Abstract
Horizontal gene transfer has occurred between organisms of all domains of life and contributed substantially to genome evolution in both prokaryotes and eukaryotes. Phylogenetic evidence suggests that eukaryotic genes horizontally transferred to bacteria provided useful new gene functions that improved metabolic plasticity and facilitated adaptation to new environments. How these eukaryotic genes evolved into functional bacterial genes is not known. Here, we have conducted a genetic screen to identify the mechanisms involved in functional activation of a eukaryotic gene after its transfer into a bacterial genome. We integrated a eukaryotic selectable marker gene cassette driven by expression elements from the red alga Porphyridium purpureum into the genome of Escherichia coli. Following growth under non-selective conditions, gene activation events were indentified by antibiotic selection. We show that gene activation in the bacterial recipient occurs at high frequency and involves two major types of spontaneous mutations: deletion and gene amplification. We further show that both mechanisms result in promoter capture and are frequently triggered by microhomology-mediated recombination. Our data suggest that horizontally transferred genes have a high probability of acquiring functionality, resulting in their maintenance if they confer a selective advantage.
INTRODUCTION
Horizontal gene transfer (HGT, sometimes also called lateral gene transfer) is the movement of genetic material between organisms other than by descent. It typically occurs by asexual or parasexual processes, and has played an important role in the evolution of both prokaryotic and eukaryotic genomes (e.g. 1–12). Genetic material can be horizontally transferred across large phylogenetic distances and between all domains of life (reviewed, e.g. in 13–18). Genome sequencing projects revealed that, especially in unicellular organisms, the contribution of HGT to the genome can be massive, and in extreme cases, may account for up to a tenth of all genes in the genome (19). The mechanisms that influence the prevalence of HGT in different groups of organisms are not well understood, but it is generally assumed that unicellularity, lack of a protected germline and natural competence for transformation (i.e. uptake of environmental DNA; 20) promote HGT and are, at least in part, responsible for the frequent presence of genes of horizontal origin in some groups of microbes.
Bacteria do not only exchange genes with other bacteria and with viruses (bacteriophages), but also acquire genes horizontally from eukaryotic source organisms. For example, a hemoglobin sequence from the bacterium Vitreoscilla is most closely related to plant leghemoglobins, and has been suggested to promote bacterial survival in oxygen-limited environments (21). Similarly, the gene for glutamine synthetase II in the root nodule bacterium Bradyrhizobium japonicum was reported to resemble plant glutamine synthetase genes, suggesting that it also originated from horizontal transfer (22). A eukaryotic version of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was found in the gut bacterium Escherichia coli (in addition to the typical bacterial enzyme), and proposed to provide increased metabolic plasticity (23). Other examples of eukaryote-to-prokaryote HGT that likely possess functional significance include aminoacyl–tRNA synthetase genes linked to bacterial antibiotic resistance (24), two components of the eukaryotic actin cytoskeleton that are found in the cyanobacterium Microcystis aeruginosa where they may play a role in cell stabilization under osmotic stress (25), and a plant pathogenesis-related proteins in the potato pathogen Streptomyces scabies (and other Streptomyces species; 26).
While it is conceivable that many functional HGT events confer an immediate selective advantage to the recipient organism, the integration of horizontally transferred genes into cellular regulatory interaction networks can take millions of years (27). An additional mechanistic barrier to eukaryote-to-prokaryote HGT is posed by the incompatibility of the gene expression mechanisms: eukaryotic expression elements will not normally function in a prokaryotic systems and vice versa. The barriers to functional HGT can be manifold, and, in addition to uptake and integration of the foreign DNA (28), include, at the level of gene expression, promoter incompatibility, presence of introns (29), incompatibility of translation initiation signals and differences in codon usage. Promoter recruitment has been suggested to represent a major mechanism driving transcriptional activation of previously silent genes (30,31). All previous experimental evolution studies on transcriptional activation of silent genes in bacterial genomes involved continuous selection pressure to isolate mutants (32–34). How and how fast spontaneous gene activation can occur in the absence of selection is largely unknown.
Here, we have investigated the molecular mechanisms that lead to functional activation of horizontally transferred eukaryotic genes in prokaryotic genomes. To mimic eukaryote-to-prokaryote HGT, we integrated a selectable marker gene cassette that is under the control of expression elements from the eukaryotic red alga Porphyridium purpureum into the genome of the bacterium E. coli, and then conducted large-scale genetic screens for acquisition of function. Our results reveal transcriptional activation of the eukaryotic gene in E. coli in the absence of selection. Activation occurs at high frequency and within approximately eight generations. Molecular analysis of the gene activation events uncovered two distinct molecular mechanisms that result in acquisition of function: deletion and gene amplification.
MATERIALS AND METHODS
Bacterial culture and integration of a eukaryotic-type marker gene into the E. coli genome
The wild-type-like E. coli strain K-12 MG1655 was grown at 37°C in liquid or agar-solidified Luria-Bertani (LB) medium (yeast extract 5 g/l, tryptone 10 g/l, NaCl 10 g/l, pH 7.5). As eukaryotic-type selectable marker gene, a ble-GFP fusion gene driven by the tubulin promoter and terminator from the red alga Porphyridium purpureum (35) was used. The lacZ gene in the E. coli genome was chosen as neutral site to insert the eukaryotic gene along with a selectable marker gene (nptII) for transformation. To construct a transformation vector for genomic integration by homologous recombination, the sequences flanking lacZ, the eukaryotic marker gene and a prokaryotic nptII expression cassette (Figure 1A) were amplified by polymerase chain reaction (PCR) using E. coli cells, plasmid pZL19 (35) and plasmid pHK20 (36), respectively, as template. The three PCR products were inserted into the SalI/EcoRI sites of the pEX-K4 backbone by recombination cloning using the In-Fusion® HD Cloning Kit (Clontech, Mountain View, CA, USA), resulting in vector pZL26. The MfeI/EcoRI restriction fragment released from pZL26 was used for chromosomal integration via homologous recombination. Transgene integration into the E. coli chromosome was performed employing previously described methods (37). Briefly, plasmid pKD46 encoding the phage lambda Red recombination system was introduced into E. coli by electroporation. Subsequently, E. coli cells harboring pKD46 were electroporated with the MfeI/EcoRI restriction fragment excised from pZL26. Successful integration events into the chromosome of E. coli (Ec-ZL26 strains) were selected on LB medium with kanamycin. The sequences of all primers used in this study are listed in Supplementary Table S1.
