Abstract
The liver-specific microRNA, miR-122, is an essential host factor for replication of the hepatitis C virus (HCV). miR-122 stabilizes the positive-strand HCV RNA genome and promotes its synthesis by binding two sites (S1 and S2) near its 5′ end in association with Ago2. Ago2 is essential for both host factor activities, but whether other host proteins are involved is unknown. Using an unbiased quantitative proteomics screen, we identified the TNRC6 protein paralogs, TNRC6B and TNRC6C, as functionally important but redundant components of the miR-122/Ago2 host factor complex. Doubly depleting TNRC6B and TNRC6C proteins reduced HCV replication in human hepatoma cells, dampening miR-122 stimulation of viral RNA synthesis without reducing the stability or translational activity of the viral RNA. TNRC6B/C were required for optimal miR-122 host factor activity only when S1 was able to bind miR-122, and restricted replication when S1 was mutated and only S2 bound by miR-122. TNRC6B/C preferentially associated with S1, and TNRC6B/C depletion enhanced Ago2 association at S2. Collectively, these data suggest a model in which TNRC6B/C regulate the assembly of miR-122/Ago complexes on HCV RNA, preferentially directing miR-122/Ago2 to S1 while restricting its association with S2, thereby fine-tuning the spatial organization of miR-122/Ago2 complexes on the viral genome.
INTRODUCTION
The hepatitis C virus (HCV) is an hepatotropic, positive-strand RNA virus classified within the family Flaviviridae. Despite the availability of effective antiviral therapies, it remains a major causative agent of chronic liver disease and hepatocellular carcinoma worldwide (1). Its single-stranded, positive-sense RNA genome is ∼9.6 kb in length, with structured 5′ and 3′ terminal untranslated regions (UTRs). The 5′UTR contains an internal ribosome entry site (IRES) that mediates the cap-independent translation of a single large open reading frame encoding both structural and nonstructural viral proteins (2). The liver-specific microRNA (miRNA), miR-122, is an essential pro-viral host factor for HCV that binds to the 5′UTR in association with argonaute 2 (Ago2) protein (3–6). Mutational studies indicate that the binding of miR-122 to HCV RNA involves base-pairing of its seed sequence at two closely spaced sites (S1 and S2) in the RNA upstream of the IRES and close to its 5′ end, with additional 3′ supplemental base-pairing (Figure 1A) (3,4,7–9).
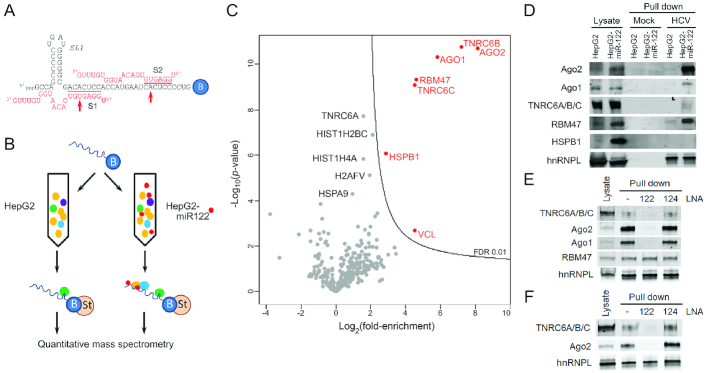
Figure 1.
TNRC6 proteins bind to HCV RNA in a miR-122-dependent fashion. (A) RNA bait used in pull-down reactions, showing base-pair interactions between miR-122 and the HCV 5′ UTR terminus. Seed sequence interactions at S1 and S2 are highlighted. Red arrows denote positions of p6m mutants. B = 3′ biotin tag. (B). Experimental scheme: the 3′ biotinylated RNA bait was bound to streptavidin beads and incubated with HepG2 or HepG2-miR-122 lysates. Proteins associated with the RNA bait were isolated and subjected to quantitative mass spectrometry. (C) Volcano plot of quantitative mass spectrometry results showing the 324 proteins that were confidently identified in (B) plotted according to their degree of enrichment in the pull-down products from the HepG2-miR122 versus miR-122-deficident HepG2 lysate: log2 (fold-enrichment) versus probability, -log10(P-value). The contour line indicates the enrichment FDR of 0.01. Proteins enriched with an FDR < 0.01 are labeled in red, whereas other proteins of potential interest are labeled in black. (D) Immunoblot confirmation of mass spectrometry results. Protein samples from lysates, mock pull-down (beads only) and RNA bait pull-downs were resolved by SDS-PAGE and blotted with indicated antibodies. TNRC6 proteins were detected with a pan-paralog antibody recognizing TNRC6A, 6B and 6C. hnRNPL binds RNA in a miR-122-independent fashion and was included as a control. (E) Pull-down assays were carried out as in (D) in HepG2-miR-122 lysate with or without prior addition of miR-122 LNA or control miR-124 LNA antagomirs. The anti-miR-122 oligo blocked the pull-down of Ago2, Ago1 and TNRC6 proteins but not RBM47. (F) Pull-down assays as in (E) were carried out with Huh-7.5 cell lysate.
The pro-viral host factor activity of miR-122 is unusual. miRNAs are small ∼22-nt RNAs that typically regulate eukaryotic gene expression post-transcriptionally, and that usually bind regions of imperfect complementarity in the 3′UTR of cellular mRNAs (10,11). miRNA silencing of gene expression requires loading of the ‘guide strand’ of a miRNA duplex into an Ago protein to form a miRNA-induced silencing complex (RISC). miRNAs target the binding of RISC complexes to specific mRNAs, resulting in translational repression and/or destabilization of the mRNA. In addition to Ago proteins, TNRC6A (trinucleotide-repeat containing gene 6A protein, otherwise known as GW182) and its two paralogs, TNRC6B and TNRC6C, are important for miRNA-induced gene silencing and serve scaffolding functions within RISC (12–15). The three TNRC6 proteins are members of the GW protein family, so named because they are rich in glycine-tryptophan repeats. The tryptophan-rich N-terminal Ago-binding domain of the TNRC6 proteins interacts with Ago (15–17), while the C-terminal domain, referred to as the ‘silencing domain’, coordinates downstream silencing processes, including interactions with poly(A)-binding protein and recruitment of the CCR4/NOT deadenylase complex, resulting in accelerated mRNA decay and/or suppression of cap-dependent translation (18–20).
In contrast to these canonical miRNA actions, the binding of miR-122 to the HCV genome stimulates replication of the virus. The mechanisms by which this occurs remain incompletely understood, but appear to be multiple (21,22). First, miR-122 stabilizes the positive-strand RNA genome by protecting its 5′ end from degradation mediated by the cytoplasmic 5′ exoribonuclease XRN1 (6,22–24) and the cellular pyrophosphatases, Dom3Z and DUSP11 (25,26). In addition, several studies suggest that miR-122 promotes HCV IRES activity (4,5,9,27,28), although this remains controversial. Increases in viral protein synthesis observed after transfecting infected cells with a duplex miR-122 mimic may result from stabilization of the viral RNA template (29,30), but two recent studies suggest that miR-122 may enhance IRES activity by shaping the folding of the 5′UTR (31,32). On the other hand, metabolic labeling experiments show such miR-122 supplementation leads to increases in viral RNA synthesis prior to detectable increases in viral protein synthesis (29). This suggests a direct effect on viral transcription, due possibly to changes in the circularization status of the positive-strand genome, or stabilization of the secondary structure of the negative-strand RNA (21,31). Other small RNAs can partially substitute for the binding of miR-122 in cell culture studies of HCV replication (32,33), but antagomirs that sequester endogenous miR-122 have potent antiviral effects in infected chimpanzees and human subjects, attesting to the importance of miR-122 as a pro-viral host factor (34,35).
Whatever the underlying mechanism(s) may be, the capacity of miR-122 to stimulate HCV RNA replication requires the involvement of an argonaute protein, primarily Ago2 (5,6,29), suggesting the formation of a RISC-like complex at the 5′ end of HCV RNA. Whether other host proteins are involved in this complex remains uncertain, however, including specifically proteins involved in miRNA silencing. Here, we describe the use of quantitative mass spectrometry to identify miR-122-dependent protein binding to a synthetic RNA bait representing the 5′ end of the HCV genome. We show that all three TNRC6 proteins, important components of the cellular RISC complex, bind to the 5′end of HCV RNA in a miR-122-dependent fashion and contribute functionally to replication of the viral genome by spatially regulating binding of miR-122/Ago2 to the 5′UTR.
MATERIALS AND METHODS
Cells and reagents
HepG2, HepG2-miR-122 (36) (obtained from Dr Matthew Evans, Icahn School of Medicine at Mount Sinai), Huh-7.5 and Huh-7.5-ΔmiR-122 (37) cells (obtained from Dr Charles Rice, Rockefeller University) were maintained in DMEM medium with 10% fetal bovine serum. Sofosbuvir (2′-deoxy-2′-α-fluoro-β-C-methyluridine-5′-monophosphate) was purchased from ChemScene, LLC (Monmouth Junction, NJ). Final dilutions contained 0.1% DMSO. Cell viability was assessed with WST-1 reagent (MilliporeSigma, Burlington, MA) using the manufacturer’s suggested protocol.
