Abstract
Determining transcriptional and translational regulatory elements in GC-rich Streptomyces genomes is essential to elucidating the complex regulatory networks that govern secondary metabolite biosynthetic gene cluster (BGC) expression. However, information about such regulatory elements has been limited for Streptomyces genomes. To address this limitation, a high-quality genome sequence of β-lactam antibiotic-producing Streptomyces clavuligerus ATCC 27 064 is completed, which contains 7163 newly annotated genes. This provides a fundamental reference genome sequence to integrate multiple genome-scale data types, including dRNA-Seq, RNA-Seq and ribosome profiling. Data integration results in the precise determination of 2659 transcription start sites which reveal transcriptional and translational regulatory elements, including −10 and −35 promoter components specific to sigma (σ) factors, and 5′-untranslated region as a determinant for translation efficiency regulation. Particularly, sequence analysis of a wide diversity of the −35 components enables us to predict potential σ-factor regulons, along with various spacer lengths between the −10 and −35 elements. At last, the primary transcriptome landscape of the β-lactam biosynthetic pathway is analyzed, suggesting temporal changes in metabolism for the synthesis of secondary metabolites driven by transcriptional regulation. This comprehensive genetic information provides a versatile genetic resource for rational engineering of secondary metabolite BGCs in Streptomyces.
INTRODUCTION
Streptomyces are Gram-positive soil bacteria harboring high GC-content chromosomes and are members of the largest genus of actinobacteria with over 900 described species (1,2). They have been prominent industrial strains given their ability to produce secondary metabolites, including antibiotics, immunosuppressants, antiparastics, antifungals and other value-added biochemical (3). Such secondary metabolites are typically synthesized via a multi-step conversion of precursor molecules, such as CoA pool and amino acids, by multi-enzyme complexes encoded in secondary metabolite biosynthetic gene clusters (BGCs) (4). Individual Streptomyces species generally encode more than thirty BGCs in their genomes, which have a vast potential to produce a diverse array of the secondary metabolites. However, their functions were found to be mostly ‘silent’ under laboratory growth conditions (5). Discovering novel bioactive compounds by activating silent BGCs in Streptomyces is therefore of major interest, motivated, in part, by the rapid rise in antibiotic-resistant pathogens. However, activation of the silent BGCs is limited by the lack of regulatory information leading to their expression. Such information is foundational to designing expression hosts for BGCs.
In recent decades, several high-throughput techniques have been developed and applied to a broad range of species to overcome the lack of regulatory information. For example, integration of differential RNA-Seq (dRNA-Seq), RNA-Seq and ribosome profiling data from Streptomyces coelicolor revealed 3570 transcription start sites (TSSs) with small RNAs, genome-wide promoter architecture, differentially expressed genes (DEGs) and translational buffering for genes encoding secondary metabolism (6). These vast amounts of genetic resources could be employed for practical applications of Streptomyces. For instance, native gene expressions were optimized by the introduction of precise promoter sequences and 5′-untranslated region (5′-UTR) for transcriptional and translational regulation (7). Also, dRNA-Seq of S. tsukubaensis NRRL18488 at two different time points enabled the identification of 8914 TSSs, including TSSs of the immunosuppressant FK506 biosynthetic gene cluster (8). Despite other regulatory network studies (9,10), our knowledge is still limited by the lack of data. Another important industrial strain, Streptomyces clavuligerus ATCC 27064 is used for the production of β-lactamase inhibitor clavulanic acid (11) and β-lactam antibiotic cephamycin C (12). Information on transcriptional and translational regulatory elements in the GC-rich S. clavuligerus genome is not available to understand the regulatory networks governing BGC activities. Here, we obtained the high-quality genome sequence of S. clavuligerus ATCC 27 064, and determined genome-wide TSSs. Then, RNA-Seq and ribosome profiling were additionally exploited to reveal fundamental regulatory elements for transcription and translation. This comprehensive analysis facilitates a new understanding of the regulation of BGC expression and thus accelerates rational strain engineering for the production of bioactive compounds.
MATERIALS AND METHODS
Strains and cell growth
Streptomyces clavuligerus ATCC 27064 cells were inoculated from its 20% glycerol stock of spores to 50 ml of R5− liquid complex medium with 8 g of glass beads (3 ± 0.3 mm diameter) in 250 ml of a baffled flask and grown at 30°C, 250 rpm. R5− medium consists of 103 g/l sucrose, 0.25 g/l K2SO4, 10.12 g/l MgCl2·6H2O, 10 gl/l glucose, 0.1 g/l casamino acids, 5 g/l yeast extract, 5.73 g/l TES (pH 7.2), 0.08 mg/l ZnCl2, 0.4 mg/l FeCl3·6H2O, 0.02 mg/l CuCl2·2H2O, 0.02 mg/l MnCl2·4H2O, 0.02 mg/l Na2B4O7·10H2O and 0.02 mg/l (NH4)6Mo7O24·4H2O. The grown mycelium was diluted 1:100 and then transferred to the fresh R5− medium for the main culture. The main culture for DNA and RNA samples was grown at the same condition as described above. To stall the ribosome and form the cross-linking for the ribosome profiling samples, thiostrepton (Sigma) was added to cultures to a final concentration of 20 μM, which is comparable to the method of a previous study on S. coelicolor and high sensitivity to the drug (6,13–14). The cultures were subsequently incubated for 5 min at 30°C before harvesting. All main cultures except for genome sequencing were prepared for biological duplicates.
Genome sequencing library preparation and high-throughput sequencing
The harvested cells from the main culture were centrifuged and resuspended in 1 ml of fresh R5− medium. The resuspended cells were frozen with liquid nitrogen, and then lysed by grinding using mortar and pestle. The lysate was then centrifuged at 4°C, 3000 g for 10 min, and the supernatant was collected. The genomic DNA was prepared using genomic DNA extraction kit (Promega) as manufacturer's protocol. The extracted genomic DNA was used for construction of both PacBio genome sequencing library and Illumina short read sequencing library. For PacBio genome sequencing library, 5 μg of genomic DNA was used as input of the SMRTbell™ Template Prep Kit (Pacific Biosciences) and the library was constructed as manufacturer's protocol. After removing fragments smaller than 20 kb by the Blue Pippin Size selection system (Sage Science), the library was sequenced using the PacBio RS II sequencing platform (Pacific Biosciences). For Illumina short read sequencing library, the genomic DNA was fragmented to ∼200 bp using Covaris (Covaris Inc.) with the following condition; Power 175, Duty factor 20%; C. burst 20; Time 20 s; Cycle 8 times. Then, the library was constructed from fragmented genomic DNA by using TruSeq DNA Sample preparation protocol (Illumina). The library was sequenced on the MiSeq v2 instrument (Illumina) with 50 bp read recipe.
De novo genome assembly and genome annotation
De novo genome assembly was conducted using the hierarchical genome assembly process workflow (HGAP, Version 2.3) (15). Then, we found total 91 conflicts between two results by mapping of Illumina sequencing reads to the assembled contigs from PacBio sequencing. Conflicts showed more than 72% frequency for Illumina reads were corrected as Illumina results and others were unchanged as PacBio results, which were verified by Sanger sequencing. Illumina reads were assembled to make the contigs by CLC genomics workbench (CLC bio) and mapped to the two assembled PacBio contigs to expand the sequence of both ends. By using the complete sequence as input, genes were annotated by latest updated version of NCBI Prokaryotic Genome Annotation Process (16). Secondary metabolite BGCs were predicted from the annotated information by antiSMASH 4.0 (17), and combined with the BGCs obtained from previous studies (18).