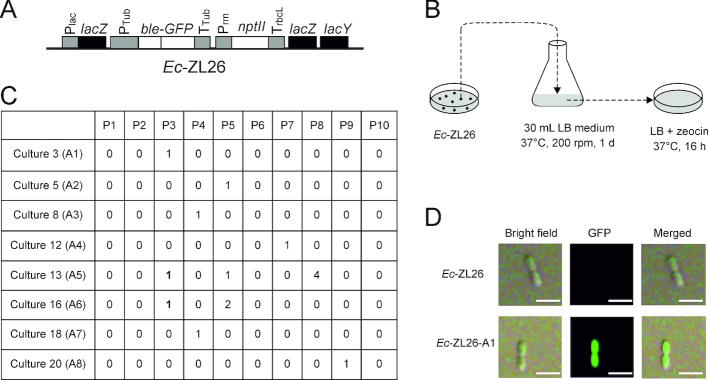
Figure 1.
Genetic screen for functional activation of a eukaryotic gene following horizontal transfer into a prokaryote (Escherichia coli). (A) Schematic map of the eukaryotic ble-GFP gene transferred to the lacZ locus in the E. coli genome by selection for kanamycin resistance (conferred by the nptII gene). The ble-GFP fusion gene is driven by the tubulin gene promoter (PTub) and terminator (TTub) from the red alga Porphyridium purpureum (35) and confers resistance to the antibiotic zeocin. Ec-ZL26 denotes the E. coli strain harboring the construct in its lacZ locus. Plac, lac operon promoter; Prrn, rRNA operon promoter from the tobacco plastid genome; TrbcL, 3′ UTR from the tobacco rbcL gene. Endogenous bacterial genes are represented as black boxes, coding regions of transgenes as white boxes and promoters and terminators (driving transgenes and the lac operon) as gray boxes. (B) Selection for functional activation of the ble-GFP gene. A large-scale genetic screen for functional activation of the ble gene in bacteria was conducted by exposing cultures of Ec-ZL26 to stringent selection for ble-mediated resistance to zeocin. The 30 ml LB medium inoculated with a single colony of Ec-ZL26 was cultivated for 1 day, followed by plating of equal aliquots onto 10 LB plates with zeocin for overnight selection of resistant colonies. The selection process was repeated 30 times (i.e. with 30 independent cultures of 30 ml each). In total, approximate 1012E. coli cells were subjected to selection on 300 plates. (C) Zeocin-resistant colonies appeared on selection plates from 8 of the 30 cultures. P, number of the selection plate; A, gene activation event; 1 (in bold), the colony representing the initially characterized activation event from cultures 13 and 16. (D) Analysis of GFP accumulation in a dividing zeocin-resistant cell from activation event Ec-ZL26-A1 by confocal laser-scanning microscopy. Scale bars: 1 μm.
Selection for functional activation of the eukaryotic ble gene in E. coli
A genetic screen for functional activation of the transferred ble gene in the E. coli genome was conducted by exposing bacterial cultures of strain Ec-ZL26 to stringent selection for ble-mediated resistance to zeocin on agar-solidified LB medium (yeast extract 5 g/l, tryptone 10 g/l, NaCl 5 g/l, pH 7.5) supplemented with 25 μg/ml zeocin. To this end, volumes of 30 ml liquid LB medium were inoculated with a single colony of Ec-ZL26 and cultivated at 37°C on a rotary shaker (at 180 rpm) for 1 day. Aliquots of the resulting bacterial culture were plated on 10 agar plates with LB medium containing 25 μg/ml zeocin and incubated at 37°C overnight to identify resistant colonies. The screen was repeated 30 times (i.e. with 30 cultures of 30 ml each).
Analysis of GFP fluorescence
GFP fluorescence in E. coli colonies growing on agar plates was detected by scanning the plates with an Amersham Typhoon RGB Biomolecular Imager (GE Healthcare, Buckinghamshire, UK) using the LD488 laser and the Cy2 525BP20 filter. Fluorescence in single E. coli cells was analyzed with a confocal laser-scanning microscope (TCS SP5; Leica, Wetzlar, Germany) using an argon laser for excitation (488 nm) and a 500–510 nm filter for detection of GFP fluorescence.
Isolation of nucleic acids, Southern blot analysis, real-time quantitative reverse-transcription PCR (qRT-PCR), and bioinformatic analyses
DNA was extracted from E. coli cultures with the GenElute™ Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA). RNA was extracted from log-phase E. coli cultures using the TRIzol™ Max™ Bacterial RNA Isolation Kit (Invitrogen, Carlsbad, CA, USA). For Southern blot analysis, samples of total bacterial DNA were digested with appropriate restriction enzymes, separated by electrophoresis in 1% agarose gels, and transferred onto Hybond XL nylon membranes (GE Healthcare) by capillary blotting. For preparation of hybridization probes, the ble coding sequence was amplified by PCR with gene-specific primers (Supplementary Table S1), purified by agarose gel electrophoresis and labeled with [α-32P]dCTP by random priming (Megaprime™ DNA Labeling System; GE Healthcare). Hybridization was performed at 65°C using standard protocols. The intensities of hybridizing bands were quantified with the Image Lab 6.0 software (Bio-Rad, Munich, Germany).