Plasmids
pHJ3-5/GLuc2A and the related S1-, S2- and S1-S2-p6m mutants, pH77s.3/Gluc2A, pJFH1-QL/Gluc2A and the HCV mini-genome plasmid, pHCVdc-GLuc, have been described previously (6,9,38). Plasmids expressing wild-type or mutant TNRC6B polypeptides (pYFP-T6B-wt and pYFP-T6B-mut) were kindly provided by Dr Gunter Meister (University of Regensburg, Germany). Plasmids expressing the 5′ and 3′ fragments of TNRC6B (pTNRC6B-5′F and pTNRC6B-3′F) were prepared by cloning the corresponding fragments from pFLAG/HA-TNRC6B plasmid (Addgene #10979) into pcDNA3 (Invitrogen). RNA transcripts were synthesized in vitro using the MegaScript T7 kit (Ambion, Foster City, CA) following the manufacturer’s protocol.
Transfections
siRNA pools targeting TNRC6A, 6B and 6C, a control (siCtrl) siRNA pool (Dharmacon, Lafayette, CO) and an additional single control siRNA (siCtrl2, Sigma-Aldrich) were transfected into cells with Lipofectamine RNAiMax reagent (Invitrogen, Carlsbad, CA) at 20 nM final concentration. miRNAs were synthesized by Dharmacon and transfected as miRNA/miRNA* duplexes at 50 nM final concentration, as described (4). In vitro transcribed HCV RNA (1.25 μg) was transfected into 1 × 106 Huh-7.5 cells using the TransIT mRNA kit (Mirus Bio, Madison, WI).
Biotinylated RNA pull-down
Commercially synthesized oligoribonucleotide representing nts 1–47 of the H77S strain HCV genome conjugated to biotin at its 3′ end (10 pmol per reaction) (Dharmacon) was heated at 75°C for 5 min, then cooled to room temperature. The annealed RNA bait was bound to magnetic streptavidin T1 beads (Invitrogen) following the manufacturer’s protocol, then incubated with HepG2, HepG2-miR-122 or Huh-7.5 cytoplasmic lysate for 1 h at 4°C. About 100 pmol of anti-miR-122 or anti-miR-124 locked nucleic acid oligonucleotide (Invitrogen, Carlsbad, CA) was added to the lysate where indicated. Proteins bound to the beads were digested by trypsin and subjected to mass spectrometry, or eluted with sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, resolved by SDS-PAGE and subjected to immunblot.
Label-free quantification (LFQ) of proteins by LC-MS/MS
Proteins were digested by trypsin on beads (39), and the desalted peptide mixtures were dissolved in 30 μl 0.1% formic acid (Thermo-Fisher), of which 5 μl peptides were injected and analyzed using an ultra2D nanoLC system (Eksigent Technologies, Dublin, CA) coupled to a Velos LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Peptides were first loaded onto a 2 mm × 0.5 mm reverse-phase (RP) C18 trap column (Eksigent) at a flow rate of 1 μl/min, then eluted and fractionated on a 25 cm C18 RP column (360 μm × 75 μm × 3 μm) with a 90 min gradient of 5–40% buffer B (ACN and 0.1% formic acid) at a constant flow rate of 250 nl/min. The Velos LTQ Orbitrap was operated in the positive-ion mode with a data-dependent automatic switch between survey Full-MS scan (m/z 300–1800, (externally calibrated to a mass accuracy of <5 ppm and a resolution of 60 000 at m/z 400) and CID MS/MS acquisition of the top 15 most intense ions.
Mass spectrometry data analysis
Mass spectral processing and peptide identification were carried out on the Andromeda search engine using MaxQuant software (Version 1.5.3.17, Max Planck Institute of Biochemistry, Munich, Germany) and the human UniProt database. All searches were conducted with a defined modification of cysteine carbamidomethylation, with methionine oxidation and protein amino-terminal acetylation as dynamic modifications. Peptides were confidently identified using a target-decoy approach with a peptide false discovery rate (FDR) of 1% and a protein FDR of 5%. A minimum peptide length of 7 amino acids was required, a maximum of two missed cleavages was allowed, initial mass deviation for precursor ion was set up to 7 ppm and the maximum allowed mass deviation for fragment ions was 0.5 Da. Data processing and statistical analysis were carried out using Perseus software (Version 1.5.5.1, Max Planck Institute of Biochemistry) (40). Protein quantification was performed on triple biological replicate runs, each with three technical replicates (Supplementary Figure S2 and Supplementary Table S1). Two-sample t-test statistics were used to assess the significance of differences in means of the label-free quantification (LFQ) intensities of peptides present in pull-down samples from HepG2 versus HepG2-miR-122 lysates, with an FDR <1% considered significant.
Real-time RT-PCR
To quantify HCV RNA, cDNA was produced by reverse transcription of RNA using random primers and SuperScript III Reverse Transcriptase (Invitrogen). qPCR analysis was carried out using iTaq SYBR Green Supermix with the CFX96 System (Bio-Rad, Hercules, CA). HCV RNA abundance was determined by reference to a standard curve using polymerase chain reaction (PCR) primers targeting the HCV 5′UTR (5′-catggcgttagtatgagtgtcgt-3′ and 5′-ccctatcaggcagtaccacaa-3′) and normalized to the abundance of β-actin mRNA where indicated (primers: 5′-gtcaccggagtccatcacg-3′ and 5′-gacccagatcatgtttgagacc-3′). HCV mini-genome RNA was amplified by primers: 5′-cacgcccaagatgaagaagt-3′ and 5′-gaacccaggaatctcaggaatg-3′.
Immunoblots
Immunoblotting was carried out using standard methods with the following monoclonal antibodies (mAbs): mouse anti-β-actin (AC-74, 1:2000, Sigma-Aldrich, St. Louis, MO), rat anti-Ago2 (11A9, 1:1000, Sigma-Aldrich), mouse anti-hnRNPL (4D11, 1:2000, Millipore), rabbit anti-RBM47 (EPR9658, 1:1000, Abcam, Cambridge, United Kingdom), mouse anti-HSPB1 (G3.1, 1:1000, Invitrogen) and rat anti-TNRC6 proteins (7A9, 1:10, kindly provided by Dr Gunter Meister, University of Regensburg, Germany). Protein bands were visualized with an Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE).
Gaussia luciferase (GLuc) HCV replication assay
TNRC6 proteins were depleted by transfecting Huh-7.5 cells with specific siRNAs or scrambled siCtrl, followed 48 h later by re-transfection with HJ3-5/GLuc RNA. Cell culture supernatant fluids were collected at intervals over 72–96 h and cells refed with fresh media. Secreted GLuc activity was measured as described (6).
HCV RNA stability assay
Huh-7.5 cells stably infected with HJ3-5 virus were transfected with siRNAs targeting TNRC6 or siCtrl siRNA, then 48 h later treated with 10 μM sofosbuvir. Cells were harvested at 0, 4, 8 and 12 h post-treatment, and residual HCV RNA quantified by reverse transcription and quantitative polymerase chain reaction (RT-qPCR) and normalized to β-actin mRNA abundance.
HCV translation assay
Huh-7.5 cells were transfected with siRNAs targeting TNRC6B/C or scrambled siCtrl, then 48 h later re-transfected with pHCVdc-GLuc mini-genome RNA. GLuc activities were measured in supernatant fluids at 6 and 24 h, and relative reporter RNA abundance determined in cell lysates at 24 h by RT-qPCR.
Nascent HCV RNA synthesis assay
Nascent RNA transcripts were quantified using the Click-iT Nascent RNA Capture Kit (Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol with some modifications. Cells were pulsed with 0.5 mM 5-ethynyl uridine (5-EU) for 8 h to incorporate 5-EU into newly synthesized RNA. Cells were then washed with phosphate-buffered saline (PBS) and total RNA extracted using the RNeasy Mini Kit (Qiagen). Five microgram of total RNA and 0.5 mM biotin azide were used in a copper-catalyzed click reaction to conjugate biotin to 5-EU-labeled RNA. Following precipitation, the RNA was dissolved in 50 μl of RNase-free water. Biotin-conjugated 5-EU-labeled RNA was isolated from 3 μg total RNA on streptavidin magnetic beads and used as a template for quantitative RT-PCR (see above). Nascent HCV RNA was normalized to the total amount of HCV RNA used to capture nascent RNA transcripts on streptavidin magnetic beads, and is expressed as ‘nascent/total HCV RNA’.
