Abstract
Background
The 5′-nucleotidase, cytosolic II gene (NT5C2, cN-II) is associated with disorders characterized by psychiatric and psychomotor disturbances. Common psychiatric risk alleles at the NT5C2 locus reduce expression of this gene in the fetal and adult brain, but downstream biological risk mechanisms remain elusive.
Methods
Distribution of the NT5C2 protein in the human dorsolateral prefrontal cortex and cortical human neural progenitor cells (hNPCs) was determined using immunostaining, publicly available expression data, and reverse transcriptase quantitative polymerase chain reaction. Phosphorylation quantification of adenosine monophosphate-activated protein kinase (AMPK) alpha (Thr172) and ribosomal protein S6 (Ser235/Ser236) was performed using Western blotting to infer the degree of activation of AMPK signaling and the rate of protein translation. Knockdowns were induced in hNPCs and Drosophila melanogaster using RNA interference. Transcriptomic profiling of hNPCs was performed using microarrays, and motility behavior was assessed in flies using the climbing assay.
Results
Expression of NT5C2 was higher during neurodevelopment and was neuronally enriched in the adult human cortex. Knockdown in hNPCs affected AMPK signaling, a major nutrient-sensing mechanism involved in energy homeostasis, and protein translation. Transcriptional changes implicated in protein translation were observed in knockdown hNPCs, and expression changes to genes related to AMPK signaling and protein translation were confirmed using reverse transcriptase quantitative polymerase chain reaction. The knockdown in Drosophila was associated with drastic climbing impairment.
Conclusions
We provide an extensive neurobiological characterization of the psychiatric risk gene NT5C2, describing its previously unknown role in the regulation of AMPK signaling and protein translation in neural stem cells and its association with Drosophila melanogaster motility behavior.
Keywords: AMP-activated protein kinase (AMPK), Drosophila melanogaster, Functional genetics, Neural stem cells, Psychiatric disorders, Ribosomal protein S6 (RPS6)
The 5′-nucleotidase, cytosolic II gene (NT5C2, cN-II) encodes a phosphatase associated with disorders characterized by psychiatric and psychomotor disturbances, including schizophrenia 1, 2, 3, 4, Parkinson’s disease (5), and spastic paraplegia (6). The NT5C2 enzyme cleaves purinergic monophosphate nucleotides and has a particularly high affinity for adenosine monophosphate (AMP) (7). These energetic molecules are required for the extensive transcriptional programming governing cell maintenance, proliferation, migration, and differentiation during neurodevelopment 8, 9, 10, 11 and have been previously implicated in adult brain function and psychiatric and psychomotor disturbances 12, 13, 14.
We previously showed that common psychiatric risk variants at the NT5C2 locus are associated with reduced neurological expression of this gene in population control subjects and in the fetus (3). As a key regulator of intracellular AMP, we hypothesize that NT5C2 modulates the AMP-activated protein kinase (AMPK), a major energy homeostasis regulator 15, 16. AMPK signaling has been previously associated with psychiatric disorders 17, 18, 19, 20, NT5C2 function in muscle fibers (21), and highly energy consuming processes such as protein translation 22, 23, 24, 25, 26, 27 and motility behavior 17, 28, 29. However, the underlying gene regulatory networks that mediate the effect of NT5C2 on AMPK signaling in the context of psychiatric disorders, and the relevant cell types and developmental time points, remain unclear.
In this study, we investigated NT5C2 expression, function, and protein distribution in the human brain and human neural progenitor cells (hNPCs); its role in the regulation of AMPK signaling and protein translation; and its association with climbing behavior in Drosophila melanogaster. First, to extend our previous work, we identified the major cell types expressing NT5C2 in the adult brain, which showed that NT5C2 protein is more expressed in neurons relative to glial cells. Second, we gathered complementary evidence that this gene is more expressed and therefore likely to play a functional role during neurodevelopment. Third, we investigated the effects of NT5C2 loss-of-function on the phosphorylation of AMPK alpha (Thr172) and ribosomal protein S6 (RPS6) (Ser235/Ser236) in hNPCs, suggesting a regulatory role in AMPK signaling and protein translation. Finally, based on evidence from genetic studies implicating NT5C2 in psychomotor disturbances, we tested the effect of a loss-of-function on climbing behavior using D. melanogaster as model organism, confirming a role in motility behavior. The present study provides an extensive neurobiological characterization of NT5C2, describing its hitherto unknown relationship with AMPK signaling and protein translation in neural stem cells and a role in motility behavior in the fly. Ultimately, these data demonstrate biological mechanisms associated with NT5C2 that may explain its association with psychiatric disorders.
Methods and Materials
See Supplemental Methods and Materials for further details.
Brain Samples
To identify cell type–specific expression of NT5C2 in the adult cortex, we obtained samples from unaffected control subjects from the Medical Research Council London Neurodegenerative Disease Brain Bank, Institute of Psychiatry, Psychology & Neuroscience, King’s College London (UK Human Tissue Authority license #12293).
Immunohistochemistry and Cytochemistry
Brain sections were deparaffinized and submitted to antigen retrieval and autofluorescence removal protocols (Supplemental Methods and Materials). hNPCs were fixed and processed as previously described (30). The following primary antibodies were used: NT5C2 monoclonal antibody (M02-3C1) (Abnova, Taipei, Taiwan), ionized calcium-binding adapter molecule 1 (IBA1) (Menarini Diagnostics Ltd., Winnersh, United Kingdom), glial fibrillary acidic protein (GFAP) (Agilent, Santa Clara, CA), microtubule-associated protein 2 (MAP2) (Abcam, Cambridge, United Kingdom), parvalbumin (PARVALB) (Abcam), and beta III tubulin (Abcam). Fluorescently labeled secondary antibodies included goat or rabbit Alexa 488, 568, and 633 antibodies (Thermo Fisher Scientific, Waltham, MA).
Cell Lines
We used hNPCs from the CTX0E16 neural stem cell line (31) or from human induced pluripotent stem cells from an unaffected control subject (30) and human embryonic kidney 293T (HEK293T) cells to identify the subcellular distribution and function of NT5C2. The CTX0E16 neural cell line (31) was obtained from ReNeuron Ltd. (Bridgend, United Kingdom) under a Material Transfer Agreement. All lines were derived and maintained as described in the Supplemental Methods and Materials and elsewhere 30, 31.
RNA and Protein Isolation and Quantification
To identify gene and protein expression and phosphorylation differences in knockdown or overexpression cultures, we isolated total RNA or protein using TRI Reagent or RIPA Buffer supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific), respectively. Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR), RNA quality control, and Western blotting details are available in the Supplemental Methods and Materials. Primary antibodies for Western blotting included AMPK alpha (D6) and phospho-AMPK alpha (Thr172) (Santa Cruz Biotechnology, Dallas, TX) and total RPS6 (54d2) and phospho-RPS6 (Ser235/Ser236) (Cell Signaling Technology, Danvers, MA).
Fly Stocks and Climbing Assay
We used Gal4-upstream activated sequences (UAS) (32) to knock down CG32549 in specific tissues by crossing a CG32549-RNA interference (RNAi line) (knockdown: v30079) with UAS lines where Gal4 expression (and thus knockdown) is driven throughout the body (ACT5C: BL4414), in neurons and neural progenitor cells (ELAV: BL8765), or in gut (GUT: DGRC113094). Crosses were submitted to the negative geotaxis assay (33), a cost-effective test that has been previously used to investigate the association between genes and motility 34, 35, 36. Climbing success was calculated as percentage of flies per tube that climbed over an arbitrary mark, and survival was determined as percentage of flies alive 17 to 20 days after emergence, out of starting flasks containing 20 flies (n = 4+ flasks per condition).
