Summary
Defects in mitotic spindle orientation (MSO) disrupt the organization of stem cell niches impacting tissue morphogenesis and homeostasis. Mutations in centrosome genes reduce MSO fidelity, leading to tissue dysplasia and causing several diseases such as microcephaly, dwarfism, and cancer. Whether these mutations perturb spindle orientation solely by affecting astral microtubule nucleation or whether centrosome proteins have more direct functions in regulating MSO is unknown. To investigate this question, we analyzed the consequences of deregulating Plk4 (the master centriole duplication kinase) activity in Drosophila asymmetrically dividing neural stem cells. We found that Plk4 functions upstream of MSO control, orchestrating centriole symmetry breaking and consequently centrosome positioning. Mechanistically, we show that Plk4 acts through Spd2 phosphorylation, which induces centriole release from the apical cortex. Overall, this work not only reveals a role for Plk4 in regulating centrosome function but also links the centrosome biogenesis machinery with the MSO apparatus.
Keywords: centrosomes, spindle orientation, symmetry breaking, centrosome positioning, Plk4, Spd2
Graphical Abstract
Highlights
-
•
Drosophila Plk4 mutant NSCs show defects in centriole asymmetry and spindle positioning
-
•
Apical centriole anchoring requires the PCM protein Spd-2 and the APC/C activator Fzr
-
•
Movement of the centriole toward the basal side of the cell requires Plk4 activity
-
•
At the mother centriole, Plk4 phosphorylates Spd2 to trigger PCM shedding and Fzr loss
Mitotic spindle orientation is tightly regulated during development and adulthood to maintain tissue organization and homeostasis. Spindle orientation requires the coordination between centrosomes and cortical cues. Gambarotto et al. report that the centrosome components Plk4 and Spd2 regulate centrosome asymmetry in interphase to influence spindle positioning in mitosis.
Introduction
Drosophila neural stem cells (NSCs; also called neuroblasts [NBs]) repeatedly divide asymmetrically to self-renew and to generate a committed progenitor, the ganglion mother cell (GMC). During interphase, a robust mechanism of centriole asymmetry controls mitotic spindle orientation (MSO) in the following mitosis (Rebollo et al., 2007, Rusan and Peifer, 2007) so that GMCs are always born at the same position relative to the NB (Figure 1A). Defects in polarity establishment or mutations in centrosome genes, which disrupt spindle positioning, interfere with asymmetric cell division and generate tumors (Basto et al., 2008, Basto et al., 2006, Castellanos et al., 2008, Caussinus and Gonzalez, 2005), highlighting the importance of regulated stem cell division.
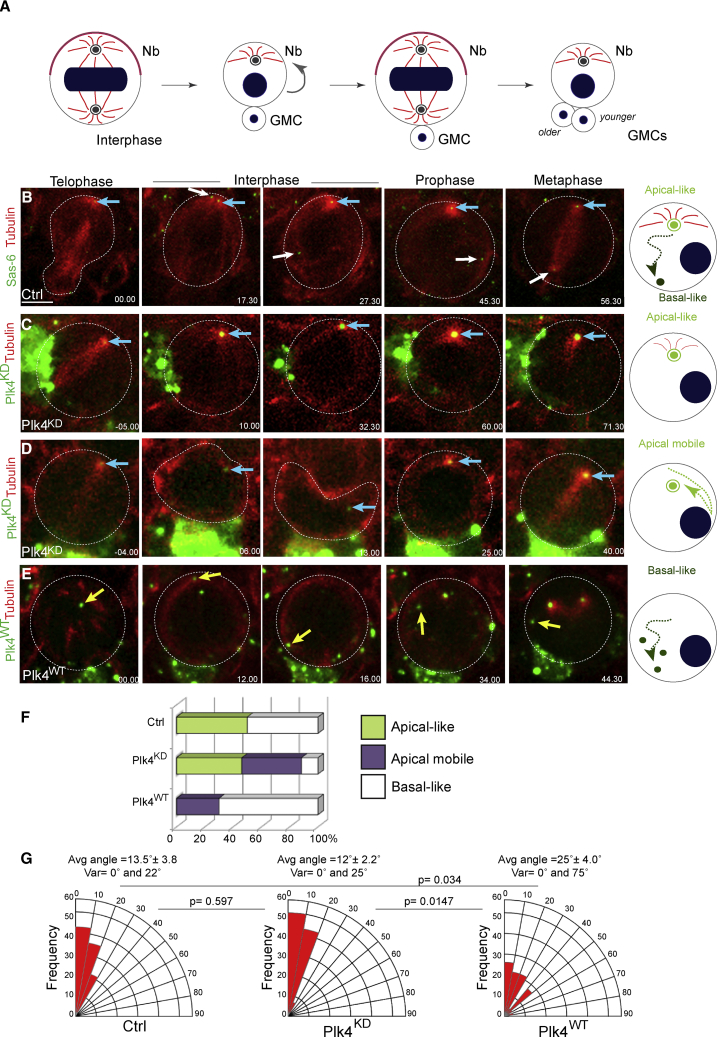
Figure 1.
Plk4 Regulates Centriole Dynamics in Interphase, Impacting Spindle Orientation
(A) Schematic drawing representing two consecutive cell cycles of a Drosophila NB depicting centrosome behavior.
(B–E) Images from time-lapse movies of Ctrl (B), Plk4KD (C and D), and Plk4WT (E) larval NBs. Tubulin in red. RFP-Sas-6 (B), GFP-Plk4KD (C and D), and GFP-Plk4WT (E) in green. See also Figures S1 and S2. The blue arrow denotes the centrosome (or centriole in the case of Plk4KD) inherited by the NB at the end of mitosis in the first column but, in all other images, marks the centriole that was localized at the apical cortex (apical centriole) after disengagement. White arrows point to the centriole that moves basally in Ctrl NBs. The yellow arrow points to the centrosome positioned at the spindle pole at the end of mitosis in Plk4WT NBs. Time, minutes. Scale, 4 μm. Diagrams on the right illustrate centriole behavior in early interphase.
(F) Graph shows the percentage of centriole behavior categories during interphase in the indicated genotypes. Centriole behavior was categorized as apical-like in (B) or (C), apical-mobile-like in (D), when the centrosome moved laterally even if remained localized within the apical hemisphere, or basal-like in (E).
(G) Quantification of the angle between two consecutive mitoses in Ctrl, Plk4KD and Plk4WT. Statistical significance (SS) was assessed by unpaired t test.
The stereotypical asymmetric centriole behavior in NBs described previously (Rebollo et al., 2007, Rusan and Peifer, 2007) largely contributes to the fidelity of asymmetric cell divisions. Within a centrosome, centrioles have different ages, and they can be structurally and/or functionally different (Conduit et al., 2015). This asymmetry is strongly visible during mitotic exit, just after disengagement of the mother-daughter centriole pair. The daughter or younger centriole retains microtubule (MT) nucleation activity, forming an aster that anchors the centriole to the apical cell cortex (hereafter called the apical centriole) (Rebollo et al., 2007, Rusan and Peifer, 2007, Conduit et al., 2010, Januschke and Gonzalez, 2010). In contrast, the mother or older centriole becomes inactivated and loses MT nucleation capacity, resulting in displacement away from the apical cortex toward the basal side (hence, referred to as basal centriole). Thus, the daughter centriole is retained in the NB, while the mother centriole is inherited by the GMC (Conduit et al., 2010, Januschke et al., 2011).
The discrepancy in the ability to nucleate MTs by the two centrioles can be explained by differences in pericentriolar material (PCM) retention (Conduit et al., 2010, Januschke et al., 2013). Maintenance of Polo kinase at the daughter centriole is crucial in retaining its PCM. Both positive and negative regulatory mechanisms control Polo localization on centrioles (Januschke et al., 2013, Ramdas Nair et al., 2016, Lerit and Rusan, 2013, Singh et al., 2014). The basal centriole is inactivated through “PCM shedding,” consisting of the rapid downregulation of the PCM (Rebollo et al., 2007, Rusan and Peifer, 2007), which is mediated by Plp and Bld10 (Drosophila orthologs of Pericentrin and Cep135, respectively) (Lerit and Rusan, 2013, Singh et al., 2014).
Although past studies have been instrumental in dissecting the molecular machinery responsible for asymmetric centriole behavior and the consequent impact on the MSO, the mechanisms responsible for centriole asymmetry establishment are not fully understood. Here, we uncover a function for Polo-like kinase 4 (Plk4), the master regulator of centriole duplication, in the establishment of this asymmetry. We show that Spd2 is a Plk4 substrate and that Spd2 phosphorylation triggers a basal-like centriole behavior. Furthermore, we found that the centriolar protein Fzr (the anaphase promoting complex [APC/C] activator and the ortholog of Cdh1 in vertebrates) functions as a positive apical centriole retention factor in NBs. Our work provides evidence of a role for centrosome proteins in orchestrating centrosome asymmetry and MSO.
Results
Altering Plk4 Activity Perturbs the Centrosome Asymmetry Cycle and Causes Spindle Positioning Defects
Drosophila NBs display a robust pattern of centriole asymmetry, which controls MSO and ensures that GMCs are always born at the same position relative to the NB (Figure 1A) (Rebollo et al., 2007, Rusan and Peifer, 2007) as confirmed by our time-lapse analysis of wild-type (WT) NBs (referred to as Control [Ctrl]) (n=20 Ctrl NBs from 8 brains) (Figures 1B and 1F; Video S1).
While analyzing the division of NBs expressing a GFP-tagged kinase-dead version of Plk4 (referred to as Plk4KD; Figure S1) in the Plk4 mutant (Plk4mut) background, we observed an unexpected behavior. The unduplicated centriole was preferentially maintained at the apical side in 88.5% of Plk4KD NBs (n = 23 out of 26 NBs from 7 brains). Of these, 46.2% contained an immobile centriole (Figures 1C and 1F; Video S2A) that remained apically anchored throughout interphase. In 46.3% of Plk4KD NBs, this centriole was maintained at an apical position even if it presented increased mobility. In this situation, the centriole movement was restricted to the apical hemisphere, the centriole moved from one side of the cell to the other (hereafter, referred to as apical mobile) (Figures 1D and 1F; Video S2A). Interestingly, in the example of Figure 1D and Video S2C, even if the apical cortex was deformed by a neighboring cell, the centriole was maintained at an apical position with little variation from one cell cycle to the following one. In the remaining 11.5% Plk4KD NBs, centrioles moved toward the basal side of the cell as described for basal centriole behavior (referred to as basal-like) (n = 3 out of 26 NBs from 7 brains) (data not shown; Figure 1F). Importantly, even if containing a single centriole, all Plk4KD NBs assembled a bipolar spindle. Initially, MT nucleation was noticed in the pole that contains the centriole; but rapidly, a bipolar array was generated, and cells invariably divided in a bipolar manner, similar to cells that lack centrioles (Basto et al., 2006, Moutinho-Pereira et al., 2009). These observations were very surprising because it is expected that as the unduplicated centriole ages, it should show a basal-like behavior, as shown by older mother centrioles (Figure S2A). The retention of the unduplicated centriole in NBs was also described in Plk4mut NBs where a significant number of NBs contain a single centriole (Bettencourt-Dias et al., 2005).
To ascertain the behavior of an unduplicated centriole that was generated using a different centriole duplication mutant background, we counted centrioles in NBs of Sas-4mut brains and compared these with Plk4KD and Plk4mut brains. Drosophila harboring mutations in key centriole genes are viable because of maternally provided centriole assembly factors that ensure centriole duplication during early embryogenesis when centrosomes are essential (Stevens et al., 2007, Basto et al., 2006, Riparbelli and Callaini, 2011, Bettencourt-Dias et al., 2005, Blachon et al., 2008). As development proceeds, centriole duplication ends around stage 15 or 16 (Basto et al., 2006) however, centrioles born prior to this stage are stably maintained. As cells continue to proliferate and increase in number throughout development, centrioles are detected in only a small number of cells. Indeed, a single centriole was detected in only 2.2% ± 1.4% of Sas-4mut NBs (n = 305 NBs from 7 brains) (Figure S2C). Moreover, we could not find centrioles in dividing NBs by live imaging of Sas-4mut brains (Figure S2D). In contrast, a single centriole was detected in 38.9% ± 5.6% of Plk4mut and 62.4% ± 15.4% of Plk4KD NBs (n = 187 NBs from 5 Plk4mut brains and n = 132 NBs from 5 Plk4KD brains) (Figure S2C). Importantly, the Plk4 mutant used in our study is a hypomorph (Bettencourt-Dias et al., 2005), explaining why some Plk4mut NBs contain centrioles at this developmental stage. Additionally, Plk4KD over-expression in the Plk4mut background might lead to a partial stabilization of the endogenous Plk4 protein, resulting in supernumerary centrosomes, which were detected at a low frequency (4.0% ± 5.2%) (Figure S2C). The presence of extra centrosomes, even in a small subset of cells, might contribute to an increased number of Plk4KD NBs with centrioles. Thus, our analysis of the behaviors of single centrioles in Plk4KD NBs suggests that loss of Plk4 activity promotes the apical cortical anchoring of an unduplicated centriole.
We next examined the consequence of over-expressing active Plk4 (referred to as a Plk4WT) on centriole positioning in NBs (Figure S1A). Over-expression of Plk4 generated a large number of NBs with extra centrosomes (78.1%; n = 274 NBs from 6 brains) (Figure S2C) (Basto et al., 2008, Habedanck et al., 2005, Marthiens et al., 2013). Surprisingly, however, and in contrast to Plk4KD NBs, centrioles in the majority of Plk4WT NBs displayed basal-like movement (70%; n = 14 of 20 NBs from 7 brains) (Figures 1E and 1F; Video S2C). Additionally, these centrioles did not produce detectable interphase MT asters, similarly to basal centrioles in Ctrl NBs. In the remaining 30% of Plk4WT NBs, centrioles presented apical mobile behavior with either a reduced MT aster (10%) or without a noticeable MT aster (20%) (n = 2 and n = 4 of 20 NBs from 7 brains, respectively). The lack of an apical centriole in Plk4WT NBs was unforeseen as at least one of the young centrioles is predicted to maintain an apical localization during interphase.
Since the asymmetric centriole cycle controls MSO over several NB divisions (Rebollo et al., 2007, Rusan and Peifer, 2007), we investigated the consequences of Plk4KD or Plk4WT expression in MSO. We filmed Ctrl, Plk4KD, and Plk4WT NBs over at least two consecutive mitoses and found that spindles in Plk4KD NBs maintained a fixed position, similar to Ctrl NBs (average angle = 13.5° ± 3.8°; variation between 0° and 22°; n = 15 NBs from 8 brains for Plk4KD and average angle = 12.0° ± 2.2°; variation between 0° and 25°; n = 14 NBs from 3 brains for Ctrl; p = not significant (ns); Figure 1G). In contrast, Plk4WT NBs displayed a more variable MSO through consecutive cycles (average angle = 25.0° ± 4.0°; variation between 0° and 75°; n = 23 from 10 brains; p = 0.026; Figure 1G). Characterization of MSO relative to the polarity axis using atypical protein kinase C (aPKC) as a marker (Lee et al., 2006, Homem and Knoblich, 2012) showed that in Ctrl NBs, spindles were oriented along the polarity axis (average angle = 9.9° ± 0.7°; variation between 2.3° and 45°; n = 48 NBs from 4 brains) (Figures S3A and S3B). In Plk4KD, the average angle was slightly increased to 15.3° ± 1.2° (variation between 5.4° and 60.9°; n = 62 NBs from 5 brains; p = 0.013) showing that even if the unduplicated centriole tends to maintain apical localization during interphase, mitotic spindles do not orient as correctly as Ctrl NBs. Importantly, in Plk4WT NBs, the average angle was increased to 23.5° ± 2.1° (variation between 3.9° and 89.6°; n = 96 NBs from 7 brains; p < 0.0001) (Figures S3A and S3B), confirming that increased Plk4 activity influences apical centrosome positioning in interphase and, thus, MSO. Defects in MSO can lead to defects in asymmetric cell division leading to the generation of two NBs instead of one NB and one GMC (Albertson and Doe, 2003, Basto et al., 2006, Basto et al., 2008, Castellanos et al., 2008, Homem and Knoblich, 2012). Using Dead pan (Dpn) (San-Juan and Baonza, 2011) as an NB marker, we determined the average number of NBs in the central brain lobe of Ctrl brains (48.9 ± 0.9; n = 9 brain lobes), similarly to previous studies (Basto et al., 2008, Gogendeau et al., 2015). Importantly, an increase in the number of NBs was noticed in Plk4WT central brain (55.2 ± 1.0; n = 9 brain lobes; p = 0.0003). Taken together, our results indicate that Plk4 activity must be tightly regulated not only to ensure the formation of a single procentriole per mother but also to control MSO and asymmetric cell division in NBs.
