Abstract
Colorectal cancer (CRC) is one of the leading causes of cancer-associated mortality worldwide. The prognosis of patients with CRC at an advanced stage is poor. Biomarkers currently used in clinical practice, including carcinoembryonic antigen (CEA) and cancer antigen (CA) 19-9, lack sufficient sensitivity and specificity for early diagnosis and prediction, therefore there remains a requirement to improve the prognosis of patients with CRC. Long non-coding RNAs (lncRNAs) have been revealed to serve fundamental roles in various pathophysiological processes, including cancer initiation and progression. The present study investigated the expression and clinical significance of the lncRNA nuclear factor-κB interacting long non-coding RNA (NKILA) in CRC. It was identified that NKILA was downregulated in six CRC cell lines and tissues (n=173). Low NKILA expression was significantly associated with a poor differentiation grade, larger tumor size and advanced Tumor-Node-Metastases stages. Further statistical analyses revealed that low NKILA expression predicted poor overall survival (OS) rate and progression-free survival (PFS) rate. In addition, low NKILA expression was determined as an independent risk factor for poor OS and PFS. Furthermore, NKILA exhibited a relatively high sensitivity and specificity compared with CEA and CA19-9 in the early diagnosis of CRC. The serum level of NKILA was positively correlated with the level in tissues. In addition, a decreased NKILA level in serum was revealed to be partially restored post-operatively. In conclusion, low NKILA expression has been demonstrated to accelerate CRC progression and NKILA may be a potential novel biomarker in early diagnosis and prognosis of patients with CRC.
Keywords: nuclear factor-κB interacting long non-coding RNA, colorectal cancer, early diagnosis, prognosis
Introduction
Colorectal cancer (CRC) is the fifth most commonly diagnosed cancer type and the fifth most common cause of cancer-associated mortality in China in 2015 (1). While invasive types of CRC that have not compromised regional lymph nodes (stages I–II) exhibit relatively good prognoses with the current treatment strategies available and are curable in 73% of cases, the disease progression is fast and untreated tumors rapidly disseminate to lymph nodes (stage III) and metastasize to distant sites (stage IV) (2). Patients with advanced CRC stages exhibit a significantly lower survival rate. Therefore, more diagnostic biomarkers with sufficient sensitivity and specificity are essential for improving the prognosis, particularly for patients with advanced-stage CRC. In addition, prognostic markers are required to identify patients with cancer who are at high risk of metastatic relapse and are therefore potential candidates for adjuvant systemic therapy. Additionally, prognostic factors can define the effects of tumor characteristics on patient outcome (3). Therefore, reliable diagnostic and prognostic markers, analyzed in non-invasively obtained surrogate samples, may exhibit vast clinical potential.
Long non-coding RNAs (lncRNAs) are defined as capped transcripts >200 nucleotides, which coincides with the cut-off for a number of RNA extraction protocols (4–6). Numerous lncRNAs have been annotated in different species and tissues (7). Notably, lncRNAs are often expressed in a disease-, tissue- or developmental stage-specific manner, which makes these molecules attractive therapeutic targets and indicates specific lncRNA functions in development and disease (8–10). These features of lncRNAs also make them potential diagnostic and prognostic biomarkers for patients with cancer (11). Furthermore, a number of studies have suggested that detection of lncRNAs provides a novel and promising early diagnostic option for cancer screening (12–16). Nuclear factor (NF)-κB interacting long non-coding RNA (NKILA) was first reported to suppress the progression of breast cancer via its binding to NF-κB/inhibitor (I)κB and directly masking phosphorylation motifs of IκB, which inhibits IκB kinase-induced IκB phosphorylation and NF-κB activation (17). Further studies have demonstrated that NKILA suppresses the progression of malignant melanoma (18), non-small cell lung cancer (19), esophageal squamous cell carcinoma (20) and laryngeal cancer (21). Notably, NF-κB has been recognized as a critical factor in the initiation and progression of CRC (22–24). However, to the best of our knowledge, the role of NKILA in CRC remains to be defined.
The present study investigated the function of NKILA in CRC. NKILA was demonstrated to be downregulated in CRC cell lines and tissues. Loss of NKILA was revealed to be associated with the clinical progression of CRC. Furthermore, decreased NKILA expression has identified to predict poor survival and serve as an independent CRC prognostic factor. In addition, NKILA downregulation was confirmed to be a promising diagnostic biomarker with sufficient sensitivity and specificity for patients with early CRC. In summary, the current study provides insights into NKILA and demonstrates a role of this lncRNA in early CRC diagnosis and prognosis.
Materials and methods
Study subjects
CRC cases were recruited at the time of diagnosis among patients treated at the Central Hospital of Weihai (Weihai, China). Only histologically confirmed new CRC cases that had not previously been diagnosed for cancer were included in the study. In addition, none of the involved patients had received anticancer treatments, including chemo-, radio- or targeted therapy. Written informed consent was obtained from each patient. The present study was approved by the Ethics Committee of the Central Hospital of Weihai (Weihai, China).