Statistical Analysis
To infer statistical differences between more than two independent groups, we used one-way analysis of variances followed by Tukey post hoc tests (for normally distributed values) or Kruskal-Wallis tests followed by Dunn’s tests (for non–normally distributed values). To infer differences between two groups, we performed two-way independent parametric t tests. Multiple testing correction was applied as described in Results. Microarray expression data were analyzed using linear regressions (Supplemental Methods and Materials). Gene overlap significance was calculated in R (R Foundation for Statistical Computing, Vienna, Austria; https://www.R-project.org) using Fisher’s exact test (GeneOverlap package). Statistical analyses were performed in R and SPSS version 24 (IBM Corp., Armonk, NY).
Results
Expression of NT5C2 Is Enriched in Neurons in the Adult Brain
To extend our previous work and investigate the relationship between NT5C2 expression and psychiatric disorders, we investigated which cell types express this gene in the adult brain. We examined single-cell RNA-sequencing data from the mouse cortex (37), which revealed cell type–specific profiles of NT5C2 expression (Kruskal-Wallis test, H4 = 52.44, p < .001). Post hoc analyses confirmed that NT5C2 was more frequently observed in pyramidal neurons (95th percentile = 3 [counts]) than astrocytes (95th percentile = 1, Dunn’s post hoc test, pcorrected < .001) and in interneurons (95th percentile = 3) than microglia (95th percentile = 2.05, pcorrected < .01) or astrocytes (95th percentile = 1, pcorrected < .001) (Supplemental Figure S1).
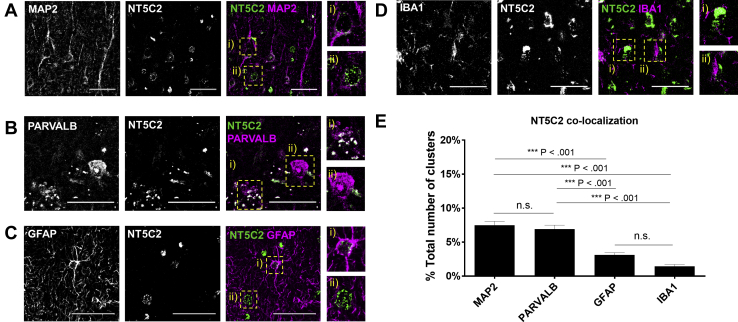
To investigate correlates with the human cortex, we performed a series of immunocolocalization experiments using postmortem brains. Initially, we investigated the specificity of an antibody for NT5C2 by probing HEK293T cells and CTX0E16 hNPCs overexpressing myc-NT5C2 (Supplemental Figures S2, S4). While immunolabeling of endogenous expression in heterologous systems may produce different results owing to the existence of tissue-specific isoforms, these findings corroborate the suitability of this antibody for our aims. This antibody was used to investigate the distribution of NT5C2 in the prefrontal cortex using standard immunoperoxidase staining with 3,3′-diaminobenzidine as chromogen (Supplemental Figure S3). Preliminary analysis of NT5C2 immunostaining using Nissl counterstain (to reveal cell morphology) suggested that NT5C2 was present in neurons, glia, and the surrounding neuropil. However, we noted that not all putative glial cells expressed NT5C2 (red arrows in Supplemental Figure S3), corroborating a previous observation by the Human Protein Atlas that expression is lower in these cells (38). To confirm this, we investigated the cell type–specific expression profile of NT5C2 in the cortex by quantifying immunocolocalization of this protein with markers of mature neurons (MAP2), a subclass of gamma-aminobutyric acid interneurons (PARVALB), astrocytes (GFAP), and microglia (IBA1) (Figure 1A–E). These cells were selected based on the wealth of evidence implicating them in the pathophysiology of psychiatric disorders 39, 40, 41, 42. Colocalization quantification was performed using a semiautomated ImageJ (National Institute of Mental Health, Bethesda, MD) macro 43, 44 (Supplemental Methods and Materials), which revealed cell type–specific profiles of NT5C2 expression (one-way analysis of variance, F3,44 = 39.12, p < .001, n = 4 unaffected control individuals, 4 technical replicates each, 20 fields of view per technical replicate). Colocalization occurred more frequently with neuron and interneuron markers than nonneuronal markers (Tukey post hoc tests: MAP2 [7.5% (cluster colocalization relative to all clusters in image) ± 2.0% (SD)], PARVALB [6.9% ± 2.1%], GFAP [3.13% ± 1.1%], IBA1 [1.4% ± 0.93%]; p < .001 for all comparisons except MAP2 vs. PARVALB and GFAP vs. IBA1 [p > .05]) (Figure 1E). NT5C2 expression at the transcript and protein levels appeared to be more highly expressed in neurons within the adult brain, consistent with recent observations that risk variants implicated in schizophrenia are enriched for neuronal genes (40).
Figure 1.
Distribution of the psychiatric risk protein NT5C2 in dorsolateral prefrontal cortex sections of postmortem unaffected individuals. Colocalization of NT5C2 staining with (A) microtubule-associated protein 2 (MAP2) (neuronal marker), (B) parvalbumin (PARVALB) (interneuron marker), (C) glial fibrillary acidic protein (GFAP) (glial marker), and (D) ionized calcium-binding adapter molecule 1 (IBA1) (microglia marker). Scale bars = 50 μm. (E) Quantification of the colocalization of NT5C2 with markers from panels (A–D) revealed a cell type–specific expression profile of NT5C2 in the mature cortex (one-way analysis of variance, Tukey pairwise comparisons: ***p < .001 for all comparisons). n.s., not significant.
