Abstract
Directed differentiation of human pluripotent stem cells (hPSCs) into functional insulin-producing cells (IPCs) holds great promise for cell therapy for diabetic patients. Despite recent advances in developing beta cell differentiation protocols, it is becoming clear that the hPSC-derived beta-like cells are functionally immature, and the efficiencies of differentiation can be variable depending on the hPSC lines used. Therefore, advanced methodologies are highly desirable for the development and refinement of beta cell differentiation protocols from hPSCs. In this report, we first derived and validated a Pdx1-mRFP/insulin-hrGFP dual-reporter cell line from MRC5-iPSCs. Then, using this dual-reporter cell line, we developed and optimized an in vitro beta cell differentiation protocol through real-time monitoring expression of Pdx1 and insulin. We demonstrated that DNA demethylation could increase the efficiency of beta cell differentiation. Furthermore, three-dimensional induction not only significantly increased the efficiency of pancreatic progenitor specification and the yield of IPCs, but also produced more mature IPCs. The current study indicates that this dual-reporter cell line is of great value for developing and optimizing the beta cell differentiation protocols. It will facilitate the development of novel protocols for generating IPCs from hPSCs and the investigation of beta cell differentiation mechanisms.
Keywords: Induced pluripotent stem cells, Pdx1/insulin dual-reporter, real-time monitoring, beta cell differentiation, diabetes
Introduction
Establishment of induced pluripotent stem cells (iPSCs) holds great promise for generating replacement cells and tissues that could be used for disease modeling, drug development, and regenerative medicine [1-3]. Moreover, generation of iPSCs from human somatic cells enables production of patient-specific cells for autologous transplantation. Recent research has shown the possibility to generate cells that would otherwise be difficult to obtain, like neurons, retinal cells, or cardiomyocytes, from pluripotent stem cells (PSCs) through development of strategies [4-6]. These cells have also been transplanted into animal models, and in some cases have demonstrated beneficial effects such as functional motor improvement in non-human primate model of Parkinson’s disease [4], improved vision after transplantation of retinal tissue into the end-stage retinal-degeneration model [5], or regeneration of infarcted non-human primate hearts [6].
One of the most rapidly growing diseases that may be treatable by stem-cell-derivatives is diabetes, affecting more than 400 million people worldwide according to the World Health Organization. Arising from autoimmune destruction of the pancreatic beta cells, type 1 diabetes (T1D) manifests itself when less than 10-20% of functional beta cells remain in the islets [7]. Type 2 diabetes is characterized by the development of beta cell dysfunction and the progressive reduction of beta cell mass via reduced proliferation and increased apoptosis [8]. Diabetic patients, particularly those suffering from T1D, could potentially be cured through transplantation of beta cells. However, its large scale application is hampered by a shortage of beta cells for transplantation and requirement of life-long immune suppression [9]. PSCs, with their potential to differentiate into a diverse array of specialized cells, provide high hopes for curing diabetes through beta cell replacement therapy. Tremendous progress has been made to derive insulin-producing cells (IPCs) from PSCs [10-12]. Research on pancreatic development in model organisms identified genes and signals important for the pancreatic lineage, and these have been effectively employed to derive beta cells in vitro from human PSCs (hPSCs) [13,14].
In general, IPCs can be acquired from both human ESCs (hESCs) and iPSCs (hiPSCs) using similar differentiation protocols [15,16]. However, significant variation in the efficiencies of differentiation has been observed between different hPSC lines, with some lines more readily differentiating into a particular cell type than others [17-19]. This variation might occur due to differences in genetic, epigenetic, and cell cycle patterns [20,21]. Owing to these differences in differentiation propensity, directed differentiation protocols often need labor-intensive and time-consuming optimization for specific hPSC lines. Thus, generation of hPSC lines reporting expression of key genes for beta cell development has the potential to greatly facilitate future efforts aimed at improving and characterizing the differentiation of hPSC towards beta cells. In this report, we describe the generation and validation of a Pdx1-mRFP/insulin-hrGFP dual-reporter cell line in a hiPSC line derived from MRC5 cells. Furthermore, we demonstrate that this dual-reporter cell line is of great value for optimizing the differentiation protocols since it allows real-time monitoring expression of the key beta cell molecular markers during beta cell differentiation.
Materials and methods
Cell culture
Insulinoma INS-1 cells were cultured as previously described [22]. MRC5- iPSCs were generated and fully characterized by our laboratory. MRC5-iPSCs were cultured on inactivated CF1 mouse embryonic fibroblasts (MEFs) (China Infrastructure of Cell Line Resources)-coated 6-well plates with iPSC medium: DMEM/F12 supplemented with 20% KSR, 1% glutamax, 1% non-essential amino acids (NEAAs), 1% penicillin/streptomycin (P/S), 0.1 mM β-mercaptoethanol, and 10 ng/ml bFGF (Gibco). The medium was changed every day.
Reverse transcription-polymerase chain reaction (RT-PCR) and quantitative PCR (qPCR)
Total RNA was purified with TRIzol reagent (Invitrogen) and digested with DNase to remove genomic DNA contamination. 1 µg of total RNA was used for reverse transcription reaction with Superscript IV first-strand synthesis system (Invitrogen), according to the manufacturer’s instructions. PCR was performed with Taq DNA polymerase (Invitrogen). The PCR conditions were as follows: initial denaturation at 94°C for 3 min followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. qPCR reactions were performed in triplicate on an ABI 7900HT (Applied Biosystem, AB) with SYBR Green PCR Master Mix (AB). Expression data were normalized relative to GAPDH transcript level. The fold change for each gene was calculated using the 2-ΔΔCt method. Results were confirmed using cDNA from at least three independent experiments. The qPCR conditions were as follows: initial denaturation at 95°C for 1 min followed by 40 cycles of 5 s at 95°C, 10 s at 60°C, and 15s at 72°C. Primer sequences are shown in Table 1.
Table 1.