Two different groups of patients with sporadic CRC were enrolled in the present study. The first group included 173 patients who underwent surgical resection between January 2011 and May 2013. Adjacent tumor tissues of the patients in the first group were at least 1 cm away from the tumor tissues. The second group consisted of 70 patients with CRC diagnosed at an early stage [Tumor-Node-Metastasis (TNM) stage I] (25), 70 patients with adenoma polyps treated between March 2016 and April 2017, and 70 healthy volunteers. Harvest of the tissues was achieved through surgical resection or colonoscopy. Pre- and postoperative peripheral blood samples of the patients in the second group, including the 70 patients with CRC diagnosed at an early stage and the 70 patients with adenoma polyps, were collected 3 days pre- and 2 weeks postoperatively, respectively. A total of 20 cases were randomly selected from the 70 volunteers and their peripheral blood samples were also collected. The obtained specimens were collected immediately after resection, frozen and stored in liquid nitrogen until used. Diagnosis of the patients was confirmed by pathology. The values of carcinoembryonic antigen (CEA) and cancer antigen (CA)19-9 used in this study were obtained in the medical records.
Patients were followed-up for a mean period of 40.4 months (range, 3–60 months). Overall survival (OS) was defined as the time interval between the date of diagnosis and the end of the follow-up or the date at which the patient succumbed to mortality due to CRC. Progression-free survival (PFS) was defined as the interval between the date of surgery and recurrence; if recurrence was not diagnosed, patients were censored on the date of mortality or the last follow-up.
Cell lines and cell culture
Six human CRC cell lines (HT29, RKO, LOVO, DLD1, SW480 and HCT116) and a human intestinal epithelial cell line (HIEC-6) were obtained from the Chinese Type Culture Collection, Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin sodium and 100 mg/ml streptomycin sulphate, at 37°C in a humidified air atmosphere containing 5% CO2. Cells were used for experiments when they were in the logarithmic growth phase.
RNA extraction and complementary DNA (cDNA) synthesis
Total RNA from HIEC-6, CRC cells (HT29, RKO, LOVO, DLD1, SW480 and HCT116), tumor adjacent tissue samples, adenomas, tumor samples and blood plasma was isolated using a MirVana isolation kit (Thermo Fisher Scientific, Inc.). The RNA concentration was determined by measuring the optical density (OD) using a NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, Inc.). Only samples with an OD260/OD280 ratio of 1.8–2.0 were utilized for further analysis. Total RNA was reverse transcribed into first-strand cDNA using SuperScript III® (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Briefly, the samples were incubated for 5 min at 25°C followed by 60 min at 42°C, then the reaction was terminated by healing at 70°C for 5 min. The obtained cDNA was stored at −20°C.
Quantitative polymerase chain reaction (qPCR)
qPCR was performed using the ABI PRISM 7000 Fluorescent Quantitative PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. SYBR Green qPCR master mix (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used for qPCR. Briefly, reactions were loaded into a 96-well plate in duplicate and incubated at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 1 min and extension at 70°C for 1 min. The results were normalized to GAPDH expression levels. The PCR primers were as follows: NKILA, forward, 5′-GGGGTACCAGACCCGGCACCCGCGCAA-3′ and reverse, 5′-CGGGATCCCCAGTTAAATTGAGATATACTTACAC-3′; and GAPDH, forward, 5′-CGCTCTCTGCTCCTCCTGTTC-3′ and reverse, 5′-ATCCGTTGACTCCGACCTTCAC-3′. All qPCR reactions were performed in triplicate. Relative quantification of gene expression was calculated and normalized using the 2−ΔΔCq method (26).
Statistical analysis
All statistical analyses were performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). Data are presented as the mean ± standard deviation. χ2 test or Student's t-test were used to compare the differences between two independent groups where appropriate. One-way analysis of variance and Dunnett's t-test was used to examine associations among three groups. Kaplan-Meier analysis was used to generate survival curves followed by a log-rank test to determine the association between NKILA expression and clinical outcomes. The effects of variables on survival were determined by univariate and multivariate Cox proportional hazards analysis. Receiver operating characteristic (ROC) curves were plotted and the area under the curve (AUC) was calculated to assess the specificity and sensitivity of distinguishing patients with CRC from healthy controls. The correlation of NKILA expression levels in serum and tissues was analyzed by Spearman's rank correlation coefficient. Wilcoxon signed-rank test was used to compare the serum levels in patients with CRC between pre- and postoperative time points. P<0.05 was considered to indicate a statistically significant difference.