dRNA-Seq library preparation and high-throughput sequencing
The harvested cells were washed with polysome buffer composed of 20 mM Tris–HCl (pH7.5), 140 mM NaCl and 5 mM MgCl2. Then, the cells were resuspended with 500 μl of lysis buffer composed of 0.3 M Sodium acetate (pH 5.2), 10 mM ethylenediaminetetraacetic acid (EDTA) and 1% Triton X-100. The resuspended cells were frozen with liquid nitrogen, and then lysed by grinding using mortar and pestle. The powdered cells were thawed and separated by centrifugation at 4°C, 3000 g for 10 min, and the supernatant was collected as the lysate. The samples at each sampling time point were immediately washed and lysed as described above, and the collected lysates were stored at −80°C, and after all sampling was carried out, all samples from each time point were thawed on ice for RNA extraction. The RNA samples were purified from the lysate by phenol–chloroform-isoamyl alcohol (Sigma), and precipitated by ice-cold 100% ethanol. This RNA samples were used for further library constructions for dRNA-Seq and RNA-Seq. To remove genomic DNA from the extracted RNA samples, the same amount of RNA samples of four different time points was pooled and treated by DNase I (2 μl of DNase I (NEB), 5 μl of 10 × DNase I buffer and 1 μl of SUPERase-In RNase Inhibitor (20 U/μl, Thermo scientific), and DEPC-treated water (Thermo scientific) in 50 μl reaction volume). After DNase I treatment by incubation at 37°C for 1 h, the RNA samples were purified by phenol–chloroform-isoamyl alcohol (Sigma) and precipitated by ice-cold 100% ethanol. Integrity of DNase-treated RNAs were confirmed by gel electrophoresis. Then, rRNAs were removed from DNase I treated RNA sample by using Ribo-Zero rRNA Removal Kit for Bacteria (Epicentre). The rRNA-removed RNA samples were purified by the ethanol precipitation. About 1.4 μg of rRNA-removed RNA was split into two samples for two different libraries that were incubated in 1 × RNA 5′-polyphosphatase reaction buffer and 1 U of SUPERase-In at 37°C for 1 h with (TAP+) or without (TAP−) 1 U of RNA 5′-polyphosphatase (tobacco acid pyrophosphatase, TAP). As TAP cleaves 5′-triphosphate of the primary transcript to make 5′-monophosphate, it was expected that TAP+ samples contain both primary and processed transcripts, and TAP− samples contain only processed transcripts. After ethanol precipitation, 5 pmol of 5′ RNA adapter was ligated to RNA with T4 RNA ligase in 1 × RNA ligase buffer (Thermo scientific) by incubation at 37°C for 90 min. Then, 1.2 × of AMPure XP beads (Beckman Coulter) were added to the solution and purified twice to select the proper size of libraries with a ligated adapter. The first cDNA strand synthesis by reverse transcription was done by Superscript III Reverse Transcriptase (Thermo Scientific) with a reverse transcription primer according to the manufacturer's instruction, followed by adding 0.8 × of AMPure XP beads and purifying twice to collect the synthesized DNA. The second strand of cDNA was synthesized using Phusion High-Fidelity DNA Polymerase (Thermo Scientific) with indexed primers. The reaction was monitored on a CFX96 Real-Time PCR Detection System (Bio-Rad) and stopped at the beginning of the saturation point. The amplified sample was purified by adding 0.8 × of AMPure XP beads to obtain the final library. The constructed libraries were sequenced using the 100-bp single-end read recipe by an Illumina HiSeq 2000 platform (Illumina).
Determination of TSSs
The sequencing results were de-multiplexed and processed by CLC genomics workbench. Total 12 938 538–13 489 136 raw reads were generated for each replicate, and trimmed by their quality (Quality score: 0.05, maximum ambiguous nucleotides: 2), their adapter sequence (Action: Remove adapter, mismatch cost: 2, gap cost: 3, internal match minimum score: 9, end match minimum score: 9), and length (>15 bp). The trimming steps yielded 81.67–86.70% of the raw reads. Total 4 607 972–7 483 560 (43.56–66.72%) reads with average read length of 100.86–100.88 bp were uniquely mapped to the completed genome (mismatch cost: 2, insertion cost: 3, deletion cost: 3, length fraction: 0.9, similarity cost: 0.9 and ignore non-specific match), which were corresponding to 54- to 88-fold coverage. The mapped information was exported as bam file format, and 5′-end of mapped reads at each genomic position were counted as TSS peak raw count. TSSs of S. clavuligerus were determined using custom perl scripts and manually curated as described previously (6). Genomic position of 5′-ends of all mapped reads from both TAP+/− libraries were called and counted as the peak intensity of each position. To determine TSS, the peak with maximum intensity among the peaks of a sub-cluster was selected. First, peaks apart from less than 100 bp were clustered. Then, adjacent peaks with <10 standard deviation of their position in the same cluster were additionally sub-clustered. Low intensity peaks less than four counts were discarded to select the enriched peaks. As the 5′-end of the processed RNAs are expected to abundantly locate the downstream of the TSS, peaks showing more than 2-fold intensity at the position in TAP+ than TAP− condition were selected. Finally, peaks that were not presented in both duplicates were removed. Assigned peaks were further manual curated according to RNA-Seq profiles and peak intensities to determine the peak as a TSS. The −10 motif was searched by MEME (P < 0.05) among the extracted sequences from −25 to +1 bp relative to the position of TSS for the different gene groups of the TSS category, COG function, spacer length and the selection of a potential regulon of sigma factors. The −35 motif was searched by MEME (oops, P < 0.05; or zoops) among the extracted sequences from −40 to −25 bp relative position of TSS. The obtained motif sequence were aligned relative to the middle position of conserved motif, and the 19 bp extracted sequence (N9 (1 bp)N9) was used as input for Weblogo 3 (19). For consensus promoter motifs of potential common regulons including the sigma (σ) factor, the promoter list was selected by FIMO (input query: the extracted sequences from −40 to −25 bp relative position of σ factor TSS; input database: the extracted sequences from −40 to −25 bp relative position of all TSS (n = 2659); P-value < 0.001). The SD motif was searched by MEME (oops, P < 0.05) among the extracted sequences from 25 bp upstream of a start codon. The obtained motif sequences were aligned relative to the second ‘G’ position of ‘GGAG’, and the 18 bp extracted sequence (N8GGAGN6) was used as input for Weblogo 3.
RNA-Seq library preparation and high-throughput sequencing
RNA-seq libraries were constructed with TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) according to the manufacturer's instructions. The library was sequenced on the HiSeq2000 platform using the 100-bp single-end read recipe. The sequencing results were de-multiplexed and processed by CLC genomics workbench. Total 12 420 952–15 701 669 raw reads were generated for each replicate, and trimmed by their quality (Quality score: 0.05, maximum ambiguous nucleotides: 2) and length (>15 bp). Total 7,193,337 (52.19% of raw reads) to 12,929,000 (95.58% of raw reads) reads with average read length of 100.84–100.88 bp were uniquely mapped to the completed genome (mismatch cost: 2, insertion cost: 3, deletion cost: 3, length fraction: 0.9, similarity cost: 0.9 and ignore non-specific match) which were corresponding to 85- to 153-fold coverage, respectively. The mapped information was exported as bam file format, and the number of mapped reads at each genomic position were counted as the read count. The RNA expression value of each gene was the sum of the number of reads within the gene, and normalized to DESeq2 value obtained from DESeq2 package in R (20).
Ribosome profiling library preparation and high-throughput sequencing
The mycelium was collected by centrifugation at 4°C for 10 min at 3000 g and the cell pellet was washed with 2 ml polysome buffer with 20 μM thiostrepton. The washed pellet was resuspended in 1 ml lysis buffer with 20 μM thiostrepton. The resuspended cells were dripped into a mortar filled with liquid nitrogen and then ground with pestle. The cell debris was removed by centrifugation at 4°C for 5 min at 3000 g. The supernatant was further clarified and collected by centrifugation at 4°C for 10 min at 16 000 g. To digest RNA, 400 U of MNase (NEB), 20 μl of 10 × MNase buffer and 2 μl of 100 × bovine serum albumin were treated to the lysate at 37°C for 2 h. The samples were loaded onto illustra MicroSpin S-400 HR Columns (GE Healthcare), which were formerly washed three times with 500 μl of washing buffer composed of 50 mM Tris–HCl pH 8, 250 mM NaCl, 50 mM MgCl2, 25 mM EGTA and 1% Triton X-100. The column was centrifuged at 4°C for 2 min at 400 g, and the flow through was further purified by phenol–chloroform extraction and ethanol precipitation. rRNA was removed by using Ribo-Zero rRNA Removal Kit. The ribosome-protected RNA fragments (RPF) between 26 and 32 bp were separated by electrophoresis for 65 min at 200 V using 15% polyacrylamide TBE-urea gel (Invitrogen), and eluted in 400 μl of RNA gel extraction buffer composed of 300 mM sodium acetate pH 5.5, 1 mM EDTA and 0.25% (w/v) sodium dodecyl sulphate. The samples were frozen for 30 min at −80°C then incubated at 37°C for 4 h with gentle mixing. The eluted RNAs were isolated by ethanol precipitation and purified again with RNeasy MinElute Column (Qiagen). To dephosphorylate the samples, they were denatured for 90 s at 80°C and incubated for 1 h at 37°C with 5 ml of 10 × T4 PNK buffer (NEB), 20 U of SUPERase-In and 10 U of T4 PNK (NEB). The dephosphorylated RNAs were purified by using RNeasy MinElute Column (Qiagen). The sequencing library was constructed with NEBNext Multiplex Small RNA Library Prep Set for Illumina (NEB) according to manufacturer's instructions. The final library of 150 bp was size-selected by gel electrophoresis for 90 min at 100 V using 2% agarose gel dyed with SYBR Gold Nucleic Acid Gel Stain (Bio-Rad). The concentration of the final library was measured with Qubit 2.0 fluorometer (Invitrogen) and the size distribution was checked with Agilent 2200 TapeStation System (Agilent). The library was sequenced on the HiSeq2000 platform using the 50-bp single-end read recipe. The sequencing results were de-multiplexed and processed by CLC genomics workbench. Total 247 353 047–307 178 979 raw reads were generated for each replicate, and trimmed by their quality (Quality score: 0.05, maximum ambiguous nucleotides: 2), their adapter sequence (Action: Remove adapter, mismatch cost: 2, gap cost: 3, internal match minimum score: 3, end match minimum score: 3) and length (>15 bp). The trimming steps yielded 94.22–98.66% of the raw reads. Total 61 294 530 (19.95% of raw reads) to 89 419 567 (35.28% of raw reads) reads with average read length of 25.8–29.58 bp were uniquely mapped to the completed genome (mismatch cost: 2, insertion cost: 3, deletion cost: 3, length fraction: 0.9, similarity cost: 0.9 and ignore non-specific match) which were corresponding to 185- to 305-fold coverage, respectively. The mapped information was exported as bam file format, and the number of mapped reads at each genomic position were counted as the read count. The RPF expression value of each gene was the sum of the number of reads within the gene, and normalized to DESeq2 and RPKM value (20).