Ago2-RNA co-immunoprecipitation
Five million Huh-7.5 cells were electroporated with 5 μg HCV RNA and seeded onto a 10 cm dish. Five hours later, cells were harvested in lysis buffer [150 mM KCl, 25 mM Tris–HCl pH 7.4, 5 mM ethylenediaminetetraacetic acid (EDTA), 1% Triton X-100, 5 mM Dithiothreitol (DTT), Complete protease inhibitor cocktail (Roche, Basel, Switzerland), 100 U/ml RNaseOUT (Invitrogen)]. Lysates were centrifuged, and supernatants were incubated with murine anti-Ago2 mAb (1B1-E2H5, 1:100, MBL International, Woburn, MA) at 4°C for 2 h, followed by addition of 30 μl of Protein G Sepharose (50% Slurry, GE Healthcare, Chicago, IL) for 1 h. The Sepharose beads were washed three times in wash buffer [1× TBS, 1.2% Triton X-100, 5 mM DTT, Complete protease inhibitor cocktail (Roche), 80 U/ml RNaseOUT (Invitrogen)] and RNA extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany).
Confocal fluorescence microscopy
Huh-7.5 cells were transfected with HJ3-5 HCV RNA for 72 h and seeded into four-well glass chamber slides at 1 × 105 cells/well. Cells were fixed 24 h later for 15 min with 4% paraformaldehyde and permeabilized with 0.25% Triton X-100 in PBS for 15 min at room temperature. The cells were immunostained with murine anti-NS5A mAb (9E10, 1:1000, kindly provided by Dr Charles Rice, Rockefeller University) or J2 anti-dsRNA antibody (Scicons, Budapest), and the rat anti-TNRC6 mAb 7A9 (1:10) for 2 h at room temperature, followed by incubation with fluorescent secondary antibodies (Invitrogen). Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) to label nuclei and mounted in ProLong Gold antifade reagent (Life Technologies). Confocal microscopic images were obtained using an Olympus FV1000 Multiphoton Laser-scanning Confocal Microscope (Leica, Wetzlar, Germany) and analyzed using the open platform ImageJ software.
Statistical analysis
Statistical significance (P < 0.05 or P < 0.01) was determined by two-sided t-test or ANOVA, as described in the legends to figures. Calculations were done using Prism 6 for Mac OS X by GraphPad Software (La Jolla, CA), version 6.h.
RESULTS
TNRC6 proteins bind the 5′ terminal HCV RNA sequence in a miR-122-dependent fashion
We demonstrated previously that miR-122/Ago2 complexes bind to a biotinylated synthetic RNA bait corresponding to the 5′ terminal 47 nucleotides of the positive-strand HCV RNA genome (Figure 1A) (42). To identify other host proteins that might be present in this complex, we used this biotinylated RNA bait to pull-down proteins from lysates of HepG2 cells, or HepG2-miR-122 cells that are engineered to constitutively express miR-122 (36) (Figure 1B). Although derived from human hepatocytes, HepG2 cells do not express detectable levels of endogenous miR-122 and are therefore not permissive for HCV replication, whereas HepG2-miR-122 cells support HCV replication (36) (Supplementary Figure S1A).
Label-free quantitative mass spectrometry identified a total of 324 host proteins binding to the RNA bait in lysates from these cells, with high reproducibility between independent pull-down experiments (Supplementary Figure S2 and Supplementary Table S1). To identify those proteins with greater binding to the RNA bait in lysates from HepG2 cells expressing miR-122 versus lysates of miR-122-deficient HepG2 cells, we compared the mean LFQ intensities of peptides identified in pull-down products from each. This analysis revealed that seven proteins were significantly enriched with an FDR < 0.01 in pull-down products from HepG2-miR-122 versus HepG2 cell lysate, and thus likely to have bound the bait in a manner dependent upon miR-122 expression (Figure 1C). As expected, Ago2 was highly enriched (∼200-fold, more than any other protein) in the HepG2-miR-122 pull-down products compared to HepG2 pull-down samples (Figure 1C). Ago1 also showed significant although lesser enrichment (∼60-fold). Ago3, while present, was not significantly enriched, whereas Ago4 was not detected. Only a limited number of other proteins were significantly enriched in the HepG2-miR-122 pull-down products, among which were the TNRC6 proteins, TNRC6B and TNRC6C (Figure 1C). TNRC6A (also known as GW182) was enriched about 3-fold in the pull-down samples from HepG2-miR122 lysate, but did not achieve an FDR < 0.01 (P-value < 10−4, Supplementary Table S1). TNRC6B peptides were significantly more abundant than either TNRC6A or TNRC6C peptides in the HepG2-miR-122 sample (Supplementary Figure S1B). Since prior studies show TNRC6A mRNA transcripts are more abundant than either TNRC6B or TNRC6C transcripts in HepG2 cells (Supplementary Figure S1C), this suggests a possible preference for TNRC6B protein associating with the RNA bait. As described above, TNRC6 proteins are known to interact with Ago proteins and are important for canonical miRNA functions (17,43). The association of these proteins with an RNA bait representing the 5′ end of the HCV genome in cells expressing miR-122 thus suggests that they may also function in miR-122 stimulation of HCV replication. RNA-binding motif protein 47 (RBM47) and heat shock protein family B member 1 (HSPB1) were also enriched in the HepG2-miR-122 sample, as were several histone proteins (Figure 1C).
Immunoblotting was done to confirm the identity of proteins that were enriched in the pull-down products from lysates of HepG2 cells expressing miR-122 (Figure 1D). Whereas little if any Ago2 was detected in the HepG2 pull-down, much larger amounts of Ago2 were pulled down from the HepG2-miR-122 lysate, supporting the miR-122-dependent nature of its association with the bait. Immunoblotting also confirmed that greater amounts of Ago1, TNRC6 and RBM47 were pulled down by the RNA bait from lysates of cells expressing miR-122. More Ago2 than Ago1 was bound to the bait, consistent with the proteomics analysis (Figure 1C) and previous studies showing Ago2 is more active than Ago1 in supporting miR-122 stabilization of the RNA genome (6). Which TNRC6 paralogs were bound by the bait could not be determined due to a lack of paralog-specific antibodies (44), but TNRC6 was detected only in immunoblots of the HepG2-miR-122 pull-down products. RBM47 was also enriched in the HepG2-miR-122 pull-down, but in contrast to the Ago proteins or TNRC6, this enrichment appeared to result from an increased abundance in the HepG2-miR-122 versus HepG2 lysate (Figure 1D), suggesting an indirect effect related to miR-122 expression. Conversely, HSPB1 was not detected in either pull-down, despite a high abundance in the lysate. hnRNP L protein, which binds the 5′UTR RNA independently of miR-122 (42), was detected equally in immunoblots of the HepG2 and HepG2-miR-122 pull-down samples (and in the proteomics analysis, Supplementary Table S1).
To assess whether protein binding to the RNA bait was directly dependent on miR-122, we treated HepG2-miR-122 lysates with an anti-sense miR-122 locked nucleic acid (LNA) oligonucleotide antagomir (or an LNA targeting miR-124 as a control for specificity) prior to streptavidin pull-down. As expected, the miR-122 antagomir inhibited binding of Ago2, Ago1 and TNRC6 proteins to the RNA, whereas the miR-124 antagomir was without effect (Figure 1E). In contrast, RBM47 binding was not reduced by the miR-122 antagomir, indicating that it occurs independently of miR-122. Because Huh-7.5 cells are commonly used to study HCV replication, we carried out similar pull-down experiments using Huh-7.5 cell lysate. These cells endogenously express a moderately high abundance of miR-122. As with HepG2-miR-122 lysates, TNRC6 proteins were detected in pull-down products from Huh-7.5 cell lysate, and reduced by prior treatment with the miR-122 but not miR-124 antagomir (Figure 1F). The miR-122 antagomir blocked the binding of Ago2 and TNRC6 proteins, but not hnRNPL. Collectively, these data show TNRC6 proteins are recruited to the 5′ end of the HCV genome in a miR-122-dependent fashion.
TNRC6 proteins are important and functionally redundant in HCV replication
Next, we asked whether TNRC6 proteins have a role in HCV replication. Individual TNRC6 paralogs were depleted by transfecting Huh-7.5 cells with specific siRNAs, and replication of a transfected recombinant HCV RNA genome expressing a Gaussia princeps luciferase (GLuc) reporter (HJ3-5/GLuc) (6) monitored over 72 h. Depletion of individual TNRC6 paralogs, confirmed by real-time RT-PCR, had little effect on replication of the RNA (Figure 2B and Supplementary Figure S3A). As TNRC6 paralogs may be functionally redundant, we similarly depleted two or more of the paralogs simultaneously (Figure 2C,D and Supplementary Figure S3B). Doubly depleting either TNRC6A/B or TNRC6A/C resulted in a small (∼20%), but statistically significant (P < 0.0001) reduction in HJ3-5/GLuc2A replication, whereas depleting TNRC6B/C or TNRC6A/B/C resulted in a larger, 50% or greater reduction (Figure 2C). In similar experiments, we confirmed the importance of TNRC6 proteins in cells infected with cell-free HJ3-5 virus, showing that depletion of TNRC6B/C or TNRC6A/B/C led to a significant reduction in viral RNA abundance (Figure 2E). The HJ3/5 and HJ3-5/GLuc viruses are chimeras with the 5′ part of the genome extending to the NS2/NS3 junction derived from the genotype 1a H77 virus and the remainder derived from the genotype 2a JFH1 virus (Supplementary Figure S3C) (45). TNRC6 depletion had negative effects on replication of both parental H77S and JFH1 GLuc reporter viruses (Supplementary Figure S3D), indicating that the pro-viral function of TNRC6 is not limited to a single HCV genotype. Importantly, TNRC6 depletion did not significantly alter cell growth (Supplementary Figure S3E) or Ago2 protein abundance (Figure 2D).