NT5C2 Is Highly Expressed and Ubiquitously Distributed in hNPCs
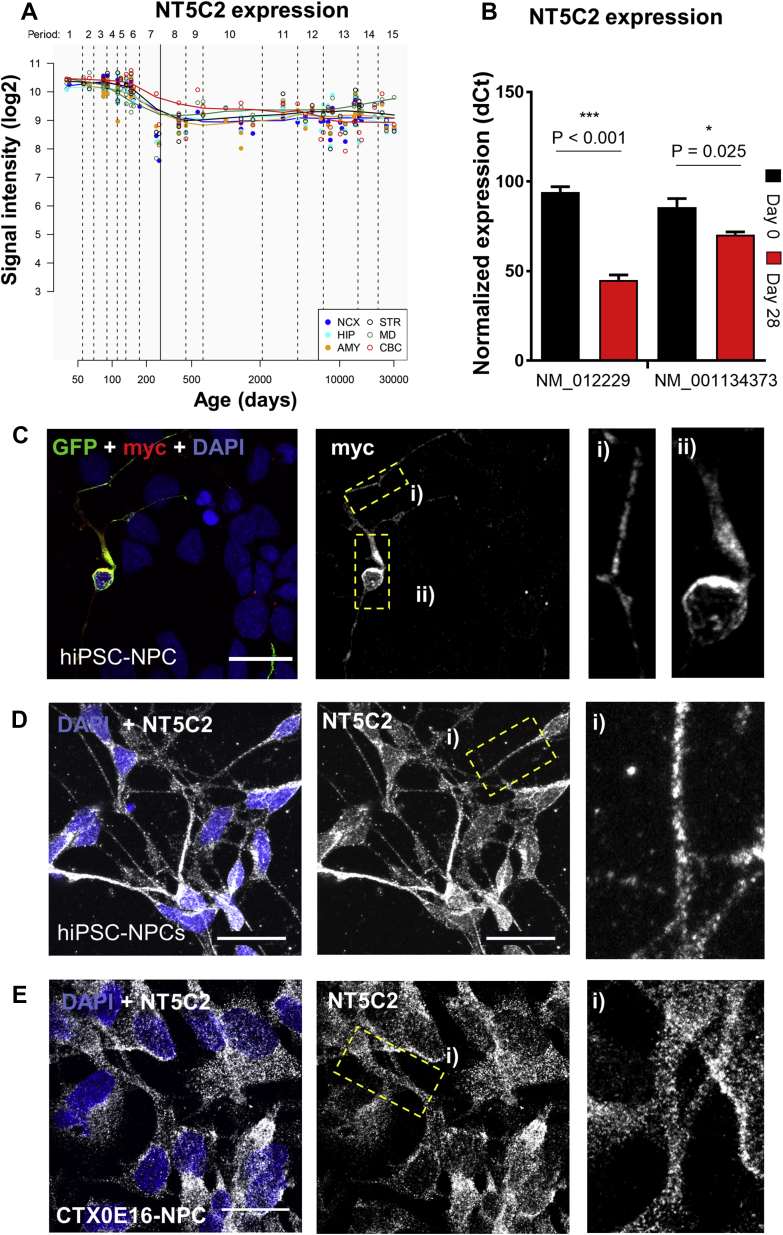
The role of NT5C2 in psychiatric disorders has been previously hypothesized to begin during neurodevelopment, a period underscored by complex processes implicated in major psychiatric disorders (45), with this risk mechanism persisting in the adult brain (3). The contribution of NT5C2 to neurobiology, however, should be greater at the developmental stage when this gene is more expressed. Thus, we investigated NT5C2 expression in the Human Brain Transcriptome atlas (46), which revealed that expression peaks during neurodevelopment (Figure 2A). Considering that established cell lines are cost-effective and easy-to-use tools to study neurodevelopment, we tested whether the CTX0E16 hNPC line 30, 31 would constitute an appropriate model. We measured expression of the NT5C2 main RefSeq transcripts (NM_012229 and NM_001134373) in these cells, specifically in hNPCs and immature neuronal cultures terminally differentiated for 28 days, which we previously showed to comprise neurons (approximately 80%) and glial cells (approximately 10%) 30, 31. The expression of NT5C2 RefSeq transcripts NM_012229 and NM_001134373 in hNPCs (day 0; NM_012229: 94.22 ± 5.85; NM_001134373: 85.67 ± 9.77) was 30% higher compared with neuronal cultures terminally differentiated for 28 days (NM_012229: 45.09 ± 5.59; NM_001134373: 70.45 ± 2.93; t tests: NM_012229, t6 = 12.14, p < .001, Bonferroni corrected p < .001; NM_001134373, t6 = 2.99, p = .0245, Bonferroni corrected p = .049). These findings are consistent with higher expression of NT5C2 at an earlier developmental stage, with persistent expression after differentiation (Figure 2B).
Figure 2.
Neurodevelopmental expression of NT5C2 and protein distribution in human neural progenitor cells (hNPCs). (A) Neurological expression of NT5C2 across human development, according to the Human Brain Transcriptome Atlas (46), showing that expression peaks during fetal development. (B) Expression of NT5C2 RefSeq transcripts NM_012229 and NM_001134373 in hNPCs (day 0) and cultures differentiated for 28 days. Expression is significantly higher at the neural progenitor state. t tests: ***p < .001, *p < .05. (C) Distribution of ectopic NT5C2 was assessed by transfecting hNPCs with plasmids encoding NT5C2-myc and enhanced green fluorescent protein (GFP), followed by immunolabeling using antibodies raised against myc or GFP (GFP was used as morphological marker). (D) Subcellular localization of endogenous NT5C2 in hNPCs derived from a human induced pluripotent stem cell (hiPSC) line and from (E) the CTX0E16 cell line. NT5C2 was ubiquitously distributed in hNPCs from both models. Scale bars = 20 μm. AMY, amygdala; CBC, cerebellar cortex; dCt, delta cycle threshold; HIP, hippocampus; MD, mediodorsal nucleus of the thalamus; NCX, neocortex; STR, striatum.
As subcellular protein distribution can inform gene function, we immunolabeled hNPCs from the CTX0E16 and human induced pluripotent stem cell lines to study NT5C2 localization. We ectopically expressed a myc-tagged NT5C2 construct in human induced pluripotent stem cell NPCs and labeled these cells using myc or NT5C2 antibodies, which revealed that myc-NT5C2 was abundantly expressed in punctate structures in the cell soma and along neurites (Figure 2C; Supplemental Figure S4). Similarly, endogenous NT5C2 was distributed in punctate structures through the cell and neurites (Figure 2D, E), suggesting that this protein is ubiquitously distributed in hNPCs, consistent with the expected distribution of a cytosolic protein.
NT5C2 Regulates the Phosphorylation of AMPK and RPS6
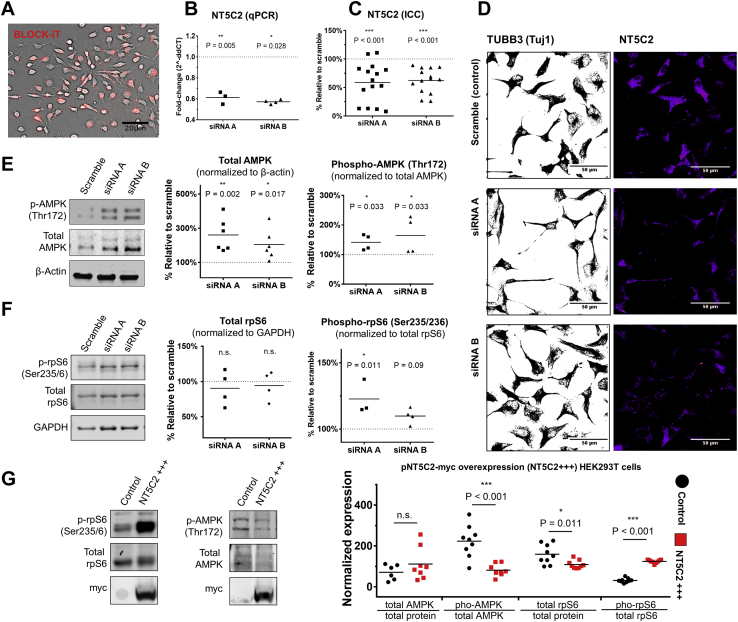
The knockdown of NT5C2 activates AMPK signaling in muscle fibers (21), and considering the relevance of AMPK to psychiatry 17, 18, 19, 20, we tested whether this also occurred in hNPCs. The knockdown in CTX0E16 hNPCs was performed using two independent small interfering RNA (siRNA) sequences, A and B. Initially, the transfection protocol efficacy was determined by assessing the uptake of fluorescently labeled oligonucleotides (BLOCK-iT; Thermo Fisher Scientific), which revealed a transfection rate of 90% ± 0.02% (n = 4 biological replicates per condition, 4 technical replicates each) (Figure 3A). We obtained knockdown cultures and confirmed reduced NT5C2 expression using RT-qPCR (linear regression to identify the effect of siRNAs on NT5C2 expression controlling for biological replicate: F2,5 = 13.9, p = .009, R2 = .92; Tukey post hoc tests against scramble [3.29 (delta cycle threshold) ± 0.23 (SD)]: siRNA A [3.79 ± 0.09], fold change = 0.71, p = .005; siRNA B [3.29 ± 0.23], fold change = 0.81, p = .028) (Figure 3B). The ability of these siRNAs to knockdown NT5C2 protein was further assessed in independent cultures (Figure 3C, D), which revealed a significant decrease in protein abundance in knockdown conditions (one-way analysis of variance, F2,41 = 12.23, p < .001; Tukey post hoc tests against scramble [100.0 ± 14.7]: siRNA A [58.8 ± 34.7], p < .001; siRNA B [62.4 ± 21.5], p < .001).