Primers for PCR and qPCR
| Genes | Accession | Forward | Reverse | Size | Tm | Cycles |
|---|---|---|---|---|---|---|
| GAPDH | NM_002046 | cgagatccctccaaaatcaagt | tgaggctgttgtcatacttctcat | 196 | 56 | 30 |
| Sox17 | NM_022454 | acgctttcatggtgtgggctaag | gtcagcgccttccacgacttg | 112 | 60 | 40 |
| Foxa2 | NM_021784 | attgctggtcgtttgttgtg | tacgtgttcatgccgttcat | 187 | 60 | 40 |
| Nkx6.1 | NM_006168 | tcttctggcccggagtgatg | ggaaaaagtgggtctcgtgtgtt | 125 | 66 | 30 |
| Ngn3 | NM_020999 | ttcgcccacaactacatc | gacagacaggtcctttcac | 282 | 56 | 30 |
| Nkx2.2 | NM_002509 | cttctacgacagcagcgaca | tgtcattgtccggtgactcg | 155 | 56 | 30 |
| Gcg | NM_002054 | aagttcccaaagagggcttg | agctgccttgtaccagcatt | 126 | 56 | 30 |
| Sst | NM_001048 | acgcaaagctggctgcaaga | gggggcgagggatcagaggt | 100 | 56 | 30 |
| Ppy | NM_002722 | aatgccacaccagagcagat | cgtaggagacagaaggtggc | 192 | 56 | 30 |
| MAFA | NM_201589 | cttcagcaaggaggaggtcatc | tctcgctctccagaatgtgcc | 119 | 60 | 30 |
| PC1 | NM_000439 | gtgacggatgctattgaggc | tgatggagatggtgtagatgct | 262 | 56 | 30 |
| PC2 | NM_002594 | tgccgaagcaagttacgact | aacttctcctgcacatcggg | 103 | 56 | 30 |
| Kir6.2 | NM_000525 | ggacccaggtggaggtaagg | ctctcggtgggcaccttctc | 103 | 56 | 30 |
| Sur1 | NM_001287174 | gaggctacttcacgtggacc | ctatctcgctgtcaggaaggc | 198 | 56 | 30 |
| Glut2 | NM_000340 | gtcactgggaccctggtttt | gtcatccagtggaacacccaa | 141 | 56 | 30 |
| Pdx1 | NM_000209 | atgaagtctaccaaagctcacgc | tctctcggtcaagttcaacatga | 201 | 60 | 40 |
| Ins | NM_000207 | acgaggcttcttctacacaccca | tgttccacaatgccacgcttc | 147 | 60 | 40 |
Immunofluorescence (IF) analysis
Cell cultures on plates or cytospins were used for IF analysis. In addition, parental iPSCs without Pdx1/insulin dual-reporter were induced in parallel using the corresponding protocols and subsequently subjected to IF analysis for beta cell markers. Briefly, cells were first fixed with 4% paraformaldehyde for 30 min at 4°C. Next, cells were permeabilized and blocked with 10% calf serum and 0.1% Triton X-100 in PBS. Subsequently, cells were stained with primary antibodies overnight at 4°C and finally incubated with fluorochrome-labeled secondary antibodies. The primary antibodies used were anti-Sox17 (R&D Systems) and anti-Foxa2 (Proteintech) for endoderm detection; and anti-Pdx1 (R&D Systems) and anti-insulin (Proteintech) for beta cell detection. The secondary antibodies used were donkey anti-mouse AF488, goat anti-rabbit AF488, and donkey anti-goat AF594 (Invitrogen). Nuclei were stained by DAPI (Sigma Aldrich). Negative staining controls were carried out by replacing the primary antibody with goat, rabbit, and mouse sera, respectively. Images were captured using Nikon Eclipse Ti-U microscope, and images were processed using Adobe Photoshop CS5.
In addition, the differentiated cells at day 34 were dissociated for preparing cytospins to calculate the percentages of Pdx1+ and Pdx1+/insulin+ cells. Nuclei were stained by DAPI. Then images were captured using Nikon Eclipse Ti-U microscope and the percentage of the cells was accounted using Adobe Photoshop CS5.
Modification of Pdx1/insulin dual-reporter vector and lentiviral production
The pTiger-Pdx1-mRFP/insulin-EGFP vector (a gift from Dr. James Johnson, University of British Columbia, Vancouver, Canada) was modified as follows [23]. First, the hPKG promoter and coding sequence for the Puromycin resistant gene was amplified by PCR from the pLKO.1 vector (Addgene) and inserted into the pTiger vector directly downstream of rat Ins1 promoter-EGFP gene using the PmeI/NotI restriction sites. Second, the coding sequence for Ins1-hrGFP (646 bp Ins1 promoter) was amplified by PCR from the pIns1-hrGFP vector (a gift from Dr. Shiwu Li, University of Florida, FL, USA), and the original rat Ins1-EGFP (410 bp Ins1 promoter) of the pTiger vector was replaced by the Ins1-hrGFP coding sequence using the NheI/PmeI restriction sites. Third, lentiviruses were produced as previously described [24]. Finally, functional expression of the new pTiger-Pdx1-mRFP/insulin-hrGFP/hPGK-Puro vector was tested and confirmed in INS-1 cells.
Generation of Pdx1/insulin dual-reporter-expressing cell lines
MRC5-derived iPSCs were first adapted to the feeder-free culture system. iPSCs were cultured on vitronectin (0.5 µg/cm2, Invitrogen)-coated 6-well plates in Essential 8 (E8) medium (Invitrogen), and the passaged cells were used for the derivation of dual-reporter cell lines. Then, iPSCs were transduced with lentiviruses carrying Pdx1/insulin dual-reporters when the cells reached 60-70% confluency. Two days after the transduction, the cells were selected using 1 µg/ml puromycin dihydrochloride (Sigma Aldrich) for 7 days. Thereafter, the cells were maintained with 0.5 µg/ml puromycin to ensure the maintenance of the transgene.
In vitro differentiation of iPSCs
iPSCs were directed through key stages of pancreatic development, including definitive endoderm (stage 1), primitive gut tube (stage 2), pancreatic progenitors (stage 3), endocrine progenitors (stage 4), and insulin-producing cells (stage 5). The following is the final protocol developed for beta cell differentiation.
Stage 1: Definitive endoderm (DE). Undifferentiated iPSCs were cultured until 50-60% confluency, washed with PBS once and treated with 100 ng/ml Activin A (R&D), 4 µM CHIR99021 (Stemgent), and 100 nM PI-103 (Selleckchem) in either medium 1 (M1) or M2 for day 1 only. For the next 2 days, the cells were cultured in either M1 or M2 supplemented with 100 ng/ml Activin A. To increase the efficiency of differentiation, iPSCs were pretreated with 2 µM 5-Azacytidine (5Aza, Sigma Aldrich) for 18 hrs before the initiation of induction. For three-dimensional (3D) induction, iPSCs were first treated with 0.5 mM EDTA and transferred onto 24-well ultra low attachment plates (Corning) in E8 medium supplemented with 1X RevitaCell supplement (RVC, Gibco) for embryonic bodies (EBs) formation. Three days later, EBs were harvested and transferred onto new wells of ultra low attachment plates for the following induction.
Stage 2: Primitive gut tube (PGT). Stage 1 cells were treated with 50 ng/ml KGF (Peprotech) for 2 days in M3.
Stage 3: Pancreatic progenitors (PP1 and PP2). Stage 2 cells were treated with 50 ng/ml KGF, 250 nM SANT1 (Sigma), 2 µM Retinoic acid (RA, Sigma), 200 nM LDN193189 (only Day 6, Selleckchem), and 500 nM PDBu (Calbiochem) in M4 for 2 days. Then the cells were exposed to 50 ng/ml KGF, 250 nM SANT1, and 2 µM RA in M4 for another 5 days.
Stage 4: Endocrine progenitors (EP). Stage 3 cells were treated with 250 nM SANT1, 100 nM RA, 1 µM XXI (Calbiochem), 10 µM Alk5i II (Calbiochem), 1 µM L-3,3’,5-Triiodothyronine (T3, Calbiochem), and 20 ng/ml Betacellulin (R&D) in M5 for 4 days.