RESULTS
High-quality genome sequence identifies 58 secondary metabolite biosynthetic gene clusters
A high-quality complete genome sequence is essential to define accurate bacterial transcription architecture and associated regulatory elements. Since the genome sequence of S. clavuligerus ATCC 27 064 has been partially completed (18,21), we sought to obtain its complete genome sequence by using two sequencing platforms; PacBio long read sequencing (22) and Illumina short read sequencing (23). As a result, we completed a high-quality genome sequence consisting of a 6.75-Mbp linear chromosome and a 1.8-Mbp mega-plasmid (Table 1), in which we filled all sequence gaps and corrected errors in the previous contig sequences (Supplementary Dataset S1). GC contents of the chromosome (72.7%) and plasmid (71.9%) were comparable with the previously established Streptomyces genomes (24).
Table 1.
General features of the completed genome of Streptomyces clavuligerus ATCC 27064
| Sequence | Total | Genome | Plasmid |
|---|---|---|---|
| Total size (bp) | 8 544 086 | 6 748 591 | 1 795 495 |
| Total gene | 7163 | 5632 | 1531 |
| Total CDS | 7076 | 5545 | 1531 |
| tRNA | 66 | 66 | 0 |
| rRNA | 18 | 18 | 0 |
| ncRNA | 3 | 3 | 0 |
| Pseudogenes | 196 | 121 | 75 |
| Coding CDS | 6880 | 5424 | 1456 |
| GC content (%) | 72.5 | 72.7 | 71.9 |
| BGC | 58 | 30 | 28 |
| CDS with putative function | 9 | 8 | 1 |
| CDS with hypothetical function | 1710 | 1161 | 549 |
| Coding density (%) | 85.3 | 85.8 | 83.4 |
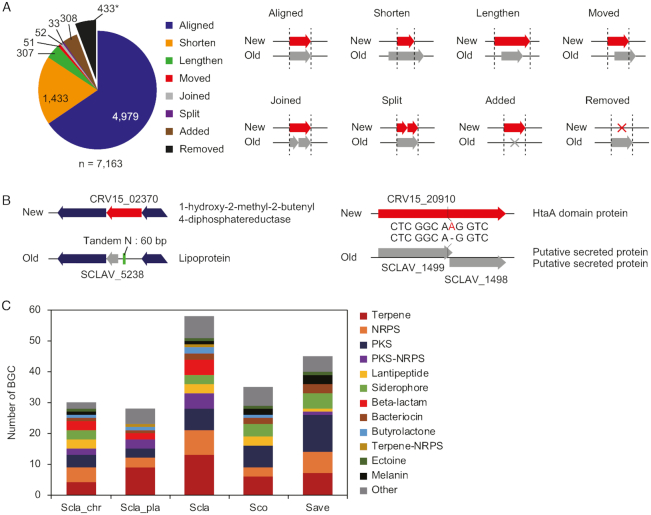
A total of 7163 genes were annotated, comprising 6,880 protein-encoding genes, 196 pseudogenes, 66 tRNAs, 18 rRNAs and 3 ncRNAs. All changes in the genome annotation were categorized into eight groups by comparing to the previous annotation (18) (Figure 1A and Supplementary Dataset S2). Although 4979 genes (69.5%) were identical in both annotations, 1433 and 307 genes were annotated as shorter and longer genes in length than those in the previous annotation. Due to the changes in both 5′ and 3′-end positions, 51 genes were assigned to the ‘moved’ category. In addition, 52 ‘joined’ genes were the merged annotation of more than two previously annotated genes, and 33 ‘split’ genes were the divided annotation of a previously annotated gene into more than two genes. A total of 308 new genes were discovered and 433 genes, previously annotated, were absent in our annotation. This new annotation allowed better prediction of gene functions (Figure 1B). For instance, CRV15_02370 was originally annotated as an unknown lipoprotein and has 60-bp of tandem N sequences at the upstream region. After correcting the tandem N sequences, this gene was re-annotated as 1-hydroxy-2-methyl-2-butenyl 4-diphosphatereductase (25). CRV15_20910 was originally annotated as two genes (SCLAV_1499 and SCLAV_1500) encoding putative secreted proteins. However, we found the absence of ‘A’ within the SCLAV_1499, which caused a frame-shift mutation. The two genes were joined into one and re-annotated as a heme-related HtaA domain protein.
Figure 1.
Completion of high-quality genome sequence of Streptomyces clavuligerus ATCC 27604. (A) Correction of genome annotation of S. clavuligerus. The 433 ‘removed’ genes were not added to the total number of genes. (B) Examples of corrected genes with functional annotations. (C) BGC distribution of three Streptomyces species. Scla_chr, the chromosome of S. clavuligerus; Scla_pla, the mega-plasmid of S. clavuligerus; Scla, total genome of S. clavuligerus; Sco, the genome of Streptomyces coelicolor; Save, the genome of Streptomyces avermitilis.
With this high-quality genome sequence and annotation, we predicted a total of 58 BGCs, including cephamycin C, clavulanic acid and 5S clavam BGCs using antiSMASH and data from previous studies (17,18). Among them, 30 and 28 BGCs were found from the chromosome and the plasmid, respectively (Supplementary Dataset S3). Note that 23 and 25 BGCs were previously predicted on the chromosome and the plasmid, respectively (18). Interestingly, holomycin and tunicamycin BGCs were newly predicted, consistent with the experimental evidence that S. clavuligerus produces the two secondary metabolites (26,27). The total number of BGCs was higher than those of other Streptomyces, such as S. coelicolor and Streptomyces avermitilis (Figure 1C) (24). Specifically, β-lactam type BGCs were unique, and the number of terpene type BGCs were much higher in S. clavuligerus. Among 14 terpene type BGCs, ten were located at the plasmid, which may be obtained in response to dynamic environmental signals during evolution (18). Consequently, a total of 1554 out of 7163 genes were assigned to the 58 BGCs from the high-quality complete genome sequence.
Transcription start sites reveal regulatory elements of individual promoters
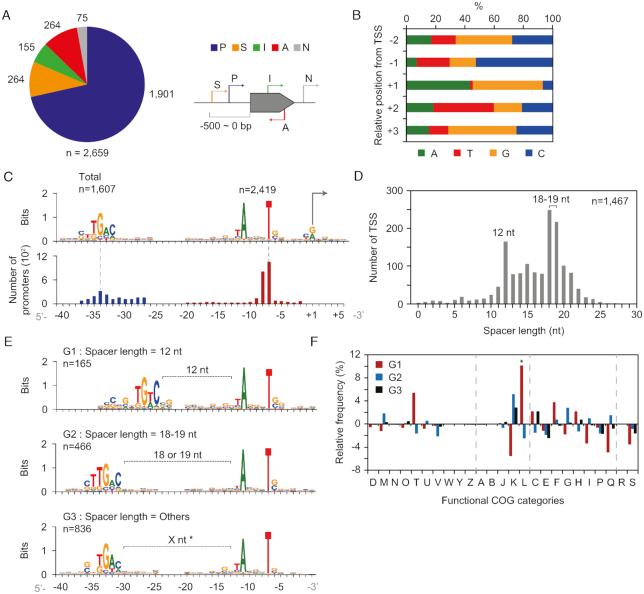
Next, we determined the genomic positions of TSSs in the complete S. clavuligerus genome sequence by using the dRNA-Seq method (10). From RNA samples obtained from the cultures grown under four different growth phases in R5− medium (Supplementary Figure S1), a total of 2,659 TSSs were determined, and subsequently classified into five categories based on their genomic positions relative to adjacent genes (Figure 2A and Supplementary Dataset S4). A total of 1901 5′-ends of sequencing reads were classified as primary TSSs (P), which have the highest peak intensity among the peaks located in the 500-bp upstream region of the associated gene, and 264 TSSs were assigned as secondary TSSs (S), which were located at the same upstream region of primary TSSs. Among the remaining TSSs, 155 internal TSSs (I) located inside of ORFs and 264 antisense TSSs (A) located inside of the opposite strand of ORFs were determined. At last, the remaining 75 TSSs were classified as intergenic TSSs (N), which were located in the intergenic region excluding 500 bp upstream regions of the associated genes.
Figure 2.
Genome-wide determination of TSSs and the architecture of their promoters. (A) Classification of determined TSSs according to their relative genomic position from the gene. P, primary; S, secondary; I, internal; A, antisense; and N, intergenic. (B) Nucleotide proportion of TSSs at relative positions. (C) The –35 and –10 motifs found from the relative position of TSSs. (D) Spacer length distribution of promoters having both –35 and –10 motifs. (E) Promoter motifs of three groups with different spacer length. G1, spacer length = 12 nt; G2, spacer length = 18 or 19 nt; and G3, all other TSSs except TSSs of G1 and G2 among the TSSs from (D). ‘X nt’ with asterisk (*) represents the variable spacer length between the G3 motifs. (F) Functional COG categories of genes from the three groups of (E). A statistical significance of ‘L’ category was indicated as an asterisk (*) (P < 0.001, chi-square test).