An MT-Independent Mechanism Contributes to Apical Centriole Maintenance in Plk4KD NBs
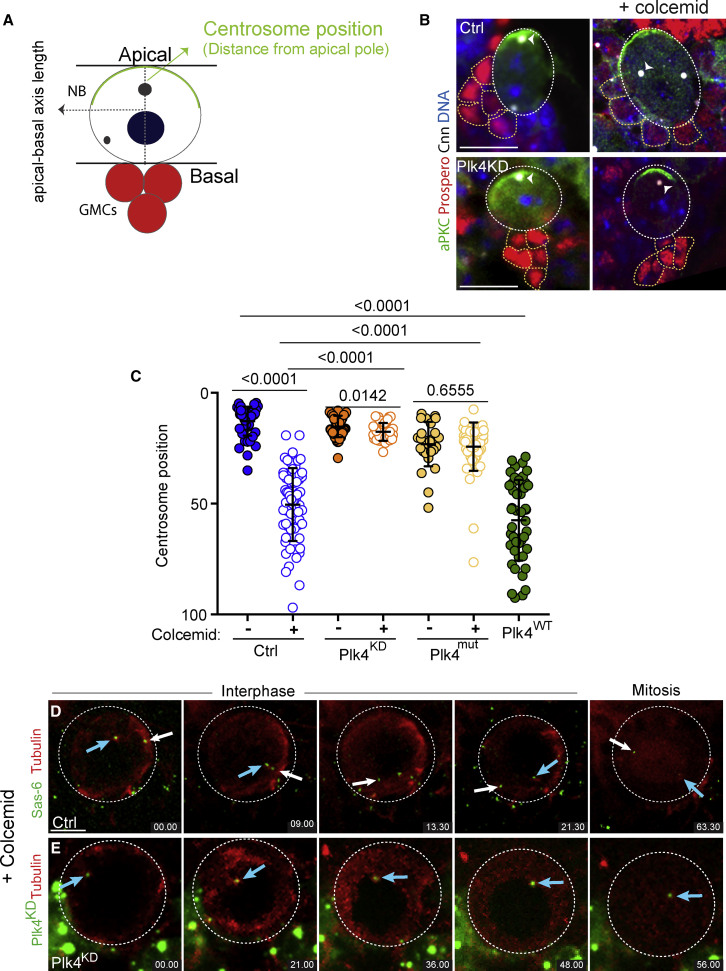
While characterizing Plk4KD NBs, we realized that the apical aster appeared to have decreased MT nucleation when compared to Ctrl NBs (Figures 1C and 1D; Videos 1 and 2), suggesting that a yet unidentified mechanism might contribute to centriole anchoring in this cell type. To further test this possibility, we treated Ctrl and Plk4KD brains with colcemid to depolymerize MTs for 1 h. After fixation, we used two different conditions to label NBs and centrosomes. First, NBs were identified by aPKC and GMCs by Prospero. In this case, only a centrosome marker (Cnn) was used (Figure 2A). In the second method, we used two centrosomal antibodies, Plp and Cnn, which is important to unambiguously identify centrosomes, in particular in centrosome mutants. NBs and accompanied GMCs were labeled with phalloidin, and their respective identity was determined by their size (Figure S3C). In Ctrl NBs treated with colcemid, centrosomes were closer to the basal axis as expected (Januschke and Gonzalez, 2010), reaching positions similar to the ones found in Plk4WT NBs (Figures 2B, 2C, S3D, and S3E). Although centrioles were further away from the apical axis in colcemid-treated Plk4KD NBs, they were not positioned closer to the basal hemisphere as observed in Ctrl NBs. This was also the case in Plk4mut NBs, suggesting that the absence of active Plk4 delays or inhibits movement toward the basal side of the cell. Consistent with these findings, time-lapse imaging revealed that centrioles in Plk4KD NBs treated with colcemid appeared less mobile than centrioles from colcemid-treated Ctrl brains (n = 16 NBs from 3 brains for each condition) (Figures 2D and 2E; Videos S3A and S3B). These results indicate that an alternative mechanism, which seems to be MT-independent, contributes to centriole apical anchoring in Plk4KD NBs.
Figure 2.
The Single Centriole of Plk4KD NBs Does Not Behave as in Ctrl NBs after MT Depolymerization
(A) Schematic drawing of NBs with a hypothetic apical basis length (dashed line). Position zero was considered the apical cortex, while position 100 at the basal cortex was determined by its connection with GMCs.
(B) Immunostaining of Ctrl (top) and Plk4KD (bottom) NBs and GMCs with (right) or without (left) colcemid treatment with antibodies against aPKC and Prospero in green and red, respectively. The centrosome is labeled with Cnn antibodies (white) and DNA in blue. The apical centrosome in Ctrl NBs was recognized by containing a higher Cnn signal (white arrowhead). Scale: Ctrl, 3 μm; and Plk4KD, 4 μm. See also Figure S3.
(C) Dot plot showing the position of centrosomes in Ctrl, Plk4KD, and Plk4mut NBs with (+) and without (−) colcemid and Plk4WT NBs (Ctrl − 12.73 ± 0.9; Ctrl + 50.4 ± 0.9; Plk4KD − 15.17 ± 0.7; Plk4KD + 17.6 ± 0.6; Plk4mut − 23.14 ± 1.9; Plk4mut + 24.27 ± 1.5; and Plk4WT − 57.5 ± 2.8). Error bars represent means ± SD from at least 3 independent experiments where at least 35 NBs were analyzed from at least 8 brains. SS was assessed by unpaired t test.
(D and E) Images from time-lapse movies of Ctrl (E) and Plk4KD (F) NBs incubated with colcemid. The blue arrow marks the apical centriole after disengagement. The white arrow marks the basal centriole in Ctrl NBs. Time, minutes. Centrosome or centriole fluorescence intensity decreases in both Ctrl and Plk4KD NBs in conditions where MTs were depolymerized. This decrease is apparent as cells re-enter the following mitosis. Their dynamics and movement were followed by increasing the intensity levels, although this is not shown in the stills. Importantly, centrosomes and centrioles remain as stable structures in conditions of MT depolymerization since they can be readily noticed in immunostaining experiments using centriole and PCM markers (Figure 2C). Scale, 4 μm.
Fzr Contributes to Anchoring of Apical Centrioles
In order to explain the apical localization of unduplicated centrioles in Plk4KDNBs, we next immunostained NBs for centriole (Asterless [Asl]) and PCM (γ-tubulin, Spd2, and Cnn) proteins and used three-dimensional structural illumination microscopy (3D SIM) to characterize potential structural changes. Of these, Spd2 localization appeared the most distinct in centrioles from Plk4KD NBs (Figure 3A; data not shown). Spd2 is the Drosophila ortholog of Cep192, a major PCM component (Dix and Raff, 2007, Conduit et al., 2014). During interphase, Spd2 appeared as a ring in Ctrl NBs (Figure 3A) (Fu and Glover, 2012, Meghini et al., 2016, Mennella et al., 2012). In Plk4KD NBs, Spd2 appeared as a larger ring, while centrioles from Plk4WT NBs displayed a smaller Spd2 diameter (Figure 3A).
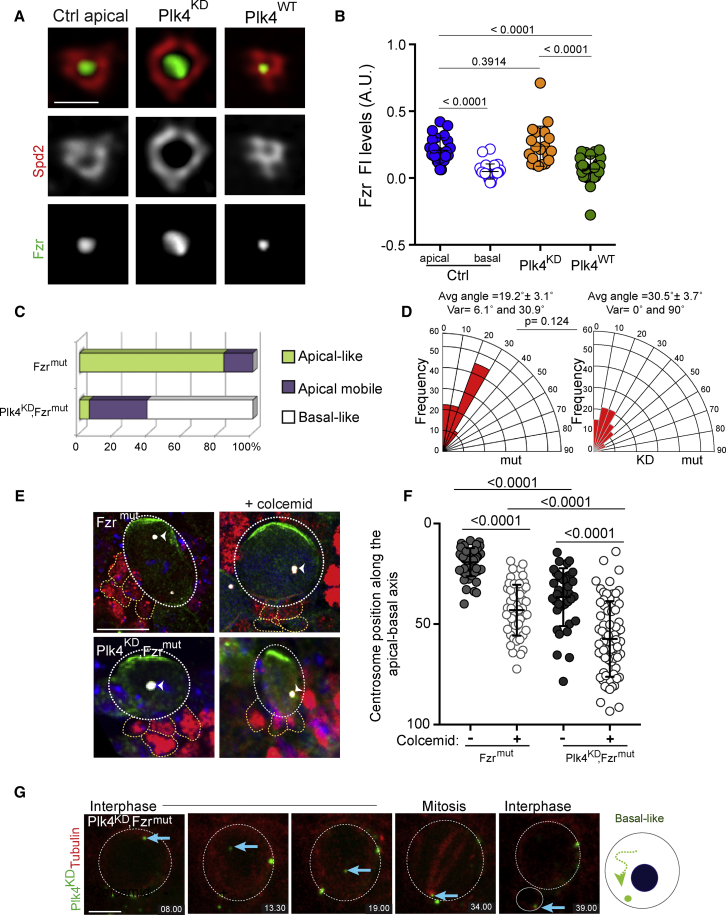
Figure 3.
Maintenance of Apical Anchoring in Plk4KD NBs Is Fzr Dependent
(A) 3D SIM images showing Spd2 (red) and Fzr (green) localization on interphase centrioles in Ctrl, Plk4KD and Plk4WT NBs. Scale, 400 nm.
(B) Dot plot showing Fzr fluorescent intensity levels at the centrosome (Ctrl apical, 0.20 ± 0.02; Ctrl basal, 0.04 ± 0.01; Plk4KD, 0.24 ± 0.03; Plk4WT, 0.07 ± 0.02). Error bars represent means ± SD from at least 3 independent experiments. SS was assessed by unpaired t test. See also Figure S4.
(C) Graph shows the percentage of centriole behavior categories in interphase (compare with Figure 1F for Ctrl and Plk4KD).
(D) Quantification of the angle between two consecutive mitoses in Fzrmut and Plk4KD, Fzrmut. SS was assessed by unpaired t test.
(E) Immunostaining of Fzrmut and Plk4KD, Fzrmut NBs and GMCs with or without colcemid treatment labeled with antibodies against aPKC and Prospero in green and red, respectively. The centrosome was labeled with Cnn antibodies (white) and DNA in blue. The apical centrosome in Fzrmut NBs was recognized by containing higher Cnn signal (white arrowhead). Scale for Ctrl, 4 μm. Note that on the Fzrmut panel with colcemid, the two centrosomes are very close to each other. See also Figure S5.
(F) Dot plot showing the position of centrosomes in Fzrmut and Fzrmut,Plk4KD NBs with (+) and without (−) colcemid (Fzrmut − 19.26 ± 0.9; Fzrmut + 43.1 ± 1.8; Fzrmut,Plk4KD − 36.6 ± 2.3; Fzrmut,Plk4KD + 57.4 ± 2.2). At least 27 NBs were analyzed for each condition from 8 different brains. Error bars represent SD. SS was assessed by unpaired t test. See also Figure S3.
(G) Images from time-lapse movies of Plk4KD, Fzrmut NBs. Tubulin (red) and GFP-Plk4KD (green). Blue arrow marks the centriole initially positioned in the NB, but later inherited by the GMC (right). Diagram illustrates centriole behavior in interphase. Time, minutes. Scale, 4 μm.
Spd2 was recently shown to recruit the APC/C activator Fizzy-related (Fzr) to the centrosome at the end of mitosis (Meghini et al., 2016). We thus investigated whether the differences in Spd2 localization in Plk4KD or Plk4WT NBs impacted Fzr levels on interphase centrioles. We generated flies expressing Red Fluorescent Protein (RFP)-Fzr (referred to as Fzr). 3D SIM images showed that Fzr occupied the internal region of the Spd2 ring (Figure 3A), as previously described (Meghini et al., 2016). In Plk4KD NBs, Spd2 formed an enlarged ring, and Fzr occupied a broader area, while in Plk4WT NBs, Fzr occupied a smaller area. Using images from confocal microscopy, we quantified Fzr fluorescent intensity levels at the centrioles of interphase NBs. Apical centrioles from Ctrl NBs and centrioles from Plk4KD NBs contained higher Fzr levels than basal centrioles from Ctrl NBs and Plk4WT centrioles (Figure 3B).
We confirmed these observations by analyzing Fzr localization using time-lapse microscopy. At the end of mitosis, Fzr was recruited to the centrosome in Ctrl NBs but exclusively remained associated with the apical centriole (Figure S4A). Fzr was also recruited to the single centriole at the end of mitosis in Plk4KD NBs and to Plk4WT centrioles (Figure S4A) however, its levels diminished on Plk4WT interphase centrioles. These results demonstrate not only that Fzr displays an asymmetric centriolar distribution in interphase NBs but also that Plk4 activity contributes to the removal of Fzr from centrioles during interphase.
To investigate whether Fzr plays a role in establishing centrosome asymmetry, we used a hypomorphic Fzr mutant (Fzrmut), which presents an overall reduction in Fzr levels (Jacobs et al., 2002). Using time-lapse microscopy of GFP-tubulin-expressing NBs in Fzrmut brains, we analyzed the position of the apical MT aster. 80% of Fzrmut NBs displayed apical centriole positioning (n = 16 out of 20 NBs from 4 brains) (Figure 3C), and a near-normal MSO was maintained over consecutive cycles (average angle = 19.2° ± 3.1°; variation between 6.1° and 30.9°; n = 9 NBs from 3 brains; Figure 3D). Fzrmut NBs displayed normal centrosome numbers (n = 70 NBs from 4 brains) (Figure S2C). Interestingly, 20% of Fzrmut NBs displayed an apical mobile behavior (Figures 3C and S4D). We also analyzed Fzrmut brains after colcemid treatment and found that centrosomes were positioned closer to the basal hemisphere (Figures 3E, 3F, S3E, and S3F). It is noteworthy that in untreated conditions, Fzrmut centrosomes were positioned further away from the apical cortex than Ctrl centrosomes (compare Figures 2C and 3F with Figures S3D and S3F). Taken together, our results suggest that Fzr may contribute to apical centrosome anchoring in NBs by acting in synergy with the MT nucleation pathway.
We next reasoned that decreasing Fzr levels in Plk4KD NBs, which have reduced MT nucleation, should induce basal-like behavior. Importantly, very few Plk4KD, Fzrmut flies reached the third instar larvae stage, suggesting that combining these two mutations perturbs development. Nevertheless, we were able to obtain larvae for these studies. Remarkably, decreasing Fzr levels in Plk4KD NBs induced movement of the unduplicated centriole toward the basal side in 61.1% of NBs (n = 11 of 18 NBs from 5 brains; p = 0.0009 compared to Plk4KD) (Figures 3C and 3G). Additionally, 33.3% of Plk4KD, Fzrmut NBs (n = 6 of 18) fell into the apical mobile category, and only one Plk4KD,Fzrmut NB maintained an apical centriole at the apical cortex during interphase (Figure 3C). Moreover, the average angle of Plk4KD, Fzrmut NBs was increased (Figure 3D) when compared to Fzrmut and, more importantly, when compared to Plk4KD NBs, (average angle = 30.5° ± 3.7°; variation between 0° and 90°; n = 32 NBs from 8 brains; p = 0.0013 relative to Plk4KD; and p = 0.124 relative to Fzrmut). Strikingly, colcemid treatment of Plk4KD, Fzrmut NBs caused centrosomes to reposition more toward the basal hemisphere (Figures 3E, 3F, S3E, and S3F). Thus, our findings suggest that Fzr participates in an MT-independent centriole apical anchoring mechanism, which is particularly active with reduced functional Plk4.
Fzr is an activator of APC/C, an E3 ubiquitin ligase that promotes mitotic exit and progression through G1 (Sivakumar and Gorbsky, 2015). Our results indicate that Fzr regulates centrosome asymmetry and, consequently, MSO. However, it remains to be investigated whether this function requires proteasome-mediated degradation of an APC/C ubiquitination target. To investigate this question, we characterized the localization of Cdc27, a core APC/C subunit, in Ctrl NBs (Huang and Raff, 2002). Although GFP-Cdc27 was slightly enriched at the centrosome at the end of mitosis, this localization was rather transient and, during interphase, GFP-Cdc27 appeared evenly distributed throughout the cytoplasm (Figure S4E). These observations reveal that APC/C is not at the centrosome at the right time to control apical anchoring. To rule out a role for APC/C in promoting apical anchoring, we tested whether inhibiting proteasomal degradation had an effect on centriole behavior by incubating Ctrl brains with Bortezomib (BZ), a proteasome inhibitor (Adams et al., 1998). Importantly, all NBs analyzed (n = 8 brains) maintained a stably anchored apical centriole, similar to controls (Figures S4F and S4G). The conditions we used increased the percentage of prometaphase-arrested NBs (41.4%; n = 7 brains) when compared to Ctrl brains incubated with DMSO (22.6%; n = 8 brains; p = 0.0433), validating the use of BZ as a proteasome inhibitor in Drosophila brains. Taken together, these results suggest that Fzr contributes to apical centriole anchoring and centrosome asymmetry independent of APC/C activity. Thus, retention of Fzr at the apical centriole constitutes a distinct mechanism that contributes to asymmetric cell division independent of MTs and APC/C activity.