Stage 5: Insulin-producing cells (IPCs). Stage 4 cells were treated with 25 nM RA, 1 µM XXI, 1 µM T3, 10 µM Alk5i II, and 20 ng/ml Betacellulin in M5 for 3 days. Then the cells were cultured in M6 supplemented with 10 µM Alk5i II and 1 µM T3 for 15 days.
Media used for directed differentiation were as follows [12,25]: M1: MCDB131 (Gibco) + 8 mM D-Glucose (Sigma) + 2.46 g/L NaHCO3 (Sigma) + 2% BSA (Calbiochem) + ITS-X (Gibco) 1:50,000 + 2 mM Glutamax (Gibco) + 0.25 mM Vitamin C (Sigma Aldrich). M2: 50% IMDM (Gibco) + 50% F12 (Gibco) + 0.1% BSA + 1% v/v chemically-defined lipid concentrate (Gibco) + 450 µM monothioglycerol (Sigma Aldrich) + ITS-X 1:200. M3: MCDB131 + 8 mM D-Glucose + 1.23 g/L NaHCO3 + 2% BSA + ITS-X 1:50,000 + 2 mM Glutamax + 0.25 mM Vitamin C. M4: MCDB131 + 8 mM D-Glucose + 1.23 g/L NaHCO3 + 2% BSA + ITS-X 1:200 + 2 mM Glutamax + 0.25 mM Vitamin C. M5: MCDB131 + 20 mM D-Glucose + 1.754 g/L NaHCO3 + 2% BSA + ITS-X 1:200 + 2 mM Glutamax + 0.25 mM Vitamin C. M6: CMRL medium 1066 (Gibco) + 10% FBS (HyClone).
Flow cytometry analysis
Differentiated cells were dissociated with trypsin (HyClone) for 5 min, followed by pipetting to separate the cells. After washing with PBS, cells were fixed and permeabilized with Fixation and Permeabilization Solution (BD Biosciences) for 20 min. After washing with Perm/Wash Buffer (BD Biosciences), cells were incubated with APC-conjugated goat anti-Sox17 (R&D Systems) and PE-conjugated mouse anti-Foxa2 antibodies (BD Biosciences) diluted in Perm/Wash Buffer for 1 hr at 4°C. After washing with PBS, cells were analyzed by flow cytometry. The corresponding isotype-matched antibodies were used as negative controls. Cells (20,000 events per sample) were acquired on a FACS Calibur flow cytometer (BD Biosciences) and data were analyzed using FCS express 4 plus software.
Insulin secretion assay
For static insulin secretion, differentiated cells at day 34 were washed 5 times and incubated for 1 hr in Krebs-Ringer bicarbonate (KRB) buffer containing 2 mM glucose, after which supernatants were collected by centrifugation. The cells were then washed 3 times and incubated in KRB buffer containing 25 mM glucose or 2 deoxy-D-glucose (2-DG) for 1 hr. After incubation, cells were removed by centrifugation and supernatants were frozen at -80°C for subsequent insulin assay. The insulin assay was performed with an Insulin ELISA kit (Alpco Diagnostics) according to the manufacturer’s instructions. Total cell protein was measured using the BCA Protein Assay Kit (Pierce Biotechnology) to normalize the amount of insulin secretion. Each experiment was repeated independently three times.
Statistical analysis
All the experiments were carried out at least three times. Data were displayed as mean ± SD. To assess statistical significance, two-tailed, unpaired Student’s t test was performed and P < 0.05 was considered significant.
Results
Pdx1/insulin dual-reporter construction and generation of dual-reporter-expressing cell lines
A pTiger-Pdx1-mRFP/insulin-EGFP vector was modified in order to generate the dual-reporter-expressing cell lines. First, the hPGK-Puromycin resistant gene was inserted into the vector to add a drug selection marker. Second, the original rat insulin 1 (Ins1)-EGFP (410 bp Ins1 promoter) of the pTiger vector was replaced by the Ins1-hrGFP (646 bp Ins1 promoter) for increasing the Ins1 promoter expression efficiency (Figure 1A). Finally, the function of the new pTiger-Pdx1-mRFP/insulin-hrGFP/hPGK-Puro vector was confirmed as demonstrated by co-expression of RFP/GFP in INS-1 cells transduced with the vector (Figure 1B).
Figure 1.
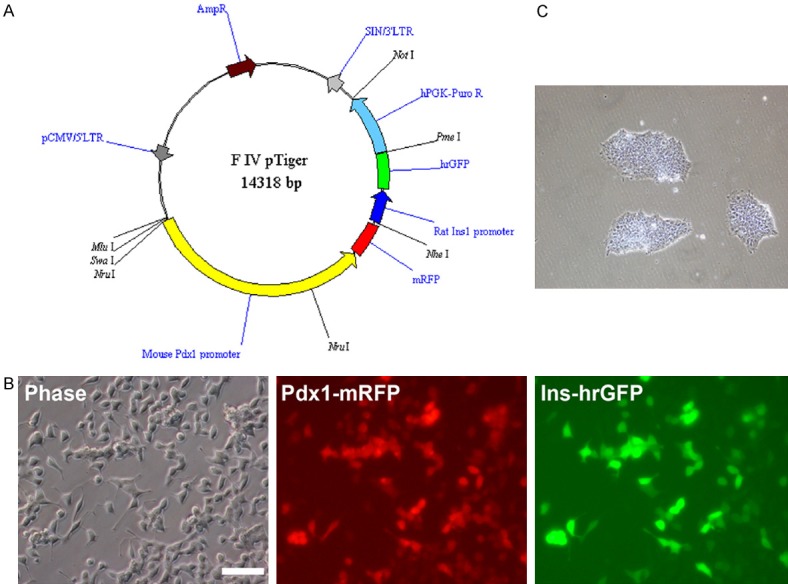
Modification of Pdx1/insulin dual-reporter vector and dual-reporter cell lines. A. Map of new dual-reporter gene expression vector. mRFP expression was driven by the mouse Pdx1 promoter, hrGFP by the rat insulin 1 promoter, and Puromycin resistant gene by the hPGK promoter. B. Functional validation of the dual-reporter vector. INS-1 cells co-expressed RFP and GFP after being infected with the dual-reporter vector. Bar is 100 µm. C. Pdx1/insulin dual-reporter-expressing cell lines. iPSCs were transduced with dual-reporters and selected using 1 µg/ml puromycin dihydrochloride for 7 days. Thereafter, the cells were maintained with 0.5 µg/ml puromycin. Bar is 200 µm.
To generate dual-reporter-expressing cell lines, iPSCs were first cultured using the feeder-free culture system to remove feeder cells. Then, the cells were transduced with lentiviruses carrying Pdx1/insulin dual-reporters. Two days after the transduction, stably transduced cells were selected using 1 µg/ml puromycin dihydrochloride. After 3-4 days of selection, the control group cells (parental iPSCs) were all dead, and the puromycin-resistant clones emerged approximately 5-7 days after selection in the transduced iPSCs group (Figure 1C). The resulting cell lines were used for the following beta cell differentiation.