Results
Low expression of NKILA is associated with clinical progression in CRC
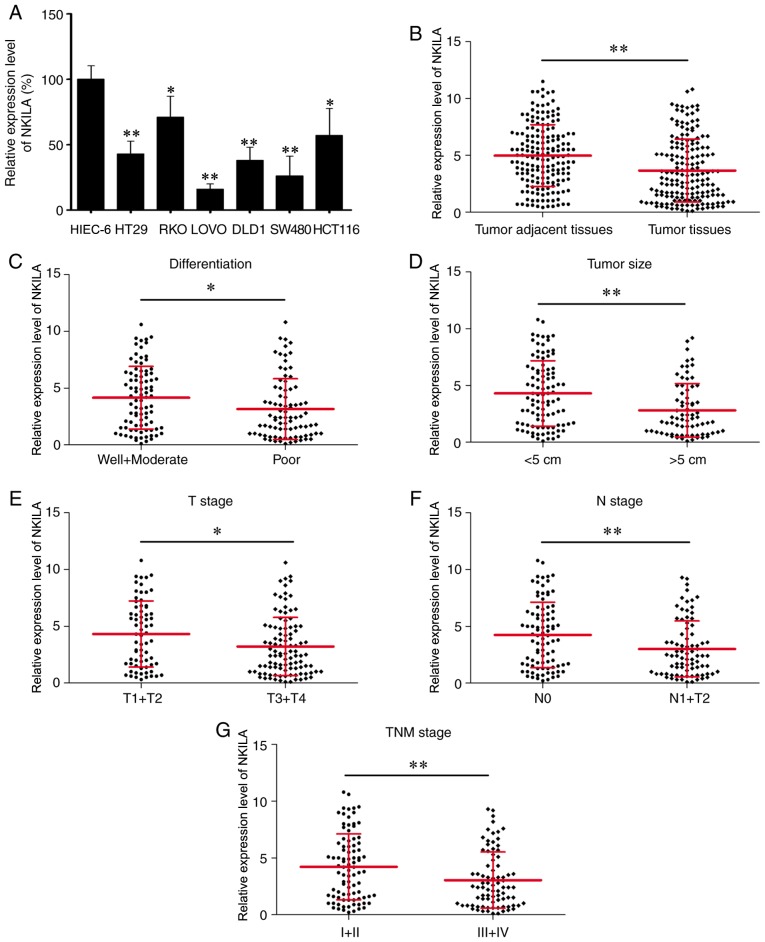
To verify the functional role of NKILA in CRC, its expression was measured in six CRC cell lines and HIEC-6 by reverse transcription-qPCR. The results demonstrated that the expression level of NKILA was lower in the six CRC cells compared with HIEC-6 (Fig. 1A). Notably, a significantly decreased expression level of NKILA was identified in CRC tissues (n=173) compared with paired tumor adjacent tissues (n=173; Fig. 1B). In addition, statistical analyses revealed that patients with poorly differentiated cancer, a larger tumor size (>5 cm), and advanced T (T3+T4), N (N1+N2) and TNM (III+IV) stages exhibited significantly lower NKILA expression levels compared with patients with well and moderately differentiated cancer, a smaller tumor size (<5 cm), and less advanced T (T1+T2), N (N0) and TNM (I+II) stages, respectively (Fig. 1C-G). Furthermore, the patients were divided into two groups, a low NKILA expression group (n=102) and a high NKILA expression group (n=71), with the mean NKILA expression level (3.7) serving as the cut-off value (patients with the exact mean value be placed in the high NKILA group). To improve understanding of the clinical significance of NKILA in CRC, the differences in the clinicopathological features between the two groups were elucidated (Table I). As a result, low NKILA expression was identified to be associated with poor differentiation grade (P=0.002), larger tumor size (>5 cm) (P=0.001), and advanced T (T3+T4) (P=0.022), N (N1+N2) (P=0.001) and TNM (III+IV) (P=0.002) stages. Overall, these findings suggested that NKILA expression was decreased in CRC and a low NKILA expression level was associated with CRC clinical progression.
Figure 1.
NKILA expression in CRC and investigation of its clinical significance. (A) Detection of NKILA expression in CRC cells and HIEC-6 by RT-qPCR. (B) RT-qPCR was performed to evaluate NKILA expression in CRC tissues (n=173) and paired tumor adjacent tissues (n=173). NKILA expression level was measured by RT-qPCR in patients with CRC with different (C) differentiation grades, (D) tumor sizes, (E) T stages, (F) N stages and (G) TNM stages. *P<0.05, **P<0.01. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CRC, colorectal cancer; NKILA, NF-κB interacting long non-coding RNA; TNM, Tumor-Node-Metastasis.
Table I.
Association between NKILA expression and clinicopathological characteristics of patients with colorectal cancer.