We also asked whether an actin-based structure could contribute to apical centriole anchoring. Indeed, actin filament nucleation at the centrosome has recently been shown to occur in certain cell types (Farina et al., 2016, Obino et al., 2016). However, even if fine sub-apical actin structures could be detected, these were not co-localizing with centrosomes, and so we think that they could not be responsible for apical centriole anchoring in Ctrl or Plk4KD NBs (Figure S5A). Further, we depolymerized filamentous actin (F-actin) using cytochalasin D. This treatment resulted in cytokinesis inhibition, confirming the effect of actin depolymerization. Interestingly, both Ctrl and Plk4KD NBs showed a modification of the MT cytoskeleton where ectopic cytoplasmic nucleation sites were noticed (Figures S5B). Nevertheless, the apical centriole remained apically localized at the cortex throughout interphase. Thus, actin does not seem to play a role in maintaining the centriole at the apical cortex during interphase.
Plk4 Phosphorylates Spd2 to Promote a Basal-like Behavior
To understand how Plk4 could promote centriole asymmetry, we next focused on Spd2 because of its unique redistribution on centrosomes in response to Plk4 activity. Measurements of Spd2 fluorescence intensity levels on interphase centrosomes in Ctrl NBs revealed that its distribution appeared asymmetric, with high levels on the apical centriole and lower levels on the basal (Figures 4A and 4B). In contrast, Spd2 levels were extremely reduced in the majority of Plk4WT centrioles, displaying basal-like values.
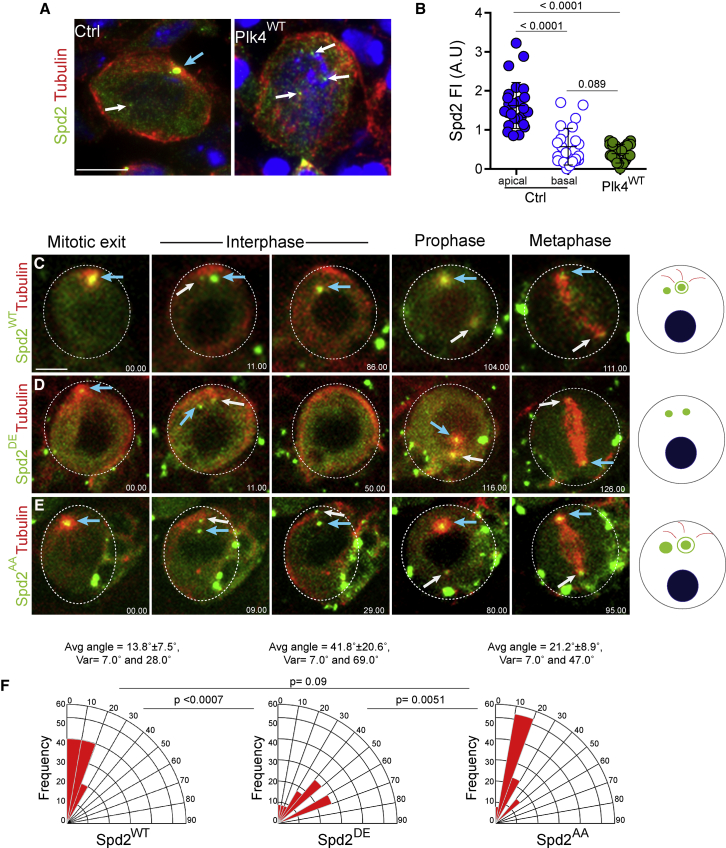
Figure 4.
Spd2 Phosphomutant NBs Display Centrosome Asymmetry and MSO Defects
(A) Ctrl and Plk4WT interphase NBs immunostained for tubulin (red) and Spd2 (green). DNA in blue. The blue arrow marks the apical centrosome in Ctrl NBs where Spd2 is detectable. White arrows denote centrosomes with low Spd2 levels. Scale, 4 μm.
(B) Dot plot showing Spd2 fluorescent intensity levels at the centrosome in the indicated genotypes (Ctrl apical, 1.6 ± 0.12; Ctrl basal, 0.6 ± 0.09; Plk4WT, 0.4 ± 0.04). Error bars represent means ± SD from at least 3 independent experiments. SS was assessed by unpaired t test.
(C–E) Images from time-lapse movies of GFP-Spd2WT (C), GFP-Spd2DE (D), and GFP-Spd2AA (E) larval NBs. Tubulin (red). See also Figures S1 and S6. Blue arrows mark the centrosome inherited by the NB at the end of mitosis. In the interphase panels, blue arrows mark the centriole that was localized at the apical cortex (apical centriole in Spd2WT) or the centrosome that was maintained at the apical hemisphere for longer periods of time after disengagement. White arrows mark the non-apical centrioles. (Right) Diagrams illustrate centriole behavior in each genotype after disengagement during early interphase. Time, minutes. Scale, 4 μm.
(F) Quantification of the angle between two consecutive mitosis in Spd2WT, Spd2DE, and Spd2AA. SS was assessed by unpaired t test.
We next explored whether Plk4 regulates centrosome asymmetry through Spd2 by testing whether Spd2 is a Plk4 substrate. We found that Plk4 phosphorylated Spd2 in vitro, and, using tandem mass spectrometry, 28 phosphorylated Ser and Thr residues within Spd2 were identified (Figures S6A–S6D). To test the functional relevance of these modifications in vivo, we generated flies expressing RFP-tagged Spd2 as either WT (Spd2WT) or phosphomutant transgenes, including phosphomimetic Spd2 harboring 28 aspartic acid or glutamic acid substitutions (Spd2DE) and a non-phosphorylatable alanine mutant (Spd2AA) (Figure S1B), all under the control of the same promoter and inserted on the same chromosome locus. The expression of Spd2 transgenes seemed to be comparable in brain extracts (Figure S6E). We then analyzed the behavior of Spd2WT and Spd2 phosphomutant-expressing NBs by time-lapse microscopy in order to observe centriole behaviors and analyze MSO over consecutive mitoses. As expected, over-expression of Spd2WT produced proper centrosome asymmetry, whereby an apical aster appeared after centriole disengagement (Figure 4C; Video S4A). As with endogenous Spd2, Spd2WT was not detected at the basal centriole soon after disengagement until just before the following mitosis, while it remained associated with the apical centrosome. Interestingly, we also observed reduced MT nucleation, suggesting that over-expression of Spd2 might change the nucleation capacity of interphase centrosomes. Importantly, MSO was maintained with relatively small variation (average angle = 13.8° ± 7.5°; variation between 7.0° and 28.0°; n = 10 NBs from 4 brains) (Figure 4F), indicating that over-expressing Spd2WT does not impact spindle positioning through consecutive cell cycles.
Despite localizing to the centrosome at the end of mitosis, Spd2DE levels decreased in a similar manner from both centrioles soon after their disengagement (Figure 4D; Video S4B). Interestingly, a fixed apical centrosome was not detected throughout most interphase, and both centrioles recruited Spd2DE with similar kinetics during late G2 and prophase. Importantly, in Spd2DE NBs, we measured a highly variable MSO over two consecutive mitoses (average angle = 41.8° ± 20.6°; variation between 7° and 69°; n = 11 NBs from 4 brains; p < 0.0007 when compared to Spd2WT) (Figure 4F).
We next examined the distribution of Spd2AA. Similar to Spd2WT and Spd2DE, Spd2AA localized to centrosomes at the end of mitosis. However, unlike Spd2WT, Spd2AA was maintained on both centrioles throughout a period of interphase (Figure 4E; Video S4C). Centrosome asymmetry was nevertheless established but much later, supporting the view that establishment of Spd2 asymmetry on centrioles relies, at least initially, on its phosphorylation state. In some NBs, the apical centriole was maintained at the apical cortex but frequently displayed an apical mobile behavior, while the other centriole also remained in the apical hemisphere (Figure 4E). Spd2AA expression caused slightly more variation in MSO than Spd2WT (average angle = 21.2° ± 8.9°; variation between 7° and 47°; n = 13 NBs from 5 brains; p = 0.09; ns, when compared to Spd2WT), but not as dramatic as Spd2DE (p = 0.0051 when compared to Spd2DE) (Figure 4F).
We investigated whether replacement of the 28 phospho-residues within Spd2 in both Spd2 phosphomutants (Spd2DE and Spd2AA) was affecting its ability to homodimerize or interact with Cnn (Galletta et al., 2016, Conduit et al., 2014). We depleted endogenous Spd2 from S2 cells by targeting its 3′ UTR and then transiently co-expressed various combinations of Spd2WT, Spd2DE, and Spd2AA tagged with either GFP or V5 (Figure S6E). Anti-GFP immunoprecipitations (IPs) from S2 cell lysates showed that V5-tagged Spd2DE and Spd2AA phosphomutants self-associate (Figure S6G). Moreover, V5-Cnn co-IPed with each of the GFP-Spd2 phosphomutants (Figure S6H), showing that the 28-amino-acid substitutions in the non-phosphorylatable and phosphomimetic Spd2 mutants do not cause protein misfolding or affect Cnn binding.
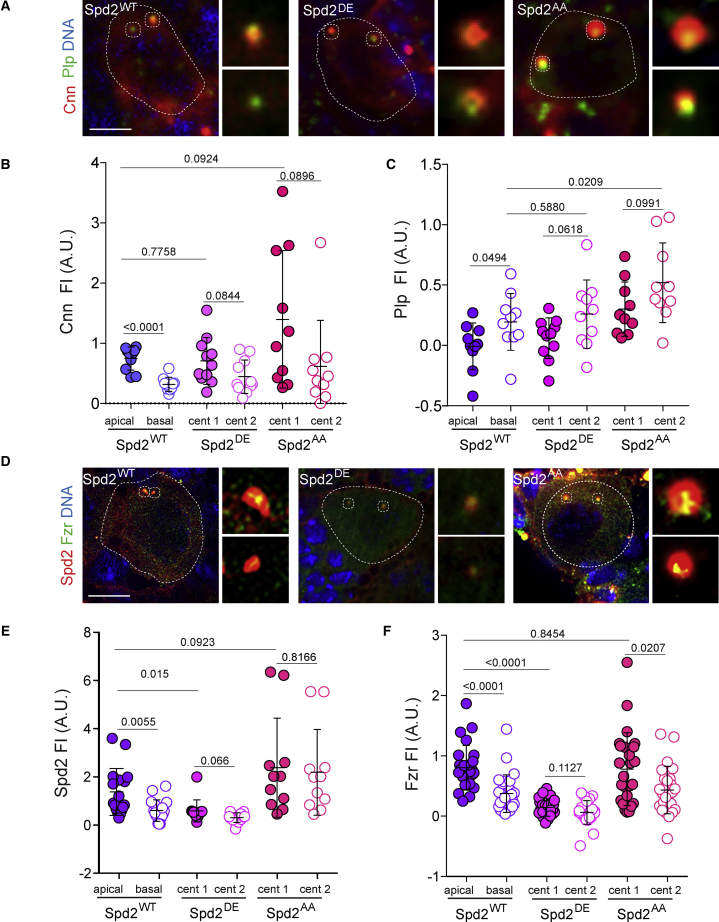
We also investigated whether Cnn centrosomal localization was influenced by the Spd2 phosphorylation state. A marked asymmetry in Cnn levels was observed between the apical and basal centrioles in Spd2WT NBs (Figures 5A and 5B). Although quite variable, Cnn was still recruited to Spd2DE and Spd2AA NBs, suggesting that Cnn is recruited to interphase centrioles regardless of the Spd2 phosphorylation status. We also measured the levels of Plp. Plp was still preferentially enriched in the basal centrioles of Spd2WT NBs and in one of the two centrioles in the Spd2 phosphomutants, even if displaying higher levels in SpdAA than in Spd2WT (Figures 5A and 5C).
Figure 5.
Spd2 Phosphomutants Influence Spd2 and Fzr Centriolar Recruitment without Impacting on Cnn or Plp
(A) Immunostaining of Spd2WT, Spd2DE, and Spd2AA early interphase NBs for Cnn (red) and Plp (green). DNA in blue. Insets show higher magnifications of each centriole. Scale, 4 μm. See also Figures S1 and S6.
(B and C) Dot plot showing Cnn and Plp fluorescent intensity levels on centrioles in the indicated genotypes (Cnn− Spd2WT: Ctrl apical 0.7 ± 0.06, Ctrl basal 0.3 ± 0.04; Spd2DE: Cent1 0.7 ± 0.1, Cent2 0.4 ± 0.08; Spd2AA: Cent1 1.4 ± 0.4, Cent2 0.6 ± 0.2 and Plp − Spd2WT: Ctrl apical − 0.01 ± 0.06, Ctrl basal 0.2 ± 0.07; Spd2DE: Cent1 0.06 ± 0.05, Cent2 0.3 ± 0.09; Spd2AA: Cent1 0.3 ± 0.07, Cent2 0.5 ± 0.1). Error bars represent means ± SD from at least 3 independent experiments. SS was assessed by unpaired t test.
(D) Images of Spd2WT, Spd2DE, and Spd2AA early interphase NBs showing Spd2 (red) and Fzr (green). DNA, blue. Insets show higher magnifications of each centriole. Scale, 4 μm.
(E and F) Dot plot showing Spd2 and Fzr fluorescent intensity levels on centrioles in the indicated genotypes. (Spd2 − Spd2WT: Ctrl apical 1.4 ± 0.2, Ctrl basal 0.6 ± 0.1; Spd2DE: Cent1 0.6 ± 0.1, Cent2 0.3 ± 0.06; Spd2AA: Cent1 2.3 ± 0.6, Cent2 2.1 ± 0.5 and Fzr- Spd2WT: Ctrl apical 0.8 ± 0.07, Ctrl basal 0.4 ± 0.06; Spd2DE: Cent1 0.1 ± 0.02, Cent2 0.06 ± 0.04; Spd2AA: Cent1 0.7 ± 0.1, Cent2 0.4 ± 0.07). Error bars represent means ± SD from at least 3 independent experiments. SS was assessed by unpaired t test.
We next measured centrosomal levels of Spd2 using anti-Spd2 antibodies (Dix and Raff, 2007). Notably, in both Spd2DE and Spd2AA NBs, Spd2 levels on centrosomes appeared more symmetric than centrioles in Spd2WT NBs (Figures 5D and 5E). In addition, Spd2 levels on Spd2AA centrosomes were comparable with Spd2WT apical centrosomes, while they were decreased in Spd2DE (Figure 5D).
Since Spd2 recruits Fzr to the centrosome (Meghini et al., 2016) and Fzr participates in centriole apical anchoring, we hypothesized that Fzr localization and/or levels might be altered in Spd2 phosphomutant NBs. Fzr showed asymmetric localization soon after centriole disengagement in Spd2WT NBs (Figures 5D and 5F; noticeable by the short distance between the two centrioles). Strikingly, Fzr levels were decreased in both centrioles of Spd2DE NBs, suggesting that Spd2 phosphorylation impacts Fzr recruitment or maintenance in interphase centrioles. Interestingly, in Spd2AA NBs, Fzr was present on both centrioles even when they were positioned far apart from one another (Figures 5D), suggesting that Fzr is maintained at both centrioles long after disengagement. Fzr asymmetry between even the two centrioles was noticeable although less pronounced than in Spd2WT NBs (Figure 5F). Our findings suggest that Plk4 phosphorylation of Spd2 downregulates Spd2 and Fzr recruitment to centrioles, thereby controlling centriole asymmetry and dynamics and, consequently, MSO in the following cell cycle.