The general features of the TSSs in S. clavuligerus genome are as follows. First, the pyrimidine-purine dinucleotide preference near TSSs was consistent with that of S. coelicolor and other bacteria (Figure 2B) (6,28). Purine nucleotide content (A/G) was strongly preferred at TSSs (91.6%), whereas +2 and −1 position from TSSs showed a high proportion of pyrimidine nucleotide content (T/C), T for 41.3% at the +2 position and C for 52.3% at the −1 position, respectively (29). Second, the −10 motif (TANNNT) was also well conserved in 91.0% of total TSSs (2419 of 2659, P < 0.05; MEME), whereas the −35 motif (NTGAC) was less conserved in 60.4% of total TSSs (1607 of 2659, P < 0.05; MEME) (Figure 2C). For TSSs having consensus −35 and −10 motifs (n = 1467), we calculated spacer length between the two motifs (Figure 2D). Promoters with an 18–19 nt spacer length were the most abundant, which is 1 or 2 nt longer than the optimal 17 nt spacer length of Escherichia coli, but similar to that of S. coelicolor (6,30). Interestingly, the second major peak was observed for the 12 nt spacer length. The −10 motifs showed no difference (i.e. TANNNT), whereas the −35 motifs of the three groups (G1 for 12 nt, G2 for 18–19 nt and G3 for all other lengths) were different according to their spacer lengths (Figure 2E). The G2 group showed the ‘TTGAC’ motif, which is one of the binding motifs of housekeeping sigma (σ) factor HrdB in S. coelicolor (31). The G1 group showed the ‘TGTC’ motif, which was previously reported in S. coelicolor and S. avermitilis for DNA damage-inducible promoters with 12 nt spacer (31). Indeed, genes in the G1 group showed specific functional enrichment related to replication, recombination, and repair functions (L, 13.5%) (Figure 2F). Moreover, −35 motifs were diverse for different categories of TSSs (Supplementary Figure S2) and COG functional categories of downstream genes (Supplementary Figure S3 and Dataset S5).
Specifically, antisense TSSs were located inside the ORF, which are highly likely to have a role in regulatory functions. They had the conserved ‘TANNNT’ −10 motif, which confirms that they are bona fide TSSs (Supplementary Figure S2). One specific feature of the promoters of antisense TSSs was a 3 nt periodic appearance of ‘G’, which is complementary to the 3 nt ‘C’ periodicity of third base of codons in GC-rich S. clavuligerus (Supplementary Figure S2). This suggests that the position of antisense TSS is closely dependent on the codon sequence context at the promoter region and the ‘ANGNTG’ motif is the only possible motif that satisfies both the 3 nt ‘G’ periodicity and conserved ‘ANNNT’ motif. Meanwhile, dispersive positions of antisense TSSs within genes were observed and there is no specific bias for the region of the codon within CDS (Supplementary Figure S4). These features might reflect the variability in the target region within CDS and its regulation mechanisms (32). Alternatively, the transcripts from antisense TSSs might involve non-functional RNAs, which are the products of transcriptional noise owing to the dependency of transcription initiation on the genomic AT content of spurious promoters in the GC-rich genome (33,34).
Promoter motifs predict the regulons regulated by a diverse set of σ-factors
The −35 and −10 motifs are the promoter elements that are mainly recognized by σ-factors. This interaction is followed by the recruitment of the RNA polymerase holo–enzyme complex to the promoter region to initiate transcription (35,36). Since the −35 motifs were diverse, the differential interaction between the −35 motifs and various σ-factors or transcription factors in a sequence-dependent manner was expected, resulting in the complex transcription regulations of morphological differentiation and synthesis of various secondary metabolites in Streptomyces (31,35). The number of σ-factors in the Streptomyces genomes were particularly large compared to other bacterial species (9). Forty one genes were annotated as σ-factors in S. clavuligerus, which were expected to play different roles in response to various environmental stimuli. Each σ-factor was expected to bind to its cognate promoter element (31). Thus, the transcription of genes with promoters having consensus −35 motif are potentially controlled by the common σ-factor.
To predict the potential regulon, all promoters were scanned for matches to the motifs of each σ-factor (P < 0.001 for FIMO; P < 0.05 for MEME) (Figure 3; Supplementary Figure S5 and Dataset S6). For example, the −35 motif of CRV15_09470 was almost identical to that of the G1 group (Figure 2E) and COG L functional group (Supplementary Figure S3). Also, the enriched gene function was highly correlated with DNA replication, recombination, and repair (Figure 3A). There are two possible interpretations; (i) Since the σ-factor usually binds to its own promoter for auto-regulation (37), the genes having the same −35 motif are the members of same regulon of CRV15_09470, or (ii) other σ-factors may bind to the promoters, including the promoter of CRV15_09470. In both cases, CRV15_09470 is considered to be involved in the regulon related to DNA replication, recombination and repair functions. The −35 motif including CRV15_24810 was ‘TCTCCGCGNGGANGG’, and the functions of some genes were related to arginine metabolism (Figure 3B). This regulon is highly likely to be related to clavulanic acid biosynthesis because one of its main precursors is arginine (38). The −35 motif (GGGAACCC) includes CRV15_08465, which is annotated as σ-factor E, and several genes in this regulon were related to peptidase and CoA-related functions (Figure 3C). This motif was also comparable to ‘GGAAC’, which is previously known to be the universal motif of the ECF family of σ-factors for stress responses (39,40). Therefore, CRV15_08465 might be related to changes in the cellular status by peptidase in response to certain stresses. Lastly, CRV15_10010, potentially encoding another σ-factor, was a member of another −35 motif group (TTGCCCCCCG). Interestingly, this regulon is composed of transporter genes (Figure 3D). The promoters of potential regulons of 18 σ-factors were also analyzed, and their consensus −35 motifs were all distinct (Supplementary Figure S5). Taken together, the promoter architecture showed the involvement of a diverse set of σ-factors in governing transcriptional regulation in S. clavuligerus.
Figure 3.
Consensus promoter motifs of four potential regulons including respective σ-factors and their sequence alignments. (A-D) The −35 and −10 motifs of the regulon including (A) CRV15_09470, (B) CRV15_024810, (C) CRV15_08465, and (D) CRV15_10010 were drawn by MEME with oops parameter. Five sequences with both low P-value and similar gene functions with each others were selected. Gene name, their TSS category, and their gene function were shown, respectively.
Transcriptome analysis determines differentially expressed genes in functional categories
Promoter elements with various σ-factors are closely connected to transcriptional initiation, which consequently affects dynamic regulation of transcription and translation. To elucidate the landscape of transcriptomic and translatomic changes under different developmental stages of S. clavuligerus (Supplementary Figure S1), RNA-Seq and ribosome profiling were performed at four different growth phases (41). A total of 7.2– 2.9 million reads and 61.3–89.4 million reads from each sequencing sample were mapped to the genome with at least 85- and 185-fold genome-wide coverage for RNA-Seq and ribosome profiling, respectively (Supplementary Figure S6). Specifically, this ribosome profiling method enables the capturing of translating ribosomes with mRNA molecules at a genome-wide scale by using deep sequencing of ribosome-protected RNA fragments (RPFs). To this end, the mycelia at different growth phases were rapidly frozen in liquid nitrogen and used to isolate the polysome fraction.
The expression levels of each gene were normalized (Supplementary Dataset S7), and hierarchically clustered to assess growth phase dependent changes in transcription. To select the DEGs, we considered the median of all BGC genes at early exponential phase as the expression cutoff value by assuming that most genes for secondary metabolism were minimally expressed at early exponential phase (6). A total of 2934 genes out of 7076 protein-coding genes were determined as DEGs (Figure 4A). The genes were divided by two large clusters, depending on the changes in expression levels from early exponential phase (E) to transition phase (T), late-exponential phase (L) and stationary phase (S), which were cluster I with an increasing pattern (n = 1452) and cluster II with a decreasing pattern (n = 1482). While housekeeping σ-factor HrdB (CRV15_05035) showed no significant expression change, 16 σ-factors were differentially expressed including WhiG (CRV15_05790), ShbA (CRV15_09955) and SigG (CRV15_27220) in cluster I, and SigF (CRV15_07985) and HrdD (CRV15_16830) in cluster II.
Figure 4.
The landscape of transcriptome and translatome. (A) Hierarchical clustering (method: Pearson, complete) of DEGs (n = 2934; expression level > median of BGC genes at early exponential phase, |fold change between at least two phases| > 2, P < 0.05, DESeq2) at four growth phases (exponential, transition, late-exponential and stationary phases). Total 16 σ genes were differentially expressed and their expression levels were indicated. (B and C) Proportion of non-differentially expressed (blue), upregulated (orange) and downregulated (gray) genes after early exponential phase that categorized by COG (B) and BGC type (C). (D) RPF/RNA distribution of protein-coding genes at four growth phases. Statistical significances were indicated for P-values of ***P < 0.001. 5% outliers were excluded. (E) Correlation between log2 RNA change and RPF/RNA change at transition phase (left), late-exponential phase (middle), and stationary phase (right) compared to exponential phase. Trend line (blue), and cutoff criteria (log2(RNA) change > 1, log2(RPF/RNA) change < −1) lines (red) were indicated. (F) Ratio of gene number involved in BGCs. Total (n = 7076) is all protein-coding genes, and Buf (n = 694) is the group of genes in yellow area of (E). Statistical significances were indicated for P-values of ***P < 0.001. 5% outliers were excluded.