| NKILA expression | ||||
|---|---|---|---|---|
| Characteristic | Total (n=173) | Low (n=102) | High (n=71) | P-value |
| Age, years | 0.216 | |||
| <65 | 98 | 53 | 44 | |
| ≥65 | 75 | 48 | 27 | |
| Sex | 0.628 | |||
| Male | 89 | 59 | 40 | |
| Female | 84 | 53 | 31 | |
| CEA, µg/ml | 0.225 | |||
| <4.5 | 71 | 38 | 33 | |
| ≥4.5 | 102 | 64 | 38 | |
| CA19-9, U/ml | 0.166 | |||
| <50 | 89 | 48 | 41 | |
| ≥50 | 84 | 54 | 30 | |
| Tumor location | 0.918 | |||
| Colon | 108 | 64 | 44 | |
| Rectum | 65 | 38 | 27 | |
| Differentiation grade | 0.002 | |||
| Well+moderate | 88 | 42 | 46 | |
| Poor | 85 | 60 | 25 | |
| Tumor size, cm | 0.001 | |||
| <5 | 99 | 48 | 51 | |
| ≥5 | 74 | 54 | 20 | |
| T stage | 0.022 | |||
| T1+T2 | 70 | 34 | 36 | |
| T3+T4 | 103 | 68 | 35 | |
| N stage | 0.001 | |||
| N0 | 91 | 43 | 48 | |
| N1+N2 | 82 | 59 | 23 | |
| M stage | 0.920 | |||
| M0 | 154 | 91 | 63 | |
| M1 | 19 | 11 | 8 | |
| TNM stage | 0.002 | |||
| I+II | 90 | 43 | 47 | |
| III+IV | 83 | 59 | 24 | |
NKILA, nuclear factor-κB interacting long non-coding RNA; CEA, carcinoembryonic antigen; CA19-9, cancer antigen 19-9; TNM, Tumor-Node-Metastasis.
Low expression of NKILA indicates poor prognosis and serves as an independent factor for poor prognosis in CRC
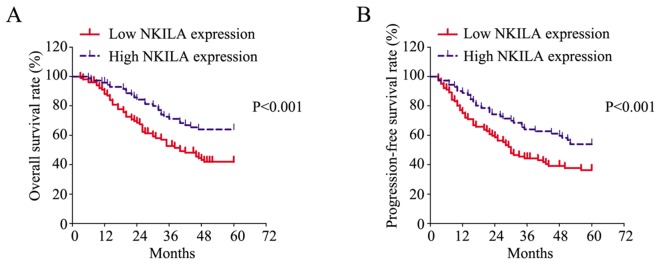
To determine the prognostic value of NKILA in CRC, Kaplan-Meier analysis was performed to evaluate the survival of patients and a log-rank test was conducted to analyze differences. As presented in Fig. 2A, patients with low NKILA expression exhibited a significantly poorer OS rate compared with patients with high NKILA expression. Furthermore, patients with low NKILA expression exhibited a significantly poorer PFS rate compared with patients with high NKILA expression (Fig. 2B).
Figure 2.
Low NKILA expression predicts poor survival in CRC. (A) Overall survival rate and (B) progression-free survival rate of patients with CRC with low or high NKILA expression. CRC, colorectal cancer; NKILA, NF-κB interacting long non-coding RNA.
In addition, the risk factors for poor CRC prognosis were statistically evaluated by univariate and multivariate Cox proportional hazards analysis. Six parameters were identified as risk factors for poor OS in CRC, including poor differentiation, advanced T (T3+T4), N (N1+N2), M (M1) and TNM (III+IV) stages, and low NKILA expression (Table II). Further examination of these factors with multivariate Cox proportional hazards analysis revealed that advanced M stage [M1; hazard ratio (HR), 4.224; 95% confidence interval (CI), 2.284–7.814; P<0.001] and low NKILA expression (HR, 0.870; 95% CI, 0.787–0.962; P=0.007) were two independent risk factors for poor OS. Consistently, six parameters, including poor differentiation, advanced T (T3+T4), N (N1+N2), M (M1) and TNM (III+IV) stages, and low NKILA expression, were also revealed as risk factors for poor PFS (Table III). Subjecting these factors to multivariate Cox proportional hazards analysis revealed that advanced M stage (M1; HR, 4.263; 95% CI, 2.382–7.630; P<0.001) and low NKILA expression (HR, 0.910; 95% CI, 0.833–0.994; P=0.036) were two independent risk factors for poor PFS. In summary, these results suggested that low NKILA expression may be a potential prognostic biomarker in CRC.
Table II.
Statistical analysis of risk factors for overall survival time of patients with colorectal cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age, years (<65 vs. ≥65) | 0.802 | 0.517–1.245 | 0.326 | |||
| Sex (male vs. female) | 1.106 | 0.655–1.575 | 0.945 | |||
| CEA level, µg/ml (<4.5 vs. ≥4.5) | 0.986 | 0.631–1.543 | 0.952 | |||
| CA19-9, U/ml (<50 vs. ≥50) | 1.192 | 0.768–1.849 | 0.434 | |||
| Tumor location (colon vs. rectum) | 1.005 | 0.639–1.580 | 0.983 | |||
| Differentiation (poor vs. well+moderate) | 1.560 | 1.003–2.425 | 0.048 | 1.476 | 0.940–2.317 | 0.091 |
| Tumor size, cm (≥5 vs. <5) | 1.100 | 0.705–1.717 | 0.675 | |||
| T stage (T1+T2 vs. T3+T4) | 1.751 | 1.096–2.797 | 0.019 | 1.064 | 0.635–1.782 | 0.814 |
| N stage (N1+N2 vs. N0) | 3.698 | 2.296–5.958 | <0.001 | 1.911 | 0.235–12.566 | 0.545 |
| M stage (M1 vs. M0) | 4.229 | 2.456–7.283 | <0.001 | 4.224 | 2.284–7.814 | <0.001 |
| TNM stage (III+IV vs. I+II) | 3.841 | 2.371–6.222 | <0.001 | 1.426 | 0.173–11.781 | 0.742 |
| NKILA expression (high vs. low) | 0.856 | 0.781–0.938 | 0.001 | 0.870 | 0.787–0.962 | 0.007 |
NKILA, NF-κB interacting long non-coding RNA; CEA, carcinoembryonic antigen; CA19-9, cancer antigen 19-9; TNM, Tumor-Node-Metastasis; CI, confidence interval; HR, hazard ratio.