Discussion
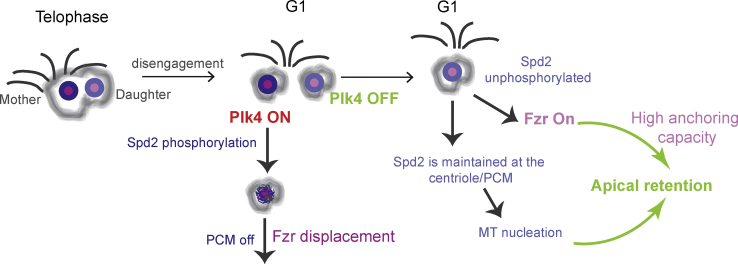
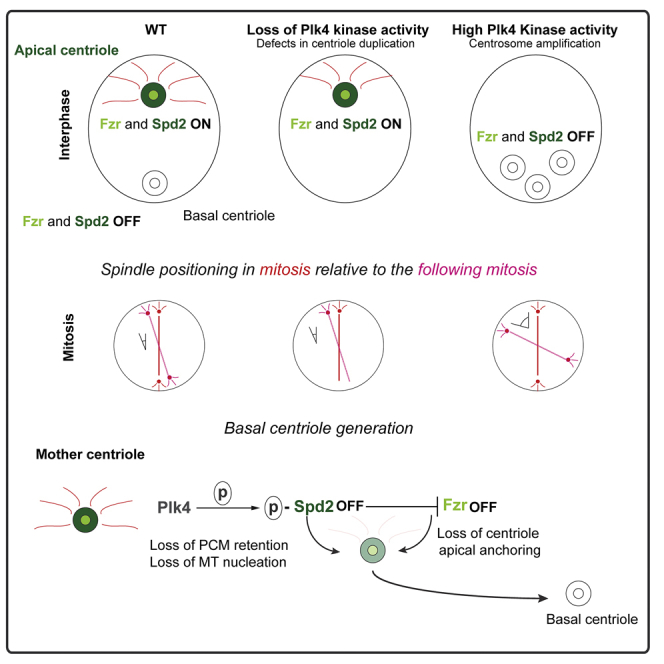
Here, we show that Plk4 plays a new role in the establishment of centrosome asymmetry and MSO in Drosophila NSCs. Our findings are consistent with a model where upon centriole disengagement at mitotic exit, the mother centriole inherits Plk4, which triggers centriole movement towards the NB basal side by disrupting microtubule-organizing center (MTOC) activity (Figure 6). Our data suggest that Plk4 needs to be removed or inactivated at the apical centriole, in order to ensure maintenance of MTOC activity. Although the model predicts an unequal distribution of Plk4 between the two NB centrioles, we are unable to test this aspect of the model. Endogenous Plk4 protein levels are extremely low and, despite several attempts to raise antibodies, we were unable to detect endogenous Plk4. Expression of GFP-tagged Plk4 under control of endogenous or weak promoters resulted in the stabilization of the protein, which invariably leads to increased activity and the unwanted supernumerary centrosomes (Basto et al., 2008, Aydogan et al., 2018).
Figure 6.
Model of Centriole Symmetry Breaking and Spindle Orientation in Drosophila NBs
At the end of mitosis, the mother-daughter centrioles of the NB disengage. The mother (or basal) centriole retains Plk4 activity, which phosphorylates Spd2, causing (1) PCM shedding and, thus, loss of MT nucleation and (2) Fzr displacement, which inhibits apical anchoring. An as yet undiscovered mechanism, Plk4 is inactive on the daughter (or apical) centriole, resulting in the stable maintenance of a centriole-bound population of non-phosphorylated Spd2, which promotes both MT nucleation and Fzr-dependent cortical anchoring.
In this model, active centriole-bound Plk4 targets Spd2, triggering Spd2 displacement and promoting, most likely, loss of additional PCM proteins from the basal centriole. It is thus conceivable that mother centrioles lacking Spd2 lose MT nucleation capacity, which induce their movement toward the basal side. It has been previously shown that asymmetric loss of PCM in the basal centriole was sufficient to trigger movement toward the basal side of the NB (Rebollo et al., 2007, Rusan and Peifer, 2007). Further, Spd2 removal also results in the loss of Fzr and consequently disables a previously undescribed second mechanism that contributes to maintaining centrosomes at the apical cortex or hemisphere. It will be important to investigate whether Spd2 mutants, which did not show defects in MSO during mitosis (Dix and Raff, 2007), display altered centriole behavior during interphase.
How Fzr promotes apical anchoring remains to be determined. Our findings support the idea that Fzr promotes centriole apical localization in an APC/C-independent manner. Although we have not identified a mechanism that displaces or inhibits Plk4 on the apical centriole, it is still possible that Fzr promotes apical anchoring by regulating, either directly or indirectly, the levels or activity of Plk4 at the apical centriole. However, we did not detect an interaction between Plk4 and Fzr in Drosophila brain extracts (D.G. and R.B., unpublished data). The observation that Fzr is absent from mitotic centrosomes in Ctrl NBs suggests that dynamic centrosome movements (typical of mitosis to allow efficient MT interactions with the cortex and chromosomes) require Fzr removal away from the centrosome. Moreover, the asymmetric maintenance of Fzr at the apical centriole, soon after disengagement, correlates with the low mobility typical of this centrosome. It is thus possible that maintenance of Fzr at the apical centriole functions as a barrier to mobility. We speculate that such an effect could be direct (Fzr itself can assemble in high-order structures to inhibit centriole mobility) or indirect (through a yet unknown interactor). Importantly, our work shows that maintenance of Spd2 at the apical centriole serves two major functions: PCM retention, MT nucleation and Fzr recruitment to inhibit mobility.
The functions of Plk4 and Spd2 uncovered and described here establish an unexpected association between the centrosome biogenesis machinery (centriole duplication and PCM recruitment) and centrosome asymmetry and spindle positioning apparatus. Additionally, we have identified a mechanism of apical centriole anchoring that appears to compensate for centriole duplication defects in NSCs by promoting apical retention. It will be interesting to investigate whether, the partial loss-of-function mutations in Plk4 described in humans, which cause microcephaly and dwarfism, support centriole retention at the apical cortex of NSCs (Martin et al., 2014, Tsutsumi et al., 2016, Shaheen et al., 2014). This might be beneficial not only by conferring the capacity to assemble primary cilia but also to ensure stem cell viability due to the presence of a centriole (Lambrus and Holland, 2017). Further, it will be important to investigate whether centrosome repositioning described during epithelial mesenchymal transition (EMT) or at mitotic exit (Burute et al., 2017, Piel et al., 2001) also relies on these mechanisms.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Spd2 | (Dix and Raff, 2007) | RRID: AB_2567456 |
| Rabbit polyclonal anti-Plp | (Martinez-Campos et al., 2004) | N/A |
| Rabbit polyclonal anti-aPKC | Santa Cruz | Cat# SC116 |
| Guinea pig polyclonal anti-Spd2 | This paper | N/A |
| Guinea pig polyclonal anti-Cnn | (Lucas and Raff, 2007) | N/A |
| Mouse monoclonal anti-α-Tubulin (clone DM1A), purified antibody | Sigma-Aldrich | Cat# T6199; RRID: AB_477583 |
| Mouse monoclonal anti-α-Tubulin (clone DM1A), ascites fluid | Sigma-Aldrich | Cat# T9026; RRID: AB_477593 |
| Mouse monoclonal anti-prospero | DSHB | MR1A; RRID: AB_528440 |
| RFP-Booster (Atto 594) | Chromotek | Cat# rba594; RRID: AB_2631390 |
| Mouse anti-GFP (clone JL-8) | Clonetech Laboratories | Cat# 632381; RRID: AB_2313808 |
| Mouse anti-HA (clone HA-7) | Sigma-Aldrich | Cat# H3663; RRID: AB_262051 |
| Mouse anti-V5 | Invitrogen | Cat# 46-0705; RRID: AB_2556564 |
| Alexa-Fluor 488 goat anti-rabbit | Thermo Fisher Scientific | A11008 |
| Alexa-Fluor 546 goat anti-mouse | Thermo Fisher Scientific | A11031 |
| Alexa-Fluor 633 goat anti-guinea pig | Thermo Fisher Scientific | A21105 |
| Fluorescent molecules | ||
| 546-conjugated Phalloidin | Thermo Fisher Scientific | A22283 |
| 647-conjugated Phalloidin | Thermo Fisher Scientific | A22287 |
| ATTO 488-Booster | ChromoTek | AD 488-21 |
| ATTO 594-Booster | ChromoTek | AD 594-21 |
| Bacterial and Virus Strains | ||
| DH5α™ Competent Cells | Thermo Fisher Scientific | Cat# 18258012 |
| One Shot™ BL21DE3 Competent Cells | Thermo Fisher Scientific | Cat# C600003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Demecolcine | Sigma-Aldrich | Cat# D7385; CAS: 477-30-5 |
| Cytochalasin D | Sigma-Aldrich | C8273 |
| Bortezomib (PS-341) | Selleck Chemicals | Cat# S1013; CAS: 179324-69-7 |
| Pfu Ultra HF DNA polymerase | Agilent Technologies Genomics | Cat# 600380 |
| Schneider’s Drosophila medium | Gibco | Cat# 21720-024 |
| Sf-900 II serum free medium | ThermoFisher | Cat# 10902096 |
| Nucleofector II | Amaxa (Lonza) | Cat# AAD-1001S |
| Fetal bovine serum | Gibco | Cat# 10500 |
| Penicillin–streptomycin | Gibco | Cat# 15140 |
| Acetic acid | VWR | Cat# 20103 295 |
| Protein-A conjugated Dynabeads | Invitrogen | Cat# 10001 D |
| Dimethyl pimelimidate dihydrochloride | Sigma-Aldrich | Cat# D8388 |
| Sodium azide (NaN3) | Fisher | Cat# S227I |
| Dithiothreitol (DTT) | Fisher | Cat# BP172 |
| Phenylmethylsophonyl fluoride (PMSF) | Sigma-Aldrich | Cat# 78830 |
| Soybean trypsin inhibitor, type II-S (SBTI) | Sigma-Aldrich | Cat# T9128 |
| SIGMAFAST protease inhibitor cocktail, EDTA-free | Sigma-Aldrich | Cat# S8830 |
| MBP-Spd2-NT1 (a.a. 1-84) | This paper | N/A |
| GST-Spd2-NT2 (a.a. 85-340) | This paper | N/A |
| GST-Spd2-M (a.a. 341-662) | This paper | N/A |
| GST-Spd2-C (a.a. 663-1146) | This paper | N/A |
| Plk4(a.a.1-317)-FLAG-His6 | (Brownlee et al., 2011) | N/A |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: UAS-GFP-Plk4KD(Plk4KD) | This paper | N/A |
| D. melanogaster: UAS-GFP-Plk4WT(Plk4WT) | This paper | N/A |
| D. melanogaster: Ubq-RFP-Fzr | This paper | N/A |
| D. melanogaster: Ubq-α-Tubulin-RFP | (Dobbelaere et al., 2008) | N/A |
| D. melanogaster: Ubq-α-Tubulin-GFP | (Dobbelaere et al., 2008) | N/A |
| D. melanogaster: Ubq-RFP-Sas-6 | (Peel et al., 2007) | N/A |
| D. melanogaster: UAS-Spd2WT | This paper | N/A |
| D. melanogaster: UAS-Spd2DE | This paper | N/A |
| D. melanogaster: UAS-Spd2AA | This paper | N/A |
| D. melanogaster: Sas-4s2214 | (Basto et al., 2006) | BDSC# 12119; FBst# 0012119 |
| D. melanogaster: FzrrapG0418 | (Jacobs et al., 2002) | BDSC# 12297 FBst# 0012297 |
| D. melanogaster: Ubq-GFP-Cdc27 | (Huang and Raff, 2002) | N/A |
| D. melanogaster: WorGAL4 | (Lee et al., 2006) | N/A |
| D. melanogaster: AseGAL4 | (Zhu et al., 2006) | N/A |
| D. melanogaster: ActGAL4 | Bloomington Drosophila Stock Center (Ahmad and Henikoff, 2001) | BDSC# 25374 FBti# 0127834 |
| D. melanogaster: GAL80ts | Bloomington Drosophila Stock Center (Davis, 2003 | BDSC# 7108 |
| D. melanogaster: Sakc06612(Plk4Mut) | (Bettencourt-Dias et al., 2005) | BDSC# 17774 FBst# 0017774 |
| D. melanogaster: wf | (Basto et al., 2006) | |
| D. melanogaster: y1w1118;PBac{y+-attp-9A}VK00020 | Bloomington Drosophila Stock Center | BDSC# 9738 FBti# 0076441 |
| Cell lines | ||
| Drosophila S2 cells | Invitrogen (Zhang et al., 2010) | Cat# R69007 |
| Oligonucleotides | ||
| Plk4KD construct generation: F 5’- GTCAAGATAG CCAACTTTGGACTGGCC-3’ and R 5’- GGCCAGT CCAAAGTTGGCTATCTTGAC-3’ |
This paper | N/A |
| Ubq-RFP-Fzr construct generation: F 5’-GGGGAC AAGTTTGTACAAAAAAGCAGGCTTC ATGTTTAGTCCCGAGTACGAGAAG-3’ and R 5’-GGGGACCACTTTGTACAA GAAAGCTGGGT CTTATCTGATATTGGCAAACAGATT-3’ |
This paper | N/A |
| UAS-RFP-Spd2 constructs generation: F 5’- cgcgcg ACTAGTGGCGGCACCGGCGGCACC ATGGACAGTAGCAGTGGAAGCCAA-3’ and R 5’-cgc gcgCCGCGGTTAAAACTAATCGGGAC-3’ |
This paper | N/A |
| RFP cloning from pURW: F 5’- cgcgcgGATATC ATGGCCTCCTCCGAGGACGTCATC-3’ and R: 5’- cgcgcgGGATCCGGCGCCGGTGGAGTGGCGGCCCTC-3’ |
This paper | N/A |
| MBP-Spd2-NT1 constructs generation: F 5’-gcgcgc GGATCCATGGACAGTAGCAGTGGAAGCCAA-3’ and R 5’- gcgcgcAAGCTTTTACTGGAGGGCAGTGCTCTTTGCTTG – 3’ |
This paper | N/A |
| GST-Spd2-NT2 constructs generation: F 5’-cgcgcg GGATCCATGCGCTTGTCCACAAACATCTCG – 3’ and R 5’ – cgcgcgCTCGAGTTATGGCTGTGGGGTCTTCTCGCCAAC – 3’ |
This paper | N/A |
| GST-Spd2-M constructs generation: F 5’ -cgcgcg GGATCCGACAATAAAACATACACTAAAACG-3’ and R 5’- cgcgcgCTCGAGTTATGTGAATCCGCTGGTGGAACTGGC - 3’ |
This paper | N/A |
| GST-Spd2-C constructs generation: F 5’ - gcgcgc GGATCCGCGAGTGGAAGACGTGGGTTGGGA - 3’ and R 5’ - cgcgcgCTCGAGTTAAAATTTAAAACTAATCGGGACACT - 3’ |
This paper | N/A |
| GFP-Spd2 constructs generation: F 5’ – cgcgGGTA CCTATGGTGAGCAAGGGCAGGAG – 3’ and 5’ – cgcg ACTAGTCTTGTACAGCTCGTCCATGCC – 3’ |
This paper | N/A |
| Spd2-UTR dsRNA synthesis: F 5’ – TAATACGACTCA CTATAGGGGTTTTCGCGTTCGCACTGCAAACTGTA ACTGTTTAAGGACAAAGCGGATTTGTTTTATTTGTG CCTGC – 3’ and R 5’ – TAATACGACTCACTATAGGG CTTTTAGGAAACAAGCG – 3’ |
This paper | N/A |
| V5-Spd2 constructs generation: F 5’ - GGGGGGATC TAGATCGGGGTACCATGggtaagcctatccctaaccctctcc tcggtctcgattctacgATGGACAGTAGCAGTGGAAGCC – 3’ and R 5’ – GGCTTCCACTGCTACTGTCCATcgtagaatcg agaccgaggagagggttagggataggcttaccCATGGTACCCCG ATCTAGATCCCCCC – 3’ |
This paper | N/A |
| V5-Cnn construct from EST: F 5’ – GGGGATCTAGATCG GGGTACCATGggtaagcctatccctaaccctctcctcggtctcgattc tacgATGGACCAGTCTAAA – 3’ and R 5’ – CGCCACTGTG CTGGATATCTTATAACTCATTCTCCATGTTTGAGCGAAC – 3’ |
This paper | N/A |
| Recombinant DNA | ||
| attB-P[acman]-Apr plasmid | DGRC | GenBank EF106980 |
| attB-pUAST-GFP-Plk4WT-SV40 | This paper | N/A |
| attB-pUAST-GFP-Plk4KD-SV40 | This paper | N/A |
| attB-pUAST-GFP-Plk4PACT-SV40 | This paper | N/A |
| pET28a-Plk4(a.a.1-317)-FLAG-His6 | (Brownlee et al., 2011) | N/A |
| pMal-C2X | Addgene | Cat# 75286 |
| pGEX-6P-2 | Addgene | Cat# 27-4598-01 |
| pMal-C2X-Spd2-NT1 (a.a. 1-84) | This paper | N/A |
| pGEX-6P-2-Spd2-NT2 (a.a. 85-340) | This paper | N/A |
| pGEX-6P-2-Spd2-M (a.a. 341-662) | This paper | N/A |
| pGEX-6P-2-Spd2-C (a.a. 663-1146) | This paper | N/A |
| pUbq-RFPNT | (Basto et al., 2008) | N/A |
| pUbq-RFPNT-Fzr | This paper | N/A |
| attB-pUAST-RFP-Spd2WT-Sv40 | This paper | N/A |
| attB-pUAST-RFP-Spd2DE-Sv40 | This paper | N/A |
| attB-pUAST-RFP-Spd2AA-Sv40 | This paper | N/A |
| pOT2 Spd2 EST (Clone ID LD24702, DGC EST Library 1.0) | DGRC | CG17286 |
| pMT/V5 His B | ThermoFisher | Cat# V412020 |
| pEGFP C1 | Addgene | Catalog #6084-1 |
| pMT/V5 His B GFP-Spd2WT | This paper | N/A |
| pMT/V5 His B GFP-Spd2AA | This paper | N/A |
| pMT/V5 His B GFP-Spd2DE | This paper | N/A |
| pMT/V5 His C GFP | (Klebba et al., 2015) | N/A |
| pMT/V5 His B V5-Spd2WT | This paper | N/A |
| pMT/V5 His B V5-Spd2AA | This paper | N/A |
| pMT/V5 His B V5-Spd2DE | This paper | N/A |
| pOT2 Cnn EST (Clone ID LD19135, DGC EST Library 1.0) | DGRC | CG4832 |
| pMT/V5 His C V5-Cnn | This paper | N/A |
| pMT/V5 His C Asl V5 | (Klebba et al., 2015) | N/A |
| Software and Algorithms | ||
| Metamorph software 7.7 | Molecular devices | N/A |
| Fiji | (Schindelin et al., 2012) | https://fiji.sc/ |
| Photoshop | Adobe | N/A |
| GraphPad Prism 7 | GraphPad Software, Inc. | N/A |
| Other | ||
| Glass-bottom 35 mm dish uncoated | MatTek Corporation | P35G-1.5-14-C |
| Membrane kit, Standard | YSI | SKU098094 |
| Voltalef oil 10S | VWR BDH Prolabo | Cat# 24627.188; CAS: 9002-83-9 |
Contact for Reagent and Resources Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Renata Basto (renata.basto@curie.fr).