For qRT-PCR analysis, extracted bacterial RNAs were treated with DNase I (Thermo Scientific, Waltham, MA, USA). First-strand cDNA was synthesized with SuperScript III Reverse Transcriptase (Invitrogen) and subsequent amplification was performed using the LightCycler 480 Real-Time PCR System (Roche Applied Science, Penzberg, Germany) and ABsolute SYBR Green ROX mix (Thermo Scientific) with rrsA as reference gene. The amplification efficiency of the primers for each gene was tested using a dilution series of cDNA as PCR template. Primer pairs with differences in the slope of the standard curve for amplification of less than 0.1 were selected. Three biological replicates were analyzed with three technical replicates each. The 2−ΔΔCT (cycle threshold) method was used to determine relative cDNA levels. Statistical evaluation was performed by analysis of variance with Microsoft Office Excel 2010.
Codon usage patterns were compared using CodonWorkbench (http://www.buba-basis.de/software/cwb/cwb.html). Relative adaptiveness of a codon is computed as the ratio between the frequency of the codon and the largest frequency among its synonymous codons. By comparing the relative adaptiveness tables of source and target organisms, the standard deviation for relative adaptiveness estimates the deviation of codon usage patterns between two organisms.
RESULTS
A genetic screen for activation of a horizontally transferred eukaryotic gene in a bacterial recipient
To investigate the molecular mechanisms that lead to functional activation of genes that were horizontally transferred from a eukaryotic donor organism into a prokaryotic recipient, we introduced an expression cassette from the eukaryotic alga Porphyridium purpureum (35) into the genome of a prokaryotic host, the bacterium E. coli. The red alga P. purpureum as the eukaryotic donor was chosen, because (i) its genome shows massive signs of HGT (19), (ii) the genome is intron-poor and (iii) its codon usage pattern is relatively similar to that of E. coli. Comparison of the codon usage patterns (http://www.buba-basis.de/software/cwb/cwb.html) revealed that the standard deviation for relative adaptiveness of all codons between P. purpureum and E. coli is 18.84, and thus even smaller than the deviation between P. purpureum and another unicellular eukaryotic model alga, the green alga Chlamydomonas reinhardtii (21.05). These properties largely eliminate confounding influences of intron possession and codon usage patterns on the probability of gene activation we aimed to determine.
The red algal expression cassette consists of the zeocin resistance gene ble translationally fused to the gene for the green fluorescent protein GFP and is driven by expression signals (promoter and terminator) taken from the tubulin gene of the red alga P. purpureum, a eukaryotic alga whose genome shows signs of exceptionally active HGT (19,35; Figure 1A). The ble-GFP cassette was integrated into the lacZ locus of the E. coli chromosome by homologous recombination and selection for a linked kanamycin resistance cassette driven by prokaryotic expression signals (Figure 1A). As expected, the resulting bacterial strain (Ec-ZL26) displayed no detectable resistance to zeocin, consistent with the eukaryotic promoter and terminator sequences not being recognized by the prokaryotic gene expression machinery of E. coli. This enabled genetic screens for activation of the ble gene by selection for the evolution of zeocin resistance (Figure 1B), which should be possible only by mutations that change the eukaryotic expression signals of the gene into expression signals that function in bacteria.
The screen was conducted by growing 30 bacterial cultures of 30 ml each (see ‘Materials and Methods’ section) followed by selection for zeocin resistance to identify putative gene activation events (Figure 1B). Eight of the 30 cultures yielded resistant colonies: six cultures gave one colony each, one culture three colonies and one culture six resistant colonies (Figure 1C). The events resulting from each of the eight culture were tentatively treated as single events and named A1–A8 (for activation events number 1–8; Figure 1C). From the two cultures that gave multiple colonies, one colony was chosen for initial characterization.
Characterization of gene activation events
Preliminary analysis of zeocin-resistant strains by confocal laser-scanning microscopy revealed intense green GFP fluorescence (Figure 1D), tentatively confirming that functional activation of the ble-GFP gene had indeed occurred.
To comparatively analyze the eight activation events, we first tested whether they result in similar levels of gene expression. When GFP fluorescence of bacterial colonies was examined (Figure 2A) and the levels of zeocin resistance were determined (Figure 2B), clear differences were observed. In general, events showing stronger GFP fluorescence also displayed higher levels of zeocin resistance (Figure 2A and B). GFP fluorescence intensities also correlated well with GFP mRNA levels as determined by qRT-PCR assays (Figure 2C).
Figure 2.