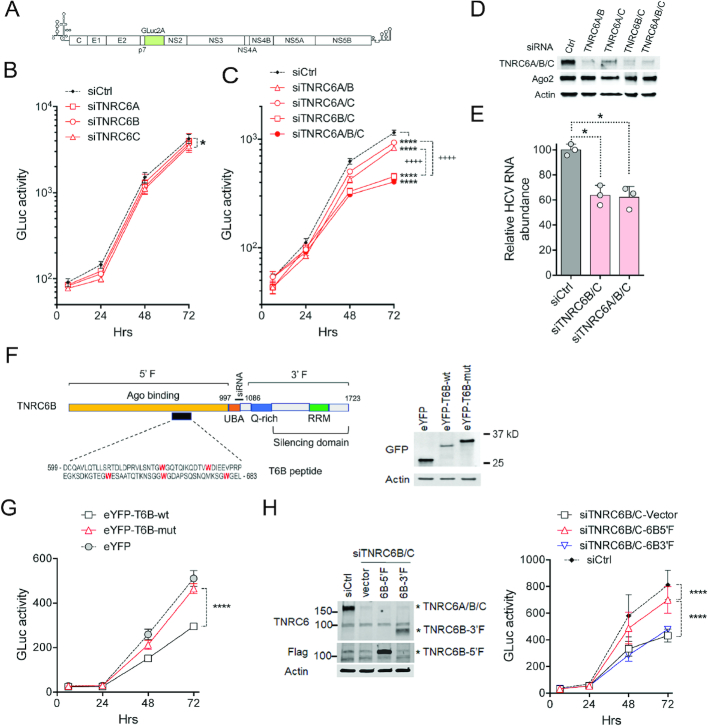
Figure 2.
TNRC6 proteins are required and functionally redundant for optimal HCV replication. (A) HJ3-5/GLuc2A reporter virus genome, showing GLuc2A insertion between p7 and NS2A coding regions. (B) HCV reporter virus replication measured by GLuc secretion in Huh-7.5 cells transfected with siRNAs depleting single TNRC6 paralogs (see Supplementary Figure S3A). Results shown are means of n = 3 technical replicates ± s.d., and are representative of two independent experiments. *TNRC6C versus Ctrl siRNAs P ≤ 0.029 at 72 h by two-way ANOVA with Sidak’s multiple comparison test. (C) Reporter virus replication in cells depleted of two or more TNRC6 proteins (see Supplementary Figure 3A). Results shown are means of three technical replicates ± s.d. and are representative of two independent experiments. ****TNRC6 versus Ctrl siRNAs, P ≤ 0.0001; ++++TNRC6B/C versus TNRC6A/B or TNRC6A/C siRNAs, at 48 and 72 h by two-way ANOVA with Sidak’s multiple comparison test. (D) Immunoblots of TNRC6 proteins 48 h after siRNA transfection. Ago2 protein was blotted for comparison. β-Actin was a loading control. (E) Cell-associated HCV RNA (normalized to β-actin mRNAabundance) 72 h after infection of TNRC6-depleted Huh-7.5 cells with cell-free HJ3-5 virus at a multiplicity of 0.1. HCV RNA abundance in siCtrl cells was arbitrarily set to 100. n = 3 from two independent experiments, one with two technical replicates (shaded symbols). *P < 0.05 versus Ctrl siRNA (adjusted P = 0.0178 for TNRC6B/C, and 0.0384 for TNRCA/B/C) by one-way ANOVA with Dunnett’s correction for multiple comparisons. (F) Left panel: Domain structure of TNRC6B with the amino-terminal fragment (5′F), carboxy-terminal fragment (3′F), T6B polypeptide segment indicated. The site targeted by siTNRC6B is also shown. Right panel: Immunoblot showing the expression of control eYFP and indicated eYFP-T6B polypeptides. (G) Reporter virus replication in Huh-7.5 cells overexpressing the indicated proteins. Results shown are means of three biological replicates ± s.d. ****P<0.0001 by two-way ANOVA with Sidak’s multiple comparison test. (H) Left panel: Immunoblots showing the levels of endogenous TNRC6A/B/C and exogenously expressed TNRC6B-5′F and 3′F. TNRC6B-5′F is not recognized by TNRC6 antibody but can be detected by Flag antibody. Right panel: Reporter virus replication in cells transfected with control siRNA or TNRC6B/C siRNAs and overexpress indicated proteins. Results shown are means of three experiments ± s.d. ****P<0.0001 by two-way ANOVA with Sidak’s multiple comparison test.
TNRC6 proteins are known to recruit the CCR4/NOT deadenylase complex, which plays an important role in miRNA-mediated gene silencing (18,20). Since the HCV RNA genome lacks a 3′ poly(A) tail, the CCR4/NOT deadenylase would not be expected to be required for TNRC6 to function in HCV replication. Consistent with this, the CCR4/NOT deadenlyase was not identified in the HCV RNA pull-down experiments (Figure 1C and Supplementary Table S1). RNAi-mediated depletion of CNOT1, an important scaffolding component in the CCR4/NOT complex, also had no effect on HCV replication (Supplementary Figure S3F).
These data suggest that TNRC6 proteins, particularly TNRC6B and TNRC6C, are required for and contribute redundantly to optimal replication of the HCV genome. The greater importance of TNRC6B and TNRC6C in replication is consistent with the greater enrichment of these paralogs versus TNRC6A in the HepG2-miR-122 pull-down experiments (Figure 1C), and provides further support for paralog-specific differences in HCV host factor activity. We thus focused on TNRC6B and TNRC6C in subsequent experiments.
HCV replication is suppressed by disrupting the interaction of TNRC6 with Ago proteins
The interaction between TNRC6 and Ago proteins is well characterized (16,17,46,47). To determine whether it is required for TNRC6 to facilitate HCV replication, we assessed the replication of the virus in Huh-7.5 cells in which the interaction was disrupted by overexpressing an Ago2-binding TNRC6 polypeptide, T6B, fused to yellow fluorescent protein (eYFP). The T6B polypeptide sequence corresponds to amino acids 599–683 of TNRC6B (Figure 2F, left), and contains a strong Ago2-binding motif in which the five tryptophan residues within the motif are important for the interaction (48). We expressed both the wild-type (T6B-wt) and a mutant (T6B-mut) version of this TNRC6 sequence in which the five tryptophan residues were substituted with alanine (Figure 2F, right). As shown in Figure 2G, overexpressing either eYFP alone or eYFP-T6B-mut had no impact on viral replication, whereas expressing the wild-type eYFP-T6B significantly suppressed viral replication. This result suggests that the interaction of TNRC6 with Ago proteins is likely to be important for HCV replication, and that the T6B peptide may act as a dominant negative suppressor of this function.
To further assess the potential requirement for a direct TNRC6–Ago interaction, we expressed the amino-terminal half of TNRC6B (6B-5′F, aa 1–997), which contains the Ago-binding domain, in cells transfected with siRNAs targeting TNRC6B/C (Figure 2H, left). Expression of this fragment partially reversed the reduction in HCV replication caused by TRNRC6B/C depletion (Figure 2H, right), providing additional support for the interaction between Ago2 and TNRC6 functionally contributing to viral replication. In contrast, expressing the complementary carboxy-terminal fragment of TNRC6B (6B-3′F, aa 1086–1723), which contains the silencing domain, had no effect on viral replication. Notably, the siRNA used for TNRC6B in this experiment targeted the region 1033–1040 (Figure 2F), such that expression of neither TNRC6B fragment was targeted by the siRNA.
TNRC6 proteins facilitate miR-122 stimulation of viral RNA synthesis but not viral RNA stability or IRES activity
To confirm that the depletion of TNRC6 suppresses HCV replication by directly reducing the viral host factor activity of miR-122, and not indirectly by globally suppressing miRNA silencing and thus altering the cellular transcriptome, we studied a previously described HCV variant, NPHV-UUGGCG (49) (previously called NPHV-delta-UUGGCG), which replicates in a miR-122-independent manner. The 5′ end of the genome of this virus has been replaced with corresponding sequence from a nonprimate hepacivirus (NPHV) in which the miR-122 seed sequence binding site was mutated to UUGGCG (Figure 3A). This virus has no miR-122-binding site near the 5′ end of its genome, yet can still replicate in Huh-7.5 cells (49). In contrast to the replication of HJ3-5 virus, which has the canonical 5′ terminal sequence of HCV and was suppressed by depletion of TNRC6 proteins, knocking down TNRC6B/C or TNRC6A/B/C did not impair and in contrast slightly increased the replication of NPHV-UUGGCG, as monitored by quantitative immunoblot analysis of viral core protein expression (Figure 3B). This result confirms that the pro-viral activity of TNRC6 proteins is linked directly to the viral host factor activity of miR-122.