In vitro differentiation of iPSCs towards beta cells
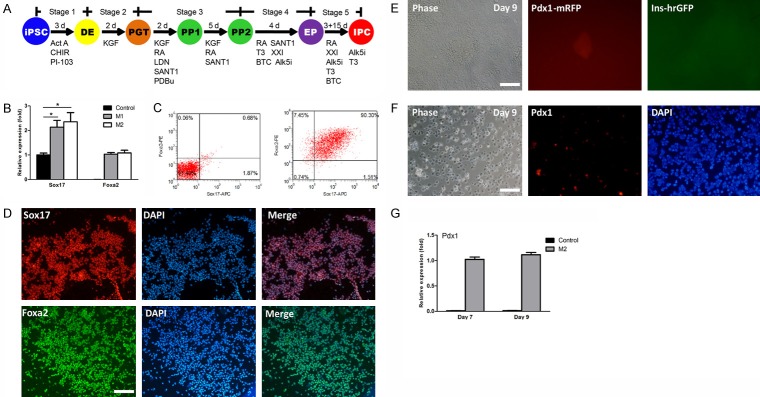
To examine the function of the Pdx1/insulin dual-reporter cell lines for real-time monitoring of cell differentiation, the cells were differentiated into beta cells by growth factors and small molecules (Figure 2A). To generate DE, the cells were first treated with Activin A, CHIR99021, and PI-103 in M1 for 1 day. Then, the cells were exposed to Activin A in M1 for another 2 days. After 3 days of treatment, qPCR analysis showed that expression levels of Sox17 and Foxa2 were significantly upregulated compared to the untreated control (Figure 2B). When continuing the induction, from day 4 on, we unexpectedly observed many cells died. Previous reports have determined that exposure to a high concentration of activin A dramatically increased cell apoptosis and reduced cell viability during the induction of DE, and optimization of induction conditions could maintain cell viability [26,27]. To increase the cell survival rates, we replaced M1 with M2 at stage 1. At day 3, qPCR analysis showed comparable expression levels of Sox17 and Foxa2 between the M2 and M1 groups (Figure 2B). Sox17 and Foxa2 double positive cells in the M2 group were around 90% as demonstrated by flow cytometry (Figure 2C). The expression of Sox17 and Foxa2 were further confirmed by IF analysis (Figure 2D). Of note, we found replacement of M1 with M2 could greatly decrease cell death which occurred previously during the induction process. These results suggest iPSCs were successfully differentiated into DE.
Figure 2.
In vitro directed differentiation from iPSCs into IPCs. A. Schematic diagram of directed differentiation from iPSCs into IPCs by growth factors and small molecules. The dual-reporter cell line was differentiated into IPCs through five stages: DE, definitive endoderm; PGT, primitive gut tube; PP, pancreatic progenitors; EP, endocrine progenitors; and IPCs, insulin-producing cells. Act A, Activin A; CHIR, CHIR99021; RA, Retinoic acid; LDN, LDN193189; PDBu, Phorbol-12,13-dibutyrate; XXI, γ-secretase inhibitor; T3, L-3,3’,5-Triiodothyronine; BTC, Betacellulin; Alk5i, Alk5 receptor inhibitor II. B. Analysis of DE gene expression by qPCR. Expression levels of Sox17 and Foxa2 were analyzed in M1, M2, and untreated control groups at the end of stage 1. Expression data are normalized to GAPDH transcript level. Each experiment was repeated independently three times. *P < 0.05. C. Flow cytometric analysis. Representative dot plots of undifferentiated iPSCs (left) and differentiated cells (Day 3) in M2 group (right) co-stained with anti-Sox17 and anti-Foxa2. Percentage in the upper right quadrant indicates the percentage of Sox17 and Foxa2 double positive cells. D. IF analysis of DE markers. Differentiated cells were stained by anti-Sox17 (1:100) and anti-Foxa2 (1:50) antibodies. Nuclei were highlighted with DAPI staining. Bar is 200 µm. E. Real-time monitoring expression of Pdx1 and insulin. The differentiated cells were checked every day under fluorescence microscope for RFP/GFP expression. Pdx1-mRFP expression was observed at day 9. Bar is 100 µm. F. IF analysis. Parental iPSCs (without dual-reporter) were induced in parallel using the corresponding protocols and stained by anti-Pdx1 (1:100) antibody at day 9. Nuclei were highlighted with DAPI staining. Bar is 100 µm. G. Analysis of Pdx1 expression by qPCR. Expression level of Pdx1 was analyzed in M2 and control groups at days 7 and 9. Expression data are normalized to GAPDH transcript level. Each experiment was repeated independently three times. *P < 0.05.
Next, PGT and PPs were sequentially induced from DE by a combined treatment of KGF, SANT1, Retinoic acid (RA), LDN193189, and PDBu, and expression of Pdx1-mRFP/insulin-hrGFP were used as indicators for monitoring the PP specification (Figure 1A). We could not observe Pdx1-mRFP expression until day 9, indicating delayed Pdx1 expression (Figure 2E), and the Pdx1 expression was confirmed by IF analysis (Figure 2F). Furthermore, the expression level of Pdx1 was not markedly increased over time as shown by qPCR analysis (Figure 2G). Considering the delayed and poor expression of Pdx1, these results suggest that the protocols still require optimization.
Effects of demethylation on beta cell differentiation
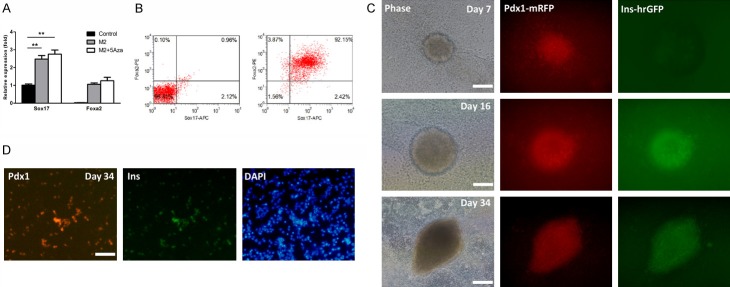
DNA methylation plays important roles on cell fate conversion [28,29]. Pretreatment with 5Aza could markedly promote the conversion of T1D patients-derived iPSCs into IPCs [29]. To efficiently derive PPs, iPSCs were pretreated with 5Aza for 18 hrs before the initiation of induction based on the above protocol. At day 3, expression levels of Sox17 and Foxa2 in the 5Aza-pretreated M2 group were significantly upregulated compared to the untreated control, but comparable to the untreated M2 group (Figure 3A). Sox17 and Foxa2 double positive cells were also around 90% in the 5Aza-pretreated M2 group as demonstrated by flow cytometry (Figure 3B), similar to the untreated M2 group. These results revealed that 5Aza pretreatment had little influence on DE induction. Next, we continued to monitor the specification of PPs. As expected, Pdx1-mRFP positive colonies emerged at day 7, two days earlier than those in the untreated M2 group, suggesting the efficient specification of PPs (Figure 3C). At day 16, we observed Pdx1-mRFP/insulin-hrGFP double positive colonies emerge, indicating the cells expressed both Pdx1 and insulin (Figure 3C). Thereafter, the Pdx1/insulin double positive colonies gradually grew over time, and the percentages of Pdx1+ and Pdx1+/insulin+ cells were 20-25% and 15-20% at day 34, respectively (Figure 3C). At day 34, we also performed IF analysis and confirmed the Pdx1 and insulin expression (Figure 3D).