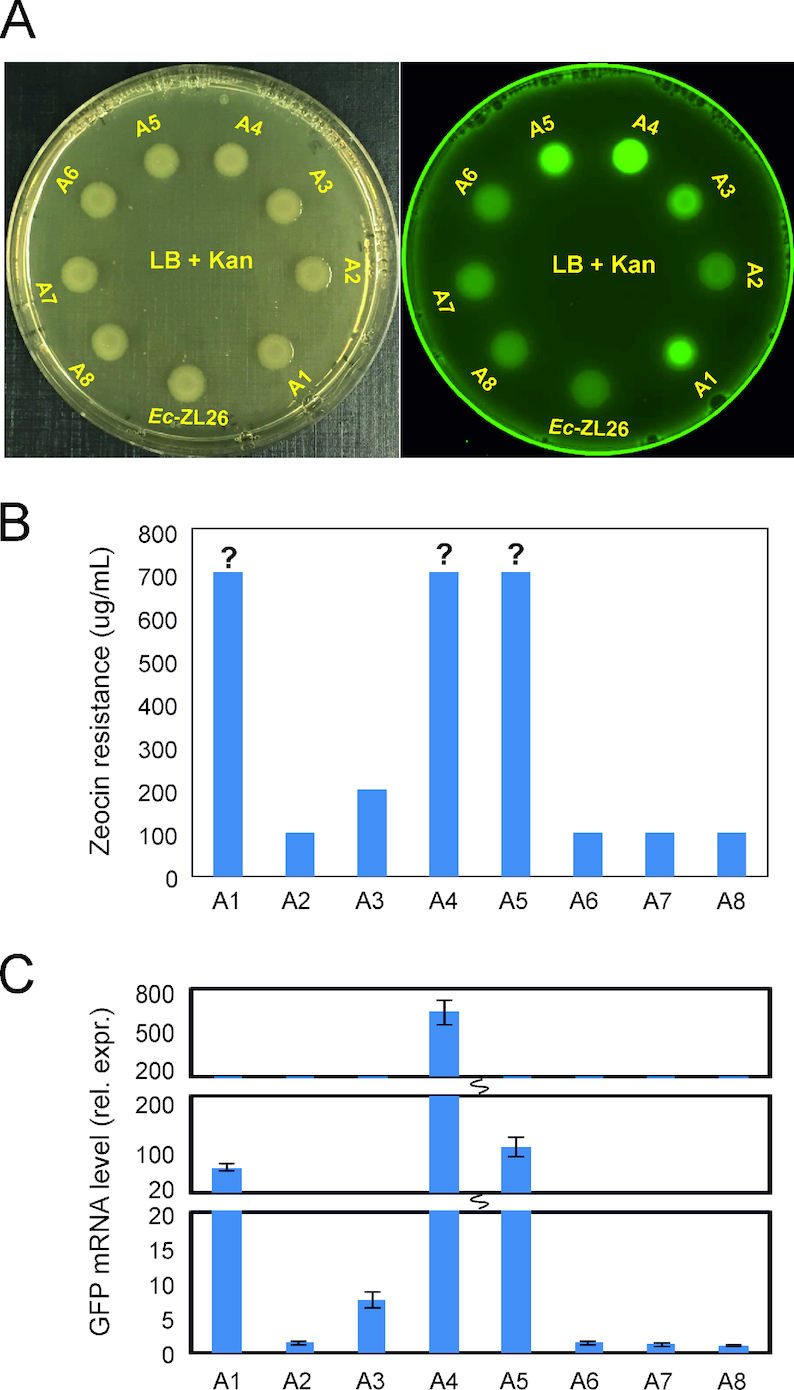
Expression levels of the ble-GFP gene differ between the activation events. (A) Analysis of GFP fluorescence of zeocin-resistant colonies. The left plate was photographed under daylight, the right plate was scanned with laser light to visualize the green fluorescence of GFP (see ‘Materials and Methods’ section). (B) Zeocin tolerance tests to determine the antibiotic resistance level by assaying growth on LB plates with zeocin concentrations between 100 and 700 μg/mL. Question marks indicate that the resistance level may be even higher, but concentrations higher than 700 μg/mL were not tested. (C) Quantification of GFP mRNA accumulation in zeocin-resistant clones by qRT-PCR. Relative expression levels (rel. expr.; normalized to Ec-ZL26-A8, the strain with the lowest GFP expression levels) are shown. Error bars represent the standard deviation from three biological replicates.
Taken together, these data suggest that, in different strains, different molecular events had occurred that led to functional activation of the eukaryotic gene in the bacterial genome.
Identification of the molecular mechanisms leading to gene activation
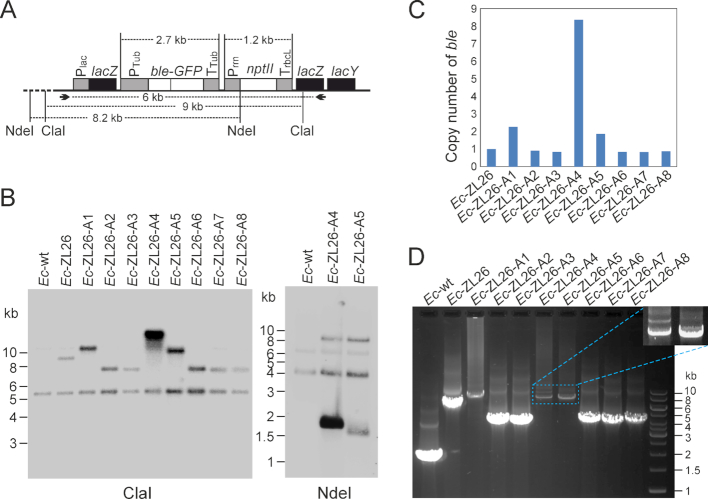
To identify the mutations and/or genomic rearrangements that resulted in gene activation, Southern blot (Figure 3A–C) and PCR analyses were performed (Figure 3D). The data revealed that at least four types of structural changes had occurred, as judged by the different sizes of the hybridizing bands (Figure 3B). Strikingly, the signal intensities of the hybridizing bands in the Southern blot analysis (relative to the reference band; Figure 3B and C) were also very different, indicating that changes in copy number of the ble-GFP gene (presumably by gene amplification) had occurred in at least some strains. Quantification of the intensities of the hybridization signals against a reference band and the fragment sizes obtained (Figure 3B) suggested that the gene copy numbers vary between one and eight, with five strains harboring one copy, two strains (A1 and A5) containing two copies and one strain (A4) even containing eight copies (Figure 3C). PCR assays of the strains with increased copy numbers yielded heterogeneous products with overall weaker amplification signals (Figure 3D), as expected for amplified loci.
Figure 3.
Identification of genomic rearrangements leading to functional activation of the eukaryotic ble-GFP gene in Escherichia coli. (A) Map of the region transferred into the E. coli genome. Restriction sites used in RFLP analyses and primers used for PCR amplification and DNA sequencing are indicated, and expected fragment sizes are given in kb. (B) Southern blot analyses of zeocin-resistant clones. Total bacterial DNA was digested with two different restriction enzymes. While ClaI does not have a recognition site in the transferred DNA, NdeI cuts once in the transgenic region (at the start codon of the nptII coding region). The blots were hybridized to a radiolabeled probe covering the ble coding region. The sizes of the restriction fragments recognized in the Ec-ZL26 strain are 9023 bp for ClaI and 8191 bp for NdeI. (C) Quantification of the bands in the Southern blot assays (with the Image Lab 6.0 software) to estimate the copy number of the ble gene in the bacterial strains with activated ble expression. The cross-hybridizing band of ∼5.1 kb (in the ClaI blot) that is also visible in the wild type was used as reference band for quantitation. (D) Gel electrophoretic analysis of PCR products of the region harboring the activated ble (obtained with the primers indicated in panel A). The magnified area shows a stronger exposure of the amplified bands corresponding to the tandem duplications in strains A4 and A5 (cf. Figure 4).