Figure 3.
Knockdown of NT5C2 in human neural progenitor cells is associated with differential phosphorylation of adenosine monophostate-activated protein kinase (AMPK) and ribosomal protein S6 (RPS6). (A) The efficiency of the small interfering RNA (siRNA) transfection was determined by uptake of BLOCK-iT, a fluorescently labeled oligonucleotide. (B)NT5C2 expression was significantly reduced in knockdown cultures (linear regressions covarying for biological replicates, Tukey post hoc tests, **p < .01, *p < .05). (C, D) siRNA treatments significantly reduce NT5C2 expression in independent human neural progenitor cell cultures at the protein level (one-way analysis of variance, Tukey post hoc tests, ***p < .001). (E) The NT5C2 knockdown was associated with increased total AMPK alpha and phosphorylated AMPK alpha (pAMPK) (Thr172) in human neural progenitor cells (Kruskal-Wallis test, Dunn’s post hoc tests, **p < .01, *p < .05). (F) The knockdown did not alter total RPS6 levels but was associated with increased phosphorylated RPS6 (prpS6) (Ser235/Ser236). siRNA A was associated with a mean 23% increase in phosphorylation (Kruskal-Wallis test, Dunn’s test, *p < .05), and siRNA B was associated with a modest 10% mean increase, which was not significant after correction (p = .09). Full blots for panels (E and F) are available in Supplemental Figure S8. (G) The overexpression of NT5C2 in human embryonic kidney 293T (HEK293T) cells causes a significant decrease in phosphorylated AMPK alpha levels and in total RPS6, and a significant increase in phosphorylated RPS6 (t test, ***p < .001, *p < .05). Full blots are available in Supplemental Figure S9. ddCt, delta-delta cycle threshold; ICC, immunocytochemistry; n.s., not significant; qPCR, quantitative polymerase chain reaction.
To test the effect of the knockdown on AMPK signaling, we measured total abundance and relative phosphorylation of AMPK alpha, a subunit of AMPK. We observed a significant effect of the knockdown on AMPK alpha abundance, which was associated with a mean 132% increase in total protein (Kruskal-Wallis test, H3 = 12.2, p < .001; Dunn’s post hoc tests against scramble [median (Mdn) = 100.0]: siRNA A [Mdn = 236.1], p = .002; siRNA B [Mdn = 182.8], p = .017) (Figure 3E). Additionally, we observed a significant effect of the knockdown on the relative phosphorylation of AMPK alpha (Thr172), with the knockdown associated with a mean 55% increase in phosphorylated AMPK, suggesting activation of this cascade (Kruskal-Wallis test, H3 = 7.65, p < .013; Dunn’s post hoc tests against scramble [Mdn = 100.0]: siRNA A [Mdn = 141.2], p = .033; siRNA B [Mdn = 160.7], p = .033) (Figure 3E).
Considering that protein translation is partly regulated by AMPK (23) and is one of the most energy-consuming cellular processes (47), we hypothesized that NT5C2 function could affect the rate of protein synthesis. To test this, we assessed the effects of the knockdown on the phosphorylation of RPS6 (Ser235/Ser236), which is routinely used as a proxy to estimate the rate of protein translation in neurons, as it correlates with mammalian target of rapamycin complex 1 activation (48). No difference was observed in total RPS6 abundance (Kruskal-Wallis test, p > .05) (Figure 3F), but we observed that the knockdown with siRNA A was associated with a mean 23% increase in phosphorylated RPS6 (Kruskal-Wallis test, H3 = 8.22, p = .002; Dunn’s post hoc test against scramble [Mdn = 100.0]: siRNA A, Mdn = 115.9, p = .012) (Figure 3F). The knockdown with siRNA B elicited a mean 10% increase in RPS6 phosphorylation, but this did not survive correction (siRNA B, Mdn = 110.2, p = .09).
We obtained complementary evidence supporting the association between NT5C2 and AMPK and RPS6 regulation using HEK293T cells. Overexpression of ectopic myc-tagged NT5C2 (NT5C2-myc) in these cells resulted in a mean 64% decrease in phosphorylated AMPK alpha (t test, control [no vector]: 223.00 [normalized expression] ± 76.99 [SD], overexpression: 81.05 ± 30.14, t15 = 4.88, p < .001, Bonferroni-adjusted [four tests] p < .001), whereas no difference in total AMPK levels was observed (p > .05) (Figure 3G). These results are consistent with our previous data and indicate that NT5C2 is a negative regulator of AMPK signaling. Subsequently, we observed a mean 28% decrease in total RPS6 abundance (t test, control: 159.10 ± 48.52, overexpression: 108.8 ± 48.52, t16 = 2.88, p = .011, pcorrected = .044) and a highly significant 300% increase in RPS6 phosphorylation (t test, control: 31.03 ± 10.66, overexpression: 124.10 ± 8.20, t16 = 20.76, p < .001, pcorrected < .001) (Figure 3G). The effect of exogenous NT5C2 on RPS6 here was opposite to what we observed in hNPCs, highlighting the complex nature of the intracellular cascades governing protein translation, which are examined in the Discussion.