Table III.
Statistical analysis of risk factors for progression-free survival time of patients with colorectal cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age, years (<65 vs. ≥65) | 0.788 | 0.524–1.187 | 0.255 | |||
| Sex (male vs. female) | 1.011 | 0.671–1.522 | 0.959 | |||
| CEA level, µg/ml (<4.5 vs. ≥4.5) | 1.012 | 0.667–1.535 | 0.957 | |||
| CA19-9, U/ml (<50 vs. ≥50) | 1.104 | 0.733–1.663 | 0.635 | |||
| Tumor location (colon vs. rectum) | 1.032 | 0.677–1.572 | 0.884 | |||
| Differentiation (poor vs. well+moderate) | 1.566 | 1.038–2.363 | 0.033 | 1.483 | 0.975–2.254 | 0.065 |
| Tumor size, cm (≥5 vs. <5) | 1.059 | 0.699–1.604 | 0.786 | |||
| T stage (T1+T2 vs. T3+T4) | 1.908 | 1.228–2.965 | 0.004 | 1.310 | 0.812–2.114 | 0.269 |
| N stage (N1+N2 vs. N0) | 3.162 | 2.055–4.868 | <0.001 | 1.566 | 0.196–12.523 | 0.673 |
| M stage (M1 vs. M0) | 4.793 | 2.843–8.081 | <0.001 | 4.263 | 2.382–7.630 | <0.001 |
| TNM stage (III+IV vs. I+II) | 3.264 | 2.114–5.040 | <0.001 | 1.445 | 0.178–11.693 | 0.730 |
| NKILA expression (high vs. low) | 0.889 | 0.820–0.964 | 0.005 | 0.910 | 0.833–0.994 | 0.036 |
NKILA, nuclear factor-κB interacting long non-coding RNA; CEA, carcinoembryonic antigen; CA19-9, cancer antigen 19-9; TNM, Tumor-Node-Metastasis; CI, confidence interval; HR, hazard ratio.
NKILA functions as a diagnostic biomarker in early CRC
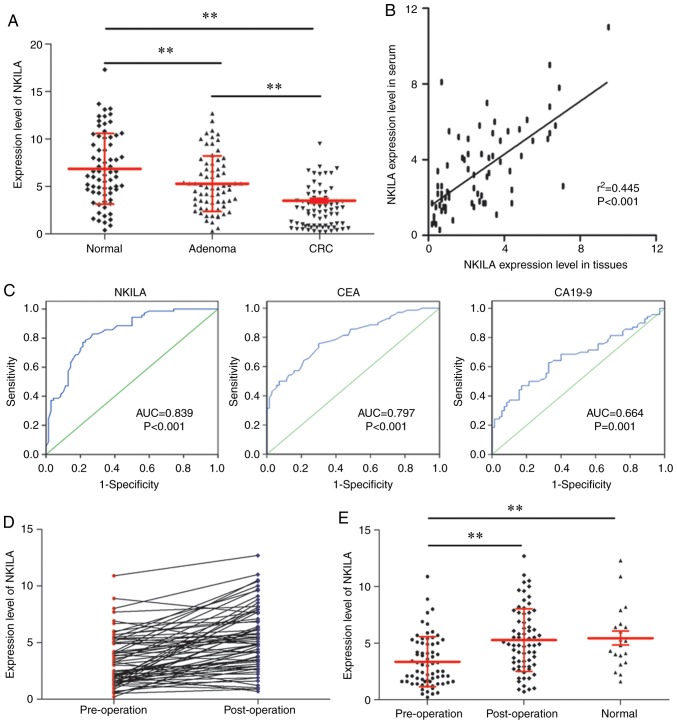
Early diagnosis serves a pivotal role in improving the prognosis of patients with CRC (27). The present study investigated the value of NKILA in the early diagnosis of CRC. By measuring the NKILA expression level in normal colorectal tissues (n=70), colorectal adenoma tissues (n=70) and early CRC tissues (TNM stage I; n=70), it was identified that the early CRC tissues exhibited a significantly lower NKILA expression level compared with the other two groups, and colorectal adenoma tissues demonstrated a significantly lower NKILA expression level compared with normal colorectal tissues. (Fig. 3A) Subsequently, the serum level of NKILA was measured for patients with early-stage CRC (n=70). Further statistical analysis determined that the serum level of NKILA was positively correlated with the corresponding NKILA level in tumor tissues (n=70; Fig. 3B) Furthermore, when comparing the specificity and sensitivity of NKILA with CEA and CA19-9 in the early diagnosis of CRC using ROC curves, NKILA exhibited a higher AUC (0.839; P<0.001) compared with CEA (AUC, 0.797; P<0.001) and CA19-9 (AUC, 0.664; P=0.001; Table IV and Fig. 3C). In addition, the expression level of NKILA was restored postoperatively (Fig. 3D), with a significant difference observed compared with the preoperative level (Fig. 3E). In summary, NKILA may function as a diagnostic marker for patients with early CRC.