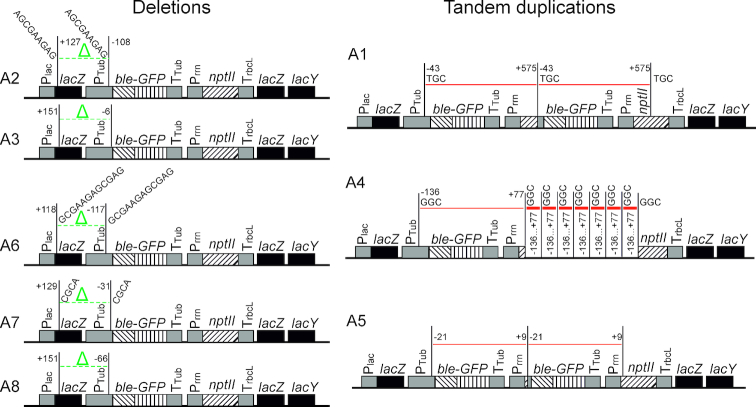
To identify the precise structure of the mutated loci, the lacZ region of all eight activation events was sequenced. Analysis of the rearrangements that had occurred revealed two distinct mechanisms of how the ble-GFP gene became a functional bacterial gene (Figure 4). Five of the eight events turned out to represent promoter capture events. The corresponding strains (A2, A3, A6, A7 and A8) harbored different deletions that all removed the eukaryotic promoter and brought the upstream lacZ promoter in close proximity to the ble-GFP coding region (Figure 4). The remaining three events (A1, A4 and A5) had retained the eukaryotic promoter in front of the ble-GFP gene, but contained additional copies of the ble-GFP coding region. The additional copies integrated downstream of the Prrn promoter driving the nptII gene (Figure 4). This led to acquisition of a prokaryotic promoter that now drives expression of ble-GFP. The unaltered structure of the original ble-GFP cassette (Figure 4) in these strains suggests that only the rearranged copy is active and results in expression of the zeocin resistance gene in strains A1, A4 and A5.
Figure 4.
Overview of the molecular rearrangements leading to functional activation of the eukaryotic ble gene in the Escherichia coli genome. Nucleotide positions at the break points of deletions (left) or duplications (right) are given and refer to the start of the coding region in which they occur (lacZ, ble or nptII), with negative numbers indicating positions upstream and positive numbers indicating positions downstream of the first nucleotide of the respective coding region. The three transgenic coding regions (ble, GFP, nptII) are distinguished by three different striping patterns. The green Δ and the green dotted lines denote deletions, the red solid lines indicate tandem duplications. The multi-copy duplications in strain A4 are schematically shown by thick red lines (representing copies 2–8). Short directly repeated sequences found at the break points are represented by their nucleotide sequences.
While strains A1 and A5 harbored only one extra copy of the ble-GFP coding region, strain A4 contained the seven additional copies estimated from quantification of the signal intensity in the Southern blot analysis (Figure 3B and C). However, the multicopy event A4 showed a smear in the Southern blot from the strongly hybridizing band into the lower molecular weight region of the gel (Figure 3B), possibly suggesting some instability of the event and occasional loss of some gene copies by homologous recombination (loop-out recombination) between the directly repeated copies.
To obtain more information about the molecular mechanisms involved in producing the identified rearrangements, the break points of the deletions and the sequences at the insertion sites of the amplified copies were investigated. In five out of eight cases, microhomologies of three to twelve basepairs were observed (Figure 4). This finding strongly suggests involvement of microhomology-mediated illegitimate recombination (38–40) in the gene activation process.
Gene activation occurs prior to selection
The gene activation events were identified by antibiotic selection (Figure 1). Since many antibiotics are known to be mutagenic (41–43), we wanted to verify that the mutations leading to activation of the ble-GFP gene are not dependent on the presence of the antibiotic. The appearance of multiple antibiotic-resistant colonies in two out of the 30 selection experiments conducted (cultures 13 and 16; Figure 1C) made it possible to distinguish between appearance of the gene-activating mutations prior to or after exposure to zeocin (Figure 5). If the mutation occurred early during growth of the bacterial culture in the absence of the antibiotic, it should give rise to more than one resistant colony in the subsequent selection for antibiotic resistance. This would be revealed by these colonies representing one and the same molecular event (Figure 5). By contrast, if appearance of the mutation depends on the action of the antibiotic, multiple events resulting from the same culture should represent independent mutations and, consequently, differ in the structure of the mutated locus in the genome (Figure 5).
Figure 5.
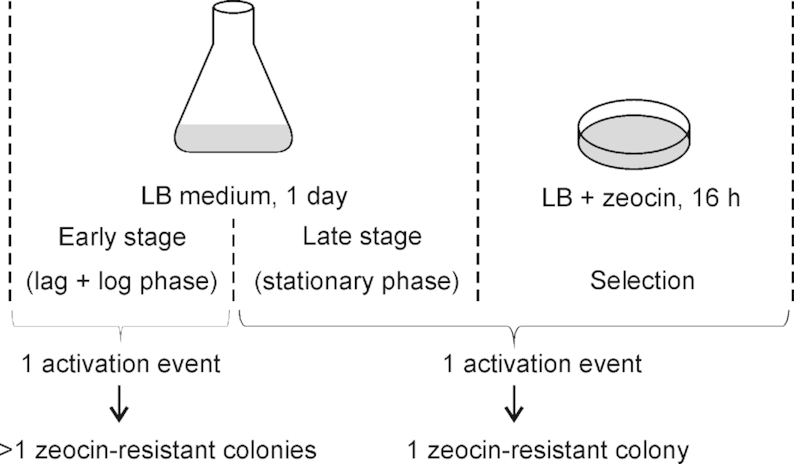
Gene activation events occur in the absence of selection. The cultivation and selection process can be divided into three phases: the early stage (lag and log phases of growth in the absence of selection), the late stage (stationary growth phase in the absence of selection) and the selection phase (growth on solid medium in the presence of the antibiotic). Only at the early stage, a single activation event can lead to more than one zeocin-resistant colony. Six and three resistant colonies were obtained from cultures 13 (A5) and 16 (A6), respectively, and all colonies from one culture showed the same molecular rearrangement, strongly suggesting that the colonies go back to one and the same gene activation event (and that this event occurred early in the absence of antibiotic selection).