NT5C2 Is Associated With Transcriptional Changes Implicated in Protein Translation
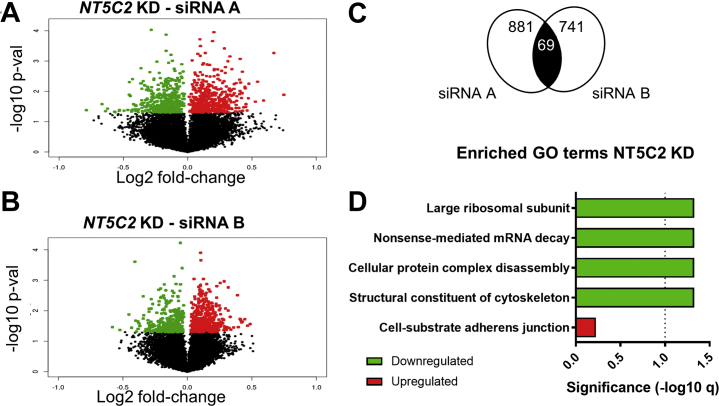
To determine the regulatory gene networks governing the effect of NT5C2 on AMPK signaling in hNPCs, we profiled the transcriptome of these cultures using microarrays (Figure 4A, B). We aimed to characterize expression differences that were shared between both siRNA treatments to reduce off-target effects associated with individual siRNAs (49). The concordant transcriptomic changes consisted of an overlap of 69 genes (Figure 4C), which is statistically unlikely to occur by chance (genes in microarray = 21,196; affected by siRNA A: 881 genes; siRNA B: 741 genes; Fisher’s exact test, p < .001, Jaccard index < 0.001, odds ratio = 2.6) (gene list in Supplemental Table S1). We observed that there was a high correlation between samples within the same siRNA groups, despite the modest number of overlapping, differentially expressed genes (Pearson’s r > .99, p < .001 for all correlations; siRNA A, n = 3 biological replicates; siRNA B and scramble, n = 4). The list of overlapping gene expression changes was subdivided by directionality of effect, and the upregulated and downregulated gene network topologies were constructed using GeneMANIA (50) (Supplemental Figure S5). This analysis revealed multiple connections between genes owing to coexpression and colocalization, corroborating their functional association. The upregulated and downregulated gene lists were analyzed for enrichment of gene ontology terms (Figure 4D; Supplemental Table S2), which revealed downregulated genes (q < .05) pertaining to the regulation of protein translation, and of the cytoskeleton [which is highly dependent on the transcriptional machinery (51)]. Furthermore, ribosomal genes, including RPL15, RPL22, RPL5, and RPL6, were downregulated, consistent with activation of AMPK signaling and inhibition of protein translation. The top upregulated gene ontology term suggested the involvement of NT5C2 in cell adhesion, but this was not significant after correction (q > .05).
Figure 4.
Transcriptional changes associated with the knockdown (KD) corroborate a role for NT5C2 on protein translation. Volcano plots indicate nominally significant transcriptomic changes elicited by (A) small interfering RNA (siRNA) A and (B) siRNA B. (C) Venn diagrams indicating the number of genes differentially regulated by siRNA A and siRNA B and the overlapping gene set, which is unlikely to occur by chance, according to Fisher’s exact test (p < .001). (D) Gene ontology (GO) terms enriched within genes concordantly, differentially expressed in both KD conditions. The line indicates the significance threshold (−log10 q < .05). mRNA, messenger RNA; p-val, p value.
We used RT-qPCR to validate a panel of gene expression changes observed in the microarray analysis (Supplemental Figure S6), which were related to protein translation and AMPK signaling, including the heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1), the proteasome 26S subunit, ATPase 4 (PSMC4), and the autophagy-related cysteine peptidase gene (ATG4B). HNRNPA1 is involved in protein translation (52), whereas ATG4B regulates AMPK signaling and energy homeostasis (53), and PSMC4 physically interacts with AMPK (54) and is involved in Parkinson’s disease (55). Considering the effects of NT5C2 in AMPK and RPS6 regulation, the transcriptional changes observed here corroborate a role for NT5C2 in protein translation, providing evidence of the gene networks involved.
Knockdown of CG32549 in D. melanogaster Is Associated With Impaired Climbing
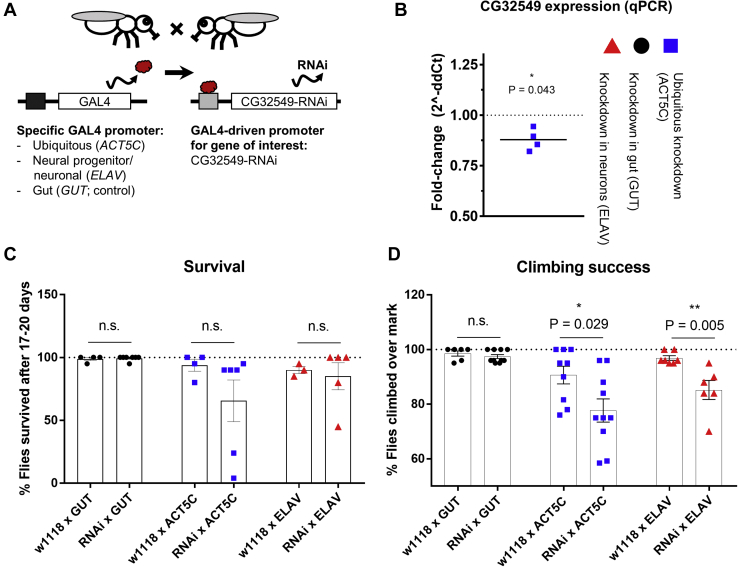
Considering the genetic link between NT5C2 and disorders associated with psychiatric and psychomotor disturbances and the importance of AMPK in energy-consuming tasks such as motility 19, 56, we investigated a potential role of the NT5C2 homologue of Drosophila in climbing. The human NT5C2 protein shares 60.5% sequence identity and 80.2% sequence similarity with CG32549 (Supplemental Figure S7), suggesting that these proteins exert the same or similar function. To investigate the role of CG32549 in motility while controlling for potential confounding effects in muscles, we generated flies using the Gal4-UAS system to obtain crosses with reduced expression of this gene ubiquitously (driven by ACT5C promoter), in neurons and neural progenitor cells (ELAV), or in gut as a control (GUT) (Figure 5A). The ubiquitous knockdown was associated with reduced CG32549 expression in the brain (UAS line [control, no RNAi cassette] = 0.042 [delta cycle threshold] ± 0.027; UAS-KD [knockdown] flies = 0.007 ± 0.004; fold change = 0.88; t test: t6 = 2.6, p = .043, n = 4) (Figure 5B). No difference in survival was observed across genotypes (UAS vs. UAS-KD lines, t tests, p > .05, n = 5 flasks on average) (Figure 5C). We observed a significant impairment in climbing success associated with the knockdown using the ELAV promoter (UAS = 96.9% ± 2.2%, UAS-KD = 85.2% ± 8.5%; t11 = 3.53, p = .005, adjusted p = .014, n = 7 per condition on average) (Figure 5D). There was also a nominally significant reduction in climbing success on knockdown of CG32549 using the ACT5C promoter (UAS = 90.6% ± 9.7%, UAS-KD = 77.7% ± 13.4%, t test: t17 = 2.4, p = .029, n = 10 per condition on average, Bonferroni (adjusted for three comparisons) p > .05). This impairment was not observed in flies with the knockdown in gut (UAS = 98.5% ± 2.3%, UAS-KD = 97.4% ± 2.3%, t test, p > .05, n = 8 per condition on average). These findings suggest there is an effect of neuronal CG32549 in D. melanogaster motility and provide an insight into the function of NT5C2 at a systems level.
Figure 5.
Knockdown of CG32549 (NT5C2 homologue) in Drosophila melanogaster using the Gal4-upstream activated sequences (UAS) system. CG32549-RNA interference (RNAi) was induced ubiquitously (ACT5C promoter), in gut (GUT), or in neural progenitor cells and neurons (ELAV). (A) Experimental design of the knockdown. (B)CG32549 was less expressed in the brain of knockdown flies (t test, *p < .05). (C) There was no difference in survival percentage between UAS lines and UAS-knockdown lines 17–20 days after emergence (t tests, p > .05). (D) Climbing success observed in UAS lines vs. UAS-knockdown lines highlight the effect of the ubiquitous and neuron-specific knockdowns on motility (t tests, *p < .05, **p < .01), an effect that was not observed in the gut-specific condition (p > .05). ddCt, delta-delta cycle threshold; n.s., not significant; qPCR, quantitative polymerase chain reaction.