Experimental Model and Subject Details
Experimental Animals
Species: Drosophila melanogaster. Flies were raised on Drosophila culture medium (0,75% agar, 3,5% cornmeal, 5% yeast, 5,5% sugar, 2,5% methyl, 1% penicillin–streptomycin, 0,4% propionic acid). Flies stocks were maintained at 18˚C in plastic vials or plastic bottles. Crosses were maintained at 25˚C in plastic vials. Brains were collected from 3rd instar larvae (which were staged as the number of days after egg laying and confirmed with developmental landmarks such as size, mouth hook and position within the tube). For all experiments except FzrrapG0418 (Fzrmut) and in combination with Plk4KD, we analyzed male and female since we did not notice any difference between the two sexes. In the case of Fzrmut, we analysed only males. In all experiments, Plk4KD, Plk4WT and Plk4PACT were recombined with the Plk4 mutant-Sakc06612 (Bettencourt-Dias et al., 2005) (BDSC#17774). Controls (Ctrl) were used accordingly to the experiments (please see Key Resources Table and below for detailed analysis).
Fly Stocks
UAS lines: UAS-GFP-Plk4KD (Plk4KD, this study), UAS-GFP-Plk4WT (Plk4WT, this study).
Reporter lines: Ubq-α-Tubulin-RFP and Ubq-α-Tubulin-GFP (Dobbelaere et al., 2008), Ubq-RFP-Sas-6 (Peel et al., 2007), Ubq-RFP-Fzr (this study), Ubq-GFP-Cdc27 (Huang and Raff, 2002).
Mutant alleles: Sakc06612 (BDSC#17774, (Bettencourt-Dias et al., 2005)), Sas-4s2214 (BDSC#12119, (Basto et al., 2006)), FzrrapG0418 (BDSC#12297, (Jacobs et al., 2002)).
Driver lines: WorGAL4 (provided by C. Doe, (Albertson et al., 2004) , AseGAL4 (provided by T. Lee, (Zhu et al., 2006), ActGAL4 (BDSC#25374), GAL80ts (BDSC#7108).wf was used as a control strain.
Cell Lines
Female Drosophila S2 cells (Zhang et al., 2010) (Invitrogen) were cultured in Sf-900 II (Life Technologies) + Pen/Strep (Gibco) and split every 3-4 days.
Method Details
Generation of Drosophila Transgenic Lines
The Plk4WT transgene was synthetized by GenScript (GenScript®, NJ, USA) in the pUC57 plasmid, using the coding DNA sequence (CDS) of Drosophila Plk4. The Plk4KD allele was generated by site-directed mutagenesis (Pfu Ultra HF DNA polymerase, 600380, Agilent Technologies Genomics) from the Plk4WT allele with primers F (5’- GTCAAGATAGCCAACTTTGGACTGGCC-3) and R (5’- GGCCAGTCCAAAGTTGGCTATCTTGAC-3’). Underlined is the triplet coding for D156, mutated into N. All Plk4 versions were initially cloned into a pDONR™/Zeo plasmid (Invitrogen, CA, USA) to have the GFP at their N-terminus, spaced by a linker of six triplets (GGCGGCACCGGCGGCACC), under the control of the UAS (X5) promoter. At the 3’ end, the SV40 sequence was also included (Figure S1). All constructs were validated through Sanger sequencing of the entire coding region. The UAS promoter was combined with the SV40 polyA sequence (ttaattgtttattgcagcttataatggttacaaataaagcaatagcatcacaaatttcacaaataaagcatttttttcactgcattctagttgtggtttgtccaaactcatcaatgtatcttatcagcggccgc), which allows for high expression and stabilization of the Plk4 sequences. To generate all Plk4 transgenic stocks, the PhiC31 integrase-mediated transgenesis system was used. The different Plk4 constructs, flanked by NotI and AscI, were cloned into a P[acman] plasmid containing the attB site (DGRC, GeNBank EF106980) and injections were performed by BestGene (BestGene Inc, CA, USA). The Bloomington stock BI9738 (y1w1118;PBac{y+-attp-9A}VK00020) was used as attP-containing docking site strain. All transgenes were inserted in the third chromosome in the insertion site 99F8.
To generate the Ubq-RFP-Fzr stock, the genomic sequence of Fzr was first amplified by PCR with primers F: (5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTTC ATGTTTAGTCCCGAGTACGAGAAG-3’) and R: (5’-GGGGACCACTTTGTACAA GAAAGCTGGGTCTTATCTGATATTGGCAAACAGATT-3’). Fzr specific sequences are underlined. The PCR product was then recombined using BP Gateway reaction (Gateway® Technology, Invitrogen, ThermoScientific) into pDONR™/Zeo plasmid. After sequence validation through Sanger sequencing, Fzr was recombined, using LR Gateway reaction in the pUbq-RFPNT vector (Basto et al., 2008), which allows for moderate expression of the tagged protein (Lee et al., 1988). BestGene performed injections in the w1118 stock. With this system the transgene insertion occurred randomly. For all experiments, stocks with transgenes inserted in the second chromosome were used.
For UAS- RFP-Spd2 stocks, the Spd2 cDNA was cloned in phase with RFP on its N-terminus with the primers F: (5’- cgcgcgACTAGTGGCGGCACCGGCGGCACC ATGGACAGTAGCAGTGGAAGCCAA) and R: (5’-cgcgcgCCGCGGTTAAAACTAATCGGGAC) and SV40 (sequence described above) at its C-terminus with the enzymes Spe1 and SacII in the Bluescript vector. RFP was cloned from the pURW vector (DGRC #1282) with primers F: (5’- cgcgcgGATATCATGGCCTCCTCCGAGGACGTCATC-3’) and R: (5’- cgcgcgGGATCCGGCGCCGGTGGAGTGGCGGCCCTC-3’) in the Bluescript vector with the enzymes EcoRV and BamHI, spaced by a linker composed of GGTGGT. Spd2 and RFP specific sequences are underlined. Restriction enzymes are in Italic. Spd2DE and Spd2AA constructs were synthetized by GeneART (ThermoFisher). In Spd2DE construct, all serines (Ser) and threonines (Thr) identified were mutated to aspartate (D) with the exception of Thr371, Thr384, Thr458, Thr493, Thr706, Thr754, which were mutated to Glutamate (E). In Spd2AA construct all 28 phosphoresidues identified were mutated to alanines (A). After fusion to RFP and SV40 and sequence validation through Sanger sequencing, all constructs comprising RFP-Spd2-SV40 was subsequently cloned in the P[acman] plasmid using AscI and Not1. Insertion site and transgenesis were performed as mentioned above for Plk4 constructs by BestGene Inc, CA, USA.
Generation of Spd2 Fragments
Spd2 fragments were amplified by PCR from the Spd2 cDNA, using the following primers: for Spd2-NT1 (a.a. 1-84) F 5’ - gcgcgcGGATCCATGGACAGTAGCAGTGGAAGCCAA - 3’ and R 5’ -gcgcgcAAGCTTTTACTGGAGGGCAGTGCTCTTTGCTTG - 3’, for Spd2-NT2 (a.a. 85-340) F 5’ - cgcgcgGGATCCATGCGCTTGTCCACAAACATCTCG – 3’ and R 5’ – cgcgcgCTCGAGTTATGGCTGTGGGGTCTTCTCGCCAAC–3’, for Spd2-M (a.a. 341-663) F 5’ – cgcgcgGGATCCGACAATAAAACATACACTAAAACG - 3’ and R 5’ - cgcgcgCTCGAGTTATGTGAATCCGCTGGTGGAACTGGC - 3’ and for Spd2-C (a.a. 664–1146) F 5’ - gcgcgcGGATCCGCGAGTGGAAGACGTGGGTTGGGA - 3’ and R 5’ - cgcgcgCTCGAGTTAAAATTTAAAACTAATCGGGACACT - 3’. Spd2 specific sequences are underlined. Restriction enzymes are in Italic. Spd2-NT1 was cloned into the pMal-C2X vector with the enzymes BamHI and HindIII, with MBP on its N-terminus. Spd2-NT2, Spd2-M and Spd2-C were cloned into the pGEX-6P-2 vector with the enzymes BamHI and XhoI, with GST on their N-terminus. After sequence validation through Sanger sequencing, Spd2 fragments were used for in vitro kinase assay.
Generation of Spd2 Phosphomutant Transgenes for S2 Experiments
Generation of the GFP-Spd2 phosphomutant transgenes was obtained as follows. GFP was amplified by PCR from the pEGFP C1 vector, flanked with KpnI and SpeI restriction sites, using the following primers: F 5’ – cgcgGGTACCTATGGTGAGCAAGGGCAGGAG – 3’ and 5’ – cgcgACTAGTCTTGTACAGCTCGTCCATGCC – 3’. GFP specific sequences are underlined. Restriction enzymes are in Italic. In the F primer, one T was added between the KpnI site and GFP sequence to maintain the frame in the pMT/V5 HisB vector. Next, Spd2 phosphomutant sequences were recovered from the Bluescript vector (already used for the generation of RFP-Spd2 stocks, see above) by digestion with the enzymes SpeI and SacII and cloned into the pMT/V5 HisB vector with GFP on their N-terminus.
The V5-Spd2 phosphomutants were generated from the GFP-Spd2 phosphomutant (in the pMT/V5 HisB vector) by a megaprimer PCR strategy. GFP sequence was replaced with V5 sequence using the megaprimers F 5’ – GGGGGGATCTAGATCGGGGTACCATGggtaagcctatccctaaccctctcctcggtctcgattctacgATGGACAGTAGCAGTGGAAGCC - 3’ and R 5’ – GGCTTCCACTGCTACTGTCCATcgtagaatcgagaccgaggagagggttagggataggcttaccCATGGTACCCCGATCTAGATCCCCCC - 3’. The V5-tag is in lower case and Spd2 specific sequences are underlined. The V5-Spd2 sequences were verified by sequencing. The V5-Cnn construct was PCR amplified from a Cnn EST (Clone ID LD19135, DGC EST Library 1.0) using primers F 5’ – GGGGATCTAGATCGGGGTACCATGggtaagcctatccctaaccctctcctcggtctcgattctacgATGGACCAGTCTAAA - 3’ and F 5’ – CGCCACTGTGCTGGATATCTTATAACTCATTCTCCATGTTTGAGCGAAC - 3’ and then inserted into the pMT/V5 HisC vector. The V5-tag is in lower case and Cnn specific sequences are underlined. Insert sequences were verified by sequencing.
Expression of Plk4 and Spd2 Transgenes
Expression of UAS-transgenes was carried out using either the UAS/GAL4 system (Brand and Perrimon, 1993) or the temporal and regional gene expression targeting (TARGET) method (McGuire et al., 2004). Worniu (Wor)GAL4 (Albertson et al., 2004) and Asense (Ase) GAL4 (Zhu et al., 2006) were used to induce transgene expression exclusively in NBs to perform live imaging experiments. WorGal4 and AseGal4 were recombined with either Ubq-α-Tubulin-RFP or Ubq-α-Tubulin-GFP transgenes (Dobbelaere et al., 2008), localized on the 2nd chromosome. As control for live imaging experiments, to analyze centriole behavior we used the Ubq-Sas-6 RFP line, which was crossed with Ubq-α-Tubulin-GFP,WorG4 or Ubq-α-Tubulin-GFP,AseGal4. As control for live imaging experiments, to analyze centriole behavior we used the Ubq-Sas-6 RFP line was crossed with Ubq-α-Tubulin-GFP, or Ubq-α-Tubulin-GFP, WorG4 or Ubq-α-Tubulin-GFP,AseGal4. All these lines showed similar centriole behavior and cell cycle timings. We referred to them as Ctrls in the text. A recombinant comprising the ActGAL4 (BDSC#25374) and GAL80ts (BDSC#7108) on the 2nd chromosome was used to induce the expression of the transgenes in a temporal manner to be used in immunostaining experiments. At 18˚C, GAL80ts binds to and inhibits the transcriptional activation domain of GAL4. The fly crosses were established and allowed for larvae to develop at either 18˚C–20˚C . Second instar larvae were then moved to 29˚C in order to inhibit the binding between GAL80ts and GAL4. This leads to the expression of the UAS-transgene within 16-18 hours. Two days after, mid third instar larval brains were dissected for immunostaining. Ctrls were performed with this stock, where the ActGal4Gal80ts recombinants were allowed to develop between 18˚C-20˚C, before being placed in the 29˚C incubator.
Drosophila genotypes and crosses
Figure 1
Control: GFP-Tub, AseGal4/Cyo-GFP; X Ubq RFP-Sas6
Plk4KD: RFP-Tub, AseGal4/Cyo-GFP; Plk4mut /TM6 Tb X +/Cyo-GFP; GFP- Plk4KD, Plk4 mut/ TM6 Tb
Plk4WT: RFP-Tub, AseGal4/Cyo-GFP; Plk4mut /TM6 Tb X +/Cyo-GFP; GFP- Plk4WT, Plk4 mut/ TM6 Tb
Crosses were maintained at 25°C
Figure 2
Control: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb
Plk4KD: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb X +/Cyo-GFP; GFP- Plk4KD, Plk4 mut/ TM6 Tb
Plk4mut: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb
Plk4WT: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb X +/Cyo-GFP; GFP- Plk4WT, Plk4 mut/ TM6 Tb
Crosses were maintained at 18 °C for 4 days after egg laying and transferred to 29°C for 48 hrs.
Control: GFP-Tub, AseGal4/Cyo-GFP; X Ubq RFP-Sas6
Plk4KD: RFP-Tub, AseGal4/Cyo-GFP; Plk4mut /TM6 Tb X +/Cyo-GFP; GFP- Plk4KD, Plk4 mut/ TM6 Tb
Crosses were maintained at 25°C
Figure 3
Control: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb
Plk4KD: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb X +/Cyo-GFP; GFP- Plk4KD, Plk4 mut/ TM6 Tb
Plk4WT: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb X +/Cyo-GFP; GFP- Plk4WT, Plk4 mut/ TM6 Tb
Crosses were maintained at 18 °C for 4 days after egg laying and transferred to 29°C for 48hrs.
Fzrmut: Fzr/Fm7 Kr-GFP; If/Cyo-GFP X GFP-Tub/Cyo-GFP
Fzrmut, Plk4KD: Fzr/Fm7 Kr-GFP; GFP- Plk4KD, Plk4 mut/ TM6 Tb X RFP-Tub, AseGal4/Cyo-GFP; Plk4mut /TM6 Tb
Crosses were maintained at 25°C
Fzrmut: Fzr/Fm7 Kr-GFP; If/Cyo-GFP
Fzrmut, Plk4KD: Fzr/Fm7 Kr-GFP; GFP- Plk4KD, Plk4 mut/ TM6 Tb X ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb
Crosses were maintained at 18 °C for 4 days after egg laying and transferred to 29°C for 48hrs.
Figure 4
Control: wf
Plk4WT: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb X +/Cyo-GFP; GFP- Plk4WT, Plk4 mut/ TM6 Tb
Crosses were maintained at 18 °C for 4 days after egg laying and transferred to 29°C for 48hrs.