Figure 3.
NKILA functions as a diagnostic biomarker for patients with early CRC. (A) NKILA expression in normal colorectal (n=70), colorectal adenoma (n=70) and early CRC tissues (n=70) measured by RT-qPCR. (B) Spearman's rank correlation coefficient analysis to evaluate NKILA expression levels in serum and tissues. (C) Receiver operating characteristic curves to investigate the diagnostic value of NKILA, CEA and CA19-9 in early-stage CRC. (D) Pre- and postoperative expression levels of NKILA detected by RT-qPCR. (E) NKILA serum levels in pre- and postoperative patients with CRC and healthy volunteers. **P<0.01. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CRC, colorectal cancer; NKILA, NF-κB interacting long non-coding RNA; AUC, area under the curve; CEA, carcinoembryonic antigen; CA19-9, cancer antigen 19-9.
Table IV.
Diagnostic value of NKILA, CES and CA19-9 as biomarkers in colorectal cancer.
| Biomarker | AUC | P-value | Cut-off value | Sensitivity | Specificity |
|---|---|---|---|---|---|
| NKILA | 0.839 | <0.001 | 4.5 | 0.829 | 0.729 |
| CEA | 0.797 | <0.001 | 3.1 | 0.757 | 0.700 |
| CA19-9 | 0.664 | 0.001 | 22.7 | 0.629 | 0.681 |
NKILA, NF-κB interacting long non-coding RNA; CEA, carcinoembryonic antigen; CA19-9, cancer antigen 19-9; AUC, area under the curve.
Discussion
The prognosis and therapeutic options for patients with CRC are associated with the stage at which they are first diagnosed (28,29). While early-stage CRC is often cured with surgery alone, more advanced or metastatic cases of CRC typically require additional adjuvant chemo- or targeted therapy, either alone or as a combined treatment (28,30–32). Therefore, early detection of CRC is important for reducing the incidence and mortality rates of the disease (33). Currently, colonoscopy is the gold-standard diagnostic test to identify colonic pathology (34,35). However, this approach is invasive, has low adherence, and is associated with potential risks and discomfort to the patient. The ideal CRC biomarker should be easily and quantitatively measured, highly specific and sensitive, as well as reliable and reproducible (36). In addition, it should be able to stratify between different risk-based populations to select patients who require a second-line test, including endoscopic and radiologic investigations. Ideally, this aim can be achieved with a noninvasive and inexpensive method, using readily available biological samples, including serum and feces (37). At present, potential molecular biomarkers for CRC diagnosis are broadly divided into the following four groups: Nucleic acids, cytokines, antibodies and proteins (38). Blood-based markers in current use, including CEA and CA19-9, are suitable for surveillance and for monitoring responses to treatment; however, they exhibit low sensitivity and specificity, ranging between 40 and 70%, and 73 and 90%, respectively, which makes them unsuitable as screening or diagnostic markers (39). Investigations of lncRNAs in CRC diagnosis are relatively rare. A previous study has demonstrated that two transcripts of lncRNA nuclear-enriched abundant gene 1 (NEAT1_v1 and NEAT1_v2) serve as biomarkers for early CRC diagnosis, with NEAT1_v2 demonstrating a 70% overall sensitivity and 96% specificity for distinguishing CRC from controls (40). The present results confirmed that NKILA expression was decreased in early CRC tissues compared with adenomas and normal tissues, and NKILA exhibited a relatively high sensitivity (82.9%) and specificity (72.9%) compared with CEA (75.7 and 70.0%, respectively) and CA19-9 (62.9 and 68.1%, respectively) for CRC diagnosis, which suggested that NKILA may serve as a biomarker for the early diagnosis of CRC.
As a NF-κB modulator, NKILA directly interacts with functional domains of signaling proteins and suppresses cancer metastasis (17). Furthermore, low NKILA expression is associated with breast cancer metastasis and poor prognosis for patients (17). Wu et al (41) reported that NKILA is upregulated by transforming growth factor-β (TGF-β) and is essential for the negative feedback regulation of the NF-κB signaling pathway, through which NKILA significantly reduces TGF-β-induced tumor metastasis by regulating the epithelial-mesenchymal transition in breast cancer. Yu et al (42) identified that NKILA expression level is associated with baicalein sensitivity in hepatocellular carcinoma by mediating IκBα phosphorylation, NF-κB nuclear translocation and NF-κB activity. In addition, reduced expression of NKILA has been identified to indicate a poor survival for patients with hepatocellular carcinoma (42). In laryngeal cancer, NKILA has been implicated in a negative feedback loop sensitizing laryngeal cancer cells to X-ray radiation via inhibition of NF-κB activation (21). Additionally, low NKILA expression was identified to be associated with a shorter OS time for patients with laryngeal cancer (21). In summary, NKILA functions as a tumor suppressor in various cancer types predominantly by interacting with NF-κB and mediating its activity.