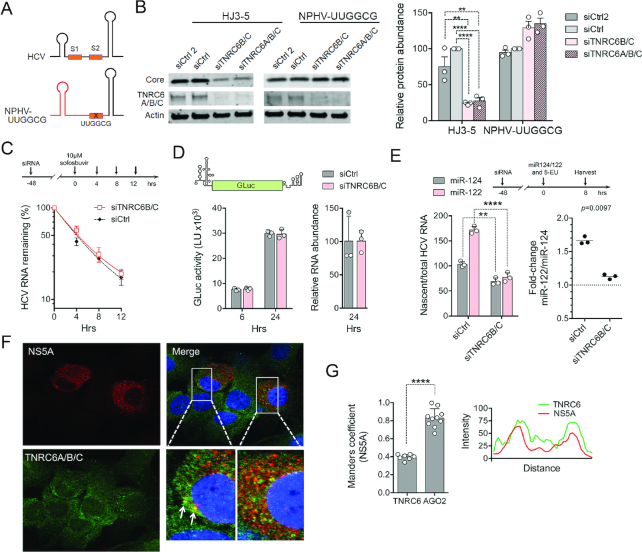
Figure 3.
TNRC6 promotes miR-122 augmented viral RNA synthesis but not HCV RNA stability or IRES activity. (A) Schematic representation of the 5′ terminal sequences of HCV and NPHV-UUGGCG viruses, with sequence from NPHV shown in red. (B) Huh7.5 cells were transfected with siCtrl (a pool of four nontargeting siRNAs), siCtrl2 (an additional single nontargeting siRNA) or the indicated TNRC6-targeting siRNAs, and 48 h later re-transfected with viral RNA. Viral and cellular protein abundance was analyzed by immunoblot after an additional 72 h. HCV core protein abundance was quantified relative to actin abundance in three independent experiments, with results shown on the right as means ± s.d. after normalizing to the siCtrl control. ****adjusted P-value < 0.0001, **adjusted P-value 0.0011–0.0019 by two-way ANOVA with Sidak’s test for multiple comparisons. (C) Positive-strand HCV RNA stability in cells following sofosbuvir arrest of viral RNA synthesis. HCV RNA level at 0 h was arbitrarily set to 100%. Data are means from three independent experiments ± s.e.m. Weighted R2 = 0.975–0.987 when fit to a single-phase decay model, with no significant difference in the decay rate, k, by extra sum-of-squares F test (P > 0.05). (D) HCV IRES activity. TNRC6-depleted Huh-7.5 cells were transfected with HCV mini-genome RNA containing GLuc sequence flanked by viral 5′ and 3′ UTRs. (Left) GLuc activities were measured in supernatant fluids, and (right) relative reporter RNA abundance determined in cell lysates (siCtrl cells arbitrarily set to 100). Data are means from two independent experiments, one with two technical replicates (shaded symbols) ± s.d. (E) Nascent HCV RNA synthesis in TNRC6-depleted cells. (Top) Stably infected Huh-7.5 cells were transfected with indicated siRNAs, and 48 h later re-transfected with miR-122, or the control miR-124, and 5-ethynyluridine (5-EU) added to the medium. Cells were harvested 8 h later for the isolation of nascent RNA. (Bottom left) The relative rate of viral RNA synthesis was calculated as nascent HCV RNA divided by total HCV RNA, with viral RNA synthesis in siCtrl control cells transfected with miR-124 arbitrarily set to 100. **P < 0.01, ****P < 0.0001 by two-way ANOVA with Sidak’s multiple comparison test. (Bottom right) miR-122-induced fold-change in viral RNA synthesis relative to that in control cells transfected with miR124. P = 0.0097 by two-sided paired t-test. Data are means of three independent experiments ± s.d. (F) Confocal fluorescence microscopy of Huh-7.5 cells infected with HJ3-5 virus and stained with antibodies to NS5A (red) and TNRC6 (green). Nuclei counterstained with DAPI (blue) in the merged image. Arrows denote puncta with co-localization of NS5A and TNRC6 proteins. (G) Quantitative analysis of confocal microscopy images. (Left) Manders’ coefficient for co-localization of TNRC6 proteins and NS5A and, for comparison, Ago2 and NS5A in similar images (see Supplementary Figure S4). ****P < 0.0001 by two-sided t-test, n = 7–10 cells. (Right) Linear trace of TNRC6 and NS5A fluorescent intensities bisecting the two puncta depicted by arrows in panel (F).
As described above, miR-122 has been implicated in several aspects of the HCV life cycle, including viral RNA stability, viral translation and viral RNA synthesis (6,27,29). We thus investigated whether TNRC6 proteins are involved in these processes. To assess stabilization of the RNA, miR-122 replete Huh-7.5 cells persistently infected with HCV were transfected with siRNAs targeting TNRC6 expression, and then treated with a potent and specific nucleoside inhibitor of the NS5B HCV polymerase to block viral RNA synthesis (6). TNRC6 depletion had no significant impact on the subsequent decay of viral RNA monitored by real-time RT-PCR (Figure 3C). To assess IRES-mediated viral translation, we transfected cells with an HCV mini-genome reporter in which the GLuc sequence is flanked by the viral 5′ and 3′ UTRs. There was no difference in secreted GLuc activity or intracellular reporter RNA abundance following depletion of TNRC6 (Figure 3D), suggesting that TNRC6 proteins do not modulate viral IRES activity.
Next, we analyzed the role of TNRC6 proteins in viral RNA synthesis. Stably infected Huh-7.5 cells were transfected with TNRC6-targeting siRNAs for 48 h, then re-transfected with duplex miR-122 or an irrelevant miRNA, miR-124, as control (Figure 3E, top). The cells were then incubated with 5-ethynyl uridine (5-EU) for 8 h to metabolically label nascent RNA, as described previously (29). 5-EU-labeled RNAs were coupled to biotin in a ‘click’ reaction, isolated on streptavidin beads and nascent HCV RNA quantified by RT-qPCR. Newly synthesized viral RNA, normalized to total HCV RNA abundance, was reduced ∼40% following depletion of TNRC6 proteins (Figure 3E, bottom left). Importantly, whereas the transfection of control cells with miR-122 resulted in a 60–70% increase in viral RNA synthesis compared to those transfected with miR-124, as described previously (29), this increase was ablated in TNRC6-depleted cells (Figure 3E, bottom right). These results were highly significant statistically in replicate experiments. Consistent with a role for TNRC6 in HCV RNA transcription, confocal fluorescent microscopy demonstrated co-localization of TNRC6 proteins with NS5A, an essential component of the HCV replication complex (Figure 3F). This was confirmed by quantitative image analysis, although co-localization was only partial and less than that observed between Ago2 and NS5A (Figure 3G and Supplementary Figure S4A). Similar results were obtained using antibody to double-stranded RNA (dsRNA) as a marker for replication complexes (Supplementary Figure S4B and C). In uninfected cells, TNRC6 proteins were diffusively distributed within the cytoplasm, with occasional foci which may be P-bodies. Taken together, these data suggest that TNRC6 proteins play an essential role in miR-122-augmented viral RNA synthesis, but not viral RNA stability or IRES-mediated translation.
TNRC6 proteins function differently when bound to HCV RNA at S1 versus S2
There are two tandem miR-122 seed sequence-binding sites (S1 and S2) at the 5′ end of the HCV genome (Figure 1A). Although both are required for optimal replication, the contributions of the individual sites differ, with mutations that ablate S1 having a greater impact on production of infectious virus (4). The requirements for miR-122 binding also differ (50). To determine whether similar differences exist in TNRC6 utilization, we studied reporter viral RNAs that contain A to U substitutions at positions 24 (S1p6m mutation) or 39 (S2p6m) that base-pair with nucleotide 6 of miR-122 in the seed sequence at S1 or S2 (Figure 1A) (4). When transfected into Huh-7.5 cells, RNA with the S1p6m mutation recruits miR-122/Ago2 primarily if not exclusively to S2, while the S2p6m RNA binds miR-122/Ago2 only at S1 (3,4). The S1p6m reporter virus (S1p6m-GLuc) was minimally competent for replication in TNRC6 replete cells, whereas S2p6m-GLuc replicated much more robustly (Figure 4A). This suggests that miR-122/Ago2 is incapable of driving robust replication of the virus when bound only at S2, consistent with the greater importance of miR-122 binding to S1 versus S2 for RNA replication noted previously (4). Surprisingly, however, TNRC6 depletion significantly boosted the replication of S1p6m-GLuc (which binds the complex at S2), while only minimally impairing the replication of S2p6m-GLuc (which binds miR-122/Ago2 at S1) (Figure 4A). Thus, eliminating complex binding at S1 reversed the pro-viral effect of TNRC6 and revealed that TNRC6 can restrict replication by interacting with miR-122/Ago2 at S2 in the absence of a competent S1 site.