To distinguish between these possibilities, the six resistant colonies selected from culture 13 and the three colonies obtained from culture 16 (Figure 1C) were analyzed by DNA sequencing. All six colonies from culture 13 represented the same event (A5) and, likewise, all three colonies from culture 16 were identical (and represented event A6; Figure 4). This result demonstrates that the gene-activating mutations arose in the absence of selection and were not caused by the mutagenic action of the antibiotic.
In our experimental setup, the bacterial cells completed approximately eight generations in the 30 ml culture during the one-day cultivation. Having isolated 15 resistant colonies (representing eight independent events) from approximately 1012 bacterial cells, this finding suggest that (i) gene activation events can occur rapidly within as little as eight generations, and (ii) the frequency of activation is approximately 10−11.
DISCUSSION
In the course of this work, we have developed a genetic screen that mimics the functional activation of eukaryotic genes in a prokaryotic cell after horizontal gene transfer. The frequent horizontal acquisition of genes from eukaryotic donors by bacterial recipients during evolution (21–26) may suggest that functional activation of eukaryotic genes in a prokaryotic environment does not pose an insurmountable obstacle to functional HGT. Our findings reported here demonstrate that transferred eukaryotic genes can quickly become functional in the genomes of bacterial recipients. This can occur in the absence of selection for gene activation and involve two distinct molecular mechanisms: (i) deletions resulting in removal of the eukaryotic promoter, or (ii) gene amplification by tandem duplications (Figure 4). In both cases, a prokaryotic promoter is captured which appears to be sufficient to achieve gene activation in the bacterial genome.
Our data show that both the lacZ promoter upstream and the rrn promoter downstream of the red algal gene can be recruited for gene activation (Figure 4). This finding may suggest that promoter type and location are not a major constraint. Given that bacterial genomes are very gene dense (and typically about 90% coding), the presence of a suitable promoter within a reasonable distance of the transferred eukaryotic gene seems unlikely to represent a serious obstacle to functional activation.
The identification of deletions as the prevailing mechanism of transcriptional gene activation may be due to the mutation bias in bacterial genomes. Previous work has revealed that deletions outweigh insertions by at least a factor of 10 in most prokaryotes (44). Promoter capture was also seen in experimental evolution experiments that used direct selection to trigger the activation of silent genes (30,31). Alternatively, random sequences can evolve into functional promoters (33), a mechanism we did not observe in our study, where gene activation was allowed to occur in the absence of selection. A recent study applying stringent selection for carbon utilization also identified gene duplication events as a mutation type that can result in activation of a previously silent bacterial gene (34).
The rapid gene activation observed in our study was surprising, given that, in a previously conducted long-term evolution experiment, it took thousands of generations to activate a previously silent citrate transporter gene present in the genome of E. coli by gene duplication and promoter capture (32). Moreover, this long-term experiment involved a continuous selection pressure for citrate utilization (32). By contrast, activation of the eukaryotic gene in our study occurred very quickly, within a day of culture and in the absence of selection for gene activation.
The presence of microhomologies at the break points of many of the observed rearrangements (Figure 4) indicates involvement of microhomology-mediated illegitimate recombination (38–40) in the generation of the mutations that lead to gene activation. Interestingly, deletions produced by microhomology-mediated illegitimate recombination and promoter capture have been demonstrated to also be involved in the functional activation of organellar (plastid and mitochondrial) genes following endosymbiotic gene transfer to the nuclear genome (39,45–47). Thus, similar molecular mechanisms seem to operate in functional gene transfer from prokaryotic to eukaryotic genomes and from eukaryotic to prokaryotic genomes.
In the screen described here, eight gene activation events were isolated from approximately 1012 bacterial cells, suggesting a gene activation frequency of ∼10−11. The eight events fell into two distinct classes, likely identifying two major mechanisms how functional activation occurs. However, the eight events all differed in the mutations that had arisen, indicating that there are many different paths to gene activation. Given this and the likely long residence time of the eukaryotic gene in the bacterial recipient genome before mutational deterioration occurs, it seems reasonable to conclude that the requirement for the horizontally transferred eukaryotic gene to evolve into a prokaryotic-type gene poses no strong barrier to functional HGT. Other constraints may more likely represent bottlenecks, including the physical transfer of the DNA (28), difference in GC content and codon usage (48–51) and/or the interruption of many eukaryotic genes by introns that require a complicated machinery for their removal (which is absent from prokaryotes). It is, however, important to note that especially some unicellular lineages of eukaryotes possess genomes that are very intron-poor, including the red algae that served as source of the eukaryotic gene cassette in this study (with only 235 introns in 8355 predicted genes; 19). Moreover, previous research has demonstrated that even presence of an unspliceable intron does not represent an unsurmountable obstacle to functional activation of a horizontally transferred gene (29). Whether or not intron-poor lineages have acted more frequently as donors in HGT is currently unknown.