Figure 3.
Effects of 5Aza on beta cell differentiation. A. Analysis of DE gene expression by qPCR. Expression levels of Sox17 and Foxa2 were analyzed in M2, M2 + 5Aza, and untreated control groups at the end of stage 1. Expression data are normalized to GAPDH transcript level. Each experiment was repeated independently three times. *P < 0.05; **P < 0.01. B. Flow cytometric analysis. Representative dot plots of undifferentiated iPSCs (left) and differentiated cells (Day 3) in M2 + 5Aza group (right) co-stained with anti-Sox17 and anti-Foxa2. Percentage in the upper right quadrant indicates the percentage of Sox17 and Foxa2 double positive cells. C. Real-time monitoring expression of Pdx1 and insulin. Pdx1-mRFP expression was first observed at day 7. Bar is 100 µm. Then Pdx1-mRFP/insulin-hrGFP double positive colonies emerged at day 16. Bar is 100 µm. And these double positive colonies gradually grew over time (Day 34). Bar is 200 µm. D. IF analysis. Parental iPSCs (without dual-reporter) were induced in parallel using the corresponding protocols and co-stained by anti-Pdx1 (1:100) and anti-insulin (1:50) antibodies at day 34. Nuclei were highlighted with DAPI staining. Bar is 100 µm.
3D induction enhances the efficiency of beta cell differentiation
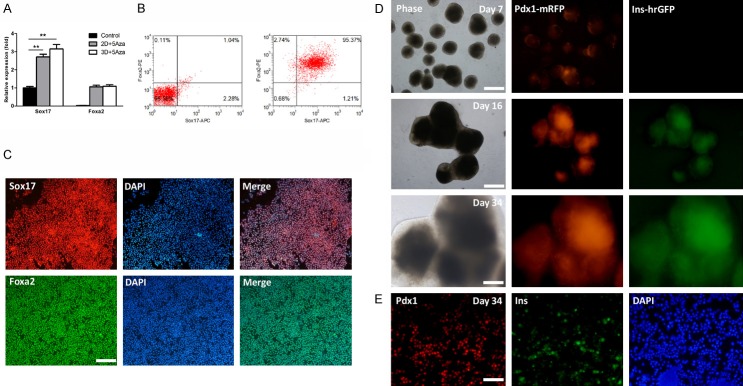
During the specification of PPs, we noticed that Pdx1-mRFP/insulin-hrGFP double positive cells tended to form clusters or 3D structures, but few scattered cells were observed. This suggests that cell-cell interactions may play pivotal roles on beta cell differentiation. Therefore, we hypothesized that the 3D induction method might further increase the efficiency of cell differentiation. In order to distinguish from the 3D induction, the above optimized protocol was designated as 2D induction thereafter. For 3D induction, we first derived embryoid bodies (EBs) from iPSCs, and RevitaCell supplement (RVC) was used to increase cell survival rates. Next, EBs were subjected to beta cell differentiation by the above optimized protocol. At the end of stage 1, expression levels of Sox17 and Foxa2 were comparable between 3D and 2D inductions (Figure 4A). The percentage of Sox17 and Foxa2 double positive cells showed a slight increase, approximately 95% in 3D induction, but no statistical significance compared to 2D induction (Figure 4B). The expression of Sox17 and Foxa2 were further confirmed by IF analysis (Figure 4C). These results reveal that 3D induction could not further increase the efficiency of DE formation. The reason for this might be that the efficiency of DE induction was already very high in 2D induction. Similar to 2D induction, Pdx1-mRFP positive colonies began to emerge at the end of stage 3, however, the efficiency increased significantly compared to 2D induction (Figure 4D). At day 16, we observed Pdx1-mRFP/insulin-hrGFP double positive colonies emerge with high efficiency (Figure 4D). These Pdx1/insulin double positive colonies rapidly grew over time, and the percentages of Pdx1+ and Pdx1+/insulin+ cells were 50-60% and 30-40% at day 34, respectively (Figure 4D). At day 34, we also performed IF analysis and confirmed the Pdx1 and insulin expression (Figure 4E).
Figure 4.
3D induction enhances the efficiency of beta cell differentiation. A. Analysis of DE gene expression by qPCR. Expression levels of Sox17 and Foxa2 were analyzed in 2D + 5Aza, 3D + 5Aza, and untreated control groups at the end of stage 1. Expression data are normalized to GAPDH transcript level. Each experiment was repeated independently three times. *P < 0.05; **P < 0.01. B. Flow cytometric analysis. Representative dot plots of undifferentiated iPSCs (left) and differentiated cells (Day 3) in 3D + 5Aza group (right) co-stained with anti-Sox17 and anti-Foxa2. Percentage in the upper right quadrant indicates the percentage of Sox17 and Foxa2 double positive cells. C. IF analysis of DE markers. Differentiated cells were stained by anti-Sox17 (1:100) and anti-Foxa2 (1:50) antibodies. Nuclei were highlighted with DAPI staining. Bar is 200 µm. D. Real-time monitoring expression of Pdx1 and insulin. Pdx1-mRFP positive colonies began to emerge at day 7 and the efficiency increased significantly. Bar is 500 µm. Pdx1-mRFP/insulin-hrGFP double positive colonies then emerged with high efficiency at day 16. Bar is 500 µm. These double positive colonies rapidly grew over time (Day 34). Bar is 200 µm. E. IF analysis. Parental iPSCs (without dual-reporter) were induced in parallel using the corresponding protocols and co-stained by anti-Pdx1 (1:100) and anti-insulin (1:50) antibodies at day 34. Nuclei were highlighted with DAPI staining. Bar is 100 µm.
To fully characterize the IPCs from 3D induction, we preformed qPCR to analyze dynamic expression of Pdx1 and insulin. The data showed that expression levels of Pdx1 and insulin were statistically higher in 3D induction than those in 2D induction at different time points (Figure 5A). Expression level of Pdx1 increased from day 7 to day 14 and maintained a high expression level up to day 34 in 3D induction (Figure 5A). Also, expression level of insulin increased greatly over time in 2D and 3D inductions, but the increase in 3D induction was more significant than that in 2D induction from day 16 to day 34 (Figure 5A). In addition, RT-PCR analysis showed that IPCs from 3D induction expressed beta cell-specific genes Nkx6.1, Ngn3, Nkx2.2, and MAFA, and beta cell function-related genes PCSK1 (PC1), PC2, Kir6.2, Sur1, and glucose transporter 2 (Glut 2), indicating the cells acquired a more mature function (Figure 5B). We also observed expression of other pancreatic islet hormone genes, i.e. glucagon (Gcg), somatostatin (Sst), and pancreatic polypeptide (Ppy) (Figure 5B), suggesting that the derivatives of 3D induction were still a mixture of different islet cell types. One of the most critical beta cell phenotypes is the capacity to synthesize and process insulin (immature beta cells), and to secrete insulin in response to glucose challenge (mature beta cells). IPCs from 2D and 3D inductions at day 34 post-directed differentiation were challenged with 25 mM glucose for 1 hr, and insulin secretion was measured by ELISA. As shown in Figure 5C, glucose-stimulated insulin secretion increased significantly in cells from 3D induction compared to those in 2D induction (P < 0.001), and there was only a slight elevated trend in cells from 2D induction (P > 0.05). To exclude the effect of osmolarity, a non-metabolizable glucose analog, 2-DG, at the same concentration as glucose (25 mM) was used as a control and failed to trigger insulin secretion. These results suggest that IPCs from 3D induction had acquired beta cell function, as they were capable of glucose-stimulated insulin secretion via a process coupling glucose sensing, uptake, and metabolism with insulin synthesis, processing, and release, consistent with the aforementioned RT-PCR analysis (Figure 5B).