Discussion
The NT5C2 gene is implicated in risk for psychiatric and neurological conditions 1, 2, 3, 5, 6, and it has been recently classified as a high confidence schizophrenia risk gene by PsychENCODE (4), but the biological mechanisms responsible for these associations remain elusive. We previously showed that psychiatric risk alleles at the NT5C2 locus are associated with reduced expression of this gene in the adult and developing brain (3). In this study, we observe that reduced NT5C2 expression is associated with differential regulation of AMPK signaling and RPS6 in hNPCs, suggesting a regulatory role in energy homeostasis and protein translation. We also observe that neurological expression of NT5C2 peaks during neurodevelopment but persists at later developmental stages, corroborating our previous hypothesis that the NT5C2 risk mechanism is an ongoing process that starts from embryonic development (3). The cell type–specific NT5C2 expression profile observed in the adult brain further revealed an enrichment for neuronal expression, suggesting that reduced NT5C2 expression in the mature cortex could be particularly detrimental to neurons. These findings are consistent with recent studies showing that schizophrenia risk variants are enriched for genes implicated in neurodevelopment and neuronal function 40, 57.
The manipulation of NT5C2 expression in hNPCs and HEK293T cells corroborates the existence of a complex regulatory network governing protein translation, with observations suggesting, at first glance, opposing effects of AMPK on the rate of protein synthesis. However, on closer inspection, we observed that our findings with HEK cells corroborate that AMPK activation inhibits protein translation by repressing the mammalian target of rapamycin complex 1 and the eukaryotic translation elongation factor 2 23, 24, 58, 59. Our findings with the hNPC model, in turn, corroborate AMPK activation leading to increased rates of protein synthesis over time, despite an initial period in which translation is halted, likely owing to a negative feedback loop (60). As a result, we observed increased abundance of AMPK alpha in the neural progenitor cell cultures, whereas no differences in expression of AMPK transcripts were detected.
We also observed that a knockdown of the NT5C2 homologue CG32549 in D. melanogaster was associated with abnormal climbing behavior, more specifically, when driven by a neuronal promoter, supporting a role for NT5C2 in motility. It is unrealistic to correlate fly motility with complex psychomotor disturbances experienced by human patients, but our results corroborate previous genetic associations between NT5C2 and diseases associated with motor symptoms 1, 2, 3, 4, 5, 6. The effect of CG32549 could be mediated by regulation of AMPK signaling and RPS6 activation, which are implicated in Drosophila motility 61, 62. A study showed that CG32549 is downregulated in a Drosophila model of seizure (28), and work by another group demonstrated that distance moved during seizure-like activity can be reduced (rescued) upon AMPK activation using the drug metformin (29).
Limitations of our study should be acknowledged. First, we obtained a modest knockdown of NT5C2 in hNPCs, which we hypothesize to be due to the proliferative nature of these cells. To support the link between NT5C2, AMPK signaling, and RPS6 activation, we overexpressed NT5C2 in HEK293T cells and confirmed the differential regulation of AMPK and RPS6. Second, we observed a modest overlap of differentially expressed genes between siRNA treatments, which we hypothesize to be due to the low specificity associated with the siRNAs. This could be overcome by using a more effective gene silencing method, such as clustered regularly interspaced short palindromic repeats (CRISPR) interference (63). Third, we quantified the rate of protein translation using relative abundance of phosphorylated RPS6, but we did not investigate overall protein synthesis using methods such as the surface sensing of translation (SUnSET) (64). We have, however, provided support for the role of NT5C2 in protein translation at the transcriptional level using our microarray and RT-qPCR data.
By exploring the role of NT5C2 in neurobiology, we observe that the study of individual risk factors for complex disorders has the potential to advance our understanding of common biological pathways contributing to disease. Functional studies using model organisms or cell culture may not entirely capture the heterogeneity and complexity of psychiatric disorders but may provide insights to accelerate the identification of novel drug targets and biomarkers for psychiatric disorders.
Acknowledgments and Disclosures
This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil (Grant No. BEX1279-13-0 [to RRRD]); National Institute for Health Research (NIHR) Maudsley Biomedical Research Centre Career Development Award (to RRRD); Medical Research Council (Grant No. G0802166 [to NJB], Skills Development Fellowship Grant No. MR/N014863/1 [to TRP], Grant No. MR/N025377/1 [to ACV], Grant No. MR/L021064/1 [to DPS]); Medical Research Council Centre for Neurodevelopmental Disorders (Grant No. MR/N026063/1); Wellcome Trust Institutional Strategic Support Fund (Grant No. 097819 [to DPS]) via the King’s Health Partners Research and Development Challenge Fund, a fund administered on behalf of King’s Health Partners by Guy’s and St Thomas’ Charity; National Alliance for Research on Schizophrenia and Depression Independent Investigator’s Award (to DPS); European Union’s Seventh Framework Programme (Grant No. FP7-HEALTH-603016 [to DPS]); and National Institutes of Health (Grant Nos. CA206488 and AI076059 [to DFN]). This study represents independent research funded in part by the NIHR-Wellcome Trust King’s Clinical Research Facility, NIHR Biomedical Research Centre at South London and Maudsley National Health Service Foundation Trust, and King’s College London. The views expressed are those of the authors and not necessarily those of the National Health Service, NIHR, or Department of Health and Social Care. Tissue samples were supplied by the London Neurodegenerative Diseases Brain Bank, which receives funding from the Medical Research Council and as part of the Brains for Dementia Research program, jointly funded by Alzheimer’s Research UK and Alzheimer’s Society.
RRRD, DPS, and NJB conceived and designed the experiments. RRRD, NDB, GAH, and M-CC performed the experiments. RRRD and TRP analyzed the data. SHL, SS, IAW, CT, GDB, ACV, IE, DFN, RMM, NJB, and TRP contributed reagents, biological material, and expertise. RRRD, TRP, and DPS wrote the article.
We thank Dr. Frank Hirth for constructive reading of the manuscript.
GDB receives funding from Eli Lilly and Company, and ACV receives funding from F. Hoffmann-La Roche AG. The other authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.biopsych.2019.03.977.