Proportions of non-differentially expressed, up-regulated, and down-regulated genes were different according to COG functional categories (Figure 4B). The genes involved in cellular transport and metabolism, such as nucleotide (F), coenzyme (H), secondary metabolites (Q), inorganic ion (P), amino acid (E) and carbohydrate (G) showed highly differential expression levels which were generally dynamic in response to the cellular growth. On the other hand, the genes involved in information storage or signal processing such as cell division (D), replication (L) and signal transduction (T) showed relatively constitutive expression levels which were essential at all growth phases. A total of 15 of 18 COG categories showed a higher proportion of cluster II genes than cluster I, indicating that most of the transcripts were decreased at later phases due to slower growth. Notably, secondary metabolism related genes (Q) showed a particularly high proportion of cluster I, which is consistent with the previous studies in various growth conditions (6). Indeed, 10 of 13 BGCs showed a higher proportion of genes upregulated at later phases (cluster I) than down-regulated (cluster II), except ectoine, siderophore and beta-lactam (Figure 4C). Most DEGs of siderophore type BGCs showed lower expression at later phases than early exponential phase, which might cause a decrease of cellular iron concentration. The opposite tendency between the expression of siderophore BGC and most of the other BGCs agrees with previous reports that iron limitation usually activates secondary metabolite production (42). Among the DEGs, translation machinery genes (J) showed mostly downregulated expression levels at later phases (cluster II) (Figure 4B).
Ribosome profiling shows significant reduction of translation efficiency at late growth phases
The ratio of RPF over RNA (RPF/RNA ratio) of all genes was calculated for four growth phases, revealing that the overall protein synthesis efficiency was significantly decreased at later phases compared to early exponential phase (Mann–Whitney U test, ***P < 0.001, two-sided) due to the limited availability of translation machinery (Figure 4D) (6). Additionally, the relationship between changes in RNA transcript levels and changes in the RPF/RNA ratio represented a global negative correlation for all three later phases compared to early exponential phase (Figure 4E). Interestingly, 694 genes showed ‘translational buffering’ (6) that had a substantially decreasing RPF/RNA ratio despite increasing RNA, indicated in the yellow box of Figure 4E. 220 of 694 (31.7%) genes were located in BGCs, which is a significantly higher ratio compared to the number of BGC genes in the total genome (1539 of 7076, 21.7%) (Figure 4F). This means that most of the genes in secondary metabolism were translationally buffered even though their transcript levels were increased compared to other genes (Figure 4B and F).
Next, we sought to integrate the two datasets with dRNA-Seq in order to determine the genome annotation of transcripts with internal TSSs. There are two possibilities for the presence of internal TSSs; (i) they are non-coding regulatory RNAs or (ii) they are actually alternative TSSs of distinct ORFs located downstream from the TSSs. The relative position of internal TSSs within the ORF was biased toward the 5′-end of the ORF, which suggests that later cases might be more abundant (Supplementary Figure S7A). Indeed, RNA read density as well as RPF read density showed increasing patterns at the downstream of internal TSSs (Supplementary Figure S7B). For example, CRV15_06880 (1,371 bp), a gene encoding sugar ABC transporter substrate-binding protein gene, had an internal TSS located at the +167 bp position (Supplementary Figure S7C). The RNA transcript and RPF profile of this gene starts at the downstream of the internal TSS, and a downstream in-frame GTG codon was found in the functional domain. Also, the ‘NTTGAC’ −35 motif, ‘TANNNT’ −10 motif, 18 nt spacer and ‘GGAGG’ SD sequence were found near this TSS. These results indicate that this internal TSS is the primary TSS of a new downstream ORF, which was previously mis-annotated. All other internal TSSs were also classified into three cases, including (i) transcript of the downstream ORF starting from the downstream location of the TSS indicating the original ORF is misannotated; (ii) transcripts of alternative downstream ORFs to the original ORF; and (iii) non-coding transcript (Supplementary Dataset S8). It is thought that the original ORF was potentially misannotated if there was no ribosome binding at the region between the start codon of the original ORF and internal TSS. To quantify the ribosome binding at the region, ribosome profiling reads at each relative position from the first nucleotide of the start codon were calculated (43). As a result, 45 of 155 internal TSSs were classified as TSSs of corrected ORFs from misannotation, 4 TSSs that did not have any possible downstream ORF were classified as the TSS of non-coding transcripts and the remaining 106 of 155 internal TSSs were classified as the TSSs of transcripts of alternative downstream ORFs. Notably, 27 of 45 transcripts of the corrected ORFs and 25 of 106 transcripts of alternative downstream ORFs were leaderless. Moreover, we predicted functional differences between original and new ORFs by pfam analysis (19). As a result, 2 of 45 corrected ORFs and 23 of 106 transcripts of alternative downstream ORFs showed different domain hits. Therefore, the integrative analysis of the transcriptome and translatome revealed a dynamic transcriptional landscape, translational buffering of secondary metabolism, novel ORFs and accurate gene annotations.
Transcription start sites define regulatory elements encoded in 5′-UTRs
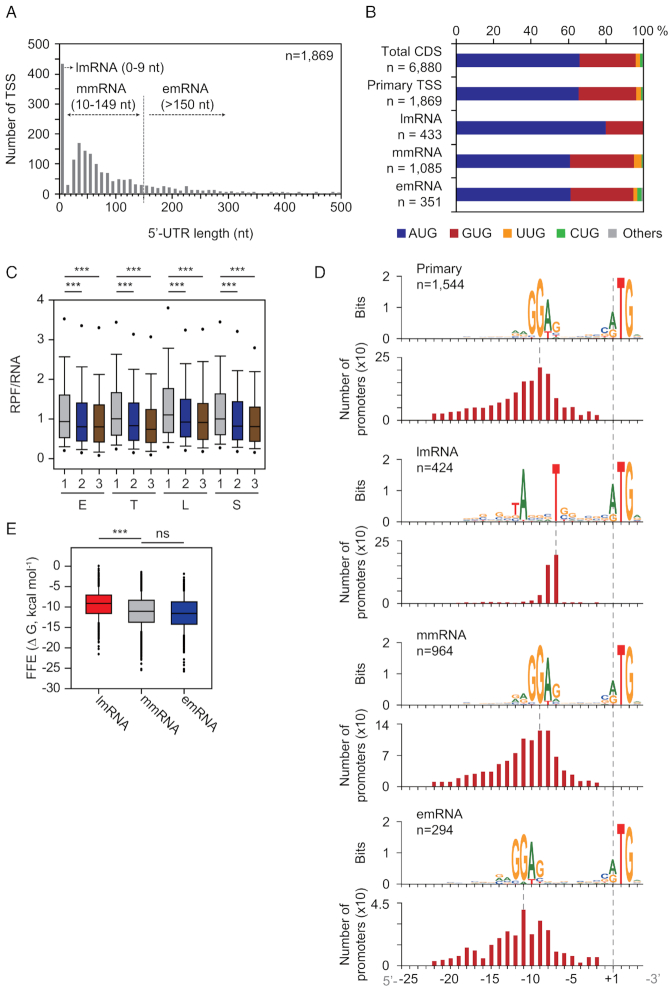
5′-UTRs encode transcriptional and translational regulatory elements such as ribosome binding sites, riboswitches and others (43,44). First, 5′-UTR length distribution was calculated from primary TSSs (n = 1869) of protein-encoding genes (Figure 5A). The median length of 5′-UTRs was 53 nt, which is comparable to the average 5′-UTR lengths of other bacteria; however, it was 9 nt longer than S. coelicolor (6,45–46). A significant portion of 5′-UTRs (n = 433) represented leaderless mRNA (lmRNA), whose 5′-UTR lengths are shorter than 9 nt. This proportion of lmRNA is 4% higher compared to other actinobacteria (19.1% lmRNA on average) (47). The absence of 5′-UTR (5′-UTR length = 0 nt) was observed in 92.8% of lmRNAs (402 of 433). Interestingly, 351 of 1869 5′-UTRs (18.8%) have lengths longer than 150 nt, indicating the presence of potential riboswitches for regulatory functions. This result led us to classify transcription units based upon 5′-UTR lengths; 0–9 nt for lmRNAs, 10–149 nt for ‘moderate-leadered mRNA’ (mmRNA, n = 1085), and longer than 150 nt for ‘extended-leadered mRNA’ (emRNA, n = 351). The lmRNA showed a higher preference of AUG start codon usage (80.0%) than other groups (60.7% for mmRNA, and 61.0% for emRNA) (Figure 5B) (43). The RPF/RNA ratio of each gene, which utilizes AUG as a start codon in total genes and lmRNA genes, suggests a higher translational initiation rate for the AUG start codon than for other start codons (Mann–Whitney U test, **P < 0.01 and ***P < 0.001, two-sided) (Figure 5C and Supplementary Figure S8).
Figure 5.