Spd2WT: If/Cyo-GFP; Spd2WT/TM6Tb X GFP-Tub, AseGal4/Cyo-GFP; Plk4mut /TM6 Tb
Spd2DE: If/Cyo-GFP; Spd2DE/TM6Tb X GFP-Tub, AseGal4/Cyo-GFP; Plk4mut /TM6 Tb
Spd2AA: If/Cyo-GFP; Spd2DE/TM6Tb X GFP-Tub, AseGal4/Cyo-GFP; Plk4mut /TM6 Tb
Crosses were maintained at 25°C
Figure 5
Spd2WT: If/Cyo-GFP; Spd2WT/TM6Tb X ActGal4, Gal80ts/Cyo-GFP; Sb/TM6Tb
Spd2DE: If/Cyo-GFP; Spd2DE/TM6Tb X ActGal4, Gal80ts /Cyo-GFP; Sb/TM6Tb
Spd2AA: If/Cyo-GFP; Spd2DE/TM6Tb X ActGal4, Gal80ts /Cyo-GFP; Sb/TM6Tb
Crosses were maintained at 18 °C for 4 days after egg laying and transferred to 29°C for 48hrs.
SFigure 2
Control: ActGal4, Gal80ts, /Cyo-GFP
Plk4KD: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb X +/Cyo-GFP; GFP- Plk4KD, Plk4 mut/ TM6 Tb
Plk4mut: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb
Plk4WT: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb X +/Cyo-GFP; GFP- Plk4WT, Plk4 mut/ TM6 Tb
Crosses were maintained at 18 °C for 4 days after egg laying and transferred to 29°C for 48hrs.
Plk4mut/TM6Tb
Sas-4mut/TM6Tb
Fzrmut/FM7 Kr-GFP
Ubq-RFP-Sas-6/Cyo-GFP; Sas-4mut/TM6Tb X Ubq-GFP-Tub/Cyo-GFP; Sas-4mut/TM6Tb
Crosses were maintained at 25°C
SFigure 3
Control: ActGal4, Gal80ts, /Cyo-GFP
Plk4KD: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb X +/Cyo-GFP; GFP- Plk4KD, Plk4 mut/ TM6 Tb
Plk4WT: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb X +/Cyo-GFP; GFP- Plk4WT, Plk4 mut/ TM6 Tb
Crosses were maintained at 18 °C for 4 days after egg laying and transferred to 29°C for 48hrs.
SFigure 4
Control: RFP-Fzr/Cyo-GFP; Plk4mut/TM6Tb X Ase-Gal4, Tub-GFP/Cyo-GFP; Plk4mut/TM6Tb
Plk4KD: RFP-Fzr/Cyo-GFP; GFP- Plk4KD, Plk4 mut X Ase-Gal4, Tub-GFP/Cyo-GFP; Plk4mut/TM6Tb
Plk4WT: RFP-Fzr/Cyo-GFP; GFP- Plk4WT, Plk4 mut X Ase-Gal4, Tub-GFP/Cyo-GFP; Plk4mut/TM6Tb
Wf
Crosses were maintained at 25°C
SFigure 5
Control: ActGal4, Gal80ts, /Cyo-GFP
Plk4KD: ActGal4, Gal80ts, /Cyo-GFP; Plk4mut /TM6 Tb X +/Cyo-GFP; GFP- Plk4KD, Plk4 mut/ TM6 Tb
Crosses were maintained at 18 °C for 4 days after egg laying and transferred to 29°C for 48 hrs.
GFP-Tub, AseGal4/Cyo-GFP; X Ubq RFP-Sas6
Crosses were maintained at 25°C
Live Imaging
WorGAL4 or AseGAL4 drivers were used to induce the transgene expression only in NBs. In general, the over-expression of any GFP-Plk4 or RFP-Spd2 proteins leads to the formation of large green/red aggregates that are frequently segregated into the GMCs. Other smaller aggregates or green particles can often been seen in NBs and GMCs. In all the movies analyzed and presented in this article, the centrosome or centriole were distinguished by its capacity to nucleate MTs, or through its position at spindle poles or association with the spindle during mitosis in the the period of the time-lapse (even if not included in the stills). In certain cases, the timeframe where it is possible to distinguish a centriole or centrosome is not included in the Figures shown, but we carefully characterize centrosome/centriole behavior and dynamics to identify the same centrosome/centriole in different time frames. Mid third instar larval brains were dissected in Schneider’s Drosophila medium (21720-024, Gibco, ThermoScientific) supplemented with 10% heat-inactivated fetal bovine serum (10500, Gibco, ThermoScientific), penicillin (100 units ml−1) and streptomycin (100 μg ml−1) (penicillin–streptomycin 15140, Gibco, ThermoScientific) (hereafter referred to as live imaging medium). Brains were placed on a glass-bottom 35 mm dish (P35G-1.5-14-C, Mat Tek Corporation, MA, USA) with 10 μl of medium, covered with a permeable membrane (Standard membrane kit, YSI, OH, USA) and sealed around the membrane borders with Voltalef oil 10S (VWR BDH Prolabo). One or two brain lobes were recorded using a Yokagawa CSU-X1 spinning head mounted on a Nikon TiE inverted microscope. The microscope was equipped with an EMCCD Evolve 512 × 512 (Photometrics, AZ, USA) and controlled by the Metamorph software 7.7 (Molecular devices). Four-dimensional z-stacks of 18–26μm at 0.75-μm intervals were acquired every 30 or 60 s using an x60, NA 1.4 oil-immersion objective. The same laser power and acquisition settings were kept for all time lapse acquisitions. Images were processed with Fiji (NIH) and Adobe Photoshop.
Immunohistochemistry and Antibodies
Third instar larval brains were dissected in PBS and fixed in 4% formaldehyde in PBS for 30 min. After fixation, brains were transferred to 45% acetic acid (diluted in water) for 15 sec and then to 60% acetic acid (diluted in water) for 3 min. Brains were then mounted onto a slide, squashed and immediately flash-frozen in liquid nitrogen, followed by a further fixation with ice-cold methanol, at -20˚C for 7 min. Next, brains were rehydrated in PBS + 0.1% Triton X-100 (T9284, Sigma), 3 times for 15 min. Once dried, brains were incubated overnight at 4˚C with the primary antibody solution diluted in PBS + 0.1% Triton, in a humid chamber. Brains were then rehydrated in PBS + 0.1% Triton, 3 times for 15 min, allowed to dry and incubated for 2h at 25˚C with the secondary antibody solution in PBS + 0.1% Triton, in the dark, in a humid chamber. Next, brains were rehydrated in PBS + 0.1% Triton, 3 times for 15 min and incubated 15 min with Hoechst 33342 (Invitrogen), at 0.5μg/ml in PBS + 0.1% Triton. Finally, once dried, brains were mounted in mounting medium (1.25% N-propyl gallate, 75% glycerol, 25% H20).
For 3D SIM imaging or confocal imaging, whole mount third instar larval brains were dissected in PBS and fixed in 4% paraformaldehyde diluted in PBS + 0.1% Triton for 30 min. Brains were then permeabilized with 3 washes in PBS + 0.3% Triton. Next, a blocking step followed, by incubation in PBS + 10% NGS for 30 min. Next, brains were incubated with primary antibody solution (PBS + 0.3% Triton), first a few hours at room temperature on gentle agitation, and then overnight at 4˚C. Brains were then washed 3 times in PBS + 0.3% Triton, followed by incubation with secondary antibody solution, overnight at 4˚C which also included a phalloidin conjugated probe to label the cell membrane. After 2 washes in PBS + 0.3% Triton (5 min each), brains were incubated with Hoechst 33342 (0.5ug/ml in PBS+0.3% Triton) for 30 min before a final wash in PBS. Finally, brains were rapidly washed 3 times in PBS. They were mounted in mounting medium as described above. For the 3D SIM analysis, the appropriate NBs in interphase were chosen based on the presence of a large intact nucleus. The apical cortex was identified by the presence of a centrosome/centriole in Ctrl and Plk4KD brains and by its position relative to GMCs. In Plk4WT NBs, centrioles are positioned towards the basal hemisphere.
Primary antibodies used: rabbit anti-Spd2 (1:500; (Dix and Raff, 2007), guinea pig anti-Cnn (1:1000, (Lucas and Raff, 2007), rabbit anti-PPHC (Plp) (Martinez-Campos et al., 2004), mouse anti-α-Tubulin (DM1α) (1:500, Sigma Aldrich), rabbit anti-aPKC (1:100, SC116, Santa Cruz) and mouse anti-Prospero (1:20, DSHB). Secondary antibodies used: Alexa Fluor 488, Alexa Fluor 546 and Alexa Fluor 647 (Molecular Probes, ThermoScientific), Phalloidin conjugated probes (R415) for F-actin labelling in red, from molecular probes (Thermo Fisher) were used to label the cell membrane, RFP and GFP Boosters (Atto 594 and Atto 488) (1:100, Chromotek).
Drug Treatments
For analysis of centrosome position in NBs, third instar brains were placed in PBS or PBS supplemented with colcemid -demecolcine (D7385, Sigma Aldrich) (50μM final concentration) for exactly 1h at 25˚C. Brains were then fixed as described above for whole mount preparations.
To inhibit the proteasome, Bortezomib (PS-341, Selleck Chemicals) was added to the live imaging medium at a concentration of 50μM, a concentration that led to mitotic arrest in NBs. Brains were fixed after 2 hours of incubation in the medium supplied with the drug. Control brains were incubated in live imaging medium with 1% DMSO. Brains were then fixed as described above for whole mount preparations.
For live imaging, third instar larval brains were dissected in live imaging medium and then incubated in live imaging medium implemented with the appropriate drug. To depolymerize MTs, demecolcine (D7385, Sigma Aldrich) was added to the medium at a concentration of 50 μM as described previously (Januschke and Gonzalez, 2010). We noticed that fluorescence intensity of centriolar tagged proteins used in this study- RFP-Sas6 and GFP-Plk4KD decreases at the centriole, indicating a possible instability of these proteins in the absence of centriolarMTs. For actin depolymerization, cytochalasin D (C8273, Sigma Aldrich) was added to the medium at a concentration of 50 μM. After dissection, brains were placed on the glass-bottom dish in 10 μl of live imaging medium with demecolcine or cytochalasin D. Brains were filmed immediately after.
Western Blot of Spd2 Transgenic Lines
Twenty third instar larval brains of each genotype were dissected in cold PBS supplied with 1% protease inhibitor cocktail (P8340, Sigma) and 1mM PMSF (P7626, Sigma) and collected in a tube. 20 ml of sample buffer was added to the brains and the tissues were stripped with the help of blunt forceps on ice to induce mechanical dissociation. Samples were boiled at 70°C for 10min. Samples were run in 10% Bis/Tris gel in MOPSSDS buffer at 180V (Np0301 and NP0001 from NuPAGE-ThermoFIsher) and transferred fir 1h at 100V in the cold using 0.2 μm NC nitrocellulose (GE Healthcare Life Science). Membranes were then blocked in PBS supplemented with 0.1% Tween20 (PBST) and 10% dried milk powder for 30 min followed by incubation O/N 4˚C with Spd2 primary antibody at 1:500 (Dix and Raff, 2007) , diluted in PBST with 3% dried milk powder. Membranes were washed 4 times for 10min in PBST and then incubated for 2h at room-temperature with a Rabbit secondary antibody conjugated to Horseradish peroxidase (HRP) (# G21234, Life Technologies) diluted in PBST with 3% dried milk powder. The secondary antibody solution was then removed and membranes washed 5 times for 10min in PBST. Finally, membranes were incubated with SuperSignalTM West Pico Chemiluminescent Substrate (34080, Termo ScientificTM) and revealed using the BioRad Chemidoc MP system. Images were analyzed using ImageLab software.
In Vitro Kinase Assays and Mass Spectrometry
Bacterially-expressed constructs of Drosophila Plk4 (amino acids 1–317) C-terminally tagged with FLAG-His6 and Drosophila Spd2 N-terminally tagged with either Glutathione S-Transferase (GST) or Maltose-binding protein (MBP) were purified on HisPur resin (ThermoFisher), glutathione resin (NEB) and amylose resin (NEB), respectively, according to manufacturer’s instructions. Prior to assay, purified proteins were resolved by SDS-PAGE and scans of the Coomassie-stained gels analyzed by densitometry (ImageJ, NIH) to determine protein purity. Total protein concentrations of the same reagents were measured by Bradford assay (BioRad). The total protein and purity measurements were used to calculate the concentration of each protein reagent. (Contaminants and proteolytic fragments are excluded by this calculation.). In vitro phosphorylation assays were performed by incubation with 100 μM ATP for 60-90 min at 24°C in reaction buffer [40 mM Na HEPES (pH 7.3), 150 mM NaCl, 5 mM MgCl2, 0.5 mM MnCl2, 1 mM DTT, 10% (by volume) glycerol]. Samples were resolved by SDS-PAGE, and proteins visualized by Coomassie staining. Phosphorylation of protein substrates was evaluated by including γ-32P-ATP in assays and, subsequently, the presence of radiolabeled substrates detected by autoradiography of dried gels. Phosphorylated residues within proteins were identified by tandem mass spectrometry (Table S1) of purified bacterially-expressed proteins phosphorylated in vitro (described above in ‘Generation of Spd2 fragments’) in the presence of non-radioactive ATP and performed at the Arizona Proteomics Consortium (University of Arizona). Samples of Spd2 were reduced (10μM dithiothreitol, 55°C, 1hr), alkylated (55mM iodoacetamide, 24°C, 45min), and trypsin digested (∼1μg trypsin, 37°C, 12hrs) in-gel, and then extracted. Peptide samples were desalted using ZipTip 0.6μL C18 resins (EMD Millipore, Billerica, MA). The peptides were then separated by HPLC on a C18 analytical column, ionized by electrospray ionization (ESI) in positive mode, and analyzed on a LTQ Orbitrap Velos (Thermo Electron Corp., San Jose, CA) mass spectrometer. All LC MS analyses were carried out in “data-dependent” mode in which the top 6 most intense precursor ions detected in the MS1 precursor scan (m/z 300-2000) were selected for fragmentation via collision induced dissociation (CID). Precursor ions were measured in the Orbitrap at a resolution of 60,000 (m/z 400) and all fragment ions were measured in the ion trap.
RNAi of Drosophila S2 Cells
Cells were plated at 50% confluency and treated with 10 μg/day dsRNA for 7 days. Cells were passaged before they reached ∼90% confluency. On day 5, cells were transfected with 2 μg of plasmid and 10 μg dsRNA using the Nucleofector II (Lonza). Transgenes were expressed by treating with 0.5 mM CuSO4 for 24 hours.
Co-Immunoprecipitation and Immunoblotting
GFP-binding protein fused to Human IgG was coupled to Protein-A conjugated Dynabeads (Invitrogen) and cross-linked to the beads with dimethyl pimelimidate dihydrochloride (Sigma). Beads were blocked overnight in PBS-0.1% Triton X-100 + 1% BSA. S2 cells were lysed in IP buffer (50 mM Tris pH 7.2, 125 mM NaCl, 1 mM EGTA, 0.5% Triton X-100, and 0.4 mM NaN3, 1 mM dithiothreitol (DTT), 1mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/mL soybean trypsin inhibitor (SBTI), and SIGMAFAST protease inhibitor cocktail (Sigma), then precleared by centrifugation at 10,000 rcf for 5 minutes at 4°C. A sample of cleared lysate was used for input blots. Beads were equilibrated in IP buffer, then 50 μL of beads were added to the remaining lysate and rocked at 4C for 30 minutes. Beads were washed 4x with IP buffer by resuspension and then harvested with a magnet between washes. Beads were transferred to a new tube during the final wash. Samples were eluted by boiling in Laemmli buffer (40 μL of 2x). Inputs and IPs were resolved by SDS-PAGE and transferred to nitrocellulose membrane, then analyzed by Western blot using mouse anti-GFP (1:3000, clone JL-8; Clontech Laboratories, Catalogue #632381), mouse anti-α-Tubulin (1:3000, clone DM1a; Sigma, Catalogue #T9026), guinea pig anti-Spd2 (1:500, polyclonal), mouse anti-HA (1:1500, Clone HA-7; Sigma, Catalogue #H3663), or mouse anti-V5 (1:1500; Invitrogen; Catalogue #46-0705) primary antibodies, followed by fluorescently-labeled goat anti-mouse (1:3000; Li-COR IRDye 800CW, Catalogue #926-32210) or donkey anti-guinea pig (1:3000; Li-COR IRDye 800 CW, Catalogue #926-32411) secondaries. Detection was performed on a LiCOR odyssey cL-X fluorescent imaging system at medium quality and 84 nm resolution.
