, Bendi S, Callen S, Fox HS, Buch S. Molecular mechanisms of long noncoding RNAs and their role in disease pathogenesis. Oncotarget. 2018;9:18648–18663. doi: 10.18632/oncotarget.24307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu M, Shi X, Wang J, Xu Y, Wei D, Zhang Y, Yang K, Wang X, Liang S, Chen X, Yang F, Sun L, Zhu X, Zhao C, Zhu L, Tang L, Zheng C, Yang Z. Association of FOXP4 gene with prostate cancer and the cumulative effects of rs4714476 and 8q24 in Chinese men. Clin Lab. 2015;61:1491–1499. doi: 10.7754/clin.lab.2015.150313. [DOI] [PubMed] [Google Scholar]
- 29.Wang N, Gu Y, Li L, Wang F, Lv P, Xiong Y, Qiu X. Circular RNA circMYO9B facilitates breast cancer cell proliferation and invasiveness via upregulating FOXP4 expression by sponging miR-4316. Arch Biochem Biophys. 2018;76:677–684. doi: 10.1016/j.abb.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 30.Ma T, Zhang J. Upregulation of FOXP4 in breast cancer promotes migration and invasion through facilitating EMT. Cancer Manag Res. 2019;11:2783–2793. doi: 10.2147/CMAR.S191641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