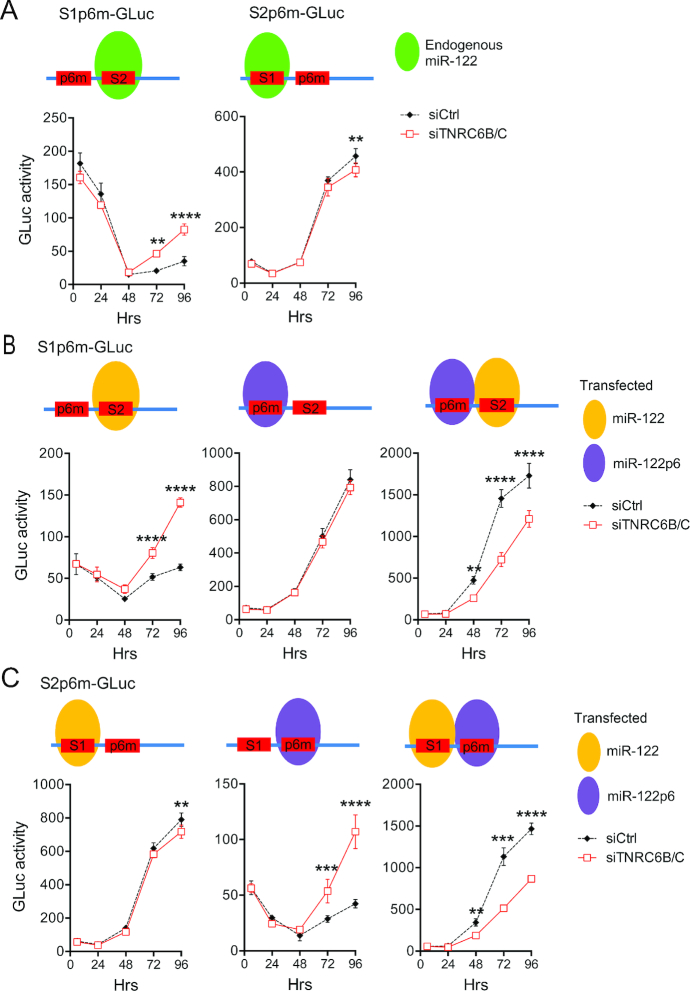
Figure 4.
TNRC6 proteins function differently when bound at S1 versus S2. (A) Huh-7.5 cells were transfected with the indicated siRNAs for 48 h and then re-transfected with S1p6m-GLuc or S2p6m-GLuc HCV RNA. Site-specific binding of endogenous miR-122/Ago2 complexes (green) to wild-type or p6 mutant (p6m) seed sequence binding sites is indicated at the top. Replication was assessed by measuring GLuc activities in supernatant fluids. (B) TNRC6B/C-depleted Huh-7.5-ΔmiR-122 cells lacking endogenous miR-122 expression were transfected with S1p6m-GLuc HCV RNA and the indicated microRNAs (miR-122, miR-122p6 or miR-122/miR-122p6 together). Binding of miR-122/Ago2 (yellow) or miR-122p6/Ago2 (magenta) complexes to the HCV RNA is shown at the top. Replication was assessed by measuring GLuc activities in supernatant fluids. (C) S2p6m-GLuc RNA replication under conditions similar to those in panel (B). In each panel, data shown represent the mean of n = 3 biological replicates ± s.d. Results were considered significant if the P-value was < 0.05 as determined by two-way ANOVA with Sidak’s multiple comparison test: **P < 0.01, ***P < 0.001, ****P < 0.0001.
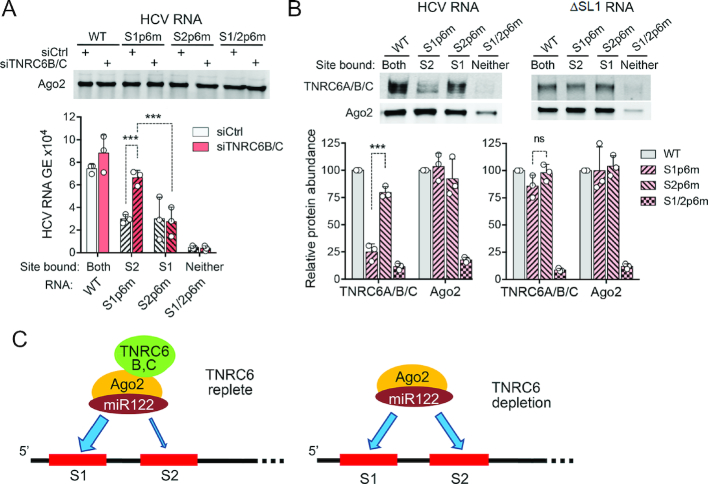
To confirm this surprising result, we carried out similar experiments in CRISPR/Cas9-edited Huh-7.5 cells lacking miR-122 expression (Huh-7.5-ΔmiR-122 cells) (37)). To specifically target miR-122 to the S1 or S2 site, or both, we co-transfected S1p6m-GLuc HCV RNA with duplex miR-122 or miR-122p6, which contains a nucleotide substitution in its seed sequence rendering it complementary to the mutant S1p6m- and S2p6m-binding sites (Figure 4B) (4). Again, we observed a striking increase in HCV replication upon depletion of TNRC6 proteins when the S2 site is predominantly bound by miR-122 (Figure 4B, left panel), and little or no effect when only the S1 site was bound by miR-122p6 (Figure 4B, middle panel). Importantly, when both sites were occupied, TNRC6 depletion reduced HCV replication (Figure 4B, right panel), as it did with virus containing the wild-type 5′UTR miR-122 interaction domain (Figure 2C and E). Similar results were obtained with a reporter virus containing the p6 mutation in the S2 site, S2p6m-GLuc (Figure 4C). Collectively, these data show that the miR-122-dependent binding of TNRC6 involves both the S1 and S2 sites within the 5′UTR, but with different functional consequences at the two sites.
TNRC6 proteins spatially regulate the binding of Ago2 to the 5′UTR
Next, we asked whether the contrasting effects of TNRC6 binding at S1 versus S2 reflect differences in miR-122/Ago2 complexes bound at these sites. We immunoprecipitated Ago2 from lysates of Huh-7.5 cells that had been electroporated with viral RNAs containing the wild-type S1 and S2 sites (‘WT’), S1p6m, S2p6m or S1/2p6m mutations (Figure 5A, top), then quantified HCV RNA associated with the Ago2 by RT-qPCR. S1/2p6m RNA, with p6 mutations at both S1 and S2, did not bind miR-122/Ago2 complexes, resulting in little viral RNA pull-down, whereas WT HCV RNA was efficiently co-precipitated with Ago2 (Figure 5A, bottom). Lesser but still substantial quantities of S1p6m and S2p6m RNAs were detected in Ago2 precipitates. Importantly, TNRC6 depletion led to a >2-fold increase in Ago2-associated S1p6m RNA (Figure 5A). A lesser, statistically insignificant increase was observed with WT RNA, whereas TNRC6 depletion had no discernible effect on Ago2 binding to S2p6m RNA. This is consistent with the increased replication of S1p6m RNA but not S2p6m RNA in TNRC6-depleted cells (Figure 4A). Thus, in the absence of TNRC6, there is a relative increase in the miR-122/Ago2 complex bound at S2, indicating that TNRC6 proteins skew the binding of miR-122/Ago2 complexes toward S1.
Figure 5.
TNRC6 binding is biased to S1 and regulates Ago2 binding at S2. (A) Ago2-associated HCV RNA in TNRC6-replete or TNRC6-depleted cells. Huh-7.5 cells were transfected with siCtrl or siTNRC6B/C, then 48 h later electroporated with wild-type (WT) or mutant HCV RNAs, and harvested 5 h later for Ago2 immunoprecipitation. (Top) Immunoblots of immunoprecipitated Ago2 protein. (Bottom) Ago2-associated HCV RNA quantified by real-time RT-qPCR. Data are means from three independent experiments ± s.d. ***P = 0.0004 by two-way ANOVA with Sidak’s multiple comparison test. (B) Biotinylated HCV or ΔSL1 RNA baits with WT or mutated miR-122 seed sequence binding sites were used to pull-down proteins from Huh-7.5 cell lysates as in Figure 1F, followed by immunoblotting for Ago2 and TNRC6 proteins. Quantification of results from three independent experiments is shown below after normalizing protein abundance to WT RNA. ***P = 0.0005 by t-test with Holm–Sidak correction for multiple comparisons. (C) Proposed model for TNRC6-mediated spatial regulation of miR-122/Ago2 binding to HCV RNA, showing skewing of complexes to S1 in TNRC6-replete cells.
Since these results indicate that TNRC6 proteins differentially regulate the binding of miR-122/Ago2 complexes to S1 versus S2, we asked whether TNRC6 proteins are also differentially associated with the two sites. To answer this question, we immunoblotted proteins that co-precipitated from Huh-7.5 cell lysates with biotinylated HCV RNA 5′ end baits similar to that used in the proteomics studies in Figure 1A, but with p6 mutations at S1 or S2. As expected, the wild-type (WT) bait efficiently pulled down both Ago2 and TNRC6 proteins, whereas less Ago2 and no detectable TNRC6 protein was associated with the S1/2p6m bait in which both sites are ablated (Figure 5B left). S1p6m and S2p6m baits pulled down similar abundances of Ago2, but S1p6m pulled down much less TNRC6 protein than S2p6m, indicating that TNRC6 proteins are preferentially associated with miR-122/Ago2 complexes bound at S1.