Transcriptional and translational elements downstream from primary TSS. (A) 5′-UTR length distribution of genes with primary TSSs. They were classified based upon their 5′-UTR lengths; lmRNA, leaderless mRNA (0–9 nt); mmRNA, moderate-leadered mRNA (10–149 nt); emRNA, extended-leadered mRNA (>150 nt). (B) Start codon usage of all protein-encoding genes (total CDS), genes with primary TSSs, and genes with different 5′-UTR lengths described in (A). (C) Log2 scale distribution of RPF level normalized by RNA level of genes having different start codons at four growth phases. E, early exponential; T, transition; L, late-exponential; S, stationary; 1, AUG (n = 4540); 2, GUG (n = 2057); and 3, other codons except AUG or GUG (n = 283). Statistical significances were indicated for P-values of ***P < 0.001. About 5% outliers were excluded. (D) 5′-UTR motifs of four different groups described in (B), and their relative position from TSSs. (E) RNA folding free energy (FFE) distribution of the extracted sequence from the first nucleotide position of start codon to the downstream 40 bp position of three different RNAs described in (A).
Next, we observed the broad appearance of the ‘RRGGAG’ motif between the −25 to −2 position from a start codon considered as the Shine-Dalgarno (SD) sequence (Figure 5D), which is complementary to the 16S rRNA 3′ tail sequence of S. clavuligerus, and also comparable to the core SD motif (GGAGG) observed in S. coelicolor (48). Most of the mmRNA and emRNA showed identical SD motifs (mmRNA: 88.8% (964 of 1085), emRNA: 83.8% (294 of 351), P < 0.05; MEME), but this motif was positioned 2–3 bp more upstream for emRNA than for mmRNA. In contrast, lmRNA showed the ‘TANNNT’ motif, which is the conserved −10 motif of the promoter as expected (97.9% (424 of 433), P < 0.05; MEME). As the interaction between fMet-tRNAfMet and 5′-AUG is the unique ribosome binding signal without SD-aSD interaction on lmRNA (49), the determinant of differential translation efficiency would be the downstream RNA structure of 5′-AUG. Interestingly, the absolute value of folding free energy of the 40 bp sequence downstream from a start codon was significantly lower for lmRNA than for mmRNA and emRNA (Mann–Whitney U test, **P < 0.001, two-sided) (Figure 5E). This suggests that lmRNAs have less structured RNA for efficient translation without 5′-UTRs. Taken together, these results provide the key regulatory elements encrypted in 5′-UTRs for transcription and translation in S. clavuligerus.
Potential regulons of β-lactam biosynthesis in S. clavuligerus
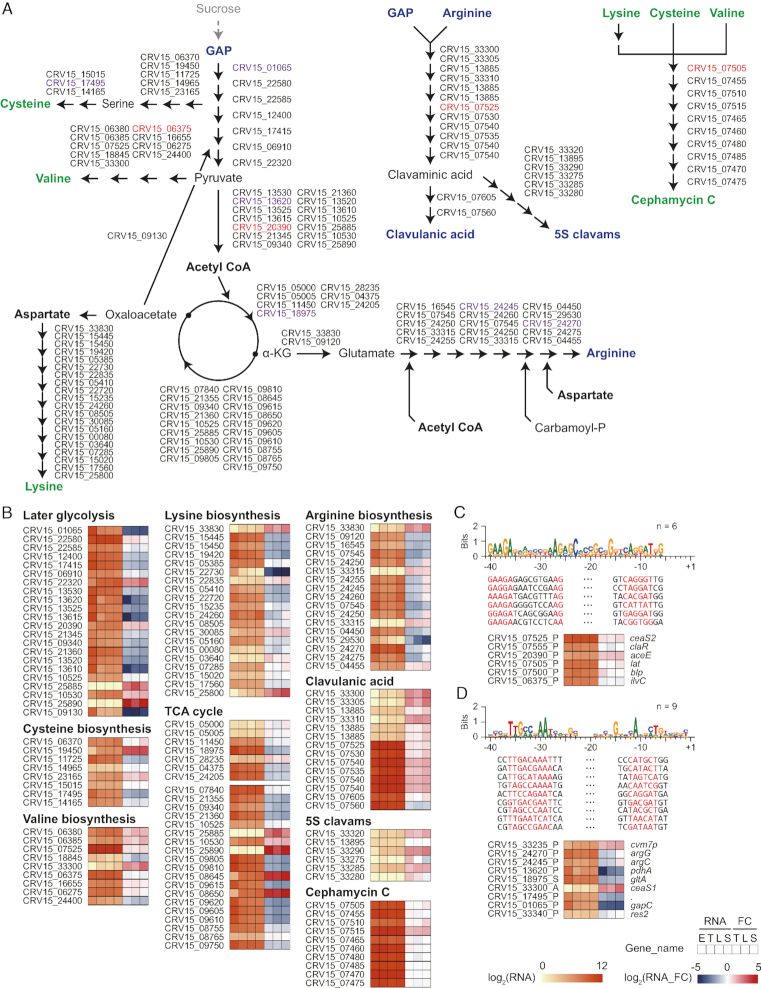
Based on the determined TSSs with expression profiles at four different growth phases, the transcriptome landscape of β-lactam biosynthetic pathway was investigated (Figure 6A and B). Overall, sucrose, which is the main carbon source of R5− medium, was converted to glyceraldehyde 3-phosphate (GAP) and arginine for precursors of clavulanic acid, and lysine, valine and cysteine for precursors of cephamycin C, respectively. For glycolysis pathway, the genes governing conversion from GAP to acetyl CoA were generally downregulated under later growth phases than early exponential phase (15 of 22 genes, P < 0.05, DESeq2). Likewise, tricarboxylic acid cycle (TCA cycle) genes for adenosine triphosphate synthesis were also downregulated through growth phases (16 of 26 genes, P < 0.05, DESeq2). These results suggest that cellular metabolism changed to precursor production for secondary metabolism rather than energy production. Interestingly, the genes involved in clavulanic acid and cephamycin C BGC showed significantly higher expression levels for all growth phases than other BGC genes (Mann–Whitney U test, P < 0.05, two-sided), even among 45 of 58 BGCs that were potentially ‘expressed’ under the culture condition of this study, which showed a higher transcription level median of at least at one growth phase than the total BGC gene median at the early exponential phase (Supplementary Figures S9-10 and Dataset S7). However, expression levels of genes related to the conversion from aspartate to lysine were downregulated at later growth phases (10 of 19 genes, P < 0.05, DESeq2), which may be the rate-limiting factor of cephamycin C biosynthesis. Moreover, the genes involved in the arginine biosynthetic pathway showed similar expression patterns that decrease at transition and late-exponential phases (13 of 17 genes, P < 0.05, DESeq2). These results suggest that a number of genes involved in the same metabolic pathways may be regulated by a common σ-factor. To find the potential common regulon, enriched −35 motifs were searched for the promoter of genes involved in β-lactam BGCs (n = 86) (Figure 6A). As a result, two motifs were significantly enriched (zoops, e < 1; MEME) (Figure 6C and D). One is the ‘GAAGANNNNNNNNAG’ motif, with two clavulanic acid BGC genes (ceaS2 and claR), two cephamycin C BGC genes (lat and blp), pyruvate dehydrogenase (aceE), and a valine biosynthesis gene (ilvC) involved (Figure 6C). They showed nearly constitutive expressions at all growth phases (|FC| < 2 for 5 of 6 genes). Another group involves genes of arginine biosynthesis (argG and argC) and 5S clavam BGC (cvm7p, ceaS1, and res2) with the ‘TTGCCNAAT’ −35 motif (Figure 6D). Seven of nine genes showed similar expression patterns decreasing at transition and late-exponential phases, reflecting common transcription regulation. In summary, the potential regulons involved in the β-lactam biosynthetic pathway were predicted by the integration of the high-quality genome, genome-wide TSSs, and their differential transcription and translation profiles.
Figure 6.
Transcriptome landscape of β-lactam biosynthetic pathway in Streptomyces clavuligerus. (A) Primary and secondary metabolism pathway of β-lactam biosynthetic pathway. Metabolites related with clavulanic acid and 5S clavams were presented as blue bold characters; metabolites related with cephamycin C were presented as green bold characters; and other important intermediates were presented as black bold. (B) Transcriptional level and the fold change of genes in each pathway were shown. RNA values are normalized log2 RPKM value, and RNA fold changes (RNA_FC) are normalized log2 ratio of RNA values of four growth phases. E, early exponential; T, transition; L, late-exponential; S, stationary. (C and D) Genes with red and purple characters in (A) were aligned in (C) and (D), respectively.
DISCUSSION
In this study, we provided (i) a high-quality genome sequence with 58 potential BGCs, (ii) a total of 2659 TSSs, (iii) transcription and translation regulatory elements and (iv) the potential regulons of β-lactam biosynthesis of S. clavuligerus ATCC 27064 by integrating dRNA-Seq, RNA-Seq and ribosome profiling. Newly annotated genes with the BGC information revealed that the S. clavuligerus genome contained an unprecedented large number of BGCs compared to other Streptomyces species (24). About half of the 58 BGCs were located in the 1.8-Mb mega-plasmid, which were likely gained by horizontal gene transfer or recombination from the chromosome (18). Also, the protein-coding genes in this plasmid were predicted to be non-essential for primary metabolism (18).