Supplementary Material
References
- 1.Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schizophrenia Working Group of the PGC Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duarte R.R.R., Troakes C., Nolan M., Srivastava D.P., Murray R.M., Bray N.J. Genome-wide significant schizophrenia risk variation on chromosome 10q24 is associated with altered cis-regulation of BORCS7, AS3MT, and NT5C2 in the human brain. Am J Med Genet B Neuropsychiatr Genet. 2016;171B:806–814. doi: 10.1002/ajmg.b.32445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D., Liu S., Warrell J., Won H., Shi X., Navarro F.C.P. Comprehensive functional genomic resource and integrative model for the human brain. Science. 2018;362:eaat8464. doi: 10.1126/science.aat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nalls M.A., Saad M., Noyce A.J., Keller M.F., Schrag A., Bestwick J.P. Genetic comorbidities in Parkinson’s disease. Hum Mol Genet. 2014;23:831–841. doi: 10.1093/hmg/ddt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Straussberg R., Onoufriadis A., Konen O., Zouabi Y., Cohen L., Lee J.Y.W. Novel homozygous missense mutation in NT5C2 underlying hereditary spastic paraplegia SPG45. Am J Med Genet A. 2017;173:3109–3113. doi: 10.1002/ajmg.a.38414. [DOI] [PubMed] [Google Scholar]
- 7.Itoh R. Enzymatic properties and physiological roles of cytosolic 5′-nucleotidase II. Curr Med Chem. 2013;20:4260–4284. doi: 10.2174/0929867311320340006. [DOI] [PubMed] [Google Scholar]
- 8.Tang Y., Illes P. Regulation of adult neural progenitor cell functions by purinergic signaling. Glia. 2017;65:213–230. doi: 10.1002/glia.23056. [DOI] [PubMed] [Google Scholar]
- 9.Rapaport E., Garcia-Blanco M.A., Zamecnik P.C. Regulation of DNA replication in S phase nuclei by ATP and ADP pools. Proc Natl Acad Sci U S A. 1979;76:1643–1647. doi: 10.1073/pnas.76.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tornroth-Horsefield S., Neutze R. Opening and closing the metabolite gate. Proc Natl Acad Sci U S A. 2008;105:19565–19566. doi: 10.1073/pnas.0810654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffman G.E., Schrode N., Flaherty E., Brennand K.J. New considerations for hiPSC-based models of neuropsychiatric disorders. Mol Psychiatry. 2019;24:49–66. doi: 10.1038/s41380-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Gil M., Camici M., Allegrini S., Pesi R., Petrotto E., Tozzi M.G. Emerging role of purine metabolizing enzymes in brain function and tumors. Int J Mol Sci. 2018;19:E3598. doi: 10.3390/ijms19113598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheffer A., Castillo A.R.G., Correa-Velloso J., Goncalves M.C.B., Naaldijk Y., Nascimento I.C. Purinergic system in psychiatric diseases. Mol Psychiatry. 2018;23:94–106. doi: 10.1038/mp.2017.188. [DOI] [PubMed] [Google Scholar]
- 14.Rioult-Pedotti M.S., Pekanovic A., Atiemo C.O., Marshall J., Luft A.R. Dopamine promotes motor cortex plasticity and motor skill learning via PLC activation. PloS One. 2015;10:e0124986. doi: 10.1371/journal.pone.0124986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omar B., Zmuda-Trzebiatowska E., Manganiello V., Goransson O., Degerman E. Regulation of AMP-activated protein kinase by cAMP in adipocytes: Roles for phosphodiesterases, protein kinase B, protein kinase A, Epac and lipolysis. Cell Signal. 2009;21:760–766. doi: 10.1016/j.cellsig.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin W., Mu J., Birnbaum M.J. Role of AMP-activated protein kinase in cyclic AMP-dependent lipolysis In 3T3-L1 adipocytes. J Biol Chem. 2003;278:43074–43080. doi: 10.1074/jbc.M308484200. [DOI] [PubMed] [Google Scholar]
- 17.Perera N.D., Turner B.J. AMPK signalling and defective energy metabolism in amyotrophic lateral sclerosis. Neurochem Res. 2016;41:544–553. doi: 10.1007/s11064-015-1665-3. [DOI] [PubMed] [Google Scholar]
- 18.Ronnett G.V., Ramamurthy S., Kleman A.M., Landree L.E., Aja S. AMPK in the brain: Its roles in energy balance and neuroprotection. J Neurochem. 2009;109:17–23. doi: 10.1111/j.1471-4159.2009.05916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosso P., Fioramonti M., Fracassi A., Marangoni M., Taglietti V., Siteni S., Segatto M. AMPK in the central nervous system: Physiological roles and pathological implications. Res Rep Biol. 2016;7:1–13. [Google Scholar]
- 20.Yuan S.Y., Liu J., Zhou J., Lu W., Zhou H.Y., Long L.H. AMPK mediates glucocorticoids stress-induced downregulation of the glucocorticoid receptor in cultured rat prefrontal cortical astrocytes. PloS One. 2016;11:e0159513. doi: 10.1371/journal.pone.0159513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulkarni S.S., Karlsson H.K., Szekeres F., Chibalin A.V., Krook A., Zierath J.R. Suppression of 5′-nucleotidase enzymes promotes AMP-activated protein kinase (AMPK) phosphorylation and metabolism in human and mouse skeletal muscle. J Biol Chem. 2011;286:34567–34574. doi: 10.1074/jbc.M111.268292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan A.Y., Soltys C.L., Young M.E., Proud C.G., Dyck J.R. Activation of AMP-activated protein kinase inhibits protein synthesis associated with hypertrophy in the cardiac myocyte. J Biol Chem. 2004;279:32771–32779. doi: 10.1074/jbc.M403528200. [DOI] [PubMed] [Google Scholar]
- 23.Reiter A.K., Bolster D.R., Crozier S.J., Kimball S.R., Jefferson L.S. AMPK represses TOP mRNA translation but not global protein synthesis in liver. Biochem Biophys Res Commun. 2008;374:345–350. doi: 10.1016/j.bbrc.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolster D.R., Crozier S.J., Kimball S.R., Jefferson L.S. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J Biol Chem. 2002;277:23977–23980. doi: 10.1074/jbc.C200171200. [DOI] [PubMed] [Google Scholar]
- 25.Kenney J.W., Sorokina O., Genheden M., Sorokin A., Armstrong J.D., Proud C.G. Dynamics of elongation factor 2 kinase regulation in cortical neurons in response to synaptic activity. J Neurosci. 2015;35:3034–3047. doi: 10.1523/JNEUROSCI.2866-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma T., Chen Y., Vingtdeux V., Zhao H., Viollet B., Marambaud P. Inhibition of AMP-activated protein kinase signaling alleviates impairments in hippocampal synaptic plasticity induced by amyloid beta. J Neurosci. 2014;34:12230–12238. doi: 10.1523/JNEUROSCI.1694-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinmetz A.B., Stern S.A., Kohtz A.S., Descalzi G., Alberini C.M. Insulin-like growth factor II targets the mTOR pathway to reverse autism-like phenotypes in mice. J Neurosci. 2018;38:1015–1029. doi: 10.1523/JNEUROSCI.2010-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin W.H., Giachello C.N., Baines R.A. Seizure control through genetic and pharmacological manipulation of Pumilio in Drosophila: A key component of neuronal homeostasis. Dis Model Mech. 2017;10:141–150. doi: 10.1242/dmm.027045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stone B., Burke B., Pathakamuri J., Coleman J., Kuebler D. A low-cost method for analyzing seizure-like activity and movement in Drosophila. J Vis Exp. 2014;84 doi: 10.3791/51460. e51460–e51460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deans P.J., Raval P., Sellers K.J., Gatford N.J., Halai S., Duarte R.R.R. Psychosis risk candidate ZNF804A localizes to synapses and regulates neurite formation and dendritic spine structure. Biol Psychiatry. 2017;82:49–61. doi: 10.1016/j.biopsych.2016.