Figure 5.
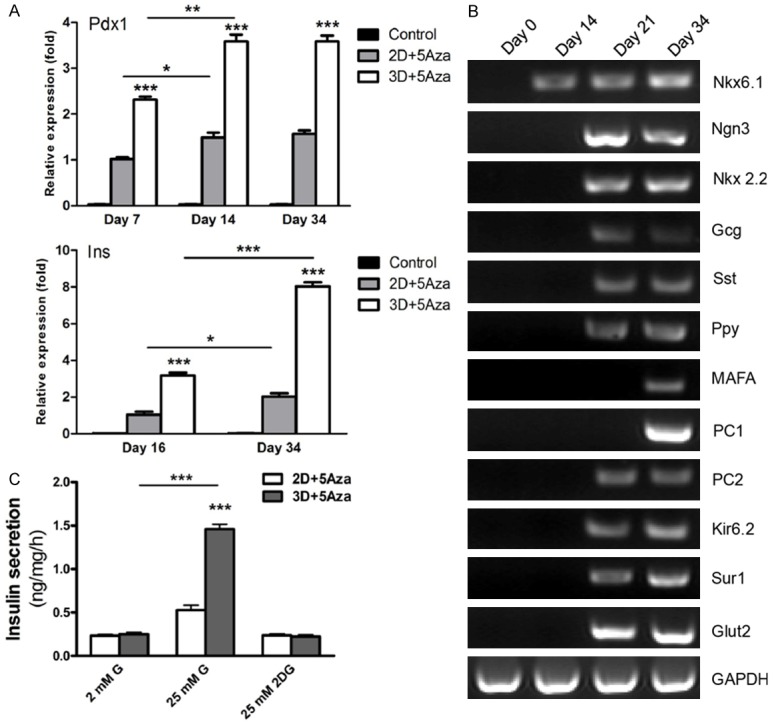
Characterization of IPCs. A. Analysis of Pdx1 and insulin expression by qPCR. Expression levels of Pdx1 and insulin were analyzed in 2D + 5Aza, 3D + 5Aza, and untreated control groups. Expression data are normalized to GAPDH transcript level. Each experiment was repeated independently three times. *P < 0.05; **P < 0.01; ***P < 0.001. B. Beta cell-related gene expression by RT-PCR. Total RNA was extracted from differentiated cells at days 0, 14, 21, and 34 following beta cell differentiation. C. ELISA for insulin secretion. The differentiated cells from 2D and 3D inductions at day 34 were analyzed for secretion of insulin upon exposure to 2 and 25 mM glucose or 25 mM 2DG. Secreted insulin in the culture medium was assayed by ELISA and expressed as amount per mg of total protein. Data represent the means ± SD of triplicate wells. Each experiment was repeated independently three times. ***P < 0.001.
Discussion
Pancreatic beta cell replacement therapy is considered one of the most promising approaches for treating and curing T1D. Directed differentiation of hPSCs into functional IPCs holds great promise for cell replacement therapy for patients suffering from diabetes. Despite recent advances in the development of beta cell differentiation protocols from hPSCs, it is becoming clear that the hPSC-derived beta-like cells have shortcomings compared to human beta cells [30,31], and the efficiencies of differentiation can be variable depending on the hPSC lines used [19,32,33]. Therefore, advanced methodologies are highly desirable for assisting efforts to develop and refine beta cell differentiation protocols from hPSCs. Pdx1 and insulin are key beta cell specific markers at different stages of beta cell development. Pdx1, a homeodomain transcription factor, plays crucial roles in early embryonic pancreatic formation, specification of different endocrine lineages, and later maturation of beta cell function [34,35]. Pdx1 expression is observed as early as embryonic day 8.5 (E8.5) in mouse [36] and around gestational week 4 (G4w) in human [37]. In rodents, Pdx1 is required for early embryonic development of the pancreas, and null mice do not develop a pancreas and die within a few days after birth [38,39]. Pdx1 is also required for the subsequent differentiation of the pancreatic lineage. During the formation of endocrine cells, elevated expression level of Pdx1 is pivotal for the commitment and differentiation of beta cells [40,41]. In adulthood, Pdx1 ultimately becomes restricted to beta and delta cells in the islets, and it maintains beta cell identity and function through the regulation of genes involved in glucose homeostasis [42]. During pancreas morphogenesis, at E10, a small number of insulin-expressing cells first arise in rodents, most of them co-expressing glucagon (primary transition) [43]. However, these few early scattered cells that produce both insulin and glucagon will not contribute to the mature endocrine compartment [44]. At approximately E13-15, the fully differentiated insulin-expressing beta cells appear during the secondary transition. In humans, the first endocrine cells that appear in pancreas are insulin-expressing cells in G7.5w, and they remain the most prevalent endocrine cell type during the first trimester [37,45]. The vast majority of alpha, beta, and delta-cells in the fetal human pancreas appear to express a single hormone [45]. However, there are a considerable number of bi-hormonal cells that express both insulin and glucagon (20-40% of alpha and beta-cells) between G9w and G16w, and they significantly decline thereafter [45,46]. These bi-hormonal cells are rarely found by the end of the late stage of prenatal pancreas development [47]. Therefore, monitoring expression of Pdx1 and insulin will greatly aid the establishment of beta cell differentiation protocols. This idea is supported by recent studies that genetically modified PSCs reporting expression of specific genes are of great value for differentiation protocol optimization and for the purification of relevant cell populations from heterogeneous cell cultures, such as Pdx1-GFP reporter mouse iPSCs, Nkx6.1 hiPSC lines, and INSGFP/w hESCs [48-50].
In the current study, we constructed a novel highly efficient Pdx1-mRFP/insulin-hrGFP dual reporter vector where the expression of RFP and GFP were driven by Pdx1 and insulin promoters, respectively. The function of the dual-reporter vector was validated in INS-1 cells as demonstrated by co-expression of RFP and GFP in these cells. Finally, we transduced iPSCs with the dual-reporter vector and generated a dual-reporter iPSC line.