Genome-wide TSS information facilitated the precise investigation of global transcription and translation regulatory elements in the S. clavuligerus genome. Base composition at the TSS region was comparable to other species in that purine was rich at +1, and pyrimidine was rich at –1 and +2. It was consistent with the structural study that the base stacking interaction between purine base at the –1 position of non-template DNA and purine base at the +1 position of RNA is one of the essential factors for transcription initiation with the abundance of pyrimidine base at the +2 position of RNA (29). Although a higher proportion of cytidine than thymine at the –1 position is reasonable due to the high GC content, it is notable that the proportion of thymine was higher than cytidine at the +2 position. This might be the outcome of additional interactions between the base composition and transcription initiation complex. Another distinct transcriptional element was the abundance of the ‘TGTC’ –35 motif with 12-bp spacer length that was related to DNA replication, recombination and repair functions. Since those functions are essential for maintaining self-replicable life, it is reasonable to have distinct motifs and the cognate σ-factor for tight regulation. For example, the CRV15_09470 gene contained the –35 motif in the promoter region and was predicted to encode the ECF family σ-factor having the σ2 and σ4 domain by Pfam (50). Remarkably, an additional SnoaL_2 domain was predicted at the C-terminal region, which was also reported in σJ of Mycobacterium tuberculosis (51). This domain was structurally similar to the domain in polyketide cyclase, expoxide hydrolase and ketosteroid isomerases. Structural and biochemical features of σJ suggested that the domain could modulate the conformation of σJ in the absence of a cognate anti-σ factor, resulting in changes of the DNA binding specificity in response to the external signals (51). Therefore, the σ-factor may bind to the distinct motif due to its unique structure in response to certain cellular signals for tight regulation of essential gene expression.
In addition to the ‘TGTC’ motif, a wide variety of –35 motifs were observed, which may be important determinants for various σ-factor interactions with the cognate promoters. As the σ4 domain directly binds to the –35 element, the structural difference of the σ4 domain among σ-factors could generate different DNA binding specificity (52). However, it is ambiguous to assign one cognate σ-factor to one cognate –35 motif due to the flexibility of σ-factor binding to the promoter sequences (53). For example, the –35 motif of the housekeeping HrdB regulon of S. coelicolor revealed by ChIP-seq was not enriched as one specific sequence (9). Other determinants such as the extended –10 element, UP element and –10 element may compensate for the weak interaction of the –35 motif with various σ-factors (54). Also, additional transcription factors may cooperate with σ-factors to alter their binding interaction (36). Even though extensive autoregulation of σ-factors to their own promoters was observed (37), other σ-factors may bind to the promoter instead of the cognate σ-factor itself, such as ShbA binding to the hrdB promoter in Streptomyces griseus (55). Nevertheless, the promoters with similar –35 and –10 motifs have the potential to be regulated by identical σ-factors. Therefore, the investigation of potential common regulons regulated by a σ-factor could be a valuable resource to understand the complex transcriptional regulatory network of secondary metabolism.
Next, the global analysis of the 5′-UTRs revealed the abundance of lmRNA in S. clavuligerus. lmRNAs were universally observed among broad species, and their proportion was conserved among phylogenetically close species (49). Thus, they are considered as an evolutionarily ancient translation mode due to the lack of translational machinery such as 5′-UTR and r-proteins (49), but also as a regulatory system for some cases such as kasugamycin resistance (56). Neither functional enrichment nor a remarkable trend of RNA, RPF, and RPF/RNA ratios were observed. Instead, lmRNA was less structured at the start codon region than mmRNA or emRNA, suggesting that the 5′-UTR might be obtained for efficient translational regulation during evolution.
At last, the transcriptome landscape of the β-lactam biosynthetic pathway provided an understanding of the metabolic changes and potential common regulons. According to differential transcription levels of the genes in our culture conditions, lysine and arginine biosynthesis were downregulated at later growth phases, which might be the rate-limiting step of cephamycin C and clavulanic acid biosynthesis, respectively. Among the second potential common regulon genes (Figure 6D), argC and argG are the first genes of two polycistronic transcription units, which are argCJBDF and argGH (57). They were known to be negatively regulated by ArgR binding to ARG box, which is located at the promoter including the –35 motif region (57). However, other genes in the same regulon such as pdhA and gltA were not known as ArgR targets. Thus, other σ-factors may regulate those genes instead of ArgR.
In conclusion, the integration of a high-quality genome sequence with 58 potential BGCs and transcriptome landscape revealed the genome-wide transcriptional and translational regulatory elements of S. clavuligerus ATCC 27064. This information will improve the understanding of complex regulatory networks in Streptomyces BGCs. Furthermore, detailed studies of diverse promoter elements, σ-factors, and their regulons will establish the foundation of rational engineering for the activation of silent BGCs.
DATA AVAILABILITY
Genome sequencing raw reads can be found at the National Center for Biotechnology Information as BioProject PRJNA414136. All other raw sequencing reads generated in this study can be found in the GEO under the accession number of GSE128216.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: B.K.C. designed and supervised the project. N.L., Y.L. and W.R. performed genome sequencing, sequence correction and validation. N.L. and Y.J. performed dRNA-Seq, RNA-Seq and ribosome profiling. S.H., Y.J. and S.C. analyzed data. S.H., S.C., B.O.P. and B.K.C. wrote the manuscript. All authors read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Novo Nordisk Foundation [NNF10CC1016517 to B.O.P.]; Intelligent Synthetic Biology Center of Global Frontier Project [2011–0031957 to B.-K.C.]; Bio & Medical Technology Development Program [2018M3A9F3079664 to B.-K.C.] through the National Research Foundation of Korea (NRF) funded by the Korean government (MSIT). Funding for open access charge: Novo Nordisk Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kim J.N., Kim Y., Jeong Y., Roe J.H., Kim B.G., Cho B.K.. Comparative genomics reveals the core and accessory genomes of Streptomyces species. J. Microbiol. Biotechnol. 2015; 25:1599–1605. [DOI] [PubMed] [Google Scholar]
- 2. Jones S.E., Elliot M.A.. Streptomyces Exploration: competition, volatile communication and new bacterial behaviours. Trends. Microbiol. 2017; 25:522–531. [DOI] [PubMed] [Google Scholar]
- 3. Procopio R.E., Silva I.R., Martins M.K., Azevedo J.L., Araujo J.M.. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012; 16:466–471. [DOI] [PubMed] [Google Scholar]
- 4. Weber T., Charusanti P., Musiol-Kroll E.M., Jiang X., Tong Y., Kim H.U., Lee S.Y.. Metabolic engineering of antibiotic factories: new tools for antibiotic production in actinomycetes. Trends. Biotechnol. 2015; 33:15–26. [DOI] [PubMed] [Google Scholar]
- 5. Hwang K.S., Kim H.U., Charusanti P., Palsson B.O., Lee S.Y.. Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol. Adv. 2014; 32:255–268. [DOI] [PubMed] [Google Scholar]
- 6. Jeong Y., Kim J.N., Kim M.W., Bucca G., Cho S., Yoon Y.J., Kim B.G., Roe J.H., Kim S.C., Smith C.P. et al.. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2). Nat. Commun. 2016; 7:11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yi J.S., Kim M.W., Kim M., Jeong Y., Kim E.J., Cho B.K., Kim B.G.. A novel approach for gene expression optimization through native Promoter and 5′ UTR combinations based on RNA-seq, Ribo-seq, and TSS-seq of Streptomyces coelicolor. ACS. Synth. Biol. 2017; 6:555–565. [DOI] [PubMed] [Google Scholar]
- 8. Bauer J.S., Fillinger S., Forstner K., Herbig A., Jones A.C., Flinspach K., Sharma C., Gross H., Nieselt K., Apel A.K.. dRNA-seq transcriptional profiling of the FK506 biosynthetic gene cluster in Streptomyces tsukubaensis NRRL18488 and general analysis of the transcriptome. RNA. Biol. 2017; 14:1617–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smidova K., Zikova A., Pospisil J., Schwarz M., Bobek J., Vohradsky J.. DNA mapping and kinetic modeling of the HrdB regulon in Streptomyces coelicolor. Nucleic Acids Res. 2019; 47:621–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Romero D.A., Hasan A.H., Lin Y.F., Kime L., Ruiz-Larrabeiti O., Urem M., Bucca G., Mamanova L., Laing E.E., van Wezel G.P. et al.. A comparison of key aspects of gene regulation in Streptomyces coelicolor and Escherichia coli using nucleotide-resolution transcription maps produced in parallel by global and differential RNA sequencing. Mol. Microbiol. 2014; 94:963–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown A.G., Butterworth D., Cole M., Hanscomb G., Hood J.D., Reading C., Rolinson G.N.. Naturally-occurring beta-lactamase inhibitors with antibacterial activity. J. Antibiot. 1976; 29:668–669. [DOI] [PubMed] [Google Scholar]