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson G.W., Deans P.J., Taylor R.D., Raval P., Chen D., Lowder H. Characterisation of neurons derived from a cortical human neural stem cell line CTX0E16. Stem Cell Res Ther. 2015;6:149. doi: 10.1186/s13287-015-0136-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 33.Gargano J.W., Martin I., Bhandari P., Grotewiel M.S. Rapid iterative negative geotaxis (RING): A new method for assessing age-related locomotor decline in Drosophila. Exp Gerontol. 2005;40:386–395. doi: 10.1016/j.exger.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Madabattula S.T., Strautman J.C., Bysice A.M., O’Sullivan J.A., Androschuk A., Rosenfelt C. Quantitative analysis of climbing defects in a Drosophila model of neurodegenerative disorders. J Vis Exp. 2015;100:e52741. doi: 10.3791/52741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calcagno B., Eyles D., van Alphen B., van Swinderen B. Transient activation of dopaminergic neurons during development modulates visual responsiveness, locomotion and brain activity in a dopamine ontogeny model of schizophrenia. Transl Psychiatry. 2013;3:e206. doi: 10.1038/tp.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhandari P., Kendler K.S., Bettinger J.C., Davies A.G., Grotewiel M. An assay for evoked locomotor behavior in Drosophila reveals a role for integrins in ethanol sensitivity and rapid ethanol tolerance. Alcohol Clin Exp Res. 2009;33:1794–1805. doi: 10.1111/j.1530-0277.2009.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeisel A., Munoz-Manchado A.B., Codeluppi S., Lonnerberg P., La Manno G., Jureus A. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 38.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez-Burgos G., Cho R.Y., Lewis D.A. Alterations in cortical network oscillations and parvalbumin neurons in schizophrenia. Biol Psychiatry. 2015;77:1031–1040. doi: 10.1016/j.biopsych.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skene N.G., Bryois J., Bakken T.E., Breen G., Crowley J.J., Gaspar H.A. Genetic identification of brain cell types underlying schizophrenia. Nat Genet. 2018;50:825–833. doi: 10.1038/s41588-018-0129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mondelli V., Vernon A.C., Turkheimer F., Dazzan P., Pariante C.M. Brain microglia in psychiatric disorders. Lancet Psychiatry. 2017;4:563–572. doi: 10.1016/S2215-0366(17)30101-3. [DOI] [PubMed] [Google Scholar]
- 42.Yamamuro K., Kimoto S., Rosen K.M., Kishimoto T., Makinodan M. Potential primary roles of glial cells in the mechanisms of psychiatric disorders. Front Cell Neurosci. 2015;9 doi: 10.3389/fncel.2015.00154. 154–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Notter T., Panzanelli P., Pfister S., Mircsof D., Fritschy J.M. A protocol for concurrent high-quality immunohistochemical and biochemical analyses in adult mouse central nervous system. Eur J Neurosci. 2014;39:165–175. doi: 10.1111/ejn.12447. [DOI] [PubMed] [Google Scholar]
- 44.Notter T., Coughlin J.M., Gschwind T., Weber-Stadlbauer U., Wang Y., Kassiou M. Translational evaluation of translocator protein as a marker of neuroinflammation in schizophrenia. Mol Psychiatry. 2018;23:323–334. doi: 10.1038/mp.2016.248. [DOI] [PubMed] [Google Scholar]
- 45.Thapar A., Cooper M., Rutter M. Neurodevelopmental disorders. Lancet Psychiatry. 2017;4:339–346. doi: 10.1016/S2215-0366(16)30376-5. [DOI] [PubMed] [Google Scholar]
- 46.Kang H.J., Kawasawa Y.I., Cheng F., Zhu Y., Xu X., Li M. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindqvist L.M., Tandoc K., Topisirovic I., Furic L. Cross-talk between protein synthesis, energy metabolism and autophagy in cancer. Curr Opin Genet Dev. 2018;48:104–111. doi: 10.1016/j.gde.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biever A., Valjent E., Puighermanal E. Ribosomal protein S6 phosphorylation in the nervous system: from regulation to function. Front Mol Neurosci. 2015;8:75. doi: 10.3389/fnmol.2015.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hill M.J., Jeffries A.R., Dobson R.J., Price J., Bray N.J. Knockdown of the psychosis susceptibility gene ZNF804A alters expression of genes involved in cell adhesion. Hum Mol Genet. 2012;21:1018–1024. doi: 10.1093/hmg/ddr532. [DOI] [PubMed] [Google Scholar]
- 50.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silva R.C., Sattlegger E., Castilho B.A. Perturbations in actin dynamics reconfigure protein complexes that modulate GCN2 activity and promote an eIF2 response. J Cell Sci. 2016;129:4521–4533. doi: 10.1242/jcs.194738. [DOI] [PubMed] [Google Scholar]
- 52.Kim H.J., Kim N.C., Wang Y.D., Scarborough E.A., Moore J., Diaz Z. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu P.F., Hsu C.J., Tsai W.L., Cheng J.S., Chen J.J., Huang I.F. Ablation of ATG4B suppressed autophagy and activated AMPK for cell cycle arrest in cancer cells. Cell Physiol Biochem. 2017;44:728–740. doi: 10.1159/000485286. [DOI] [PubMed] [Google Scholar]
- 54.Ewing R.M., Chu P., Elisma F., Li H., Taylor P., Climie S. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molochnikov L., Rabey J.M., Dobronevsky E., Bonucelli U., Ceravolo R., Frosini D. A molecular signature in blood identifies early Parkinson’s disease. Mol Neurodegener. 2012;7:26. doi: 10.1186/1750-1326-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorgensen S.B., Richter E.A., Wojtaszewski J.F. Role of AMPK in skeletal muscle metabolic regulation and adaptation in relation to exercise. J Physiol. 2006;574:17–31. doi: 10.1113/jphysiol.2006.109942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Owen M.J., O’Donovan M.C. Schizophrenia and the neurodevelopmental continuum: evidence from genomics. World Psychiatry. 2017;16:227–235. doi: 10.1002/wps.20440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saha A.K., Xu X.J., Lawson E., Deoliveira R., Brandon A.E., Kraegen E.W. Downregulation of AMPK accompanies leucine- and glucose-induced increases in protein synthesis and insulin resistance in rat skeletal muscle. Diabetes. 2010;59:2426–2434. doi: 10.2337/db09-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L., Cash T.P., Jones R.G., Keith B., Thompson C.B., Simon M.C. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dreyer H.C., Fujita S., Cadenas J.G., Chinkes D.L., Volpi E., Rasmussen B.B. Resistance exercise increases AMPK activity and reduces 4E-BP1 phosphorylation and protein synthesis in human skeletal muscle. J Physiol. 2006;576:613–624. doi: 10.1113/jphysiol.2006.113175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ng C.H., Guan M.S., Koh C., Ouyang X., Yu F., Tan E.K. AMP kinase activation mitigates dopaminergic dysfunction and mitochondrial abnormalities in Drosophila models of Parkinson’s disease. J Neurosci. 2012;32:14311–14317. doi: 10.1523/JNEUROSCI.0499-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim M., Semple I., Kim B., Kiers A., Nam S., Park H.W. Drosophila Gyf/GRB10 interacting GYF protein is an autophagy regulator that controls neuron and muscle homeostasis. Autophagy. 2015;11:1358–1372. doi: 10.1080/15548627.2015.1063766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng Y., Shen W., Zhang J., Yang B., Liu Y.N., Qi H. CRISPR interference-based specific and efficient gene inactivation in the brain. Nat Neurosci. 2018;21:447–454. doi: 10.1038/s41593-018-0077-5. [DOI] [PubMed] [Google Scholar]
- 64.Goodman C.A., Hornberger T.A. Measuring protein synthesis with SUnSET: A valid alternative to traditional techniques? Exerc Sport Sci Rev. 2013;41:107–115. doi: 10.1097/JES.0b013e3182798a95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.