In addition to transcriptional activation by promoter capture, expression of a eukaryotic gene in a prokaryotic recipient organism also requires appropriate signals for translation initiation that reside in the 5′ untranslated region (5′ UTR). The Shine-Dalgarno (SD) sequence provides the most common signal for translation initiation by providing a ribosome-binding site that engages in a base-pairing interaction with the 3′ end of the 16S rRNA (52). However, SD-independent translation initiation (promoted by the absence of mRNA secondary structure around the initiation codon; 53) is also widespread in many prokaryotes. The promoter and 5′ UTR sequence from the P. purpureum tubulin gene used for construction of the ble-GFP cassette contains a sequence five nucleotides upstream of the start codon that resembles an SD sequence (AGAGG). We propose that this sequence motif may act as an SD-like sequence and facilitates translation to levels that are sufficient to make the gene functional after transcriptional activation by promoter capture. Since the sequence of the SD element and its distance from the start codon are variable, and moreover, translation initiation can also occur in an SD-independent manner (53), it seems reasonable to assume that translation does not pose a strong barrier to functional HGT.
In summary, our work reported here has identified genetic mechanisms that enable a horizontally transferred eukaryotic gene to become functional in its new prokaryotic environment. The relatively high frequency at which this occurs suggests that horizontally transferred genes have a high probability of acquiring functionality, if they confer a selective advantage to the recipient cell.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Daniel Karcher for discussion and Hai He (both MPI-MP) for providing the bacterial strain and help with genomic integration of transgene cassettes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Max Planck Society; European Research Council (ERC) under the European Union's Horizon 2020 Research and Innovation Programme (ERC-ADG-2014) [669982 to R.B.]. Funding for open access charge: Max-Planck-Gesellschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1. Vaughn J.C., Mason M.T., Sper-Whitis G.L., Kuhlman P., Palmer J.D.. Fungal origin by horizontal gene transfer of a plant mitochondrial group I intron in the chimeric coxI gene of Peperomia. J. Mol. Evol. 1995; 41:563–572. [DOI] [PubMed] [Google Scholar]
- 2. Bergthorsson U., Adams K.L., Thomason B., Palmer J.D.. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003; 424:197–201. [DOI] [PubMed] [Google Scholar]
- 3. Lindell D., Sullivan M.B., Johnson Z.I., Tolonen A.C., Rohwer F., Chisholm S.W.. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:11013–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Denker E., Bapteste E., Le Guyader H., Manuel M., Rabet N.. Horizontal gene transfer and the evolution of cnidarian stinging cells. Curr. Biol. 2008; 18:R858–R859. [DOI] [PubMed] [Google Scholar]
- 5. Gladyshev E.A., Meselson M., Arkhipova I.R.. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008; 320:1210–1213. [DOI] [PubMed] [Google Scholar]
- 6. Stegemann S., Keuthe M., Greiner S., Bock R.. Horizontal transfer of chloroplast genomes between plant species. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:2434–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schönknecht G., Chen W.-H., Ternes C.M., Barbier G.G., Shrestha R.P., Stanke M., Bräutigam A., Baker B.J., Banfield J.F., Garavito R.M. et al.. Gene transfer from bacteria and Archaea facilitated evolution of an extremophilic eukaryote. Science. 2013; 339:1207–1210. [DOI] [PubMed] [Google Scholar]
- 8. Li F.-W., Villarreal J.C., Kelly S., Rothfels C.J., Melkonian M., Frangedakis E., Ruhsam M., Sigel E.M., Der J.P., Pittermann J. et al.. Horizontal transfer of an adaptive chimeric photoreceptor from bryophytes to ferns. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:6672–6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Metcalf J.A., Funkhouser-Jones L.J., Brileya K., Reysenbach A.-L., Bordenstein S.R.. Antibacterial gene transfer across the tree of life. Elife. 2014; 3:4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kyndt T., Quispe D., Zhai H., Jarret R., Ghislain M., Liu Q., Gheysen G., Kreuze J.F.. The genome of cultivated sweet potato contains Agrobacterium T-DNAs with expressed genes: an example of a naturally transgenic food crop. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marsit S., Mena A., Bigey F., Sauvage F.-X., Couloux A., Guy J., Legras J.-L., Barrio E., Dequin S., Galeote V.. Evolutionary advantage conferred by an eukaryote-to-eukaryote gene transfer event in wine yeasts. Mol. Biol. Evol. 2015; 32:1695–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ropars J., de la Vega R.C.R., López-Villavicencio M., Gouzy J., Sallet E., Dumas E., Lacoste S., Debuchy R., Dupont J., Brance A. et al.. Adaptive horizontal gene transfers between multiple cheese-associated fungi. Curr. Biol. 2015; 25:2562–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Keeling P.J., Palmer J.D.. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 2008; 9:605–618. [DOI] [PubMed] [Google Scholar]
- 14. Bock R. The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci. 2010; 15:11–22. [DOI] [PubMed] [Google Scholar]
- 15. Zhaxybayeva O., Doolitle W.F.. Lateral gene transfer. Curr. Biol. 2011; 21:R242–R246. [DOI] [PubMed] [Google Scholar]
- 16. Overballe-Petersen S., Willerslev E.. Horizontal transfer of short and degraded DNA has evolutionary implications for microbes and eukaryotic sexual reproduction. Bioessays. 2014; 36:1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Soucy S.M., Huang J., Gogarten J.P.. Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 2015; 16:472–482. [DOI] [PubMed] [Google Scholar]
- 18. Husnik F., McCutcheon J.P.. Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 2018; 16:67–79. [DOI] [PubMed] [Google Scholar]
- 19. Bhattacharya D., Price D.C., Chan C.X., Qiu H., Rose N., Ball S., Weber A.P.M., Arias M.C., Henrissat B., Coutinho P.M. et al.. Genome of the red alga Porphyridium purpureum. Nat. Commun. 2013; 4:1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blokesch M. Natural competence for transformation. Curr. Biol. 2016; 26:R1119–R1136. [DOI] [PubMed] [Google Scholar]