To investigate the functionality of dual-reporter cell lines for real-time monitoring of cell differentiation, the cells were directed to differentiate into beta cells by growth factors and small molecules. During the DE induction, we found remarkable cell loss after 3-day induction, which might be due to activin A-induced cell apoptosis and reduced cell viability [26,27]. After optimization of the basal induction medium at stage 1, the cell survival rates were dramatically increased. Then the cells were directed into PGT and PPs by a combined treatment of KGF, SANT1, RA, LDN193189, and PDBu. The delayed RFP expression indicated Pdx1 expression was inefficient and the protocol required further refinement. When pretreated with 5Aza before the initiation of induction, the cells expressed RFP two days earlier than before, suggesting the efficient specification of PPs. This result is consistent with reports that epigenetic regulation plays important roles in cell differentiation [28,29]. In the process of optimizing the protocols, we observed RFP and RFP/GFP positive cells tended to aggregate 3D structures. Therefore, we speculated that 3D induction would further enhance the efficiency of beta cell differentiation. As expected, 3D induction not only significantly increased the efficiency of PP specification, but also enhanced the yield of IPCs, compared to 2D induction. The proliferation rates of Pdx1/insulin double positive cells in 3D induction were also faster than those in 2D induction. Furthermore, the differentiated cells of 3D induction were more mature and capable of glucose-stimulated insulin secretion. Several reports showed that the 3D differentiation culture system was superior to the traditional 2D culture system [10,11]. The 3D induction system could increase the efficiency of cell differentiation and produce more mature IPCs [12,29], which is in line with our results. The reason for this improvement is that, during embryogenesis, the developing cells are arranged in 3D clusters, which support cell-cell paracrine signaling [51,52]. 3D differentiation of hiPSCs has also been used to derive functionally and morphologically superior tissues, such as cerebral organoids [51] and liver buds [52].
Taken together, our results demonstrate that Pdx1/insulin dual-reporter hiPSC lines are a valuable resource for implementing and optimizing differentiation protocols towards the pancreatic lineage. Using the dual-reporter line, we established a novel and efficient beta cell differentiation protocol from MRC5-dervied iPSCs by simultaneously real-time monitoring the expression of Pdx1 and insulin, which demonstrated the advantages of time-saving and cost-effectiveness. Furthermore, the dual-reporter cell line will also be a useful tool for investigating the mechanisms of beta cell differentiation by isolating different populations of Pdx1 and Pdx1/insulin positive cells. We hope this study will facilitate the development of novel protocols for generating IPCs from hPSCs.
Acknowledgements
This project was supported by the National Science and Technology Major Project (2018ZX0801012B and 2016ZX08006002-004) and the National Science and Technology Support Program (2015BA108B03, H. Y.).
Disclosure of conflict of interest
None.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Hallett PJ, Deleidi M, Astradsson A, Smith GA, Cooper O, Osborn TM, Sundberg M, Moore MA, Perez-Torres E, Brownell AL, Schumacher JM, Spealman RD, Isacson O. Successful function of autologous iPSC-derived dopamine neurons following transplantation in a non-human primate model of Parkinson’s disease. Cell Stem Cell. 2015;16:269–274. doi: 10.1016/j.stem.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandai M, Fujii M, Hashiguchi T, Sunagawa GA, Ito SI, Sun J, Kaneko J, Sho J, Yamada C, Takahashi M. iPSC-derived retina transplants improve vision in rd1 End-stage retinal-degeneration mice. Stem Cell Reports. 2017;8:1112–1113. doi: 10.1016/j.stemcr.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiba Y, Gomibuchi T, Seto T, Wada Y, Ichimura H, Tanaka Y, Ogasawara T, Okada K, Shiba N, Sakamoto K, Ido D, Shiina T, Ohkura M, Nakai J, Uno N, Kazuki Y, Oshimura M, Minami I, Ikeda U. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature. 2016;538:388–391. doi: 10.1038/nature19815. [DOI] [PubMed] [Google Scholar]
- 7.Gillespie KM. Type 1 diabetes: pathogenesis and prevention. CMAJ. 2006;175:165–170. doi: 10.1503/cmaj.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joost HG. Pathogenesis, risk assessment and prevention of type 2 diabetes mellitus. Obes Facts. 2008;1:128–137. doi: 10.1159/000137673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atkinson MA, Eisenbarth GS. Type 1 diabetes: new perspectives on disease pathogenesis and treatment. Lancet. 2001;358:221–229. doi: 10.1016/S0140-6736(01)05415-0. [DOI] [PubMed] [Google Scholar]
- 10.D’Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, Baetge EE. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 11.Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, Agulnick AD, D’Amour KA, Carpenter MK, Baetge EE. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 12.Pagliuca FW, Millman JR, Gurtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA. Generation of functional human pancreatic beta cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 14.Hebrok M, Kim SK, St Jacques B, McMahon AP, Melton DA. Regulation of pancreas development by hedgehog signaling. Development. 2000;127:4905–4913. doi: 10.1242/dev.127.22.4905. [DOI] [PubMed] [Google Scholar]
- 15.Maehr R, Chen S, Snitow M, Ludwig T, Yagasaki L, Goland R, Leibel RL, Melton DA. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci U S A. 2009;106:15768–15773. doi: 10.1073/pnas.0906894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Borowiak M, Fox JL, Maehr R, Osafune K, Davidow L, Lam K, Peng LF, Schreiber SL, Rubin LL, Melton D. A small molecule that directs differentiation of human ESCs into the pancreatic lineage. Nat Chem Biol. 2009;5:258–265. doi: 10.1038/nchembio.154. [DOI] [PubMed] [Google Scholar]
- 17.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, Miller J, Schlaeger T, Daley GQ, Feinberg AP. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, Khvorostov I, Ott V, Grunstein M, Lavon N, Benvenisty N, Croce CM, Clark AT, Baxter T, Pyle AD, Teitell MA, Pelegrini M, Plath K, Lowry WE. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, Melton DA. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 20.Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, Gnirke A, Eggan K, Meissner A. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chetty S, Pagliuca FW, Honore C, Kweudjeu A, Rezania A, Melton DA. A simple tool to improve pluripotent stem cell differentiation. Nat Methods. 2013;10:553–556. doi: 10.1038/nmeth.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao LZ, Tang DQ, Horb ME, Li SW, Yang LJ. High glucose is necessary for complete maturation of Pdx1-VP16-expressing hepatic cells into functional insulin-producing cells. Diabetes. 2004;53:3168–3178. doi: 10.2337/diabetes.53.12.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szabat M, Luciani DS, Piret JM, Johnson JD. Maturation of adult beta-cells revealed using a Pdx1/insulin dual-reporter lentivirus. Endocrinology. 2009;150:1627–1635. doi: 10.1210/en.2008-1224. [DOI] [PubMed] [Google Scholar]