Synthesis of Spd2 dsRNA
dsRNA was synthesized by in vitro T7 transcription using PCR product amplified from Spd2 EST (CG17286, Clone ID LD24702, DGC EST Library 1.0) with the primers Spd2-UTR-For: 5′ – TAATACGACTCACTATAGGGGTTTTCGCGTTCGCACTGCAAACTGTAACTGTTTAAGGACAAAGCGGATTTGTTTTATTTGTGCCTGC and Spd2-UTR-Rev: 5′ – TAATACGACTCACTATAGGGCTTTTAGGAAACAAGCG.
Image Acquisition
Images of squashed preparations were collected with a x100 objective on a Leica DM6B epifluorescence microscope, this microscope was equipped with an ORCA-Flash4.0 V2 Digital CMOS camera (Hamamatsu) and controlled by the Metamorph software 7.7 (Molecular devices). Images of whole mount brain lobes were acquired on a Nikon A1R inverted TiE confocal microscope with a 40X 1.3NA or a X60 1.4 NA objective in NIS Element software. For the characterization of Spd2 and Fzr localization in interphase centrioles, images were acquired with a N-SIM Nikon microscope in 3D SIM mode before image reconstruction using the NIS Elements software (Gustafsson et al., 2008). The system is equipped with an APO TIRF SR 100x 1.49NA oil immersion, a laser illumination (488nm 200mW, 561nm 100mW, 640nm 100mW) and an EMCCD DU-897 Andor camera. Images were acquired with the following protocol, a Z stack (0.12 micron steps) was acquired. Images were then reconstructed using Nikon elements software.
Quantification and Statistical Analysis
Characterization of Centriole Behavior
The characterization of centriole movement was performed on raw data taking into account the Z-stacks required to follow centriole behavior in x, y and z. Z-stacks were used to generate projections to follow the centriole behavior over time. In Ctrl cells, after mitosis, the two centrioles display stereotypic movements as described previously (Rebollo et al., 2007, Rusan and Peifer, 2007) and this behavior was taken for the basis of characterization of centriole behavior in the different genotypes. Centrioles were considered “Apical” when behaving as Ctrl apical centrioles- maintaining a stable and fixed position closer to the apical cell cortex throughout interphase. The apical cell cortex was established by the position of the large aster, at the end of telophase, typical of the asymmetric spindle in NBs. Centrioles were categorized as apical mobile when they were maintained within the apical hemisphere (top half of the cell), Centrioles that moved away from the apical cortex and were positioned towards the basal hemisphere (bottom half of the cell) were considered as “Basal-like” centrioles. Frequently, they displayed a random jumping movement). In certain cases, the NB moved and the neighboring cells were used as landmarks to position the apical cortex at different time points. . For the characterization of centriole behavior in the different Plk4 transgenes, 20 NBs from 3 Ctrl brains, 26 NBs from 7 Plk4KD brains and 20 NBs from 7 Plk4WT brains were analyzed. In the same type of analysis in the Fzrmut background, 12 NBs from 3 Fzrmut brains and 18 NBs from 5 Plk4KD,Fzrmut brains were analyzed.
Stabilization and Tracking Centriole Dynamics in Time-lapse Movies
To stabilize the full set of images that comprise each time-lapse movie, we used the “SetLandmark” and “StabilizeMovie” plugins from ImageJ. With the “SetLandmark” we defined a landmark on each frame to identify the apical and basal axis of each NB cell. Subsequently, translations and rotations were applied to align the center of the previously established landmarks at the center of the image. Centrosome/centriole tracking was performed with the “Tracking” plugins from ImageJ. In each time frame the centriole was identified and labelled manually in order to allow tracking of their movement. After this step, we used “PrintTracking” from imageJ to draw the tracking of each centrosome/centriole.
Analysis of Spindle Orientation after Two Consecutive Mitosis
Analysis of spindle orientation was performed on time-lapse movies where two consecutive mitosis of the same NB could be identified. In both mitosis spindle axis was determined at early anaphase. The angle between the two spindles was measured using the “Angle tool” from Fiji (NIH).
Analysis of Spindle Orientation in Mitotic NBs
Analysis of spindle orientation was performed on metaphase or anaphase NBs that contained a clear aPKC crescent. The angle between the spindle and the middle of the aPKC crescent was determined using the “Angle tool” from Fiji (NIH).
Analysis of Centrosome Position in NBs
We only analyzed NBs were a clear group of GMCs could be seen to identify the position of the apical cortex at the opposite side of the GMCs. First, a line was drawn from the apical cortex to the basal to measure the apical-basal axis length with the Fiji (NIH) “Straight line” tool. Then a second line, positioned at the same position than the first line (at the apical side), was drawn till it reached the centrosome, labelled with two centrosome markers (only one is shown in the Figure). The two values for each NB (apical-basal axis length and centrosome distance from the apical cortex) were plotted on an excel sheet. Centrosome position was then calculated by dividing the second measure by the first one.
Characterization of Centrosomal Protein Levels in Interphase Centrioles
Quantification of the levels of centrosome proteins in interphase were performed in at least ten NBs for each genotype from 3 independent experiments. For each set of immunostaining experiments, the same acquisition settings were kept for all conditions. Images were analyzed with Fiji (NIH) and quantifications done as follows. The fluorescence mean grey value (Fc) was measured by drawing the area occupied by the protein of interest with the freehand tool in a single Z plane, representing the center of the centrosome. The cytoplasmic fluorescence (Fcy) was measured by drawing a round area as big as about one-fourth of the cell size and then subtracted from Fc to obtain the net fluorescence of the centrosome (Vc=Fc-Fcy). The signal of the background (Fbkg) outside the cell was also measured to obtain the net fluorescence of the cytoplasm (Vcy=Fcy-Fbkg). Finally, centrosome enrichment was measured as ratio between Vc and Vcy (Vc/Vcy).
Statistical Analysis
Statistical analysis was performed with GraphPad Prism 7. A two-tailed unpaired t-test was used to assess statistical differences. All details of statistical analyses, including n and p values, are found in the text and Figures. All results are presented as mean ± SD.
Acknowledgments
We dedicate this work to the memory of Giuseppe Boccia. We thank J. Raff, Y. Bellaiche, T. Lee, C. Doe, the Bloomington Stock Center, and BestGene for stocks and reagents; L. Sengmanivong, V. Fraisier, and L. Leconte of the NIC at the IC for valuable help with microscopy; P. Maiuri and M. Rujano for help with data analysis; V. Marthiens, D. Vargas, S. Gemble, O. Goudiam, A. Echard, M. Piel, A. Audibert, J. Azimzadeh, M. H. Verlhac, M. Bornens, J. Raff, and C. Gonzalez for discussions and/or critical comments on the manuscript. Financial support from an ERC starting grant CentroStemCancer 242598, a FRM installation and ATIP grants, the Institut Curie and CNRS. G.C.R. is grateful for support from NCI P30CA23074 and NIGMS R01 GM110166 and GM126035. D.G. was supported by PhD fellowships from IC and FRM. The Basto laboratory is a member of the Labex CelTisPhyBio
Author Contributions
D.G. and R.B. conceived the project, analyzed the data, and, together with G.C.R., wrote the manuscript. D.G. performed most of the experimental procedures. C.P. generated tools and most of the transgenic fly stocks. J.M.R, D.W.B., and G.C.R. conceived and performed kinase assays, mass spectrometry, and S2 cell experiments. D.G. generated Plk4WT transgenic fly stocks. A.S. helped with the maintenance of fly stocks, crosses, and dissection, and D.-B. and V.R. identified the methodology used to track centrosome or centriole behavior in time-lapse movies and the representation of their dynamics. Y.K. shared unpublished data. A.G. and M.N. helped R.B. to set up the live imaging of some fly strains. R.B. supervised the project.
Declaration of Interests
The authors declare no competing interests.
Published: May 23, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.devcel.2019.04.036.
Contributor Information
Gregory C. Rogers, Email: gcrogers@email.arizona.edu.
Renata Basto, Email: renata.basto@curie.fr.
Supplemental Information
References
- Adams J., Behnke M., Chen S., Cruickshank A.A., Dick L.R., Grenier L., Klunder J.M., Ma Y.T., Plamondon L., Stein R.L. Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorg. Med. Chem. Lett. 1998;8:333–338. doi: 10.1016/s0960-894x(98)00029-8. [DOI] [PubMed] [Google Scholar]; Adams, J., Behnke, M., Chen, S., Cruickshank, A.A., Dick, L.R., Grenier, L., Klunder, J.M., Ma, Y.T., Plamondon, L., and Stein, R.L. (1998). Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorg. Med. Chem. Lett. 8, 333-338. [DOI] [PubMed]
- Ahmad K., Henikoff S. Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell. 2001;104:839–847. doi: 10.1016/s0092-8674(01)00281-1. [DOI] [PubMed] [Google Scholar]; Ahmad, K., and Henikoff, S. (2001). Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell 104, 839-847. [DOI] [PubMed]
- Albertson R., Chabu C., Sheehan A., Doe C.Q. Scribble protein domain mapping reveals a multistep localization mechanism and domains necessary for establishing cortical polarity. J. Cell Sci. 2004;117:6061–6070. doi: 10.1242/jcs.01525. [DOI] [PubMed] [Google Scholar]; Albertson, R., Chabu, C., Sheehan, A., and Doe, C.Q. (2004). Scribble protein domain mapping reveals a multistep localization mechanism and domains necessary for establishing cortical polarity. J. Cell Sci. 117, 6061-6070. [DOI] [PubMed]
- Albertson R., Doe C.Q. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat. Cell Biol. 2003;5:166–170. doi: 10.1038/ncb922. [DOI] [PubMed] [Google Scholar]; Albertson, R., and Doe, C.Q. (2003). Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat. Cell Biol. 5, 166-170. [DOI] [PubMed]
- Aydogan M.G., Wainman A., Saurya S., Steinacker T.L., Caballe A., Novak Z.A., Baumbach J., Muschalik N., Raff J.W. A homeostatic clock sets daughter centriole size in flies. J. Cell Biol. 2018;217:1233–1248. doi: 10.1083/jcb.201801014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Aydogan, M.G., Wainman, A., Saurya, S., Steinacker, T.L., Caballe, A., Novak, Z.A., Baumbach, J., Muschalik, N., and Raff, J.W. (2018). A homeostatic clock sets daughter centriole size in flies. J. Cell Biol. 217, 1233-1248. [DOI] [PMC free article] [PubMed]
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., Raff J.W. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]; Basto, R., Brunk, K., Vinadogrova, T., Peel, N., Franz, A., Khodjakov, A., and Raff, J.W. (2008). Centrosome amplification can initiate tumorigenesis in flies. Cell 133, 1032-1042. [DOI] [PMC free article] [PubMed]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., Raff J.W. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]; Basto, R., Lau, J., Vinogradova, T., Gardiol, A., Woods, C.G., Khodjakov, A., and Raff, J.W. (2006). Flies without centrioles. Cell 125, 1375-1386. [DOI] [PubMed]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., Glover D.M. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]; Bettencourt-Dias, M., Rodrigues-Martins, A., Carpenter, L., Riparbelli, M., Lehmann, L., Gatt, M.K., Carmo, N., Balloux, F., Callaini, G., and Glover, D.M. (2005). SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15, 2199-2207. [DOI] [PubMed]
- Blachon S., Gopalakrishnan J., Omori Y., Polyanovsky A., Church A., Nicastro D., Malicki J., Avidor-Reiss T. Drosophila asterless and vertebrate Cep152 are orthologs essential for centriole duplication. Genetics. 2008;180:2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]; Blachon, S., Gopalakrishnan, J., Omori, Y., Polyanovsky, A., Church, A., Nicastro, D., Malicki, J., and Avidor-Reiss, T. (2008). Drosophila asterless and vertebrate Cep152 are orthologs essential for centriole duplication. Genetics 180, 2081-2094. [DOI] [PMC free article] [PubMed]
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]; Brand, A.H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415. [DOI] [PubMed]
- Brownlee C.W., Klebba J.E., Buster D.W., Rogers G.C. The protein phosphatase 2A regulatory subunit Twins stabilizes Plk4 to induce centriole amplification. J. Cell Biol. 2011;195:231–243. doi: 10.1083/jcb.201107086. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brownlee, C.W., Klebba, J.E., Buster, D.W., and Rogers, G.C. (2011). The protein phosphatase 2A regulatory subunit Twins stabilizes Plk4 to induce centriole amplification. J. Cell Biol. 195, 231-243. [DOI] [PMC free article] [PubMed]
- Burute M., Prioux M., Blin G., Truchet S., Letort G., Tseng Q., Bessy T., Lowell S., Young J., Filhol O. Polarity reversal by centrosome repositioning primes cell scattering during epithelial-to-mesenchymal transition. Dev. Cell. 2017;40:168–184. doi: 10.1016/j.devcel.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Burute, M., Prioux, M., Blin, G., Truchet, S., Letort, G., Tseng, Q., Bessy, T., Lowell, S., Young, J., Filhol, O., et al. (2017). Polarity reversal by centrosome repositioning primes cell scattering during epithelial-to-mesenchymal transition. Dev. Cell 40, 168-184. [DOI] [PMC free article] [PubMed]
- Castellanos E., Dominguez P., Gonzalez C. Centrosome dysfunction in Drosophila neural stem cells causes tumors that are not due to genome instability. Curr. Biol. 2008;18:1209–1214. doi: 10.1016/j.cub.2008.07.029. [DOI] [PubMed] [Google Scholar]; Castellanos, E., Dominguez, P., and Gonzalez, C. (2008). Centrosome dysfunction in Drosophila neural stem cells causes tumors that are not due to genome instability. Curr. Biol. 18, 1209-1214. [DOI] [PubMed]
- Caussinus E., Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 2005;37:1125–1129. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]; Caussinus, E., and Gonzalez, C. (2005). Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 37, 1125-1129. [DOI] [PubMed]
- Conduit P.T., Brunk K., Dobbelaere J., Dix C.I., Lucas E.P., Raff J.W. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr. Biol. 2010;20:2178–2186. doi: 10.1016/j.cub.2010.11.011. [DOI] [PubMed] [Google Scholar]; Conduit, P.T., Brunk, K., Dobbelaere, J., Dix, C.I., Lucas, E.P., and Raff, J.W. (2010). Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr. Biol. 20, 2178-2186. [DOI] [PubMed]
- Conduit P.T., Richens J.H., Wainman A., Holder J., Vicente C.C., Pratt M.B., Dix C.I., Novak Z.A., Dobbie I.M., Schermelleh L. A molecular mechanism of mitotic centrosome assembly in Drosophila. Elife. 2014;3:e03399. doi: 10.7554/eLife.03399. [DOI] [PMC free article] [PubMed] [Google Scholar]; Conduit, P.T., Richens, J.H., Wainman, A., Holder, J., Vicente, C.C., Pratt, M.B., Dix, C.I., Novak, Z.A., Dobbie, I.M., Schermelleh, L., et al. (2014). A molecular mechanism of mitotic centrosome assembly in Drosophila. Elife 3, e03399. [DOI] [PMC free article] [PubMed]
- Conduit P.T., Wainman A., Raff J.W. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 2015;16:611–624. doi: 10.1038/nrm4062. [DOI] [PubMed] [Google Scholar]; Conduit, P.T., Wainman, A., and Raff, J.W. (2015). Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 16, 611-624. [DOI] [PubMed]
- Davis, R. (2003). RE: P{tubP-GAL80ts} construct and insertions. Type to CENTER, B. S.