- 12. Ward J.M., Hodgson J.E.. The biosynthetic genes for clavulanic acid and cephamycin production occur as a ‘super-cluster’ in three Streptomyces. FEMS. Microbiol. Lett. 1993; 110:239–242. [DOI] [PubMed] [Google Scholar]
- 13. Vecchione J.J., Alexander B. Jr, Sello J.K.. Two distinct major facilitator superfamily drug efflux pumps mediate chloramphenicol resistance in Streptomyces coelicolor. Antimicrob. Agents Chemother. 2009; 53:4673–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcia-Dominguez M., Martin J.F., Mahro B., Demain A.L., Liras P.. Efficient plasmid transformation of the beta-lactam producer Streptomyces clavuligerus. Appl. Environ. Microbiol. 1987; 53:1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chin C.S., Alexander D.H., Marks P., Klammer A.A., Drake J., Heiner C., Clum A., Copeland A., Huddleston J., Eichler E.E. et al.. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013; 10:563–569. [DOI] [PubMed] [Google Scholar]
- 16. Haft D.H., DiCuccio M., Badretdin A., Brover V., Chetvernin V., O’Neill K., Li W., Chitsaz F., Derbyshire M.K., Gonzales N.R. et al.. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018; 46:D851–D860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blin K., Wolf T., Chevrette M.G., Lu X., Schwalen C.J., Kautsar S.A., Suarez Duran H.G., de Los Santos E.L.C., Kim H.U., Nave M. et al.. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017; 45:W36–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Medema M.H., Trefzer A., Kovalchuk A., van den Berg M., Muller U., Heijne W., Wu L., Alam M.T., Ronning C.M., Nierman W.C. et al.. The sequence of a 1.8-mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome. Biol. Evol. 2010; 2:212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Song J.Y., Jeong H., Yu D.S., Fischbach M.A., Park H.S., Kim J.J., Seo J.S., Jensen S.E., Oh T.K., Lee K.J. et al.. Draft genome sequence of Streptomyces clavuligerus NRRL 3585, a producer of diverse secondary metabolites. J. Bacteriol. 2010; 192:6317–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B. et al.. Real-time DNA sequencing from single polymerase molecules. Science. 2009; 323:133–138. [DOI] [PubMed] [Google Scholar]
- 23. Bentley D.R., Balasubramanian S., Swerdlow H.P., Smith G.P., Milton J., Brown C.G., Hall K.P., Evers D.J., Barnes C.L., Bignell H.R. et al.. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008; 456:53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nett M., Ikeda H., Moore B.S.. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009; 26:1362–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ludwiczuk A., Skalicka-Woźniak K., Georgiev M.I.. Badal S, Delgoda R. Terpenoids. Pharmacognosy. 2017; Boston: Academic Press; 233–266. [Google Scholar]
- 26. Li B., Walsh C.T.. Identification of the gene cluster for the dithiolopyrrolone antibiotic holomycin in Streptomyces clavuligerus. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:19731–19735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen W., Qu D., Zhai L., Tao M., Wang Y., Lin S., Price N.P., Deng Z.. Characterization of the tunicamycin gene cluster unveiling unique steps involved in its biosynthesis. Protein Cell. 2010; 1:1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cortes T., Schubert O.T., Rose G., Arnvig K.B., Comas I., Aebersold R., Young D.B.. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 2013; 5:1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Basu R.S., Warner B.A., Molodtsov V., Pupov D., Esyunina D., Fernandez-Tornero C., Kulbachinskiy A., Murakami K.S.. Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J. Biol. Chem. 2014; 289:24549–24559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Typas A., Hengge R.. Role of the spacer between the −35 and −10 regions in σs promoter selectivity in Escherichia coli. Mol. Microbiol. 2005; 59:1037–1051. [DOI] [PubMed] [Google Scholar]
- 31. Touzain F., Schbath S., Debled-Rennesson I., Aigle B., Kucherov G., Leblond P.. SIGffRid: a tool to search for sigma factor binding sites in bacterial genomes using comparative approach and biologically driven statistics. BMC Bioinformatics. 2008; 9:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thomason M.K., Storz G.. Bacterial antisense RNAs: how many are there, and what are they doing. Annu. Rev. Genet. 2010; 44:167–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Llorens-Rico V., Cano J., Kamminga T., Gil R., Latorre A., Chen W.H., Bork P., Glass J.I., Serrano L., Lluch-Senar M.. Bacterial antisense RNAs are mainly the product of transcriptional noise. Sci. Adv. 2016; 2:e1501363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raghavan R., Sloan D.B., Ochman H.. Antisense transcription is pervasive but rarely conserved in enteric bacteria. Mbio. 2012; 3:e00156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun D., Liu C., Zhu J., Liu W.. Connecting metabolic pathways: sigma factors in Streptomyces spp. Front. Microbiol. 2017; 8:2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Browning D.F., Busby S.J.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. [DOI] [PubMed] [Google Scholar]
- 37. Chauhan R., Ravi J., Datta P., Chen T., Schnappinger D., Bassler K.E., Balazsi G., Gennaro M.L.. Reconstruction and topological characterization of the sigma factor regulatory network of Mycobacterium tuberculosis. Nat. Commun. 2016; 7:11062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jensen S.E. Biosynthesis of clavam metabolites. J. Ind. Microbiol. Biotechnol. 2012; 39:1407–1419. [DOI] [PubMed] [Google Scholar]
- 39. Sachdeva P., Misra R., Tyagi A.K., Singh Y.. The sigma factors of Mycobacterium tuberculosis: regulation of the regulators. FEBS. J. 2010; 277:605–626. [DOI] [PubMed] [Google Scholar]
- 40. Sauviac L., Philippe H., Phok K., Bruand C.. An extracytoplasmic function sigma factor acts as a general stress response regulator in Sinorhizobium meliloti. J. Bacteriol. 2007; 189:4204–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ingolia N.T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014; 15:205–213. [DOI] [PubMed] [Google Scholar]
- 42. Locatelli F.M., Goo K.S., Ulanova D.. Effects of trace metal ions on secondary metabolism and the morphological development of streptomycetes. Metallomics. 2016; 8:469–480. [DOI] [PubMed] [Google Scholar]
- 43. Moll I., Grill S., Gualerzi C.O., Blasi U.. Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control. Mol. Microbiol. 2002; 43:239–246. [DOI] [PubMed] [Google Scholar]
- 44. Winkler W.C., Breaker R.R.. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005; 59:487–517. [DOI] [PubMed] [Google Scholar]
- 45. Irnov I., Sharma C.M., Vogel J., Winkler W.C.. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 2010; 38:6637–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kroger C., Dillon S.C., Cameron A.D., Papenfort K., Sivasankaran S.K., Hokamp K., Chao Y., Sittka A., Hebrard M., Handler K. et al.. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E1277–E1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zheng X., Hu G.Q., She Z.S., Zhu H.. Leaderless genes in bacteria: clue to the evolution of translation initiation mechanisms in prokaryotes. BMC Genomics. 2011; 12:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rudolph M.M., Vockenhuber M.P., Suess B.. Synthetic riboswitches for the conditional control of gene expression in Streptomyces coelicolor. Microbiology. 2013; 159:1416–1422. [DOI] [PubMed] [Google Scholar]
- 49. Beck H.J., Moll I.. Leaderless mRNAs in the spotlight: ancient but not outdated. Microbiol. Spectr. 2018; 6:doi:10.1128/microbiolspec.RWR-0016-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A. et al.. The Pfam protein families database in 2019. Nucleic Acids Res. 2019; 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goutam K., Gupta A.K., Gopal B.. The fused SnoaL_2 domain in the Mycobacterium tuberculosis sigma factor sigmaJ modulates promoter recognition. Nucleic Acids Res. 2017; 45:9760–9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Feklistov A., Sharon B.D., Darst S.A., Gross C.A.. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 2014; 68:357–376. [DOI] [PubMed] [Google Scholar]
- 53. Hook-Barnard I.G., Hinton D.M.. Transcription initiation by mix and match elements: flexibility for polymerase binding to bacterial promoters. Gene. Regul. Syst. Bio. 2007; 1:275–293. [PMC free article] [PubMed] [Google Scholar]
- 54. Guzina J., Djordjevic M.. Promoter recognition by extracytoplasmic function sigma factors: analyzing DNA and protein interaction motifs. J. Bacteriol. 2016; 198:1927–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Otani H., Higo A., Nanamiya H., Horinouchi S., Ohnishi Y.. An alternative sigma factor governs the principal sigma factor in Streptomyces griseus. Mol. Microbiol. 2013; 87:1223–1236. [DOI] [PubMed] [Google Scholar]
- 56. Schluenzen F., Takemoto C., Wilson D.N., Kaminishi T., Harms J.M., Hanawa-Suetsugu K., Szaflarski W., Kawazoe M., Shirouzu M., Nierhaus K.H. et al.. The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat. Struct. Mol. Biol. 2006; 13:871–878. [DOI] [PubMed] [Google Scholar]
- 57. Rodriguez-Garcia A., de la Fuente A., Perez-Redondo R., Martin J.F., Liras P.. Characterization and expression of the arginine biosynthesis gene cluster of Streptomyces clavuligerus. J. Mol. Microbiol. Biotechnol. 2000; 2:543–550. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequencing raw reads can be found at the National Center for Biotechnology Information as BioProject PRJNA414136. All other raw sequencing reads generated in this study can be found in the GEO under the accession number of GSE128216.