- 21. Wakabayashi S., Matsubara H., Webster D.A.. Primary sequence of a dimeric bacterial haemoglobin from Vitreoscilla. Nature. 1986; 322:481–483. [DOI] [PubMed] [Google Scholar]
- 22. Carlson T.A., Chelm B.K.. Apparent eukaryotic origin of glutamine synthetase II from the bacterium Bradyrhizobium japonicum. Nature. 1986; 322:658–570. [Google Scholar]
- 23. Doolittle R.F., Feng D.F., Anderson K.L., Alberro M.R.. A naturally occurring horizontal gene transfer from a eukaryote to a prokaryote. J. Mol. Evol. 1990; 31:383–388. [DOI] [PubMed] [Google Scholar]
- 24. Brown J.R., Zhang J., Hodgson J.E.. A bacterial antibiotic resistance gene with eukaryotic origins. Curr. Biol. 1998; 8:R365–R367. [DOI] [PubMed] [Google Scholar]
- 25. Guljamow A., Jenke-Kodama H., Saumweber H., Quillardet P., Frangeul L., Castets A.M., Bouchier C., de Marsac N.T., Dittmann E.. Horizontal gene transfer of two cytoskeletal elements from a eukaryote to a cyanobacterium. Curr. Biol. 2007; 17:R757–R759. [DOI] [PubMed] [Google Scholar]
- 26. Armijos-Jaramillo V., Santander-Gordón D., Soria R., Pazmiño-Betancourth M., Echeverría M.C.. A whole genome analysis reveals the presence of a plant PR1 sequence in the potato pathogen Streptomyces scabies and other Streptomyces species. Mol. Phylogenet. Evol. 2016; 114:346–352. [DOI] [PubMed] [Google Scholar]
- 27. Lercher M.J., Pál C.. Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol. Biol. Evol. 2007; 25:559–567. [DOI] [PubMed] [Google Scholar]
- 28. Popa O., Dagan T.. Trends and barriers to lateral gene transfer in prokaryotes. Curr. Opin. Microbiol. 2011; 14:615–623. [DOI] [PubMed] [Google Scholar]
- 29. Fuentes I., Karcher D., Bock R.. Experimental reconstruction of the functional transfer of intron-containing plastid genes to the nucleus. Curr. Biol. 2012; 22:763–771. [DOI] [PubMed] [Google Scholar]
- 30. Matus-Garcia M., Nijveen H., van Passel M.W.. Promoter propagation in prokaryotes. Nucleic Acids Res. 2012; 40:10032–10040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oren Y., Smith M.B., Johns N.I., Kaplan Zeevi M., Biran D., Ron E.Z., Corander J., Wang H.H., Alm E.J., Pupko T.. Transfer of noncoding DNA drives regulatory rewiring in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:16112–16117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blount Z.D., Barrick J.E., Davidson C.J., Lenski R.E.. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012; 489:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yona A.H., Alm E.J., Gore J.. Random sequences rapidly evolve into de novo promoters. Nat. Commun. 2018; 9:1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van Hofwegen D.J., Hovde C.J., Minnich S.A.. Rapid evolution of citrate utilization by Escherichia coli by direct selection sequires citT and dctA. J. Bacteriol. 2016; 198:1022–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Z., Bock R.. Replication of bacterial plasmids in the nucleus of the red alga Porphyridium purpureum. Nat. Commun. 2018; 9:3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuroda H., Maliga P.. Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res. 2001; 29:970–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chan C.Y., Kiechle M., Manivasakam P., Schiestl R.H.. Ionizing radiation and restriction enzymes induce microhomology-mediated illegitimate recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2007; 35:5051–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stegemann S., Bock R.. Experimental reconstruction of functional gene transfer from the tobacco plastid genome to the nucleus. Plant Cell. 2006; 18:2869–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hazkani-Covo E., Covo S.. Numt-mediated double-strand break repair mitigates deletions during primate genome evolution. PLoS Genet. 2008; 4:e1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Balashov S., Humayun M.Z.. Mistranslation induced by streptomycin provokes a RecABC/RuvABC-dependent mutator phenotype in Escherichia coli cells. J. Mol. Biol. 2002; 315:513–527. [DOI] [PubMed] [Google Scholar]
- 42. Ramotar D., Wang H.. Protective mechanisms against the antitumor agent bleomycin: lessons from Saccharomyces cerevisiae. Curr. Genet. 2003; 43:213–224. [DOI] [PubMed] [Google Scholar]
- 43. MacLean R.C., Torres-Barceló C., Moxon R.. Evaluating evolutionary models of stress-induced mutagenesis in bacteria. Nat. Rev. Genet. 2013; 14:221–227. [DOI] [PubMed] [Google Scholar]
- 44. Kuo C.H., Ochman H.. Deletional bias across the three domains of life. Genome Biol. Evol. 2009; 1:145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bock R., Timmis J.N.. Reconstructing evolution: gene transfer from plastids to the nucleus. Bioessays. 2008; 30:556–566. [DOI] [PubMed] [Google Scholar]
- 46. Lloyd A.H., Timmis J.N.. The origin and characterization of new nuclear genes originating from a cytoplasmic organellar genome. Mol. Biol. Evol. 2011; 28:2019–2028. [DOI] [PubMed] [Google Scholar]
- 47. Bock R. Witnessing genome evolution: experimental reconstruction of endosymbiotic and horizontal gene transfer. Annu. Rev. Genet. 2017; 51:1–22. [DOI] [PubMed] [Google Scholar]
- 48. Kudla G., Murray A.W., Tollervey D., Plotkin J.B.. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009; 324:255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boel G., Letso R., Neely H., Price W.N., Wong K.-H., Su M., Luff J.D., Valecha M., Everett J.K., Acton T.B. et al.. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature. 2016; 529:358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barahimipour R., Strenkert D., Neupert J., Schroda M., Merchant S.S., Bock R.. Dissecting the contributions of GC content and codon usage to gene expression in the model alga Chlamydomonas reinhardtii. Plant J. 2015; 84:704–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hanson G., Coller J.. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018; 19:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999; 234:187–208. [DOI] [PubMed] [Google Scholar]
- 53. Scharff L.B., Childs L., Walther D., Bock R.. Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet. 2011; 7:e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.