- Dix C.I., Raff J.W. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 2007;17:1759–1764. doi: 10.1016/j.cub.2007.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dix, C.I., and Raff, J.W. (2007). Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 17, 1759-1764. [DOI] [PMC free article] [PubMed]
- Dobbelaere J., Josué F., Suijkerbuijk S., Baum B., Tapon N., Raff J. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dobbelaere, J., Josue, F., Suijkerbuijk, S., Baum, B., Tapon, N., and Raff, J. (2008). A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6, e224. [DOI] [PMC free article] [PubMed]
- Farina F., Gaillard J., Guérin C., Couté Y., Sillibourne J., Blanchoin L., Théry M. The centrosome is an actin-organizing centre. Nat. Cell Biol. 2016;18:65–75. doi: 10.1038/ncb3285. [DOI] [PMC free article] [PubMed] [Google Scholar]; Farina, F., Gaillard, J., Guerin, C., Coute, Y., Sillibourne, J., Blanchoin, L., and Thery, M. (2016). The centrosome is an actin-organizing centre. Nat. Cell Biol. 18, 65-75. [DOI] [PMC free article] [PubMed]
- Fu J., Glover D.M. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012;2:120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fu, J., and Glover, D.M. (2012). Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2, 120104. [DOI] [PMC free article] [PubMed]
- Galletta B.J., Fagerstrom C.J., Schoborg T.A., Mclamarrah T.A., Ryniawec J.M., Buster D.W., Slep K.C., Rogers G.C., Rusan N.M. A centrosome interactome provides insight into organelle assembly and reveals a non-duplication role for Plk4. Nat. Commun. 2016;7:12476. doi: 10.1038/ncomms12476. [DOI] [PMC free article] [PubMed] [Google Scholar]; Galletta, B.J., Fagerstrom, C.J., Schoborg, T.A., Mclamarrah, T.A., Ryniawec, J.M., Buster, D.W., Slep, K.C., Rogers, G.C., and Rusan, N.M. (2016). A centrosome interactome provides insight into organelle assembly and reveals a non-duplication role for Plk4. Nat. Commun. 7, 12476. [DOI] [PMC free article] [PubMed]
- Gogendeau D., Siudeja K., Gambarotto D., Pennetier C., Bardin A.J., Basto R. Aneuploidy causes premature differentiation of neural and intestinal stem cells. Nat. Commun. 2015;6:8894. doi: 10.1038/ncomms9894. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gogendeau, D., Siudeja, K., Gambarotto, D., Pennetier, C., Bardin, A.J., and Basto, R. (2015). Aneuploidy causes premature differentiation of neural and intestinal stem cells. Nat. Commun.. 6, 8894. [DOI] [PMC free article] [PubMed]
- Gustafsson M.G., Shao L., Carlton P.M., Wang C.J., Golubovskaya I.N., Cande W.Z., Agard D.A., Sedat J.W. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gustafsson, M.G., Shao, L., Carlton, P.M., Wang, C.J., Golubovskaya, I.N., Cande, W.Z., Agard, D.A., and Sedat, J.W. (2008). Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94, 4957-4970. [DOI] [PMC free article] [PubMed]
- Habedanck R., Stierhof Y.D., Wilkinson C.J., Nigg E.A. The polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]; Habedanck, R., Stierhof, Y.D., Wilkinson, C.J., and Nigg, E.A. (2005). The polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7, 1140-1146. [DOI] [PubMed]
- Homem C.C., Knoblich J.A. Drosophila neuroblasts: a model for stem cell biology. Development. 2012;139:4297–4310. doi: 10.1242/dev.080515. [DOI] [PubMed] [Google Scholar]; Homem, C.C., and Knoblich, J.A. (2012). Drosophila neuroblasts: a model for stem cell biology. Development 139, 4297-4310. [DOI] [PubMed]
- Huang J.Y., Raff J.W. The dynamic localisation of the Drosophila APC/C: evidence for the existence of multiple complexes that perform distinct functions and are differentially localised. J. Cell Sci. 2002;115:2847–2856. doi: 10.1242/jcs.115.14.2847. [DOI] [PubMed] [Google Scholar]; Huang, J.Y., and Raff, J.W. (2002). The dynamic localisation of the Drosophila APC/C: evidence for the existence of multiple complexes that perform distinct functions and are differentially localised. J. Cell Sci. 115, 2847-2856. [DOI] [PubMed]
- Jacobs H., Richter D., Venkatesh T., Lehner C. Completion of mitosis requires neither fzr/rap nor fzr2, a male germline-specific Drosophila Cdh1 homolog. Curr. Biol. 2002;12:1435–1441. doi: 10.1016/s0960-9822(02)01074-6. [DOI] [PubMed] [Google Scholar]; Jacobs, H., Richter, D., Venkatesh, T., and Lehner, C. (2002). Completion of mitosis requires neither fzr/rap nor fzr2, a male germline-specific Drosophila Cdh1 homolog. Curr. Biol. 12, 1435-1441. [DOI] [PubMed]
- Januschke J., Gonzalez C. The interphase microtubule aster is a determinant of asymmetric division orientation in Drosophila neuroblasts. J. Cell Biol. 2010;188:693–706. doi: 10.1083/jcb.200905024. [DOI] [PMC free article] [PubMed] [Google Scholar]; Januschke, J., and Gonzalez, C. (2010). The interphase microtubule aster is a determinant of asymmetric division orientation in Drosophila neuroblasts. J. Cell Biol. 188, 693-706. [DOI] [PMC free article] [PubMed]
- Januschke J., Llamazares S., Reina J., Gonzalez C. Drosophila neuroblasts retain the daughter centrosome. Nat. Commun. 2011;2:243. doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]; Januschke, J., Llamazares, S., Reina, J., and Gonzalez, C. (2011). Drosophila neuroblasts retain the daughter centrosome. Nat. Commun.. 2, 243. [DOI] [PMC free article] [PubMed]
- Januschke J., Reina J., Llamazares S., Bertran T., Rossi F., Roig J., Gonzalez C. Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nat. Cell Biol. 2013;15:241–248. doi: 10.1038/ncb2671. [DOI] [PubMed] [Google Scholar]; Januschke, J., Reina, J., Llamazares, S., Bertran, T., Rossi, F., Roig, J., and Gonzalez, C. (2013). Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nat. Cell Biol. 15, 241-248. [DOI] [PubMed]
- Klebba J.E., Buster D.W., Mclamarrah T.A., Rusan N.M., Rogers G.C. Autoinhibition and relief mechanism for Polo-like kinase 4. Proc. Natl. Acad. Sci. USA. 2015;112:E657–E666. doi: 10.1073/pnas.1417967112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Klebba, J.E., Buster, D.W., Mclamarrah, T.A., Rusan, N.M., and Rogers, G.C. (2015). Autoinhibition and relief mechanism for Polo-like kinase 4. Proc. Natl. Acad. Sci. USA 112, E657-E666. [DOI] [PMC free article] [PubMed]
- Lambrus B.G., Holland A.J. A new mode of mitotic surveillance. Trends Cell Biol. 2017;27:314–321. doi: 10.1016/j.tcb.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lambrus, B.G., and Holland, A.J. (2017). A new mode of mitotic surveillance. Trends Cell Biol. 27, 314-321. [DOI] [PMC free article] [PubMed]
- Lee C.Y., Robinson K.J., Doe C.Q. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]; Lee, C.Y., Robinson, K.J., and Doe, C.Q. (2006). Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature 439, 594-598. [DOI] [PubMed]
- Lee H.S., Simon J.A., Lis J.T. Structure and expression of ubiquitin genes of Drosophila melanogaster. Mol. Cell. Biol. 1988;8:4727–4735. doi: 10.1128/mcb.8.11.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee, H.S., Simon, J.A., and Lis, J.T. (1988). Structure and expression of ubiquitin genes of Drosophila melanogaster. Mol. Cell. Biol. 8, 4727-4735. [DOI] [PMC free article] [PubMed]
- Lerit D.A., Rusan N.M. PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J. Cell Biol. 2013;202:1013–1022. doi: 10.1083/jcb.201303141. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lerit, D.A., and Rusan, N.M. (2013). PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J. Cell Biol. 202, 1013-1022. [DOI] [PMC free article] [PubMed]
- Lucas E.P., Raff J.W. Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J. Cell Biol. 2007;178:725–732. doi: 10.1083/jcb.200704081. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lucas, E.P., and Raff, J.W. (2007). Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J. Cell Biol. 178, 725-732. [DOI] [PMC free article] [PubMed]
- Marthiens V., Rujano M.A., Pennetier C., Tessier S., Paul-Gilloteaux P., Basto R. Centrosome amplification causes microcephaly. Nat. Cell Biol. 2013;15:731–740. doi: 10.1038/ncb2746. [DOI] [PubMed] [Google Scholar]; Marthiens, V., Rujano, M.A., Pennetier, C., Tessier, S., Paul-Gilloteaux, P., and Basto, R. (2013). Centrosome amplification causes microcephaly. Nat. Cell Biol. 15, 731-740. [DOI] [PubMed]
- Martin C.A., Ahmad I., Klingseisen A., Hussain M.S., Bicknell L.S., Leitch A., Nürnberg G., Toliat M.R., Murray J.E., Hunt D. Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat. Genet. 2014;46:1283–1292. doi: 10.1038/ng.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]; Martin, C.A., Ahmad, I., Klingseisen, A., Hussain, M.S., Bicknell, L.S., Leitch, A., Nurnberg, G., Toliat, M.R., Murray, J.E., Hunt, D., et al. (2014). Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat. Genet. 46, 1283-1292. [DOI] [PMC free article] [PubMed]
- Martinez-Campos M., Basto R., Baker J., Kernan M., Raff J.W. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 2004;165:673–683. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]; Martinez-Campos, M., Basto, R., Baker, J., Kernan, M., and Raff, J.W. (2004). The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165, 673-683. [DOI] [PMC free article] [PubMed]
- Mcguire S.E., Mao Z., Davis R.L. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]; Mcguire, S.E., Mao, Z., and Davis, R.L. (2004). Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE 2004, pl6. [DOI] [PubMed]
- Meghini F., Martins T., Tait X., Fujimitsu K., Yamano H., Glover D.M., Kimata Y. Targeting of Fzr/Cdh1 for timely activation of the APC/C at the centrosome during mitotic exit. Nat. Commun. 2016;7:12607. doi: 10.1038/ncomms12607. [DOI] [PMC free article] [PubMed] [Google Scholar]; Meghini, F., Martins, T., Tait, X., Fujimitsu, K., Yamano, H., Glover, D.M., and Kimata, Y. (2016). Targeting of Fzr/Cdh1 for timely activation of the APC/C at the centrosome during mitotic exit. Nat. Commun.. 7, 12607. [DOI] [PMC free article] [PubMed]
- Mennella V., Keszthelyi B., Mcdonald K.L., Chhun B., Kan F., Rogers G.C., Huang B., Agard D.A. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 2012;14:1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mennella, V., Keszthelyi, B., Mcdonald, K.L., Chhun, B., Kan, F., Rogers, G.C., Huang, B., and Agard, D.A. (2012). Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14, 1159-1168. [DOI] [PMC free article] [PubMed]
- Moutinho-Pereira S., Debec A., Maiato H. Microtubule cytoskeleton remodeling by acentriolar microtubule-organizing centers at the entry and exit from mitosis in Drosophila somatic cells. Mol. Biol. Cell. 2009;20:2796–2808. doi: 10.1091/mbc.E09-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Moutinho-Pereira, S., Debec, A., and Maiato, H. (2009). Microtubule cytoskeleton remodeling by acentriolar microtubule-organizing centers at the entry and exit from mitosis in Drosophila somatic cells. Mol. Biol. Cell 20, 2796-2808. [DOI] [PMC free article] [PubMed]
- Obino D., Farina F., Malbec O., Sáez P.J., Maurin M., Gaillard J., Dingli F., Loew D., Gautreau A., Yuseff M.I. Actin nucleation at the centrosome controls lymphocyte polarity. Nat. Commun. 2016;7:10969. doi: 10.1038/ncomms10969. [DOI] [PMC free article] [PubMed] [Google Scholar]; Obino, D., Farina, F., Malbec, O., Saez, P.J., Maurin, M., Gaillard, J., Dingli, F., Loew, D., Gautreau, A., Yuseff, M.I., et al. (2016). Actin nucleation at the centrosome controls lymphocyte polarity. Nat. Commun.. 7, 10969. [DOI] [PMC free article] [PubMed]
- Peel N., Stevens N.R., Basto R., Raff J.W. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]; Peel, N., Stevens, N.R., Basto, R., and Raff, J.W. (2007). Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 17, 834-843. [DOI] [PMC free article] [PubMed]
- Piel M., Nordberg J., Euteneuer U., Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]; Piel, M., Nordberg, J., Euteneuer, U., and Bornens, M. (2001). Centrosome-dependent exit of cytokinesis in animal cells. Science 291, 1550-1553. [DOI] [PubMed]
- Ramdas Nair A., Singh P., Salvador Garcia D., Rodriguez-Crespo D., Egger B., Cabernard C. The microcephaly-associated protein Wdr62/CG7337 is required to maintain centrosome asymmetry in Drosophila neuroblasts. Cell Rep. 2016;14:1100–1113. doi: 10.1016/j.celrep.2015.12.097. [DOI] [PubMed] [Google Scholar]; Ramdas Nair, A., Singh, P., Salvador Garcia, D., Rodriguez-Crespo, D., Egger, B., and Cabernard, C. (2016). The microcephaly-associated protein Wdr62/CG7337 is required to maintain centrosome asymmetry in Drosophila neuroblasts. Cell Rep.. 14, 1100-1113. [DOI] [PubMed]
- Rebollo E., Sampaio P., Januschke J., Llamazares S., Varmark H., González C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]; Rebollo, E., Sampaio, P., Januschke, J., Llamazares, S., Varmark, H., and Gonzalez, C. (2007). Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell 12, 467-474. [DOI] [PubMed]
- Riparbelli M.G., Callaini G. Male gametogenesis without centrioles. Dev. Biol. 2011;349:427–439. doi: 10.1016/j.ydbio.2010.10.021. [DOI] [PubMed] [Google Scholar]; Riparbelli, M.G., and Callaini, G. (2011). Male gametogenesis without centrioles. Dev. Biol. 349, 427-439. [DOI] [PubMed]
- Rusan N.M., Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rusan, N.M., and Peifer, M. (2007). A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 177, 13-20. [DOI] [PMC free article] [PubMed]
- San-Juán B.P., Baonza A. The bHLH factor deadpan is a direct target of Notch signaling and regulates neuroblast self-renewal in Drosophila. Dev. Biol. 2011;352:70–82. doi: 10.1016/j.ydbio.2011.01.019. [DOI] [PubMed] [Google Scholar]; San-Juan, B.P., and Baonza, A. (2011). The bHLH factor deadpan is a direct target of Notch signaling and regulates neuroblast self-renewal in Drosophila. Dev. Biol. 352, 70-82. [DOI] [PubMed]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. [DOI] [PMC free article] [PubMed]
- Shaheen R., Al Tala S., Almoisheer A., Alkuraya F.S. Mutation in PLK4, encoding a master regulator of centriole formation, defines a novel locus for primordial dwarfism. J. Med. Genet. 2014;51:814–816. doi: 10.1136/jmedgenet-2014-102790. [DOI] [PubMed] [Google Scholar]; Shaheen, R., Al Tala, S., Almoisheer, A., and Alkuraya, F.S. (2014). Mutation in PLK4, encoding a master regulator of centriole formation, defines a novel locus for primordial dwarfism. J. Med. Genet. 51, 814-816. [DOI] [PubMed]
- Singh P., Ramdas Nair A., Cabernard C. The centriolar protein Bld10/Cep135 is required to establish centrosome asymmetry in Drosophila neuroblasts. Curr. Biol. 2014;24:1548–1555. doi: 10.1016/j.cub.2014.05.050. [DOI] [PubMed] [Google Scholar]; Singh, P., Ramdas Nair, A., and Cabernard, C. (2014). The centriolar protein Bld10/Cep135 is required to establish centrosome asymmetry in Drosophila neuroblasts. Curr. Biol. 24, 1548-1555. [DOI] [PubMed]
- Sivakumar S., Gorbsky G.J. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 2015;16:82–94. doi: 10.1038/nrm3934. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sivakumar, S., and Gorbsky, G.J. (2015). Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 16, 82-94. [DOI] [PMC free article] [PubMed]
- Stevens N.R., Raposo A.A., Basto R., St Johnston D., Raff J.W. From stem cell to embryo without centrioles. Curr. Biol. 2007;17:1498–1503. doi: 10.1016/j.cub.2007.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stevens, N.R., Raposo, A.A., Basto, R., St Johnston, D., and Raff, J.W. (2007). From stem cell to embryo without centrioles. Curr. Biol. 17, 1498-1503. [DOI] [PMC free article] [PubMed]
- Tsutsumi M., Yokoi S., Miya F., Miyata M., Kato M., Okamoto N., Tsunoda T., Yamasaki M., Kanemura Y., Kosaki K. Novel compound heterozygous variants in PLK4 identified in a patient with autosomal recessive microcephaly and chorioretinopathy. Eur. J. Hum. Genet. 2016;24:1702–1706. doi: 10.1038/ejhg.2016.119. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tsutsumi, M., Yokoi, S., Miya, F., Miyata, M., Kato, M., Okamoto, N., Tsunoda, T., Yamasaki, M., Kanemura, Y., Kosaki, K., et al. (2016). Novel compound heterozygous variants in PLK4 identified in a patient with autosomal recessive microcephaly and chorioretinopathy. Eur. J. Hum. Genet. 24, 1702-1706. [DOI] [PMC free article] [PubMed]
- Zhang Y., Malone J.H., Powell S.K., Periwal V., Spana E., Macalpine D.M., Oliver B. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 2010;8:e1000320. doi: 10.1371/journal.pbio.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang, Y., Malone, J.H., Powell, S.K., Periwal, V., Spana, E., Macalpine, D.M., and Oliver, B. (2010). Expression in aneuploid Drosophila S2 cells. PLoS Biol. 8, e1000320. [DOI] [PMC free article] [PubMed]
- Zhu S., Lin S., Kao C.F., Awasaki T., Chiang A.S., Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]; Zhu, S., Lin, S., Kao, C.F., Awasaki, T., Chiang, A.S., and Lee, T. (2006). Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell 127, 409-422. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.