- 24.Curran MA, Nolan GP. Recombinant feline immunodeficiency virus vectors. Preparation and use. Methods Mol Med. 2002;69:335–350. doi: 10.1385/1-59259-141-8:335. [DOI] [PubMed] [Google Scholar]
- 25.Loh KM, Ang LT, Zhang J, Kumar V, Ang J, Auyeong JQ, Lee KL, Choo SH, Lim CY, Nichane M, Tan J, Noghabi MS, Azzola L, Ng ES, Durruthy-Durruthy J, Sebastiano V, Poellinger L, Elefanty AG, Stanley EG, Chen Q, Prabhakar S, Weissman IL, Lim B. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell. 2014;14:237–252. doi: 10.1016/j.stem.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawton BR, Sosa JA, Roman S, Krause DS. Effect of a matrigel sandwich on endodermal differentiation of human embryonic stem cells. Stem Cell Rev. 2013;9:578–585. doi: 10.1007/s12015-013-9447-2. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, Barron MR, Hou L, Soerens AG, Yu J, Palecek SP, Lyons GE, Thomson JA, Herron TJ, Jalife J, Kamp TJ. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ Res. 2012;111:1125–1136. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pennarossa G, Maffei S, Campagnol M, Tarantini L, Gandolfi F, Brevini TA. Brief demethylation step allows the conversion of adult human skin fibroblasts into insulin-secreting cells. Proc Natl Acad Sci U S A. 2013;110:8948–8953. doi: 10.1073/pnas.1220637110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manzar GS, Kim EM, Zavazava N. Demethylation of induced pluripotent stem cells from type 1 diabetic patients enhances differentiation into functional pancreatic beta cells. J Biol Chem. 2017;292:14066–14079. doi: 10.1074/jbc.M117.784280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kushner JA, MacDonald PE, Atkinson MA. Stem cells to insulin secreting cells: two steps forward and now a time to pause? Cell Stem Cell. 2014;15:535–536. doi: 10.1016/j.stem.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Johnson JD. The quest to make fully functional human pancreatic beta cells from embryonic stem cells: climbing a mountain in the clouds. Diabetologia. 2016;59:2047–2057. doi: 10.1007/s00125-016-4059-4. [DOI] [PubMed] [Google Scholar]
- 32.Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L, Rodolfa CT, Dimos JT, Mikkilineni S, MacDermott AB, Woolf CJ, Henderson CE, Wichterle H, Eggan K. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishizawa M, Chonabayashi K, Nomura M, Tanaka A, Nakamura M, Inagaki A, Nishikawa M, Takei I, Oishi A, Tanabe K, Ohnuki M, Yokota H, Koyanagi-Aoi M, Okita K, Watanabe A, Takaori-Kondo A, Yamanaka S, Yoshida Y. Epigenetic variation between human induced pluripotent stem cell lines is an indicator of differentiation capacity. Cell Stem Cell. 2016;19:341–354. doi: 10.1016/j.stem.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 34.Shih HP, Wang A, Sander M. Pancreas organogenesis: from lineage determination to morphogenesis. Annu Rev Cell Dev Biol. 2013;29:81–105. doi: 10.1146/annurev-cellbio-101512-122405. [DOI] [PubMed] [Google Scholar]
- 35.Edlund H. Pancreatic organogenesis--developmental mechanisms and implications for therapy. Nat Rev Genet. 2002;3:524–532. doi: 10.1038/nrg841. [DOI] [PubMed] [Google Scholar]
- 36.Miki R, Yoshida T, Murata K, Oki S, Kume K, Kume S. Fate maps of ventral and dorsal pancreatic progenitor cells in early somite stage mouse embryos. Mech Dev. 2012;128:597–609. doi: 10.1016/j.mod.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Jennings RE, Berry AA, Kirkwood-Wilson R, Roberts NA, Hearn T, Salisbury RJ, Blaylock J, Piper Hanley K, Hanley NA. Development of the human pancreas from foregut to endocrine commitment. Diabetes. 2013;62:3514–3522. doi: 10.2337/db12-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 39.Holland AM, Hale MA, Kagami H, Hammer RE, MacDonald RJ. Experimental control of pancreatic development and maintenance. Proc Natl Acad Sci U S A. 2002;99:12236–12241. doi: 10.1073/pnas.192255099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernardo AS, Hay CW, Docherty K. Pancreatic transcription factors and their role in the birth, life and survival of the pancreatic beta cell. Mol Cell Endocrinol. 2008;294:1–9. doi: 10.1016/j.mce.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Gao N, LeLay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22:3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao T, McKenna B, Li C, Reichert M, Nguyen J, Singh T, Yang C, Pannikar A, Doliba N, Zhang T, Stoffers DA, Edlund H, Matschinsky F, Stein R, Stanger BZ. Pdx1 maintains beta cell identity and function by repressing an alpha cell program. Cell Metab. 2014;19:259–271. doi: 10.1016/j.cmet.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teitelman G, Alpert S, Polak JM, Martinez A, Hanahan D. Precursor cells of mouse endocrine pancreas coexpress insulin, glucagon and the neuronal proteins tyrosine hydroxylase and neuropeptide Y, but not pancreatic polypeptide. Development. 1993;118:1031–1039. doi: 10.1242/dev.118.4.1031. [DOI] [PubMed] [Google Scholar]
- 44.Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127:2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 45.Riedel MJ, Asadi A, Wang R, Ao Z, Warnock GL, Kieffer TJ. Immunohistochemical characterisation of cells co-producing insulin and glucagon in the developing human pancreas. Diabetologia. 2012;55:372–381. doi: 10.1007/s00125-011-2344-9. [DOI] [PubMed] [Google Scholar]
- 46.Jeon J, Correa-Medina M, Ricordi C, Edlund H, Diez JA. Endocrine cell clustering during human pancreas development. J Histochem Cytochem. 2009;57:811–824. doi: 10.1369/jhc.2009.953307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meier JJ, Kohler CU, Alkhatib B, Sergi C, Junker T, Klein HH, Schmidt WE, Fritsch H. Beta-cell development and turnover during prenatal life in humans. Eur J Endocrinol. 2010;162:559–568. doi: 10.1530/EJE-09-1053. [DOI] [PubMed] [Google Scholar]
- 48.Micallef SJ, Li X, Schiesser JV, Hirst CE, Yu QC, Lim SM, Nostro MC, Elliott DA, Sarangi F, Harrison LC, Keller G, Elefanty AG, Stanley EG. INS(GFP/w) human embryonic stem cells facilitate isolation of in vitro derived insulin-producing cells. Diabetologia. 2012;55:694–706. doi: 10.1007/s00125-011-2379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta SK, Wesolowska-Andersen A, Ringgaard AK, Jaiswal H, Song L, Hastoy B, Ingvorsen C, Taheri-Ghahfarokhi A, Magnusson B, Maresca M, Jensen RR, Beer NL, Fels JJ, Grunnet LG, Thomas MK, Gloyn AL, Hicks R, McCarthy MI, Hansson M, Honore C. NKX6.1 induced pluripotent stem cell reporter lines for isolation and analysis of functionally relevant neuronal and pancreas populations. Stem Cell Res. 2018;29:220–231. doi: 10.1016/j.scr.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 50.Porciuncula A, Kumar A, Rodriguez S, Atari M, Arana M, Martin F, Soria B, Prosper F, Verfaillie C, Barajas M. Pancreatic differentiation of Pdx1-GFP reporter mouse induced pluripotent stem cells. Differentiation. 2016;92:249–256. doi: 10.1016/j.diff.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 51.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, Aoyama S, Adachi Y, Taniguchi H. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]