This result is surprising, considering that TNRC6 can pre-assemble with Ago2 before binding to a target RNA (17,47). However, the S1 site differs from S2 in that the bases pairing with the miRNA seed sequence and those forming 3′ supplemental base pair interactions are separated by a stem–loop structure (SL1, Figure 1A). To test whether this stem–loop structure might contribute to the differential binding of TNRC6, we deleted it from the biotin-RNA baits (ΔSL1 mutants) and repeated the pull-down analysis (Figure 5B, right). The ΔSL1 RNA bait showed no preference for TNRC6 binding at S1 versus S2, as the ΔSL1-S1p6m and ΔSL1-S2p6m baits pulled down similar amounts of TNRC6 proteins. This suggests that the SL1 stem–loop is important for the preferential association of TNRC6 at the S1 miR-122 interaction site.
DISCUSSION
Strong genetic evidence indicate that miR-122/Ago2 complexes must bind to both the S1 and S2 sites near the 5′ end of the HCV genome for optimal viral replication, and that both seed sequence and 3′ supplemental base-pair interactions of miR-122 with the viral RNA are important for binding at both sites (4,7,9). However, the two sites show subtle differences in their requirements for miR-122 binding, with S1 failing to recruit a minority 21-nt miR-122 species that binds to S2 (50). We show here that TNRC6 proteins (all three paralogs, but especially TNRC6B and TNRC6C) associate with the 5′UTR of HCV in a miR-122-dependent manner (Figure 1), and that they are functionally important components of the miR-122/Ago2 complex assembling on the viral RNA (Figure 2). Earlier reports have hinted at the involvement of TNRC6 proteins in HCV replication. Bukong et al. (51) reported that siRNA-mediated depletion of GW182 (TNRC6A) negatively impacted replication of the virus, reducing levels of viral RNA and protein within infected cells. A similar observation was made by Pager et al. (52). By contrast, we found little effect on replication after depleting TNRC6A only, and noted substantial inhibition only after dual depletion of TNRC6B and TNRC6C (discussed in greater detail below). Roberts et al. (28) showed that dually depleting TNRC6A and TNRC6B reduced miR-122 stimulated translation of a reporter protein placed under control of the HCV 5′UTR, but were unable to conclude that TNRC6 proteins specifically regulated the miR-122 effect on its translation. They did not assess the impact of TNRC6 protein depletion on viral replication. None of these reports demonstrated that TNRC6 proteins are recruited to the viral RNA in a miR-122 dependent fashion, as we show here.
While bound at both S1 and S2, TNRC6 is preferentially bound at the S1 site (Figure 5B). Experiments with mutant viral RNAs, in which either S1 or S2 was rendered unable to bind miR-122, indicate that TNRC6 has a strong, pro-viral function only when both sites are active, and that TNRC6 surprisingly restricts replication when only S2 can bind miR-122 (Figure 4). These results provide strong, albeit indirect, evidence for simultaneous occupancy of both sites in single viral RNA molecules, consistent with a previous report suggesting cooperative binding of the miRNA at the two sites (53). Similar experiments suggested that the binding of miR-122/Ago2 complexes to S2 is increased in cells depleted of TNRC6B/C (Figure 5A), leading us to suggest a model in which TNRC6 scaffolds the optimal assembly of miR-122/Ago2 complexes at the 5′ end of the genome, preferentially directing complex formation to S1 where it is more active in promoting amplification of the genome (Figure 5C). When TNRC6 is depleted, this preference for the S1 site is disrupted and binding is increased at S2. Although the overall recruitment of miR-122/Ago2 to the RNA is not significantly changed, its spatial distribution is sub-optimal, resulting in reduced HCV replication. Thus, TNRC6 proteins fine-tune the distribution of miR-122/Ago2 complex at the 5′ terminus of HCV RNA to promote maximal viral replication.
TNRC6 family proteins show relatively low sequence conservation, but are characterized by a conserved domain structure that includes the amino-terminal GW-rich region that interacts with Ago proteins and the carboxy-terminal silencing domain (Figure 2F). Recent structural studies show that the amino-terminal Ago-binding domain of TNRC6B is highly disordered, and that the presence of multiple tryptophan-binding pockets in the PIWI domain of Ago2 allows for a variety of structurally diverse interactions between these proteins (17). Thus, Ago2–TNRC6 interactions may differ both structurally and kinetically at S1 and S2, potentially explaining the differences we observed in TNRC6 function at the two sites. Our data suggest that the SL1 stem–loop (Figure 1A) is important for the differential binding of TNRC6 at the S1 versus S2 sites (Figure 5B). How the stem–loop may mediate this difference in TNRC6 binding is uncertain in the absence of a high-resolution structural model of the HCV-Ago2/miR-122/TNRC6 complex. However, TNRC6 is a large protein (∼182 kD), and it is possible that one or more cellular proteins binding to the stem–loop might interfere sterically with TNRC6 binding at the adjacent S2 site. Alternatively, it is possible that the stem–loop contributes directly to TNRC6 binding at S1.
TNRC6 paralogs function redundantly in miRNA-mediated gene silencing (54–56), and our results suggest TNRC6 proteins also function redundantly in HCV replication. However, TNRC6A seems to be less important than TNRC6B and TNRC6C, based both on the degree of enrichment in the pull-down experiments (Figure 1C and Supplementary Figure S1B,C) and on the results of depletion studies (Figure 2C). This may relate to the presence of a nuclear localization signal in TNRC6A that is not conserved in TNRC6B or TNRC6C (57). Additionally, TNRC6A binds importin-α, whereas TNRC6B and TNRC6C do not (44).
miR-122 has been reported to be involved in multiple facets of the HCV life cycle, including viral RNA stability, IRES activity and viral RNA synthesis (6,22,27–29), and some evidence suggest that miR-122 can rebalance the engagement of viral genomes in translation versus replication (29). TNRC6 depletion reduced the rate of HCV nascent RNA synthesis in Huh-7.5 cells, and also blunted the ability of miR-122 to stimulate viral RNA synthesis (Figure 3E). In contrast, we found no effect on activity of the HCV IRES (Figure 3D) or stability of the viral genome (Figure 3C). Taken collectively, these data suggest that TNRC6 promotes HCV replication by optimizing miR-122-dependent viral RNA synthesis (29). Partial co-localization of TNRC6 proteins with replicase complexes is also consistent with a role in RNA replication (Figure 3F,G and Supplementary Figure S4B,C). Recent studies by Sheu-Gruttadauria and MacRae (17) indicate that the interaction of TNRC6B with Ago2 drives the formation of phase-separated droplets, both in vitro and in cells, condensing RISC complexes and their target RNAs with effector proteins and promoting target RNA processing. It is intriguing to consider that TNRC6 might function similarly in HCV replication, with phase-separated TNRC6B-Ago2 droplets sequestering viral RNA from the cytosol and promoting the assembly of HCV replication complexes.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (41) partner repository with the dataset identifier PXD013054.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Ian MacRae and Dr Luca Gebert for helpful discussions; Dr Matthew Evans, Dr Gunter Meister and Dr Charles Rice for kindly providing reagents; the Michael Hooker Microscopy Facility of the University of North Carolina for technical support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01-AI095690 to S.M.L.; U19-AI109965 to S.M.L., X.C.; T32-AI007151 to E.R.S. (in part)]. Funding for open access charge: NIH R01-AI095690.
Conflict of interest statement. None declared.