The present study confirmed a low expression level of NKILA in CRC, and low NKILA expression was identified to be significantly associated with a poor differentiation grade, larger tumor size (>5 cm), and advanced T (T3+T4), N (N1+N2) and TNM (III+IV) stages. Therefore, it was hypothesized that NKILA may also function as a tumor suppressor in CRC. Due to the heterogeneity of CRC, the benefits from adjuvant chemotherapy for patients with stage II and III CRC may vary to a large extent (33). Therefore, identifying molecular prognostic markers, which are capable of identifying patients who are more likely to benefit from adjuvant chemotherapy, may improve the prognosis and assist in the selection of appropriate therapy and subsequently improve outcomes (33). The current study revealed that NKILA was associated with poor OS and PFS rates in CRC, and NKILA expression was recognized as an independent risk factor for poor OS and PFS. Therefore, NKILA detection may serve as a useful tool for stratifying patients with different risks for metastasis and recurrence.
In conclusion, NKILA may be a potential diagnostic biomarker in early CRC. In addition, NKILA may serve as a novel prognostic marker and therapeutic target in CRC. However, the detailed mechanisms of NKILA-induced suppression of CRC progression were not investigated in the present study and further confirmation of the current results requires more evidence from prospective multi-center studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YZ conducted the statistical analyses. PJ, XH and JS collected the samples, clinical information and evaluated the expression levels of NKILA. WB designed the study, conducted the statistical analysis and wrote the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Central Hospital of Weihai (Weihai, China). Written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinicrope FA, Okamoto K, Kasi PM, Kawakami H. Molecular biomarkers in the personalized treatment of colorectal cancer. Clin Gastroenterol Hepatol. 2016;14:651–658. doi: 10.1016/j.cgh.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: Evolutionary, regulatory and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta. 2010;1799:597–615. doi: 10.1016/j.bbagrm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mele M, Rinn JL. ‘Cat's cradling’ the 3D genome by the act of LncRNA transcription. Mol Cell. 2016;62:657–664. doi: 10.1016/j.molcel.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, Mattick JS. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA. 2009;15:2013–2027. doi: 10.1261/rna.1705309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25:135–141. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- 10.Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: LncRNAs in cancer. J Clin Invest. 2016;126:2775–2782. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slaby O. Non-coding RNAs as biomarkers for colorectal cancer screening and early detection. Adv Exp Med Biol. 2016;937:153–170. doi: 10.1007/978-3-319-42059-2_8. [DOI] [PubMed] [Google Scholar]
- 13.Li Q, Li N, Lao Y, Lin W, Jiang G, Wei N, Wang C, Liu K, Wu J. Variable levels of long noncoding RNA expression in dna mismatch repair-proficient early-stage colon cancer. Dig Dis Sci. 2017;62:1235–1245. doi: 10.1007/s10620-017-4465-6. [DOI] [PubMed] [Google Scholar]
- 14.Sawaki K, Kanda M, Kodera Y. Review of recent efforts to discover biomarkers for early detection, monitoring, prognosis, and prediction of treatment responses of patients with gastric cancer. Expert Rev Gastroenterol Hepatol. 2018;12:657–670. doi: 10.1080/17474124.2018.1489233. [DOI] [PubMed] [Google Scholar]
- 15.Lu Q, Yu T, Ou X, Cao D, Xie T, Chen X. Potential lncRNA diagnostic biomarkers for early gastric cancer. Mol Med Rep. 2017;16:9545–9552. doi: 10.3892/mmr.2017.7770. [DOI] [PubMed] [Google Scholar]
- 16.Shang C, Zhu W, Liu T, Wang W, Huang G, Huang J, Zhao P, Zhao Y, Yao S. Characterization of long non-coding RNA expression profiles in lymph node metastasis of early-stage cervical cancer. Oncol Rep. 2016;35:3185–3197. doi: 10.3892/or.2016.4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Bian D, Gao C, Bao K, Song G. The long non-coding RNA NKILA inhibits the invasion-metastasis cascade of malignant melanoma via the regulation of NF-κB. Am J Cancer Res. 2017;7:28–40. [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Z, Li Y, Wang J, Che Y, Sun S, Huang J, Chen Z, He J. Long non-coding RNA NKILA inhibits migration and invasion of non-small cell lung cancer via NF-κB/Snail pathway. J Exp Clin Cancer Res. 2017;36:54. doi: 10.1186/s13046-017-0518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Z, Chen Z, Li Y, Wang J, Zhang Z, Che Y, Huang J, Sun S, Mao S, Lei Y, et al. TGF-β-induced NKILA inhibits ESCC cell migration and invasion through NF-κB/MMP14 signaling. J Mol Med (Berl) 2018;96:301–313. doi: 10.1007/s00109-018-1621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang T, Li S, Liu J, Yin D, Yang X, Tang Q. lncRNA-NKILA/NF-κB feedback loop modulates laryngeal cancer cell proliferation, invasion, and radioresistance. Cancer Med. 2018;7:2048–2063. doi: 10.1002/cam4.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaiopoulos AG, Athanasoula KCh, Papavassiliou AG. NF-κB in colorectal cancer. J Mol Med (Berl) 2013;91:1029–1037. doi: 10.1007/s00109-013-1045-x. [DOI] [PubMed] [Google Scholar]
- 23.Pasparakis M. Role of NF-κB in epithelial biology. Immunol Rev. 2012;246:346–358. doi: 10.1111/j.1600-065X.2012.01109.x. [DOI] [PubMed] [Google Scholar]
- 24.Sunami Y, Wirth T. Intestinal carcinogenesis: IKK can go all the way. J Clin Invest. 2011;121:2551–2553. doi: 10.1172/JCI58454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Maida M, Macaluso FS, Ianiro G, Mangiola F, Sinagra E, Hold G, Maida C, Cammarota G, Gasbarrini A, Scarpulla G. Screening of colorectal cancer: Present and future. Expert Rev Anticancer Ther. 2017;17:1131–1146. doi: 10.1080/14737140.2017.1392243. [DOI] [PubMed] [Google Scholar]
- 28.Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies JM, Goldberg RM. Treatment of metastatic colorectal cancer. Semin Oncol. 2011;38:552–560. doi: 10.1053/j.seminoncol.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Pohl M, Schmiegel W. Colorectal cancer-personalized, stage-adjusted tumour therapy. Dtsch Med Wochenschr. 2013;138:1790–1795. doi: 10.1055/s-0033-1343343. (In German) [DOI] [PubMed] [Google Scholar]
- 31.Beretta GD, Milesi L, Pessi MA, Mosconi S, Labianca R. Adjuvant treatment of colorectal cancer. Surg Oncol. 2004;13:63–73. doi: 10.1016/j.suronc.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Nesbitt C, Glendinning RJ, Byrne C, Poston GJ. Factors that influence treatment strategies in advanced colorectal cancer. Eur J Surg Oncol. 2007;33(Suppl 2):S88–S94. doi: 10.1016/j.ejso.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen HT, Duong HQ. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy. Oncol Lett. 2018;16:9–18. doi: 10.3892/ol.2018.8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heiss JA, Brenner H. Epigenome-wide discovery and evaluation of leukocyte DNA methylation markers for the detection of colorectal cancer in a screening setting. Clin Epigenetics. 2017;9:24. doi: 10.1186/s13148-017-0322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vatandoost N, Ghanbari J, Mojaver M, Avan A, Ghayour-Mobarhan M, Nedaeinia R, Salehi R. Early detection of colorectal cancer: From conventional methods to novel biomarkers. J Cancer Res Clin Oncol. 2016;142:341–351. doi: 10.1007/s00432-015-1928-z. [DOI] [PubMed] [Google Scholar]
- 36.Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, Goel A. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766–1774. doi: 10.1158/1055-9965.EPI-10-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pellino G, Gallo G, Pallante P, Capasso R, De Stefano A, Maretto I, Malapelle U, Qiu S, Nikolaou S, Barina A, et al. Noninvasive biomarkers of colorectal cancer: Role in diagnosis and personalised treatment perspectives. Gastroenterol Res Pract. 2018;2018:2397863. doi: 10.1155/2018/2397863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikolaou S, Qiu S, Fiorentino F, Rasheed S, Tekkis P, Kontovounisios C. Systematic review of blood diagnostic markers in colorectal cancer. Tech Coloproctol. 2018;22:481–498. doi: 10.1007/s10151-018-1820-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young GP, Pedersen SK, Mansfield S, Murray DH, Baker RT, Rabbitt P, Byrne S, Bambacas L, Hollington P, Symonds EL. A cross-sectional study comparing a blood test for methylated BCAT1 and IKZF1 tumor-derived DNA with CEA for detection of recurrent colorectal cancer. Cancer Med. 2016;5:2763–2772. doi: 10.1002/cam4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y, Yang L, Zhao J, Li C, Nie J, Liu F, Zhuo C, Zheng Y, Li B, Wang Z, Xu Y. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol Cancer. 2015;14:191. doi: 10.1186/s12943-015-0455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu W, Chen F, Cui X, Yang L, Chen J, Zhao J, Huang D, Liu J, Yang L, Zeng J, et al. LncRNA NKILA suppresses TGF-β-induced epithelial-mesenchymal transition by blocking NF-κB Signaling in breast cancer. Int J Cancer. 2018;143:2213–2224. doi: 10.1002/ijc.31605. [DOI] [PubMed] [Google Scholar]
- 42.Yu X, Tang W, Yang Y, Tang L, Dai R, Pu B, Feng C, Xia J. Long noncoding RNA NKILA enhances the anti-cancer effects of baicalein in hepatocellular carcinoma via the regulation of NF-κB signaling. Chem Biol Interact. 2018;285:48–58. doi: 10.1016/j.cbi.2018.02.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.