REFERENCES
- 1. Perz J.F., Armstrong G.L., Farrington L.A., Hutin Y.J., Bell B.P.. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J. Hepatol. 2006; 45:529–538. [DOI] [PubMed] [Google Scholar]
- 2. Honda M., Beard M.R., Ping L.H., Lemon S.M.. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 1999; 73:1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P.. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005; 309:1577–1581. [DOI] [PubMed] [Google Scholar]
- 4. Jangra R.K., Yi M., Lemon S.M.. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J. Virol. 2010; 84:6615–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wilson J.A., Zhang C., Huys A., Richardson C.D.. Human Ago2 is required for efficient microRNA 122 regulation of hepatitis C virus RNA accumulation and translation. J. Virol. 2011; 85:2342–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shimakami T., Yamane D., Jangra R.K., Kempf B.J., Spaniel C., Barton D.J., Lemon S.M.. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jopling C.L., Schutz S., Sarnow P.. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008; 4:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Machlin E.S., Sarnow P., Sagan S.M.. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:3193–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shimakami T., Yamane D., Welsch C., Hensley L., Jangra R.K., Lemon S.M.. Base pairing between hepatitis C virus RNA and microRNA 122 3′ of Its seed sequence Is essential for genome stabilization and production of infectious virus. J. Virol. 2012; 86:7372–7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fabian M.R., Sonenberg N., Filipowicz W.. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010; 79:351–379. [DOI] [PubMed] [Google Scholar]
- 11. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu J., Rivas F.V., Wohlschlegel J., Yates J.R., Parker R., Hannon G.J.. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005; 7:1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baillat D., Shiekhattar R.. Functional dissection of the human TNRC6 (GW182-related) family of proteins. Mol. Cell Biol. 2009; 29:4144–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen C.Y., Zheng D., Xia Z., Shyu A.B.. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 2009; 16:1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eulalio A., Huntzinger E., Izaurralde E.. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008; 15:346–353. [DOI] [PubMed] [Google Scholar]
- 16. Pfaff J., Hennig J., Herzog F., Aebersold R., Sattler M., Niessing D., Meister G.. Structural features of Argonaute-GW182 protein interactions. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E3770–E3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sheu-Gruttadauria J., MacRae I.J.. Phase transitions in the assembly and function of human miRISC. Cell. 2018; 173:946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Braun J.E., Huntzinger E., Fauser M., Izaurralde E.. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell. 2011; 44:120–133. [DOI] [PubMed] [Google Scholar]
- 19. Huntzinger E., Izaurralde E.. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011; 12:99–110. [DOI] [PubMed] [Google Scholar]
- 20. Huntzinger E., Kuzuoglu-Öztürk D., Braun J.E., Eulalio A., Wohlbold L., Izaurralde E.. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res. 2013; 41:978–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y., Yamane D., Masaki T., Lemon S.M.. The yin and yang of hepatitis C: synthesis and decay of hepatitis C virus RNA. Nat. Rev. Microbiol. 2015; 13:544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Y., Masaki T., Yamane D., McGivern D.R., Lemon S.M.. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1881–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Y., Yamane D., Lemon S.M.. Dissecting the roles of the 5′ exoribonucleases Xrn1 and Xrn2 in restricting hepatitis C virus replication. J. Virol. 2015; 89:4857–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sedano C.D., Sarnow P.. Hepatitis C virus subverts liver-specific miR-122 to protect the viral genome from exoribonuclease Xrn2. Cell Host Microbe. 2014; 16:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Amador-Cañizares Y., Bernier A., Wilson J.A., Sagan S.M.. miR-122 does not impact recognition of the HCV genome by innate sensors of RNA but rather protects the 5′ end from the cellular pyrophosphatases, DOM3Z and DUSP11. Nucleic Acids Res. 2018; 46:5139–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kincaid R.P., Lam V.L., Chirayil R.P., Randall G., Sullivan C.S.. RNA triphosphatase DUSP11 enables exonuclease XRN-mediated restriction of hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:8197–8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Henke J.I., Goergen D., Zheng J., Song Y., Schuttler C.G., Fehr C., Junemann C., Niepmann M.. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008; 27:3300–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roberts A.P., Lewis A.P., Jopling C.L.. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011; 39:7716–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Masaki T., Arend K.C., Li Y., Yamane D., McGivern D.R., Kato T., Wakita T., Moorman N.J., Lemon S.M.. miR-122 stimulates hepatitis C virus RNA synthesis by altering the balance of viral RNAs engaged in replication versus translation. Cell Host Microbe. 2015; 17:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thibault P.A., Huys A., Amador-Canizares Y., Gailius J.E., Pinel D.E., Wilson J.A.. Regulation of hepatitis C virus genome replication by Xrn1 and microRNA-122 binding to individual sites in the 5′ untranslated region. J. Virol. 2015; 89:6294–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schult P., Roth H., Adams R.L., Mas C., Imbert L., Orlik C., Ruggieri A., Pyle A.M., Lohmann V.. microRNA-122 amplifies hepatitis C virus translation by shaping the structure of the internal ribosomal entry site. Nat. Commun. 2018; 9:2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Amador-Canizares Y., Panigrahi M., Huys A., Kunden R.D., Adams H.M., Schinold M.J., Wilson J.A.. miR-122, small RNA annealing and sequence mutations alter the predicted structure of the Hepatitis C virus 5′ UTR RNA to stabilize and promote viral RNA accumulation. Nucleic Acids Res. 2018; 46:9776–9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Y.P., Gottwein J.M., Scheel T.K., Jensen T.B., Bukh J.. MicroRNA-122 antagonism against hepatitis C virus genotypes 1–6 and reduced efficacy by host RNA insertion or mutations in the HCV 5′ UTR. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:4991–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lanford R.E., Hildebrandt-Eriksen E.S., Petri A., Persson R., Lindow M., Munk M.E., Kauppinen S., Orum H.. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010; 327:198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y. et al.. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013; 368:1685–1694. [DOI] [PubMed] [Google Scholar]
- 36. Narbus C.M., Israelow B., Sourisseau M., Michta M.L., Hopcraft S.E., Zeiner G.M., Evans M.J.. HepG2 cells expressing microRNA miR-122 support the entire hepatitis C virus life cycle. J. Virol. 2011; 85:12087–12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luna J.M., Scheel T.K., Danino T., Shaw K.S., Mele A., Fak J.J., Nishiuchi E., Takacs C.N., Catanese M.T., de Jong Y.P. et al.. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015; 160:1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamane D., McGivern D.R., Wauthier E., Yi M., Madden V.J., Welsch C., Antes I., Wen Y., Chugh P.E., McGee C.E. et al.. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat. Med. 2014; 20:927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang L., Wrobel J.A., Xie L., Li D., Zurlo G., Shen H., Yang P., Wang Z., Peng Y., Gunawardena H.P. et al.. Novel RNA-Affinity proteogenomics dissects tumor heterogeneity for revealing personalized markers in precision prognosis of cancer. Cell Chem. Biol. 2018; 25:619–633. [DOI] [PubMed] [Google Scholar]
- 40. Cox J., Mann M.. Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 2011; 80:273–299. [DOI] [PubMed] [Google Scholar]
- 41. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M. et al.. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li Y., Masaki T., Shimakami T., Lemon S.M.. hnRNP L and NF90 interact with hepatitis C virus 5′ terminal untranslated RNA and promote efficient replication. J. Virol. 2014; 88:7199–7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jonas S., Izaurralde E.. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015; 16:421–433. [DOI] [PubMed] [Google Scholar]
- 44. Schraivogel D., Schindler S.G., Danner J., Kremmer E., Pfaff J., Hannus S., Depping R., Meister G.. Importin-β facilitates nuclear import of human GW proteins and balances cytoplasmic gene silencing protein levels. Nucleic Acids Res. 2015; 43:7447–7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ma Y., Yates J., Liang Y., Lemon S.M., Yi M.. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 2008; 82:7624–7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schirle N.T., MacRae I.J.. The crystal structure of human Argonaute2. Science. 2012; 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Elkayam E., Faehnle C.R., Morales M., Sun J., Li H., Joshua-Tor L.. Multivalent recruitment of human argonaute by GW182. Mol. Cell. 2017; 67:646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hauptmann J., Schraivogel D., Bruckmann A., Manickavel S., Jakob L., Eichner N., Pfaff J., Urban M., Sprunck S., Hafner M. et al.. Biochemical isolation of Argonaute protein complexes by Ago-APP. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:11841–11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu Y., Scheel T.K.H., Luna J.M., Chung H., Nishiuchi E., Scull M.A., Echeverría N., Ricardo-Lax I., Kapoor A., Lipkin W.I. et al.. miRNA independent hepacivirus variants suggest a strong evolutionary pressure to maintain miR-122 dependence. PLoS Pathog. 2017; 13:e1006694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamane D., Selitsky S.R., Shimakami T., Li Y., Zhou M., Honda M., Sethupathy P., Lemon S.M.. Differential hepatitis C virus RNA target site selection and host factor activities of naturally occurring miR-122 3′ variants. Nucleic Acids Res. 2017; 45:4743–4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bukong T.N., Hou W., Kodys K., Szabo G.. Ethanol facilitates hepatitis C virus replication via up-regulation of GW182 and heat shock protein 90 in human hepatoma cells. Hepatology. 2013; 57:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pager C.T., Schutz S., Abraham T.M., Luo G., Sarnow P.. Modulation of hepatitis C virus RNA abundance and virus release by dispersion of processing bodies and enrichment of stress granules. Virology. 2013; 435:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nieder-Röhrmann A., Dünnes N., Gerresheim G.K., Shalamova L.A., Herchenröther A., Niepmann M.. Cooperative enhancement of translation by two adjacent microRNA-122/Argonaute 2 complexes binding to the 5′ untranslated region of hepatitis C virus RNA. J. Gen. Virol. 2017; 98:212–224. [DOI] [PubMed] [Google Scholar]
- 54. Lazzaretti D., Tournier I., Izaurralde E.. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA. 2009; 15:1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zipprich J.T., Bhattacharyya S., Mathys H., Filipowicz W.. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009; 15:781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Takimoto K., Wakiyama M., Yokoyama S.. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA. 2009; 15:1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nishi K., Nishi A., Nagasawa T., Ui-Tei K.. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA. 2013; 19:17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (41) partner repository with the dataset identifier PXD013054.